Abstract
Aurora kinase B (AKB), a Ser/Thr kinase that plays a crucial role in mitosis, is overexpressed in several cancers. Clinical inhibitors targeting AKB bind to the active DFG “in” conformation of the kinase. It would be beneficial, however, to understand if AKB is susceptible to type II kinase inhibitors that bind to the inactive, DFG “out” conformation, since type II inhibitors achieve higher kinome selectivity and higher potency in vivo. The DFG “out” conformation of AKB is not yet experimentally determined which makes the design of type II inhibitors exceedingly difficult. An alternate approach is to simulate the DFG “out” conformation from the experimentally determined DFG “in” conformation using atomistic molecular dynamics (MD) simulation. In this work, we employed metadynamics (MTD) approach to simulate the DFG “out” conformation of AKB by choosing the appropriate collective variables. We examined structural changes during the DFG-flip and determined the interactions crucial to stabilize the kinase in active and inactive states. Interestingly, the MTD approach also identified a unique transition state (DFG “up”), which can be targeted by small molecule inhibitors. Structural insights about these conformations is essential for structure-guided design of next-generation AKB inhibitors. This work also emphasizes the usefulness of MTD simulations in predicting macromolecular conformational changes at reduced computational costs.
Keywords: Aurora kinase B, DFG-in conformation, DFG-out conformation, metadynamics
INTRODUCTION
Protein kinases are key regulators in several signaling cascades (1-3). They are responsible for catalyzing the transfer of the γ-phosphate of ATP to downstream substrates by switching from an inactive “off” state to an active “on” state (4,5). Their deregulation is linked to several diseases, in particular cancers (6-8). The cell cycle kinase, Aurora kinase B (AKB), has been associated with a variety of malignancies. AKB is a serine-threonine kinase that is part of the chromosomal passenger complex (CPC), a protein complex crucial in mitosis for chromosomal condensation, ensuring proper microtubule-chromosome attachments, spindle assembly check point, and cytokinesis (9,10). AKB is the only enzymatic unit of this protein complex.
Overexpression of AKB is reported in several cancers including lung cancer (11), breast cancer (12), colorectal cancer (13), prostate cancer (14,15), acute myeloid leukemia, and acute lymphatic leukemia (16). There are several studies relating genomic instability with increased Aurora B activity, establishing it as a therapeutic target in cancer therapy. These studies provide important insight into the use of AKB inhibitors in combination therapy. Barasertib (AZD1152), an Aurora B inhibitor, sensitizes cancer cells to chemotherapeutic agents such as paclitaxel (17) and cytarabine (18). AKB inhibition can lower radiation resistance in gynecological cancers (19) and also sensitizes colorectal cancer to radiotherapy (20). Reports also suggest that AKB inhibitors enhance the efficacy of vemurafenib (17). These results suggest that AKB inhibitors have a dual-use capacity in oncology by (1) directly impairing cell division and (2) synergizing with other treatments including radiotherapy, chemotherapy, and targeted therapy. The dual-use capacity of AKB inhibitors represents an attractive treatment approach that has yet to be exploited clinically.
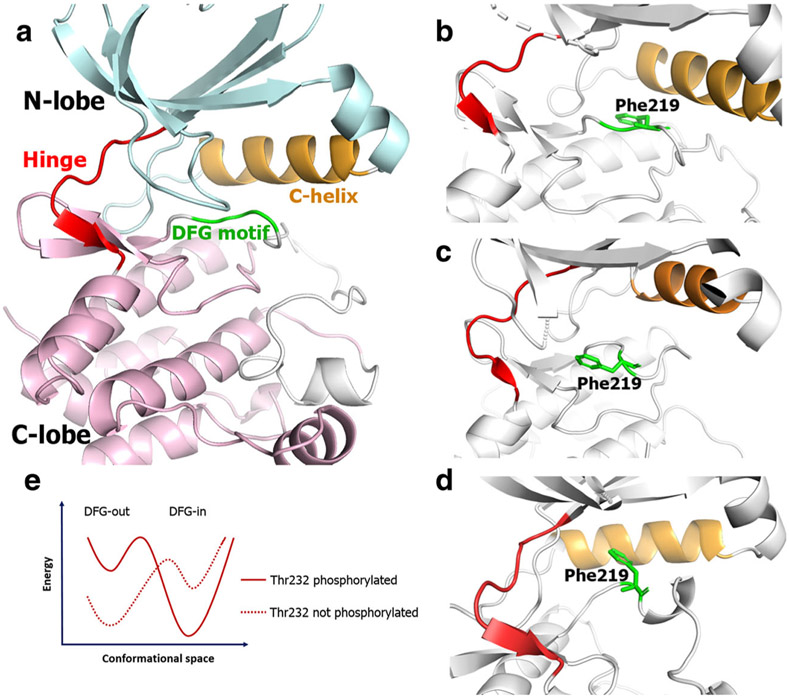
Structurally, the AKB kinase domain is split between an N-lobe and C-lobe, and at the interface of these lobes is the ATP binding pocket. Within the ATP binding pocket is a motif of three highly conserved residues: Asp-Phe-Gly (DFG). In the active state, the DFG motif assumes an “in” conformation and, in the inactive state, the DFG motif adopts an “out” conformation (21). This nomenclature is based on the conformation adopted by phenylalanine. In the DFG “in” conformation, the phenylalanine points into the cavity adjacent to the DFG residues and towards the αC-helix (Fig. 1b), which is called the “allosteric” or “back” pocket. In the DFG “out” conformation, the phenylalanine points away from the αC-helix (Fig. 1c). The nomenclature can also be defined based on the aspartate residue preceding phenylalanine—it points into the ATP-binding site in the DFG “in” conformation and away from it facing the αC-helix in DFG “out.” These structural motifs are conserved within the human kinome. Kinase inhibitors binding to both DFG “in” and “out” conformations have been reported (22). However, for AKB, only DFG “in” inhibitors (type I inhibitors) are reported (23). The most well-known AKB inhibitor is barasertib, which is currently under clinical investigation. Barasertib exhibits ATP-competitive inhibition, and the co-crystal structure of barasertib in AKB is in the DFG “in” conformation (24). A few other compounds, such as chiauranib, also inhibit AKB but are non-selective, multi-kinase inhibitors (25). The limited success of targeting the various conformations of AKB may stem from a lack of sufficient structural information required for drug discovery efforts. Since kinases adopt various biologically relevant conformations between maximally and minimally active states, knowledge of the structural configuration of the target protein is of vital importance for structure-based drug design. In the case of AKB, only the active state is crystallized (24,26,27), while the inactive and other less active states are yet to be solved. Thus, studies providing structural insights for other AKB conformations are crucial for drug discovery efforts targeting AKB.
Fig. 1.
Schematic representation of general kinase structure (a). The orientation of Phe219 residue in the DFG “in” (b), DFG “out” (c), and DFG “up” (d) conformations. Hypothesis for the DFG “in” to “out” transition (e)
In its fully active state, AKB adopts the DFG “in” conformation, which is the thermodynamic endpoint of two events: phosphorylation of Thr232 and binding of Inner Centromere Protein (INCENP) (9). In the absence of these two events, the thermodynamic endpoint is the DFG “out” conformation. We hypothesized that the experimentally known DFG “in” conformation, without INCENP and Thr232 phosphorylated, would transition the kinase to the DFG “out” conformation during molecular dynamic simulations (Fig. 1e). One approach to achieve this is through unbiased molecular dynamics simulations (28). Prior reports on molecular dynamics of other kinases suggest the need of several microseconds to simulate the DFG-flip (29). Such long scale simulations are challenged by the limitations of computational power. This problem was addressed by employing several enhanced sampling methods including, but not limited to, replica exchange molecular dynamics (REMD) (30), Markov state models (31), Bayesian clustering (32), and Metadynamics (33). In this study, we used metadynamics (MTD) (34,35) to simulate the DFG “out” conformation of AKB from the DFG “in” conformation and characterized key structural changes in the kinase during the DFG-flip. We also discuss a unique DFG “up” conformation in AKB, which is reported in an Aurora A triple-mutant (36) and on the possibility of an Src-like inactive state in AKB. To the best of our knowledge, this is the first molecular dynamics study exploring the conformational space of AKB.
METHODS
System Building for MD Runs.
The human and Xenopus laevis AKB amino acid sequences were retrieved from Uniprot database. Sequence alignment was performed to determine the non-conserved residues. The Xenopus laevis Aurora kinase B crystal structure (PDB ID: 4C2V, 1.49 Å) was retrieved from RCSB Protein Data Bank. The non-conserved residues were computationally mutated using the Pymol Mutagenesis tool. The structure thus obtained was minimized in a solvent box using GROMACS simulation package. The protein structures were prepared with the Protein Preparation Wizard in Maestro 2017-2 suite obtained through Desmond academic license. The missing hydrogens were added, bond orders were assigned, and the protein was minimized using OPLS3e force field. Simulation system was prepared using system builder wizard. The proteins and protein-ligand complexes were centered in an orthorhombic box with the edges 10 Å away from the protein in all directions. The box was solvated with simple point charge (SPC) waters. NaCl counter ions were added to neutralize and maintain the system at physiological pH of 0.15 M. The systems were equilibrated at 300 K for 1 ns in NVT ensembles using Nose-Hoover chain thermostat, followed by another 1 ns equilibration at 300 K in NPT ensemble using Martyna-Tobias-Klein barostat.
Metadynamics Simulations.
The equilibrated systems were submitted to metadynamics using the GPU accelerated Desmond software run on NVIDIA Quadro P5000 graphic card installed on a system with Intel Xeon processor. A combination of two collective variables (CVs) that describe the movement around the DFG loop was defined (see “RESULTS” for the CVs’ definitions). For the distance CVs, the Gaussian width was set to 0.05 Å and distance was capped at 30 Å to prevent the protein from being pushed to biologically irrelevant conformations. A Gaussian width of 2.5° was set for the angular CV. The starting height of the Gaussian potential was set to 0.03 kcal/mol, and the Gaussians were deposited every 0.09 ps. The simulations were performed at 300 K and 1.01325 bar pressure. RESPA integrator was used with a time step of 2.0 fs. For coulombic interactions, short range cutoff radius was defined at 9 Å. No positional restraints were specified for any of the atoms. The trajectory frames were recorded at an interval of 100 ps. The simulations were visualized in Maestro suite. Analysis of the trajectory was performed using Simulation Event Analysis of Maestro and Visual Molecular Dynamics.
Molecular Docking.
Molecular docking was performed using the commercially obtained Schrödinger license in Maestro 2017-2 suite. The protein for docking was prepared using the protein preparation wizard as described above. The docking grid was generated using the “Receptor grid generation” module. The binding pocket residues were manually specified which includes the hinge region, DFG loop, glycine-rich loop, and αC-helix. A grid was generated with the centroid of these residues as the center of the grid with no constraints. The ligand was prepared using the LigPrep module. Docking studies were performed using Glide module in Xtreme Precision (XP) mode and with flexible ligand sampling. The per-residue interaction scores were calculated for all the binding pocket residues and the residues within 20 Å of the grid center. The results were analyzed using XP Visualizer.
RESULTS
Preliminary Studies.
Visualization of crystal structure (PDB ID: 4C2V) revealed the kinase in the DFG “in” state, with Asp218 pointing towards the ATP binding pocket and Phe219 oppositely oriented. Sequence alignment of X. laevis AKB with human AKB revealed 87% similarity (Figure S1), with only two residues not conserved in the ATP binding site. These residues were computationally mutated to the corresponding human AKB residues. This structure was energy minimized and used as the starting point for the studies. This structure does not contain phosphorylation on Thr248 and is not bound to INCENP. Also, the DFG-Asp (Asp218) is not protonated.
Optimization of Metadynamics Parameters
In an unbiased simulation performed for 1 μs to explore the possibility of a DFG-flip, we observed no significant change in kinase conformation, indicating the need of long-scale simulations or enhanced sampling. We employed metadynamics, a tool successfully applied in biophysics and structural biology to study protein conformational changes with excellent results (37-39). To optimize the best CVs and time scales, several metadynamics simulations were performed with different combinations of CVs for different lengths of time. We determined that the DFG-flip is best observed with a combination two CVs: (1) the distance between the center of mass of DFG-Phe side chain and COM of Gln129 amide and (2) the angle defined by COM of phenyl ring of Phe219, terminal oxygen on the side chain of Asp218, and oxygen of the backbone carbonyl group of Lys215 (Figure S2). We also determined that a 200-ns metadynamics run with these CVs is optimal to observe a DFG conformational change.
Structural Changes During the DFG Flip
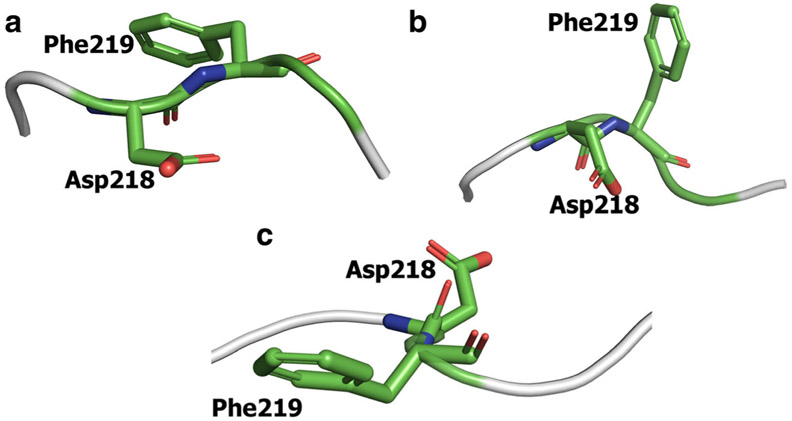
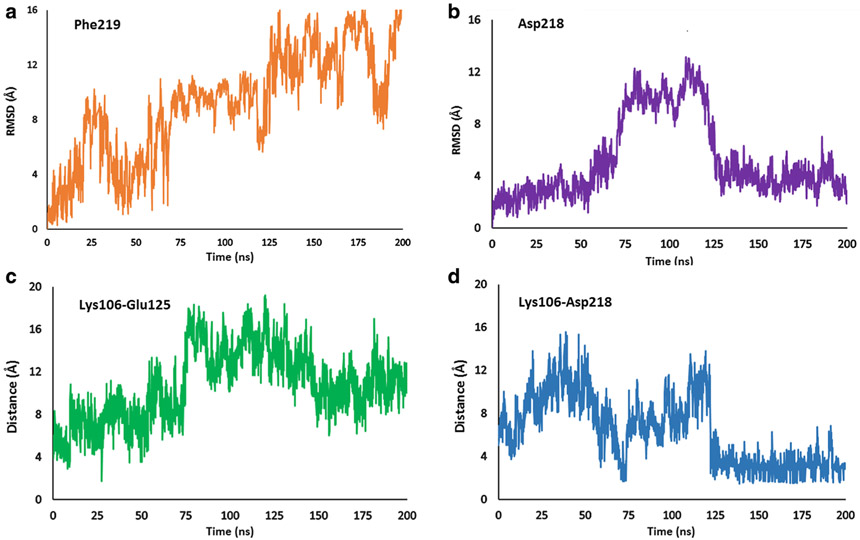
Visualization of a 200-ns metadynamics run with the abovementioned CVs exhibited complete transition of DFG motif from the “in” to “out” state. As expected, major structural changes during this transition are associated with Phe219 and Asp218. In the starting DFG “in” structure, the phenyl ring of Phe219 is oriented towards the αC-helix in the N-lobe (Fig. 2a). As the simulation proceeds, this ring gradually moves away from the αC-helix and eventually points in the direction opposite to its starting position (Fig. 2c), which corresponds to the DFG “out” conformation. The RMSD plot shows that the Phe219 side chain adopts three major conformations (Fig. 4a). Initially, Phe219 points into the allosteric cavity. It briefly enters the DFG “up” conformation from 20 to 35 ns, before coming to initial orientation pointing the αC-helix. As the simulation proceeds, the residue enters the DFG “up” conformation (Fig. 2b) and is stable in this conformation from 70 to 125 ns. From the DFG “up” state, the residue flips 90° pointing away from the allosteric pocket. A simultaneous change during this transition is the movement of Asp218. The terminal carboxyl of aspartate initially orients away from the αC-helix (Fig. 2a) and ends up in the opposite direction pointing towards the αC-helix (Fig. 2c). The RMSD plot of the terminal carboxyl of Asp218 exhibits three distinct positions of the residue (Fig. 4b). Over the course of 70 ns, the aspartate residue is close to its initial conformation pointing away from the allosteric pocket. From approximately 70–125 ns, the carboxyl enters the allosteric pocket and subsequently stabilizes towards the end of the simulation. Comparison of the initial and final structures reveals that the aspartate and phenylalanine residues of DFG swap positions during the simulation. This suggests that the metadynamics simulation successfully predicts the DFG-flip with the selected CV set and simulation parameters. The phi and psi angles of the Asp218 and Phe219 are shown in Table S1.
Fig. 2..
Orientation of Asp218 and Phe219 residues in the starting DFG “in” conformation (a) and the simulated DFG “out” conformation (b). Phe219 points towards the αC-helix in DFG “in” conformation while Asp218 points in the opposite direction. These two residues swap their positions in the DFG “out” conformation. The DFG-flip occurs through DFG “up” conformation in which Phe219 points towards the N-lobe while Asp218 points opposite to the allosteric pocket (c)
Fig. 4.
a RMSD plot of Phe219. b RMSD plot of Asp218. c Lys106 Glu125 salt-bridge distance. d Lys106-Asp218 salt bridge distance
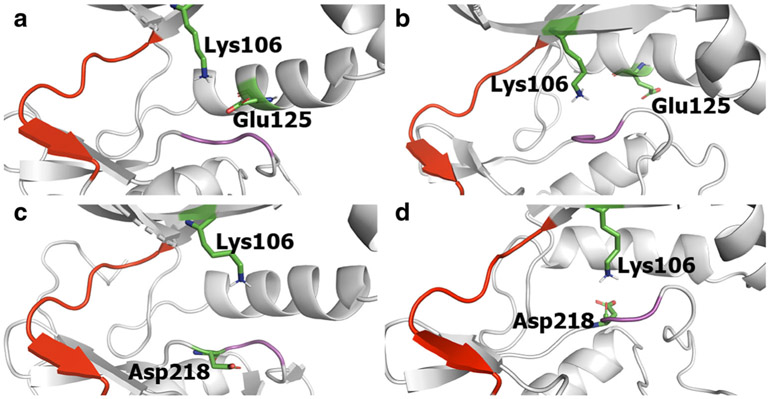
We further examined residues involved in salt-bridge interactions. Lys106 in the N-lobe is conserved across the human kinome (40,41) and is important for catalysis (42). In the DFG “in” conformation, the terminal amine of Lys106 and terminal carboxyl of Glu125 form a salt-bridge at a distance of 4.39 Å (Fig. 3a). This distance, plotted in Fig. 4c, changes as the simulation proceeds. As the kinase moves away from the DFG “in” state, the distance between these two residues increases, reaching up to 16 Å in the DFG “out” conformation (Fig. 3b).
Fig. 3.
Schematic representations depicting the proximity of Lys106 and Glu125 and of Lys106 and Asp218 in DFG “in” and “out” conformations. In the DFG “in” state, Lys106 and Glu125 are close to each other forming a salt bridge (a), which is broken in the DFG “out” state (b). Lys106 and Asp218 are far away in the DFG “in” state (c) and they move closer to each other forming a salt bridge that stabilizes the DFG “out” state (d). The hinge region is depicted in red and the DFG loop in pink
Another significant salt-bridge forms between Asp218 of the DFG motif and Lys106. In the DFG “in” conformation, these residues are 6–8 Å apart (Fig. 3c), which increases up to 15 Å as the simulation progresses. At the end of the simulation, the kinase enters the DFG “out” conformation and Asp218 and Lys106 are in close proximity (Fig. 3d). The RMSD plot (Fig. 4d) shows that the Lys106-Asp218 distance fluctuates between 2 and 4 Å.
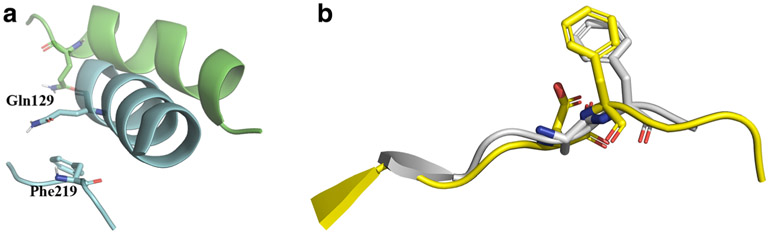
We also observed that the αC-helix moves away from the DFG motif. In the DFG “in” conformation, Gln129 is located 3.75 Å above the phenyl ring of Phe219. Also, the side chain of Gln129 is oriented such that the terminal amide points towards the ATP-binding site. As the simulation proceeds, the αC-helix moves up to 3.5–4 Å away from the DFG motif. The side chain of Gln129 also rotates finally pointing towards the solvent front.
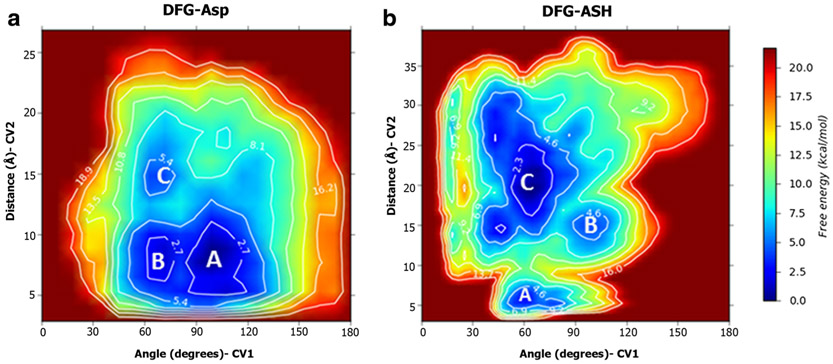
To quantify the energy difference of the conformations, we generated a free energy plot (FEP) as a function of the two CVs (Fig. 6a). We observed three minima. Two minima correspond to the DFG “in” and DFG “out” conformations, while the third minimum corresponds to the intermediate DFG “up” state, discussed in the next sub-section. These three energy wells are depicted in blue. While the DFG “in” and DFG “up” conformations have similar free energies, the free energy of the DFG “out” conformation is nearly 4 kcal/mol higher than that of the DFG “in” conformation.
Fig. 6.
2D free energy surfaces for the DFG-Asp (a) and DFG-ASH (b) ensembles, as a function of the collective variables. The DFG “in,” “up,” and “out” conformations are labelled A, B, and C respectively
DFG-Flip Occurs Through the DFG “Up” Conformation
We observed through our simulation that AKB resides in the DFG “up” conformation for a significant time during the transition from DFG “in” to “out.” In this conformation, the phenylalanine is oriented in an “up” position, which is halfway between the DFG “in” and “out” conformations (Fig. 2c). This conformation is similar to the experimentally determined AKA and AKB DFG “up” crystal structures, with Phe219 standing upright towards the N-lobe and Asp218 pointing into the allosteric pocket. The overlay of the simulated and experimentally determined DFG “up” structures is depicted in Fig. 5b.
Fig. 5.
a αC-helix moves away from the DFG motif during the flip. The side chain of Gln129 which points the ATP-binding site in the “in” state, moves away facing the solvent front to facilitate the rotation of Phe219. The “in” conformation is represented in blue ribbons and the “out” conformation in green. The DFG motif in the “in” conformation is shown for reference. b Overlay of the DFG loop of the simulated (yellow ribbon) and crystallized (gray ribbons, PDB ID: 4AF3) DFG “up” conformations
The RMSD plots and salt-bridge distance plots also suggest this conformation. In the RMSD plots of Asp218 and Phe219, the abrupt increase in the RMSD between 70 and 120 ns corresponds to the DFG “up” conformation. The free energy plot of the simulation generated as a function of the CVs shows a local minimum for the DFG “up” conformation (Fig. 6a). Unlike the DFG “in” and “out” conformations, we did not observe any salt-bridge interactions in the DFG “up” conformation. However, Phe219 is involved in hydrophobic contacts with Ile126, Leu152 and Leu140 (Figure S3).
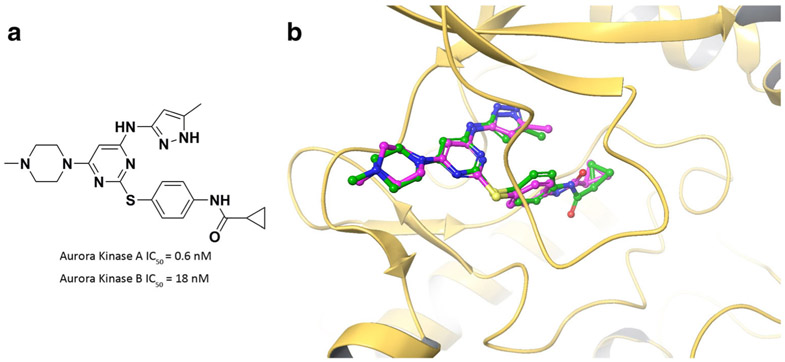
The DFG “up” conformation was previously reported in a co-crystal structure of AKB with an Aurora kinase A (AKA) inhibitor, VX-680 (PDB ID: 4AFE). VX-680 is a potent AKA inhibitor (IC50 = 0.6 nM) but is also active on AKB (IC50 = 18 nM) (43). To confirm the accuracy of our DFG “up” model, we docked VX-680 into AKB. Comparison of the crystallized pose and the docked pose exhibits perfect overlap of the ligand (Fig. 7b). This result supports that the DFG “up” model from our simulation predicts the crystallized binding pose of VX-680.
Fig. 7.
a Chemical structure of VX-680. b Overlap of docked pose (green) and crystallized pose (purple) of VX-680 in AKB DFG “up” conformation
Presence of Src-Like Inactive Conformation
An intermediate state adopted by kinases when transitioning from the DFG “in” to “out” conformation is the Src-like inactive state. In this conformation, the DFG-Asp faces away from the allosteric pocket and the DFG-Phe points towards the allosteric pocket. However, the conserved glutamate in the αC-helix points away from the DFG motif assuming a “classical-like” inactive state. In our metadynamics studies, we did not observe AKB adopting such a conformation.
Role of the Protonation State of DFG-Asp in the Flip
In some kinases like Abl and EGFR, the protonation state of DFG-Asp is known to influence the conformation adopted by the kinase (29,44). In order to examine the role of the protonation state of the DFG-Asp in AKB, we performed a metadynamics run with the AKB DFG “in” with the residues protonated (DFG-ASH). The simulation was performed for 200 ns with other parameters, such as CVs, and equilibration criteria consistent with the non-protonated simulation (DFG-Asp). We observed that the DFG-flip occurs in both cases, with Asp218 and Phe219 swapping their positions. Both transitions occur through a DFG “up” conformation. Free energy plots of the DFG-ASH system demonstrate that the DFG “out” conformation has 7 kcal/mol less energy compared to the DFG “in” state (Fig. 6b).
DISCUSSION
In this work, we have studied the conformational change of the DFG motif of Aurora kinase B. We used the DFG-in conformation as the starting structure. AKB is activated upon the occurrence of two events: (1) INCENP binds at the N-terminal of AKB, and (2) Thr248 is phosphorylated. Thus, to facilitate the transition to the inactive DFG “out” conformation, INCENP was removed from the starting structure and Thr248 was de-phosphorylated. Using unbiased simulations is not feasible, since the DFG-flip in other kinases, like EGFR, are reported to take several microseconds (29). Thus, in this work, we used an enhanced sampling method called metadynamics. A key factor deciding the success of a metadynamics run is the choice of collective variables (CV). A CV is a set of reaction co-ordinates used to enhance fluctuation and to which the Gaussian potential is added (45). Another important parameter of a metadynamics run is the length of the simulation. Running the simulation for longer times will force the protein to biologically irrelevant conformations (46).
The trajectories of the simulations were visualized focusing on the DFG motif. The rotation of Asp218 and Phe219 in opposite directions to swap their positions indicated that our simulation could predict the DFG flip successfully. In the active state, the terminal carboxyl group of Asp218 faces the ATP binding pocket. This conformation enables chelation to the Mg2+ ion. We observed that when the kinase is in DFG “in” state, the Lys106-Glu125 salt-bridge is intact. These two residues are conserved in many kinases and this salt-bridge is a key feature of several active kinases. A second salt-bridge was observed between Lys106 and Asp218. This interaction was very strong throughout the DFG “out” conformation of the kinase, indicating stabilization of the inactive state.
In the DFG “in” state, Phe219 and Gln129 are in close proximity to each other (Fig. 5a). This impedes the rotation of Phe219 by creating a steric clash, thus stabilizing the kinase in the DFG “in” state. During the simulation, as the kinase exits the DFG “in” state, the αC-helix moves away from the DFG loop. Also, the side chain of Gln129, which was initially facing the ATP binding pocket, orients itself to the solvent front. This movement could be occurring to evade the steric clash of Phe219, while rotating, with Gln129.
Kinases adopt several intermediate or transition states between the “in” and “out” conformations (22). One such conformation is the Src-like inactive state, which is also reported in other kinases including Abl (47), BTK (48), and EGFR (49). Kinases that adopt the Src-like inactive state engage in a strong salt-bridge between the conserved glutamate in the αC-helix and an arginine present two residues down the C-terminus of the DFG motif. This salt-bridge stabilizes the kinase in the Src-like inactive conformation. However, AKB lacks positively charged residues like arginine or lysine next to the DFG motif. Instead, it has a serine at this position, which cannot form a salt-bridge and consequently does not favor stabilization of the kinase in the Src-like inactive conformation.
A second intermediate state reported in kinases is the DFG “up” conformation. In one study, a triple mutant of Aurora kinase A (AKA) was crystallized in this state (36). In another study, an AKA inhibitor was co-crystallized with human AKB. Examining the crystal structure reveals the kinase to be adopting a DFG “up” conformation. In our simulation, we observed this conformation with Asp218 pointing away from the ATP-binding pocket and Phe219 halfway through its rotation. The kinase resides in this state for significant time before completing the flip to the DFG “out” state. Our simulation indicates that the DFG “up” conformation could be the transition state between DFG “in” and “out” conformations.
In the starting structure, the kinase is not bound to INCENP and Thr232 is not phosphorylated. In such conditions, DFG “out” is the thermodynamic endpoint and is expected to be at a lower energy than DFG “in.” However, the free energy diagram of the simulation showed the DFG “in” conformation at a higher energy compared to DFG “out.” We examined the influence of the protonation state of Asp218 to address this question. The role of the protonation state of DFG-Asp in kinase activation and drug binding was first reported by Kuriyan and Shaw (50). The group experimentally demonstrated a pH dependence of imatinib binding to Abl, which was lost upon DFG-Asp mutation. This supported that protonated DFG-Asp favors “out” over “in” enabling imatinib binding. Meng et al. further quantitatively explained this effect in solvent explicit free energy simulations (44). However, in similar studies examining insulin receptor kinase (51) and c-Src, the protonation state of DFG-Asp did not significantly affect the DFG-flip.
In the simulation with the DFG-Asp protonated, we observed the DFG “out” state to be at lower energy compared to DFG “in.” This indicates that the protonation state of the DFG-Asp determines the energetics of the DFG “in” to “out” conformational change. When DFG-Asp is protonated, the DFG “in” conformation is not energetically favored. On the other hand, when DFG is not protonated, the DFG “in” conformation predominates. This observation is justified by the fact that Asp218 engages Mg2+ in the active DFG “in” state. For this interaction to occur, Asp218 must be ionized to chelate the magnesium cation.
Structural insights from this work will be useful for the discovery of next generation AKB inhibitors. Kinase inhibitors have been historically classified as type I or II, based on binding modes. Type I bind to the active form of the kinase and are ATP competitive. However, type II inhibitors bind to the inactive form of the kinase and display non-competitive enzyme kinetics. This binding mode was discovered when imatinib was co-crystallized with the Abl kinase (52). Inhibitors of this class occupy both the ATP pocket and an adjacent “back” pocket created by the flip of the DFG motif from “in” to “out” and the movement of the αC-helix. Type II inhibitors are more selective and potent, which improves drug-likeness and clinical utility.
Drug discovery efforts for AKB have targeted the active conformation of the kinase. The limited success of these projects can stem from a lack of structural information to help target the inactive, type II conformation. To further study the conformational space of AKB, we opted for an alternative approach of computationally simulating other possible conformations of the kinase, which can be targeted for drug discovery. The reported structural characterizations will be useful for designing next generation inhibitors. For instance, the flip of Phe219 creates a back pocket and new inhibitors can be generated with functional groups that exploit this pocket. Also, the Asp218 side chain points into the newly formed allosteric pocket and new inhibitors can be designed to engage the side chain. Similarly, Lys106, which was interacting with Glu125 in the DFG “in” conformation, interacts with Asp218. The orientation of these residues provides important direction for structure guided discovery of type II AKB inhibitors. Type II inhibitors have the advantage of (1) exploiting a less-conserved back pocket and (2) are ATP non-competitive. We demonstrated the ability of our DFG “up” model to accurately predict the binding pose of VX-680. Next-generation AKB inhibitors can be screened against our model to identify new candidates with improved properties for clinical development.
CONCLUSION
In conclusion, using metadynamics, we simulated the DFG “out” conformation of AKB from the experimentally determined DFG “in” conformation. We also characterized the structural changes during the DFG-flip of AKB, which are consistent with previous kinase structural research. Further, we predict that the transition from “in” to “out” occurs through a previously reported DFG “up” state. This widens the spectrum of AKB conformations that could be targeted in drug discovery projects. Our studies also reveal the crucial role of protonation of the DFG-Asp residue, which directly contributes to catalytic activity. In an unprotonated state, the DFG “in” conformation is favored and the kinase is catalytically active. Conversely, in the protonated state, the DFG “out” conformation is favored and the kinase is not catalytically active. The results of this study will be useful in the design of next generation AKB inhibitors.
Supplementary Material
Acknowledgments
FUNDING INFORMATION
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number “P20 GM109005.” BF was supported by the UAMS Seeds of Science Grant, the UAMS COP Seed Grant, and a grant from the American Thyroid Association (ATA/Thyca). HL was supported by NIH grants (NIH 1R01CA194094 and 1R01CA197178). MB was supported by Inglewood Scholars Program and NIA/NIH grant 2P01AG012411-17A1 (W.S.T. Griffin, P.I.)
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1208/s12248-019-0399-6) contains supplementary material, which is available to authorized users.
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
REFERENCES
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. [DOI] [PubMed] [Google Scholar]
- 2.Nolen B, Taylor S, Ghosh G. Review regulation of protein kinases: controlling activity through activation segment conformation. Mol Cell. 2004;15:661–75. [DOI] [PubMed] [Google Scholar]
- 3.Hunter T. A thousand and one protein kinases Cell. Elsevier; 1987;50:823–9. [DOI] [PubMed] [Google Scholar]
- 4.Kornev AP, Haste NM, Taylor SS, Ten Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–58. [DOI] [PubMed] [Google Scholar]
- 6.Endicott JA, Noble MEM, Johnson LN. The structural basis for control of eukaryotic protein kinases. Annu Rev Biochem. 2012;81:587–613. [DOI] [PubMed] [Google Scholar]
- 7.Cohen P. The origins of protein phosphorylation. Nat Cell Biol. 2002;4:E127–30. [DOI] [PubMed] [Google Scholar]
- 8.Dar AC, Shokat KM. The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem. 2011;80:769–95. [DOI] [PubMed] [Google Scholar]
- 9.Vader G, Medema RH, Lens SMA. The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol. 2006;173:833–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol. 2012;13:789–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeshita M, Koga T, Takayama K, Ijichi K, Yano T, Maehara Y, et al. Aurora-B overexpression is correlated with aneuploidy and poor prognosis in non-small cell lung cancer. Lung Cancer. 2013;80:85–90. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Jiang C, Li H, Lv F, Li X, Qian X, et al. Elevated Aurora B expression contributes to chemoresistance and poor prognosis in breast cancer. Int J Clin Exp Pathol. 2015;8:751–7. [PMC free article] [PubMed] [Google Scholar]
- 13.Tuncel H, Shimamoto F, Kaneko Guangying Qi H, Aoki E, Jikihara H, Nakai S, et al. Nuclear Aurora B and cytoplasmic Survivin expression is involved in lymph node metastasis of colorectal cancer. Oncol Lett. 2012;3:1109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fadri-Moskwik M, Weiderhold KN, Deeraksa A, Chuang C, Pan J, Lin S-H, et al. Aurora B is regulated by acetylation/deacetylation during mitosis in prostate cancer cells. FASEB J. 2012;26:4057–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chieffi P, Cozzolino L, Kisslinger A, Libertini S, Staibano S, Mansueto G, et al. Aurora B expression directly correlates with prostate cancer malignancy and influence prostate cell proliferation. Prostate. 2006;66:326–33. [DOI] [PubMed] [Google Scholar]
- 16.Hartsink-Segers SA, Zwaan CM, Exalto C, Luijendijk MWJ, Calvert VS, Petricoin EF, et al. Aurora kinases in childhood acute leukemia: the promise of aurora B as therapeutic target. Leukemia. 2013;27:560–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porcelli L, Guida G, Quatrale AE, Cocco T, Sidella L, Maida I, et al. Aurora kinase B inhibition reduces the proliferation of metastatic melanoma cells and enhances the response to chemotherapy. J Transl Med. 2015;13:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamauchi T, Uzui K, Shigemi H, Negoro E, Yoshida A, Ueda T. Aurora B inhibitor barasertib and cytarabine exert a greater-than-additive cytotoxicity in acute myeloid leukemia cells. Cancer Sci. 2013;104:926–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marampon F, Gravina GL, Popov VM, Scarsella L, Festuccia C, La Verghetta ME, et al. Close correlation between MEK/ERK and Aurora-B signaling pathways in sustaining tumorigenic potential and radioresistance of gynecological cancer cell lines. Int J Oncol. 2014;44:285–94. [DOI] [PubMed] [Google Scholar]
- 20.Wu X, Liu W, Cao Q, Chen C, Chen Z, Xu Z, et al. Inhibition of Aurora B by CCT137690 sensitizes colorectal cells to radiotherapy. J Exp Clin Cancer Res. 2014;33:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treiber Dk, Shah NP. Ins and outs of kinase DFG Motifs. Chem Biol. 2013;20:745–6. [DOI] [PubMed] [Google Scholar]
- 22.Roskoski R. Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacol Res. 2016;103:26–48. [DOI] [PubMed] [Google Scholar]
- 23.Tang A, Gao K, Chu L, Zhang R, Yang J, Zheng J. Aurora kinases: novel therapy targets in cancers. Oncotarget. 2017;8:23937–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sessa F, Villa F. Structure of Aurora B-INCENP in complex with barasertib reveals a potential transinhibitory mechanism. Acta Crystallogr F Struct Biol Commun. 2014;70:294–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y, Shan S, Li Z-B, Xin L-J, Pan D-S, Yang Q-J, et al. CS2164, a novel multi-target inhibitor against tumor angiogenesis, mitosis and chronic inflammation with anti-tumor potency. Cancer Sci. 2017;108:469–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sessa F, Mapelli M, Ciferri C, Tarricone C, Areces LB, Schneider TR, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–91. [DOI] [PubMed] [Google Scholar]
- 27.Sini P, Gurtler U, Zahn SK, Baumann C, Rudolph D, Baumgartinger R, et al. Pharmacological profile of BI 847325, an orally bioavailable, ATP-competitive inhibitor of MEK and Aurora kinases. Mol Cancer Ther. 2016;15:2388–98. [DOI] [PubMed] [Google Scholar]
- 28.Shaw DE, Maragakis P, Lindorff-Larsen K, Piana S, Dror RO, Eastwood MP, et al. Atomic-level characterization of the structural dynamics of proteins. Science. 2010;330:341–6. [DOI] [PubMed] [Google Scholar]
- 29.Shan Y, Arkhipov A, Kim ET, Pan AC, Shaw DE. Transitions to catalytically inactive conformations in EGFR kinase. Proc Natl Acad Sci USA. 2013;110:7270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–51. [Google Scholar]
- 31.Meng Y, Gao C, Clawson DK, Atwell S, Russell M, Vieth M, et al. Predicting the conformational variability of Abl tyrosine kinase using molecular dynamics simulations and Markov state models. J Chem Theory Comput. 2018;14:2721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y, Li X, Ma W, Dong Z. Conformational transition pathways of epidermal growth factor receptor kinase domain from multiple molecular dynamics simulations and Bayesian clustering. J Chem Theory Comput. 2014;10:3503–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Niu H, Li Y, Chu H, Shen H, Zhang D, et al. Mechanistic insight into the functional transition of the enzyme guanylate kinase induced by a single mutation. Sci Rep. 2015;5:8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laio A, Parrinello M. Escaping free-energy minima. Proc Natl Acad Sci USA. 2002;99:12562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tribello GA, Ceriotti M, Parrinello M. A self-learning algorithm for biased molecular dynamics. Proc Natl Acad Sci USA. 2010;107:17509–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dodson CA, Kosmopoulou M, Richards MW, Atrash B, Bavetsias V, Blagg J, et al. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem J. 2010;427:19–28. [DOI] [PubMed] [Google Scholar]
- 37.Bian Y, Zhang J, Wang J, Wang W. On the accuracy of metadynamics and its variations in a protein folding process. Mol Simul. 2015;41:752–63. [Google Scholar]
- 38.Park J, McDonald JJ, Petter RC, Houk KN. Molecular dynamics analysis of binding of kinase inhibitors to WT EGFR and the T790M Mutant. J Chem Theory Comput. 2016;12:2066–78. [DOI] [PubMed] [Google Scholar]
- 39.Callegari D, Lodola A, Pala D, Rivara S, Mor M, Rizzi A, et al. Metadynamics simulations distinguish short- and long-residence-time inhibitors of cyclin-dependent kinase 8. J Chem Inf Model. 2017;57:159–69. [DOI] [PubMed] [Google Scholar]
- 40.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knight JDR, Qian B, Baker D, Kothary R. Conservation, variability and the modeling of active protein kinases Fugmann S, editor. PLoS One. 2007;2:e982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carrera AC, Alexandrov K, Roberts TM. The conserved lysine of the catalytic domain of protein kinases is actively involved in the phosphotransfer reaction and not required for anchoring ATP. Proc Natl Acad Sci USA. 1993;90:442–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10:262–7. [DOI] [PubMed] [Google Scholar]
- 44.Meng Y, Lin Y, Roux B. Computational study of the “DFG-Flip” conformational transition in c-Abl and c-Src tyrosine kinases. J Phys Chem B. 2015;119:1443–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ensing B, De Vivo M, Liu Z, Moore P, Klein ML. Metadynamics as a tool for exploring free energy landscapes of chemical reactions. Acc Chem Res. 2006;39(2):73–81. [DOI] [PubMed] [Google Scholar]
- 46.Barducci A, Bussi G, Parrinello M. Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys Rev Lett. 2008;100:020603. [DOI] [PubMed] [Google Scholar]
- 47.Levinson nM, Kuchment O, Shen K, Young MA, Koldobskiy M, Karplus M, et al. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sultan MM, Denny RA, Unwalla R, Lovering F, Pande VS. Millisecond dynamics of BTK reveal kinome-wide conformational plasticity within the apo kinase domain. Sci Rep. 2017;7:15604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–49. [DOI] [PubMed] [Google Scholar]
- 50.Shan Y, Seeliger MA, Eastwood MP, Frank F, Xu H, Jensen MØ, et al. A conserved protonation-dependent switch controls drug binding in the Abl kinase. Proc Natl Acad Sci USA. 2009;106:139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vashisth H, Maragliano L, Abrams CF. DFG-flip in the insulin receptor kinase is facilitated by a helical intermediate state of the activation loop. Biophys J. 2012;102:1979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Structural mechanism for STI-571 inhibition of Abelson tyrosine kinase. Science. 2000;289:1938–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.