Abstract
Although some previous studies have examined epigenomic alterations in lung adenocarcinomas, correlations between epigenomic events and genomic driver mutations have not been fully elucidated. Single-CpG resolution genome-wide DNA methylation analysis with the Infinium HumanMethylation27 BeadChip was performed using 162 paired samples of adjacent normal lung tissue (N) and the corresponding tumorous tissue (T) from patients with lung adenocarcinomas. Correlations between DNA methylation data on the one hand and clinicopathological parameters and genomic driver mutations, i.e. mutations of EGFR, KRAS, BRAF and HER2 and fusions involving ALK, RET and ROS1, were examined. DNA methylation levels in 12 629 probes from N samples were significantly correlated with recurrence-free survival. Principal component analysis revealed that distinct DNA methylation profiles at the precancerous N stage tended not to induce specific genomic driver aberrations. Most of the genes showing significant DNA methylation alterations during transition from N to T were shared by two or more driver aberration groups. After small interfering RNA knockdown of ZNF132, which showed DNA hypermethylation only in the pan-negative group and was correlated with vascular invasion, the proliferation, apoptosis and migration of cancer cell lines were examined. ZNF132 knockdown led to increased cell migration ability, rather than increased cell growth or reduced apoptosis. We concluded that DNA hypermethylation of the ZNF132 gene participates in the clinicopathological aggressiveness of ‘pan-negative’ lung adenocarcinomas. In addition, DNA methylation alterations at the precancerous stage may determine tumor aggressiveness, and such alterations that accumulate after driver mutation may additionally modify clinicopathological features through alterations of gene expression.
Methylome analysis has revealed that DNA methylation alterations at the precancerous stage may determine the aggressiveness of lung adenocarcinomas and that such alterations accumulating after driver mutation, e.g. epigenetic silencing of ZNF132, additionally modify the clinicopathological features.
Introduction
It is well known that epigenomic alterations play a key role in carcinogenesis in various human organs (1–3). Aberrant DNA methylation is one of the most important epigenomic alterations resulting in chromosomal instability and altered expression of tumor-related genes (4). In the context of lung adenocarcinoma, various groups including The Cancer Genome Atlas (TCGA) (5) have reported the results of methylome analysis using appropriate screening tools, such as the Infinium assay (6), for large numbers of human tissue samples (7–9). For example, we have shown that epigenomic clustering of lung adenocarcinomas based on their genome-wide DNA methylation profiles is significantly correlated with carcinogenetic background factors (10) such as cigarette smoking and chronic obstructive pulmonary disease (11). In addition, as in cancers of other organs (12–14), it has been revealed that the DNA methylation profiles of lung adenocarcinomas are significantly correlated with clinicopathological aggressiveness and poorer prognosis (15,16).
On the other hand, various genomic alterations, such as mutations of the EGFR, KRAS, BRAF and HER2 genes and fusions involving the ALK, RET and ROS1 genes are known to be mutually exclusive driver aberrations that are amenable to molecular targeted therapies (5,17). Genomic driver aberrations are known to be essential for growth, but alone they are insufficient for malignant progression of cancers through metastasis, for which molecular alterations other than driver aberrations, such as epigenomic lesions, are important (18). Therefore, in individual patients, epigenomic analyses and driver mutation screening are necessary to explain the clinicopathological diversity of lung adenocarcinomas.
As DNA methylation alterations are observed even in non-cancerous lung tissue of patients with lung adenocarcinomas, which have already be exposed to carcinogenetic factors such as cigarette smoking and chronic obstructive pulmonary disease (11,19), we have focused on the correlations between DNA methylation profiles at the precancerous stage and subsequent driver mutations. In addition, during multistage carcinogenesis, epigenomic aberrations may further accumulate in the established cancers even after driver mutations have occurred. Since genomic polymorphism is known to affect DNA methylation profiles (20), the relationship between the driver mutation profile and the DNA methylation profile that accumulates in the later stage of carcinogenesis has also been a focus of interest. In the present study, based on the results of the Infinium assay using 364 lung tissue samples from patients who had undergone screening of genomic driver aberrations, we examined the significance of DNA methylation profiles at the early and later stages of multistage lung adenocarcinogenesis.
Materials and methods
Patients and tissue samples
We employed 162 samples of adjacent normal lung tissue (N) and 162 samples of the corresponding tumorous tissue (T) obtained from patients with primary lung adenocarcinomas who underwent lung resection at the National Cancer Center Hospital, Japan. All of the patients had undergone complete resection and none had received any preoperative treatment or adjuvant therapy after surgery. The patients comprised 85 males and 77 females, and their median age was 60 years (range, 30–76 years). Histological diagnosis was based on the World Health Organization classification (21), and the follow-up period ranged from 65 to 4904 days (mean, 1653 days) after surgery. Recurrence was diagnosed by clinicians on the basis of physical examination and imaging modalities such as computed tomography, magnetic resonance imaging and positron emission tomography, and was sometimes confirmed histopathologically by biopsy. The clinicopathological parameters of the 162 patients are summarized in Supplementary Table 1, available at Carcinogenesis Online. A proportion of this cohort had also been included in our previous studies focusing on the correlation with smoking history (11) and recurrence-related genes (16).
All of the 162 cases were screened for EGFR, KRAS, BRAF and HER2 hot spot mutations by the high resolution melt method, and for EML4- and KIF5B-ALK, KIF5B- and CCDC6-RET, and CD74-, EZR- and SLC34A2-ROS1 fusions by reverse transcription (RT)–PCR, as described previously (17).
Tissue samples were taken immediately after surgery, and then frozen and stored in liquid nitrogen at the National Cancer Center Biobank, Japan, until analysis in accordance with the ‘Japanese Society of Pathology Guidelines for the handling of pathological tissue samples for genomic research’ (22). This study was approved by the Ethics Committees of Keio University School of Medicine and the National Cancer Center, Tokyo, Japan, and was performed in accordance with the Declaration of Helsinki. All patients included in this study provided written informed consent for use of their materials and data.
Infinium assay
Genomic DNA was extracted from all tissue samples and cell lines using a QIAamp DNA Mini kit (Qiagen, Hilden, Germany) or phenol–chloroform extraction followed by dialysis. Five-hundred-nanogram aliquots of DNA were subjected to bisulfite conversion using an EZ DNA Methylation-Gold Kit (Zymo Research, Irvine, CA). DNA methylation status at 27 578 CpG loci was examined at single-CpG resolution using the Infinium HumanMethylation27 Bead Array (Illumina, San Diego, CA). After hybridization, the specifically hybridized DNA was fluorescence-labeled by a single-base extension reaction and detected using a BeadScan reader (Illumina) in accordance with the manufacturer’s protocols. The data were then assembled using GenomeStudio methylation software (Illumina). At each CpG site, GenomeStudio provided the so-called β-value, which is the ratio of the fluorescence signal measured using a methylated probe relative to the sum of those measured using methylated and unmethylated probes. These data were deposited in the Integrative Disease Omics Database (iDOx DB, https://gemdbj.ncc.go.jp/omics/). In accordance with a previous study (23), the M-value was calculated by logit transformation of the β-value and used for further statistical analysis.
Gene ontology enrichment analysis
Gene ontology (GO) enrichment analysis was conducted using genes showing DNA methylation alterations during lung adenocarcinogenesis by the GeneGO MetaCore™ software package (version 6.7) (Thomson Reuters, NY), an analytical tool based on a proprietary manually curated database. Such genes were considered significantly enriched in protein classes for which the P value was less than 0.0001.
Quantitative analysis of DNA methylation using pyrosequencing
The PCR and sequencing primers for the promoter regions of the TRIM58 and ZNF132 genes were designed using PSQ Assay Design Software Version 1.0.6 (Biotage, Uppsala, Sweden). To overcome any PCR bias, we optimized the PCR conditions: 0, 50 and 100% of the fully methylated control DNA (Epitect methylated human control DNA, QIAGEN) was used as a template, as described previously (24), and the linearity of the measured values and their consistency with the theoretical values were confirmed. As a result of this optimization experiment, both PCR reactions were performed using HotStarTaq DNA polymerase (QIAGEN). The further optimized PCR conditions for each primer set are summarized in Supplementary Table 2, available at Carcinogenesis Online. The biotinylated PCR product was captured on streptavidin-coated beads, and quantitative sequencing was performed on a PyroMark Q24 (QIAGEN) using the Pyro Gold Reagents (QIAGEN). The experiment was conducted in triplicate and the mean DNA methylation levels for each set of three experiments were used as quantitative values.
In addition to the exact Infinium probe CpG sites, i.e. cg20855565 (position 1) for the TRIM58 gene and cg13877915 (position 1) for the ZNF132 gene, DNA methylation data for neighboring CpG sites (positions 2 and 3) within the promoter regions were obtained for both the TRIM58 and ZNF132 genes by pyrosequencing, as shown in Supplementary Table 2, available at Carcinogenesis Online. Then mean values for positions 1–3 were considered to be the DNA methylation levels (%) of the promoter region for each gene obtained by pyrosequencing.
Cell lines
The lung adenocarcinoma cell lines PC-9 (25), H1975 (26) and A549 (27) and a hepatocellular carcinoma cell line JHH-7 (28) were purchased from Immuno-Biological Laboratories (Fujioka, Japan), the American Type Culture Collection (Manassas, VA) and Japanese Collection of Research Bioresources (Osaka, Japan), and authenticated based on short tandem repeat analysis by Japanese Collection of Research Bioresources Cell Bank (certification number, KBN0734 and KBN0752) in June and October 2020. PC-9, H1975 and A549 were maintained in RPMI 1640 medium (Gibco-Thermo Fisher Scientific, Waltham, MA) and JHH-7 was maintained in William’s E medium (Gibco-Thermo Fisher Scientific), supplemented with 10% fetal bovine serum, under 95% air and 5% CO2 at 37°C.
5-Aza-2′-deoxycytidine (5-aza-dC) treatment of cell lines
A549 and JHH-7 cells were seeded at a density of 9 × 105 cells per 15-cm dish on day 0 and then allowed to attach for 24 h. Then, 5-aza-dC (Sigma–Aldrich, St. Louis, MO) was added to a final concentration of 1 µM. Cells were passaged at a subculture ratio of 1:2 on day 3. At 24 h after replacing, 5-aza-dC was added again to the same final concentration. Genomic DNA and total RNA were extracted from both cell lines on day 3 or 6.
Quantitative real-time RT–PCR analysis
Total RNA was extracted from the cell lines using TRIzol reagent (Life Technologies, Carlsbad, CA). cDNA was then synthesized from the total RNA with random primers using Super-Script IV Reverse Transcriptase (Thermo Fisher Scientific) and preamplified using TaqMan PreAmp Master Mix (Thermo Fisher Scientific). Levels of mRNA expression for ZNF132 were analyzed using a custom TaqMan expression Assay probe (Hs01036387_m1, Thermo Fisher Scientific) on the 7500 Fast Real-Time PCR System (Thermo Fisher Scientific) employing the relative standard curve method. Experiments were performed in triplicate, and the mean value was used as the CT value. The CT value was normalized to that of GAPDH (Hs02758991_g1, Thermo Fisher Scientific) in the same sample.
Transfection with small interfering RNA
H1975 and PC-9 cells were seeded in 96-well plates at a concentration of 1 × 104 cells/well. When the cells had reached about 60% confluence, the medium was replaced with Opti-MEM® I Reduced Serum Medium (Thermo Fisher Scientific). The cells were then transfected with either the negative control siRNA (siNC) or a ZNF132-specific small interfering RNA (siRNA) (s15162 and s15163) (Thermo Fisher Scientific) using Lipofectamine™ RNAiMAX reagent (Thermo Fisher Scientific). At 48 h after transfection, the levels of expression of mRNAs for ZNF132 were determined by quantitative real-time RT–PCR analysis, using GAPDH as the reference gene. The transfected cells were then subjected to cell proliferation, apoptosis and cell migration assays.
Western blotting
Cultured cells were lysed in radioimmunoprecipitation assay buffer containing proteinase inhibitors (sc-24948, Santa Cruz Biotechnology), and loaded on 12.5% polyacrylamide gel. Proteins were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. The membranes were immunoblotted with antihuman ZNF132 rabbit polyclonal antibody (PA5-41096, 1:500 dilution, Invirtogen, Carlsbad, CA) or antihuman β-actin rabbit monoclonal antibody (13E5, 1:1000 dilution, Cell Signaling Technology, Danvers, MA) and reacted with horseradish peroxidase-conjugated goat antibody (antirabbit IgG, 7074, Cell Signaling Technology), followed by incubation with SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific).
MTS cell viability assay
The 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell viability assay was performed as described previously (29). Briefly, 48 h after transfection with negative control or ZNF132-specific siRNAs, cells were treated with CellTiter 96 Aqueous One Solution Reagent (Promega, Madison, WI). After 1 h of treatment, the optical density was measured at 490 nm on a GloMax®-Multi+ Detection System Glomax (Promega). Results were presented as the mean ± standard deviation for three separate determinations.
Apoptosis assay
The apoptosis assay was performed as described previously (29). Briefly, 48 h after transfection with negative control or ZNF132-specific siRNAs, cells were treated with a Caspase-Glo 3/7 assay kit (Promega). After 1 h of incubation, the luminescent signal was measured on a GloMax-Multi+ Detection System Glomax (Promega). Results were presented as the mean ± standard deviation for three separate determinations.
Cell migration assay
Cell migration was determined using 24-well transwell chambers with an 8-µm pore polycarbonate filter (Corning, Corning, NY). Forty-eight hours after transfection with the negative control or ZNF132-specific siRNAs, 5 × 104 H1975 and PC-9 cells were seeded onto the upper-side transwells in 100 µl of serum-free medium, and 500 µl of the complete medium was added to the lower chamber. The cells were incubated to allow migration for 48 h at 37°C and 5% CO2. At the end of the assay, the non-motile cells on the top surface of the inserts were removed with cotton swabs. Cells that had passed through the polycarbonate membrane were fixed with 10% formalin and stained with 0.5% crystal violet to visualize the attached cells. The crystal violet was eluted with 10% acetic acid and the optical density was measured at 600 nm on a GloMax Multi Detection System (Promega). Results were presented as the mean ± standard deviation for three separate determinations.
Statistics
In the Infinium assay, all CpG sites on chromosomes X and Y were excluded to avoid any gender-specific methylation bias. The call proportions (P values for detection of signals above the background <0.01) for 42 probes in all of the tissue samples examined were less than 90%. As such a low proportion may have been attributable to polymorphism at the probe CpG sites, these 42 probes were excluded from the present assay as described previously (30,31), leaving a final total of 26 444 autosomal CpG sites.
When the false discovery rate (FDR) of q was less than 0.01, differences in the M-values of Infinium probes between sample groups were considered significant. The receiver operating characteristic curve was generated and the Youden index (32) was calculated in order to discriminate recurrence-positive patients from negative patients. Recurrence-free survival curves were generated by the Kaplan–Meier method, and differences were compared by the log-rank test. The DNA methylation profiles were analyzed using principal component analysis. Correlations between DNA methylation levels and clinicopathological parameters were examined using variance between groups (analysis of variance) and Welch’s t-test at a significance level of FDR <0.5. Subsequently, multivariate analyses were performed using logistic regression. The mRNA expression levels, optical density assessed by MTS and cell migration assays, and luminescence for the apoptosis assay were examined by Welch’s t-test at a significance level of P < 0.05. All statistical analyses were performed using programming language R.
Results
Prognostic impacts of DNA methylation profiles
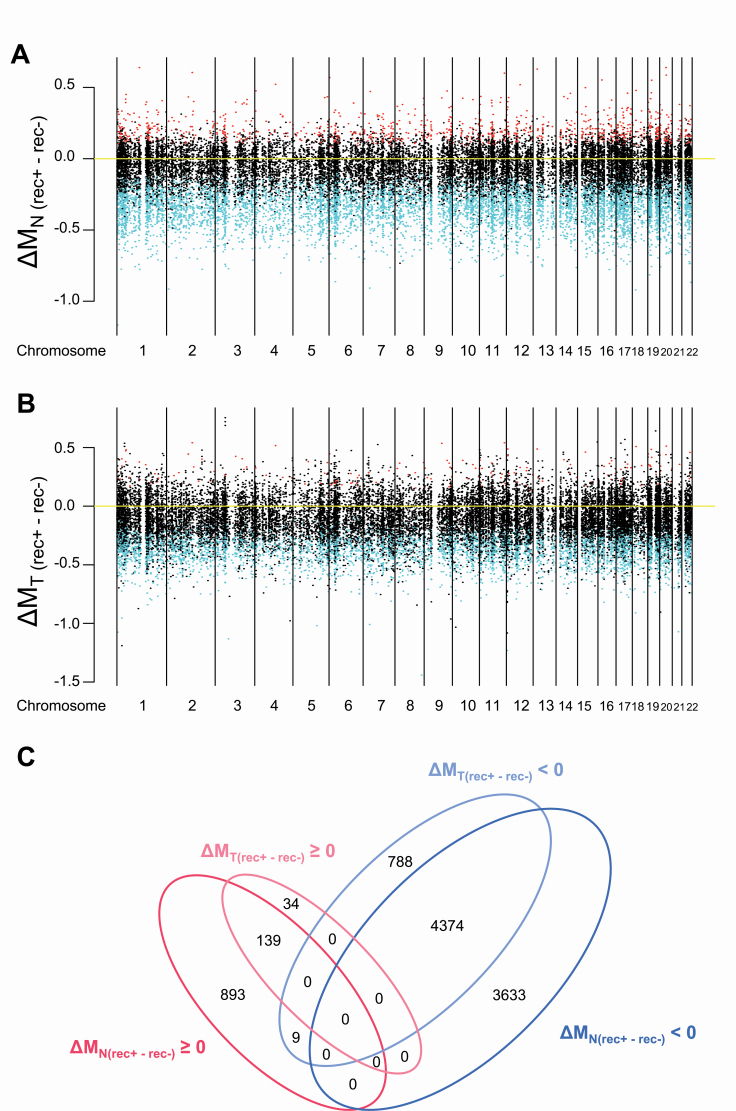
Differences in the M-value for each of the 26 444 autosomal probe CpG sites in 162 N and 162 T samples were examined between recurrence-negative cases (n = 39) and recurrence-positive cases (n = 123) (FDR <0.01). In N samples, 9048 probes, designed for 7132 genes, showed significant differences between patients who were positive and negative for recurrence (Supplementary Table 3A, available at Carcinogenesis Online), indicating that a distinct DNA methylation profile with a prognostic impact had already been established at the precancerous N stage. As shown in Figure 1A, 1041 probes (red dots) and 8007 probes (blue dots) showed DNA hypermethylation and DNA hypomethylation in recurrence-positive patients relative to recurrence-negative patients, respectively.
Figure 1.
Correlations between DNA methylation profiles and recurrence in samples of adjacent normal lung tissue (N) (A) and the corresponding tumorous tissue (T) (B) from patients with lung adenocarcinomas. All 26 444 probes are shown along the chromosome. Red and blue indicate probes showing significantly higher and lower DNA methylation levels observed in recurrence-positive (rec+) patients when compared with those in recurrence-negative (rec−) patients, respectively (FDR <0.01). (C) A Venn diagram summarizing the number of probes showing significantly higher and lower DNA methylation levels observed in recurrence-positive (rec+) patients relative to those in recurrence-negative (rec−) patients in panels (A) and (B). ΔM (rec+ − rec−) ≧ 0 means DNA hypermethylation (shown in red) and ΔM (rec+ − rec−) < 0 means DNA hypomethylation (shown in blue) in rec+ patients relative to rec− patients. Most of the DNA methylation alterations in T samples were shared with Ns and the number of probes showing DNA methylation alterations unique to Ts was limited.
In T samples, 5344 probes, designed for 4477 genes, showed significant differences between recurrence-positive and recurrence-negative patients (Supplementary Table 3B, available at Carcinogenesis Online). As shown in Figure 1B, 173 probes (red dots) and 5171 probes (blue dots) showed DNA hypermethylation and DNA hypomethylation in recurrence-positive patients relative to recurrence-negative patients, respectively. Among the 5344 probes showing a significant correlation with recurrence in T samples, 4513 also revealed such a correlation even in N samples.
A Venn diagram summarizing the number of signatures from Figure 1A and B is shown as Figure 1C. This indicates that most of the DNA methylation alterations in T samples were shared with Ns and that the number of probes showing DNA methylation alterations unique to Ts was limited, suggesting that DNA methylation profiles at the precancerous N stages were inherited by the tumors themselves.
In addition, we compared the M-values of the present 162 N and 162 T samples with those of 32 samples of normal control lung tissue (C) obtained from patients without lung cancers, who may not have been exposed to lung carcinogenetic factors [the C samples had been analyzed and published in our previous paper (11)]. Representative probes showing ordered differences in M-values from C to N, and then to T samples (P < 0.05 by Jonckheere–Terpstra trend test), are shown in Supplementary Figure 1, available at Carcinogenesis Online. Supplementary Figure 1, available at Carcinogenesis Online again indicates that DNA methylation alterations in N samples relative to control C samples were inherited by or strengthened in T samples.
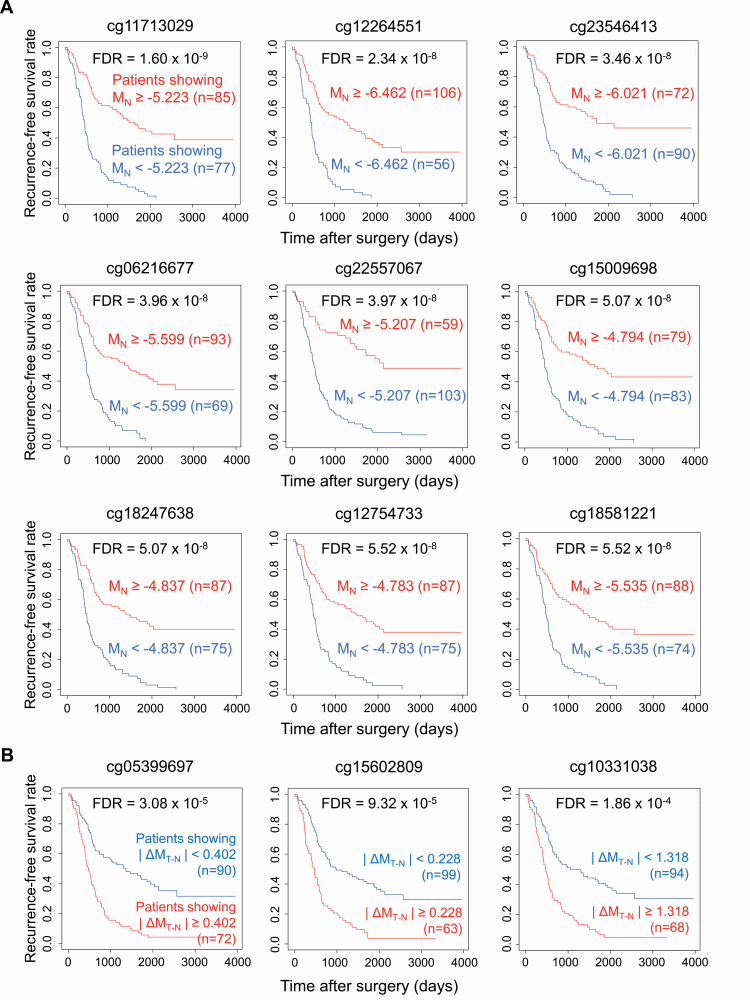
The prognostic impact of DNA methylation alterations at the precancerous N stage was further examined. Receiver operating characteristic curves were generated for all 26 444 probe CpG sites using MN values. Youden indices were calculated to discriminate recurrence-positive from recurrence-negative patients. Recurrence-free survival curves for patients belonging to groups with higher (MN ≥ Youden index) and lower (MN < Youden index) M-values were generated by the Kaplan–Meier method. For 12 629 probes, designed for 8984 genes, the log-rank test showed that recurrence-free survival differed significantly between patients showing higher and lower DNA methylation levels in N samples (FDR <0.01) (Supplementary Table 4A, available at Carcinogenesis Online): the Kaplan–Meier curves for nine representative probes are shown in Figure 2A.
Figure 2.
Prognostic impact of DNA methylation profiles at the precancerous stage (A) and DNA methylation alterations during progression from the precancerous stage to established cancers (B). (A) Representative Kaplan–Meier survival curves of patients showing higher (MN ≥ Youden index based on receiver operating characteristic curves discriminating recurrence-positive patients from recurrence-negative patients) and lower (MN < Youden index) DNA methylation levels in adjacent normal lung tissue (N) samples. (B) Representative Kaplan–Meier survival curves of patients showing larger (|ΔMT–N| ≥ |Youden index|) and smaller (|ΔMT–N| < |Youden index|) differences in DNA methylation levels between N and the corresponding tumorous tissue (T) samples.
Next, to examine the significance of DNA methylation alterations during the N to T transition stage, receiver operating characteristic curves were generated for all 26 444 probe CpG sites using the ΔMT–N values. Recurrence-free survival curves for patients belonging to groups showing larger (|ΔMT–N| ≥ |Youden index|) and smaller (|ΔMT–N| < |Youden index|) differences were generated by the Kaplan–Meier method. For only 130 probes, designed for 124 genes, the log-rank test revealed that recurrence-free survival differed significantly between patients showing larger and smaller differences during the N to T transition (FDR <0.01) (Supplementary Table 4B, available at Carcinogenesis Online): the Kaplan–Meier curves for three representative probes are shown in Figure 2B.
Correlations between DNA methylation profiles and genomic driver aberrations
M-values for all 26 444 probes in all 162 N samples were analyzed using principal component analysis.
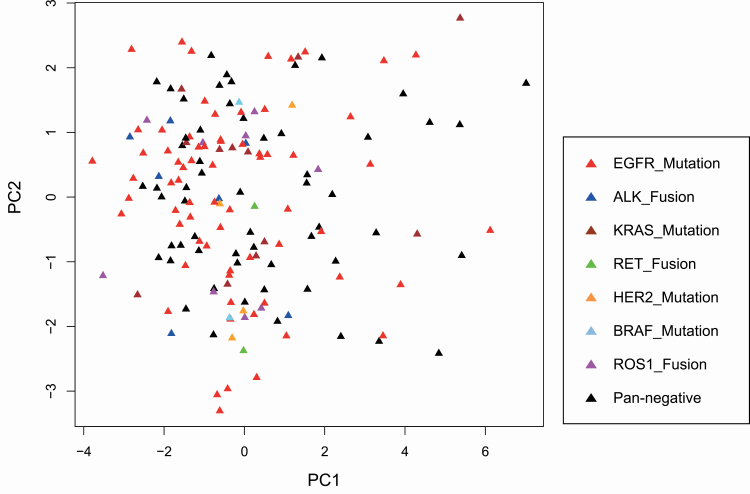
The patients showing each genomic driver aberration, i.e. mutations of the EGFR, KRAS, BRAF and HER2 genes and fusions including the ALK, ROS1 and RET genes, were scattered on the plot (Figure 3), indicating that the distinct DNA methylation profiles at the precancerous N stage did not appear to induce any specific genomic driver aberrations.
Figure 3.
Principal component analysis using DNA methylation levels of all 26 444 probes in adjacent normal lung tissue samples from patients with lung adenocarcinomas harboring EGFR mutation (red) (n = 68), ALK fusion (blue) (n = 7), KRAS mutation (brown) (n = 12), ROS1 fusion (pink) (n = 9), HER2 mutation (orange) (n = 4), BRAF mutation (light blue) (n = 2), RET fusion (green) (n = 2) and none (pan-negative cases, black) (n = 58). The horizontal and vertical axes show the first (PC1) and second (PC2) principal components, respectively.
Next, the correlation between the DNA methylation profile and genomic driver aberrations was examined during the N to T transition. In patients with EGFR mutation (n = 68), ALK fusion (n = 7), KRAS mutation (n = 12), ROS1 fusion (n = 9) and HER2 mutation (n = 4) and pan-negative patients (n = 58), 9583, 599, 792, 470, 102 and 8907 probes showed significant differences in DNA methylation levels between N and T samples, respectively (FDR <0.01) (Supplementary Table 5, available at Carcinogenesis Online). Since the number of patients was so small, patients with BRAF mutation (n = 2) and RET fusion (n = 2) were excluded from further statistical analysis. Among these aberrantly methylated probes, DNA methylation alterations on 7648 probes were shared by two or more driver aberration groups (Supplementary Table 6, available at Carcinogenesis Online). The numbers of probes shared by two to six driver aberration groups are summarized in Supplementary Table 7, available at Carcinogenesis Online. On the other hand, only 1948, 18, 4, 4, 3 and 1297 probes showed significant differences between N and T samples that were specific to the EGFR mutation-positive, ALK fusion-positive, KRAS mutation-positive, ROS1 fusion-positive, HER2 mutation-positive and pan-negative groups, respectively (Supplementary Table 7, available at Carcinogenesis Online).
DNA methylation alterations shared by multiple genomic driver aberration groups
Among the 7648 probes aberrantly methylated during the N to T transition that were shared by two or more driver aberration groups, 1404 probes were located within the CpG islands around the transcription start site, i.e. the region from the transcription start site to 1500 bp upstream of it, the 5′ untranslated region and the first exon or the first intron of the 1116 genes based on the UCSC Genome Browser (http://genome.ucsc.edu/). Among 1116 genes, the levels of mRNA expression were inversely correlated with the levels of DNA methylation for 197 genes (259 probes) in lung cancers deposited in the TCGA database (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) using the MethHC pipeline (http://MethHC.mbc.nctu.edu.tw) (Pearson correlation coefficient [r] < −0.2 and P < 0.05) (Supplementary Table 8A, available at Carcinogenesis Online).
These 197 genes were evaluated for protein function by enrichment analysis using the MetaCore software, and compared with the protein function distribution of genes within the GeneGo database. Genes showing DNA methylation alterations shared by multiple driver aberration groups were clearly overrepresented by ‘transcriptional factors’ (P = 3.734 × 10−13) and ‘enzyme’ (P = 2.016 × 10−6) (Table 1). In addition, known tumor-related genes, e.g. CDH1, CDH2 and CLDN4, which participate in cell adhesion and epithelial–mesenchymal transition, and FZD2, SOX2 and PAX7, which participate in Wnt/β-catenin signaling, have been included in Supplementary Table 8A, available at Carcinogenesis Online.
Table 1.
Gene ontology enrichment analysis of the 197 genes commonly showing significant differences in DNA methylation levels between non-cancerous and cancerous tissue specimens in two or more driver aberration groups, for which DNA methylation alterations would potentially result in mRNA expression alterations (listed in Supplementary Table 8A, available at Carcinogenesis Online), according to protein function using MetaCore software
| Protein class | r | n | R | N | Expected | Ratio | P |
|---|---|---|---|---|---|---|---|
| Transcription factors | 32 | 220 | 1254 | 40 677 | 6.782 | 4.718 | 3.734E−13 |
| Enzymes | 36 | 220 | 2869 | 40 677 | 15.52 | 2.32 | 2.016E−06 |
| Receptors | 19 | 220 | 1700 | 40 677 | 9.194 | 2.066 | 2.367E−03 |
| Kinases | 9 | 220 | 685 | 40 677 | 3.705 | 2.429 | 1.279E−02 |
| Ligands | 5 | 220 | 562 | 40 677 | 3.04 | 1.645 | 1.898E−01 |
| Phosphatases | 2 | 220 | 242 | 40 677 | 1.309 | 1.528 | 3.769E−01 |
| Proteases | 4 | 220 | 623 | 40 677 | 3.369 | 1.187 | 4.358E−01 |
| Other | 115 | 220 | 32 813 | 40 677 | 177.5 | 0.648 | 1.534E−21 |
r, number of objects from the present data set for a given protein class; n, total number of objects from the present data set; R, number of background objects from the database for a given class; N, total number of background objects from the database; Expected, mean value for hypergeometric distribution (n × R/40 677); Ratio, the ratio of actual/expected. If the ratio is more than 1, P values less than 0.0001 mean significant enrichment, and these are bold.
Genomic driver aberration-specific occurrence of DNA methylation alterations
Among the aforementioned 1948, 18, 4, 4, 3 and 1297 probes showing EGFR mutation-specific, ALK fusion-specific, KRAS mutation-specific, ROS1 fusion-specific, HER2 mutation-specific and pan-negativity-specific DNA methylation alterations, respectively, levels of mRNA expression were inversely correlated with DNA methylation levels for only 41 EGFR-specific genes (43 probes) and 26 pan-negative-specific genes (29 probes) (67 genes in total), based on the TCGA database (r < −0.2 and P < 0.05) (Supplementary Table 8B, available at Carcinogenesis Online).
Among the 67 genes, the DNA methylation alterations of 66 genes occurring during the N to T transition (ΔMT–N) were significantly correlated with clinicopathological parameters reflecting tumor aggressiveness, i.e. a larger tumor size, pleural invasion, lymphatic invasion, vascular invasion, intrapulmonary metastasis and recurrence (FDR <0.5) (Supplementary Table 9, available at Carcinogenesis Online).
Since gender, age and smoking status are known to be associated with DNA methylation (33), multivariate analysis of these parameters in addition to the DNA methylation status (ΔMT–N) of the genes included in Supplementary Table 9, available at Carcinogenesis Online, were performed. The DNA methylation status of multiple genes was significantly correlated with a larger tumor size, pleural invasion, lymphatic invasion and intrapulmonary metastasis, independently from gender, age and smoking status (Supplementary Table 10, available at Carcinogenesis Online).
Technical verification of DNA methylation levels in the promoter region of the TRIM58 and ZNF132 genes
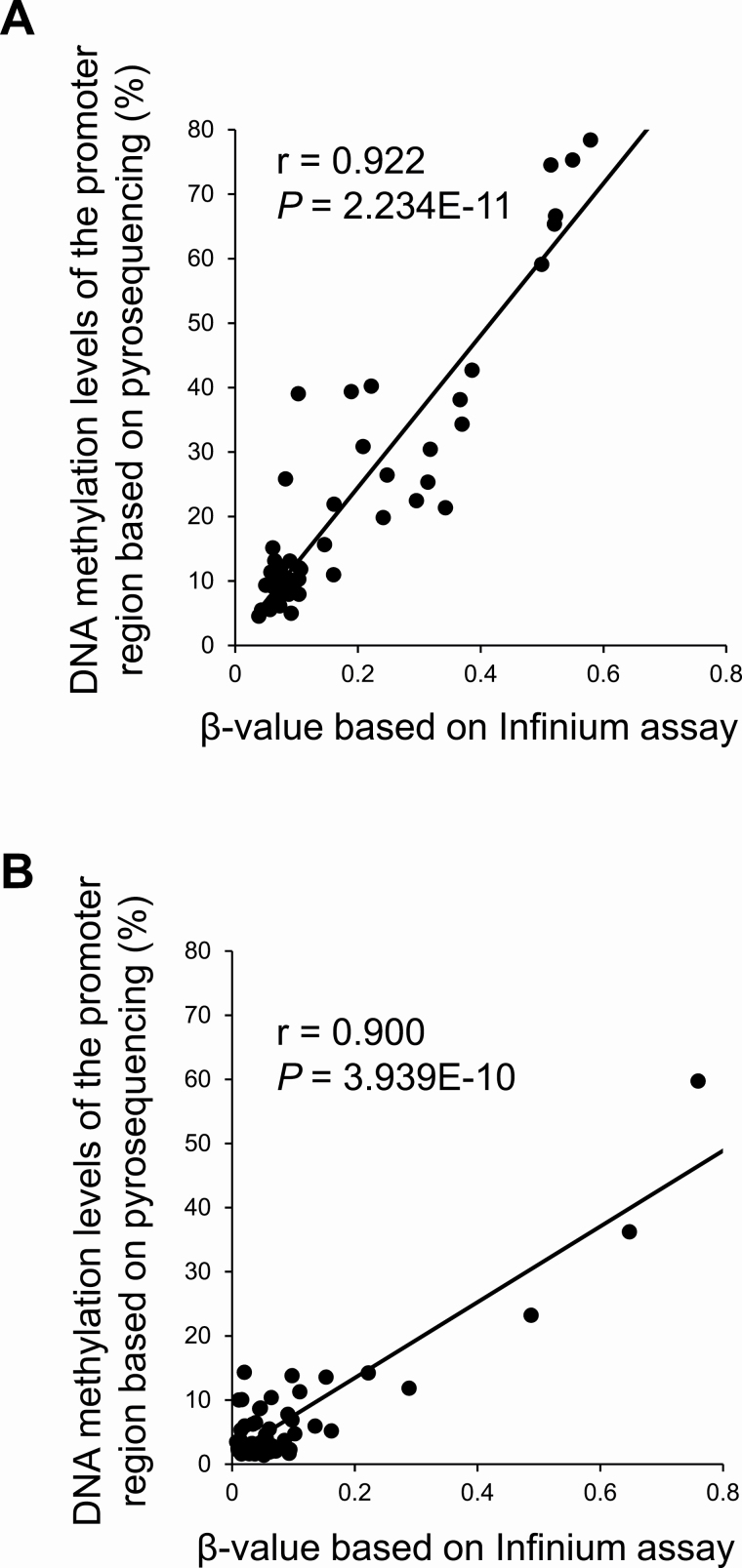
Using pyrosequencing, technical verification was performed for representative genes, i.e. the TRIM58 gene, for which our Infinium assay had demonstrated DNA hypermethylation shared by multiple genomic driver aberration groups and an inverse correlation with its mRNA expression had been evident in the TCGA database (Supplementary Table 8A, available at Carcinogenesis Online), and the ZNF132 gene, for which DNA hypermethylation based on our Infinium assay had been observed only in the ‘pan-negative’ group and was significantly correlated with vascular invasion (Supplementary Table 9, available at Carcinogenesis Online). Since tissue samples from the present 162 patients were exhausted after the Infinium assay, 26 paired N and their corresponding T samples in another cohort [validation cohort (n = 52)] of lung adenocarcinomas were subjected to pyrosequencing. The β-values of cg20855565 and cg13877915 for the TRIM58 and ZNF132 genes based on the Infinium assay and DNA methylation levels in the promoter regions of the TRIM58 and ZNF132 genes based on pyrosequencing were significantly correlated with each other in the validation cohort (Pearson’s correlation coefficient r = −0.922 and 0.900, and P = 2.234 × 10−11 and 3.939 × 10−10, respectively) (Figure 4), indicating that the Infinium data had been successfully verified using pyrosequencing.
Figure 4.
Pyrosequencing verification of the β-values of the TRIM58 (A) and ZN132 (B) genes based on the Infinium assay. Since tissue samples from the present 162 patients were exhausted after the Infinium assay, pyrosequencing and the Infinium assay were performed using 26 paired samples of adjacent normal lung tissue and their corresponding cancerous tissues in another cohort [validation cohort (n = 52)] of lung adenocarcinomas. The β-values of cg20855565 and cg13877915 for the TRIM58 and ZNF132 genes and the DNA methylation levels in the promoter regions based on pyrosequencing were significantly correlated with each other in the validation cohort (Pearson’s correlation coefficient r = −0.922 and 0.900, and P = 2.234 × 10−11 and 3.939 × 10−10, respectively), indicating that the Infinium data had been successfully verified using pyrosequencing.
Silencing of ZNF132 due to DNA hypermethylation
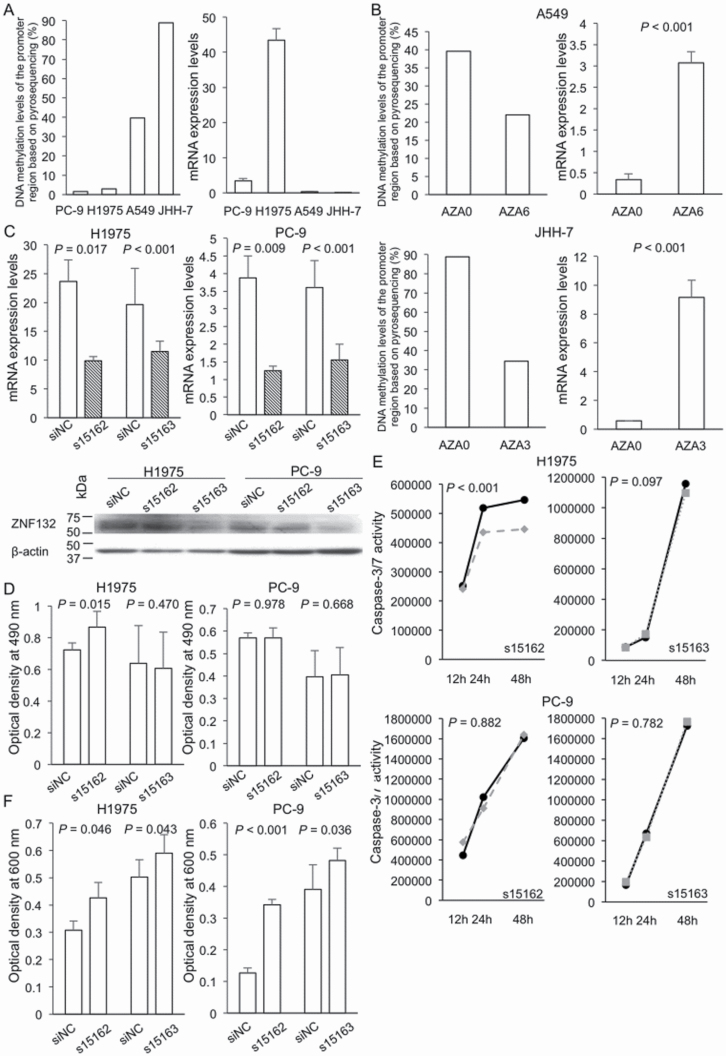
We further focused on the ZNF132 gene, for which DNA hypermethylation during N to T transition (ΔMT–N) was significantly correlated with vascular invasion (Supplementary Table 9, available at Carcinogenesis Online). The levels of DNA methylation and mRNA expression of ZNF132 in PC-9, H1975, A549 and JHH-7 cells are shown in Figure 5A. In the top two cell lines showing the highest DNA methylation levels, A549 and JHH-7, the level of ZNF132 mRNA expression was low. These two cell lines were then subjected to 5-aza-dC treatment. This led to a marked reduction of DNA methylation and restored the expression level of ZNF132 mRNA (Figure 5B), indicating that ZNF132 had been silenced due to DNA hypermethylation in lung adenocarcinomas.
Figure 5.
Treatment with 5-aza-2′-deoxycytidine (5-aza-dC) and ZNF132 gene knockdown experiments in lung adenocarcinoma cell lines. (A) DNA methylation levels of the promoter region based on pyrosequencing and mRNA expression levels based on RT–PCR analysis of ZNF132 in PC-9, H1975, A549 and JHH-7 cells. A549 and JHH-7 cells showing higher DNA methylation levels were subjected to 5-aza-dC treatment and H1975 and PC-9 showing higher mRNA expression levels were subjected to knockdown experiments. (B) DNA methylation and mRNA expression levels of ZNF132 on day 3 or 6 (AZA3 or AZA6) of 5-aza-dC treatment and untreated control cells (AZA0). After 5-aza-dC treatment, reduction of DNA methylation levels and restoration of the mRNA expression levels (9.1- and 15.8-fold increase) were observed in A549 and JHH-7 cells, respectively. (C) Knockdown of ZNF132 by transfection with a ZNF132-specific siRNA (dashed bar) and a negative control siRNA (siNC) (clear bar) in H1975 and PC-9 cells. mRNA and protein expression levels were reduced in both cell lines. (D) Effects of ZNF132 knockdown on cell proliferation. Forty-eight hours after transfection of H1975 and PC-9 cells with siNC and siZNF132, the MTS assay was performed. (E) Effects of ZNF132 knockdown on apoptosis. Twelve, 24 and 48 h after transfection with siNC (solid line) and siZNF132, s15162 (broken line) and s15163 (dotted line), caspase-3/7 activity was evaluated in H1975 and PC-9 cells. (F) Effects of ZNF132 knockdown on cell migration ability. Forty-eight hours after transfection of H1975 and PC-9 cells with siNC and siZNF132, transwell assay was performed. Although cell growth and apoptosis were altered only in H1975 using s15162 (panels D and E), cell migration ability was significantly enhanced by reduction of ZNF132 in both H1975 and PC-9 using both s15162 and s15163 (panel F).
Significance of ZNF132 in proliferation, apoptosis and migration of lung adenocarcinoma cells
Knockdown of ZNF132 using siRNA transfection was performed in the top two cell lines showing the highest mRNA expression levels, H1975 and PC-9. After transfection, marked reduction of ZNF132 expression was confirmed by quantitative real-time RT–PCR and western blotting (Figure 5C and Supplementary Figure 2, available at Carcinogenesis Online). Although an increase of cell growth and a decrease of caspase-3/7 activities were observed only in s15162-treated H1975 cells (Figure 5D and E, respectively), cell migration ability was significantly enhanced by knockdown of ZNF132 using both s15162 and s15163 in both H1975 and PC-9 cells (Figure 5F and Supplementary Figure 3, available at Carcinogenesis Online).
Discussion
Here, we have reported the results of the Infinium assay for 324 samples of lung tissue (162 N and 162 T samples) along with the genomic driver aberration data for T samples. According to the field cancerization concept (34), N samples obtained from patients with lung adenocarcinomas, which can be exposed to carcinogenetic factors such as cigarette smoking and chronic persistent inflammation during chronic obstructive pulmonary disease (10), are considered to be at precancerous stages. Therefore, we first focused on the DNA methylation profiles of N samples.
More probes for N samples than for T samples showed significant correlation with recurrence after surgery, reflecting the poorer prognosis associated with the former (Figure 1 and Supplementary Table 3, available at Carcinogenesis Online). In fact, a significant association between the level of DNA methylation at each of the CpG sites in N samples with recurrence-free survival rate had been confirmed by long-term follow-up (Figure 2A). Moreover, in T samples, most of the probes showing significant correlation with recurrence also showed such a correlation even from the N stages (Figure 1C and Supplementary Table 3, available at Carcinogenesis Online), indicating that DNA methylation profiles at precancerous N stages may already determine the aggressiveness of established cancers developing in the same individuals. However, this conclusion has a limitation in that DNA methylation profiles of N samples may be affected by some paracrine factors derived from adjacent established tumors.
On the other hand, the number of probes showing a correlation between DNA methylation alterations during the N to T transition (ΔMT–N) and recurrence-free survival was very limited (Supplementary Table 4B, available at Carcinogenesis Online), suggesting that many passenger DNA methylation alterations may occur, at least partly due to genomic alterations such as copy number alterations, during progression from the precancerous N stage to established Ts. The finding that the number of probes showing DNA methylation alterations significantly associated with recurrence in T samples was smaller than that in N samples (Figure 1) may be attributable to the fact that passenger DNA methylation alterations can mask any clinicopathologically significant DNA methylation profiles in T samples.
Next, we focused on the interaction between epigenomic and genomic alterations during multistage lung adenocarcinogenesis. Our principal component analysis clearly revealed that patients with each type of genomic driver aberration were individually scattered on the scattergram (Figure 3), indicating that, in general, preceding distinct DNA methylation profiles at the precancerous N stage may not induce specific genomic driver mutations, although this conclusion has some limitation due to the very limited sample size. Since, in general, DNA methylation alterations are more dependent on environmental factors than genomic aberrations (35), it is feasible that DNA methylation profiles are generally independent of mutation profiles.
During multistage carcinogenesis, DNA methylation alterations are further accumulated in established cancers even after a driver mutation has arisen. The majority of DNA methylation alterations (on the 7648 probes) occurring during transition from N to T, potentially after driver mutation, were shared in two or more genomic driver aberration groups (Supplementary Table 7, available at Carcinogenesis Online), indicating that, in general, each genomic driver mutation does not induce a specific DNA methylation profile. DNA methylation alterations again occur independently, regardless of the driver aberration profile. This conclusion again has some limitation as it was based on results from a very limited sample size.
We considered that such commonly shared DNA methylation alterations in established cancers with various driver mutations might shed light on the significance of epigenomic events in the later steps of multistage carcinogenesis. Commonly shared DNA methylation alterations which could potentially result in expression abnormalities were enriched among ‘transcriptional factors’ (Table 1). It is feasible that such factors affect the transcriptional regulation of various downstream genes including tumor-related genes. In addition, some known tumor-related genes themselves have shown commonly shared DNA methylation alterations (Supplementary Table 8A, available at Carcinogenesis Online). These findings suggest that DNA methylation alterations in the later stages may further participate in the progression of cancers.
On the other hand, the number of genes showing DNA methylation alterations, which are specific to some mutation groups and could potentially result in expression abnormalities, was very small, i.e. only 67. However, such alterations were frequently correlated with tumor aggressiveness (Supplementary Table 9, available at Carcinogenesis Online), indicating that DNA methylation alterations in the later steps of carcinogenesis may further modify the clinicopathological diversity of lung adenocarcinomas.
For example, DNA hypermethylation of ZNF132 during progression from the precancerous stage to established lung adenocarcinoma was significantly correlated with vascular invasion. Although epigenomic silencing of ZNF132 has been reported in prostatic adenocarcinoma (36) and esophageal squamous cell carcinoma (37), its participation in lung cancer has never been confirmed previously. In addition to the inverse correlation between DNA methylation and mRNA expression in the TCGA database, our 5-aza-dC treatment revealed that ZNF132 is silenced by DNA methylation. Our knockdown study indicated that silencing of ZNF132 may participate in the malignant progression of lung adenocarcinomas, especially that of pan-negative cancers that have not yet been well studied, through an increase of cell migration ability, which can result in invasiveness and potential for metastasis, rather than increased cell proliferation or reduced apoptosis.
In summary, our study has shown that DNA methylation alterations at the precancerous stage, which precedes genomic driver aberration, largely determine the clinicopathological aggressiveness and outcome of lung adenocarcinomas. In general, such distinct DNA methylation alterations at the precancerous stage do not induce specific genomic driver mutations, and each driver mutation tends not to induce specific DNA methylation profiles. Moreover, DNA methylation alterations occurring after driver mutation at the later stages of multistage carcinogenesis again contribute to the further progression and clinicopathological diversity of lung adenocarcinomas by altering the expression of the affected genes.
Supplementary Material
Glossary
Abbreviations
- 5-aza-dC
5-aza-2′-deoxycytidine
- FDR
false discovery rate
- GO
gene ontology
- N
adjacent normal lung tissue
- RT
reverse transcription
- siRNA
small interfering RNA
- T
tumorous tissue
- TCGA
The Cancer Genome Atlas
Funding
This work was supported by the Japan Society for the Promotion of Science under grant KAKENHI JP19K07444; the Japan Agency for Medical Research and Development under grants 19ak0101043h0105 and JP19ae0101020.
Conflict of Interest Statement: None declared.
References
- 1. Ilango, S. et al. (2020) Epigenetic alterations in cancer. Front. Biosci. (Landmark Ed.), 25, 1058–1109. [DOI] [PubMed] [Google Scholar]
- 2. Jones, P.A. et al. (2016) Targeting the cancer epigenome for therapy. Nat. Rev. Genet., 17, 630–641. [DOI] [PubMed] [Google Scholar]
- 3. Baylin, S.B. et al. (2016) Epigenetic determinants of cancer. Cold Spring Harb. Perspect. Biol., 8, a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kanai, Y. et al. (2014) Multilayer-omics analyses of human cancers: exploration of biomarkers and drug targets based on the activities of the International Human Epigenome Consortium. Front. Genet., 5, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cancer Genome Atlas Research, N. (2014) Comprehensive molecular profiling of lung adenocarcinoma. Nature, 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bibikova, M. et al. (2009) Genome-wide DNA methylation profiling using Infinium® assay. Epigenomics, 1, 177–200. [DOI] [PubMed] [Google Scholar]
- 7. Husni, R.E. et al. (2019) DNA hypomethylation-related overexpression of SFN, GORASP2 and ZYG11A is a novel prognostic biomarker for early stage lung adenocarcinoma. Oncotarget, 10, 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quek, K. et al. (2017) DNA methylation intratumor heterogeneity in localized lung adenocarcinomas. Oncotarget, 8, 21994–22002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bjaanaes, M.M. et al. (2016) Genome-wide DNA methylation analyses in lung adenocarcinomas: association with EGFR, KRAS and TP53 mutation status, gene expression and prognosis. Mol. Oncol., 10, 330–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Santoro, A. et al. (2019) Tobacco smoking: risk to develop addiction, chronic obstructive pulmonary disease, and lung cancer. Recent Pat. Anticancer Drug Discov., 14, 39–52. [DOI] [PubMed] [Google Scholar]
- 11. Sato, T. et al. (2014) Epigenetic clustering of lung adenocarcinomas based on DNA methylation profiles in adjacent lung tissue: its correlation with smoking history and chronic obstructive pulmonary disease. Int. J. Cancer, 135, 319–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makabe, T. et al. (2019) Genome-wide DNA methylation profile of early-onset endometrial cancer: its correlation with genetic aberrations and comparison with late-onset endometrial cancer. Carcinogenesis, 40, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yamanoi, K. et al. (2015) Epigenetic clustering of gastric carcinomas based on DNA methylation profiles at the precancerous stage: its correlation with tumor aggressiveness and patient outcome. Carcinogenesis, 36, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Arai, E. et al. (2012) Single-CpG-resolution methylome analysis identifies clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Carcinogenesis, 33, 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robles, A.I. et al. (2015) An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J. Thorac. Oncol., 10, 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sato, T. et al. (2013) DNA methylation profiles at precancerous stages associated with recurrence of lung adenocarcinoma. PLoS One, 8, e59444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saito, M. et al. (2018) Treatment of lung adenocarcinoma by molecular-targeted therapy and immunotherapy. Surg. Today, 48, 1–8. [DOI] [PubMed] [Google Scholar]
- 18. Matano, M. et al. (2015) Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat. Med., 21, 256–262. [DOI] [PubMed] [Google Scholar]
- 19. Eguchi, K. et al. (1997) DNA hypermethylation at the D17S5 locus in non-small cell lung cancers: its association with smoking history. Cancer Res., 57, 4913–4915. [PubMed] [Google Scholar]
- 20. Arai, E. et al. (2018) Epigenome mapping of human normal purified hepatocytes: personal epigenome variation and genome-epigenome correlation. Epigenomics, 10, 955–979. [DOI] [PubMed] [Google Scholar]
- 21. Travis, W.D. et al. (2015) Adenocarcinoma. In Travis, W.D et al. (eds) WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart. 4th edn IARC, Lyon, France, pp. 26–43. [Google Scholar]
- 22. Kanai, Y. et al. (2018) The Japanese Society of Pathology Guidelines on the handling of pathological tissue samples for genomic research: standard operating procedures based on empirical analyses. Pathol. Int., 68, 63–90. [DOI] [PubMed] [Google Scholar]
- 23. Du, P. et al. (2010) Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform., 11, 587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fujimoto, M. et al. (2020) Establishment of diagnostic criteria for upper urinary tract urothelial carcinoma based on genome-wide DNA methylation analysis. Epigenetics, 4, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kinjo, M. et al. (1979) Thromboplastic and fibrinolytic activities of cultured human cancer cell lines. Br. J. Cancer, 39, 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Phelps, R.M. et al. (1996) NCI-Navy Medical Oncology Branch cell line data base. J. Cell. Biochem. Suppl., 24, 32–91. [DOI] [PubMed] [Google Scholar]
- 27. Giard, D.J. et al. (1973) In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J. Natl. Cancer Inst., 51, 1417–1423. [DOI] [PubMed] [Google Scholar]
- 28. Fujise, K (1990) Integration of hepatitis B virus DNA into cells of six established human hepatocellular carcinoma cell lines. Hepatogastroenterology, 37, 457–460. [PubMed] [Google Scholar]
- 29. Arai, E. et al. (2015) Alterations of the spindle checkpoint pathway in clinicopathologically aggressive CpG island methylator phenotype clear cell renal cell carcinomas. Int. J. Cancer, 137, 2589–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ohara, K. et al. (2017) Genes involved in development and differentiation are commonly methylated in cancers derived from multiple organs: a single-institutional methylome analysis using 1007 tissue specimens. Carcinogenesis, 38, 241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuramoto, J. et al. (2017) Genome-wide DNA methylation analysis during non-alcoholic steatohepatitis-related multistage hepatocarcinogenesis: comparison with hepatitis virus-related carcinogenesis. Carcinogenesis, 38, 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fluss, R. et al. (2005) Estimation of the Youden Index and its associated cutoff point. Biom. J., 47, 458–472. [DOI] [PubMed] [Google Scholar]
- 33. Reid, M.E. et al. (2008) Molecular epidemiology to better predict lung cancer risk. Clin. Lung Cancer, 9, 149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Curtius, K. et al. (2018) An evolutionary perspective on field cancerization. Nat. Rev. Cancer, 18, 19–32. [DOI] [PubMed] [Google Scholar]
- 35. Martin, E.M. et al. (2018) Environmental influences on the epigenome: exposure-associated DNA methylation in human populations. Annu. Rev. Public Health, 39, 309–333. [DOI] [PubMed] [Google Scholar]
- 36. Abildgaard, M.O. et al. (2012) Downregulation of zinc finger protein 132 in prostate cancer is associated with aberrant promoter hypermethylation and poor prognosis. Int. J. Cancer, 130, 885–895. [DOI] [PubMed] [Google Scholar]
- 37. Jiang, D. et al. (2018) Epigenetic silencing of ZNF132 mediated by methylation-sensitive Sp1 binding promotes cancer progression in esophageal squamous cell carcinoma. Cell Death Dis., 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.