Abstract
Prostate cancer is the second most common type of cancer and the second leading cause of cancer death in American men. RAD9 stabilizes the genome, but prostate cancer cells and tumors often have high quantities of the protein. Reduction of RAD9 level within prostate cancer cells decreases tumorigenicity of nude mouse xenographs and metastasis phenotypes in culture, indicating that RAD9 overproduction is essential for the disease. In prostate cancer DU145 cells, CpG hypermethylation in a transcription suppressor site of RAD9 intron 2 causes high-level gene expression. Herein, we demonstrate that DNA methyltransferases DNMT1 and DNMT3B are highly abundant in prostate cancer cells DU145, CWR22, LNCaP and PC-3; yet, these DNMTs bind primarily to the transcription suppressor in DU145, the only cells where methylation is critical for RAD9 regulation. For DU145 cells, DNMT1 or DNMT3B shRNA reduced RAD9 level and tumorigenicity, and RAD9 ectopic expression restored this latter activity in the DNMT knockdown cells. High levels of RAD9, DNMT1, DNMT3B and RAD9 transcription suppressor hypermethylation were significantly correlated in prostate tumors, and not in normal prostate tissues. Based on these results, we propose a novel model where RAD9 is regulated epigenetically by DNMT1 and DNMT3B, via targeted hypermethylation, and that consequent RAD9 overproduction promotes prostate tumorigenesis.
DNMT1/DNMT3B-mediated, targeted RAD9 hypermethylation in human prostate cancer cells increases RAD9 level and mouse xenograph tumorigenesis. Excessive RAD9, DNMT1, DNMT3B and RAD9 hypermethylation are significantly correlated in cancer but not normal prostate tissue, indicating disease relevance.
Introduction
RAD9 regulates fundamental cellular activities, including maintenance of genomic integrity, DNA repair, cell cycle checkpoints, apoptosis, and transcriptional transactivation of specific target genes (1,2). There is an optimum RAD9 level required for proper function; too much or too little can have deleterious consequences (3). Rad9 null mouse embryonic stem cells are hypersensitive to numerous DNA damaging agents (4) and display defects in homologous recombination repair (5), mismatch repair (6), nucleotide excision repair (7) and base excision repair (8). RAD9 also participates in alternative non-homologous end joining (9), through microhomology-mediated DNA strand annealing (10). Rad9 null cells demonstrate high frequencies of spontaneous mutations and chromosomal aberrations (4). Cells that are Rad9 null or overproduce the protein can undergo apoptosis (11,12).
RAD9 has oncogene and tumor suppressor activities, dependent at least in part on concentration of the protein. High levels are associated with cancers of breast, lung and thyroid (3). Furthermore, RAD9 overproduction is causally linked to prostate tumorigenesis and metastatic phenotypes, as RAD9 knockdown or knockout can neutralize these cancer-related characteristics (2,3,13,14). In contrast, Rad9 null exacerbates carcinogen-induced skin cancer (15).
Gene expression can be regulated by epigenetic mechanisms, not involving nucleotide sequence changes. Most common epigenetic alterations include methylation of cytosine carbon-5 within CpG, and post-translational modification of histones, such as methylation of H3 at lys4, lys9 or lys27 (16). Epigenomic status is highly regulated and frequently critical for biological processes. Cancer cells in general are hypomethylated genome-wide yet hypermethylated at certain gene promoters (17,18). This affects carcinogenesis, as hypermethylation can silence tumor suppressor genes (18), whereas hypomethylation can lead to chromosome instability (19), a hallmark of cancer.
In mammalian cells, CpG methylation is performed by DNA methyltransferases (DNMTs) (20). DNMT1 is a maintenance methyltransferase that methylates cytosines in hemimethylated CpG dinucleotides (21). DNMT3 is a family containing DNMT3A and DNMT3B, capable of methylating, de novo, unmethylated CpG, and also stably maintaining methylated sites (22). DNMT3L is another member, whose function is confined mainly to gametogenesis. The protein does not demonstrate DNA methyltransferase activity but can modulate DNMT3A and DNMT3B functions (23).
RAD9 expression can be controlled by DNA methylation. Cheng et al. (24) identified a suppressor of transcription site in human RAD9 intron 2 that regulates expression by differential methylation in breast cancer cells. When the site is hypomethylated Sp3 binds and decreases transcription. In contrast, when the intron 2 site is highly methylated transcription increases as Sp3 does not bind. Zhu et al. (13) found that hypermethylation of CpG sites within this RAD9 location in prostate cancer DU145 cells is responsible for abnormally high levels of the protein. Nevertheless, little is known about the mechanism of RAD9 methylation, or how widespread it is with respect to prostate cancer.
The work herein is a follow-up to our previously published studies of the relationship between RAD9 expression and prostate carcinogenesis (13). We showed high quantities of RAD9 in prostate cancer cell lines compared to the ‘normal’ controls, abundant methylation at the RAD9 intron 2 transcription suppressor site in DU145 cells, that 5-AzaC and hence methylation regulates RAD9 expression in DU145, and that one prostate cancer cell line, PC-3, in contrast, had amplification of the RAD9 DNA sequence, which was the likely underlying mechanism of RAD9 overproduction in that instance. In addition, we reported high levels of RAD9 protein staining in prostate tumor sections, compared to normal, non-cancerous prostate tissues, and that decreasing RAD9 quantity in the cell lines reduced their ability to form tumors in mouse xenographs. In the present investigation, we demonstrate that prostate cancer cells have high levels and activity of DNMT1 and DNMT3B. We show that although these enzymes are abundant in four prostate cancer cell lines, relative to nontumorigenic controls, they bind and hypermethylate the RAD9 transcription suppressor only in DU145. We provide evidence that this hypermethylation is important to achieve the high levels of RAD9 needed for tumorigenicity of DU145 cells in mouse xenographs. We also show that human prostate tumors, compared to non-cancer specimen, frequently contain extensively methylated CpG sites within the RAD9 suppressor and high levels of RAD9, DNMT1 and DNMT3B proteins. Our results suggest hypermethylation is a prevalent mechanism for upregulating oncogenic RAD9 expression in prostate tumors.
Materials and methods
Cells, culture conditions
Human prostate cancer cells CWR22, DU145, LNCaP and PC-3, as well as normal prostate epithelial cells (PrEC) were grown as described (13). Construction and growth of a PC-3 derivative knocked out for RAD9 by CRISPR/cas9 (crRAD9) was also reported (2). Immortalized, non-tumorigenic RWPE-1 cells were grown in serum-free medium containing bovine pituitary extract and human epidermal growth factor (Life Technologies). All cells were cultured at 37°C in 5% CO2. Cell lines are tested before use for Mycoplasma contamination using Venor TM GEM Mycoplasma Detection Kit (Sigma–Aldrich). All cell lines were purchased between the years 2006 and 2011 from ATCC, which authenticate CWR22, PC-3 and LNCaP by the STR method, DU145 and RWPE-1 by STR and the isozyme analysis method, and PrEC by Pan-Cytokeratin (+), TE-7 (−) assessment, as well as observation of an epithelial, packed cuboidal morphology. Cells were expanded, aliquoted and frozen in multiple vials upon receipt. After thawing, cells were passaged for no longer than 6 months before a fresh vial was used.
RNAi, plasmid construction, DNA transfection into cells
pSUPER.retro.puro RNAi vector and transfection of cells to knockdown RAD9 expression are described (13). We employed a similar strategy to knockdown DNMT1 and DNMT3B expression. Two DNMT1 (5′-CTGACACCCTGCATGCGGG-3′ and 5′-GCAGTTCAACACCCTCATC-3′) and three DNMT3B (5′-AACAAGACTCGAAGACGCA-3′, 5′-AGGCTGAAAGATGACGGAT-3′ and 5′-ATCGACCTCACAGACGACA-3′) siRNA target sequences, designed and synthesized by Oligoengine Inc. were used. pZeoSV2(+)-RAD9 (25) was used to express RAD9 in cells.
Quantitative reverse transcription–PCR, western Blotting
Quantitative reverse transcription–PCR (qRT–PCR) was performed as described (13). cDNA was used to amplify DNMT1 and DNMT3B by PCR. DNA primers were 5′-GCCCTGCCAAACGGAAACCTCA-3′, 5′-TCCCCGGCCTCGTCATAACTCTCC-3′ for DNMT1 and 5′-CTCCGACTCGCCCCCAATCCTG-3′, 5′-CACGGGGTTTTTCCTGCCACAAGA-3′ for DNMT3B. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) served as an internal reference gene. DNA primers for GAPDH were purchased (Super Array, Inc., UNiGene no. Hs.544577, RefSeq Accession no. NM_002046.2). Relative quantification of DNMT1 and DNMT3B to GAPDH RNA abundance was analyzed by the comparative threshold cycle (Ct) method (26).
Western blotting was conducted as reported (13). Blots were probed with monoclonal mouse anti-human DNMT1, DNMT3B or RAD9 antibody (Novus Biologicals; BD Biosciences) and goat anti-human actin antibody (Santa Cruz Biotechnology, Inc.), followed by secondary antibody conjugated with horseradish peroxidase. Enhanced chemiluminescence western blotting substrate (Pierce, Inc.) was used to detect protein bands. Uncropped gel images are in Supplementary Figure 1, available at Carcinogenesis Online.
In vivo complex of methylation assay
In vivo complex of methylation (ICM) assay kits were used to measure complexes of DNMT1 and DNMT3B with genomic DNA in vivo, following manufacturer’s instructions (Methylation, LTD.) and published methods (27). Slot blots were probed by standard western procedures, using antibodies against DNMT1 and DNMT3B (Methylation, LTD., or Novus Biologicals). Immune complexes were detected using enhanced chemiluminescence.
Chromatin immunoprecipitation
DNMT1 and DNMT3B binding to the RAD9 transcription suppressor was assessed by chromatin immunoprecipitation (ChIP), using a kit (no. 17–295) according to the manufacturer (Upstate Biotechnology, Inc., now part of EMD Millipore) and as published (28,29). ChIP grade monoclonal DNMT1 or DNMT3B antibody (Novus Biologicals), normal rat IgG (negative control; Novus Biologicals) or no antibody (negative control) were used. Isolated DNA was examined by PCR with a primer pair spanning RAD9 +293 to +559, an intron 2 region containing the transcription suppressor. Primers were MF1, 5′-GGCAAAGTTTCAGTTTCTTAGTCTGG-3′; MB1, 5′-CCCAGCCCTCTGGCTGCTTCTGCTC-3. PCR products were analyzed on 2% agarose gels.
Bisulfite sequencing
Methylation status of RAD9 CpG sites in cells and tissues was determined by sodium bisulfite sequencing, as described (13). Primer pairs were as follows: (i) MF2, 5′-GGGGTAGTATGAAGTGTTTGGTTA-3′; MB3, 5′-CCTCCAAAAATTCCAAATAAAACT-3′. Amplifies RAD9 −8 to +182, including the first and second exon and first intron; (ii) MF3, 5′-AGTTTTATTTGGAATTTTTGGAGG-3′; MB2, 5′- CCAAACTAAAAAACTAAAACTTTAC-3′. Amplifies +159 to +318, including the second intron 5′ of RAD9. (iii) MF1, 5′-GGTAAAGTTTTAGTTTTTTAGTTTGG-3′; MB1, 5′- CCCAACCCTCTAACTACTTCTACTC-3. Amplifies RAD9 +293 to +559, including the rest of intron 2, containing the transcription suppressor bearing 9 CpG sites (13,24).
Tumor development by human prostate cells injected into mice
Human prostate cancer cells were tested for ability to form tumors in mice, as described (13). Four week old Nu/Nu male mice (Harlan Sprague-Dawley, Inc.) were injected with cells into their backs subcutis. Tumors were assessed starting 2 to 3 weeks, post-injection. Tumor size was measured every 2 weeks with a vernier caliper by two investigators, unaware of animal identities during assessment. Tumor volume was calculated as the average of the two sets of measurements. At study conclusion, cancerous growths were harvested and stored in 10% formaldehyde. Tumor sections were stained with hematoxylin and eosin to assess histology, and with antibodies against human epithelial cell-specific markers, cytokeratin 5, 18 and 19 (Sigma), and RAD9 (BD Biosciences) via the ABC kit (Vector Laboratories) to confirm human origin.
Human prostate tissue
Human prostate specimens were obtained from Imgenex Corp., US Biomax, and the Columbia University Molecular Pathology Shared Resource of the Herbert Irving Comprehensive Cancer Center, as thin section arrays on slides. There were 22 normal prostate tissues and 17 prostate adenocarcinomas, representing stages II, III and IV from different patients. Available information did not include patient identifiers, race or ethnicity, surgical techniques employed, neo-adjuvant treatment or patient outcomes.
Immunohistochemical staining of prostate tissues, data quantification
Immunostaining of prostate paraffin-embedded tissue array slides was performed as described (13). Mouse monoclonal anti-human RAD9 (BD Biosciences), DNMT1 or DNMT3B (Novus Biologicals) primary antibody, and biotin-conjugated secondary antibody were used. Meyer’s hematoxylin was employed for counterstaining.
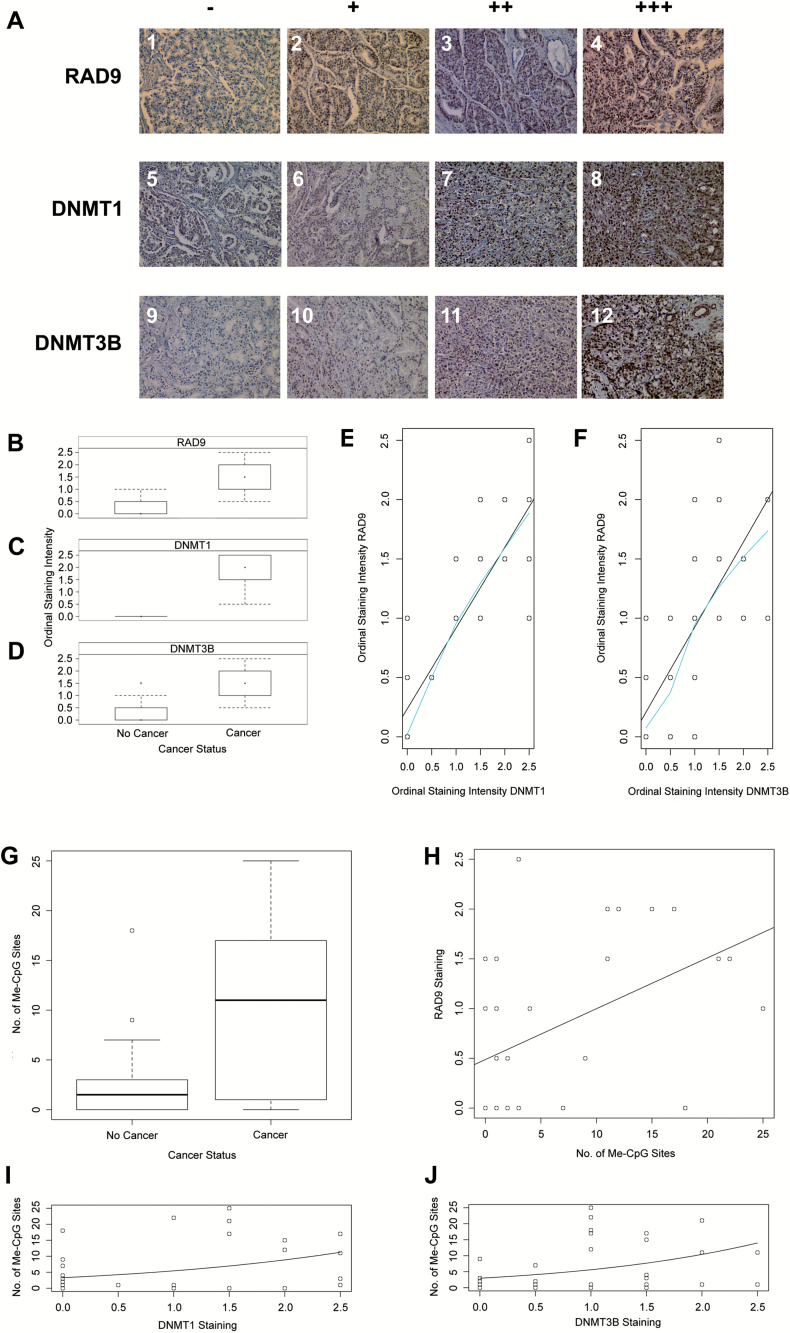
Immunostaining for DNMT1, DNMT3B and RAD9 proteins was quantified in categorical and ordinal terms for analyses, because tissue staining intensity is scored on a scale from − to +++ (Figure 3A; Tables 1 and 2), and there is some degree of subjectivity in the values chosen. For categorical data analysis, samples with a ‘partial +’ or above are assigned a ‘1’, and all others are assigned a ‘0’. For ordinal data analysis, samples are assigned a number between 0 and 3 corresponding to their average number of pluses. For example, a sample scored +/++ is assigned 1.5 staining intensity.
Figure 3.
High RAD9, DNMT1 and DNMT3B protein abundance, and elevated number of methylated CpG sites within the RAD9 intron 2 suppressor region of human prostate tissues are significantly correlated in cancer but not in normal prostate tissue. (A) Prostate tissue thin sections were immunostained for RAD9, DNMT1 and DNMT3B proteins. Blue, negative; Brown, positive. No detectable stain (−), weak stain (+), strong stain (++), very intense stain (+++). This illustrates the criteria used to assign staining intensity for the three proteins of interest in prostate tissue. Employing these guidelines, we found dependence of the ordinal intensity of RAD9, DNMT1 and DNMT3B protein staining on cancer status of human prostate tissue. Box and whisker plots of the dependence of ordinal intensity of (B) RAD9, (C) DNMT1 and (D) DNMT3B protein staining on whether human prostate tissue is non-cancerous or cancerous are indicated. Data from Tables 1 and 2 were used for these analyses. (E, F) We found that the intensity of immunohistochemical staining of human prostate tissues for RAD9 depends upon that of DNMT1 and DNMT3B, as illustrated by these plots, respectively. Data from Tables 1 and 2 were used for these analyses. (G) We found that CpG sites within the RAD9 transcription suppressor region of intron 2 are more highly methylated in cancer than in non-cancer human prostate tissue. Box and whisker plots illustrate dependence of the total number of methylated CpG sites (Me-CpG) in the RAD9 transcription suppressor region on the cancer status of prostate tissue. There are 9 CpG sites per clone, and 12 clones per patient, for a total of 108 potential methylation sites. Data from Tables 1 and 2 were used for these analyses. (H) There is dependence of RAD9 protein staining on the total number of methylated CpG (Me-CpG) sites in the RAD9 intron 2 transcription suppressor region of human prostate tissue. Line indicates best fit from linear regression. Data from Tables 1–3 were used for these analyses. Dependence of the total number of methylated CpG (Me-CpG) sites in the RAD9 intron 2 transcription suppressor on (I) DNMT1 and (J) DNMT3B protein staining in human prostate tissues are shown. Lines indicate best fit from logistic regression. Data from Tables 1–3 were used for these analyses.
Table 1.
Immunohistochemical staining of RAD9, DNMT1 and DNMT3B proteins in non-cancer human prostate tissues
| Sample number | Non-cancer prostate tissue | RAD9 staininga | DNMT1 staininga | DNMT3B staininga |
|---|---|---|---|---|
| 1 | 141N | − | − | − |
| 2 | 142N | −/+, partial + | −/+ | −/+ |
| 3 | 143N | −/+, partial + | − | − |
| 4 | 320N | − | − | − |
| 5 | 321N | − | − | − |
| 6 | 322N | − | −/+ | −/+ |
| 7 | 323N | − | − | − |
| 8 | N1 | − | − | − |
| 9 | N2 | − | − | − |
| 10 | N3 | − | − | − |
| 11 | N4 | − | − | − |
| 12 | N5 | −/+, partial + | − | +/++ |
| 13 | N6 | −, partial −/+ | −/+ | −/+ |
| 14 | N7 | − | −, partial −/+ | − |
| 15 | N8 | − | − | − |
| 16 | N9 | − | −/+ | −, partial + |
| 17 | N10 | − | − | + |
| 18 | N11 | −/+ | − | − |
| 19 | N12 | − | − | −/+ |
| 20 | N13 | −/+ | − | + |
| 21 | N14 | − | − | −/+ |
| 22 | N15 | − | − | − |
| Percent positiveb | 13.6 (3/22) | 0 (0/22) | 18.8 (4/22) |
aImmunostaining for RAD9, DNMT1 and DNMT3B proteins designated ‘—’ (negative), ‘+’ (weak), ‘++’ (strong), ‘−/+’ (borderline weakly positive) or ‘+/++’ (borderline weak to strongly positive). When ‘partial’ is indicated, the first designation is the predominant staining intensity and only a small portion of the tissue stains with the second indication of intensity.
bBorderline weakly positive (−/+) is considered negative for the Percent Positive calculation.
Table 2.
Immunohistochemical staining of RAD9, DNMT1 and DNMT3B protein levels in cancerous human prostate tissues
| Sample number | Cancer prostate tissuea | RAD9 stainingb | DNMT1 stainingb | DNMT3B stainingb |
|---|---|---|---|---|
| 1 | PR803E7-T IV | ++/+++ | ++/+++ | +/++ |
| 2 | 375-T IV | ++ | ++/+++ | ++/+++ |
| 3 | 376-T IV | ++ | ++/+++ | +/++ |
| 4 | PR803G2-T IV | +/++ | ++/+++ | ++ |
| 5 | 362-T IV | + | +/++ | −/+ |
| 6 | 363-T IV | +/++ | + | −/+, partial+ |
| 7 | 377-T III | −/+, partial+ | +/++ | + |
| 8 | 378-T III | ++ | +/++ | + |
| 9 | PR208K6/7 III | +/++ | ++/+++ | +/++ |
| 10 | 364-T III | −/+ | −/+ | −/+ |
| 11 | 365-T III | +/++ | ++ | +/++ |
| 12 | 371-T III | +/++ | +/++ | ++ |
| 13 | 372-T III | + | + | ++ |
| 14 | 370-T II-III | + | ++/+++ | ++/+++ |
| 15 | J7,8 II | −/+ | + | + |
| 16 | 1125 IV | ++ | ++ | +/++ |
| 17 | 1126III | ++ | ++ | + |
| Percent positivec | 88.2 (15/17) | 94.1 (16/17) | 88.2 (15/17) |
aRoman numerals (II, II-III, III, IV) indicate stage of cancer tissue.
bImmunostaining for RAD9, DNMT1 and DNMT3B proteins designated ‘−’ (negative), ‘+’ (weak), ‘++’ (strong), ‘-/+’ (borderline weakly positive), ‘+/++’ (borderline weak to strongly positive) or ‘++/+++’ (borderline strong to intensely positive). When ‘partial’ is indicated, the first designation is the predominant staining intensity and only a small portion of the tissue stains with the second indication of intensity.
cBorderline weakly positive (−/+) is considered negative for the Percent Positive calculation.
Immunocytochemistry staining of cells
PC-3 cells expressing normal levels of endogenous RAD9 or reduced levels (shRAD9, or crRAD9) were grown in 4-chamber slides. After 24 h of plating, cells were fixed in 3.7% paraformaldehyde for 20 min, washed 3 times in PBS and permeabilized with 0.2% Triton X-100 for 5 min. Cells were washed 3 times in PBS and nonspecific binding was blocked in 5% goat normal serum in PBS (blocking buffer) for 1 h. Cells were labeled with mouse monoclonal anti-RAD9 antibody (611324; BD Biosciences) diluted in blocking buffer (1:500) overnight at 40°C. The primary antibody was discarded and cells were washed 3 times with PBS. Subsequently, cells were incubated with ImmPRESS goat anti-mouse IgG polymer, peroxidase (MP-7452; Vector Laboratories) for 30 min. The cells were washed 2 times with PBS and color was developed by DAB (3,3′-diaminobenzidine) peroxidase substrate kit (SK-4100; Vector Laboratories) following the manufacturer’s protocol. Slides were rinsed under tap water, and nuclei were counterstained with Mayer’s hematoxylin. The slides were covered with glass coverslips using Cytoseal 60 solution and examined under an Echo Revolve microscope (Discover Echo, Inc) using a 60X oil immersion objective.
Statistical analysis of PCR and tumor volume data
PCR and tumor volume data were analyzed by ANOVA (30) corrected for multiple comparisons by Westfall’s method (31). All P-values and effect sizes are in Supplementary Table 1, available at Carcinogenesis Online. PCR data were analyzed as ΔCt = (ΔCexp − ΔCctrl), rather than as relative concentrations, because we found the former quantity yielded a linear qqplot, indicating normality (32), whereas the later quantity did not. Concentrations are plotted relative to a reference. Tumor volume was analyzed as log transformed values, logvol = log2(volume + 1), to achieve normality, and for plotting, back-transformed as volume = 2logvol − 1. Significance codes are ‘***’, ≤ 0.001; 0.001< ‘**’ ≤ 0.01; 0.01< ‘*’ ≤ 0.05; ‘ns ’ > 0.05.
Statistical analysis of tissue immunostaining and CpG methylation data
All P-values and effect sizes are in Supplementary Table 2, available at Carcinogenesis Online. Dependence of protein staining on cancer status (Figure 3B–D) is analyzed two ways: (i) with staining expressed as a categorical variable, it is analyzed by the Fisher exact test (30); (ii) with staining expressed as an ordinal variable, it is analyzed by the Wilcoxon method (30). The dependence of RAD9 on DNMT1 and DNMT3B staining (Figure 3E and F) is analyzed as a categorical variable by the Fisher exact test and as an ordinal variable by the Spearman correlation (30). The straight and smoothed lines in Figure 3E and F are linear regression and Loess curves (30), respectively. Dependence of the number of RAD9 intron 2 transcription suppressor methylated cytosines on cancer status (Figure 3G) is analyzed as a fraction of the 108 (9 CpG sites × 12 clones) potential sites by analysis of deviance with a logistic transformation and a quasibinomial error model (30). Dependence of RAD9 staining on RAD9 methylation (Figure 3H) is analyzed by linear regression (effect size, slope) and Spearman correlation. Dependence of the number (or fraction) of RAD9 transcription suppressor methylated CpG sites on DNMT1 and DNMT3B staining (Figures 3I and J) is estimated by logistic regression with a quasibinomial error model (30) and by Spearman correlation. Effect size for logistic regression is the coefficient of the ordinal staining in the logistic regression equation. Significance codes are as above.
Software
Microsoft Excel is the graphing software used to produce Figures 1C, D, E, I, and J, and 2E, F, G. Calculations were performed and graphs drawn (except as noted above) using R (30) and the following R packages: Calculations were performed using stats (30). Multiple comparisons were performed using multcomp (31). Figure 3B–D were drawn using lattice (33). Figure 3E–J was drawn using graphics (30).
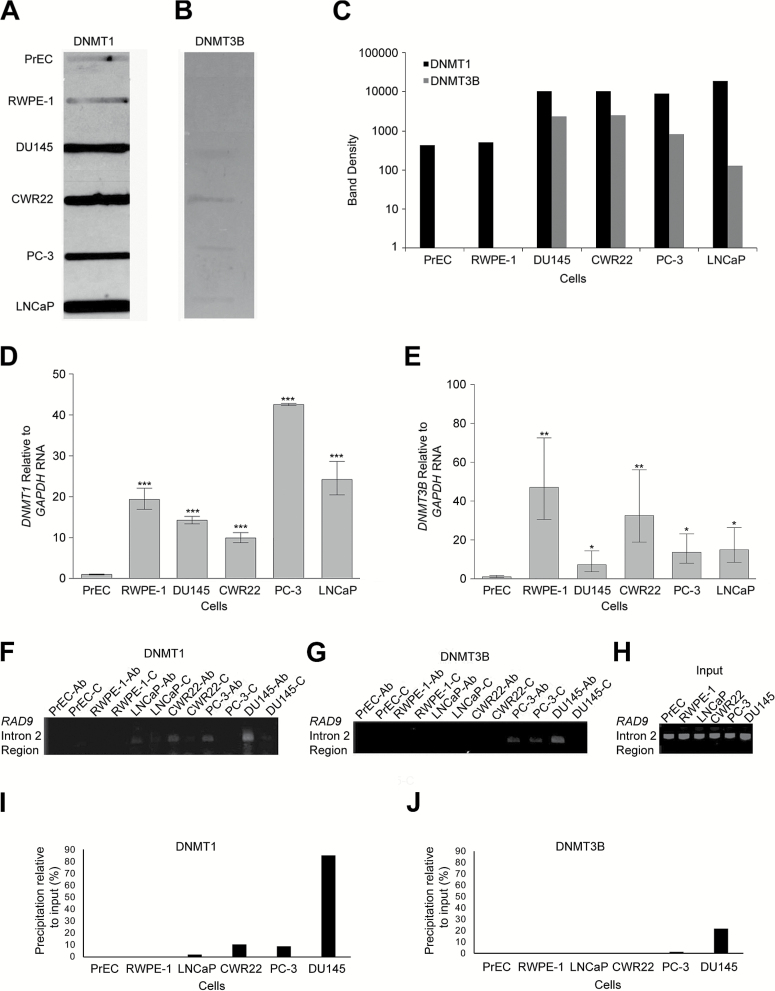
Figure 1.
DNMT1 and DNMT3B DNA methyltransferase levels are high in multiple prostate cancer cell lines, relative to non-cancer prostate cells. However, both enzymes bind the transcription suppressor region of RAD9 intron 2 primarily in DU145, the only cells tested where methylation regulates RAD9 expression. The in vivo complex of methylation (ICM) assay was used to measure complexes of (A) DNMT1 and (B) DNMT3B with aza-dC adducts in genomic DNA, formed in vivo, within PrEC, RWPE-1, DU145, CWR22, PC-3 and LNCaP cells. (C) Slot blot band densities were quantified. ICM was performed three times and a representative slot blot as well as slot band density profiles are presented. Quantitative RT–PCR was used to measure (D) DNMT1 and (E) DNMT3B RNA abundance relative to GAPDH in each of the cell lines, and values were normalized to results from PrEC. Experiments were performed in triplicate, and average values are presented. Bars indicate standard error of the mean. p values: ‘***’, ≤ 0.001; 0.001< ‘**’ ≤ 0.01; 0.01< ‘*’ ≤ 0.05. Chromatin immunoprecipitation (ChIP) was used to determine binding of (F) DNMT1 and (G) DNMT3B to the transcription suppressor domain of RAD9 intron 2. Studies were performed with PrEC, RWPE-1, LNCaP, CWR22, PC-3 and DU145 cells. ‘Ab’ indicates that DNMT1 or DNMT3B antibodies were used for the assay, and ‘C’ indicates use of IgG control antibodies. (H) Input DNA is indicated. Band densities derived from ChIP with either (I) DNMT1 or (J) DNMT3B antibodies, minus background ChIP using IgG antibodies, relative to input DNA band densities are presented. Representative data from three independent trials are shown.
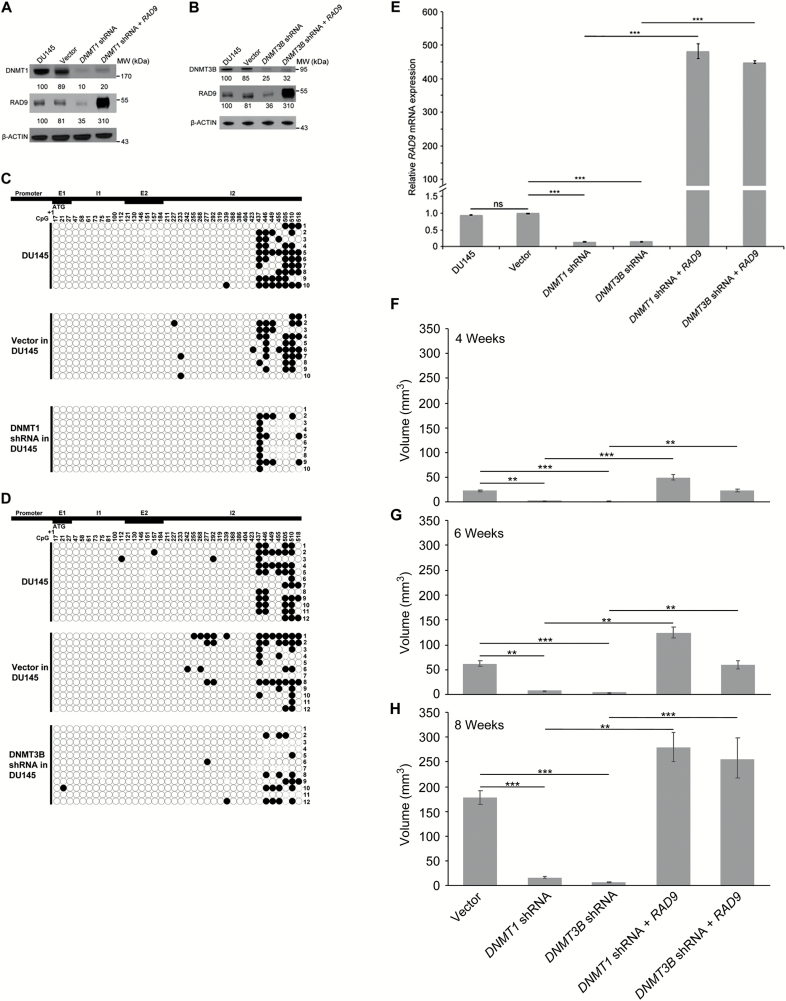
Figure 2.
Knock down of DNMT1 or DNMT3B expression in DU145 cells reduces RAD9 protein abundance, methylation of CpG sites within the transcription suppressor domain of RAD9 intron 2 and tumorigenic capability in mouse xenographs. (A) Western blot analysis of DNMT1, RAD9 and β-ACTIN control protein levels in DU145 cells, those cells containing an insertless vector, a vector encoding DNMT1 shRNA, or the latter additionally expressing RAD9. DNMT1 and RAD9 band densities, relative to β-ACTIN, in DU145 cells was designated 100, and relative band densities in the other cells normalized to within DU145 are indicated. (B) Western blot analysis of DNMT3B, RAD9 and β-ACTIN control protein levels in DU145 cells, those cells containing an insertless vector, with a vector encoding DNMT3B shRNA, or the latter additionally expressing RAD9. Comparable to A, DNMT3B and RAD9 band densities, relative to β-ACTIN, in DU145 cells was designated 100, and relative band densities in the other cells normalized to within DU145 are indicated. (C) Bisulfite sequencing was used to determine methylation of CpG sites 437 to 518 within the transcription suppressor domain of RAD9 intron 2. Ten clones were examined from DU145 cells, those containing an insertless vector, or DU145 containing a vector encoding DNMT1 shRNA. (D) Twelve clones were examined from DU145 cells, those containing an insertless vector, or DU145 containing a vector encoding DNMT3B shRNA. Black dot indicates a methylated CpG site. +1, ATG start of translation; E1, exon 1; I1, intron 1; E2, exon 2; I2, intron 2. (E) Effect of DNMT1 and DNMT3B knockdown on RAD9 RNA abundance was detected by qRT–PCR, and results reflected findings at the protein level; abundance of RAD9 in the knockdown cells after ectopic expression is also indicated. RAD9 levels were calculated relative to GAPDH RNA levels in respective cells. Experiments were performed in triplicate. Values normalized against DU145 with insertless vector. P-values: ‘***’, ≤ 0.001; ‘ns’ > 0.1. The tumorigenic potential of the cells was also assessed. Human cells were injected subcutaneously into the backs of nude mice, and growths at injection sites were monitored from 4 to 8 weeks post-injection: (F) 4 weeks; (G) 6 weeks; (H) 8 weeks. Each column represents the mean tumor volume at 6 to 12 injection sites. Bars indicate standard error of the mean. P-values: ‘***’, ≤ 0.001; 0.001< ‘**’ ≤ 0.01.
Results
Prostate cancer cells overproducing RAD9 have high levels of DNMT1 and DNMT3B
We demonstrated previously that human prostate cancer cells CWR22, DU145, LNCaP and PC-3 have high levels of RAD9, relative to non-cancer prostate PrEC cells (3). RWPE-1, immortalized non-tumorigenic prostate cells, have an intermediate level of RAD9 (Supplementary Figure 2A, available at Carcinogenesis Online). We showed that a suppressor of transcription within RAD9 intron 2 in DU145 is hypermethylated, and that is responsible for high levels of RAD9 expression in those cells (13). To examine whether DNMT1 and DNMT3B play a role in hypermethylating RAD9 intron 2 in DU145, we used the highly sensitive ICM assay first to quantify the association of these DNMTs in vivo to DNA aza-dC adducts in the total genome (27). There are much higher amounts of bound DNMT1 in all four prostate cancer cell lines, compared to PrEC and RWPE-1 (Figure 1A). DNMT3B, a protein generally less abundant than DNMT1, is also comparatively more plentiful in CWR22, DU145, LNCaP and PC-3 (Figure 1B). Band densities in the slot blots are illustrated in Figure 1C. The ICM assay is a method to determine activity of the DNMTs in the context of the entire genome. Hence, the activity in the other cancer cell lines compared to DU145 is not low, and lower activity therefore does not explain lower methylation of the intron 2 site in the CWR22, LNCaP and PC-3 cell lines.
We performed qRT–PCR to corroborate these findings at the RNA level. We found that DNMT1 (Figure 1D) and DNMT3B (Figure 1E) RNA are more abundant in the four cancer cell populations and in RWPE-1, relative to PrEC.
Differential binding of DNMT1 and DNMT3B to the RAD9 transcription suppressor
We found that global genomic binding of DNMT1 and DNMT3B to aza-dC adducts, as well as corresponding RNA levels for these proteins, was high in CWR22, DU145, LNCaP and PC-3, relative to PrEC. RWPE-1 cells also have elevated levels. However, only DU145 cells are highly methylated at CpG residues within the RAD9 transcription suppressor (Supplementary Figure 2B, available at Carcinogenesis Online) (13). To test whether differential binding of DNMT1 and DNMT3B in these cells could explain differences in DNMT levels versus CpG methylation, we performed ChIP. As per Figures 1F–J, DNMT1 and DNMT3B bind more abundantly to the suppressor in DU145 than in the other cells. The lower ChIP activity in the cancer cell lines other than DU145 is consistent with previous studies where we showed some methylation at the intron 2 site in CWR22, LNCaP and PC-3 but not as much as DU145 (13). This result indicates that although DNMT1 and DNMT3B are plentiful in all cancer cells tested, CpG sites in the RAD9 suppressor are likely hypermethylated only in DU145 due to greater RAD9-DNMT1/3B interactions in those cells.
DNMT1 and DNMT3B shRNA reduce methylated sites within the RAD9 transcription suppressor and RAD9 protein abundance in DU145
To demonstrate that DNMT1 and DNMT3B are responsible for RAD9 hypermethylation and high expression in DU145, we stably transfected these cells with DNMT shRNAs, or related insertless vectors. Then, we examined DNMT1, DNMT3B and RAD9 expression. Relative to DU145 cells or those with insertless vector, DNMT1 shRNA reduces DNMT1 and RAD9 protein abundance (Figure 2A). DNMT3B shRNA reduces DNMT3B and RAD9 levels, compared to controls (Figure 2B). Thus, DNMT1 and DNMT3B can regulate RAD9 protein quantities. Additionally, as per Figure 2A and B (lane 4 of each gel), we examined DU145 cells wherein DNMT1 and DNMT3B expression were knocked down, respectively, while ectopically overexpressing RAD9. These cells were used for tumorigenesis studies described later (Figure 2F–H).
To test whether DNMT1 and DNMT3B shRNA reduce methylation of RAD9 suppressor CpG sequences in DU145, we performed bisulfite sequencing of that region in cells used for Figure 2A and B. As per Figure 2C, of 10 clones sequenced, each with 9 CpG sites in the RAD9 transcription suppressor, DU145 cells have 46 sites methylated, insertless vector has 34, and cells knocked down for DNMT1 expression have 17. As per Figure 2D where 12 clones per cell population were examined, DU145 had 44 CpG sites methylated, vector control had 34, but DU145 with DNMT3B knockdown had only 18. In summary, relative to controls, DU145 cells with reduced RAD9 and DNMT1 or DNMT3B protein have fewer methylated cytosines in the RAD9 transcription suppressor. When DNMT1 or DNMT3B expression is knocked down by shRNA, RAD9 RNA levels, measured by qRT–PCR, are significantly reduced relative to DU145 cells with insertless vector (Figure 2E). Ectopic expression of RAD9 in DNMT1 or DNMT3B knockdown cells restores high abundance of RAD9 RNA, as predicted.
Knockdown of DNMT1 or DNMT3B expression reduces tumorigenicity of DU145 cells, which is restored by ectopic RAD9 expression
Elevated levels of RAD9 within DU145 cells cause tumorigenesis, as demonstrated by grafting cells into mice (13). DU145 cells with high levels of RAD9 can form tumors, whereas DU145 cells with reduced, knocked down levels of RAD9 have diminished capacity to do so. We tested whether DNMT1 and DNMT3B regulation of RAD9 is important for tumorigenesis, using DU145 cells stably transfected with insertless vector, or made to produce DNMT1 or DNMT3B shRNA, alone or in combination with ectopic RAD9 expression (Figure 2A and B). Each cell population was injected at 6 to 12 sites, subcutaneously into the backs of nude mice, and tumor growth was measured as a function of time. Between 4 and 8 weeks, post-injection, there were significantly larger tumor volumes at sites injected with DU145 cells bearing insertless vector, versus DU145 producing shRNA against DNMT1 or DNMT3B (Figure 2F–H). However, ectopic expression of RAD9 (albeit at levels higher then parental DU145) increased tumorigenicity of DU145 cells knocked down for either DNMT1 or DNMT3B (Figure 2F–H), thus rescuing the tumorigenic phenotype noted for controls. Effect sizes and P-values for these results are listed in Supplementary Table 1, available at Carcinogenesis Online. These studies indicate that DNMT1 and DNMT3B are needed for the tumorigenic activity of DU145 cells, secondary to their ability to regulate RAD9 protein abundance. In support, we determined that knockdown of DNMT1 or DNMT3B in PC-3 cells, where RAD9 is not regulated by methylation of intron 2, had no effect on RAD9 protein abundance, RAD9 suppressor site methylation or tumorigenicity (data not shown).
Immunohistochemical staining of human prostate tissues for RAD9, DNMT1 and DNMT3B
To expand upon our initial findings and further the translational relevance of the work, we examined prostate specimens immunohistochemically for RAD9, DNMT1 and DNMT3B. Validation for RAD9 staining is demonstrated in Supplementary Figure 3A, available at Carcinogenesis Online, where immunocytochemical staining for RAD9 in PC-3 cells is very high, but dramatically diminished in shRAD9 knockdown or crRAD9 knockout derivatives, or when anti-RAD9 is not applied, as predicted. A western blot illustrated the quantities of RAD9 in those cell populations (Supplementary Figure 3B, available at Carcinogenesis Online), and results commensurate with the immunocytochemical staining data. Figure 3A displays tissue sections considered negative (−), weak (+), strong (++) or very intense (+++) for immunostaining each of these proteins. Table 1 lists results for 22 non-cancer human prostate samples. Of note, when tissue sections showed regional differences in staining for the same protein, multiple levels of staining are listed. As per Table 1, undetectable or very weak levels of all three proteins were observed, consistent with our published data (13), as well as with cell-based results herein that examined DNMT1 and DNMT3B RNA levels and protein activity (Figures 1A–E). Table 2 lists immunohistochemical results for 17 prostate tumor sections. Relative to non-cancer samples, a much higher percentage of tissues was positive for all three proteins. When protein was detected, staining intensity was much greater in the cancer cohort. Moreover, unlike many non-cancer specimens, none of the cancer tissues demonstrated a completely negative stain for any of the proteins assayed. Abundance levels of all three proteins tended to be consistent within each sample [e.g. PR803E7-T V had regions that stain strongly (++) for RAD9, DNMT1 and DNMT3B. In contrast, 364-T III stained weakly (−/+) for all three]. Statistical analyses of the relationships of DNMT1, DNMT3B and RAD9 protein levels with respect to each other and prostate cancer, as well as methylation of the RAD9 transcription suppressor, are presented at the end of the Results section. Published reports by others also indicate low levels of DNMT1 and DNMT3B in non-cancer prostate cells and tissues. Furthermore, there is a strong, established correlation between higher levels as well as activities of these proteins, and advanced stage of prostate cancer (34,35).
RAD9 transcription suppressor CpG methylation in non-cancer and cancerous prostate tissue
We demonstrated that a high percentage of prostate cancer tissue has elevated levels of RAD9, DNMT1 and DNMT3B, relative to non-cancer controls (Tables 1 and 2) (13). To assess whether DNA methylation might play a role in RAD9 regulation within prostate tumors, we examined methylation status of nine CpG sites in the RAD9 transcription suppressor of 17 prostate cancer and 22 non-cancer tissues. We evaluated these sites in 12 independent samples from each of the tissue specimens, via bisulfite sequencing (Table 3). In non-cancer tissues, 57 of 2,376 (2.4%) transcription suppressor CpG sites are methylated. Eight tissues had no detectable methylation (8 of 22 specimen; 36.4%), and most positives contained one to four modified CpG. Tissues N10, N11 and N12 were exceptions, with 18, 9 and 7 modified sites, respectively. In stark contrast, 158 of 1,836 (8.6%) potentially modifiable sites in the cancer specimens were methylated, a 3.6-fold increase relative to the average of non-cancer specimen. Only three of the cancer tissues (3 of 17; 17.6%) had none of these CpG sites methylated.
Table 3.
CpG methylation in the transcription suppressor region of RAD9 intron 2 in non-cancer and cancerous prostate tissues
| Sample number | Non-cancer prostate tissue | Number of methylated CpG sitesa | Sample number | Cancer prostate tissueb | Number of methylated CpG sitesa |
|---|---|---|---|---|---|
| 1 | 141N | 0 | 1 | PR803E7-T IV | 3 |
| 2 | 142N | 0 | 2 | 375-T IV | 11 |
| 3 | 143N | 0 | 3 | 376-T IV | 17 |
| 4 | 320N | 3 | 4 | PR803G2-T IV | 11 |
| 5 | 321N | 0 | 5 | 362-T IV | 0 |
| 6 | 322N | 0 | 6 | 363-T IV | 22 |
| 7 | 323N | 2 | 7 | 377-T III | 25 |
| 8 | N1 | 0 | 8 | 378-T III | 17 |
| 9 | N2 | 0 | 9 | PR208K6/7 III | 1 |
| 10 | N3 | 2 | 10 | 364-T III | 1 |
| 11 | N4 | 3 | 11 | 365-T III | 0 |
| 12 | N5 | 4 | 12 | 371-T III | 21 |
| 13 | N6 | 2 | 13 | 372-T III | 1 |
| 14 | N7 | 1 | 14 | 370-T II-III | 1 |
| 15 | N8 | 2 | 15 | J7,8 II | 0 |
| 16 | N9 | 0 | 16 | 1125 IV | 15 |
| 17 | N10 | 18 | 17 | 1126III | 12 |
| 18 | N11 | 9 | |||
| 19 | N12 | 7 | |||
| 20 | N13 | 1 | |||
| 21 | N14 | 1 | |||
| 22 | N15 | 2 | |||
| Total number of methylated CpG sitesc | 57 | 158 | |||
| Percent methylated CpG sitesd | 2.4 (57/2376) | 8.6 (158/1836) |
aThis indicates number of methylated CpG sites in the RAD9 intron 2 transcription suppressor region and reflects the number within 9 potentially methylated sites examined in 12 clones per sample.
bRoman numerals (II, II-III, III, IV) indicate stage of cancer tissue.
cThe total number of methylated RAD9 intron 2 transcription suppressor CpG sites detected for non-cancer and cancer prostate tissues is indicated.
dPercent methylated CpG sites are calculated by total number of CpG methylated sites detected for non-cancer or cancer tissues, divided by the number of sites that could be methylated (9 per intron 2 suppressor region X 12 clones per sample tested X 22 samples examined for non-cancer or X 17 for cancer tissues), times 100.
Statistical analyses of relationships among DNMT1, DNMT3B, RAD9, RAD9 suppressor CpG methylation and cancer in human prostate tissue
Relationships among levels of DNMT1, DNMT3B and RAD9 proteins (Tables 1 and 2), frequency of RAD9 transcription suppressor CpG methylation (Table 3) and prostate cancer were analyzed statistically to assess significance. Figure 3B through 3J illustrates pairwise relationships. Supplementary Table 2, available at Carcinogenesis Online, lists effect sizes and P-values of these relationships. Analyses revealed that abundance of RAD9, DNMT1 and DNMT3B is greater in cancerous than in noncancerous prostate tissue (Tables 1 and 2; Supplementary Table 2, available at Carcinogenesis Online; Figures 3B–D). Amounts of these proteins are positively correlated with each other (Figures 3E and F; Supplementary Table 2, available at Carcinogenesis Online). In particular, RAD9 abundance is positively correlated with that of DNMT1 and DNMT3B, consistent with DNMT1 and DNMT3B increasing expression of RAD9 by methylating the transcription suppressor of the gene in a high percentage of prostate tumors. RAD9 suppressor is methylated to a greater extent in cancerous rather than in non-cancerous prostate tissue (Figure 3G; Table 3; Supplementary Table 2, available at Carcinogenesis Online), again suggesting that methylation of this region is important for prostate cancer. RAD9 protein level is positively correlated with frequency of RAD9 suppressor methylation (Figure 3H; Supplementary Table 2, available at Carcinogenesis Online) as well. Further, number of Me-CpG sites is proportional to DNMT1 and DNMT3B protein levels (Figure 3I and J; Supplementary Table 2, available at Carcinogenesis Online). These results all support the model that RAD9 expression is modulated by methylation of the transcription suppression site by DNMT1 and DNMT3B, and that event is critical for prostate carcinogenesis.
Discussion
RAD9 is aberrantly overexpressed in a large fraction of human prostate cancer cell lines and is critical for their ability to form tumors as a mouse xenograph, and demonstrate metastasis related phenotypes in vitro, such as anchorage independent growth, anoikis resistance, and rapid cell migration (13,14). Investigations with breast tumors and MCF-7 cancer cells, where RAD9 expression is high, showed transcription factor Sp1/3 sites in intron 2 of the gene are hypermethylated and that enhances RAD9 transcription (24). The same methylation-based mechanism occurs in prostate cancer DU145 cells, although they are not any more aggressive at forming tumors than other human prostate cancer cell lines where RAD9 quantities are also high but by a different mechanism (13). About 45% of prostate tumors have high RAD9 levels, whereas only 4% of non-cancer prostate specimen demonstrate low yet detectable amounts of the protein. Herein, we report that prostate cancer tissues had, on average, 3.6-fold more RAD9 Sp1/3-interacting transcription suppressor CpG sites hypermethylated than noncancer tissues and, concomitantly, higher levels of RAD9 protein. These results provide evidence that RAD9 is regulated by methylation in some prostate tissues to potentiate cancer.
DNA methyltransferases are a family of enzymes that methylate cytosines of CpG dinucleotides (20). We addressed whether DNMT1 and DNMT3B were critical for RAD9 intron 2 transcription suppressor site methylation. Initially, we looked at four prostate cancer cell lines, PC-3, CWR22 and LNCaP, as well as DU145 in which we already demonstrated the latter is the only cell line amongst those cited where CpG sequences are hypermethylated and critical for RAD9 regulation. Unexpectedly, we found all four cell populations, as well as non-tumorigenic, immortalized RWPE-1 cells, had high levels of both DNA methyltransferases, compared to PrEC. However, ChIP analyses indicated only the RAD9 suppressor site within DU145 cells bound DNMT1 and DNMT3B abundantly in vivo. This is consistent with previous findings indicating that some methylation at the intron 2 site is found in all the prostate cancer cell lines examined but the most was detected in DU145 where RAD9 is regulated by hypermethylation (13). Several factors can influence DNA methyltransferase activity and could be responsible for the cell line differences, despite all demonstrating high quantities of DNMT1 and DNMT3B. The major de novo methylation enzymes DNMT3A and DNMT3B form complexes and cooperate with maintenance methylation enzyme DNMT1, which leads to spreading of methylation in the genome (36). Chromatin structure affects localization and activity of DNMT1 (37) and DNMT3B (38). In addition, DNA methyltransferases localize more frequently to transcribed regions of the genome, and presence of two or more of these DNMTs is strongly associated with sequences targeted for DNA hypermethylation (38,39). Therefore, although not yet experimentally addressed, unique cell line–specific features of DNMT protein complex formation or chromatin structure near RAD9 might dictate the differential extent of methylation observed. Moreover, our findings indicate that there is a difference between DU145 and the other cancer cell lines in the way they regulate RAD9 expression. For example, CWR22 and DU145 have the same activity levels of the DNMTs but there is a large difference in ChIP at intron 2. The RAD9 gene is amplified in PC-3, which could explain the high abundance of the protein in this cell line. However, the mechanism of aberrantly high RAD9 expression in CWR22 or LNCaP cells has not been defined. Perhaps alterations in DNA regulatory elements, other than the transcription suppressor site in RAD9 intron 2, or hyperactive transcription factor activity drive the high RAD9 levels observed. This also needs to be determined.
We found that the activity of DNMT1 was much higher than that of DNMT3B in DU145 cells. Nevertheless, the shRNA of each of the DNMTs caused similar knockdown of RAD9. Further, knockdown of an individual DNMT by shRNA is able to substantially decrease RAD9 level even though the other DNMT is present. The reason for this is not clear. The differential extent of each DNMT knockdown by their respective shRNAs (10-fold for DNMT1 versus 4-fold for DNMT3B) might play a role, especially if total DNA methyltransferase activity is influenced by individual DNMT function as well as their interactions. Also, the question arises as to whether DNMT1 and DNMT3B can methylate specific CpG sites, and if one site is more critical than another for RAD9 expression. In this regard, it is interesting to note that we found CpG position 437 within the RAD9 transcription suppressor region methylated in 9 of 10 sites when DNMT1 is knocked down and not methylated in 12 of 12 sites when DNMT3B is knocked down.
Decreased RAD9 expression by RNAi reduces or eliminates the ability of prostate cancer cells to form tumors when injected subcutaneously into mice (13). We show that knockdown of DNMT1 or DNMT3B also reduces tumor formation by DU145 cells. This is due to the resulting downregulation of RAD9, as ectopic expression of only that gene in DU145 with reduced expression of either DNMT1 or DNMT3B restores ability of the cells to more frequently form tumors. This is also supported by studies indicating that DNMT1 or DNMT3B knockdown in PC-3 cells, which have high amounts of RAD9 not due to a methylation-related mechanism, do not show reduced tumorigenicity in mouse xenographs, relative to control cells expressing wild type levels of these two DNMTs (data not shown). This latter finding, together with our demonstration that DNMT1 and DNMT3B primarily bind to the transcription suppressor region of RAD9 in DU145 and not in PC-3, suggests that these DNA methyltransferases directly regulate RAD9 transcription. Furthermore, the finding that hypermethylation of CpG mediated by DNMT1 and DNMT3B regulates RAD9 appears not just confined to DU145 cells, as we show that a high percentage of prostate tumor tissues, relative to non-cancer controls, have high levels of RAD9, DNMT1, DNMT3B and hypermethylated RAD9 transcription suppressor.
These findings suggest that DNA methyltransferases could serve as therapeutic targets for some prostate cancer patients, and such investigations have been reported (40). 5-Azacitidine inhibits DNMT-mediated methylation and has shown promise as an anti-tumor agent in both in vivo preclinical models (41,42) and early stage clinical trials (43) for this disease. Given our demonstration that hypermethylation regulates overproduction of RAD9, and that in turn promotes prostate carcinogenesis, 5-azacitidine might be mediating reported anti-carcinogenic effects at least in part through control of RAD9 abundance.
Although it is clear that high levels of RAD9 cause prostate tumorigenesis, the underlying mechanism is not well understood. RAD9 participates in activities that influence genomic stability, including apoptosis, cell cycle checkpoints and multiple DNA repair processes (2). High levels of the protein could alter these activities and serve as the driving force to trigger carcinogenesis, including metastasis. RAD9 also functions as a transcription factor, with a select set of target genes, and some have roles in carcinogenesis (1,2,5,25). Furthermore, Wen et al. (44) demonstrated that RAD9 suppresses the epithelial-mesenchymal transition through inhibition of SLUG transcription, so negative regulation of gene transcription could be another mechanism to evaluate in terms of tumor development.
In summary, we provide evidence for a novel mechanism of RAD9 regulation and function in prostate cancer. DNMT1 and DNMT3B hypermethylate CpG sites within RAD9 intron 2 in prostate cancer cells, inhibiting a transcription suppressor domain, and increasing RAD9 expression. This dysregulates activities important for genomic stability and cancer suppression (8,26,44). High levels of RAD9 promote genomic instability and cancer through inappropriate transcriptional control of RAD9 target genes, or by mechanisms unrelated to transcription. RAD9 physically interacts with proteins, such as members of the base excision repair machinery, stimulates their activity (45) and consequently alters genome stability, but effects of RAD9 overproduction are not known. High levels of RAD9 promote phosphorylation-mediated activation of AKT, critical for anoikis resistance (14). Moreover, high levels of RAD9 influence radioresistance by increasing ITGB1 abundance through stabilizing the protein (14,46). Determining the complete list of functions driven by high levels of RAD9 that influence genomic stability, promote prostate cancer and enhance metastasis is critical for understanding the molecular basis of the disease. In terms of RAD9 regulation by DNA methylation, there are other reports where epigenetic status affects cancer-related gene expression, including for prostate cancer, and the relationship to RAD9 should be investigated (47). Identifying critical upstream and downstream network elements could lead to novel, anti-cancer strategies to diagnose and treat patients with prostate cancer (48,49).
Funding
National Institutes of Health (CA130536 to H.B.L.). This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant Number UL1 TR001873. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This research was also funded in part through the National Institutes of Health/National Cancer Institute Cancer Center Support Grant P30CA013696 and used the Molecular Pathology Shared Resource.
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations
- CRISPR
clustered regularly interspaced short palindromic repeats
- DNMTs
DNA methyltransferases
- PrEC
prostate epithelial cells
- qRT–PCR
quantitative reverse transcription–PCR
- SP1/3
Specific Protein 1/3
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- ICM
in vivo complex of methylation
- ChIP
chromatin immunoprecipitation
References
- 1. Lieberman, HB et al. (2017) p53 and RAD9, the DNA damage response, and regulation of transcription networks. Radiat. Res., 187, 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Broustas, CG et al. (2019) RAD9A promotes metastatic phenotypes through transcriptional regulation of anterior gradient 2 (AGR2). Carcinogenesis, 40, 164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lieberman, HB et al. (2011) The role of RAD9 in tumorigenesis. J. Mol. Cell Biol., 3, 39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hopkins, KM et al. (2004) Deletion of mouse rad9 causes abnormal cellular responses to DNA damage, genomic instability, and embryonic lethality. Mol. Cell. Biol., 24, 7235–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pandita, RK et al. (2006) Mammalian Rad9 plays a role in telomere stability, S- and G2-phase-specific cell survival, and homologous recombinational repair. Mol. Cell. Biol., 26, 1850–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He, W et al. (2008) Rad9 plays an important role in DNA mismatch repair through physical interaction with MLH1. Nucleic Acids Res., 36, 6406–6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li, T et al. (2013) Checkpoint protein Rad9 plays an important role in nucleotide excision repair. DNA Repair (Amst)., 12, 284–292. [DOI] [PubMed] [Google Scholar]
- 8. Panigrahi, SK et al. (2015) Regulation of NEIL1 protein abundance by RAD9 is important for efficient base excision repair. Nucleic Acids Res., 43, 4531–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai, FL et al. (2014) The checkpoint clamp protein Rad9 facilitates DNA-end resection and prevents alternative non-homologous end joining. Cell Cycle, 13, 3460–3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Crespan, E et al. (2012) Microhomology-mediated DNA strand annealing and elongation by human DNA polymerases λ and β on normal and repetitive DNA sequences. Nucleic Acids Res., 40, 5577–5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komatsu, K et al. (2000) Human homologue of S. pombe Rad9 interacts with BCL-2/BCL-xL and promotes apoptosis. Nat. Cell Biol., 2, 1–6. [DOI] [PubMed] [Google Scholar]
- 12. Zhu, A et al. (2005) Differential impact of mouse Rad9 deletion on ionizing radiation-induced bystander effects. Radiat. Res., 164, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu, A et al. (2008) Rad9 has a functional role in human prostate carcinogenesis. Cancer Res., 68, 1267–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Broustas, CG et al. (2012) Rad9 protein contributes to prostate tumor progression by promoting cell migration and anoikis resistance. J. Biol. Chem., 287, 41324–41333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hu, Z et al. (2008) Targeted deletion of Rad9 in mouse skin keratinocytes enhances genotoxin-induced tumor development. Cancer Res., 68, 5552–5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dolinoy, DC et al. (2007) Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr. Res., 61(5 Pt 2), 30R–37R. [DOI] [PubMed] [Google Scholar]
- 17. Feinberg, AP et al. (1983) Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature, 301, 89–92. [DOI] [PubMed] [Google Scholar]
- 18. Rhee, I et al. (2002) DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature, 416, 552–556. [DOI] [PubMed] [Google Scholar]
- 19. Gaudet, F et al. (2003) Induction of tumors in mice by genomic hypomethylation. Science, 300, 489–492. [DOI] [PubMed] [Google Scholar]
- 20. Lyko, F (2018) The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet., 19, 81–92. [DOI] [PubMed] [Google Scholar]
- 21. Hermann, A et al. (2004) The Dnmt1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem., 279, 48350–48359. [DOI] [PubMed] [Google Scholar]
- 22. Chen, T et al. (2003) Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol., 23, 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wienholz, BL et al. (2010) DNMT3L modulates significant and distinct flanking sequence preference for DNA methylation by DNMT3A and DNMT3B in vivo. PLoS Genet., 6, e1001106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng, CK et al. (2005) The cell cycle checkpoint gene Rad9 is a novel oncogene activated by 11q13 amplification and DNA methylation in breast cancer. Cancer Res., 65, 8646–8654. [DOI] [PubMed] [Google Scholar]
- 25. Yin, Y et al. (2004) Human RAD9 checkpoint control/proapoptotic protein can activate transcription of p21. Proc. Natl. Acad. Sci. U. S. A., 101, 8864–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shao, G et al. (2006) Epigenetic inactivation of Betaig-h3 gene in human cancer cells. Cancer Res., 66, 4566–4573. [DOI] [PubMed] [Google Scholar]
- 27. Liu, K et al. (2003) Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol. Cell. Biol., 23, 2709–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu, CH et al. (2012) Nickel-induced epithelial-mesenchymal transition by reactive oxygen species generation and E-cadherin promoter hypermethylation. J. Biol. Chem., 287, 25292–25302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paschos, K et al. (2012) BIM promoter directly targeted by EBNA3C in polycomb-mediated repression by EBV. Nucleic Acids Res., 40, 7233–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. R Core Team. (2017) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 31. Bretz, F et al. (2010) Multiple Comparisons Using R. CRC Press, Boca Raton, FL. [Google Scholar]
- 32. Crawley, M (2007) The R Book. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 33. Sarkar, D (2008) Lattice: Multivariate Data Visualization with R. Springer, New York, pp. 268. [Google Scholar]
- 34. Benbrahim-Tallaa, L et al. (2007) Tumor suppressor gene inactivation during cadmium-induced malignant transformation of human prostate cells correlates with overexpression of de novo DNA methyltransferase. Environ. Health Perspect., 115, 1454–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gravina, GL et al. (2013) Increased levels of DNA methyltransferases are associated with the tumorigenic capacity of prostate cancer cells. Oncol. Rep., 29, 1189–1195. [DOI] [PubMed] [Google Scholar]
- 36. Kim, GD et al. (2002) Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. Embo. J., 21, 4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robertson, AK et al. (2004) Effects of chromatin structure on the enzymatic and DNA binding functions of DNA methyltransferases DNMT1 and Dnmt3a in vitro. Biochem. Biophys. Res. Commun., 322, 110–118. [DOI] [PubMed] [Google Scholar]
- 38. Baubec, T et al. (2015) Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature, 520, 243–247. [DOI] [PubMed] [Google Scholar]
- 39. Jin, B et al. (2012) Linking DNA methyltransferases to epigenetic marks and nucleosome structure genome-wide in human tumor cells. Cell Rep., 2, 1411–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baumgart, SJ et al. (2017) Exploiting epigenetic alterations in prostate cancer. Int J Mol Sci., 18, E1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McCabe, MT et al. (2006) Inhibition of DNA methyltransferase activity prevents tumorigenesis in a mouse model of prostate cancer. Cancer Res., 66, 385–392. [DOI] [PubMed] [Google Scholar]
- 42. Gravina, GL et al. (2010) 5-Azacitidine restores and amplifies the bicalutamide response on preclinical models of androgen receptor expressing or deficient prostate tumors. Prostate, 70, 1166–1178. [DOI] [PubMed] [Google Scholar]
- 43. Singal, R et al. (2015) Phase I/II study of azacitidine, docetaxel, and prednisone in patients with metastatic castration-resistant prostate cancer previously treated with docetaxel-based therapy. Clin. Genitourin. Cancer, 13, 22–31. [DOI] [PubMed] [Google Scholar]
- 44. Wen, FC et al. (2014) hRAD9 functions as a tumor suppressor by inducing p21-dependent senescence and suppressing epithelial-mesenchymal transition through inhibition of Slug transcription. Carcinogenesis, 35, 1481–1490. [DOI] [PubMed] [Google Scholar]
- 45. Helt, CE et al. (2005) Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle, 4, 529–532. [DOI] [PubMed] [Google Scholar]
- 46. Broustas, CG et al. (2014) RAD9 enhances radioresistance of human prostate cancer cells through regulation of ITGB1 protein levels. Prostate, 74, 1359–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Massie, CE et al. (2017) The importance of DNA methylation in prostate cancer development. J. Steroid Biochem. Mol. Biol., 166, 1–15. [DOI] [PubMed] [Google Scholar]
- 48. Brien, GL et al. (2016) Exploiting the epigenome to control cancer-promoting gene-expression programs. Cancer Cell, 29, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lieberman, HB et al. (2018) Prostate cancer: unmet clinical needs and RAD9 as a candidate biomarker for patient management. Transl. Cancer Res., 7(Suppl 6), S651–S661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.