Abstract
LGR4 and LGR5 encode two homologous receptors with critical yet distinct roles in organ development and adult stem cell survival. Both receptors are co-expressed in intestinal crypt stem cells, bind to R-spondins (RSPOs) with high affinity, and potentiate Wnt/β-catenin signaling, presumably by the same mechanism – forming RSPO-bridged complexes with the E3 ligases RNF43 and ZNRF3 to inhibit ubiquitylation of Wnt receptors. However, direct evidence for RSPO-bound full-length LGR5 interacting with the E3 ligases in whole cells has not been reported, and only LGR4 is essential for the self-renewal of intestinal stem cells. Here, we examined the mechanisms of action of LGR4 and LGR5 in parallel using co-immunoprecipitation, proximity ligation, competition binding, and time-resolved FRET assays in whole cells. Full-length LGR4 formed a tight complex with ZNRF3 and RNF43 even without RSPO, whereas LGR5 did not interact with either E3 ligase with or without RSPO. Domain-swapping experiments with LGR4 and LGR5 revealed that the seven transmembrane domain of LGR4 conferred interaction with the E3 ligases. Native LGR4 and LGR5 existed as dimers on the cell surface, and LGR5 interacted with both FZD and LRP6 of the Wnt signalosome to enhance LRP6 phosphorylation and potentiate Wnt–β-catenin signaling. These findings provide a molecular basis for the weaker activity of LGR5 in potentiation of Wnt signaling that may underlie the distinct roles of LGR4 and LGR5 in organ development as well as the self-renewal and fitness of adult stem cells.
One-Sentence Summary:
LGR5 does not inhibit E3 ligases RNF43 and ZNRF3 to potentiate Wnt signaling.
Editor’s Summary:
Mechanistic differences between LGR4 and LGR5
The receptor LGR4 promotes Wnt signaling in response to the binding of R-spondin ligands (RSPOs) both by inhibiting the activities of the E3 ligases RNF43 and ZNRF3 and by directly stimulating the Wnt signalosome through the scaffold protein IQGAP1. It has been proposed that LGR5 stimulates Wnt signaling through the same mechanism as LGR4 despite the observation that LGR4 and LGR5 are not functionally equivalent in vivo. By analyzing the interactions of LGR4 and LGR5 with RSPOs, components of the Wnt signalosome, and RNF43 and ZNRF3 in cultured cells, Park et al. found that LGR5 did not bind to RNF43 or ZNRF3 regardless of the presence of RSPOs but did promote Wnt signaling through IQGAP1. They also found that LGR4 interacted with RNF43 or ZNRF3 regardless of the presence of RSPOs. These findings demonstrate that LGR4 and LGR5 potentiate Wnt signaling through distinct mechanisms.
Introduction
LGR5 is a well-established marker of adult stem cells in multiple epithelial tissues, including the gastrointestinal tract, liver, and skin (1, 2). LGR5 and its two closely related homologues, LGR4 and LGR6 (~50% amino acid identity), consist of a large extracellular domain (ECD) containing 17 leucine-rich repeats, a seven-transmembrane (7TM) domain similar to those of the rhodopsin family of G protein–coupled receptors, and an intracellular tail with potential phosphorylation sites (3, 4). In the gastrointestinal tract and skin, expression of LGR5 and LGR6 is limited to the stem cells, whereas LGR4 is expressed in both stem cells and proliferating progenitor cells (1, 5–7). Absence of LGR5 in the intestine affects only stem cell fitness with no apparent effect on stem cell self-renewal, but inducible removal of LGR4 leads to an immediate halt in cell proliferation in the crypts and collapse of the intestine (8–10), suggesting that LGR4 and LGR5 have distinct, non-equivalent functions in intestinal stem cells. Several studies have demonstrated that the four R-Spondins (RSPO1–4) function as high-affinity ligands of LGR4–6 to potentiate Wnt signaling (8, 11, 12). All four RSPOs contain a furin-like domain that binds to LGRs and is essential for the potentiation of Wnt signaling and a thrombospondin domain (TSP) that binds to the extracellular matrix and enhances the potencies of the RSPOs (13, 14). Mechanistically, LGR4-RSPO forms a trimeric complex with ZNRF3, an E3 ligase that ubiquitylates the Wnt receptor Frizzled (FZD) proteins for degradation (15, 16). When part of this trimeric complex, ZNFR3 is not able to ubiquitylate FZDs for degradation, leading to increased abundance of FZDs and, therefore, enhanced Wnt signaling (15, 16). LGR4 also interacts with the Wnt signalosome by binding to the scaffold protein IQGAP1 and potentiating Wnt–β-catenin signaling directly (8, 17).
For LGR5, crystal structure analysis showed that the extracellular domain (LGR5ECD) formed a 2:2 dimer with the furin domain of RSPO, whereas LGR5ECD and RSPO formed a 1:1:1 trimer with the extracellular domain of RNF43, a ZNRF3 homolog (18–20). These structural data prompted the model that RSPO-LGR5, like RSPO-LGR4, potentiates Wnt–β-catenin signaling by forming a trimeric LGR5-RSPO-E3 ligase complex that inhibits the ligase activity to increase Wnt receptor abundance (19–22). However, LGR5-RSPO-E3 ligase trimer formation has never been demonstrated with full-length LGR5 and E3 ligases in whole cells. Here, we examined LGR4 and LGR5 side-by-side in multiple assays and found that LGR5, unlike LGR4, did not interact with the E3 ligases in response to RSPO. Instead, full-length LGR5 interacted with both FZD and LRP6 of the Wnt signalosome and potentiated Wnt signaling in an IQGAP1-dependent manner.
Results
LGR5 potentiates Wnt–β-catenin signaling in response to RPSO1–3 but not in response to RSPO4
LGR5 binds all four RSPOs with high affinity and potentiates Wnt–β-catenin signaling in cultured cells in response to RSPO1–4 (8, 11, 23). Some studies have also reported that overexpression of LGR5 inhibits Wnt–β-catenin signaling in certain cancer cell lines (24, 25). A major caveat to these findings is that effects mediated by LGR4 could not be ruled out because most of these studies were carried out in cell lines that express LGR4 endogenously or in HEK293T cells with transient knockdown of endogenous LGR4 (8, 11, 18, 23, 25). To clarify the specific activities pf LGR5, we previously used a CRISPR/Cas9-genereated LGR4 knockout cell line derived from the Wnt–β-catenin reporter HEK293T-STF (Super TOPFlash) cell line (26, 27). This cell line (STF-LGR4KO) showed near total loss of response to RSPO1 and RSPO4 with partial response to RSPO2 and RSPO3 (27). From STF-LGR4KO cells, we generated cell lines that stably express Myc-tagged full-length LGR5 (STF-4KO-LGR5) or empty vector control (STF-4KO-vect). Expression of LGR5 in STF-4KO-LGR5 cells was validated by live cell staining with the LGR5-specific antibody 8F2 (8, 28), and binding of purified RSPO2-derived ligand with STF-4KO-vect cells was used as a negative control (27) (fig. S1, A to D).
We first compared the activities of LGR-dependent RSPO1 and RSPO4 in STF-4KO-LGR5 and STF-4KO-vect cells side-by-side using the TOPFlash assay (26). STF-4KO-vect cells showed no response to either RSPO, as expected, whereas STF-4KO-LGR5 cells showed a dose-dependent response to RSPO1 with high potency but little response to RSPO4 (Fig. 1A). RSPO2 and RSPO3 showed activity in STF-4KO-vect cells, and their potency was much higher in STF-4KO-LGR5 cells (Fig. 1B). However, the response of STF-4KO-LGR5 cells to RSPO4 was not restored even at high concentrations of RSPO4 (Fig. 1A) despite RSPO4’s high binding affinity to LGR5 (11, 20). This was distinct from parental HEK293T-STF or HEK293T cells overexpressing LGR4, which responded to all four RSPOs with RSPO4 showing approximately half of the efficacy (27, 29).
Fig. 1. LGR5 potentiates Wnt–β-catenin signaling without increasing Wnt receptor level.
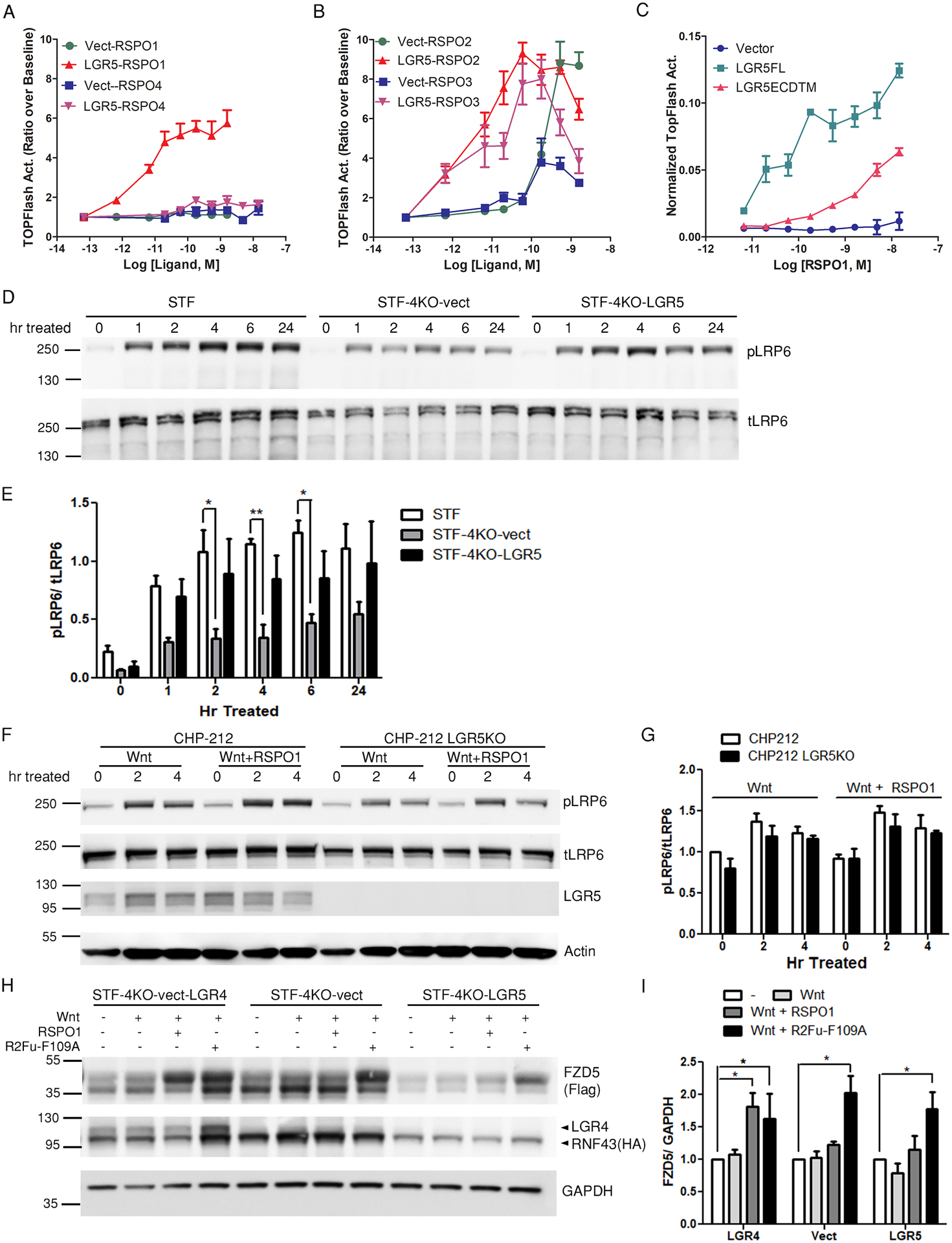
(A and B) TOPFlash activity of Super TOPFlash LGR4 knockout cells stably transfected with empty vector (STF-4KO-vect) or Myc-LGR5 construct (STF-4KO-LGR5) after stimulation with RSPO1 or RSPO4 (A) or with RSPO2 or RSPO3 (B) at the indicated concentrations in Wnt3a-conditioned media (CM). TOPFlash activity was normalized to baseline before RSPO addition in Wnt3a CM. (C) Normalized TOPFlash activity of Super TOPFlash LGR4 knockout (STF-LGR4KO) cells transiently transfected with empty vector, Myc-LGR5FL, or Myc-LGR5ECDTM after stimulation with the indicated concentrations of RSPO1 in Wnt3a CM. The TOPFlash activities are representatives of 6–10 independent experiments, and error bars show SEM of 2–3 technical repeats. (D and E) Western blot (D) and quantification (E) of phosphorylated LRP6 (pLRP6) and total LRP6 (tLRP6) in parental STF (endogenous LGR4 expression), STF-4KO-vect, and STF-4KO-LGR5 cell lines after treatment with Wnt3A CM + RSPO1. n = 3 independent experiments. Data represent mean and standard deviation (SD) of the quantified pLRP6 normalized to tLRP6. Two-way Anova Bonferroni posttest was performed, and * represents P < 0.05 against STF cells. (F and G) Western blot (F) and quantification (G) of pLRP6 and tLRP6 in CHP-212 and CHP-212 LGR5KO cells after treatment with Wnt3a CM only or Wnt3a CM + RSPO1. Actin is a loading control. Data represent mean and standard deviation (SD) of the quantified pLRP6 normalized to tLRP6. n = 3 independent experiments. Two-way Anova Bonferroni posttest was performed against non-treated cells, and no significance was found. (H and I) Western blot (H) and quantification (I) of RNF43 and FZD5 in STF-4KO-LGR5 cells co-expressing HA-RNF43 and FLAG-FZD5 (LGR5 overexpression) and STF-4KO-vect cells coexpressing HA-tagged LGR4,RNF43 and FZD5 (LGR4 overexpression) HA-RNF43 and FLAG-FZD5 after Wnt3a CM, Wnt3a CM + RSPO1, or Wnt3a CM + R2Fu-F109A treatment. STF-4KO-vect cells (no LGR4 or LGR5) co-expressing HA-RNF43 and FLAG-FZD5 are a negative control. GAPDH is a loading control. n = 3–4 independent experiments. Data represent mean and SD of FZD/GAPDH normalized to the untreated condition of each cell line. One-way Anova Dunnett’s multiple comparison test against the untreated cells was performed, and * represents P < 0.05.
We previously reported an RSPO2 furin domain mutant (designated R2Fu-F109A) that had normal binding affinity to ZNRF3 but lost binding to LGR4 and LGR5 due to a mutation (F109A) in the LGR-binding domain (27). We compared the activities of the wild-type RSPO2 furin domain (R2Fu-WT) and R2Fu-F109A in STF-4KO-LGR5 cells and STF-4KO-vect cells, and the data showed that R2Fu-WT was more active than R2Fu-F109A in LGR5-expressing cells but not in the vector control cells (fig. S2, A and B), providing further evidence that LGR5 could enhance Wnt signaling in response to RSPO. We then asked if the 7TM domain of LGR5 was essential for responding to RSPO by comparing the activity of full-length LGR5 with that of LGR5ECD fused to the single transmembrane (TM) domain of CD4 (LGR5ECDTM) using transient transfection in STF-LGR4KO cells. RSPO1 showed higher potency and efficacy in cells expressing full-length LGR5 cells compared to cells expressing LGR5ECDTM (Fig. 1C). Also, cells transfected with full-length LGR5 but not LGR5ECDTM showed an increase in basal Wnt signaling activity (Fig. 1C) even though both proteins were produced in similar amounts (fig. S3). These data indicate that the 7TM domain of LGR5 was essential for its full activity in potentiating Wnt signaling, whereas the ECD alone anchored to cell membrane had partial response to high concentrations of RSPO1. Overall, these data show that LGR5, similar to LGR4, could mediate RSPO-induced potentiation of Wnt–β-catenin signaling in a 7TM-dependent manner, except that LGR5 was not activated by RSPO4 whereas LGR4 was.
RSPO1-LGR5 enhances LRP6 phosphorylation without increasing FZD abundance
To understand the mechanism of LGR5-mediated potentiation of Wnt–β-catenin signaling and because previous studies had provided inconclusive results in cells with endogenous expression of LGR4 (11, 25), we first checked if RSPO1-LGR5 enhanced the phosphorylation of LRP5 and LRP6 (LRP5/6), which is a critical early step in the activation of Wnt–β-catenin signaling, in the absence of LGR4. Parental STF, STF-4KO-vect, and STF-4KO-LGR5 cells were stimulated with Wnt3a-conditioned media (Wnt3a CM) plus RSPO1, and LRP6 phosphorylation was followed for 24 hours. Although total LRP6 did not change during the time-course experiment, phosphorylated LRP6 (pLRP6) increased upon Wnt3a + RSPO1 treatment for all three cell lines tested (Fig. 1, D and E). Parental STF cells (expressing LGR4 endogenously) showed a strong and prolonged increase in pLRP6 that lasted 24 hours, and STF-4KO-LGR5 cells exhibited similar trends as parental STF cells (Fig. 1, D and E). On the other hand, the amounts of pLRP6 in STF-4KO-vect cells also increased upon Wnt3a + RSPO1 stimulation and was maintained throughout 24 hour treatment (Fig. 1, D and E), but pLRP6 amounts in STF-4KO-vect cells were significantly lower than those in the parental STF cells at the 2, 4, and 6 hour time points. Because RSPO1 had no activity in STF-4KO-vect cells in the TOPFlash assay at the concentrations tested (Fig. 1A), the data suggest that RSPO1-LGR5 enhanced LRP6 phosphorylation.
In order to confirm that the RSPO1-LGR5–induced increase in LRP6 phosphorylation was not limited to recombinant LGR5, we employed two neuroblastoma cancer cell lines (SK-NAS and CHP-212) that expresses LGR5 endogenously but do not express LGR4 (table S1) (30). In CHP-212 cells, the addition of RSPO1 enhanced Wnt3a-induced LRP6 phosphorylation, and this effect was attenuated when LGR5 was knocked out (Fig. 1, F and G). In SK-NAS cells, which were confirmed to produce high amounts of LGR5 by immunocytochemistry (ICC) (fig. S4A), the addition of RSPO1 also further increased LRP6 phosphorylation compared to stimulation with Wnt3a alone (fig. S4B). Because RSPO1 is totally dependent on LGR4 or LGR5 to enhance Wnt signaling (14, 27), these results confirmed that endogenous LGR5 could enhance LRP6 phosphorylation in response to RSPO1 stimulation.
RSPO-LGR4 inhibits RNF43 and ZNRF3 (RNF43/ZNRF3), thus reducing the degradation of the Wnt co-receptor FZD and enhancing LRP6 phosphorylation (15). We directly tested whether RSPO-LGR5 would also increase FZD abundance in response to RSPO1 when co-expressed with E3 ligases. STF-4KO cells expressing vector, LGR4, or LGR5 were co-transfected with FLAG-tagged FZD5 (FLAG-FZD5) and HA-tagged RNF43 (HA-RNF43), and then treated with vehicle, Wnt3a, Wnt3a+RSPO1, or Wnt3a+R2Fu-F109A, which inhibits E3 ligase activity independently of LGR (27). Although we used the pan-FZD antibody, 18R5, in subsequent experiments to detect endogenous FZDs using the pan-FZD antibody, for these experiments we used recombinant, tagged FZD5 because 18R5 does not detect denatured proteins, and the change in the endogenous FZD levels upon treatments would likely not be observable in the presence of an excess of recombinant RNF43. RSPO1 significantly increased the amount of FLAG-FZD5 in cells expressing LGR4 as expected (Fig. 1, H and I) but not in cells expressing LGR5 or in vector control cells (Fig. 1, H and I). As a positive control, R2Fu-F109A significantly increased FZD5 abundance in all three cell lines, consistent with its LGR-independent activity. Together with the TOPFlash data, these results suggest that both LGR4 and LGR5 could enhance LRP6 phosphorylation to potentiate Wnt–β-catenin signaling in response to RSPO; however, RSPO1-LGR5 appeared to achieve this function without preventing the degradation of FZD.
Full-length LGR5 does not interact with ZNRF3 or RNF43 in response to RSPO
RSPO-LGR4 interacts directly with ZNRF3 and inhibits the ligase’s activity (15). Because RSPO1-LGR5 did not block RNF43 activity (Fig. 1, H and I), we compared LGR4 and LGR5 side-by-side in interaction with ZNRF3 in the absence and presence of RSPOs by co-immunoprecipitation and proximity ligation assay (PLA). HEK293T cells were co-transfected with a Myc-tagged form of ZNRF3 missing its RING domain (Myc-ZNRF3ΔR) and HA-tagged full-length LGR4 or LGR5 (HA-LGR4FL or HA-LGR5FL). Use of ZNRF3ΔR was necessary because overexpression of full-length ZNRF3 is toxic to the cells, and deletion of the RING domain in ZNRF3 has no effect on ZNRF3’s interaction with RSPO-LGR4 (15). LGR4FL was readily pulled down by immunoprecipiation of ZNRF3 even in the absence of RSPO, whereas little if any LGR5FL was pulled down by immunoprecipitation of ZNRF3 with or without RSPO (Fig. 2A). The amounts of LGR4, LGR5, and ZNRF3 in the cells were similar (Fig. 2A). Similar results were obtained RNF43 immunoprecipitates were blotted for LGR5 (fig. S5). However, a small amount of LGR5ECDTM was pulled down by RNF43 in the presence of RSPO1 (fig. S5), suggesting that RSPO1 induced LGR5ECDTM-RNF43 complex formation, which may explain the partial response of LGR5ECDTM to RSPO1 in the TOPFlash assay (Fig. 1C).
Fig. 2. LGR5 does not interact with ZNRF3 or RNF43.
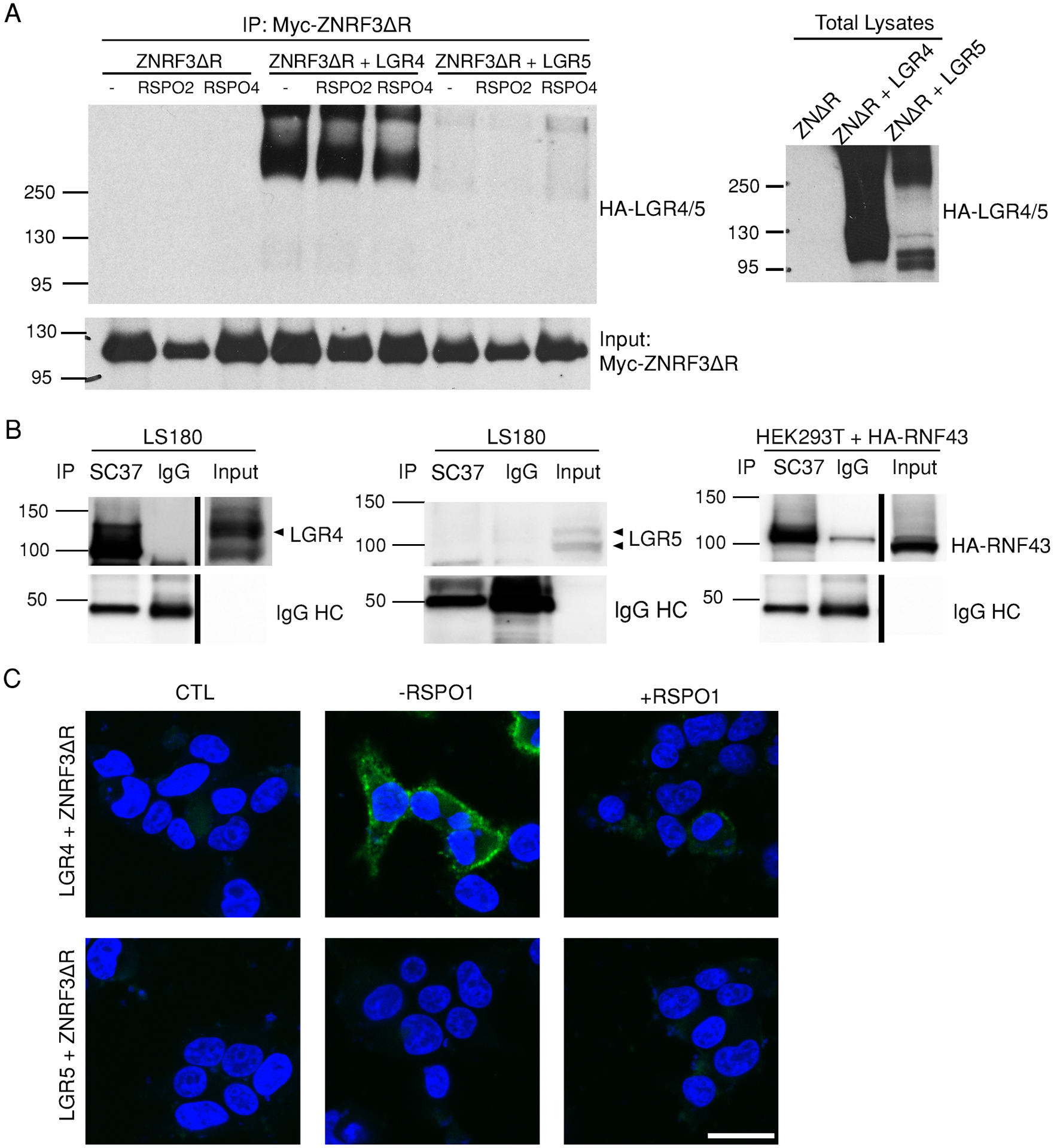
(A) Co-immunoprecipitation of LGR4 and LGR5 with experiments with ZNRF3ΔR. Myc immunoprecipitates from HEK293T cells expressing Myc-ZNRF3ΔR only or co-transfected with HA-tagged full-length LGR4 or LGR5 in the presence or absence of RSPO2 and RSPO4 as indicated were blotted for HA and Myc. Total lysates were also probed with the HA antibody. Blot is representative of 5 independent experiments. (B) Co-immunoprecipitation of LGR4 and LGR5 with RNF43. SC37 immunoprecipitates (endogenous RNF43) from LS180 cells were blotted for LGR4 and LGR5, and RNF43 immunoprecipitates from HEK293T cells transiently transfected with HA-tagged RNF43 were blotted for HA. IgG was used as a negative control. Blots are representative of 4–6 independent experiments. (C) Confocal microscopy images of PLA (green) between LGR4 or LGR5 using the antibodies 8D2 and 8F2, respectively and Myc-ZNRF3ΔR in the absence or presence of RSPO1 in HEK293T cells transiently co-expressing Myc-ZNRF3ΔR and either LGR4 or LGR5. CTL is the negative control without primary antibody in the absence of RSPO1. Nuclei were stained with TO-PRO-3(blue). Images are representative of 2 independent experiments. Scale bar, 25 μm.
We then probed the interactions of LGR4 and LGR5 with RNF43 using the colon cancer cell line LS180, which expresses LGR4, LGR5, and RNF43 endogenously at moderate to high levels (table S1). An RNF43-specific antibody named SC37 (31) was produced and validated to bind to native RNF43 with high affinity (fig. S6). Immunoprecipitation with SC37 pulled down LGR4 but not LGR5 in LS180 cells (Fig. 2B; fig. S13, A–D), confirming that only LGR4 interacted with RNF43 endogenously; detection of RNF43 in LS180 cells by immunoblotting could not be carried out due to the lack of an antibody that detects denatured RNF43, but the effectiveness of SC37 for immunoprecipiation was validated using cells expressing recombinant HA-tagged RNF43 (Fig. 2B; fig. S13, A–D).
Next, we explored interactions between ZNRF3ΔR and full-length LGR4 and LGR5 using PLA, which, compared to immunoprecipitation, is more sensitive and specific for the detection of proteins that are in close proximity (32–34). In this assay, HEK293T cells were co-transfected with Myc-ZNRF3ΔR and HA-LGR4FL or HA-LGR5FL. Myc-ZNRF3ΔR was detected with a mouse antibody recognizing the Myc tag, LGR4 with the in-house generated rat-human chimeric antibody 8D2 (35), and LGR5 with the rat-human chimeric antibody 8F2 (8, 28). 8D2 and 8F2 bind to the ECD of native LGR4 and LGR5, respectively, without interfering with RSPO function (fig. S7A) (28). Strong PLA signals were observed between ZNRF3ΔR and LGR4FL with or without RSPO1 (Fig. 2C), suggesting close proximity between LGR4 and ZNRF3 even in the absence of RSPO. The LGR4-ZNRF3 PLA signal appeared to decrease following RSPO1 treatment, which may be due to altered orientation of the proteins and increased distance between LGR4 and ZNRF3 to cause less ligation of the probes following RSPO1 binding. This phenomenon of decreased PLA signal with RSPO1 was also observed when LGR4 was labeled with an antibody recognizing an HA tag at the N-terminus (fig. S7B). In contrast, no PLA signal was detected between HA-LGR5FL and ZNRF3ΔR with or without RSPO1, using either 8F2 (Fig. 2C) or the HA antibody (fig. S7B). Confocal immunofluorescence microscopy confirmed that both LGR4 and LGR5 were on the cell surface (fig. S7C). Together with the co-immunoprecipitation results, these data indicate that full-length LGR5 did not interact with ZNRF3 or RNF43 in whole cell assays with or without RSPO, whereas LGR4 clearly did so even without RSPO.
Next, we carried out competition assays to examine the interactions between RSPO-LGR4 or RSPO-LGR5 and ZNRF3 in live cells using the ligand R2Fu-F109A, which binds to ZNRF3 with high affinity but does not bind to LGR4 or LGR5 (27). We also generated an RSPO4-furin-Fc fusion mutant protein (designated R4Fu-Q65R) with a mutation (Q65R) in the RNF43/ZNRF3 binding domain that retains high-affinity binding to LGR4 and LGR5 but no binding to ZNRF3 (20). Because RSPO1 and RSPO4 have little affinity to ZNRF3 alone, we reasoned that if LGR4, RSPO, and ZNRF3 form a trimer, then R2Fu-F109A’s binding to ZNRF3 could be displaced by RSPO1 and RSPO4 in cells that produce both LGR4 and ZNRF3, but not in cells that produce only ZNRF3. On the other hand, RSPO1 and RSPO4 would not be able to displace the binding of R2Fu-F109A to ZNRF3 in cells that make both LGR5 and ZNRF3 if RSPO-LGR5 did not form a trimer with ZNRF3. To this end, HEK293T cells were transfected with ZNRF3ΔR alone or with LGR4FL or LGR5ΔC and incubated with a sub-saturating concentration of R2Fu-F109A plus various concentrations of recombinant RSPO1–4. Here, instead of LGR5FL, we used LGR5ΔC, which lacks the C-terminal tail but remains fully functional, because its expression causes high receptor abundance on the cell surface due to impaired endocytosis (36, 37), as confirmed by RSPO binding (fig. S8). In contrast, LGR5FL constantly internalizes and recycles such that its surface availability at a given time point is less than that of LGR5ΔC (fig. S8). Through saturation binding analysis of R2Fu-F109A, we confirmed that all three transfections produced ZNRF3ΔR in similar amounts (Fig. 3A). When tested with binding of R4Fu-Q65R, ZNRF3ΔR+LGR4 and ZNRF3ΔR+LGR5ΔC cells displayed specific binding at similar levels (Fig. 3B), indicating that LGR4 and LGR5ΔC had similar receptor density on the cell surface. ZNRF3ΔR cells, as expected, showed negligible binding of R4Fu-Q65R (Fig. 3B). In competition assays, neither RSPO1 nor RSPO4 inhibited the binding of R2Fu-F109A to cells expressing ZNRF3ΔR alone, whereas RSPO2 competed with R2Fu-F109A for binding to ZNFR3 with high affinity, consistent with RSPO2 binding to ZNRF3 without LGR (Fig. 3C). Both RSPO1 and RSPO4 were able to compete R2Fu-F109A off ZNRF3ΔR-LGR4 with almost the same affinity as RSPO3 (Fig. 3D), suggesting that RSPO1 and RSPO4 were able to occupy the RSPO binding site of ZNRF3 when LGR4 was present. In contrast, in cells with ZNRF3ΔR and LGR5, only RSPO2 competed off R2Fu-109A (Fig. 3E), as in cells expressing ZNRF3ΔR alone. These results suggest that RSPO1- or RSPO4-bound LGR5 was not able to engage with ZNRF3 on the cell membrane to occupy the RSPO binding site on ZNRF3. Together, these data in live cells provide further support that RSPO-LGR5 did not form heterotrimers with the E3 ligases, whereas LGR4 clearly did.
Fig. 3. LGR5 does not enable binding of RSPO1 or RSPO4 to ZNRF3.
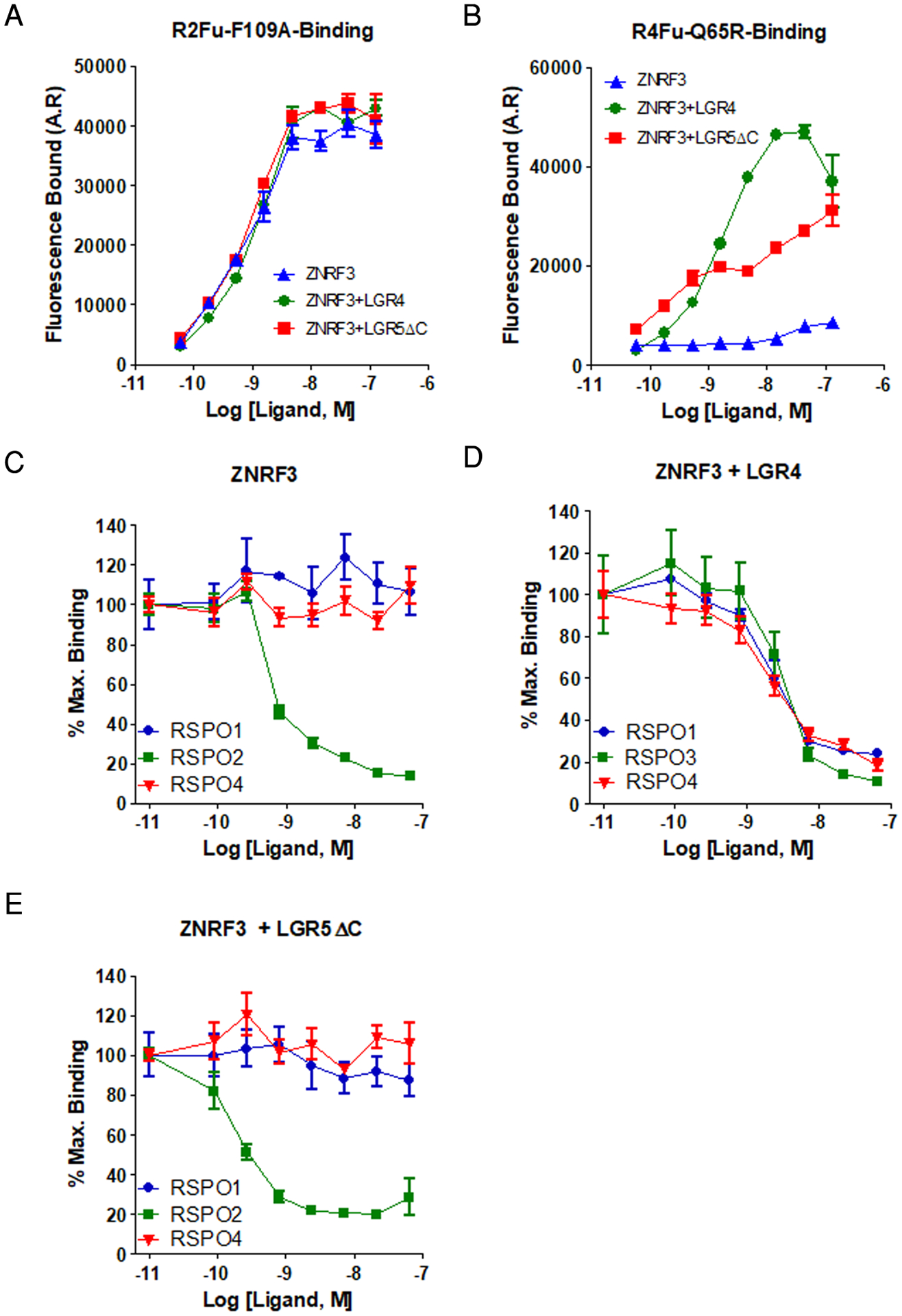
(A and B) Saturation binding analyses of R2Fu-F109A (A) and R4Fu-Q65R (B) to HEK293T cells transfected with ZNRF3ΔR, ZNRF3ΔR + LGR4FL, or ZNRF3ΔR + LGR5ΔC. (C to E) Competition binding curves for R2Fu-F109A with increasing concentrations of recombinant RSPO1, RSP2, or RSPO4 to HEK293T cells transfected with ZNRF3ΔR alone (C), ZNRF3ΔR + LGR4FL (D), and ZNRF3ΔR + LGR5ΔC (E). All error bars are SEM from 2–3 technical replicates, and the curves are representative of 3–5 independent experiments.
The 7TM domain of LGR4 confers interaction with the E3 ligase
LGR4 and LGR5 are highly similar in overall protein sequence and domain organization with 54% and 49% sequence identity in the ECD and 7TM regions, respectively. We asked if the ability to interact with the E3 ligases could be conferred by exchanging the ECD or 7TM domain of the two receptors. Two chimeric receptors, LGR4ECD-5TM and LGR5ECD-4TM were constructed with LGR4ECD fused to LGR5–7TM, and LGR5ECD to LGR4–7TM, respectively. The switch was at the ECD-7TM juncture of each receptor, and both chimeric receptors showed expression and RSPO binding similar to those of wild-type LGR4 and LGR5 (fig. S9A). TOPFlash assay also showed similar response to RSPO1 in potency and efficacy (fig. S9, B and C), suggesting that both chimeric receptors were functional. We then performed co-immunoprecipitation assays for ZNRF3ΔR with all four receptors in parallel and found that LGR5ECD-4TM gained interaction with ZNRF3, whereas LGR4ECD-5TM showed weaker co-immunoprecipitation with ZNRF3 compared to wild-type LGR4 (Fig. 4A), suggesting that the 7TM domain of LGR4 drove the interaction with ZNRF3. Of note, LGR4ECD-5TM still showed weak co-immunoprecipitation signal with ZNRF3ΔR (Fig. 4A), implying the ECD of LGR4 may have slight direct interaction with ZNRF3. We then tested if LGR5ECD-4TM could protect FZD from E3 ligase–mediated degradation given its interaction with ZNRF3. LGR5ECD-4TM clearly increased FZD abundance in response to RSPO1 (Fig. 4B). These results indicate that the 7TM region of LGR4 was largely responsible for RSPO-LGR4 interacting with ZNRF3 and inhibiting its activity. The results are also consistent with our finding that LGR4 binding to the ZNRF3 did not require RSPO (Fig. 2A) because the 7TM region was not involved in RSPO binding.
Fig. 4. The 7TM domain of LGR4 confers interaction with ZNRF3, and the LGR4-ECD does not antagonize RSPO activity.
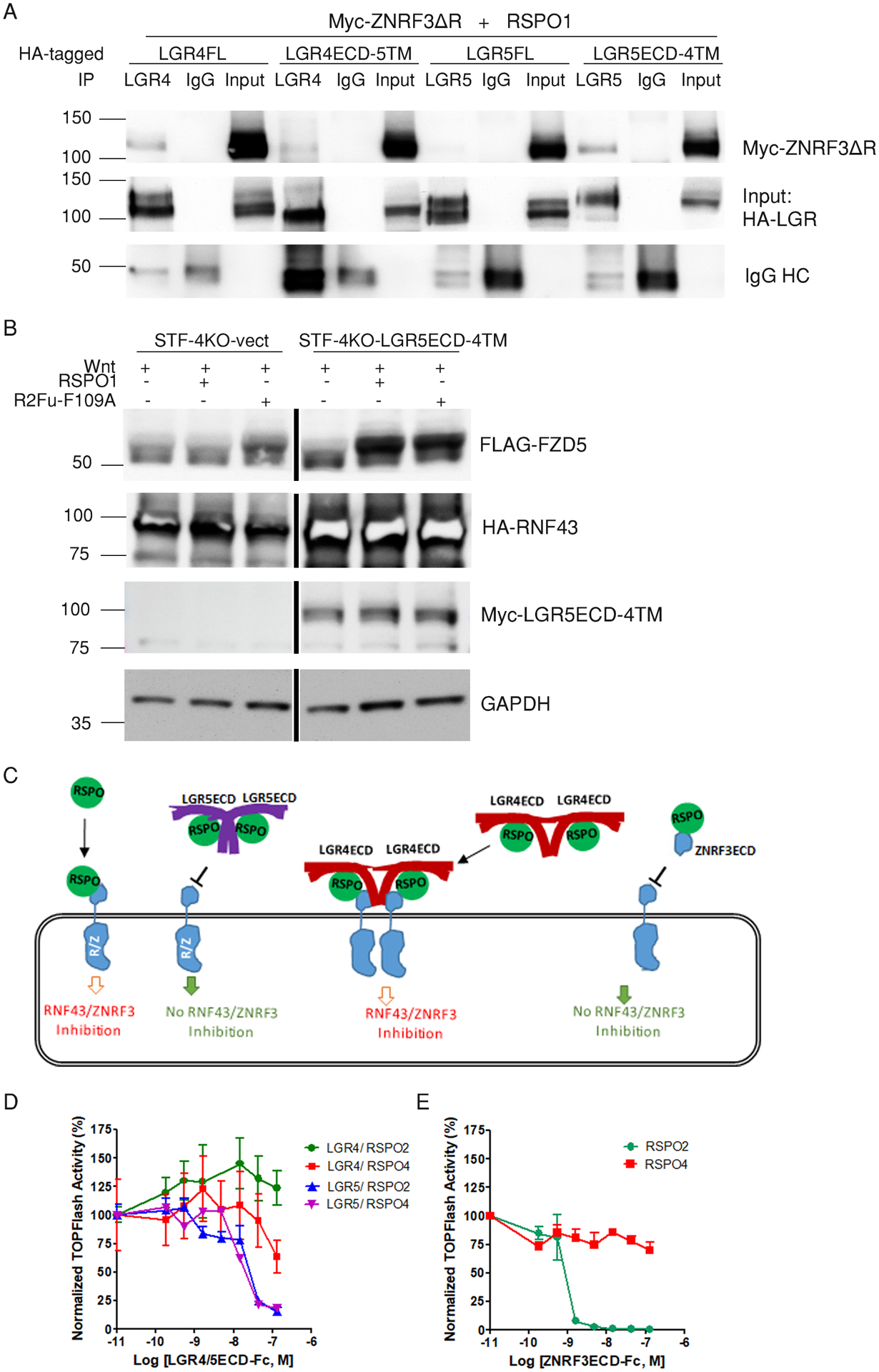
(A) Co-immunoprecipitation of ZNRF3ΔR with LGR4 and LGR5 full-length and ECD constructs. Western blotting for Myc and HA in LGR4 and LGR5 immunoprecipitates (8D2 or 8F2 antibody, respectively) from HEK293T cells co-transfected with Myc-ZNRF3ΔR and HA-tagged LGR4-FL, LGR4ECD-5TM, LGR5FL, or LGR5ECD-4TM as indicated in the presence of 1 μg/mL RSPO1. IgG was used as a negative control. Blot is representative of 3 independent experiments. (B) Western blotting for FLAG, HA, and Myc in STF-LGR4KO cells transiently transfected with vector or Myc-tagged LGR5ECD-4TM, HA-tagged RNF43, and FLAG-tagged FZD5. Cells were treated with Wnt3a CM, Wnt3a CM+RSPO1, or Wnt3a CM+R2Fu-F109A overnight. GAPDH is a loading control. Blot is representative of 2 independent experiments. (C) A schematic model illustrating the distinct interactions among RSPO-LGRECDs and RNF43/ZNRF3. RSPO and LGR5ECD form a 2:2 dimer that can no longer bind to RNF43/ZNRF3 due to steric hindrance of the LGR5ECD in trans. RSPO and LGR4ECD form a 2:2 dimer that remains capable of binding to RNF43/ZNRF3. RSPO and ZNRF3ECD forms a 1:1 dimer that cannot bind to native ZNRF3 or RNF43. (D and E) TOPFlash results for the effect of LGR4ECD-Fc and LGR5ECD-Fc (D) and ZNRF3ECD-Fc (E) on the activity of 0.01 μg/mL RSPO2 or 0.2 μg/mL RSPO4 in 293-STF cells. The TOPFlash activities are representatives of 5 independent experiments with the error bars representing SEM of 2–3 technical repeats.
Soluble LGR5ECD, but not LGR4ECD, antagonizes RSPO-induced potentiation of Wnt–β-catenin signaling
We then asked if there is intrinsic difference between the ECDs of LGR4 and LGR5. Crystal structures show RSPO-LGR5ECD forms 2:2 dimers that cannot bind to RNF43ECD or ZNRF3ECD (18, 21), whereas crystal structures of RSPO-LGR4ECD showed it as 1:1 dimers that could still bind to RNF43-ECD or ZNRF3ECD (38–40). This would suggest that purified, soluble LGR5ECD would act as antagonist of RSPOs but LGR4ECD would not (Fig. 4C). In fact, Yan et al. showed that His-tagged LGR5ECD antagonized the Wnt signaling potentiation activities of all RSPOs (41). We compared the effect of purified, soluble LGR4ECD-Fc and LGR5ECD-Fc in parallel on RSPO-induced activity in the TOPFlash assay. Among the four RSPOs, RSPO1 and RSPO4 (RSPO1/4) are similar in that they are completely dependent on LGR proteins to potentiate Wnt signaling and have relatively low binding affinities to the E3 ligases. On the other hand, RSPO2 and RSPO3 (RSPO2/3) can potentiate Wnt signaling independent of LGR and have high binding affinities to E3 ligases (14, 27). Because we expect similar Wnt functional activities from RSPO1/4 or RSPO2/3, we employed RSPO2 and RSPO4 to represent the two classes. LGR5ECD-Fc inhibited the activity of RSPO2 and RSPO4 in a dose-dependent fashion (Fig. 4D), consistent with published results (41). LGR4ECD-Fc, in contrast, had no antagonist activity (Fig. 4D). In fact, it even slightly enhanced the activity of RSPO2 (Fig. 4D). We also tested the effect of ZNRF3ECD-Fc and found that it could block the activity of RSPO2 but not RSPO4 (Fig. 4E), consistent with the lack of binding of RSPO4 to ZNRF3 itself (20, 42). These results are consistent with our finding of only RSPO-bound LGR4 still capable of interacting with the ZNRF3 (Fig. 3, C and D).
RSPOs interact with full-length LGR4 and LGR5 in cells as 2:2 dimers or higher oligomers
Because RSPO-LGR5ECD was consistently found as 2:2 dimers in crystal structures, we asked if RSPO bound to full-length LGR4 and LGR5 as 2:2 dimers or higher oligomers in whole cells. A previous study reported that LGR4 and LGR5 exist as dimers when overexpressed in whole cells (43). We also routinely observed that recombinant full-length LGR4 and LGR5 run as dimers in SDS-PAGE even under denaturing conditions, whereas their ECDs fused to the TM domain of CD4 run largely as monomers (fig. S3, A andB). We first examined if endogenous LGR4 and LGR5 also exist as dimers by SDS-PAGE analysis. In a cancer cell line (OV90) with high endogenous LGR4 expression (table S1), endogenous LGR4 was detected in both dimeric and monomeric forms (Fig. 5A). Treatment with the reducing agent β-mercaptoethanol (BME) or RSPO1 had no effect on the relative amounts monomer and dimer (Fig. 5A). To assay endogenous LGR5 in cells, we used LoVo cells because they highly express LGR5 and produce LGR5 that is readily detectable by western blotting (44). No dimer of LGR5 was detected unless the cells were pre-treated with the chemical crosslinker bis(sulfosuccinimidyl)suberate (BS3) (Fig. 5B), suggesting that LGR5 dimers in LoVo cells could not withstand SDS gel separation. The effect of BS3 on LGR5 was also validated using a stable HEK293 cell line expressing Myc-tagged LGR5 (Fig. 5C).
Fig. 5. Both LGR4 and LGR5 exist primarily as dimers with or without ligand binding.
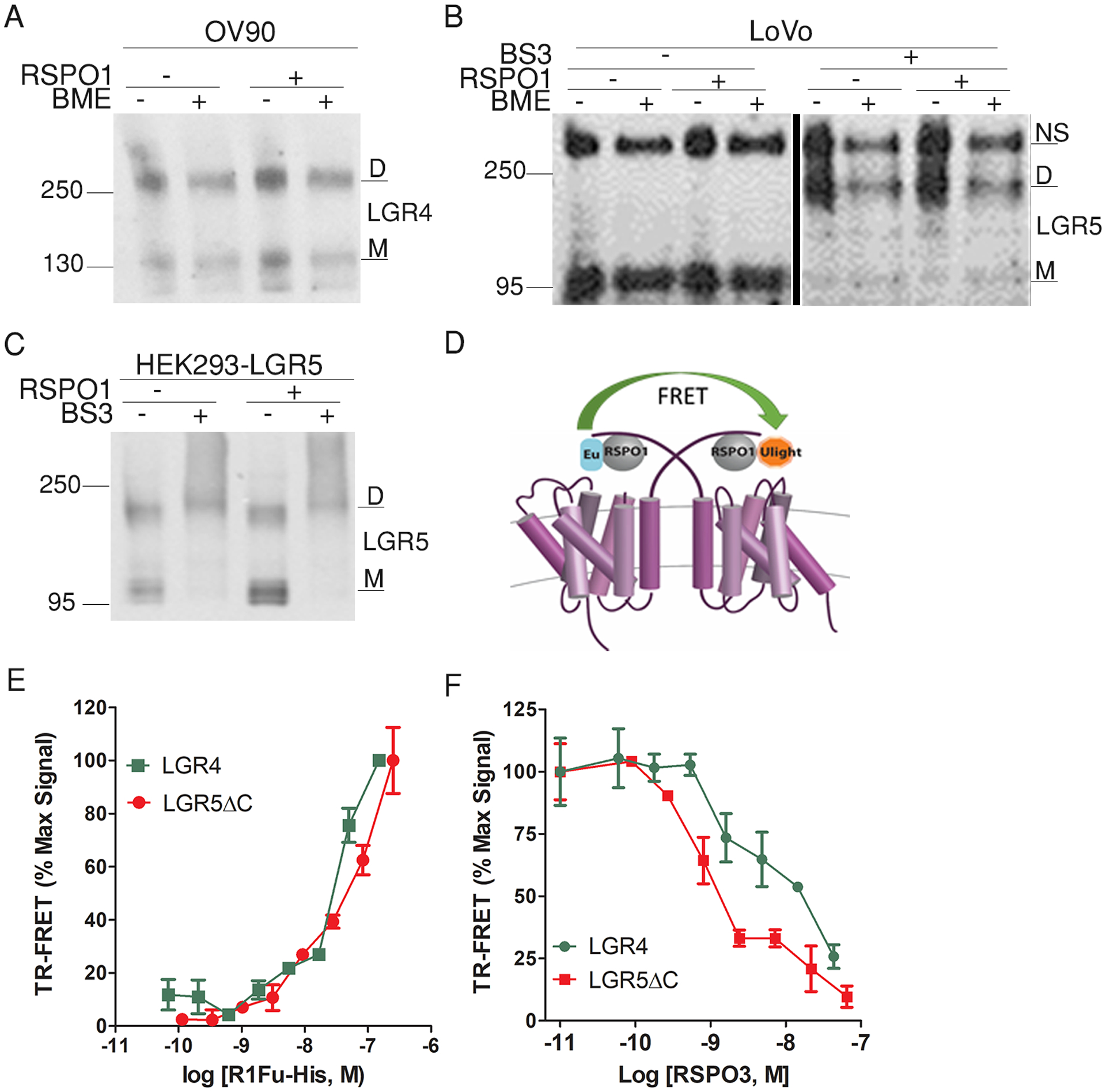
(A) Western blot for endogenous LGR4 in extracts from OV90 cells with and without the addition of β-mercaptoethanol (BME) to the samples. (B) Western blot for endogenous LGR5 in extracts from LoVo cells with and without BS3 cross-linker pretreatment and with or without or BME added to the samples. (C) Western blot for recombinant LGR5 in HEK293-LGR5 cells with or without BS3 pre-treatment. Blots are representative of 3 independent experiments. (D) Schematic illustration of fluorescence resonance energy transfer (FRET) between two RSPO ligands bound to LGR4 or LGR5. A 1:1 mixture of Europium (Eu)- and Ulight-labelled His tag–specific antibodies were added to the cells. FRET would occur if two ligands were within 10 nm of one another. (E) Baseline-corrected time-resolved FRET (TR-FRET) signal of R1Fu-His bound to HEK293-LGR4 or HEK293-LGR5ΔC cells with increasing concentrations of the ligand. (F) Baseline-corrected TR-FRET signal of LGR4 and LGR5ΔC bound R1Fu-His molecules in the presence of increasing concentrations of unlabeled RSPO3. TR-FRET curves are representatives of 3 independent experiments, and error bars are SEM of 2 technical repeats.
Next we tested if RSPO ligands bound to the receptors as dimers using time-resolved Fluorescence assay (TR-FRET), which is commonly used to detect G protein–coupled receptor (GPCR) oligomerization (45). If each monomer of LGR4 and LGR5 dimers bound one molecule of RSPO, FRET may occur between the two bound ligands that were labeled with either donor or recipient dyes (Fig. 5D). We used purified RSPO1 furin domain with a His tag at the C-terminus (R1Fu-His) and Europium (Eu)-labeled His antibody as the donor dye and Ulight-labeled His antibody as the recipient. TR-FRET was easily detected between R1Fu-His and HA-LGR4 or HA-LGR5FL using Eu-labeled His antibody and Alexa647-labeled antibody against HA-tag (fig. S10). With increasing concentrations of R1Fu-His, strong FRET signal between two ligands was detected in a dose-dependent fashion in both LGR4- and LGR5ΔC-expressing cells (Fig. 5E); LGR5ΔC was used instead of full-length LGR5 here to increase receptor density on the cell surface. Furthermore, the FRET signal was completely competed off by full-length, untagged RSPO3 with the expected affinity (Fig. 5F), indicating the FRET signal was specific to RSPO binding. Taken together, the TR-FRET and SDS gel data suggested that both LGR4 and LGR5 existed as dimers on the cell surface in the absence of RSPO and bound to RSPO with a 2:2 dimer architecture.
LGR5 interacts with both FZD and LRP6 in the Wnt signalosome
Previously, we used immunoprecipitation, FRET, and co-internalization assays to show that LGR5 interacts with LRP6 of the Wnt signalosome (36). Given the increased sensitivity and specificity of PLA compared to these other techniques, we re-examined if RSPO-LGR5 could interact with both FZD and LRP in the Wnt signalosome using PLA. For this assay, STF-4KO-LGR5 cells were transiently transfected with HA-LRP6 while FZD was provided by endogenous expression. Then, we used Myc and a pan-FZD antibody (18R5) (46) to label LGR5 and endogenous FZD, respectively, or the LGR5 antibody 8F2 and HA antibody to label LGR5 and LRP6, respectively. The primary antibody incubation with and without sub-saturating amounts of recombinant RSPO1 was performed on ice to prevent internalization of LGR5 and to exclusively detect interactions at the cell surface. PLA signal was detected between LGR5 and endogenous FZD without and with RSPO1, and a stronger PLA signal was detected between LGR5 and LRP6 even without RSPO1 (Fig. 6A). Immunofluorescence confocal microscopy also confirmed co-expression and co-localization of LGR5 with both FZD and LRP6 (fig. S11). We also asked if interaction between LGR5 and FZD could be detected with co-immunoprecipiation analysis. Immunoprecipitation of FZD5 pulled down LGR5 when the two receptors were co-expressed (Fig. 6B). Together, these results provided further evidence that reinforced previous findings of LGR5 interacting directly with Wnt signalosome (8, 36).
Fig. 6. LGR5 interacts with both FZD and LRP6 of the Wnt signalosome and promotes signaling IQGAP1-dependently.
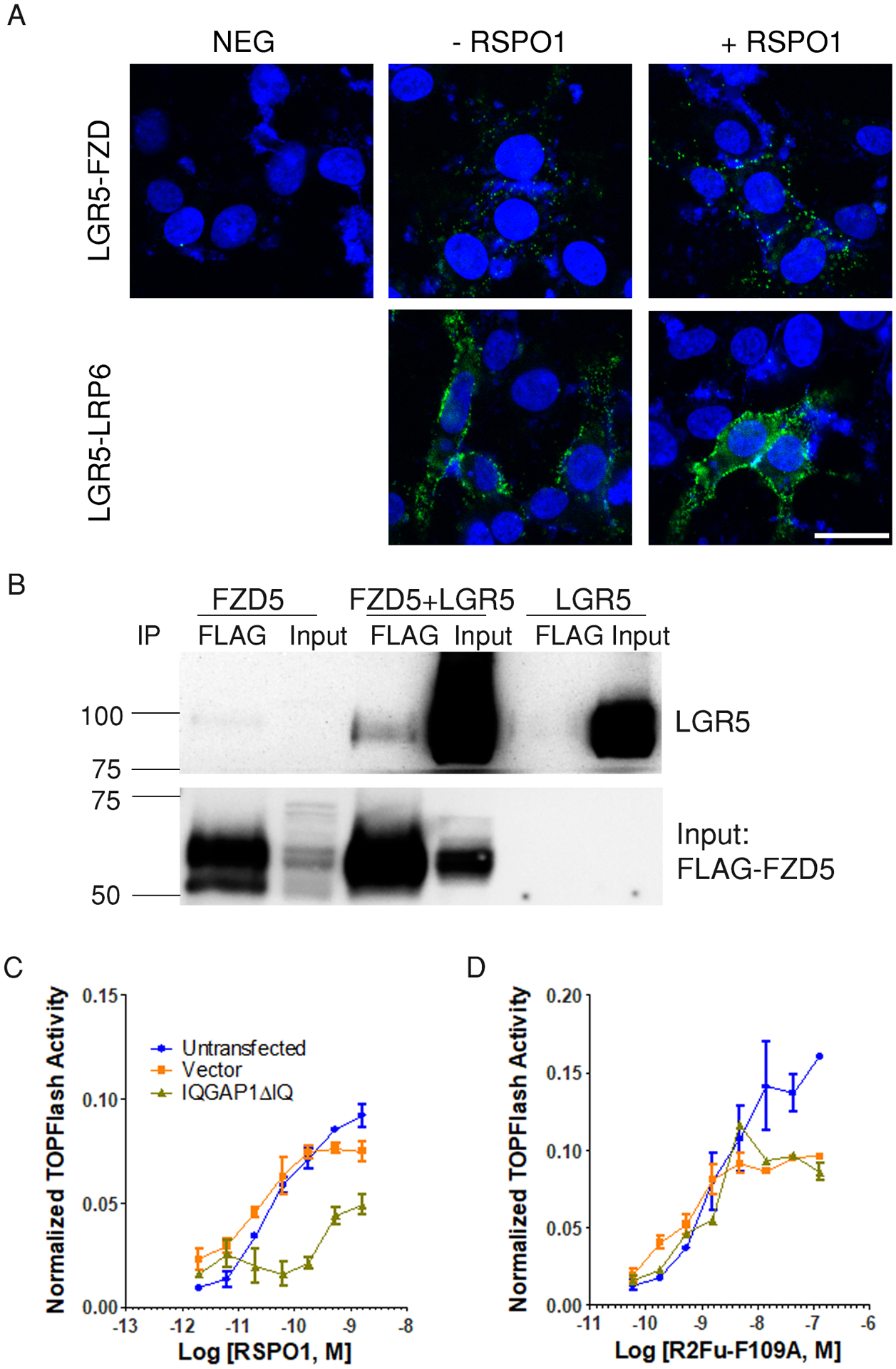
(A) Confocal microscopy images of PLA between LGR5 and endogenous FZD and between LGR5 and HA-LRP6 in the absence or presence of RSPO1 during primary antibody incubation in STF-4KO-LGR5 cells transiently transfected with HA-LRP6. CTL is negative control without primary antibody. Nuclei were stained with TO-PRO-3 (blue). Images are representative of 2 independent experiments. Scale bar, 25 μm. (B) Coimmunoprecipitation of LGR5 with FZD. Western blot for LGR5 and FLAG in FLAG immunoprecipitates from HEK293T cells overexpressing FLAG-FZD5, FLAG-FZD5 + LGR5, or LGR5. Blot is representative of 2 independent experiments. (C and D) Normalized TOPFlash activity of STF-4KO-LGR5 cells transfected with vector or IQGAP1ΔIQ (dominant negative IQGAP1) stimulated with RSPO1 (C) or R2Fu-F109A (D). Curves are representatives of 3 independent experiments and error bars represent SEM of 2 technical repeats.
Previously, we showed that LGR5 interacted with the scaffold protein IQGAP1 to enhance cell-cell adhesion in the absence of RSPO and that LGR4 interacted with the Wnt signalosome through IQGAP1 and enhanced the interaction between IQGAP1 with Dishevelled (DVL), thus promoting LRP6 phosphorylation and Wnt signaling (17, 44). Because RSPO-LGR5 did not show interaction with RNF43 or ZNRF3, we reasoned that LGR5 may function through IQGAP1 to potentiate Wnt–β-catenin signaling. Expression of a dominant negative form of IQGAP1 inhibited RSPO1-induced activity in STF-4KO-LGR5 cells but had no effect on signaling induced by R2Fu-F109A, which is LGR-independent (Fig. 6, C and D; fig. S12), suggesting that RSPO-LGR5–mediated potentiation of Wnt signaling depended on IQGAP1.
Discussion
LGR5 and the closely related receptor LGR4 are co-produced in intestinal crypt stem cells, and both have been shown bind to RSPOs with high affinity and potentiate Wnt signaling in response to RSPO (8, 11). Mechanistically, LGR4 was suggested to potentiate Wnt signaling primarily by enabling or enhancing RSPO-mediated inhibition of RNF43/ZNRF3 through the formation of LGR4-RSPO-RNF43/ZNRF3 heterotrimers (15). Given the high homology between LGR4 and LGR5, it has been widely assumed that LGR5 mediates RSPO-induced potentiation of Wnt signaling in the same way as does LGR4 (19, 20, 22). However, LGR4 and LGR5 clearly do not have interchangeable functions, because only LGR4 is essential for the self-renewal of crypt stem cells (8, 10), and direct evidence of RSPO-LGR5 interacting with the E3 ligases in whole cells has not been reported. Here we present multiple lines of evidence showing that full-length LGR5 did not form a complex with the RNF43 or ZNRF3 in whole cells with or without RSPOs. Instead, RSPO-LGR5 interacted with the Wnt signalosome directly and promoted Wnt–β-catenin signaling through an IQGAP1-dependent mechanism. LGR4, on the other hand, interacted with RNF43/ZNRF3 even in the absence of RSPO, and the LGR4-E3 ligase interaction depended on the 7TM domain of LGR4. We also found that both LGR4 and LGR5 existed as dimers in whole cells and that RSPO bound to LGR4 and LGR5 in 2:2 dimers. Based on these data, we propose distinct structural models for LGR4 and LGR5: LGR4 pre-assembles with RNF43/ZNRF3 as 2:2 dimer, and binding of RSPO to the complex leads to inhibition of E3 ligase activity (Fig. 7A). LGR4 dimers not in complex with E3 ligase would also be able to enhance Wnt signaling through IQGAP1 in response to RSPO (17). LGR5 exists as a homodimer in complex with the Wnt signalosome, and binding of RSPO to LGR5 enhances LRP phosphorylation through an IQGAP1-dependent mechanism (Fig. 7B). These models, we argue, are more congruent with existing data on the structure and function of LGR4 and LGR5 in Wnt signaling.
Fig. 7. A schematic model illustrating the mechanistic differences between LGR4 and LGR5 in mediating RSPO-induced potentiation of Wnt–β-catenin signaling.
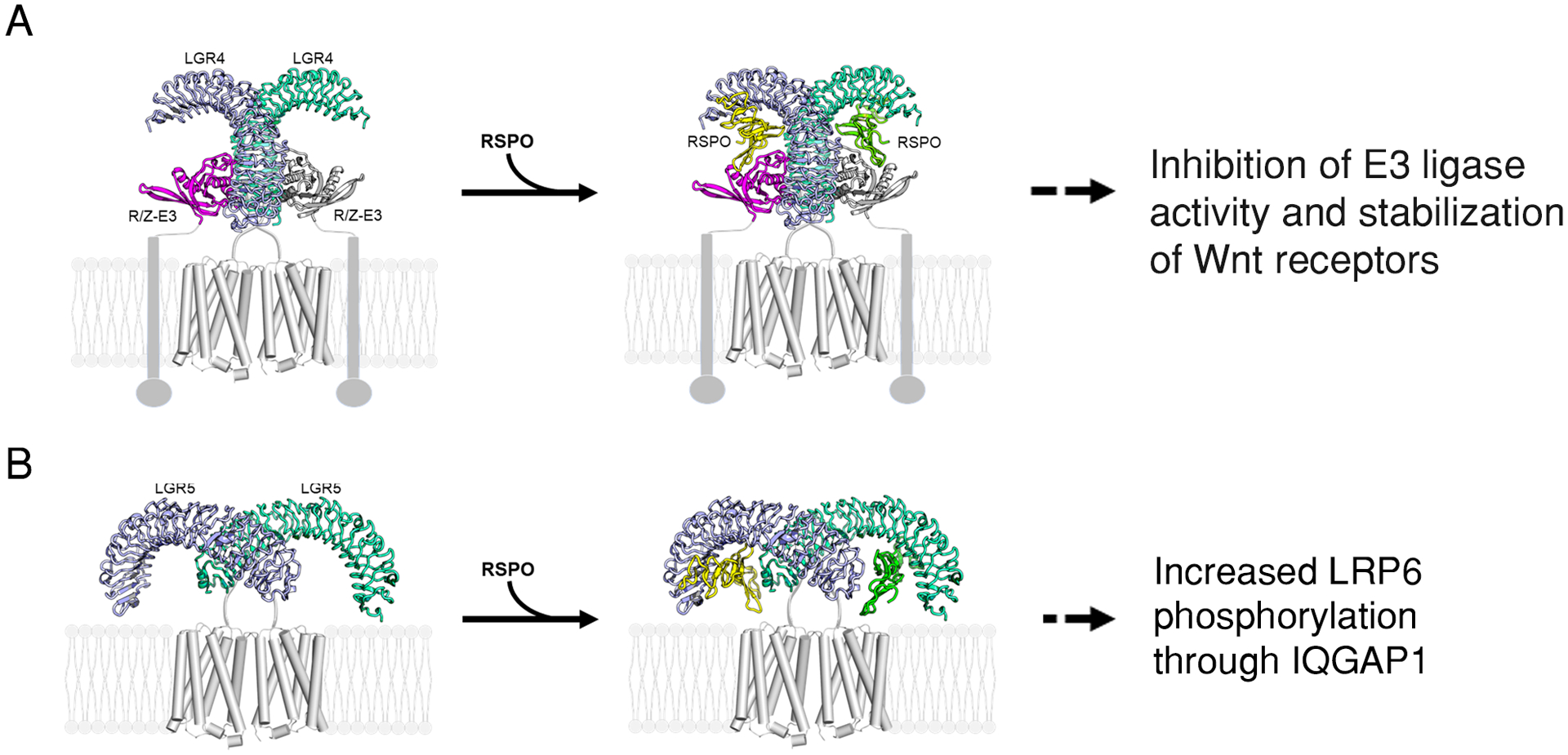
(A). LGR4 binds to the E3 ligases RNF43 and ZNRF3 (R/F-E3) as a 2:2 dimer without inhibiting ligase activity. Upon RSPO binding and trimer formation, E3 ligase activity is lost. LGR4 dimers that are not engaged with E3 ligases can also enhance LRP6 phosphorylation through direct interaction with the Wnt signalosome and IQGAP1 (not shown). (B) LGR5 exists as a homodimer that is not bound to an E3 ligase but instead interacts with Wnt receptors (not shown). Through an unknown mechanism, RSPO binding to LGR5 enhances LRP6 phosphorylation in an IQGAP1-dependent manner (not shown). LGR4-RSPO was modeled after PDB structures 4KT1 and 4KNG, and LGR5-RSPO was modeled after 4UFR, and the 7TM was modeled after 4OR2.
The prevailing model of RSPO-LGR4/5 functioning in Wnt signaling is that LGR4 and LGR5 both serve as high-affinity receptors of RSPOs to recruit RSPOs to the membrane so that the RSPO-LGR complex can bind to and inhibit the function of ZRNF3/RNF43 (20–22, 47). However, this mechanism for LGR5 is inconsistent with multiple observations. Free RSPO1 and RSPO4 have little to no affinity for binding to ZNRF3/RNF43, and so they could not enhance Wnt signaling without LGR (14, 20, 27). In the presence of LGR4 or LGR5, RSPO1 potentiates Wnt–β-catenin signaling at similar efficacy with potency within an order of magnitude of those values for RSPO2 and RSPO3 (8, 11, 20, 29). RSPO4 shows high potency and half-efficacy when combined with LGR4 (27, 29), but nearly no activity with LGR5 (Fig. 1). If LGR4/5 function simply as recruiting receptors, then LGR4 must be able to alter conformations of RSPO1 and RSPO4 to enable them to bind to the E3 ligases, whereas LGR5 could only alter that of RSPO1. However, structural data reveal that RSPO1 bound to LGR5ECD had no increase in binding affinity to ZNRF3 (20, 47). Also, RSPO2 bound to LGR5ECD actually showed a decrease in binding affinity to ZNRF3 when compared to free RSPO2 (850 vs 25 nM) (20). Moreover, when LGR5ECD and RSPO furin domains were co-crystalized without RNF43/ZNRF3ECD, they invariably formed a 2:2 dimer that would not be able to bind to the E3 ligase (18, 20). This would predict that purified, soluble LGR5ECD would act as an RSPO antagonist, which is what it did in our experiments (Fig. 4D) (41). Furthermore, native LGR5 existed as dimers in whole cells, and RSPO bound LGR5 in a 2:2 dimer (Fig. 5), which would prevent the complex from further interacting with the E3 ligases. In addition, if LGR4 and LGR5 just serve as recruiting receptors, the ECDs of LGR4 and LGR5 anchored to cell membrane by a single transmembrane domain in place of the 7TM domain should function just as well as the wild-type protein, yet they showed much less response to RSPO1 and RSPO4 in potency and efficacy despite the ECD’s high binding affinity and strong presence on the cell surface (Fig. 1C) (27). All these data suggest that LGR5 could not function by recruiting RSPOs to inhibit the E3 ligases. Instead, we propose that LGR5 dimerizes on its own and interacts with FZD and LRP6, both of which also exist as homodimers before Wnt ligand binding, on the cell surface (48). Upon RSPO1–3 binding, RSPO-LGR5 enhances LRP6 phosphorylation through an IQGAP1-dependent mechanism, the details of which remain to be determined. RSPO4 did not activate LGR5 to engage this pathway. This model is consistent with the structural data of RSPO-LGR5ECD and implies that RSPO-LGR5 is only a weak potentiator of Wnt signaling in vivo due to the lack of inhibition of the E3 ligases.
As for LGR4, our model that LGR4-RNF43/ZNRF3 forms a pre-assembled complex and that RSPO binding to this complex abolishes E3 ligase activity either through direct inhibition or by altering its endocytosis would explain the high potency and efficacy of RSPO1 and RSPO4 despite their poor binding to the E3 ligases. It would also explain why the 7TM domain of LGR4 was essential for high-potency activity of RSPOs because it was critical for LGR4 to interact with the E3 ligases. Both the ECD and 7TM domains of LGR4 must be able to impose a dimer structure that is compatible with RSPO-E3 ligase binding because LGR5ECD-4TM was capable of binding to the E3 ligases, RSPO-LGR4ECD-5TM still had weak interaction with ZNRF3, and purified, soluble LGR4ECD did not act as antagonist (Fig. 4). It is also supported by structural data that all RSPO1-LGR4ECD structures were observed as 1:1 or 2:2 dimers that were compatible with further binding to the E3 ligases (38–40). Co-crystal structures of LGR4ECD-RSPO with either RNF43- or ZNRF3ECD has never been reported. The dual mechanisms of LGR4 – inhibition of RNF43/ZNRF3 and enhancement of LRP6 phosphorylation – would enable LGR4 to give much stronger potentiation of Wnt signaling, which may explain why LGR4 is both necessary and sufficient for the survival of crypt stem cells (8). Of note, genetic ablation of LGR4 in crypt stem cells leads to near total loss of Wnt–β-catenin signaling, whereas loss of LGR5 has little effect (8, 49). Also, double knockout of RNF43 and ZNRF3 enables RSPO-independent self-renewal of crypt stem cells (15, 16), further implicating the importance of inhibiting the E3 ligases for stem cell survival. The lack of interaction between RNF43/ZNRF3 and RSPO-LGR5 may explain why endogenous LGR5 alone is not sufficient to support stem cell survival.
In summary, we compared the mechanisms of LGR4 and LGR5 side-by-side in multiple assays and found that RSPO-LGR5 did not interact with RNF43/ZNRF3 to potentiate Wnt–β-catenin signaling, whereas LGR4 bound to the E3 ligases even in the absence of RSPO. Instead, LGR5 interacted directly with FZD and LRP6 of the Wnt signalosome and functioned by an IQGAP1-dependent mechanism. These findings are consistent with previous reports of LGR5’s interaction with the Wnt signalosome, antagonist activity of LGR5ECD, and dimer quaternary structure on the cell surface. They also provide a molecular basis for the weak activity of LGR5 in Wnt signaling potentiation and the inability of endogenous LGR5 to support the self-renewal of crypt stem cells. Details of how RSPOs activate LGR5 to enhance Wnt signaling and how RSPOs inhibit LGR4-bound E3 ligases remain to be elucidated.
Materials and Methods
Plasmids and Cloning
Plasmids encoding HA-LGR4, Myc-LGR5FL, Myc-LGR5ECD-CD4TM, Myc-LGR5ΔC, FLAG-IQGAP1ΔIQ, HA-LRP6, FLAG-FZD5, HA-RNF43 in pIRES-puro3 were prepared as described previously (11, 17, 36, 44, 50). Myc-ZNRF3ΔR was a gift from Dr. Feng Cong (Novartis Institute for Biomedical Research) (15). HA tagged LGR5 (HA-LGR5FL), HA tagged LGR4ECD-5TM, in which LGR4ECD was fused with the 7TM of LGR5 at T542, and HA or Myc tagged LGR5ECD-4TM, in which LGR5ECD was fused with the 7TM of LGR4 at Ser561 were cloned to pIRES-puro3 using standard PCR procedures and In-Fusion HD cloning (Clontech). Also, Myc-tagged LGR5FL was cloned to pIREShyg3 vector using In-Fusion HD cloning. All plasmids were verified by sequencing.
Recombinant proteins and antibodies
Human recombinant full-length RSPO proteins (RSPO1–4) were purchased from R&D Systems, and human recombinant RSPO1 furin domain (aa 1–146, C-terminal His-tag) was from Sino Biological. For western blot and immunoprecipitation, anti-HA (Invitrogen cat #71–5500 or Cell Signaling cat #2367), anti-Myc (Cell Signaling cat #2276), anti-phosphorylated and total LRP6 (Cell Signaling cat #2568 and #3395), anti-β-actin (Cell Signaling cat #4970), anti-LGR4 (7E7) (7), anti-FLAG (Sigma cat #F7425), anti-LGR5 (Abcam cat #ab75850), anti-RNF43 (SC37) (31), and anti-GAPDH (Cell Signaling cat #2118) were used. For ICC, anti-HA-Alexa 488 (Cell Signaling cat #2350), anti-Myc-Cy3 (Sigma cat #C6594s), TO-PRO-3 Iodide (Thermo Fisher cat #T3605), and anti-human-Alexa 488 or –Alexa 555 (Invitrogen cat #A11013 and #A21433) were used. Anti-pan FZD antibody (18R5) were prepared as described before (46). For PLA and IP, anti-LGR5 (8F2) and anti-LGR4 (8D2) antibodies were produced as described (28, 35). Duolink® In Situ PLA probes (anti-mouse PLUS and MINUS, anti-rabbit PLUS and MINUS, and anti-human PLUS and MINUS) and Duolink® In Situ PLA detection reagents green were purchased from Sigma. For TR-FRET, Lance Europium-labeled anti-6X His antibody, Lance Ultra Ulight™-anti-6xHis, and ULight anti-Myc were purchased from PerkinElmer, and anti-HA-Alexa 647 were from Invitrogen.
Cell culture, transient transfection, generation of stable cell lines, and Wnt–β-catenin signaling TOPFlash reporter enzyme assay
HEK293T and STF cells were purchased from ATCC and cultured in high glucose DMEM supplemented with 10 % fetal bovine serum (FBS) and pen/strep at 37 °C with 95 % humidity and 5 % CO2. LoVo, colorectal cancer cell line, was from Dr. Shao-Cong Sun at M.D. Anderson and cultured in RPMI supplemented with 10% FBS and pen/strep. LS180, OV90, SK-NAS, and CHP-212 cells were purchased from ATCC. HEK293-LGR4, -LGR5FL, and LGR5ΔC overexpressing cells and STF-LGR4KO cells were generated and cultured as described previously (11, 27, 36), and CHP-212 LGR5KO cells were generated using CRISPR-based knockout by cloning the guide sequence to Lenti-CRISPR2 vector as described (51); the guide sequence is CAG GAG CAC ACC GAG CCG GG (corresponding to nucleotide 314–295 of human LGR5 open reading frame). HEK293F cells for protein expression were from Invitrogen and cultured in FreestyleTM 293 Expression medium at 37 °C with 95 % humidity and 5 % CO2. STF-4KO-LGR5 and –vect cells were generated by transfecting Myc-LGR5FL (hygromycin resistant) or Hyg3-vector into STF-LGR4KO cells and selecting with 200 μg/mL of hygromycin B (Gibco).
TOPFlash assays were performed as described previously (27). Briefly, 5000 cells were incubated overnight with Wnt3aCM and/or treatments indicated in each experiments. To test LGR4ECD-Fc and LGR5ECD-Fc, 293-STF cells were co-incubated overnight with 0.01 μg/mL RSPO2 or 0.2 μg/mL RSPO4 and serially diluted LGR4ECD-Fc or LGR5ECD-Fc. The effect of LGR4 antibody 8D2 in 293-STF cells were assessed by co-incubating 8D2 with fixed concentration of RSPO1 at 0.02 μg/mL. The cell viability was measured using alamar blue (Thermo Fisher), and luciferase activity was measured by FireFly Luciferase glow assay kit (Pierce).
Western blotting and immunoprecipitation
Western blotting was carried out using standard procedures as described before (27). Lysates subjected to western blotting was harvested in RIPA buffer (Pierce) with protease and phosphatase inhibitors. The lysates were reduced at 37 °C for 1 hour in sample buffer with BME to protect 7TM domains of membrane receptor proteins. For co-IP experiments, cell lysates were collected in IP lysis buffer (Pierce) supplemented with protease and phosphatase inhibitors. HEK293T cells were transiently co-transfected with Myc-ZNRF3ΔR and HA-LGR4 or HA-LGR5FL plasmids and pulled down with Myc antibody with and without 1 μg/mL of the recombinant RSPO2 or RSPO4. LGR5FL and LGR5ECD stable cell lines were transfected with HA-RNF43 and IPed with HA antibody with and without 1 μg/mL RSPO1. LS180 cell lysates and HEK293T cells transiently transfected with HA-RNF43 were IPed with RNF43 antibody (SC37) or human IgG control. HEK293T and LGR5 stable cell lines transiently transfected with FLAG-FZD5 were pulled down with anti-FLAG M2 magnetic beads (Sigma Cat #M8823). HEK293T cells transiently co-transfected with Myc-ZNRF3ΔR and HA-tagged LGR4FL, LGR4ECD-5TM, LGR5FL, or LGR5ECD-4TM were pulled down with 8D2 antibody for LGR4ECD and 8F2 antibody for LGR5ECD. Cell lysates were incubated with agarose protein A/G beads (Santa Cruz) with antibodies or antibody conjugated magnetic beads and incubated overnight at 4 °C on a rotator. The beads were washed with IP lysis buffer 3 times and reduced at 37 °C for 1 hour in 2X sample buffer with BME, and both immunoprecipitated and total lysates were subjected to western blotting.
Competition whole-cell binding
HEK293T cells were co-transfected with ZNRF3ΔR and LGR4 or LGR5ΔC as described before (27) and seeded to 96-well poly-D-lysine-coated black and clear bottom plate. After 24 hours, the cells chilled on ice were incubated with a sub-saturating concentration of R2Fu-F109A, and serially diluted recombinant RSPO1–4. After 1 hour incubation, cells were washed with cold PBS and fixed with 4.2 % PFA and blocked with 1 % bovine serum albumin (BSA) in PBS. The secondary antibody, anti-human-Alexa 555 was incubated at the room temperature for an hour followed by PBS washing. The fluorescence intensity of labeled R2Fu-F109A was measured with Tecan multiplate reader. To confirm the expression of transfected proteins, whole-cell binding of R2Fu-F109A and R4Fu-Q65R was performed as described previously (27). The fluorescence reading was normalized to the baseline where no recombinant RSPO was added.
Proximity Ligation Assay (PLA)
PLA to detect protein interaction generally followed the standard protocol provided by Sigma. To detect interaction between ZNRF3ΔR and LGR4 or LGR5, HEK293T cells were transiently co-transfected with Myc-ZNRF3ΔR and HA-LGR4 or HA-LGR5FL. Then, the transfected cells were re-seeded to 8-well poly-D-lysine coated slide (Corning) and incubated at 37 °C with 5 % CO2 and 95 % humidity for 24 hours. After 24 hours, the chilled cells were incubated with anti-human-8D2 for LGR4 and anti-human-8F2 for LGR5, and anti-mouse-Myc for ZNRF3ΔR with and without 1 μg/mL of the recombinant RSPO1. For detection of LGR5 and Wnt signalosome interactions (FZD and LRP6), STF-4KO-LGR5 cells were transfected with HA tagged LRP6 as described previously. Then, the cells, chilled on ice, were incubated with primary antibodies and with and without 1 μg/mL of the recombinant RSPO1; for LGR5 and FZD5, anti-mouse-Myc and anti-human-FZD (18R5), and for LGR5 and LRP6, anti-LGR5 (8F2, human) and anti-rabbit-HA were used. For all negative controls, no primary antibody was added to the cells. After 1.5 hour primary antibody incubation, cells were washed with PBS, fixed with 4.2 % paraformaldehyde (PFA), and permeabilized with 0.1 % saponin. Then, the cells were blocked using the provided blocking reagent from Duolink® PLA probes, and labeled with PLUS and MINUS PLA probes corresponding to the primary antibody pairs. The labeled probes were then ligated and amplified with green fluorescence. Lastly, nuclei were stained with TO-PRO-3 and mounted. Expression and localization of the transiently transfected proteins were confirmed by standard ICC protocol. The stained cells were imaged under a confocal microscope (Leica) and analyzed by Leica LAS AF Lite software.
Time Resolved-Förster Resonance Energy Transfer (TR-FRET) assay and receptor cross-linking
HEK293-LGR4 and –LGR5ΔC overexpressing cells were seeded on white PDL coated 96 well plate. The pre-chilled cells were incubated with the saturating concentration of the recombinant RSPO1 furin domain with C-terminal His-tag and varying concentration of unlabeled recombinant RSPO3. After 2 hour incubation on ice, the cells were fixed with 4.2 % PFA and blocked with 1 % BSA in PBS. 1 nM anti-Europium-His and 50 nM anti-Ulight-His were co-incubated with the cells at the room temperature for 1 hour and washed with PBS. TR-FRET signal was measured from the excitation at 317 nm and emission at 620 nm with lag time between excitation and emission 50 μs. TR-FRET reading was then percent normalized to the baseline. For cross-linking of receptors, cells were treated with BS3 (1.5 mM) at 37 °C for 30 min and then harvested for SDS-PAGE analysis with and without BME using standard procedures.
Supplementary Material
Supplementary Materials
Fig. S1. Validation of LGR5 expression in STF-4KO-LGR5 cell line.
Fig. S2. R2Fu-F109A has no LGR5-dependent activity.
Fig. S3. Full-length LGR4 and LGR5 exist as both monomers and dimers but their ECDs only form monomers.
Fig. S4. LGR5-RSPO1-induced potentiation of Wnt signaling is through phosphorylation of LRP6.
Fig. S5. LGR5FL does not interact with RNF43 with or without RSPO1.
Fig. S6. Validation of RNF43 antibody, SC37.
Fig. S7. PLA between ZNRF3ΔR and LGR4 or LGR5 shows that LGR5 does not interact with ZNRF3ΔR with or without RSPO1.
Fig. S8. LGR4 and LGR5 differ in surface distribution but bind RSPO similarly.
Fig. S9. Expression of LGR4ECD-5TM and LGR5ECD-4TM chimeras.
Fig. S10. TR-FRET assay between RSPO and LGR as a positive control.
Fig. S11. Co-localization of LGR5 with FZD and LRP6.
Fig. S12. Expression of IQGAP1ΔIQ.
Fig. S13. Images of full blots for Fig.2B.
Table S1. Published RNA-seq data for selected genes in the cancer cell lines used in the study.
Acknowledgements:
The authors would like to thank Dr. Feng Cong of Novartis for the ZNRF3 plasmid, Dr. Shao-Cong Sun of M.D. Anderson Cancer center for the LoVo cells, and Dr. Jun Song at UNC Charlotte for statistics consult.
Funding: The work was support in part by Cancer Prevention and Research Institute of Texas (RP160235 and RP170245 to Q.J.L.), National Institute of Health (R01GM102485 to Q.J.L and R01CA226894 to K.S.C.), and Gordon Endowment for Bowel Cancer Research (to Q.J.L.).
Footnotes
Competing interests: Q.J.L. and K.S.C., and the University of Texas have patents issued related to LGR4 and LGR5 signaling and patent applications files on anti-LGR4 antibodies.
Data and materials availability: All the data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, Clevers H, Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007); published online EpubOct 25 ( 10.1038/nature06196). [DOI] [PubMed] [Google Scholar]
- 2.Koo BK, Clevers H, Stem cells marked by the R-spondin receptor LGR5. Gastroenterology 147, 289–302 (2014); published online EpubAug ( 10.1053/j.gastro.2014.05.007). [DOI] [PubMed] [Google Scholar]
- 3.McDonald T, Wang R, Bailey W, Xie G, Chen F, Caskey CT, Liu Q, Identification and cloning of an orphan G protein-coupled receptor of the glycoprotein hormone receptor subfamily. Biochem Biophys Res Commun 247, 266–270 (1998); published online EpubJun 18 (S0006–291X(98)98774–5 [pii] 10.1006/bbrc.1998.8774). [DOI] [PubMed] [Google Scholar]
- 4.Hsu SY, Liang SG, Hsueh AJ, Characterization of two LGR genes homologous to gonadotropin and thyrotropin receptors with extracellular leucine-rich repeats and a G protein-coupled, seven-transmembrane region. Mol Endocrinol 12, 1830–1845 (1998); published online EpubDec ( 10.1210/mend.12.12.0211) [DOI] [PubMed] [Google Scholar]
- 5.Barker N, Clevers H, Leucine-rich repeat-containing G-protein-coupled receptors as markers of adult stem cells. Gastroenterology 138, 1681–1696 (2010); published online EpubMay (S0016–5085(10)00336–7 [pii] 10.1053/j.gastro.2010.03.002). [DOI] [PubMed] [Google Scholar]
- 6.Van Schoore G, Mendive F, Pochet R, Vassart G, Expression pattern of the orphan receptor LGR4/GPR48 gene in the mouse. Histochem Cell Biol 124, 35–50 (2005); published online EpubJul ( 10.1007/s00418-005-0002-3). [DOI] [PubMed] [Google Scholar]
- 7.Yi J, Xiong W, Gong X, Bellister S, Ellis LM, Liu Q, Analysis of LGR4 Receptor Distribution in Human and Mouse Tissues. PLoS One 8, e78144 (2013) 10.1371/journal.pone.0078144). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lau W, Barker N, Low TY, Koo BK, Li VS, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, Stange DE, van Es JE, Guardavaccaro D, Schasfoort RB, Mohri Y, Nishimori K, Mohammed S, Heck AJ, Clevers H, Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature 476, 293–297 (2011); published online EpubAug 18 ( 10.1038/nature10337). [DOI] [PubMed] [Google Scholar]
- 9.Snyder JC, Rochelle LK, Ray C, Pack TF, Bock CB, Lubkov V, Lyerly HK, Waggoner AS, Barak LS, Caron MG, Inhibiting clathrin-mediated endocytosis of the leucine-rich G protein-coupled Receptor-5 diminishes cell fitness. J Biol Chem, (2017); published online EpubMar 08 ( 10.1074/jbc.M116.756635). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mustata RC, Van Loy T, Lefort A, Libert F, Strollo S, Vassart G, Garcia MI, Lgr4 is required for Paneth cell differentiation and maintenance of intestinal stem cells ex vivo. EMBO Rep, (2011); published online EpubApr 21 (embor201152 [pii] 10.1038/embor.2011.52). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmon KS, Gong X, Lin Q, Thomas A, Liu Q, R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A 108, 11452–11457 (2011); published online EpubJul 12 (1106083108 [pii] 10.1073/pnas.1106083108). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glinka A, Dolde C, Kirsch N, Huang YL, Kazanskaya O, Ingelfinger D, Boutros M, Cruciat CM, Niehrs C, LGR4 and LGR5 are R-spondin receptors mediating Wnt/beta-catenin and Wnt/PCP signalling. EMBO Rep 12, 1055–1061 (2011); published online EpubOct ( 10.1038/embor.2011.175). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK, Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J Biol Chem 281, 13247–13257 (2006); published online EpubMay 12 (M508324200 [pii] 10.1074/jbc.M508324200). [DOI] [PubMed] [Google Scholar]
- 14.Lebensohn AM, Rohatgi R, R-spondins can potentiate WNT signaling without LGRs. eLife 7, (2018); published online EpubFeb 6 ( 10.7554/eLife.33126). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, Mao X, Ma Q, Zamponi R, Bouwmeester T, Finan PM, Kirschner MW, Porter JA, Serluca FC, Cong F, ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485, 195–200 (2012); published online EpubMay 10 ( 10.1038/nature11019). [DOI] [PubMed] [Google Scholar]
- 16.Koo BK, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJ, Maurice MM, Clevers H, Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 488, 665–669 (2012); published online EpubAug 30 ( 10.1038/nature11308). [DOI] [PubMed] [Google Scholar]
- 17.Carmon KS, Gong X, Yi J, Thomas A, Liu Q, RSPO-LGR4 functions via IQGAP1 to potentiate Wnt signaling. Proc Natl Acad Sci U S A 111, E1221–1229 (2014); published online EpubApr 1 ( 10.1073/pnas.1323106111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peng WC, de Lau W, Forneris F, Granneman JC, Huch M, Clevers H, Gros P, Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell reports 3, 1885–1892 (2013); published online EpubJun 27 ( 10.1016/j.celrep.2013.06.009). [DOI] [PubMed] [Google Scholar]
- 19.Chen PH, Chen X, Lin Z, Fang D, He X, The structural basis of R-spondin recognition by LGR5 and RNF43. Genes Dev 27, 1345–1350 (2013); published online EpubJun 15 ( 10.1101/gad.219915.113). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zebisch M, Xu Y, Krastev C, MacDonald BT, Chen M, Gilbert RJ, He X, Jones EY, Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nature communications 4, 2787 (2013) 10.1038/ncomms3787). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zebisch M, Jones EY, Crystal structure of R-spondin 2 in complex with the ectodomains of its receptors LGR5 and ZNRF3. J Struct Biol 191, 149–155 (2015); published online EpubAug ( 10.1016/j.jsb.2015.05.008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Lau W, Peng WC, Gros P, Clevers H, The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev 28, 305–316 (2014); published online EpubFeb 15 ( 10.1101/gad.235473.113). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruffner H, Sprunger J, Charlat O, Leighton-Davies J, Grosshans B, Salathe A, Zietzling S, Beck V, Therier M, Isken A, Xie Y, Zhang Y, Hao H, Shi X, Liu D, Song Q, Clay I, Hintzen G, Tchorz J, Bouchez LC, Michaud G, Finan P, Myer VE, Bouwmeester T, Porter J, Hild M, Bassilana F, Parker CN, Cong F, R-Spondin potentiates Wnt/beta-catenin signaling through orphan receptors LGR4 and LGR5. PLoS One 7, e40976 (2012) 10.1371/journal.pone.0040976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker F, Zhang HH, Odorizzi A, Burgess AW, LGR5 Is a Negative Regulator of Tumourigenicity, Antagonizes Wnt Signalling and Regulates Cell Adhesion in Colorectal Cancer Cell Lines. PLoS One 6, e22733 (2011) 10.1371/journal.pone.0022733PONE-D-11-02367 [pii]). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, Qiu S, Lu L, Zou J, Li WF, Wang O, Zhao H, Wang H, Tang J, Chen L, Xu T, Sun Z, Liao W, Luo G, Lu X, RSPO2-LGR5 signaling has tumour-suppressive activity in colorectal cancer. Nature communications 5, 3149 (2014) 10.1038/ncomms4149). [DOI] [PubMed] [Google Scholar]
- 26.Xu Q, Wang Y, Dabdoub A, Smallwood PM, Williams J, Woods C, Kelley MW, Jiang L, Tasman W, Zhang K, Nathans J, Vascular development in the retina and inner ear: control by Norrin and Frizzled-4, a high-affinity ligand-receptor pair. Cell 116, 883–895 (2004); published online EpubMar 19 ( 10.1016/s0092-8674(04)00216-8) [DOI] [PubMed] [Google Scholar]
- 27.Park S, Cui J, Yu W, Wu L, Carmon KS, Liu QJ, Differential activities and mechanisms of the four R-spondins in potentiating Wnt/beta-catenin signaling. J Biol Chem 293, 9759–9769 (2018); published online EpubJun 22 ( 10.1074/jbc.RA118.002743). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong X, Azhdarinia A, Ghosh SC, Xiong W, An Z, Liu Q, Carmon KS, LGR5-Targeted Antibody-Drug Conjugate Eradicates Gastrointestinal Tumors and Prevents Recurrence. Mol Cancer Ther 15, 1580–1590 (2016); published online EpubJul ( 10.1158/1535-7163.MCT-16-0114). [DOI] [PubMed] [Google Scholar]
- 29.Warner ML, Bell T, Pioszak AA, Engineering high-potency R-spondin adult stem cell growth factors. Mol Pharmacol 87, 410–420 (2015) 10.1124/mol.114.095133). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira GC, Chockalingam S, Melegh Z, Greenhough A, Malik S, Szemes M, Park JH, Kaidi A, Zhou L, Catchpoole D, Morgan R, Bates DO, Gabb PD, Malik K, LGR5 regulates pro-survival MEK/ERK and proliferative Wnt/beta-catenin signalling in neuroblastoma. Oncotarget 6, 40053–40067 (2015); published online EpubNov 24 ( 10.18632/oncotarget.5548). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boontanrart M, Rokkam D, Liu D, Dylla SJ, Aujay M, Novel anti-RNF43 antibodies and methods of use. Patent publication PCT WO2015/164392A2 (2015).
- 32.Gustafsdottir SM, Schallmeiner E, Fredriksson S, Gullberg M, Soderberg O, Jarvius M, Jarvius J, Howell M, Landegren U, Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem 345, 2–9 (2005); published online EpubOct 1 ( 10.1016/j.ab.2005.01.018). [DOI] [PubMed] [Google Scholar]
- 33.Gauthier T, Claude-Taupin A, Delage-Mourroux R, Boyer-Guittaut M, Hervouet E, Proximity Ligation In situ Assay is a Powerful Tool to Monitor Specific ATG Protein Interactions following Autophagy Induction. PLoS One 10, e0128701 (2015) 10.1371/journal.pone.0128701). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avin A, Levy M, Porat Z, Abramson J, Quantitative analysis of protein-protein interactions and post-translational modifications in rare immune populations. Nature communications 8, 1524 (2017); published online EpubNov 15 ( 10.1038/s41467-017-01808-6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Q, Carmon KS, Gong X, Lgr4 specific monoclonal antibodies and methods of their use. Patent Publication PCT WO2017106034A1 (2017).
- 36.Carmon KS, Lin Q, Gong X, Thomas A, Liu Q, LGR5 Interacts and Cointernalizes with Wnt Receptors To Modulate Wnt/beta-Catenin Signaling. Mol Cell Biol 32, 2054–2064 (2012); published online EpubJun ( 10.1128/MCB.00272-12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder JC, Rochelle LK, Lyerly HK, Caron MG, Barak LS, Constitutive Internalization of the Leucine-rich G Protein-coupled Receptor-5 (LGR5) to the Trans-Golgi Network. J Biol Chem 288, 10286–10297 (2013); published online EpubApr 12 ( 10.1074/jbc.M112.447540). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu K, Xu Y, Rajashankar KR, Robev D, Nikolov DB, Crystal structures of Lgr4 and its complex with R-spondin1. Structure 21, 1683–1689 (2013); published online EpubSep 3 ( 10.1016/j.str.2013.07.001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang D, Huang B, Zhang S, Yu X, Wu W, Wang X, Structural basis for R-spondin recognition by LGR4/5/6 receptors. Genes Dev 27, 1339–1344 (2013); published online EpubJun 15 ( 10.1101/gad.219360.113). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu JG, Huang C, Yang Z, Jin M, Fu P, Zhang N, Luo J, Li D, Liu M, Zhou Y, Zhu Y, Crystal structure of LGR4-Rspo1 complex: insights into the divergent mechanisms of ligand recognition by leucine-rich repeat G-protein-coupled receptors (LGRs). J Biol Chem 290, 2455–2465 (2015); published online EpubJan 23 ( 10.1074/jbc.M114.599134). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, Ootani A, Roelf K, Lee M, Yuan J, Li X, Bolen CR, Wilhelmy J, Davies PS, Ueno H, von Furstenberg RJ, Belgrader P, Ziraldo SB, Ordonez H, Henning SJ, Wong MH, Snyder MP, Weissman IL, Hsueh AJ, Mikkelsen TS, Garcia KC, Kuo CJ, Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature 545, 238–242 (2017); published online EpubMay 11 ( 10.1038/nature22313). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moad HE, Pioszak AA, Reconstitution of R-spondin:LGR4:ZNRF3 Adult Stem Cell Growth Factor Signaling Complexes with Recombinant Proteins Produced in E. coli. Biochemistry, (2013); published online EpubSep 19 ( 10.1021/bi401090h). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felce JH, Latty SL, Knox RG, Mattick SR, Lui Y, Lee SF, Klenerman D, Davis SJ, Receptor Quaternary Organization Explains G Protein-Coupled Receptor Family Structure. Cell reports 20, 2654–2665 (2017); published online EpubSep 12 ( 10.1016/j.celrep.2017.08.072). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carmon KS, Gong X, Yi J, Wu L, Thomas A, Moore CM, Masuho I, Timson DJ, Martemyanov KA, Liu QJ, LGR5 receptor promotes cell-cell adhesion in stem cells and colon cancer cells via the IQGAP1-Rac1 pathway. J Biol Chem 292, 14989–15001 (2017); published online EpubSep 8 ( 10.1074/jbc.M117.786798). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cottet M, Albizu L, Comps-Agrar L, Trinquet E, Pin JP, Mouillac B, Durroux T, Time resolved FRET strategy with fluorescent ligands to analyze receptor interactions in native tissues: application to GPCR oligomerization. Methods Mol Biol 746, 373–387 (2011) 10.1007/978-1-61779-126-0_21). [DOI] [PubMed] [Google Scholar]
- 46.Gurney A, Axelrod F, Bond CJ, Cain J, Chartier C, Donigan L, Fischer M, Chaudhari A, Ji M, Kapoun AM, Lam A, Lazetic S, Ma S, Mitra S, Park IK, Pickell K, Sato A, Satyal S, Stroud M, Tran H, Yen WC, Lewicki J, Hoey T, Wnt pathway inhibition via the targeting of Frizzled receptors results in decreased growth and tumorigenicity of human tumors. Proc Natl Acad Sci U S A 109, 11717–11722 (2012); published online EpubJul 17 ( 10.1073/pnas.1120068109). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng WC, de Lau W, Madoori PK, Forneris F, Granneman JC, Clevers H, Gros P, Structures of Wnt-Antagonist ZNRF3 and Its Complex with R-Spondin 1 and Implications for Signaling. PLoS One 8, e83110 (2013) 10.1371/journal.pone.0083110). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DeBruine ZJ, Xu HE, Melcher K, Assembly and architecture of the Wnt/beta-catenin signalosome at the membrane. British journal of pharmacology 174, 4564–4574 (2017); published online EpubDec ( 10.1111/bph.14048). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kinzel B, Pikiolek M, Orsini V, Sprunger J, Isken A, Zietzling S, Desplanches M, Dubost V, Breustedt D, Valdez R, Liu D, Theil D, Muller M, Dietrich B, Bouwmeester T, Ruffner H, Tchorz JS, Functional roles of Lgr4 and Lgr5 in embryonic gut, kidney and skin development in mice. Dev Biol 390, 181–190 (2014); published online EpubJun 15 ( 10.1016/j.ydbio.2014.03.009). [DOI] [PubMed] [Google Scholar]
- 50.Tu J, Park S, Yu W, Zhang S, Wu L, Carmon K, Liu QJ, The most common RNF43 mutant G659Vfs*41 is fully functional in inhibiting Wnt signaling and unlikely to play a role in tumorigenesis. Scientific reports 9, 18557 (2019); published online EpubDec 6 ( 10.1038/s41598-019-54931-3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanjana NE, Shalem O, Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783–784 (2014); published online EpubAug ( 10.1038/nmeth.3047). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials
Fig. S1. Validation of LGR5 expression in STF-4KO-LGR5 cell line.
Fig. S2. R2Fu-F109A has no LGR5-dependent activity.
Fig. S3. Full-length LGR4 and LGR5 exist as both monomers and dimers but their ECDs only form monomers.
Fig. S4. LGR5-RSPO1-induced potentiation of Wnt signaling is through phosphorylation of LRP6.
Fig. S5. LGR5FL does not interact with RNF43 with or without RSPO1.
Fig. S6. Validation of RNF43 antibody, SC37.
Fig. S7. PLA between ZNRF3ΔR and LGR4 or LGR5 shows that LGR5 does not interact with ZNRF3ΔR with or without RSPO1.
Fig. S8. LGR4 and LGR5 differ in surface distribution but bind RSPO similarly.
Fig. S9. Expression of LGR4ECD-5TM and LGR5ECD-4TM chimeras.
Fig. S10. TR-FRET assay between RSPO and LGR as a positive control.
Fig. S11. Co-localization of LGR5 with FZD and LRP6.
Fig. S12. Expression of IQGAP1ΔIQ.
Fig. S13. Images of full blots for Fig.2B.
Table S1. Published RNA-seq data for selected genes in the cancer cell lines used in the study.


