Abstract
Mycobacterium tuberculosis possesses a significant arsenal of strategies to combat immune defense of the host organism. Small noncoding RNAs, which constitute the largest group of regulatory RNAs, play an important role in the host–pathogen interactions and represent one of the levels of the regulation of interactions of microbial cells with their environment. The regulatory role of small RNAs in pathogenic bacteria is essential when rapid adaptation to the changing environmental conditions with further synchronization of metabolic reactions are required to ensure microbial survival and infection progression. During the past few years, eight small RNAs from M. tuberculosis have been functionally characterized, and targets for four of them have been identified. Small RNAs from M. tuberculosis and other pathogenic microorganisms were found to be one of the most important functional factors in the adaptive response to changing environmental conditions.
Keywords: Mycobacterium tuberculosis, small non-coding RNA, pathogenesis, host–pathogen interactions, adaptive response
INTRODUCTION
Tuberculosis is a globally spread infectious disease that claims almost 2 million human lives annually. Moreover, more than eight million new cases are reported every year [1]. The causative agent of this infection – Mycobacterium tuberculosis – can persist in an organism in the form of latent infection, which could reactivate upon compromised host immune status [2]. The mechanisms used by M. tuberculosis to avoid the host immune defense, to maintain its viability under stress during the intracellular persistence, and to transit from the dormant state into the active one are under scrutiny of modern immunologists, bacteriologists, and molecular biologists.
After entering a human body, M. tuberculosis interacts with the host immune cells. Notably, the host response to the mycobacteria differs from that to other bacterial infections. Mycobacteria modulate or reprogram the process of phagosome maturation using cell wall components and secreted products, which is known as the phagosome maturation arrest. Mycobacteria can suppress autophagy and cell–cell signaling in macrophages, which are the first immune cells interacting with M. tuberculosis, as well as resist the action of toxic compounds (reactive oxygen species, nitrogen oxide, and metals [3, 4]); that facilitate survival of mycobacteria inside the macrophages. Thus, M. tuberculosis uses macrophages as a main niche for the long-term persistence in the organism [5-8].
Long co-evolution of M. tuberculosis and its hosts provide the pathogen with a set of strategies to efficiently fight the host immune defense. One of them is noncoding RNAs, which form complex regulatory networks together with their targets that allow M. tuberculosis to adjust its metabolism at different stages of infection [9]. It has been established that small noncoding RNAs, which are the most numerous group of regulatory RNAs, play the most important role in the adaptation of M. tuberculosis to a parasitic life style [9].
Small noncoding RNAs (small RNAs, sRNAs) are subdivided into cis-encoded and trans-encoded antisense RNAs [10]. cis-Encoded antisense RNAs are encoded in the same locus with their target mRNAs; they are transcribed from the opposite DNA strand to ensure their complementary binding to the target. Mostly, cis-encoded sRNAs block translation via complementary binding to the ribosome-binding site (RBS) in the mRNA target. trans-Encoded antisense RNAs are especially interesting as they are encoded by individual genes and, hence, have their own promoters and terminators. The genes for these sRNAs are located at a considerable distance from the genes they regulate, and their size varies from 50 to 300 nucleotides. sRNAs genes are often localized between the protein-coding genes. sRNAs have a stable secondary structure that prevents their rapid degradation and facilitates formation of loops with unpaired nucleotide bases that form the target-binding sites (so-called seed regions). The role of sRNAs (both cis- and trans-encoded) in bacterial metabolism was characterized in detail in the review by Azhikina et al. [10]. A recently published review by Taneja and Dutta describes the search for mycobacterial sRNAs and their regulation [11]. Here, we describe the regulatory role of trans-encoded sRNAs in one of the most dangerous pathogen – M. tuberculosis.
REGULATORY MECHANISMS IMPLEMENTED BY SMALL NONCODING RNAs
As trans-encoded small RNAs (sRNAs) are only partially complementary to their targets, they can regulate more than one target. sRNAs control the expression of target RNAs via two different mechanisms: by direct RNA–RNA interaction and by binding with effector proteins (Hfq or ProQ) [9-12].
Such interactions could result in the termination of mRNA translation due to the RBS blocking, which is the most common mechanism of sRNA action [9]. The binding of sRNA with its target not only prevents ribosome binding, but most often, initiates the cleavage of the RNA-RNA duplex by RNases J and E. In the case of polycistronic mRNAs, the binding of sRNA at the 5′-region could result in the Rho-dependent termination of translation at all mRNA sites [13].
sRNAs can also activate protein translation. In some mRNAs, the 5′-untranslated region (UTR) forms a hairpin structure protecting the RBS. Interaction of the sRNA loop with the complementary mRNA sequence opens this hairpin, thus initiating translation [14]. Moreover, sRNAs can stabilize other transcripts. The binding of sRNA to a cleavage site in the target mRNA prevents the cleavage of the latter and increases the half-life of the transcript [15]. Another example of transcription activation is prevention of internal Rho-dependent termination in long 5′-UTRs of some genes [16].
Sponge RNAs is a special functional class of sRNAs that bind other sRNAs and regulate their activity. Sponge RNAs were identified in enterobacteria and Bacillus subtilis in the investigations of RNA–RNA interactions. The diversity of their origin, as well as the diversity of biological processes involving sponge RNAs, suggests that this regulatory mechanism is widespread in bacteria. Similar to other sRNAs, sponge RNAs can be encoded by individual genes, 3′- or 5′-UTRs, or spacers in other genes. Each RNA sponge has a partial complementarity to another sRNA, which is its target. The formation of the duplex sequesters the target sRNA and results in the degradation of the complex. The extent of the inhibition varies depending on the concentrations of the target sRNA and sponge RNA, which, in turn, depend on the level of the expression (see review [17] for detailed description of the mechanism of action of sponge RNAs).
sRNAs with a dual function have also been found. Such sRNAs both serve as transcription regulators, and encode small peptides, as their genes contain open reading frames. In particular, the well-known RNAIII sRNA from Staphylococcus aureus controls expression of several pathogenic genes by the classic mechanism, i.e., via binding to the target mRNA through the seed region in one of its loops. In addition, RNAIII encodes a protein δ-hemolysin, that disrupts cell membranes of the host organism [18].
Hence, sRNAs represent a powerful regulatory reservoir to provide the fast adaptive response to rapidly changing environmental conditions. Identification of such sRNAs in M. tuberculosis and elucidation of their functions are very important for understanding the role of these molecules in the progression of tuberculosis infection and in the microbe interaction with the host organism.
STRATEGIES FOR IDENTIFICATION OF sRNAs IN M. tuberculosis
To date, around 2000 noncoding regulatory RNAs have been identified in M. tuberculosis that belong to different functional classes. Approximately 560 of them are small trans-encoded RNAs. More than 20 trans-encoded sRNAs in M. tuberculosis have been confirmed experimentally by Northern blotting and/or microarray analysis, but only eight of them have been characterized in detail.
Some of sRNAs found in M. tuberculosis have been also identified in all members of the genus Mycobacterium, e.g., sRNA B11 (MTS2822). Other sRNAs, such as Mcr11 (MTS0997) and DrrS (MTS1338), were identified only in pathogenic mycobacteria. Remarkably, no homologs of the chaperon proteins Hfq and ProQ mediating sRNA interactions with the mRNA targets, have been found in M. tuberculosis [9]. Nevertheless, it was suggested that a protein mediating the functions of sRNAs could exist in M. tuberculosis cells. RNA-binding proteins CspA and CspB from the large family of cold-shock proteins are plausible candidates for this role [9, 19].
Identification of sRNAs undoubtedly attracts the researchers’ attention. Back in 2006 the sRNAPredict2 computer program predicted56 noncoding RNAs in the M. tuberculosis genome [20]. The search was based on comparative analysis of sequences of the intergenic regions in different bacterial species with consideration of the homology and characteristic features of sRNAs. However, the existence of sRNAs predicted using this approach has not been confirmed experimentally. Taking into account that only few regulatory sRNAs were discovered at that time, and most of them were from Gram-negative organisms, which differ significantly from mycobacteria, the probability of identifying mycobacterial noncoding sRNAs was rather low.
Another approach to identify sRNAs in M. tuberculosis that involved the sequencing of cDNA library of low-molecular-weight RNAs (20-75 bases) was undertaken several years later [21]. The identified sRNAs were confirmed by Northern blotting, and their genomic coordinates were determined by RACE (rapid amplification of cDNA ends). In total, five trans-encoded sRNAs were found: B11, B55, C8, F6, and G2. The first studies aimed to characterize the physiological significance of these sRNAs. In particular, C8 was classified as a structural RNA homologous to the 4.5S RNA.
Later, 34 sRNAs were found in the Mycobacterium bovis BCG cells [22]. The authors used a combination of two approaches: computer prediction based on the interspecies homology and cloning of short-length cDNAs. All identified sRNAs were confirmed by Northern blotting. From 34 sRNAs, 20 were also identified in M. tuberculosis; three of them – F6, C8, B11 – were the same as in the previous study [21].
Analysis of M. tuberculosis transcriptome by the RNA sequencing technique (RNA-seq) [23] revealed the presence of 15 sRNAs; 11 of them were confirmed by Northern blotting, including five previously discovered sRNAs [21]. Furthermore, the authors characterized in detail three sRNAs: MTS2823, MTS1338, and MTS0997 [23].
Later, a new algorithm for predicting noncoding RNAs was developed [24], which included analysis of RNA-seq data and data on the conservatism of certain genome regions produced by the method of comparative genomics. When applied, this algorithm identified 1948 noncoding RNAs in the transcriptome of exponential M. tuberculosis; 977 of them were trans-encoded sRNAs, including all previously discovered sRNAs. By introducing additional criteria (minimal distance to the nearest gene, minimal free energy of the secondary structure), the list of candidates for the trans-encoded sRNAs was reduced to 59. Analysis of 1373 predicted noncoding RNAs by microarrays [25] confirmed 258 corresponding transcripts; 22 of them were assigned to trans-encoded sRNAs. The presence of four sRNAs (#149, #161, #224, and #1096 according to the author’s designations) was additionally confirmed by Northern blotting.
Another algorithm for identification of sRNAs in M. tuberculosis, which was based on the same principle, i.e., the presence of conserved, highly expressed sequences in the bacterial transcriptome, was tested in 2016 [26]. The cDNA libraries in this case were constructed separately for different RNA fractions isolated from the microbial cultures in the logarithmic growth phase. The RNAz algorithm used in the experiment had several built-in search parameters, including conservatism of the nucleotide sequence among the members of Mycobacterium genus, conserved structure, and stability. In addition to the sRNAs identified and confirmed in the previous studies, 192 new potential sRNAs were found; 14 of them were selected for the confirmation by Northern blotting, and 13 of them passed this test.
Recently, 62 potential sRNAs from different functional categories were identified by analysis of the M. tuberculosis genome using saturated transposon mutagenesis; mutations in 7 of these sRNAs was found to cause defects in the cell growth [27]. This approach allows the researchers to identify genes required for bacterial growth that was based on the frequencies of mutations induced by the transposon insertion. The BS_finder program for predicting regulatory sRNAs based on the RNA-seq data was developed to scan the transcriptome using the sliding window approach. This approach allows finding small transcripts with significantly greater read depth encoded fully or partially in the intergenic regions and identifying their 5′ and 3′ coordinates.
Notably, all methods which were previously used for identification of sRNAs were based on the transcriptome analysis of bacteria cultivated under standard conditions in rich nutrient media. Under such conditions, many sRNAs, which could participate in the stress adaptation of bacteria, will be missed, as they are expressed at very low levels. Gerrick et al. used BS_finder to search for sRNAs in the transcriptomes of bacteria exposed to various stress factors [28]. They discovered 189 potential sRNAs; 103 of them have not been identified previously. The authors also reported that the expression levels of these potential sRNAs varied depending on the cultivation conditions. One of the sRNAs, MrsI, which was identified and confirmed for the first time, was examined in detail.
The idea of analysis of M. tuberculosis transcriptomes obtained under different growth conditions was later developed by Ami et al. [29], who analyzed RNA-seq data from M. tuberculosis cells in the logarithmic growth phase using the sliding window approach. The search revealed 65 intergenic regions related to potential trans-encoded sRNAs. The list included four sRNA that had been experimentally verified before: ncRv11147Ac, ncRv2395, ncRv11534A, and MrsI. Expression of genes for potential sRNAs was studied using analysis of DNA–protein interactions by the chromatin immunoprecipitation (ChIP) technique and massive parallel DNA sequencing (ChIP-seq) with RNAP (RNA polymerase) protein and NusA protein (component of transcriptional complex that participates in the transcription termination and antitermination). The data were obtained for M. tuberculosis cultures under various growth conditions: logarithmic and stationary phases, on dormancy, reactivation, and under stress exposure (15 states totally). It was found that the putative sRNA coding regions were significantly bound by the transcription machinery. Differential expression was observed for 24 sRNAs, confirmed earlier.
SMALL RNAs OF M. tuberculosis WITH THE DEFINED MECHANISM OF ACTION
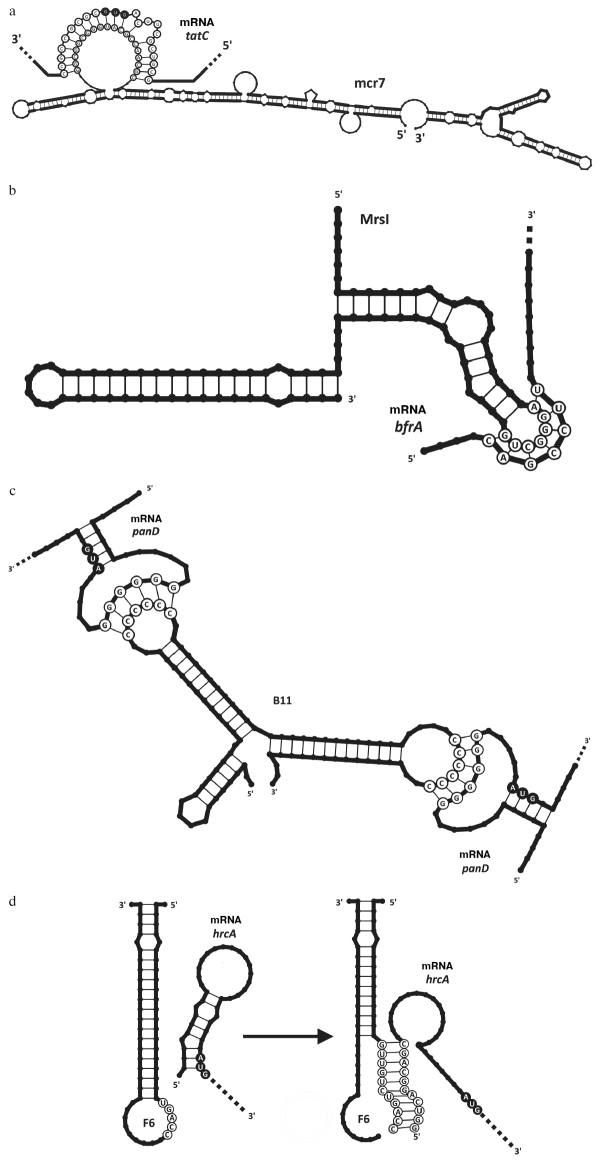
Despite numerous attempts to elucidate the functional role of M. tuberculosis sRNAs, these studies have had a very limited success for a long time. However, recent years have been marked by a series of successful investigations. By present, eight sRNAs from M. tuberculosis have been functionally characterized, and target mRNAs have been identified for four of them (figure).
Figure.
Interaction of M. tuberculosis sRNAs with mRNA targets. Start codons are marked with black circles. a) Mcr7 binds to the RBS and the first codons in tatC, resulting in the translation suppression; b) MrsI interacts with the bfrA 5′-UTR, resulting in the translation suppression; c) B11 binds to the RBS in panD, resulting in the translation suppression; d) F6 interacts with the hrcA 5′-UTR, resulting in the opening of the hairpin covering the Shine–Dalgarno sequence and translation initiation.
Mcr7 (MTB000067). Mcr7 was initially discovered in the analysis of cDNA libraries from M. bovis BCG and then confirmed by Northern blotting. Later, its homologue was found in M. tuberculosis [22]. Mcr7 is the first M. tuberculosis sRNA, whose function was established [30]. Expression of Mcr7 is regulated by the two-component PhoPR signaling system, which is essential for the virulence of mycobacteria and controls expression of approximately 2% of all M. tuberculosis genes, including the ESX-1 secretion system genes.
While being a target of the PhoP protein, Mcr7 also modulates translation of the tatC gene by binding to its mRNA (figure, a), which affects the activity of the Tat secretory complex in M. tuberculosis. The Tat complex mediates secretion of proteins with a specific signaling sequence that contains two arginine residues (twin-arginine motif), including immunodominant Ag85 complex [31] and beta-lactamase BlaC [32].
MrsI (MTB000142, ncRv11846). In 2018, Gerrick et al. published the study on the search for sRNAs typical for various stress conditions [28]. In this study, bacterial cells were subjected to one of the stress factors such as (i) lack of iron in the cultivation medium, (ii) oxidative stress, (iii) membrane damage, (iv) acidic medium pH, and (v) nutrient deficiency. The sequencing of the cDNA libraries generated from the respective RNA samples and selection of transcripts with the highest probabilities of being sRNAs followed by comparison of the expression levels of these transcripts with the expression levels in the control samples revealed 82 sRNA candidates that exhibited differential expression under at least one of the stress conditions. Thus, the cultivation in the iron-deficient medium slightly increased the content of ncRv11846, whereas membrane damage strongly upregulated expression of this sRNA. The ncRv11846 sequence is conserved among the members of Mycobacteriaceae family and microorganisms from the closely related Nocardiaceae family, and its homologue have previously been described in Mycobacterium smegmatis [33]. This sRNA was selected by the authors for detailed investigation and, as a result, was named MrsI (mycobacterial regulatory sRNA in iron) [28]. MrsI was predicted to have a stable secondary structure was with a putative binding site consisting of 6 nucleotides in the apical loop. Deletion of the gene coding for this sRNA affected the viability of bacterial cells only in the iron-deficient medium. Proteomic and transcriptomic analyses of such cells revealed elevated levels of iron-binding proteins and their transcripts. Bioinformatics analysis demonstrated the possibility of interaction between the identified transcripts and suggested MrsI seed region. One of the MrsI targets is the bfrA mRNA encoding bacterioferritin – an iron deficiency protein (figure, b). The involvement of MrsI in the regulation of iron metabolism was also supported by identification of the binding site for the transcriptional factor IdeR activated in mycobacteria under the iron deficiency conditions of [34]. Hence, the function of MrsI was found to be similar to the function of the RyhB sRNA in Escherichia coli [35]. MrsI suppresses translation of iron-binding proteins (but not affecting cell survival), thus establishing a more economical regime of iron utilization in the cells.
B11 (ncRv13660c, MTS2822, 6C). B11 was identified in 2009 by Arnvig et al. during the screening of cDNA libraries obtained for a low-molecular-weight RNA fraction from M. tuberculosis [21]. The secondary structure of B11 includes two hairpins, each containing 6 consequent cytosine residues in the loop. This sRNA is highly conserved among bacteria of the Mycobacterium genus, and its homologues have been found in the bacteria of the Streptomyces and Corynebacterium genera. Expression of B11 is induced by the oxidative stress and low medium pH [21]. Overexpression of B11 causes the death of M. tuberculosis cells and suppresses the growth of M. smegmatis cells [21].
B11 has a stable secondary structure with three loops. Two of these loops contain seed regions comprised of 6 or 7 consecutive cytosines. The complementary guanidine sequences were found in the 5′ leader sequences of many mycobacterial genes. It was shown that the presence of both loops and the conservation of the cytosine sequences in B11 are critical for its function [36]. The transcriptome analysis revealed that B11 modulates (predominantly, reduces) expression of a significant number of genes, and putative B11-binding sites were detected in the mRNAs of 47 genes. Fifteen potential targets (including panD, dnaB, espE, espF, eccA1, PE35, MTB84, and mycP1 genes) were identified among these 47 candidates, for which interaction with B11 was experimentally confirmed. In all the cases, upregulation of the B11 expression in M. smegmatis resulted in the inhibition of translation of the corresponding mRNA target. Suppression of the dnaB gene encoding replicative DNA helicase was found to be essential for the formation of the characteristic B11 overexpression phenotype. Interestingly enough, B11 interacts directly with the mRNA target (figure, c), as confirmed by the gel electrophoresis mobility shift assay (EMSA) and transposon mutagenesis [36].
Investigations of the B11 functions were continued in Mycobacterium kansasi cells [37]. The authors generated a strain with mutation in the 5′-region of the sRNA gene, which resulted in complete suppression of its expression but did not affect expression of the neighboring genes. The M. kansasii strain lacking B11 demonstrated growth defects on solid nutrient media. The mutant mycobacteria did not form biofilms, although their planktonic growth was the same as in the wild-type strain. Despite the fact that no molecular targets were identified in this study, such physiological characterization expanded the spectrum of the regulatory functions of B11.
F6 (ncRv10243, MTS194, MTB000051). F6 was discovered by sequencing of the low-molecular-weight fraction of M. tuberculosis cDNA libraries [21] and confirmed by Northern blotting. It was found in the pathogenic representatives of the Mycobacterium genus and in M. smegmatis. The attempts to characterize its physiological role revealed the upregulation of the F6 expression under conditions of oxidative stress, hypoxia, low medium pH, and macrophage infection, but the strongest upregulation was observed upon the nutrient deficiency [21, 38]. It was found that overexpression of F6 suppressed the growth of M. tuberculosis cells [21]; however, neither overexpression, nor deletion of the F6 gene affected the growth of M. smegmatis cells [21]. The deletion of F6 prevented transition of M. tuberculosis from the dormant state in the Wayne hypoxia model [38]. Analysis of the transcription profile of M. tuberculosis strain deficient by F6 under conditions of nutrient deficiency revealed the upregulated expression of four genes: Rv0440 (groEL2), Rv3418c (groES), Rv0990c, and Rv0991c. All these genes are in the same regulon under control of the HrcA (Rv2374c) transcriptional repressor, which is activated during the heat shock. The hairpin in the 5′-region of the hrcA mRNA prevents translation of this template, as it partially covers the Shine-Dalgarno sequence. The complementary sequence contains the binding site for F6, so that its interaction with F6 results in the hairpin opening, making the RBS accessible for the translation initiation (figure, d). Activation of the HrcA repressor decreases production of the GroEL/S chaperones in the cell.
Mcr11 (MTS0997, MTB000063). Mcr11 was first identified in 2010 in the transcriptome of the logarithmic-phase culture of M. bovis BCG cells by RNA-seq and confirmed by Northern blotting [22]. The gene for Mcr11 is also present in M. tuberculosis, and its expression was confirmed experimentally. No Mcr11 homologues were found in non-pathogenic mycobacteria.
It was established that the level of Mcr11 expression increased upon transition to the stationary growth phase (both in M. tuberculosis and M. bovis cells). The expression of Mcr11 was noticeably upregulated in the mouse infection model [23, 39] and in dormant M. tuberculosis cells [40]. Analysis of the expression profiles revealed that Mcr11 transcription increased under nutrient deficiency and decreased at low pH; it also depended on the level of cAMP in the cells [41]. Overexpression of Mcr11 resulted in the inhibition of M. tuberculosis growth [40]. It was shown that the Mcr11 transcription was regulated by the AbmR (ATP-binding Mcr11 regulator) protein encoded by the gene adjacent to the Mcr11 gene [42]. The promoter regions of the AbmR and Mcr11 genes partially overlap. AbmR exhibits the autoinhibiting activity by specifically binding to its promoter region in the presence of ATP. In the AbmR-deficient strain, the content of Mcr11 did not change with transition between the growth phases. It is likely that the Mcr11 expression depends on the intracellular ATP concentration and is regulated by the ATP-binding transcription factor AbmR [42]. The binding of Mcr11 with the mRNAs for the lipB (lipoate protein ligase B required for lipoate synthesis), Rv3282 (protein presumably participating in the formation of division septum), and fadA3 (β-ketoacyl coenzyme A thiolase) genes was predicted using the target-searching program TargetRNA [43]. Taking into consideration that association of these genes with lipid metabolism had been demonstrated previously, the researchers examined the growth of M. tuberculosis and M. bovis BCG cells with deleted Mcr11 gene in the fatty acid-deficient cultivation medium. It was found that the growth of the mutant strains in the medium without exogenous sources of oleate was disrupted. Furthermore, expression of the predicted target genes in the mutant strains was increased in comparison with the control wild-type strain. It was concluded that Mcr11 controls expression of three operons: (i) dlaT-Rv2216-lipB, (ii) accD5-accE5-Rv3282, and (iii) fadA3, but also could have other targets involved in the regulation of lipid metabolism in mycobacteria [44].
MTS1338 (DrrS, MTB000077). Similar to Mcr11, MTS1338, which was identified in the M. tuberculosis transcriptome by RNA-seq, is present only in the pathogenic species belonging to Mycobacterium tuberculosis complex [23]. Expression of MTS1338 increases with the transition to the stationary growth phase [23]. MTS1338 is regulated by the genes from the DosR regulon, the expression of which is upregulated upon transition to the stationary phase and under stress conditions. MTS1338 has a stable secondary structure; its half-life is 6 h [44]. Significant accumulation of MTS1338 was observed during infection (e.g., in mouse models and during infection of the macrophage cell line) [23, 39, 45]. The concentration of MTS1338 increased in response to the activation of macrophages by γ-interferon via activation of the macrophage NO synthase [45]. The content of MTS1338 in dormant M. tuberculosis cells almost reached the level of 16S rRNA [40]. MTS1338 overexpression resulted in the inhibition of growth of mycobacterial cell [23, 40]. Moreover, the strains overexpressing MTS1338 were characterized with a higher resistance to unfavorable factors; in particular, the resistance of such bacteria to the medium acidification increases noticeably [45, 46]. Transcriptome analysis of the MTS1338-overexpressing strain cultivated under normal conditions revealed the expression profile typical for the cells grown under hypoxic conditions, indicating a decrease in the translation activity [45]. Taken together, the results of the conducted studies confirm the role of MTS1338 in the pathogenicity of M. tuberculosis and its adaptation to the conditions inside the infected organism.
MTS2823 (MTB000078). This sRNA was first described in 2011 [23]. The concentration of MTS2823 in the stationary-phase culture of mycobacterial cells and during chronic infection in mice was found to be high, almost reaching the levels of ribosomal RNAs. The high expression levels of MTS2823 were also observed in the dormant state [40] and under the action of stress factors in vitro [29]. Expression of many genes encoding the central metabolism enzymes in M. tuberculosis was significantly decreased during MTS2823 overexpression [23].
MTS2823 is highly conserved among bacteria of the Mycobacterium genus; it was also found in other representatives of actinobacteria. The predicted secondary structure of MTS2823 is a double-stranded hairpin with a knot in the middle and two small loops at the ends. This structure imitates DNA during transcription and can bind RNA polymerase. The 6S sRNA from E. coli has the same structure and acts as a transcription inhibitor. MTS2823 is a functional analogue of 6S, although they have certain differences. The mechanism of MTS2823 action was investigated in M. smegmatis, which has its homologue Ms1 [47]. Ms1 binds to RNA polymerase (RNAP) prior to the binding of the latter to the sigma factors. The more RNAP is in the complex with MS1, the lower is the total transcription level in the cell. The MS1–RNAP complex is unstable and dissociates easily; hence, the efficiency of the enzyme sequestration and, consequently, of the transcription inhibition depends directly of the Ms1 concentration. Ms1 is degraded efficiently by polynucleotide phosphorylase (PNPase) during the exponential growth phase. During the stationary phase, the concentration of PNPase decreases, leading to the Ms1 accumulation and, consequently, more efficient RNAP sequestration and suppression of the total transcription. It was also found that Ms1 upregulates expression of the β- and β′-subunits of RNAP. Deletion of Ms1 caused a decrease of RNAP concentration, resulting in no dramatic upregulation of the transcription activity in the mutant. The concentration of RNAP in the wild-type M. smegmatis cells in the stationary phase remained high, but the activity of the enzyme was reduced. This allows fast transition of bacterial cells to the active transcription state under favorable conditions [47].
ncRv12659 (MTS2048). ncRv12659 was found in the M. tuberculosis transcriptome in 2011 by RNA-seq [23]. It was demonstrated that this sRNA is accumulated in the mouse infection model and during starvation [48]. Overexpression of ncRv12659 resulted in the reduced growth of bacterial cells and changes in the transcription of ~50 genes. Deletion of ncRv12659 did not reduce the survival of M. tuberculosis under normal or stress conditions, as well as during infection of the macrophage cell line [49]. No targets of this sRNA have been identified so far.
The ncRv12659 gene is encoded in the PhiRv2 prophage, which is found in only some M. tuberculosis isolates. The ncRv12659 gene is located on the opposite strand from the gene for Rv2660, which has been considered for some time as an immunogenic protein potentially important for vaccine development [50]. However, following detailed examination of the transcription profile by RNA-seq with consideration of the strand specificity and bacteria in different physiological states, it was concluded that the Rv2660 mRNA was absent in M. tuberculosis cells, which most likely indicated that the Rv2660 protein did not exist [48].
FEATURES OF HOST–PATHOGEN INTERACTION CARRIED OUT BY sRNAs
Recent studies have indicated the importance of bacterial RNAs in pathogenesis. For many pathogenic bacteria, the mechanisms involving noncoding regulatory RNAs in the manifestation of virulent properties have been investigated in detail (see recent review [51]). In particular, it was demonstrated that the ssrS sRNA activates expression of many secretory virulence factors in bacteria of the Legionella genus, and deletion of this gene results in a significant virulence attenuation. The sRNA IhtA was found to facilitate the regulation of the Chlamydia trachomatis cell cycle and transition of this microorganism from the vegetative form to the infectious one.
It was found that the sRNAs RyhB and SgrS found in a number of bacteria and investigated in detail in model organisms, are important pathogenicity factors that facilitate survival of bacterial cells during infection. It is known that SgrS regulates the transport of sugars into the cell and is important for normal bacterial growth at elevated extracellular glucose concentration [52]. Although its functions are similar in different enterobacteria (E. coli, Salmonella, Yersinia pestis, Klebsiella pneumoniae, and others), SgrS from Salmonella enterica belongs to the chromosomal cluster of pathogenicity genes and suppresses expression of secreted SopD protein, which is one of the essential virulence factors in Salmonella during mouse infection [53]. RyhB downregulates expression of iron-binding proteins, thus facilitating efficient storage of iron in bacteria in the iron-deficient environment. Association of RyhB with virulence was confirmed for Pseudomonas aeruginosa, S. enterica, E. coli EHEC, Shigella flexneri, Shigella sonnei, Shigella dysenteriae, and Yersinia pseudotuberculosis [51].
It is interesting that the so-called RNA thermometers – temperature-sensitive riboswitches – were found in such infectious agents as Y. pestis, P. aeruginosa, Vibrio cholera, and Listeria monocytogenes. After bacteria enter the host organism and the environment temperature increases to 37°C, these RNA thermometers initiate expression of virulence-associated genes.
sRNAs can be secreted into the cytoplasm of immune cells after phagocytosis of bacteria. Similar to microRNAs, they can be processed in the cytoplasm and affect metabolic processes in host cells. This idea was first proposed in 2014 and experimentally confirmed for mycobacteria [54], however, no targets for the sRNAs were identified. In 2016, Westermann et al. described secretion of the Salmonella sRNAs into the intracellular medium and their processing (similar to microRNAs) in detail [55]. At present, the interaction between the secreted bacterial transcripts and host genomes is one of the widely discussed issues. It has been found that bacterial RNAs stimulate the innate immunity of host cells by interacting with the Toll-like receptors (TLRs) [56]. microRNA-like sRNAs from bacteria could be secreted into the environment via microvesicles, which could be then transported into eukaryotic cells, causing changes in the cytokine expression profile [57]. M. tuberculosis RNA is one of the key factors of host–pathogen interactions. After phagocytosis, bacteria secrete RNA, which is recognized by receptors expressed on the inner side of the phagosome membrane (TLR8, TLR7, and TLR3) [58].
CONCLUSIONS
Small noncoding RNAs of bacteria are one of the levels of global regulation of interaction of microbial cells with the environment. sRNAs of pathogenic bacteria are especially important for rapid adaptation of these microorganisms to a changing environment and following synchronization of various metabolic reactions during host–pathogen interaction.
sRNAs were found to play a crucial role in the stress response; they can also directly regulate expression of virulence factors under certain conditions or, more often, adjust metabolic pathways in order to facilitate the survival of bacterial cells in the infected organism.
Elucidation of these processes could help in the development of more efficient approaches to the diagnostics, therapy, and prevention of dangerous infections with the participation of regulatory sRNAs. Recently, gene of the sRNA npcTB_6715 was suggested for diagnostics of tuberculosis by multiplex PCR. This highly conserved gene is present only in the Mycobacterium tuberculosis complex, which provides almost 100% specificity and a high sensitivity of the assay [59]. Moreover, it is known that the sRNA expression profile could change under the antibiotic therapy [60]. In particular, the growth of M. tuberculosis cells in the presence of isoniazid resulted in differential expression of not only protein-encoding genes, but also of sRNAs, including B55, G2, MTS1338, MTS0997, F6, etc. [61]. All these make small regulatory RNAs of M. tuberculosis and pathogenic organisms in general a crucial element of adaptive response of bacteria to changes in environmental conditions.
Abbreviations
- RBS
ribosome-binding site
- RNA-seq
RNA sequencing
- UTR
untranslated region
- sRNA
small RNA
Funding
This work was supported by the Russian Science Foundation (project no. 18-15-00332).
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.World Health Organization (2019) Geneva: Global Tuberculosis Report, 2019.
- 2.Dye C. Global epidemiology of tuberculosis. Lancet. 2006;367:938–940. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- 3.Moraco A. H., Kornfeld H. Cell death and autophagy in tuberculosis. Semin. Immunol. 2014;26:497–511. doi: 10.1016/j.smim.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat, K., and Yaseen, I. (2018) Mycobacterium tuberculosis: macrophage takeover and modulation of innate effector responses, 10.5772/intechopen.75003.
- 5.Ehrt S., Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell. Microbiol. 2009;11:1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lerner T. R., Borel S., Gutierrez M. G. The innate immune response in human tuberculosis. Cell. Microbiol. 2015;17:1277–1285. doi: 10.1111/cmi.12480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behar S. M., Divangahi M., Remold H. G. Evasion of innate immunity by Mycobacteriumtuberculosis: is death an exit strategy? Nat. Rev. Microbiol. 2010;8:668–674. doi: 10.1038/nrmicro2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zondervan N. A., van Dam J. C. J., Schaap P. J., Martins Dos Santos V. A. P., Suarez-Diez M. Regulation of three virulence strategies of Mycobacterium tuberculosis: a success story. Int. J. Mol. Sci. 2018;19:347. doi: 10.3390/ijms19020347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwenk S., Arnvig K. B. Regulatory RNA in Mycobacteriumtuberculosis, back to basics. Pathog. Dis. 2018;76:1–12. doi: 10.1093/femspd/fty035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azhikina T. L., Ignatov D. V., Salina E. G., Fursov M. V., Kaprelyants A. S. Role of small non-coding RNAs in bacterial metabolism. Biochemistry (Moscow) 2015;80:1633–1646. doi: 10.1134/S0006297915130015. [DOI] [PubMed] [Google Scholar]
- 11.Taneja S., Dutta T. On a stake-out: Mycobacterial small RNA identification and regulation. Noncoding RNA Res. 2019;4:86–95. doi: 10.1016/j.ncrna.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wagner E. G., Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. Adv. Genet. 2015;90:133–208. doi: 10.1016/bs.adgen.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Bossi L., Schwartz A., Guillemardet B., Boudvillain M., Figueroa-Bossi N. A role for Rho-dependent polarity in gene regulation by a noncoding small RNA. Genes Dev. 2012;26:1864–1873. doi: 10.1101/gad.195412.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papenfort K., Vanderpool C. K. Target activation by regulatory RNAs in bacteria. FEMS Microbiol. Rev. 2015;39:362–378. doi: 10.1093/femsre/fuv016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frohlich K. S., Papenfort K., Fekete A., Vogel J. A small RNA activates CFA synthase by isoform-specific mRNA stabilization. EMBO J. 2013;32:2963–2979. doi: 10.1038/emboj.2013.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sedlyarova N., Shamovsky I., Bharati B. K., Epshtein V., Chen J., et al. sRNA-mediated control of transcription termination in E. coli. Cell. 2016;167:111–121.e13. doi: 10.1016/j.cell.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denham E. L. The Sponge RNAs of bacteria – how to find them and their role in regulating the post-transcriptional network. Biochim. Biophys. Acta Gene Regul. Mech. 2020;1863:194565. doi: 10.1016/j.bbagrm.2020.194565. [DOI] [PubMed] [Google Scholar]
- 18.Gimpel M., Brantl S. Dual-function small regulatory RNAs in bacteria. Mol. Microbiol. 2017;103:387–397. doi: 10.1111/mmi.13558. [DOI] [PubMed] [Google Scholar]
- 19.Płociński P., Macios M., Houghton J., Niemiec E., Płocińska R., et al. Proteomic and transcriptomic experiments reveal an essential role of RNA degradosome complexes in shaping the transcriptome of Mycobacterium tuberculosis. Nucleic Acids Res. 2019;47:5892–5905. doi: 10.1093/nar/gkz251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Livny J., Brencic A., Lory S., Waldor M. K. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arnvig K. B., Young D. B. Identification of small RNAs in Mycobacterium tuberculosis. Mol. Microbiol. 2009;73:397–408. doi: 10.1111/j.1365-2958.2009.06777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DiChiara J. M., Contreras-Martinez L. M., Livny J., Smith D., McDonough K. A., Belfort M. Multiple small RNAs identified in Mycobacterium bovis BCG are also expressed in Mycobacterium tuberculosis and Mycobacterium smegmatis. Nucleic Acids Res. 2010;38:4067–4078. doi: 10.1093/nar/gkq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnvig K. B., Comas I., Thomson N. R., Houghton J., Boshoff H., et al. Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002342. doi: 10.1371/journal.ppat.1002342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miotto P., Forti F., Ambrosi A., Pellin D., Veiga D. F., et al. Genome-wide discovery of small RNAs in Mycobacterium tuberculosis. PLoS One. 2012;7:e51950. doi: 10.1371/journal.pone.0051950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellin D., Miotto P., Ambrosi A., Cirillo D. M., Di Serio C. A genome-wide identification analysis of small regulatory RNAs in Mycobacteriumtuberculosis by RNA-seq and conservation analysis. PLoS One. 2012;7:e32723. doi: 10.1371/journal.pone.0032723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang M., Fleming J., Li Z., Li C., Zhang H., et al. An automated approach for global identification of sRNA-encoding regions in RNA-Seq data from Mycobacterium tuberculosis. Acta Biochim. Biophys. Sin. 2016;48:544–553. doi: 10.1093/abbs/gmw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeJesus M., Gerrick E. R., Xu W., Park S. W., Long J. ., et al. Comprehensive essentiality analysis of the Mycobacterium tuberculosis genome via saturating transposon mutagenesis. mBio. 2017;8:e02133–16. doi: 10.1128/mBio.02133-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gerrick E. R., Barbier T., Chase M. R., Xu R., François J., et al. Small RNA profiling in Mycobacteriumtuberculosis identifies MrsI as necessary for an anticipatory iron sparing response. Proc. Natl. Acad. Sci. USA. 2018;115:6464–6469. doi: 10.1073/pnas.1718003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ami V. K. G., Balasubramanian R., Hegde S. R. Genome-wide identification of the context-dependent sRNA expression in Mycobacterium tuberculosis. BMC Genomics. 2020;21:167. doi: 10.1186/s12864-020-6573-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Solans L., Gonzalo-Asensio J., Sala C., Benjak A., Uplekar S., et al. The PhoP-dependent ncRNA Mcr7 modulates the TAT secretion system in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004183. doi: 10.1371/journal.ppat.1004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiker H. G., Harboe M. The antigen 85 complex: a major secretion product of Mycobacterium tuberculosis. Microbiol. Rev. 1992;56:648–661. doi: 10.1128/mr.56.4.648-661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores A. R., Parsons L. M., Pavelka M. S., Jr. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology. 2005;151:521–532. doi: 10.1099/mic.0.27629-0. [DOI] [PubMed] [Google Scholar]
- 33.Tsai C.-H., Baranowski C., Livny J., McDonough K. A., Wade J. T., Contreras L. M. Identification of novel sRNAs in Mycobacterial species. PLoS One. 2013;8:e79411. doi: 10.1371/journal.pone.0079411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prakash P., Yellaboina S., Ranjan A., Hasnain S. E. Computational prediction and experimental verification of novel IdeR binding sites in the upstream sequences of Mycobacterium tuberculosis open reading frames. Bioinformatics. 2005;21:2161–2166. doi: 10.1093/bioinformatics/bti375. [DOI] [PubMed] [Google Scholar]
- 35.Jacques J. F., Jang S., Prévost K., Desnoyers G., Desmarais M., et al. RyhB small RNA modulates the free intracellular iron pool and is essential for normal growth during iron limitation in Escherichia coli. Mol. Microbiol. 2006;62:1181–1190. doi: 10.1111/j.1365-2958.2006.05439.x. [DOI] [PubMed] [Google Scholar]
- 36.Mai J., Rao C., Watt J., Sun X., Lin C., Zhang L., Lui J. Mycobacterium tuberculosis 6C sRNA binds multiple mRNA targets via C-rich loops independent of RNA chaperones. Nucleic Acids Res. 2019;47:4292–4307. doi: 10.1093/nar/gkz149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Budell W. C., Germain G. A., Janisch N., McKie-Krisber Z., Jayaprakash A. D., et al. Transposon mutagenesis in Mycobacterium kansasii links a small RNA gene to colony morphology and biofilm formation and identifies 9,885 intragenic insertions that do not compromise colony outgrowth. Microbiol. Open. 2019;9:e988. doi: 10.1002/mbo3.988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houghton J., Rodgers A., Rose G., Arnvig K. B. The Mycobacterium tuberculosis sRNA F6 regulates expression of groEL/S. bioRxiv. 2020 doi: 10.1101/2020.07.15.204107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ignatov D. V., Timoshina O. Yu., Logunova N. N., Skvortsov T. A., Azhikina T. L. Expression of small RNAs of Mycobacterium tuberculosis in murine models of tuberculosis infection. Russ. J. Bioorg. Chem. 2014;40:233. doi: 10.1134/s1068162014020058. [DOI] [PubMed] [Google Scholar]
- 40.Ignatov D. ., Salina E. G., Fursov M. V., Skvortsov T. A., Azhikina T. L., Kaprelyants A. S. Dormant non-culturable Mycobacterium tuberculosis retains stable low-abundant mRNA. BMC Genomics. 2015;16:954. doi: 10.1186/s12864-015-2197-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelly S., Bishai W. R., Lamichhane G. A screen for non-coding RNA in Mycobacteriumtuberculosis reveals a cAMP-responsive RNA that is expressed during infection. Gene. 2012;500:85–92. doi: 10.1016/j.gene.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girardin R. C., Bai G., He J., Sui H., McDonough K. A. AbmR (Rv1265) is a novel transcription factor of Mycobacterium tuberculosis that regulates host cell association and expression of the non-coding small RNA Mcr11. Mol. Microbiol. 2018;110:811–830. doi: 10.1111/mmi.14126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Girardin R. C., McDonough K. A. Small RNA Mcr11 requires the transcription factor AbmR for stable expression and regulates genes involved in the central metabolism of Mycobacterium tuberculosis. Mol. Microbiol. 2019;113:504–520. doi: 10.1111/mmi.14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moores A., Riesco A. B., Schwenk S., Arnvig K. B. Expression, maturation and turnover of DrrS, an unusually stable, DosR regulated small RNA in Mycobacterium tuberculosis. PLoS One. 2017;12:e0174079. doi: 10.1371/journal.pone.0174079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salina E. G., Grigorov A., Skvortsova Y., Majorov R., Bychenko O., et al. MTS1338, a small Mycobacterium tuberculosis RNA, regulates transcriptional shifts consistent with bacterial adaptation for entering into dormancy and survival within host macrophages. Front. Cell. Infect. Microbiol. 2019;9:405. doi: 10.3389/fcimb.2019.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ostrik A. A., Salina E. G., Skvortsova Yu. V., Grigorov A. S., Bychenko O. S., et al. Small RNAs of Mycobacterium tuberculosis in adaptation to host-like stress conditions iv vitro. Appl. Biochem. Microbiol. 2020;56:381–386. [Google Scholar]
- 47.Šiková M., Janoušková M., Ramaniuk O., Páleníková P., Pospíšil J., et al. Ms1 RNA increases the amount of RNA polymerase in Mycobacterium smegmatis. Mol. Microbiol. 2019;111:354–372. doi: 10.1111/mmi.14159. [DOI] [PubMed] [Google Scholar]
- 48.Houghton J., Cortes T., Schubert O., Rose G., Rodgers A., et al. A small RNA encoded in the Rv2660c locus of Mycobacterium tuberculosis is induced during starvation and infection. PLoS One. 2013;8:e80047. doi: 10.1371/journal.pone.0080047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Houghton, J. (2015) The regulatory role of sRNAs in Mycobacterium tuberculosis: a thesis for the degree of Doctor of Philosophy, L.: The National Institute for Medical Research, p. 265.
- 50.Aagaard C., Hoang T., Dietrich J., Cardona P. J., Izzo A., et al. A multistage tuberculosis vaccine that confers efficient protection before and after exposure. Nat. Med. 2011;17:189–194. doi: 10.1038/nm.2285. [DOI] [PubMed] [Google Scholar]
- 51.Westermann A. J. Regulatory RNAs in virulence and host–microbe interactions. Microbiol. Spectrum. 2018;6:RWR–0002–2017. doi: 10.1128/microbiolspec.rwr-0002-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bobrovskyy M., Vanderpool C. K. The small RNA SgrS: roles in metabolism and pathogenesis of enteric bacteria. Front. Cell. Infect. Microbiol. 2014;4:61. doi: 10.3389/fcimb.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang X., Rossanese O. W., Brown N. F., Kujat-Choy S., Galán J. E., et al. The related effector proteins SopD and SopD2 from Salmonella enterica serovar Typhimurium contribute to virulence during systemic infection of mice. Mol. Microbiol. 2004;54:1186–1198. doi: 10.1111/j.1365-2958.2004.04344.x. [DOI] [PubMed] [Google Scholar]
- 54.Furuse Y., Finethy R., Saka H. A., Xet-Mull A. M., Sisk D. M., et al. Search for microRNAs expressed by intracellular bacterial pathogens in infected mammalian cells. PLoS One. 2014;9:e106434. doi: 10.1371/journal.pone.0106434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Westermann A. J., Förstner K. U., Amman F., Barquist L., et al. Dual RNA-seq unveils noncoding RNA functions in host–pathogen interactions. Nature. 2016;529:496–501. doi: 10.1038/nature16547. [DOI] [PubMed] [Google Scholar]
- 56.Lee J., Lee S., Kim K. K., Lim Y.-J., Choi J.-A., et al. Characterisation of genes differentially expressed in macrophages by virulent and attenuated Mycobacterium tuberculosis through RNA-Seq analysis. Sci. Rep. 2019;9:4027. doi: 10.1038/s41598-019-40814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi J.-W., Kim S. C., Hong S.-H., Lee H. J. Secretable small RNAs via outer membrane vesicles in periodontal pathogens. J. Dent. Res. 2017;96:458–466. doi: 10.1177/0022034516685071. [DOI] [PubMed] [Google Scholar]
- 58.Burkert S., Schumann R. R. RNA Sensing of Mycobacterium tuberculosis and its impact on TB vaccination strategies. Vaccines. 2020;8:67. doi: 10.3390/vaccines8010067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanniappan P., Ahmed S. A., Rajasekaram G., Marimuthu C., Ch'ng E. S., et al. RNomic identification and evaluation of npcTB_6715, a non-protein-coding RNA gene as a potential biomarker for the detection of Mycobacterium tuberculosis. J. Cell. Mol. Med. 2017;21:2276–2283. doi: 10.1111/jcmm.13148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dersch P., Khan M. A., Mühlen S., Görke B. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 2017;8:803. doi: 10.3389/fmicb.2017.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeeves R. E., Marriott A. A. N., Pullan S. T., Hatch K. A., Allnutt J. C., et al. Mycobacterium tuberculosis is resistant to isoniazid at a slow growth rate by single nucleotide polymorphisms in katG codon Ser315. PLoS One. 2015;10:e0138253. doi: 10.1371/journal.pone.0138253. [DOI] [PMC free article] [PubMed] [Google Scholar]