Abstract
Aminoacyl-RNA synthetases (aaRSs) are among the key enzymes of protein biosynthesis. They are responsible for conducting the first step in the protein biosynthesis, namely attaching amino acids to the corresponding tRNA molecules both in cytoplasm and mitochondria. More and more research demonstrates that mutations in the genes encoding aaRSs lead to the development of various neurodegenerative diseases, such as incurable Charcot–Marie–Tooth disease (CMT) and distal spinal muscular atrophy. Some mutations result in the loss of tRNA aminoacylation activity, while other mutants retain their classical enzyme activity. In the latter case, disease manifestations are associated with additional neuron-specific functions of aaRSs. At present, seven aaRSs (GlyRS, TyrRS, AlaRS, HisRS, TrpRS, MetRS, and LysRS) are known to be involved in the CMT etiology with glycyl-tRNA synthetase (GlyRS) being the most studied of them.
Keywords: GARS, aaRS, CMT, SMA, dHMN, ALS
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRSs) have been found in all domains of life. These enzymes function at the initial stage of protein biosynthesis by conjugating amino acids with the corresponding tRNAs in cytoplasm and mitochondria. A growing body of evidence indicates that mutations in the aaRS genes are associated with various neurodegenerative disorders, including the incurable neurodegenerative neuropathy known as the Charcot–Marie–Tooth disease (CMT) [1].
In 2003, it was found that mutations in the human glycyl-tRNA synthetase (GlyRS or GARS) are associated with the distal spinal muscle atrophy type V (dSMA-V) and CMT. This discovery has initiated the search for association between mutations in the aaRS genes and hereditary diseases of motor neurons [2]. By now, mutations in seven aaRSs (GlyRS, TyrRS, AlaRS, HisRS, TrpRS, MetRS, and LysRS) associated with the axonal and intermediate CMT forms have been identified [2-9].
CHARCOT–MARIE–TOOTH DISEASE
CMT is a heterogenous group of hereditary neurodegenerative disorders of the peripheral nervous system. The disease was named after the three doctors who first described it in 1886. Clinical picture of CMT can vary significantly, but the common symptoms include progressive motor dysfunction, muscle weakness, wasting of distal muscles, loss of sensation, skeletal deformations, and loss of tendon reflexes.
Population and epidemiological studies of hereditary diseases provide information on variations in the prevalence of the pathologies in populations, which indicates the necessity for investigating regional features in the occurrence of hereditary diseases.
In various populations, CMT (all types) affects from 0.1 to 41.0 individuals in 100,000 people (average, 4.0-15.0 in 100,000 people) [10].
CMT is traditionally classified into three types based on electrophysiological and neuropathological criteria as well as inheritance pattern, which can be determined from the family medical history. Type 1 CMT (CMT1) involves abnormal myelination of peripheral axons. The average nerve conduction velocity (NCV) in this case is below 35 m/s. CMT1 affects 15 individuals in 100,000 people (https://neuromuscular.wustl.edu/).
The symptoms first appear in patients of 5 to 25 years old. Typical clinical manifestations (weakness and atrophy of distal muscles and loss of touch sensation) slowly progress and are often accompanied with the development of foot deformities such as high arch (pes cavus) and foot drop in both feet. Life expectancy is not affected by this type of disease and less than 5% of affected individuals become disabled.
Type 2 or not demyelinating (axonal) CMT (CMT2) is characterized by the damage of the axon itself; the NCV remains unaffected or decreases only slightly (> 45 m/s). The prevalence of CMT2 is 7 individuals per 100,000 people (https://neuromuscular.wustl.edu/).
Clinical manifestations of CMT2 (axonal peripheral neuropathy) are similar to those of CMT1 (demyelinating peripheral neuropathy). CMT2 patients rarely experience loss of sensation and rarely become disabled.
The dominating intermediate CMT (DI-CMT) is a CMT variant with the features typical for both CMT1 and CMT2. In DI-CMT, the NCV is 35 to 45 m/s; hence, the disease is difficult to classify based on traditional electrophysiological criteria. The NCV values can vary significantly, and affected members of the same family can demonstrate NCVs typical for either axonal or demyelinating peripheral neuropathy [11-13].
As more information on the genetic features of the CMT forms became available it was suggested to use letters for designation of the CMT types based on the involved genes (e.g., CMT1A). More than 90 genes have been found, mutations in which are associated with CMT (https://neuromuscular.wustl.edu/). However, many patients demonstrate similar neuropathy features and inheritance patterns, which made the letter/number classification too cumbersome [13]. In 2018, a new CMT classification based on the CMT-related genes was introduced [14]. One of the advantages of this classification is the possibility for describing the disease in terms of inheritance patterns, neuropathy type, and genes involved [15].
The CMT diagnostics is also complicated because of the involvement of the same genes in the phenotypically different distal hereditary motor neuropathies (dHMNs) and CMT2. For this reason, it was suggested to change the classification to include dHMNs as a subcategory of CMT.
Nevertheless, the mechanism(s) of gene involvement in the CMT development are poorly studied for most of the related genes [16, 17]. Inherited peripheral neuropathies are associated with more than 75 genes with the autosomal dominant, autosomal recessive, and X-linked inheritance patterns. CMT1 with the dominant inheritance pattern is the most common and easy-to-diagnose type of disease (since the associated genes are known in 80% cases). Association of CMT2 with mutations has been shown in 25% of cases only, which might be explained by the lack of information on the genetic basis of this type of neuropathy [18].
CLASSES OF AMINOACYL-tRNA SYNTHETASES AND THEIR FUNCTIONS
Protein biosynthesis is one of the essential cell processes. The first stage of protein biosynthesis is aminoacylation reaction, i.e., formation of the ester bond between an amino acid and its specific tRNA with generation of the aminoacylated (charged) tRNAs. This reaction is catalyzed by the enzymes from the aminoacyl-tRNA synthetase (aaRS) family that includes 20 enzymes (one for each proteinogenic amino acid and the corresponding tRNA) [19].
The aminoacylation reaction occurs in two stages. At the first stage, the amino acid is activated by ATP in the aaRS active site with formation of aminoacyl-AMP. Two out of the three ATP phosphates are released in the form of inorganic pyrophosphate (PPi). At the second stage, the complex of aaRS and aminoacyl-AMP interacts with the corresponding tRNA molecule. The amino acid binds to the CAA end of tRNA, followed by the release of AMP and, later, aminoacyl-tRNA [20].
Depending on the structural properties of the catalytic domain and the tRNA-recognizing sites, all aaRSs could be divided in two classes (I and II) with each class containing ten enzymes. The classes are subdivided into three subclasses (a, b, and c) each, based on the enzyme structure [21, 22]. Most class I enzymes are monomers (except dimeric TyrRS and TrpRS); their catalytic domains are structurally conserved. Class I aaRSs contain the Rossmann fold – six parallel β-strands alternating with α-helices [19]. This class is also characterized by the presence of two conserved motifs: HIGH (His-Ile-Gly-His) and KMSKS (Lys-Met-Ser-Lys-Ser) [23]. The HIGH motif is required for positioning of the ATP adenine and enzyme interaction with the phosphates. The KMSKS motif stabilizes transition state in the aminoacylation reaction via the second Lys residue. The open state of the KMSMS motif ensures amino acid recognition and binding; the KMSKS loop closes after the aminoacyl-AMP formation [24].
Class II aaRSs are multimeric enzymes. Most of them are homodimers, although some function as tetramers (Phe-aaRS, Ala-aaRS, and bacterial Gly-tRNA synthetase) [25]. The catalytic site in these enzymes is formed by seven antiparallel β-sheets surrounded by α-helices [20].
Human aaRSs could be divided into cytoplasmic, mitochondrial, and dual-localized enzymes. Human genome contains 37 genes for aaRSs: 18 genes for cytoplasmic only aaRSs; 17 genes for mitochondrial only aaRSs, and 2 for the dual-localized enzymes (aaRSs found in both the cytoplasm and the mitochondria) [19]. All mitochondrial aaRSs are synthesized in the cytosol and then are imported to mitochondria due to the presence of the N-terminal mitochondrial targeting sequence that is cleaved off after the enzyme translocation [26]. The dual-localized enzymes are glycyl-tRNA synthetase (GARS) and lysyl-tRNA synthetase (KARS). Besides, no mitochondrial glutamyl-tRNA synthetase (QARS) has been found. Instead, it was suggested that the glutamyl-tRNA is formed when tRNAGln is mistakenly charged with glutamic acid by the mitochondrial GluRS, which is then converted into Gln by an aminoacyl-tRNA aminotransferase [19]. It must be mentioned that the mitochondrial aaRSs are designated with number 2 after the name (e.g., YARS1 for the cytoplasmic tyrosyl-tRNA synthetase and YARS2 for its mitochondrial form).
Some mammalian aaRSs function as single proteins (WARS, HARS, SARS, FARS, YARS, NARS, TARS, GARS, CARS, AARS, VARS). The other are active as components of the multimeric multi-tRNA synthetase complex (MSC). MSC consists of several aaRSs and three aaRS-interacting multifunctional proteins (AIMP1, AIMP2, and AIMP3) that act as a scaffold in the MSC assembly and participate in various signaling pathways. Although the structure and functions of MCS have been studied insufficiently, it is evident that it contributes to the systemic control and homeostasis maintenance in higher eukaryotes [24].
In additions to their main activity (aminoacylation), some aaRSs have noncanonical functions and play a role in the translation initiation, transcription regulation, apoptosis, ribosomal RNA biogenesis, angiogenesis, and cell signaling [27-32].
MUTATIONS IN AMINOACYL-tRNA SYNTHETASES
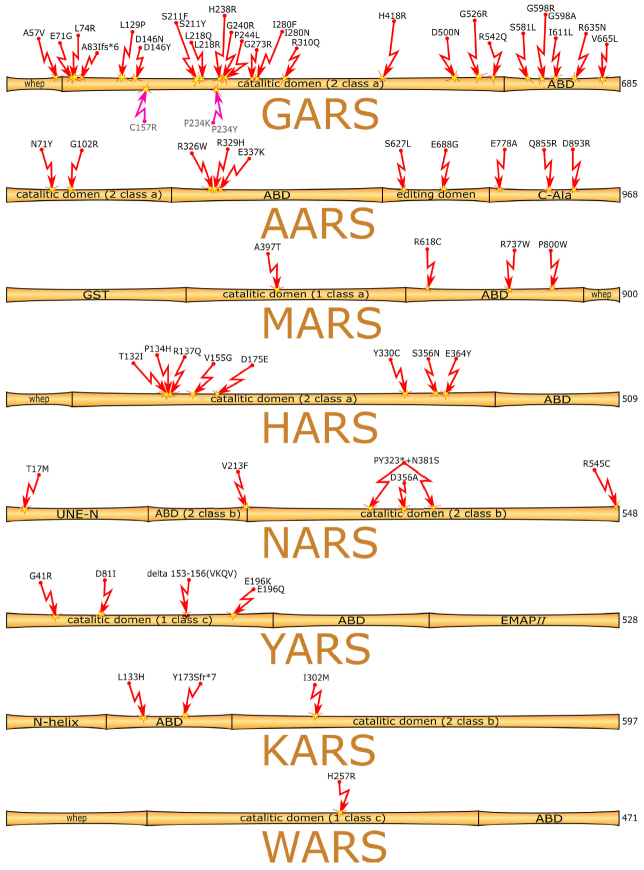
Histidyl-tRNA synthetase. Histidyl-tRNA synthetase (HisRS or HARS) is one of the seven aaRSs associated with the CMT2W also known as AD-CMTax [33]. HARS consists of the N-terminal WHEP domain, catalytic domain, and C-terminal anticodon-binding domain. All known mutations in the HARS gene are located in the catalytic domain around the active site (figure). The Arg137Gln mutation was found in 2013 in a patient with sporadic motor and sensory peripheral neuropathy characterized by the distal motor and sensory disfunctions. Although this is so far a single case of neuropathy associated with this particular mutation, its importance was confirmed by the functional studies in yeast and the observed toxic effect of this mutation on neurons in Caenorhabditis elegans expressing the mutant protein [6]. Discovery of the Arg137Gln mutation has prompted the search for other HARS mutations, resulting in identification of Thr132Ili, Pro134His, Asp175Glu, and Asp364Tyr substitutions [18]. In 2018, three more missense mutations (Val155Gly, Tyr330Cys, Ser356Asn) were found that are also located in the HARS catalytic domain. It should be noted that the evidence in favor of the Ser356Asn mutation involvement in the CMT phenotype is less convincing, since the mother of the patient with the Ser356Asn substitution was heterozygous by the same mutation but did not manifest any CMT symptoms [34].
Figure.
Domain structure of aaRSs. Red arrows, mutations associated with human neurodegenerative diseases; pink arrows, mutations found in mice (numbering according to the corresponding human protein).
Asparaginyl-tRNA synthetase. Asparaginyl-tRNA synthetase (AspRS or NARS) belongs to the subtype a of class II aaRSs. Similarly to the majority of aaRSs, there are the cytoplasmic (NARS1) and mitochondrial (NARS2) isoforms of the enzyme. NARS1 consists of three domains: unique N-terminal extension (UNE-N), anticodon-binding domain (ABD), and catalytic domain. NARS2 is composed of the ABD and catalytic domain only. Missense mutations in both NARS isoforms result in various neurodegenerative diseases, but unlike most aaRSs mutations, mutations in NARS can be located in domains other than the catalytic one (figure).
The study involving 21 families with 32 members manifesting delayed development, seizures, peripheral neuropathy, and ataxia reveal association of these disorders with some heterozygous and biallelic mutations in NARS1 [35]. In seven patients with microcephaly and delayed development from three unrelated families, the NARS1 gene contained biallelic Thr17Met, Asp356Ala, Arg545Cys mutations and frameshift mutations. Content of the NARS1 protein in these patients was decreased, the activity of NARS1 was suppressed, and the total protein synthesis was impaired [36]. The homozygous Val213Phe mutation in NARS2 causes nonsyndromic hearing loss (DFNB94), while the compound heterozygous mutations Tyr323* + Asn381Ser result in the mitochondrial respiratory chain deficiency and Leigh syndrome, (neurodegenerative disease characterized by the symmetrical lesions in the basal ganglia, thalamus, and brainstem) [37].
Tyrosyl-tRNA synthetase. Tyrosyl-tRNA synthetase (TyrRS or YARS) is a class I aaRS. Together with tryptophanyl-tRNA synthetase (WARS or TrpRS), it belongs to the subtype c enzymes, and contains the AIDQ motif typical for the ATP-binding site [38]. YARS consists of three domains: evolutionary conserved catalytic domain, ABD, and C-terminal EMAPII (endothelial monocyte-activating polypeptide II)-like domain. Unlike other aaRSs, human YARS is processed by elastase with formation of the N-terminal TyrRS (also known as mini-TyrRS) and the C-terminal EMAPII-like domain [39].
YARS is the second aaRSs, whose role in the CMT was demonstrated. There are at least five mutations in the YARS catalytic domain associated with the intermediate CMT form (DI-CMTC/AD-CMTin-YARS) (Fig. 1): Originally, heterozygous Gly41Arg and Glu196Lys mutations and Δ153-156 (VKQV) deletion were identified [3], pathogenicity of which was confirmed in the transgenic Drosophila models [40]. All three mutations were characterized in detail both in vitro and in vivo. Not all mutant enzymes displayed a decreased amino acylating activity, and none of the mutations altered the protein stability or secondary structure (Gly41Arg and Glu196Lys caused slight conformational changes) [41]. Later, the Asp81Ile mutation was found in an adult patient. According to the family medical history, the parents of this patient were healthy and carried no other CMT-associated mutations [42]. The fifth mutation in the YARS gene (Glu196Gln) was identified in a family, in which representatives of three generations had intermediate CMT type. The Glu196Gln mutation was inherited by two children from the affected mother [43].
Tryptophanyl-tRNA synthetase. Human tryptophanyl-tRNA synthetase (WARS or TrpRS) consists of the N-terminal WHEP domain, catalytic domain, and C-terminal ABD. The His257Arg mutation was first discovered in two unrelated Taiwanese families, and then in a European family in Belgium (figure). All affected individuals demonstrated phenotypical manifestations of dHMN. The His257 residue is evolutionary conserved from fish (Danio rerio) to humans [8].
Methionyl-tRNA synthetase. Methionyl-tRNA synthetase (MetRS or MARS) belongs to the subtype a of class I aaRSs. It consists of the N-terminal glutathione S-transferase (GST) domain, catalytic domain, ABD, and C-terminal WHEP domain. MARS is the only CMT-associated aaRS that does not form a dimer. Mutations in MARS rarely cause CMT [7]. There are four mutations known to be associated with the type 2U CMT (CMT2U/AD-CMTax-MARS) (figure): three of them are in the ABD and one in the catalytic domain. Mutations in the MARS gene can cause autosomal recessive spastic paraplegy [44]. The first discovered mutation (Arg618Cys) was found in two CMT2 patients from the same family (45- and 67-year-old males) [7]. The Pro800Thr mutation was identified in several unrelated Korean families [42, 45] and in a 71-year-old Japanese female [46]. CMT2U is a sensory and motor neuropathy manifestations of which rise with aging. However, two individuals with the Pro800Thr mutation from a Korean family were found that developed the disease in an early age [45]. In 2018, a new missense mutation (Arg737Trp) was identified in a 13-year-old girl. The Arg73 residue is conserved from bacteria to humans. The patient inherited this gene variant from a 60-year-old mother, who demonstrated slightly diminished leg reflexes, especially in the Achilles tendon. No cavus foot or other foot abnormalities were observed [47].
Recently, the Ala397Thr mutation was found in an 11-year-old girl with a progressing CMT that started in the early childhood. Sanger sequencing revealed absence of this mutation in the genome of the healthy mother (healthy father was not tested). The Ala397 residue is highly conserved (up to yeast) [48].
Structure modeling showed that Arg618 and Pro800 are important for the anticodon recognition and correct functioning of MARS, while Ala397 is located near the Zn2+-binding site and is essential for catalytic activity of the enzyme [48].
Mutations Arg618Cys and Arg737Trp were also found in non-symptomatic patients and patients with weakly manifested symptoms. All the patients with the Pro800Thr mutation displayed significant neurological symptoms [47].
Alanyl-tRNA synthetase. Alanyl-tRNA synthetase (AARS or AlaRS) belongs to the class II aaRSs and consists of four domains: N-terminal catalytic domain, tRNA-binding domain, editing domain, and C-terminal domain involved in oligomerization (dimerization or tetramerization). The tRNA-Ala has the unique G3: U70 base pair in the acceptor stem which determines its affinity for AARS [49]. At the same time, AASR can activate wrong amino acid because of the structural similarity between alanine and some other amino acids (e.g., serine), which requires hydrolytic editing activity of the enzyme to provide aminoacylation of the correct tRNA [19]. So far, all mutations discovered in AARS are associated with the subtype 2N CMT (CMT2N/AD-CMTax-AARS). The average age for the first manifestations of this disease is 28 years (from 2 to 60 years). The most studied mutation in the AARS is Arg329His (figure) that was discovered in 17 patients with the axonal form of CMT (CMT2) [4]. The Asn71Tyr mutation was found in 36 unrelated Taiwanese patients. It has been believed for a long time that this mutation is the major Asian mutation in CMT2N [50, 51]. Another missense heterozygous mutation, Gly102Arg, was found in the catalytic domain. All patients with this mutation demonstrated a mild axonal neuropathy; three out of four patients exhibited lower extremity hyperreflexia, indicating superimposed myelopathy [52].
Three more mutations were discovered when studying larger cohorts of the affected individuals. Two of these mutations (Arg326Trp and Glu337Lys) are located in the catalytic domain; the third one is in the editing domain (Ser627Leu) [53]. Another mutation in the AARS gene (Asp893Asn) was found in a Chinese family. The mutation had the autosomal dominant inheritance pattern. Clinical features included mild weakness and wasting of the distal muscles of the lower limb and foot deformity, without clinically detected sensory disruptions [54].
Very recently, a novel mutation (Gln855Arg) was discovered in a CMT2 patient, but its pathogenic role has not been confirmed yet (variant of uncertain significance, VUS) [55].
The Glu778Ala mutation was revealed in an Australian family. Unlike the abovementioned mutations, the substituted amino acid is not evolutionary conserved [5].
The Glu688Gly mutation in the editing domain was found in a family of Irish origin during studying four British and two Irish families with neuropathy [56].
Lysyl-tRNA synthetase. Lysyl-tRNA synthetase (LysRS or KARS) is a dual-localization aaRS of the class II, subtype b. It consists of the N-terminal helical domain, ABD, and C-terminal catalytic domain. Mutations associated with the recessive intermediate CMT type (CMTRIB) were first identified in two patients. One of these patients had the Ile302Met mutation in a heterozygous state. The studies of the pedigree of this patient indicated the autosomal dominant type of inheritance. The Leu133His substitution and Tyr173Serfs*7 frameshift were identified in another patient that were phenotypically manifested as the intermediate CMT, delayed development, self-deprecation, dysmorphia, neurinoma of the auditory nerve [9], and nonsyndromic hearing loss [57, 58]. The study published in 2019 described a patient with severe neurological and neurosensory disfunctions who had two new mutations: Pro228Leu and Phe291Val (Pro200Leu and Phe263Val in the cytoplasmic enzyme) (figure). The patient exhibited atypical optical neuropathy that has not been reported before [59].
Glycyl-tRNA synthetase. Glycyl-tRNA (GlyRS or GARS) belongs to the subtype of class II aaRSs. Similar to KARS, GARS is a dual-localized aaRS. It consists of the WHEP domain, catalytic domain, and ABD. The mitochondrial enzyme also contains targeting sequence for the import to mitochondria.
CMT is the most common hereditary peripheral neuropathy. So far, there is no effective treatment of this disease [60]. Autosomal dominant mutations in GARS cause the 2D subtype of CMT (CMT2D).
Although most of the CMT2D-related mutations are located in the catalytic domain, autosomal recessive mutations in the ABD domain have been found in patients with the mitochondrial disease phenotype. For example, a child homozygous by the Arg635Trp substitution exhibited strong neonatal cardiomyopathy and deficit of cytochrome c oxidase (the child died at 10 days of age) [61]. Another child heterozygous by the Ser581Leu and Arg542Gln mutations displayed myalgia, cardiomyopathy, periventricular lesions, and persistent elevation of blood lactate [62].
A 40-year-old patient living in the United Kingdom, but originally from Ghana experienced difficulties with writing, as he has noticed weakening of his hands since the age of 12. His symptoms worsened when he was badly beaten at the age of 34 in Ghana. When the plaster was removed three weeks later, both hands were more wasted than previously. His lower limbs were normal. Genetic studies of this patient revealed the presence of the earlier undescribed Ala57Val mutation in GARS [63].
Out of eight studied families, three families of Algerian Sephardic Jewish origin (16 patients) had the Glu526Arg mutation. The clinical phenotype consisted of a slowly progressive motor distal neuropathy that started in the hands in most patients, although four mutation carriers were still asymptomatic, two of whom were already 49 years of age [64].
The Ser211Phe mutation was discovered in a Chinese family with CMT2D patients, adding to the variety of GARS mutations observed in the Chinese population [65]. The Asp146Asn and Ser211Phe mutations located in the highly conserved regions of the catalytic domain were found in Korean patients with CMT2D. Both mutations have the dominant inheritance pattern and were found in parents, as well as in children. These mutations were missing from the databases on human genomic polymorphisms [66]; they are the first GARS mutations discovered in Koreans. Both mutations correspond to the dHMN-V phenotype. The Glu71Gly and Pro244Leu mutations have been found in the Asian patients only [2, 67].
The majority of GARS mutations result in deterioration of upper extremities with the clinical manifestations staring at puberty. Mutations in the ABD mostly affect the lower limbs with the first clinical manifestation appearing in the early age [68, 69].
Mutations in GARS are atypical for dHMN-V patients in China. However, a new autosomal dominant mutation (Leu74Arg) was recently discovered and characterized in a Chinese family with dHMN-V. An 11-year-old girl had difficulties with writing and walking on her heels but was still able to ambulate. No sensory problems or tremor were present. Another five members from three successive generations of the family (grandparent, father, aunt, elder female cousin, and younger sister) showed similar symptoms. All the above individuals with dHMN-V (but not their healthy relatives) had the Leu74Arg mutations. The mutation is absent in the polymorphism databases; it is pathogenic, as has been demonstrated in several in silico experiments. Clinical symptoms of dHMN-V include atrophy of leg muscle caused by peripheral motor neuropathy. No sensory disfunctions were registered in the family, thus excluding the CMT-related neuropathy. Hence, mutations in GARS cause purely motor neuropathies [70].
As of today, over 10 dominant mutations in GARS have been described that cause motor rather than sensory neuropathies [71].
Recently, the new Gly273Arg mutation was found in an 18-year-old young woman that had problems with ankles during running and in a 22-year-old woman with the weakness in arms and legs. This mutation was discovered by sequencing and was absent in the genomes of the patients’ relatives. The phenotype of the affected women was similar to that of other patients with the dominant GARS mutations (Glu71Gly, Leu129Pro, Asp146Asn, Ser211Phe, Leu218Gln, Pro244Leu, Ile280Phe, His418Arg, Gly526Arg, Gly598Ala). The Gly273 residue is conserved between yeast and mammals. The mutant protein (Gly327Arg) did not support yeast growth in the complementation assay, showing pathogenicity of this mutation [71]. The recently discovered recessive mutations in the catalytic domain were found to cause the multisystem developmental syndrome that includes severe growth retardation, thinning of the corpus callosum, decrease in the cerebral white matter volume, and atrophy of the brain stem, but not peripheral neuropathy [72]. Children with the recessive mutations do not present neuropathies, although it cannot be excluded that such neuropathies will develop in future.
Mutations in GARS also result in the reduction of aminoacylation activity, changes in the axon location, and alterations in the neuropilin 1 (Nrp1) pathway [3, 73, 74].
However, the role of mitochondrial GARS and its effects on the phenotypical manifestations of the disease are still poorly studied. It was demonstrated that mutations in GARS lead to the tissue-specific mitochondrial defects in neurons that develop via different mechanisms in patients with autosomal dominant and recessive mutations.
Dominant mutations in the GARS gene are associated with hereditary neuropathies, while recessive mutations cause severe childhood disorders affecting various muscles (including cardiac muscle). The mechanisms for the tissue-specific character of disease manifestation and relations between the genotype and the phenotype still remain unknown.
It was shown that GARS mutants can bind Nrp1, which is a direct antagonist of the signaling pathway essential for survival of the motor neurons. Nrp1 is a membrane-bound co-receptor of the tyrosine kinase receptor for the vascular endothelial growth factor (VEGF). It is involved in the VEGF-induced angiogenesis, axon guidance, and cell survival, migration, and metastasis [74]. Aberrant interaction between the GARS mutants and Nrp1 prevents VEGF binding to Nrp1. A decrease in the Nrp1 level exacerbates neuropathy, while upregulation of the VEGF expression restores motor functions. Although this suggests the pathological effects of GARS mutant proteins secreted by the neurons, it does not explain all the pathologies associated with the mutations in GARS.
Downregulation or complete absence of GARS lead to the decline in the mitochondrial translation [75]. Suppression of GARS biosynthesis with siRNAs results in the reduction of mitochondrial translations in neurons and myoblasts (but not in fibroblasts), which suggests that mutant GARS proteins cause tissue-specific defects in the mitochondrial translation because of the loss of function.
Besides, the observed changes in the mitochondrial metabolism cannot be fully explained by the defects in the cytoplasmic and mitochondrial translation and suggest existence of additional noncanonical GARS functions in neurons [76, 77].
GARS colocalizes with the mitochondrial RNA granules that act as the centers of post-translational processing and biogenesis of mitochondrial ribosomes, as well as a platform for RNA maturation, ribosome assembly, and translation initiation. This fact confirms the possibility of the effects of the mutant forms on these processes. However, RNA granules are not typical for neurons and cannot explain the tissue-specificity of the pathological effects of mutations in GARS [75].
RNA sequencing and comparative proteomic studies have shown that both dominant and recessive GARS mutations lead to the significant alterations in the mitochondrial transcriptome and proteome, including changes in the respiratory chain subunits, Krebs cycle enzymes, and mitochondrial transport proteins. Moreover, the recessive GARS mutations noticeably affect the metabolism of fatty acid. These data correlate well with the fact that patients with recessive GARS mutations display exercise intolerance, as well as myopathy and cardiomyopathy commonly observed in the tissues with defective fatty acid oxidation.
The neuropathy-associated GARS mutations are accompanied by the decrease in the content of the vesicle-associated membrane protein B (VAPB) and changes in the mitochondrial and cellular calcium homeostasis, which might explain tissue specificity of their clinical manifestations. VAPB is a component of a structural complex that binds to the plasma membrane and membranes of the endoplasmic reticulum (ER) and mitochondria. Modulation of the VAPB activity is manifested by the changes in the mitochondrial-ER contacts and can affect calcium metabolism in these organelles [78].
More than 20 GARS mutations related to various neurodegenerative diseases have been described (figure), Glu71GLy, Leu129Pro, and GLy240Arg being the most strongly associated with the diseases. Only few mutations affect aminoacylation insignificantly. Mice with one copy of the GARS gene (and therefore, 50% GARS activity) present no symptoms [79]. Moreover, overexpression of the wild-type GARS in the CMT2D mice does not cure neuropathy [80], as the disease is caused by impairments of the GARS functions other than aminoacylation, as suggested earlier.
Most class II aaRSs function as dimers (including GARS). All CMT2D-associated mutations are located in the region of subunit contact [81]. Some of these mutations lead to the conformational opening and exposure of the protein surfaces normally inaccessible to the solvent [74, 82], which might result in attainment of the ability for binding of an unusual partners. Thus, the wild-type GARS binds Nrp1 very poorly (with the binding constant of 1 mM), while binding constant of the Leu129Pro mutant is 29.8-66.3 nM [74, 82].
Mutations in GARS result in the its subcellular redistribution and can lead to the loss of additional functions at the cellular level, even if the aminoacylating activity is retained [83].
Despite the fact CMT is a commonly occurring disease, it is rarely reported in the sub-Saharan Africa. Mutations resulting in CMTD2 have been found in very few families, mostly of Caucasian ancestry. Until recently, no CMTD2 cases have been registered in Africa. Recently, an African family was found in Mali, whose members presented the CMT phenotype due to the novel GARS mutation [84]. Two out of five siblings were found to be affected (35-year-old male and 19-year-old female); the symptoms started in their teenage years with muscle weakness and atrophy in hands. Later, distal involvement of the lower limbs was noticed. The affected individuals experienced insignificant loss of sensation. Genetic studies revealed the novel Ser211Tyr mutation in GARS. The same mutation was identified in the 58-year-old mother, who presented no symptoms [84]. More phenotypical and genetic studies in Africa might provide additional information that will help to distinguish between the CMT2D and distal spinal muscular atrophy type V (dSMA-V).
Spinal muscle atrophy (SMA) is a disease characterized by the symmetric proximal muscle weakness and atrophy, resulting in the progressive degeneration and loss of neurons in the ventral horns of the spinal cord. Unlike CMT patients, most individuals with SMA carry deletions in the chromosome 5; however, 5% of SMA cases are associated with other mutations. These patients often exhibit the damage of proximal muscles, including respiratory system dysfunctions and type V distal SMA (dSMA-V). Both CMT2D and dSMA-V are caused by heterogenous mutations in GARS. Multiple cases of childhood SMA related to GARS mutation have been described recently [68, 69, 85].
GARS is the most commonly analyzed gene in the genetic screening of patients with suspected CMT2D and dSMA-V. However, these tests are conducted mainly in adults, while in newborns and babies, the diseases associated with mutations in GARS can be easily missed.
Very recently, three patients with mutations in the GARS gene were found to developed early neuropathy with respiratory distress that mimicked the childhood SMA [86]. All three patients were born full-term, without visible pathologies. Two of the them were from the Hispanic families. The mother of the third patient was French Canadian, and the farther was Caucasian. No hereditary neurodegenerative diseases have been reported in the patients’ families. The first manifestations of neuropathy were observed 1.5 to 2 months after birth in two patients and within the first days after birth in the third patient. The first two patients had the Ile280Asn mutation in the catalytic domain, while the third child had the Gly598Arg substitution in the ABD. Both Ile280 and Gly598 are conserved in eukaryotes (from nematodes to humans). Another mutation at position 598 (Gly598Ala) has been described before. This substitution results in CMT and distal SMA in children and newborns [68, 69]. It is believed that mutations in the ABD disturb the enzymatic function of GARS more than mutations in any other domain.
All three patients presented the symptoms of SMA and associated dysfunctions and had problems with sensation, which is typical of CMT2D. Based on the pronounced weakness of distal muscles and breathing problems, it is likely that these patients had SMARD1 (spinal muscular atrophy with respiratory distress type 1).
Making correct diagnosis is essential, since many degenerative diseases have similar symptoms [87, 88]. Genomic screening in combination with analysis of clinical manifestations might be helpful in the revision of the existing disease classification. Thus, previously, GARS mutations had been linked to CMT2D and dSMA-V only; at present, they are also associated with early manifestations of SMA.
Yeasts are commonly used for verifying the ability of mutant forms of human GARS to restore cell growth in the complementation assay that uses haloid yeast strain deficient in the GARS1 gene. The GARS gene can be introduced into the cells with the URA3 plasmid [89]. Expression of the full-size enzyme or its truncated form lacking the mitochondrial targeting signal and the WHEP domain (GARS ΔMTSΔWHEP) restored rapid yeast growth on the selective medium that caused spontaneous loss of the URA3 vector. This proved that human GARS complemented the loss of the endogenous GARS1 locus [72, 90]. Expression of the GARS ΔMTSΔWHEP Ile280Asn mutant did not restore the growth of yeast cells, indicating the loss of function by the mutant enzyme [86].
Another neurodegenerative disease that should be mentioned is amyotrophic lateral sclerosis (ALS), which is a progressive neurological disorder characterized by the damage of motor neurons in the brain and spinal cord. More than 30 ALS-associated genes have been identified [91]. Previously, no association between the defects in the GARS gene and motor neuron disorders had been considered. However, genetic screening of a 70-year-old female with the classical bulbar ALS revealed the presence of the heterozygous GARS mutation Val665Leu in the cytoplasmic enzyme (Val719Leu in its mitochondrial form), that most probably was the cause of the disease. No mutations in the ALS-associated genes were found [92]. The discovered mutation has not been described before. It is located in the ABD, mutations in which are known to cause the most pronounced symptoms.
More than ten mutations in GARS have been associated with the development of CMT and dHMN [93], and this number continues to increase. We believe that GARS should be added to the list of genes that are sequenced in ALS because of the newly discovered association of this gene with various neurodegenerative diseases.
In addition to the above-described GARS-related disorders, there are cases of the multisystem developmental syndrome. Genome sequencing of these patients revealed the presence of the heterozygous Glu83Ilefs*6 frameshift and Arg310Gln mutation. In vivo and in vitro experiments showed that both mutations resulted in the GARS loss of function via reduction in the protein level (frameshift) or decrease in the aminoacylating activity (missense mutation).
A girl born prematurely at 36-weeks gestation with a birth weight of 1.52 kg increased the weight poorly and showed cephalofacial disproportion, low-normal bone mineralization, and loss of hearing. No infections were found. She continued to suffer from atopic dermatitis and recurrent rhinitis. Developmentally, she rolled at 4 months, sat at 16 months, started to hold a pencil and feed herself at 5 years, and walked at 6 years. Her verbal skills included 20-25 single words with poor intelligibility. At the age of 2 years and 9 months, her weight was 7.6 kg, height was 72 cm, and head circumference was 43 cm [72]. She did not show signs of lower motor neuron disorder or neuropathy and her sensation to temperature was normal. To reveal the possible cause of the observed symptoms, her genome and genomes of her healthy parents were sequenced. The sequencing identified two mutations in the child’s GARS gene: heterozygous Glu83Ilefs*6 frameshift and Arg310Gln mutation. The mother had the Glu83Ilefs*6 frameshift in a heterozygous form, and the father had heterozygous Arg310Gln substitution. The frameshift affected both the core domain and the ABD, while substitution Arg31 (residue conserved between worms and humans) affected the core domain only. The frameshift resulted in the emergence of premature stop codon and, consequently, cause a decrease in the content of the full-size GARS (synthesized from the second allele). The Arg310Gln substitution resulted in the loss of the aminoacylating activity (by ~99%); the mutant protein did not restore yeast growth in the yeast complementation assay [72]. The presence of both mutations led to multiple dysfunctions typical for the defective aaRSs. Interestingly, none of the three examined individuals with the Arg310Gln mutation presented CMT symptoms.
CONCLUSION
Correct diagnosis is one of prerequisites for successful treatment. The discovery that mutations in GARS might results in the development of diseases other than CMT and dSMA (e.g., ALS, SMA, and multisystem developmental syndrome) prompts the scientists to add the GARS gene to the list of genes tested for the presence of mutations involved in the development of neurodegenerative disorders. In recent years, new methods for the treatment of neuropathies (especially, their early stages in children) using antisense oligonucleotides have been under development. However, antisense oligonucleotides and even more advanced adenoviral vectors should be used only after the underlying causes of the disease (chromosomal deletion, mutations in aaRS genes, etc.) has been elucidated.
The molecular mechanisms of selective neuronal susceptibility in neurodegenerative diseases might be related to the newly acquired activities of the incorrectly folded proteins (aaRSs) interacting with the signaling proteins in a particular type of cells. Studying these additional activities of aaRSs may be helpful in understanding the mechanisms of development of neurodegenerative disorders and searching for new approaches and effective drugs to treat these diseases.
Abbreviations
- aaRS
aminoacyl-tRNA synthetase
- AlaRS
alanyl-tRNA synthetase
- AsnRS
asparaginyl-tRNA synthetase
- CMT
Charcot–Marie–Tooth disease
- dHMN
distal hereditary motor neuropathy
- GARS or GlyRS
glycyl-tRNA synthetase
- HisRS
histidyl-tRNA synthetase
- LysRS
lysyl-tRNA synthetase
- MetRS
methionyl-tRNA synthetase
- NCV
nerve conduction velocity
- SMA
spinal muscular atrophy
- TrpRS
tryptophanyl-tRNA synthetase
- TyrRS
tyrosyl-tRNA synthetase
Funding
This work was financially supported by the Russian Foundation for Basic Research (project no. 19-34-90135).
Ethics declarations
The authors declare no conflict of interest in financial or any other sphere. This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Skre H. Genetic and clinical aspects of Charcot–Marie–Tooth’s disease. Clin. Genet. 1974;6:9–118. doi: 10.1111/j.1399-0004.1974.tb00638.x. [DOI] [PubMed] [Google Scholar]
- 2.Antonellis A., Ellsworth R. E., Sambuughin N., Puls I., Abel A., et al. Glycyl tRNA synthetase mutations in Charcot–Marie–Tooth disease type 2D and distal spinal muscular atrophy type V. Am. J. Hum. Genet. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordanova A., Irobi J., Thomas F. P., Dijck P. V., Meerschaert K., et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot–Marie–Tooth neuropathy. Nat. Genet. 2006;38:197–202. doi: 10.1038/ng1727. [DOI] [PubMed] [Google Scholar]
- 4.Latour P., Thauvin-Robinet C., Baudelet-Méry C., Soichot P., Cusin V., et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot–Marie–Tooth disease. Am. J. Hum. Genet. 2010;86:77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McLaughlin H. M., Sakaguchi R., Giblin W., NISC Comparative Sequencing Program. Wilson T. E., et al. A recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with Charcot–Marie–Tooth disease type 2N (CMT2N) Hum. Mutat. 2012;33:244–253. doi: 10.1002/humu.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vester A., Velez-Ruiz G., McLaughlin H. M., NISC Comparative Sequencing Program. Lupski J. R., et al. A loss-of-function variant in the human histidyl-tRNA synthetase (HARS) gene is neurotoxic in vivo. Hum. Mutat. 2013;34:191–199. doi: 10.1002/humu.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez M., McLaughlin H., Houlden H., Guo M., Yo-Tsen L., et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatry. 2013;84:1247–1249. doi: 10.1136/jnnp-2013-305049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai P. C., Soong B. W., Mademan I., Huang Y. H., Liu C. R., et al. A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain. 2017;140:1252–1266. doi: 10.1093/brain/awx058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McLaughlin H. M., Sakaguchi R., Liu C., Igarashi T., Pehlivan D., et al. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. Am. J. Hum. Genet. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gur’eva, P. I. (2014) Epidemiological and clinical-genetic characteristics of Charcot–Marie–Tooth disease in the Republic of Sakha (Yakutia). Diss. Cand. Med. Sci., KrasGMU im. prof. Voino-Yasenetsky, Ministry of Health of the Russian Federetion, p. 109.
- 11.Pareyson D., Marchesi C. Diagnosis, natural history, and management of Charcot–Marie–Tooth disease. Lancet Neurol. 2009;8:654–667. doi: 10.1016/S1474-4422(09)70110-3. [DOI] [PubMed] [Google Scholar]
- 12.Jordanova A., Thomas F. P., Guergueltcheva V., Tournev I., Gondim F. A., et al. Dominant intermediate Charcot–Marie–Tooth type C maps to chromosome 1p34-p35. Am. J. Hum. Genet. 2003;73:1423–1430. doi: 10.1086/379792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toriello, H. V. (2009) Thrombocytopenia Absent Radius Syndrome (Adam, M. P., Ardinger, H. H., Pagon, R. A., et al., eds.) GeneReviews, University of Washington, Seattle, 1993-2019.
- 14.Magy L., Mathis S., Le Masson G., Goizet C., Tazir M., Vallat J. M. Updating the classification of inherited neuropathies: Results of an international survey. Neurology. 2018;90:870–876. doi: 10.1212/WNL.0000000000005074. [DOI] [PubMed] [Google Scholar]
- 15.Bansagi B., Griffin H., Whittaker R. G., Antoniadi T., Evangelista T., et al. Genetic heterogeneity of motor neuropathies. Neurology. 2017;88:1226–1234. doi: 10.1212/WNL.0000000000003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossor A. M., Polke J. M., Houlden H., Reilly M. M. Clinical implications of genetic advances in Charcot–Marie–Tooth disease. Nat. Rev. Neurol. 2013;9:562–571. doi: 10.1038/nrneurol.2013.179. [DOI] [PubMed] [Google Scholar]
- 17.Gutmann L., Shy M. Update on Charcot–Marie–Tooth disease. Curr. Opin. Neurol. 2015;28:462–467. doi: 10.1097/WCO.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 18.Brozkova S. D., Deconinck T., Griffin L. B., Ferbert A., Haberlova J., et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain. 2015;138:2161–2172. doi: 10.1093/brain/awv158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei N., Zhang Q., Yang X. L. Neurodegenerative Charcot–Marie–Tooth disease as a case study to decipher novel functions of aminoacyl-tRNA synthetases. J. Biol. Chem. 2019;294:5321–5339. doi: 10.1074/jbc.REV118.002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rajendran V., Kalita P., Shukla H., Kumar A., Tripathi T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018;111:400–414. doi: 10.1016/j.ijbiomac.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 21.Chaliotis A., Vlastaridis P., Mossialos D., Ibba M., Becker H. D., et al. The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 2017;45:1059–1068. doi: 10.1093/nar/gkw1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf Y. I., Aravind L., Grishin N. V., Koonin E. V. Evolution of aminoacyl-tRNA synthetases – analysis of unique domain architectures and phylogenetic trees reveals a complex history of horizontal gene transfer events. Genome Res. 1999;9:689–710. [PubMed] [Google Scholar]
- 23.Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 24.Kwon N. H., Fox P. L., Kim S. Aminoacyl-tRNA synthetases as therapeutic targets. Nat. Rev. Drug Discov. 2019;18:629–650. doi: 10.1038/s41573-019-0026-3. [DOI] [PubMed] [Google Scholar]
- 25.Banik S. D., Nandi N. Mechanism of the activation step of the aminoacylation reaction: a significant difference between class I and class II synthetases. J. Biomol. Struct. Dynam. 2012;30:701–715. doi: 10.1080/07391102.2012.689701. [DOI] [PubMed] [Google Scholar]
- 26.Boczonadi V., Jennings M. J., Horvath R. The role of tRNA synthetases in neurological and neuromuscular disorders. FEBS Lett. 2018;592:703–717. doi: 10.1002/1873-3468.12962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown M. V., Reader J. S., Tzima E. Mammalian aminoacyl-tRNA synthetases: cell signaling functions of the protein translation machinery. Vasc. Pharmacol. 2010;52:21–26. doi: 10.1016/j.vph.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Bhatt T. K., Kapil C., Khan S., Jairajpuri M. A., Sharma V., et al. A genomic glimpse of aminoacyl-tRNA synthetases in malaria parasite Plasmodium falciparum. BMC Genom. 2009;10:644. doi: 10.1186/1471-2164-10-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo M., Yang X. L., Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat. Rev. Mol. Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo M., Schimmel P., Yang X. L. Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett. 2010;584:434–442. doi: 10.1016/j.febslet.2009.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andreev D. E., Hirnet J., Terenin I. M., Dmitriev S. E., Niepmann M., Shatsky I. N. Glycyl-tRNA synthetase specifically binds to the poliovirus IRES to activate translation initiation. Nucleic Acids Res. 2012;40:5602–5614. doi: 10.1093/nar/gks182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikonova E. Y., Mihaylina A. O., Nemchinova M. S., Garber M. B., Nikonov O. S. Glycyl-tRNA synthetase as a potential universal regulator of translation initiation at IRES-I. Molek. Biol. 2018;52:10–18. doi: 10.7868/S0026898418010020. [DOI] [PubMed] [Google Scholar]
- 33.Blocquel D., Sun L., Matuszek Z., Li S., Weber T., et al. CMT disease severity correlates with mutation-induced open conformation of histidyl-tRNA synthetase, not aminoacylation loss, in patient cells. Proc. Natl. Acad. Sci. USA. 2019;116:19440–19448. doi: 10.1073/pnas.1908288116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott J. A., Meyer-Schuman R., Lupo V., Feely S., Mademan I., et al. Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy. Hum. Mutat. 2018;39:415–432. doi: 10.1002/humu.23380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salpietro V., Harripaul R., Badalato L., Walia J., Francklyn C. S., et al. Denovo and bi-allelic pathogenic variants in NARS1 cause neurodevelopmental delay due to toxic gain-of-function and partial loss-of-function effects. Am. J. Hum. Genet. 2020;107:311–324. doi: 10.1016/j.ajhg.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang L., Li Z., Sievert D., Smith D., Mendes M. I., et al. Loss of NARS1 impairs progenitor proliferation in cortical brain organoids and leads to microcephaly. Nat. Commun. 2020;11:4038. doi: 10.1038/s41467-020-17454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon M., Richard E. M., Wang X., Shahzad M., Huang V. H., et al. Mutations of human NARS2, encoding the mitochondrial asparaginyl-tRNA synthetase, cause nonsyndromic deafness and Leigh syndrome. PLoS Genet. 2015;11:e1005097. doi: 10.1371/journal.pgen.1005097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barros-Álvarez X., Kerchner K. M., Koh C. Y., Turley S., Pardon E., et al. Leishmania donovani tyrosyl-tRNA synthetase structure in complex with a tyrosyl adenylate analog and comparisons with human and protozoan counterparts. Biochimie. 2017;138:124–136. doi: 10.1016/j.biochi.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatt T. K., Khan S., Dwivedi V. P., Banday M. M., Sharma A., et al. Malaria parasite tyrosyl-tRNA synthetase secretion triggers pro-inflammatory responses. Nat. Commun. 2011;2:530. doi: 10.1038/ncomms1522. [DOI] [PubMed] [Google Scholar]
- 40.Storkebaum E., Leitão-Gonçalves R., Godenschwege T., Nangle L., Mejia M., et al. Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot–Marie–Tooth neuropathy. Proc. Natl. Acad. Sci. USA. 2009;106:11782–11787. doi: 10.1073/pnas.0905339106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blocquel D., Li S., Wei N., Daub H., Sajish M., et al. Alternative stable conformation capable of protein misinteraction links tRNA synthetase to peripheral neuropathy. Nucleic Acids Res. 2017;45:8091–8104. doi: 10.1093/nar/gkx455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hyun Y. S., Park H. J., Heo S. H., Yoon B. R., Nam S. H., et al. Rare variants in methionyl- and tyrosyl-tRNA synthetase genes in late-onset autosomal dominant Charcot–Marie–Tooth neuropathy. Clin. Genet. 2014;86:592–594. doi: 10.1111/cge.12327. [DOI] [PubMed] [Google Scholar]
- 43.Gonzaga-Jauregui C., Harel T., Gambin T., Kousi M., Griffin L. B., et al. Exome sequence analysis suggests that genetic burden contributes to phenotypic variability and complex neuropathy. Cell Rep. 2015;12:1169–1183. doi: 10.1016/j.celrep.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadchouel A., Wieland T., Griese M., Baruffini E., Lorenz-Depiereux B., et al. Biallelic mutations of methionyl-tRNA synthetase cause a specific type of pulmonary alveolar proteinosis prevalent on Réunion Island. Am. J. Hum. Genet. 2015;96:826–831. doi: 10.1016/j.ajhg.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nam S. H., Hong Y. B., Hyun Y. S., Nam D., Kwak G., et al. Identification of genetic causes of inherited peripheral neuropathies by targeted gene panel sequencing. Mol. Cells. 2016;39:382–388. doi: 10.14348/molcells.2016.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirano M., Oka N., Hashiguchi A., Ueno S., Sakamoto H., et al. Histopathological features of a patient with Charcot–Marie–Tooth disease type 2U/AD-CMTax-MARS. J. Peripher. Nerv. Syst. 2016;21:370–374. doi: 10.1111/jns.12193. [DOI] [PubMed] [Google Scholar]
- 47.Sagi-Dain L., Shemer L., Zelnik N., Zoabi Y., Orit S., et al. Whole-exome sequencing reveals a novel missense mutation in the MARS gene related to a rare Charcot–Marie–Tooth neuropathy type 2U. J. Peripher. Nerv. Syst. 2018;23:138–142. doi: 10.1111/jns.12264. [DOI] [PubMed] [Google Scholar]
- 48.Gillespie M. K., McMillan H. J., Kernohan K. D., Pena I. A., Meyer-Schuman R., et al. A novel mutation in MARS in a patient with Charcot–Marie–Tooth disease, axonal, type 2U with congenital onset. J. Neuromusc. Diseases. 2019;6:333–339. doi: 10.3233/JND-190404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naganuma M., Sekine S., Fukunaga R., Yokoyama S. Unique protein architecture of alanyl-tRNA synthetase for aminoacylation, editing, and dimerization. Proc. Natl. Acad. Sci. USA. 2009;106:8489–8494. doi: 10.1073/pnas.0901572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin K. P., Soong B. W., Yang C. C., Huang L. W., Chang M. H., et al. The mutational spectrum in a cohort of Charcot–Marie–Tooth disease type 2 among the Han Chinese in Taiwan. PLoS One. 2011;6:e29393. doi: 10.1371/journal.pone.0029393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tatsumi Y., Matsumoto N., Iibe N., Watanabe N., Torii T., et al. CMT type 2N disease-associated AARS mutant inhibits neurite growth that can be reversed by valproic acid. Neurosci. Res. 2019;139:69–78. doi: 10.1016/j.neures.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 52.Motley W. W., Griffin L. B., Mademan I., Baets J., De Vriendt E., et al. A novel AARS mutation in a family with dominant myeloneuropathy. Neurology. 2015;84:2040–2047. doi: 10.1212/WNL.0000000000001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weterman M., Kuo M., Kenter S. B., Gordillo S., Karjosukarso D. W., et al. Hypermorphic and hypomorphic AARS alleles in patients with CMT2N expand clinical and molecular heterogeneities. Hum. Mol. Genet. 2018;27:4036–4050. doi: 10.1093/hmg/ddy290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhao Z., Hashiguchi A., Hu J., Sakiyama Y., Okamoto Y., et al. Alanyl-tRNA synthetase mutation in a family with dominant distal hereditary motor neuropathy. Neurology. 2012;78:1644–1649. doi: 10.1212/WNL.0b013e3182574f8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee A. J., Nam D. E., Choi Y. J., Nam S. H., Choi B. O., Chung K. W. Alanyl-tRNA synthetase 1 (AARS1) gene mutation in a family with intermediate Charcot–Marie–Tooth neuropathy. Genes Genom. 2020;42:663–672. doi: 10.1007/s13258-020-00933-9. [DOI] [PubMed] [Google Scholar]
- 56.Bansagi B., Antoniadi T., Burton-Jones S., Murphy S. M., McHugh J., et al. Genotype/phenotype correlations in AARS-related neuropathy in a cohort of patients from the United Kingdom and Ireland. J. Neurol. 2015;262:1899–1908. doi: 10.1007/s00415-015-7778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Santos-Cortez R. L., Lee K., Azeem Z., Antonellis P. J., Pollock L. M., et al. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am. J. Hum. Genet. 2013;93:132–140. doi: 10.1016/j.ajhg.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Zhou J. B., Zeng Q. Y., Wu S., Xue M. Q., et al. Hearing impairment-associated KARS mutations lead to defects in aminoacylation of both cytoplasmic and mitochondrial tRNALys. Sci. China Life Sci. 2020;63:1227–1239. doi: 10.1007/s11427-019-1619-x. [DOI] [PubMed] [Google Scholar]
- 59.Scheidecker S., Bär S., Stoetzel C., Geoffroy V., Lannes B., et al. Mutations in KARS cause a severe neurological and neurosensory disease with optic neuropathy. Hum. Mutat. 2019;40:1826–1840. doi: 10.1002/humu.23799. [DOI] [PubMed] [Google Scholar]
- 60.Patzkó A., Shy M. E. Update on Charcot–Marie–Tooth disease. Curr. Neurol. Neurosci. Rep. 2011;11:78–88. doi: 10.1007/s11910-010-0158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taylor R. W., Pyle A., Griffin H., Blakely E. L., Duff J., et al. Use of whole-exome sequencing to determine the genetic basis of multiple mitochondrial respiratory chain complex deficiencies. JAMA. 2014;312:68–77. doi: 10.1001/jama.2014.7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McMillan H. J., Schwartzentruber J., Smith A., Lee S., Chakraborty P., et al. Compound heterozygous mutations in glycyl-tRNA synthetase are a proposed cause of systemic mitochondrial disease. BMC Med. Genet. 2014;15:36. doi: 10.1186/1471-2350-15-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohkamm B., Reilly M. M., Lochmüller H., Schlotter-Weigel B., Barisic N., et al. Further evidence for genetic heterogeneity of distal HMN type V, CMT2 with predominant hand involvement and Silver syndrome. J. Neurol. Sci. 2007;263:100–106. doi: 10.1016/j.jns.2007.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dubourg O., Azzedine H., Yaou R. B., Pouget J., Barois A., et al. The G526R glycyl-tRNA synthetase gene mutation in distal hereditary motor neuropathy type V. Neurology. 2006;66:1721–1726. doi: 10.1212/01.wnl.0000218304.02715.04. [DOI] [PubMed] [Google Scholar]
- 65.Sun A., Liu X., Zheng M., Sun Q., Huang Y., Fan D. A novel mutation of the glycyl-tRNA synthetase (GARS) gene associated with Charcot–Marie–Tooth type 2D in a Chinese family. Neurol. Res. 2015;37:782–787. doi: 10.1179/1743132815Y.0000000055. [DOI] [PubMed] [Google Scholar]
- 66.Lee H. J., Park J., Nakhro K., Park J. M., Hur Y. M., et al. Two novel mutations of GARS in Korean families with distal hereditary motor neuropathy type V. J. Peripher. Nerv. Syst. 2012;17:418–421. doi: 10.1111/j.1529-8027.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- 67.Abe A., Hayasaka K. The GARS gene is rarely mutated in Japanese patients with Charcot–Marie–Tooth neuropathy. J. Hum. Genet. 2009;54:310–312. doi: 10.1038/jhg.2009.25. [DOI] [PubMed] [Google Scholar]
- 68.James P. A., Cader M. Z., Muntoni F., Childs A. M., Crow Y. J., Talbot K. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology. 2006;67:1710–1712. doi: 10.1212/01.wnl.0000242619.52335.bc. [DOI] [PubMed] [Google Scholar]
- 69.Eskuri J. M., Stanley C. M., Moore S. A., Mathews K. D. Infantile onset CMT2D/dSMA V in monozygotic twins due to a mutation in the anticodon-binding domain of GARS. J. Peripher. Nerv. Syst. 2012;17:132–134. doi: 10.1111/j.1529-8027.2012.00370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu X., Chen B., Tang H., Li W., Fu Y., Zhang Z., Yan Y. Novel Mutation of GARS in a Chinese family with distal hereditary motor neuropathy type V. Front. Neurol. 2018;9:571. doi: 10.3389/fneur.2018.00571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee D. C., Meyer-Schuman R., Bacon C., Shy M. E., Antonellis A., Scherer S. S. A recurrent GARS mutation causes distal hereditary motor neuropathy. J. Peripher. Nerv. Syst. 2019;24:320–323. doi: 10.1111/jns.12353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oprescu S. N., Chepa-Lotrea X., Takase R., Golas G., Markello T. C., et al. Compound heterozygosity for loss-of-function GARS variants results in a multisystem developmental syndrome that includes severe growth retardation. Hum. Mutat. 2017;38:1412–1420. doi: 10.1002/humu.23287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Griffin L. B., Sakaguchi R., McGuigan D., Gonzalez M. A., Searby C., et al. Impaired function is a common feature of neuropathy-associated glycyl-tRNA synthetase mutations. Hum. Mutat. 2014;35:1363–1371. doi: 10.1002/humu.22681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He W., Bai G., Zhou H., Wei N., White N. M., et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526:710–714. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boczonadi V., Meyer K., Gonczarowska-Jorge H., Griffin H., Roos A., et al. Mutations in glycyl-tRNA synthetase impair mitochondrial metabolism in neurons. Hum. Mol. Genet. 2018;27:2187–2204. doi: 10.1093/hmg/ddy127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo M., Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat. Chem. Biol. 2013;9:145–153. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pang Y. L., Poruri K., Martinis S. A. tRNA synthetase: tRNA aminoacylation and beyond. Wiley Interdiscip. Rev. RNA. 2014;5:461–480. doi: 10.1002/wrna.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spaulding E. L., Sleigh J. N., Morelli K. H., Pinter M. J., Burgess R. W., Seburn K. L. Synaptic deficits at neuromuscular junctions in two mouse models of Charcot–Marie–Tooth type 2d. J. Neurosci. 2016;36:3254–3267. doi: 10.1523/JNEUROSCI.1762-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Seburn K. L., Nangle L. A., Cox G. A., Schimmel P., Burgess R. W. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot–Marie–Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- 80.Motley W. W., Seburn K. L., Nawaz M. H., Miers K. E., Cheng J., et al. Charcot–Marie–Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet. 2011;7:e1002399. doi: 10.1371/journal.pgen.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nangle L. A., Zhang W., Xie W., Yang X. L., Schimmel P. Charcot–Marie–Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc. Natl. Acad. Sci. USA. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.He W., Zhang H. M., Chong Y. E., Guo M., Marshall A. G., Yang X. L. Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc. Natl. Acad. Sci. USA. 2011;108:12307–12312. doi: 10.1073/pnas.1104293108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cader M. Z., Ren J., James P. A., Bird L. E., Talbot K., Stammers D. K. Crystal structure of human wildtype and S581L-mutant glycyl-tRNA synthetase, an enzyme underlying distal spinal muscular atrophy. FEBS Lett. 2007;581:2959–2964. doi: 10.1016/j.febslet.2007.05.046. [DOI] [PubMed] [Google Scholar]
- 84.Yalcouyé A., Diallo S. H., Coulibaly T., Cissé L., Diallo S., et al. A novel mutation in the GARS gene in a Malian family with Charcot–Marie–Tooth disease. Mol. Genet. Genom. Med. 2019;7:e00782. doi: 10.1002/mgg3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liao Y. C., Liu Y. T., Tsai P. C., Chang C. C., Huang Y. H., et al. Two novel denovo GARS mutations cause early-onset axonal Charcot–Marie–Tooth disease. PLoS One. 2015;10:e0133423. doi: 10.1371/journal.pone.0133423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Markovitz R., Ghosh R., Kuo M. E., Hong W., Lim J., et al. GARS-related disease in infantile spinal muscular atrophy: implications for diagnosis and treatment. Am. J. Med. Genet. A. 2020;182:1167–1176. doi: 10.1002/ajmg.a.61544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Darras B. T. Non-5q spinal muscular atrophies: the alphanumeric soup thickens. Neurology. 2011;77:312–314. doi: 10.1212/WNL.0b013e3182267bd8. [DOI] [PubMed] [Google Scholar]
- 88.Peeters K., Chamova T., Jordanova A. Clinical and genetic diversity of SMN1-negative proximal spinal muscular atrophies. Brain J. Neurol. 2014;137:2879–2896. doi: 10.1093/brain/awu169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turner R. J., Lovato M., Schimmel P. One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. J. Biol. Chem. 2000;275:27681–27688. doi: 10.1074/jbc.M003416200. [DOI] [PubMed] [Google Scholar]
- 90.Chien C.-I., Chen Y.-W., Wu Y.-H., Chang C.-Y., Wang T.-L., Wang C. C. Functional substitution of a eukaryotic glycyl-tRNA synthetase with an evolutionarily unrelated bacterial cognate enzyme. PLoS One. 2014;9:e94659. doi: 10.1371/journal.pone.0094659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Brown R. H., Al-Chalabi A. Amyotrophic lateral sclerosis. New Eng. J. Med. 2017;377:162–172. doi: 10.1056/NEJMra1603471. [DOI] [PubMed] [Google Scholar]
- 92.Corcia P., Brulard C., Beltran S., Marouillat S., Bakkouche S. E., et al. Typical bulbar ALS can be linked to GARS mutation. Amyotroph. Lateral Scler. Frontotemporal Degener. 2019;20:275–277. doi: 10.1080/21678421.2018.1556699. [DOI] [PubMed] [Google Scholar]
- 93.Rossor A. M., Kalmar B., Greensmith L., Reilly M. M. The distal hereditary motor neuropathies. J. Neurol. Neurosurg. Psychiatry. 2012;83:6–14. doi: 10.1136/jnnp-2011-300952. [DOI] [PubMed] [Google Scholar]