Abstract
Background:
Traumatic injury can lead to a compromised intestinal epithelial barrier, decreased gut perfusion, and inflammation. While recent studies indicate that the gut microbiome (GM) is altered early following traumatic injury, the impact of GM changes on clinical outcomes remains unknown. Our objective of this follow-up study was to determine if the GM is associated with clinical outcomes in critically injured patients.
Methods:
We conducted a prospective, observational study in adult patients (n=67) sustaining severe injury admitted to a Level I Trauma Center. Fecal specimens were collected on admission to the Emergency Department (ED), and microbial DNA from all samples was analyzed using the QIIME pipeline and compared against the Greengenes database. Alpha and β-diversity were estimated using the observed species metrics, and analyzed with t-tests and permutational analysis of variance (PERMANOVA) for overall significance, with post-hoc pairwise analyses.
Results:
Our patient population consisted of 63% males with a mean age of 44 years old. 78% of the patients suffered blunt trauma with 22% undergoing penetrating injuries. The mean body mass index (BMI) was 26.9 kg/m2. Significant differences in admission β-diversity were noted by hospital LOS, ICU LOS, number of days on the ventilator, infections, and ARDS (P<0.05). Beta-diversity on admission differed in patients that died compared to patients that lived (mean time to death=8 days). There were also significantly less OTUs in samples from patients who died versus those who survived. A number of species were enriched in the GM of injured patients who died, which included some traditionally probiotic species such as Akkermansia muciniphilia, Oxalobacter formigenes, and eubacterium biforme; p<0.05.
Conclusion:
GM diversity on admission in severely injured patients is predictive of a variety of clinically important outcomes. While our study does not address causality, the GM of trauma patients may provide valuable diagnostic and therapeutic targets for the care of injured patients.
Level of Evidence:
Level III
Study Type:
Prognostic and Epidemiological
Keywords: trauma, injury, gut microbiome, dysbiosis, clinical outcomes
Background:
The microbiome is the ecological community of commensal, symbiotic and pathogenic microorganisms that share our body space with a composition that differs according to body location. Given the relationship to the external environment, the gut functions as an immune organ in conjunction with the gut microbiome (GM) which influences the development and maintenance of the immune system and, subsequently inflammation 1, 2. Gut microbes protect against transiently invading pathogens by providing tonic stimulation to the innate immune system via toll-like receptor (TLR) signaling that increases intestinal motility, reinforces epithelial integrity, and produces metabolites 3, 4.
While alterations in the diversity of the human GM have recently been implicated in a number of disease states, relatively little is still known in the context of traumatic injury 5–8.
Trauma can influence the gut in a number of ways whether by directly injuring the gut, interrupting the brain-gut axis, or systemically via global hypoperfusion or the inflammatory response. The impact on the gut following traumatic injury in turn alters the GM and causes dysbiosis, or an imbalance or altered distribution of gut microbes 9. Rapid dysbiosis has also been demonstrated in critical illness with worsening during prolonged hospitalization and has also been attributed to septic complications 10, 11.
Although reductions in intestinal microbial diversity have been linked to increased mortality in critically ill patients, less defined is the impact of trauma on the intestinal microbial community. There is a distinct dysbiosis in trauma patients likely related to this population’s susceptibility to complications such as multiple organ dysfunction syndrome, hospital acquired infection, and systemic inflammatory response 12, 13. Several early clinical studies in small patient populations have demonstrated phylogenetic changes among gut microbial populations following traumatic and burn injuries, yet these studies have lacked power to include clinically relevant outcomes 14–17. Preclinical data from various injury models including polytrauma, burn injury, traumatic brain injury (TBI) and spinal cord injury (SCI) also support the concept that traumatic injury alters the GM which impacts outcomes 16, 18–23. Larger clinical studies are needed to address this gap and better understand microbial changes that occur in the gut following injury, and their relationship to outcomes.
To this end, we conducted a prospective, observational cohort study of severely injured patients. The aim of this study was to characterize differences in gut microbial communities at admission in trauma patients, and to begin to elucidate the potential impact on clinical outcomes. We hypothesized that the diversity and composition of the GM would be differentially altered depending on clinical course of the patient. Specifically, we examined whether patients who experienced infectious complications, lengthy hospital and ICU stays, or mortality had distinct GM characteristics upon admission to the emergency department.
Methods:
Approval was obtained from the University of Texas Health San Antonio Institutional Review Board to conduct this study. Adult patients (age ≥ 18 years; n=67) sustaining a severe injury from blunt or penetrating trauma admitted to University Hospital (UH), a Level 1 Trauma Center in San Antonio, Texas were enrolled prospectively from 2015 to 2016. Enrollment criteria included age ≥ 18 years, an estimated Injury Severity Score (ISS) >15, ground transport to UH from the scene, and admission to the UH Surgical Intensive Care Unit (SICU). Exclusion criteria included prisoners, age < 18 years, pregnancy, and patients transferred from outside hospitals. Patients were initially enrolled under a waiver of consent on admission to the UH Emergency Department (ED). Consent to participate and continue the study was obtained from the patient or a legal authorized representative as soon as possible following admission. Healthy volunteers (n=13) were also enrolled for comparison purposes, but not included in any statistical analysis included herein.
Sample collection
This is a follow-up study to a previous study by our group, and both collection and data processing were performed in a similar manner 9. Fecal specimens were collected on admission to the UH Emergency Department (Day 0) by rectal swab (COPAN, California, USA) on routine trauma evaluation. All fecal samples were stored at −80°C within 20 minutes of sampled collection for DNA isolation at a later time. Extensive demographic, injury, clinical and outcome data were prospectively collected on all patients. Of note, only three of the 67 patients enrolled received antibiotics prior to the rectal exam performed on secondary survey.
Gut microbiome analysis
Microbial DNA was isolated from all fecal samples using the QIAGEN QIAamp® DNA Stool Mini Kit (QIAGEN, Hilden, Germany). DNA was quantified using the Thermo Scientific NanoDrop 1000 Spectrophotometer. Extracted genomic DNA was then used to amplify the V1-V2 variable region of the 16S rRNA genes with custom-designed primers (F27/R355) using PCR. The Forward Bosshard sequence was AGAGTTTGATCMTGGCTCAG (27F) and the Reverse Bosshard sequence was GCTGCCTCCCGTAGGAGT (355R) with the amplicon size of V1-V2 about 340bp (355–27). Libraries for all samples were prepared and sequenced by Paired-end sequencing (2 × 300 bp) using the Illumina MiSeq platform.
Subsequently, raw data were processed through the software package Quantitative Insights Into Microbial Ecology (QIIME). A mean of 164,813 pair-end raw reads (median of 165,738 pair-end raw reads) per sample were generated with read length of 301bps. Raw sequences were quality trimmed by removing reads shorter than 200 bases, resulting in a median quality score of 36 for forward reads, and 30 for reverse reads. The operational taxonomic units (OTU) were clustered based on at 97% similarity. Taxonomic classifications were made using the QIIME-formatted Greengenes (gg_13_8) 16S rRNA gene database according to standard phylogenetic methods. The OTU table was further filtered by removing OTUs found in only one sample. Rarefaction was performed to a depth of 28000 base pairs, which allowed inclusion of all samples.
Beta (β) diversity, or the inter-population diversity (the microbial diversity between patients at each time point), was estimated by constructing principal coordinate analysis plots for the following β-diversity measures: weighted and unweighted UniFrac distances, Bray-Curtis, and Jaccard Indices using QIIME. These ß diversity measures plot the three largest variances of the whole data set against each other in 3 axes, on order to allow clustering of different samples/groups as a way to measure similarity/dissimilarity between groups. Mortality was the primary outcome. Due to the available sample size, secondary outcomes and risk factor data were not analyzed in a continuous fashion, but rather categorized according to the following criteria: BMI- underweight (<18.5kg/m2), normal (18.5–24.9 kg/m2), overweight (25–29.9 kg/m2), obese (>30 kg/m2); hospital length of stay (LOS)- low (0–7 days), medium (8–14 days), high (15–30 days), profound (>30 days), and died; intensive care unit LOS- low (0–3 days), medium (4–7 days), high (8–14 days), profound (>14 days), and died; and ventilator days- low (0–3 days), medium (4–7 days), high (>7days); presence of documented infection (pneumonia, urinary tract infection, bacteremia, wound infection, etc. as defined by positive cultures); and acute respiratory distress syndrome (ARDS; as documented and confirmed by P:F ratio ≤300). Statistical analysis of these measures were performed with a permutational analysis of variance (PERMANOVA) for overall significance, with post-hoc pairwise PERMANOVAs run to assess differences across groups.
Alpha (α) diversity, or the intra-population diversity (microbial diversity within individual patients at each time point), was estimated by calculating the number of observed OTUs (richness), evenness of OTU abundance, and diversity using the Faith_PD and Shannon Diversity Indices. Two way ANOVA with Tukey’s multiple comparisons was used to perform statistical analyses α-diversity. For all parameters comparing survivors and non-survivors (e.g., demographics and phylogeny) D’Agostino and Pearson normality tests were performed. Data that was normally distributed are presented as mean ± SEM, while non-normally distributed data is presented as median ± IQR. Significance was performed with t-tests or Mann-Whitney tests with Welch’s correction as needed (when variance was not equal). Alpha < 0.05 was considered significant for all analyses. QIIME, STAMP and GraphPad Prism were used for the visualization and the statistics of the comparative metagenomics data sets.
Results:
Patient characteristics
Patient characteristics shown in Table 1 reveal that the patients (n = 67) were predominantly male with a mean age of 45 years and an average ISS of 21. The majority of the patients suffered blunt trauma with only 22% sustaining penetrating injuries. The mean body mass index (BMI) was 27.0 ±0.7 kg/m2. The average transport time from the scene of injury to the emergency department was 29 minutes, and was significantly faster in patients who died versus those who survived. Patients who died were older (p = 0.04) and more severely injured as indicated by a greater ISS (p = 0.0001), but not shock index. There was no difference in sex or BMI in patients who lived or died.
Table 1.
Patient characteristics of the total injured population and those that survived vs died.
| Total (n=67) | Survived (n=59) | Died (n=8) | p value | |
|---|---|---|---|---|
| Mean Age (years) | 45 (2.3) | 44 (2.5) | 58 (7.3) | 0.04 |
| # Males | 44 (67%) | 38 (64%) | 6 (75%) | 0.70 |
| Mean BMI (kg/m2) | 27.0 (0.7) | 27.2 (0.7) | 25.5 (1.3) | 0.44 |
| # Smokers | 19 (28%) | 18 (31%) | 1 (13%) | 0.42 |
| Mean ISS | 20.9 (1.2) | 19.8 (1.2) | 33.2 (3.9) | 0.0019 |
| Median Shock Index (beats/min/mmHg) | 0.80 (0.66–1.21) | 0.80 (0.67–1.20) | 0.93 (0.58–1.83) | 0.79 |
| Blunt Mechanism | 52 (78%) | 46 (78%) | 6 (75%) | 1.00 |
| Penetrating Mechanism | 15 (22%) | 13 (22%) | 2 (25%) | 1.00 |
| Transport Time (min) | 29 | 29 | 18 | 0.0001 |
| LOS (days) | 20.3 (7.0–24.8) | 13 (8–25.0) | 6 (2.5–24.0) | 0.24 |
| ICU LOS (days) | 6.0 (4.0–16.3) | 6.0 (4.0–14.0) | 6.0 (2.5–24.0) | 0.95 |
| Mean Ventilator (days) | 4.0 (3.0–14.0) | 6.0 (3.0–13.8) | 3.0 (1.0–19.0) | 0.26 |
| Diagnosis of infection | 29 (43%) | 26 (44%) | 3 (38%) | 1.00 |
| Diagnosis of ARDS | 31 (46%) | 27 (46%) | 4 (50%) | 1.00 |
α-diversity
Largely, the Evenness, Faith_PD, and Shannon indices of α-diversity did not reveal differences in most all of the different demographic and outcome variables. It was shown that operational taxonomic units (OTUs) greatly increased in aging populations (Figure 1), which is unlikely related to trauma, as this has been shown previously in healthy individuals 24. However, we saw an increase in the OTUs from fecal swabs of patients who survived (n=59) compared to those who died (n=8; p=0.049).
Figure 1.
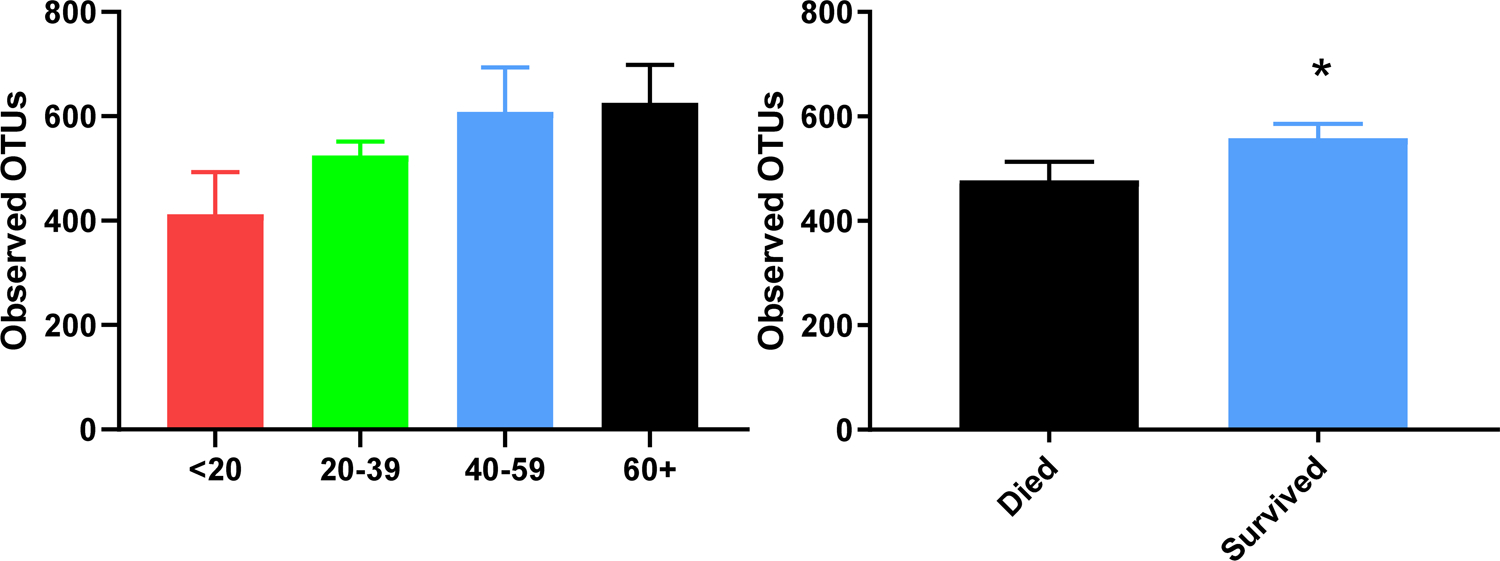
Operational Taxonomic Units (OTUs) demonstrating alpha diversity for chosen patient characteristics. Statistical analyses reveal that OTUs increase with age (A) and in patients who died (B). *p < 0.05.
β- diversity
Significance levels for all β-diversity values are shown in Table 2, and The Principle Components Analysis (PCA) plots for all of these variables are shown in Figures 2 and 3, as well as Supplemental Figures 1–5. As previously reported, ISS significantly influenced β-diversity whereas the mechanism of injury (i.e., blunt vs. penetrating trauma) did not 9. Bray Curtis dissimilarity was significantly clustered based on BMI (p=0.025, Figure 2) and gender (p=0.0005, Supplemental Figure 1), but not smoking (p=0.298).
Table 2.
β-diversity significance levels found via PERMANOVA.
| β-diversity measure | BMI (kg/m2) | Sex (M/F) | Smoking (Y/N) | Hospital LOS (days) | ICU LOS (days) | Ventilator Days | Infection (Y/N) | ARDS (Y/N) | MOF (Y/N) | Mortality (Y/N) |
|---|---|---|---|---|---|---|---|---|---|---|
| Bray-Curtis | p= 0.025 |
p= 0.005 |
p= 0.298 |
p= 0.074 |
p= 0.019 |
p= 0.033 |
p= 0.123 |
p= 0.030 |
p= 0.311 |
p= 0.026 |
| Jaccard | p= 0.207 |
p= 0.001 |
p= 0.087 |
p= 0.089 |
p= 0.002 |
p= 0.006 |
p= 0.089 |
p= 0.029 |
p= 0.076 |
p= 0.016 |
| Unweighted Unifrac | p= 0.605 |
p= 0.027 |
p= 0.228 |
p= 0.11 |
p= 0.027 |
p= 0.505 |
p= 0.039 |
p= 0.360 |
p= 0.150 |
p= 0.053 |
| Weighted Unifrac | p= 0.583 |
p= 0.119 |
p= 0.098 |
p= 0.007 |
p= 0.009 |
p= 0.101 |
p= 0.205 |
p= 0.074 |
p= 0.510 |
p= 0.010 |
Figure 2.
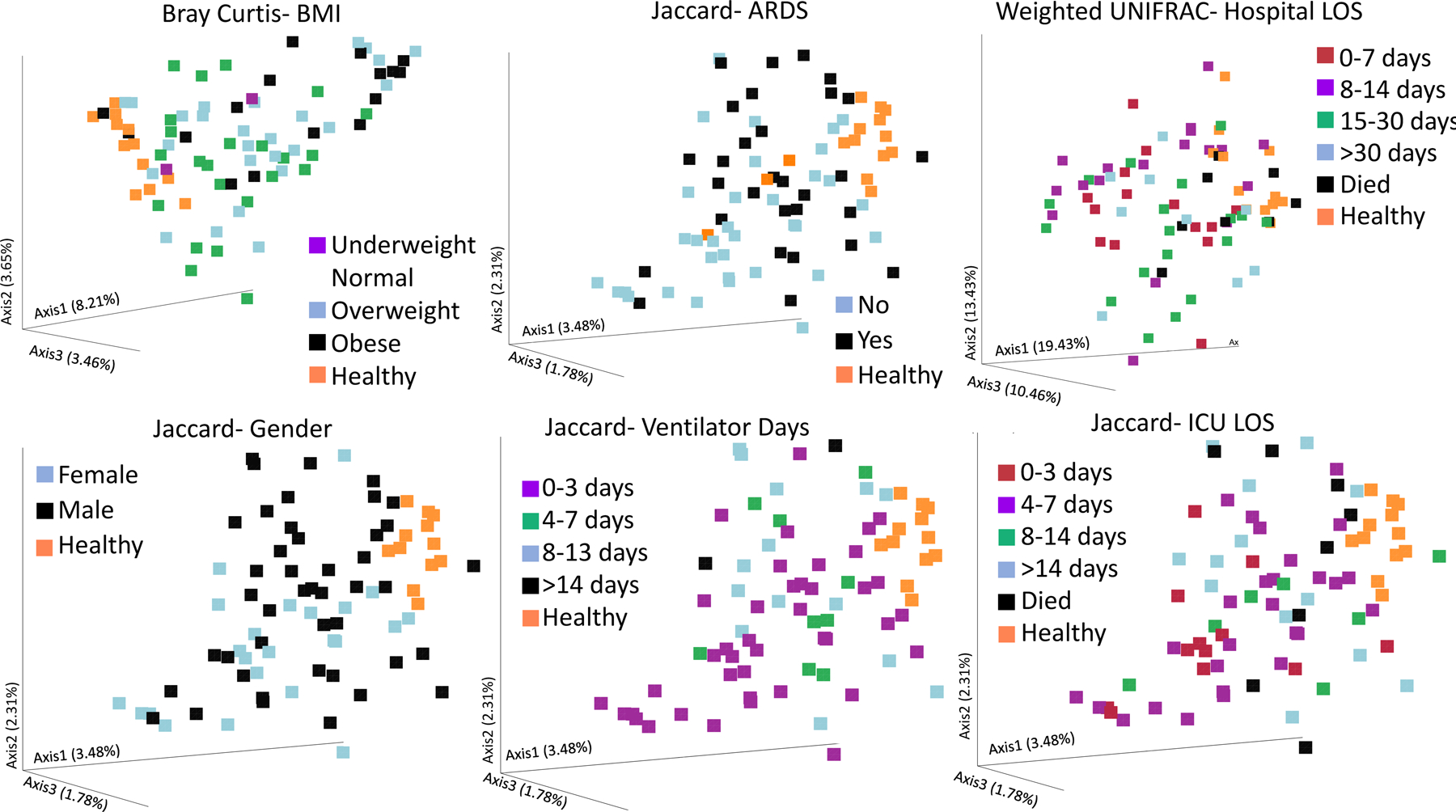
Significant findings in beta diversity measures for chosen patient characteristics. Bray Curtis measures clustered different depending on BMI (A), while the weighted Unifrac clustered patients based upon their hospital length of stay (B). The Jaccard similarity index was able to significantly cluster patients based on whether they experienced acute respiratory distress syndrome (C), Gender (D), ventilator days (E), and ICU LOS (F). p < 0.05 for all plots.
Figure 3.
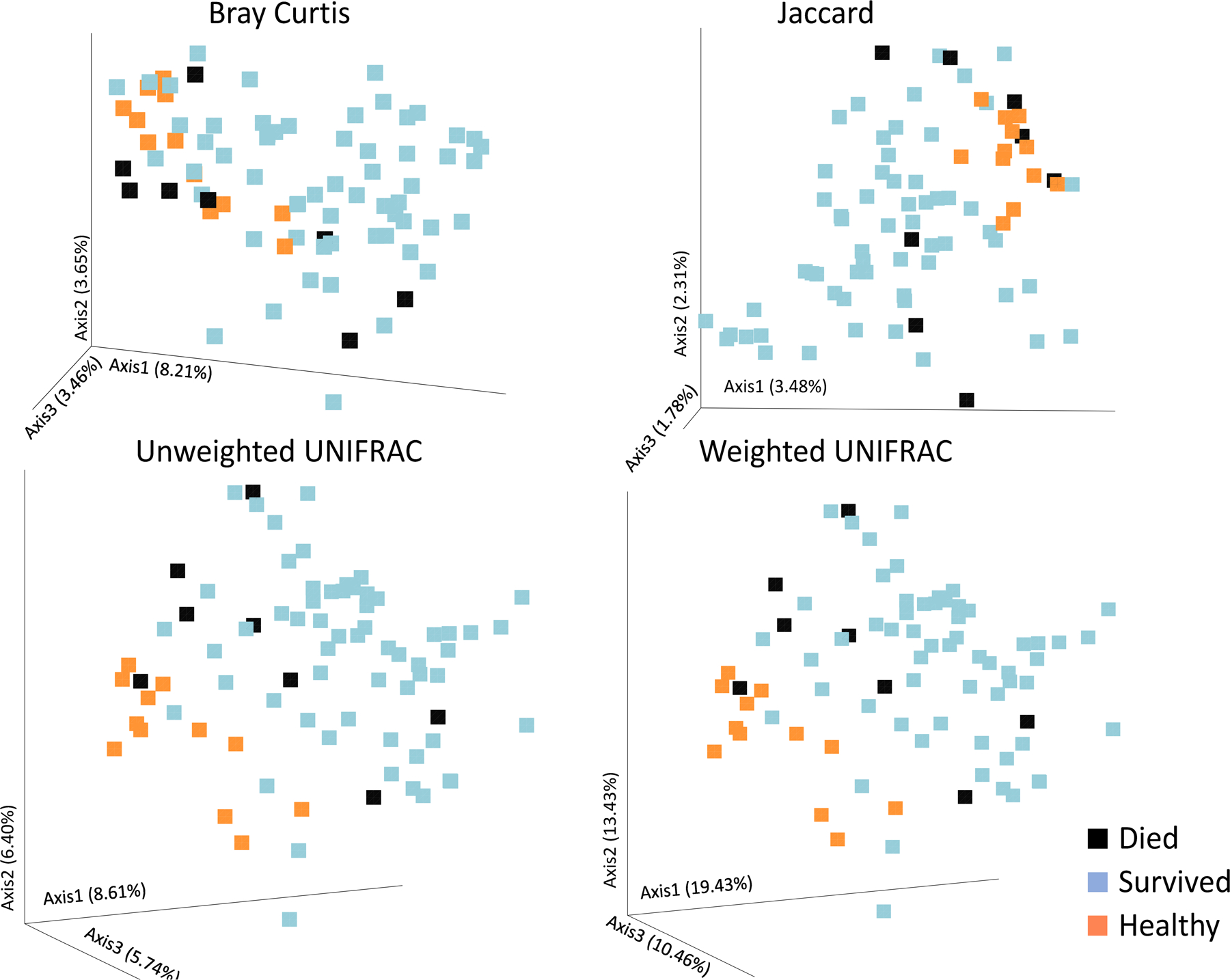
Beta diversity plots for mortality. Patients that lived showed significantly different β-diversity than those that died for all β-diversity measures: Bray Curtis (A), Jaccard (B), Unweighted UNIFRAC (C), and weighted UNIFRAC (D). p < 0.05 for all plots.
Figure 2 also reveals that many other clinically important endpoints and risk factors were significantly clustered for different β-diversity measures. For example, the weighted UNIFRAC (taking into account the relative number of different species), but not the unweighted UNIFRAC was significantly clustered based on overall hospital LOS (p=0.007). Moreover, every single β-diversity measure analyzed revealed substantial clustering based on the ICU LOS (Figure 2, Supplemental Figure 2). Interestingly, in general, more severe outcomes tended to cluster in space on PCA plots more closely to those of healthy controls than outcomes generally considered to be preferred. For example, in the case of ICU LOS (Figure 2), death (black squares), and profound ICU LOS over 14 days (blue squares) cluster more closely to healthy controls (orange squares) than lower LOS (Figure 2, Supplemental Figure 3), however the healthy controls were not included in the statistical analysis.
In terms of pulmonary function, the most relevant β-diversity measure was the Jaccard similarity index (Figure 2, Supplemental Figure 4). For this measure, patients that developed ARDS at some point in their hospital stay were clustered in a significantly different manner on the PCA than those that did not experience ARDS. Moreover, the number of days on a ventilator was also clustered based on whether patients spent 0–3, 4–7, 8–13, or over 14 days on a ventilator (Figure 2, Supplemental Figure 4). Surprisingly, only one of the four β-diversity measures (Unweighted UNIFRAC) was able to distinguish patients based on the presence of infection, perhaps owing to the mixed bag of infectious complications included (Supplemental Figure 5).
Figure 3 shows the PCA plots for all for β-diversity measures separating patients that died versus those that survived. Every single measure examined was successful in identifying differences in admission microbiome between patients that died and those that did not (p ≤ 0.05). It was surprising that all measures found such significant differences in β-diversity given the small sample size of the group that died (n=8) and that the mean time to death was 8 days. Moreover, the GM from patients who died tended to cluster more closely to the GM from healthy volunteer samples, rather than surviving patients. The statistical analysis, however, only included data from trauma patients and not healthy volunteers, so it is not known whether this difference was statistically significant.
All phylogenetic differences found associated with mortality are shown in Figure 4 and in the Supplemental Figures 6–9. Patients that survived had GM compositions with a significantly lower relative abundance of the Firmicutes phylum (p = 0.0049). These patients had a tendency to contain more Proteobacteria, however the difference did not reach statistical significance (p = 0.24). Largely the difference in Firmicutes can be attributed to members of the Order Clostridiales (p = 0.0024) and the Family Ruminococcaceae (p = 0.0017, Supplemental Figure 6). Increases in the Genus Prevotella (p = 0.024) and Corynebacterium (p = 0.055) were also seen in patients who survived versus those who died (Supplemental Figure 7, 8). Lastly, we were surprised to see that there was an increase in the gut microbial composition of traditional probiotic bacteria in patients who died versus those who survived. Specifically, Eubacterium biforme, Ruminococcus flavefaciens, Akkermansia muciniphila, and Oxalobacter formigenes (p = 0.037, 0.010, 0.0004, and <0.0001, respectively) were all increased in the admission fecal swabs of patients that died (Figure 4B).
Figure 4.
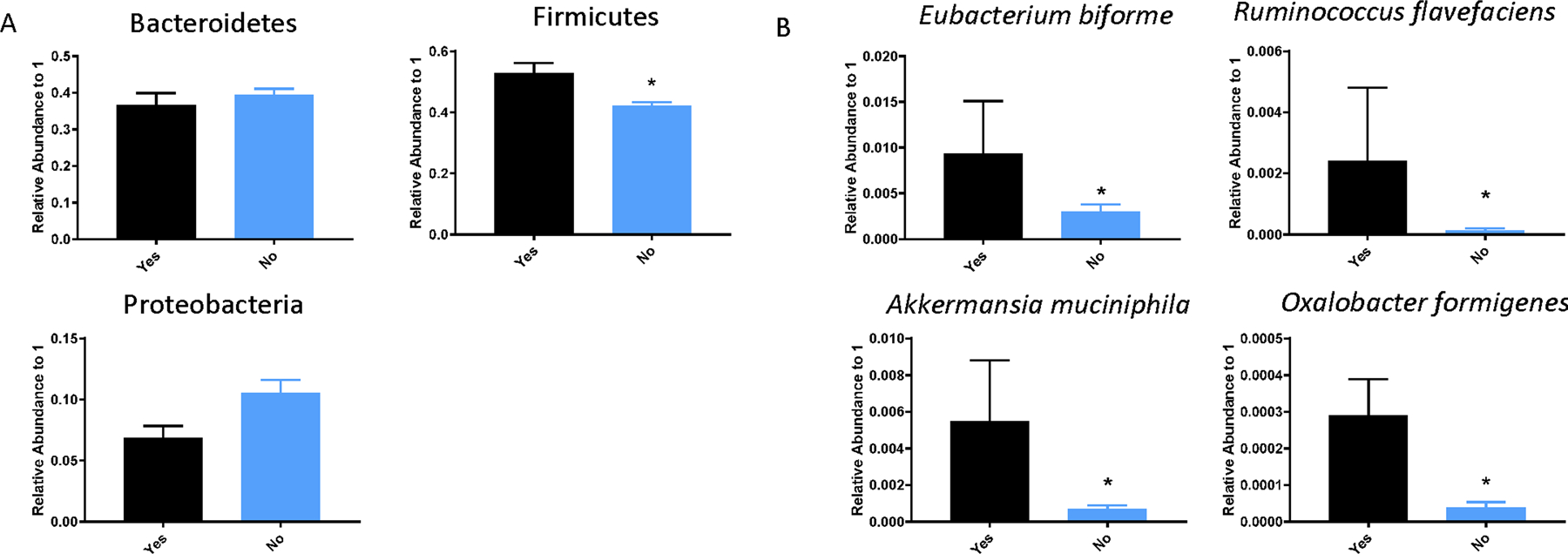
Phylogenetic differences associated with survival. (A) At the phylum level, levels of Bacteroidetes was unchanged, while levels of Firmicutes was significantly enriched in patients that died versus those that did not. (B) At the species level, there were 4 specific species (Eubacterium biforme, Ruminococcus flavefaciens, Akkermansia muciniphila, and Oxalobacter formigenes) that were enriched in the gut microbiota of patients that died versus those that did not (P<0.05).
Discussion:
From an evolutionary standpoint, it is logical to conclude that the ecological community of commensal, symbiotic and pathogenic microorganisms that share our body space has a great influence on homeostasis. What has become apparent in the past decade is that the GM can greatly influence pathogenic states. Once thought to be more influential in metabolic syndromes such as obesity and diabetes, the GM has been implicated in a wide variety of conditions, both acute and chronic. Herein, we describe that admission rectal swabs of severely injured patients can categorize the GM based on a number of clinically important outcomes to include hospital and ICU LOS, ventilator days, and mortality. Moreover, to our knowledge, this is the first report to show that specific bacteria are altered upon admission in trauma in patients that succumbed to their injury.
The importance of the microbiome has been demonstrated in many conditions (e.g. surgery), however, the specific role of the microbiome in trauma patients has been limited to demonstration that the microbiome changes with time post-injury 25. Our previous study on this data set also suggested that changes may be linked to perfusion, as the amounts of blood products administered also impacts the bacterial composition 9. That study utilized healthy volunteers as a comparison, which was limited by the fact that healthy volunteers may be demographically different than trauma patients. While we show the data for healthy controls here on PCA plots, our analyses only included admission samples. The average pre-hospital transport time for these patients was less than 30 minutes, and rectal swabs occurred within 30 minutes of admission. Thus, the differences observed have occurred within 1 hour of traumatic injury. While this does not address the presumably massive effects of clinical care (drugs, fluids, etc.) on the GM, this highlights the potential for the admission microbiome to hold powerful prognostic value on endpoints such as mortality and LOS. Indeed, the GM has been described as one of the key tenets that will facilitate precision medicine in becoming a reality 26.
One challenge in microbiome research is the rapidly evolving bioinformatics used in the interpretation of the data. It is very likely that realizing the diagnostic value of different β-diversity values will require a deep understanding of the nuances between these values (i.e., Unifrac vs. Bray Curtis). The data presented herein show that the Jaccard index was very efficient at categorizing patients based on how long the needed a ventilator and whether they developed ARDS. This highlights the possibility that if a patient presents with an injury pattern that causes concern over pulmonary function, targeted methods to probe the Jaccard index may be warranted. The nuanced differences between these variables is also worth considering. For example, while Bray-Curtis and Jaccard indices are used to quantify compositional dissimilarity, Jaccard is based on metric space. Similarly, while UniFrac is a distance metric comparing communities, weighted UniFrac is considered more quantitative due to the fact that it takes into account abundances of various species 27.
To further emphasize the importance of the bioinformatic processing, setting variables such as the sampling depth (rarefaction) or OTU similarity used can drastically influence findings, and efforts should be made to, at minimum, report how this processing is done, if not to standardize how data is treated within the trauma field. Moreover, while we did not employ false discovery rate methods, we do report 21 unique bacterial reads in the phylogenic results (to include related microbes such as Clostridia and Clostridiales) which would, mathematically, lead to 1.05 false discoveries. This type of statistical check will become very important when larger mutli center datasets are available. Still, we believe that the wealth of information that comes from analyzing the microbiome holds untapped valuable information.
Despite our small sample size (n = 8 patients who died) we found robust differences in patients who died after trauma versus those who survived. Interestingly, all β-diversity variables were clustered differently depending on survival, and those who died tended to cluster more closely with healthy individuals. Moreover, we found a variety of bacterial populations that were different in the patients that died, including down to the species level. Surprisingly, four of the species that were identified to be enriched in patients who died (Eubacterium biforme, Ruminococcus flavefaciens, Akkermansia muciniphila, and Oxalobacter formigenes) are commercially available as probiotics. Probiotic species have been proposed to prevent mortality in, for example necrotizing enterocolitis 28. While the implication of these findings are unclear, we propose that there is possibly a compensatory response in the patients who died, which was ultimately unsuccessful. Our data also suggest the possibility that an early alteration in gut microbial communities after traumatic injury may convey a protective benefit to patients and play an integral part in survival and outcome.
There are several limitations of the current study worth noting. This study represents a single center study and is representative of the population in the South Texas and urban San Antonio area. Given the substantial variability in the GM of healthy individuals, especially in regards to diet, larger-multicenter trials interrogating the microbiome are warranted. Also, the findings herein are purely associative and causation between the GM and outcomes cannot be inferred. Additionally, the use of antibiotics most certainly affects the GM, which represents a confounder that will likely occur in all trauma trials. Only three patients received antibiotics in the prehospital setting en route to the hospital, which drastically alter the GM 29. Similarly, only one timepoint was examined. However, these admission samples were largely taken before administration of antibiotics or resuscitation products, and thus the iatrogenic influences on the GM were not examined herein. Lastly, while the patient cohort was not large enough to identify significant differences in several α-diversity parameters that may ultimately prove informative.
In conclusion, the current report represents the largest report on the GM of trauma patients to date. Mortality due to traumatic injury is associated with a GM that includes fewer unique organisms, but does contain a higher number of several different commercially available probiotic species. Moreover, all β-diversity parameters probed were able to categorize patients according to whether or not they succumbed to their injury. While there were some of these diversity calculations that also were able to cluster other important outcomes, the specific associations are not as robust as the ones between mortality and all β-diversity parameters. Taken together, the GM holds great promise for diagnostic and therapeutic targets in traumatic injury, and further clinical and preclinical studies are needed.
Supplementary Material
Supplemental Figure 1. Significant β-diversity plots according to Gender.
Supplemental Figure 2. Significant β-diversity plots according to Ventilator days and ARDS.
Supplemental Figure 3. Significant β-diversity plots according to hospital length of stay (LOS).
Supplemental Figure 4. Significant β-diversity plots according to ICU length of stay (LOS).
Supplemental Figure 5. Significant β-diversity plots according to infection.
Supplemental Figure 6. Significant changes in phylogeny within the phylum Firmicutes.
Supplemental Figure 7. Significant changes in phylogeny within the phylum Bacteroidetes.
Supplemental Figure 9. Significant changes in phylogeny within the phylum Proteobacteria.
Supplemental Figure 8. Significant changes in phylogeny within the phylum Actinobacteria.
Acknowledgments and Source of Funding:
These findings were presented in part at the Military Health System Research Symposium August 18–22, 2019. Kissimmee, Florida and will be presented at the 78th Annual Meeting of the American Association for the Surgery of Trauma and the Clinical Congress of Acute Care Surgery, Dallas, TX . The project described was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2 TR001118. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the Department of the Army and the Department of Defense. Support was also received by the University of Texas Health San Antonio Military Health Institute and the Bob Kelso Endowment awarded to the University of Texas Health San Antonio Department of Surgery. The authors would like to thank the following individuals for their support: Basil A. Pruitt, Jr., Dawn Garcia and Korri S. Weldon for 16S sequencing sample processing and data generation.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to disclose.
Data from this paper, in varying forms, have been presented and will be presented at: The 2019 Military Health System Research Symposium, August 19–22, 2019, Kissimmee, FL
The 78th Annual Meeting of the American Association for the Surgery of Trauma and the Clinical Congress of Acute Care September 18–21, Dallas, TX, 2019.
Conflicts of Interest:
No competing financial interests exist.
References:
- 1.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. [DOI] [PubMed] [Google Scholar]
- 4.Ivanov II, Honda K Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12(4):496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491(7422):119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8(9):523–531. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, Iwakura Y, Oshima K, Morita H, Hattori M, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. [DOI] [PubMed] [Google Scholar]
- 8.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicholson SE, Burmeister DM, Johnson TR, Zou Y, Lai Z, Scroggins S, DeRosa M, Jonas RB, Merrill DR, Zhu C, et al. A prospective study in severely injured patients reveals an altered gut microbiome is associated with transfusion volume. J Trauma Acute Care Surg. 2019;86(4):573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pham TA, Lawley TD. Emerging insights on intestinal dysbiosis during bacterial infections. Curr Opin Microbiol. 2014;17:67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimizu K, Ogura H, Hamasaki T, Goto M, Tasaki O, Asahara T, Nomoto K, Morotomi M, Matsushima A, Kuwagata Y, et al. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Dig Dis Sci. 2011;56(4):1171–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moore FA, Sauaia A, Moore EE, Haenel JB, Burch JM, Lezotte DC. Postinjury multiple organ failure: a bimodal phenomenon. J Trauma. 1996;40(4):501–510; discussion 510–502. [DOI] [PubMed] [Google Scholar]
- 13.Papia G, McLellan BA, El-Helou P, Louie M, Rachlis A, Szalai JP, Simor AE. Infection in hospitalized trauma patients: incidence, risk factors, and complications. J Trauma. 1999;47(5):923–927. [DOI] [PubMed] [Google Scholar]
- 14.Howard BM, Kornblith LZ, Christie SA, Conroy AS, Nelson MF, Campion EM, Callcut RA, Calfee CS, Lamere BJ, Fadrosh DW, et al. Characterizing the gut microbiome in trauma: significant changes in microbial diversity occur early after severe injury. Trauma Surg Acute Care Open. 2017;2(1):e000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayakawa M, Asahara T, Henzan N, Murakami H, Yamamoto H, Mukai N, Minami Y, Sugano M, Kubota N, Uegaki S, et al. Dramatic changes of the gut flora immediately after severe and sudden insults. Dig Dis Sci. 2011;56(8):2361–2365. [DOI] [PubMed] [Google Scholar]
- 16.Earley ZM, Akhtar S, Green SJ, Naqib A, Khan O, Cannon AR, Hammer AM, Morris NL, Li X, Eberhardt JM, et al. Burn Injury Alters the Intestinal Microbiome and Increases Gut Permeability and Bacterial Translocation. PLoS One. 2015;10(7):e0129996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu K, Ogura H, Asahara T, Nomoto K, Matsushima A, Hayakawa K, Ikegawa H, Tasaki O, Kuwagata Y, Shimazu T. Gut microbiota and environment in patients with major burns - A preliminary report. Burns. 2014. [DOI] [PubMed]
- 18.Nicholson SE, Merrill D, Zhu C, Burmeister DM, Zou Y, Lai Z, Darlington DN, Lewis AM, Newton L, Scroggins S, et al. Polytrauma independent of therapeutic intervention alters the gastrointestinal microbiome. Am J Surg. 2018. [DOI] [PubMed]
- 19.Houlden A, Goldrick M, Brough D, Vizi ES, Lenart N, Martinecz B, Roberts IS, Denes A. Brain injury induces specific changes in the caecal microbiota of mice via altered autonomic activity and mucoprotein production. Brain Behav Immun. 2016;57:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang G, Sun K, Yin S, Jiang B, Chen Y, Gong Y, Chen Y, Yang Z, Chen J, Yuan Z, et al. Burn Injury Leads to Increase in Relative Abundance of Opportunistic Pathogens in the Rat Gastrointestinal Microbiome. Front Microbiol. 2017;8:1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson SE, Watts LT, Burmeister DM, Merrill D, Scroggins S, Zou Y, Lai Z, Grandhi R, Lewis AM, Newton LM, et al. Moderate Traumatic Brain Injury Alters the Gastrointestinal Microbiome in a Time-Dependent Manner. Shock. 2018. [DOI] [PubMed]
- 22.Waligora-Dupriet AJ, Lafleur S, Charrueau C, Choisy C, Cynober L, Butel MJ, Moinard C. Head injury profoundly affects gut microbiota homeostasis: Results of a pilot study. Nutrition. 2018;45:104–107. [DOI] [PubMed] [Google Scholar]
- 23.Kigerl KA, Hall JC, Wang L, Mo X, Yu Z, Popovich PG. Gut dysbiosis impairs recovery after spinal cord injury. J Exp Med. 2016;213(12):2603–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Cuesta-Zuluaga J, Kelley ST, Chen Y, Escobar JS, Mueller NT, Ley RE, McDonald D, Huang S, Swafford AD, Knight R, et al. Age- and Sex-Dependent Patterns of Gut Microbial Diversity in Human Adults. mSystems. 2019;4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alverdy JC. Microbiome Medicine: This Changes Everything. J Am Coll Surg. 2018;226(5):719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riscuta G, Xi D, Pierre-Victor D, Starke-Reed P, Khalsa J, Duffy L. Diet, Microbiome, and Epigenetics in the Era of Precision Medicine. Methods Mol Biol. 2018;1856:141–156. [DOI] [PubMed] [Google Scholar]
- 27.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. Isme j. 2011;5(2):169–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang HY, Chen JH, Chang JH, Lin HC, Lin CY, Peng CC. Multiple strains probiotics appear to be the most effective probiotics in the prevention of necrotizing enterocolitis and mortality: An updated meta-analysis. PLoS One. 2017;12(2):e0171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iizumi T, Battaglia T, Ruiz V, Perez Perez GI. Gut Microbiome and Antibiotics. Arch Med Res. 2017;48(8):727–734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Significant β-diversity plots according to Gender.
Supplemental Figure 2. Significant β-diversity plots according to Ventilator days and ARDS.
Supplemental Figure 3. Significant β-diversity plots according to hospital length of stay (LOS).
Supplemental Figure 4. Significant β-diversity plots according to ICU length of stay (LOS).
Supplemental Figure 5. Significant β-diversity plots according to infection.
Supplemental Figure 6. Significant changes in phylogeny within the phylum Firmicutes.
Supplemental Figure 7. Significant changes in phylogeny within the phylum Bacteroidetes.
Supplemental Figure 9. Significant changes in phylogeny within the phylum Proteobacteria.
Supplemental Figure 8. Significant changes in phylogeny within the phylum Actinobacteria.


