Abstract
The COVID-19 situation had escalated into an unprecedented global crisis in just a few weeks. On the 30th of January 2020, World Health Organization officially declared the COVID-19 epidemic as a public health emergency of international concern. The confirmed cases were reported to exceed 105,856,046 globally, with the death toll of above 2,311,048, according to the dashboard from Johns Hopkins University on the 7th of February, 2021, though the actual figures may be much higher. Conserved regions of the South Asian strains were used to construct a phylogenetic tree to find evolutionary relationships among the novel virus. Off target similarities were searched with other microorganisms that have been previously reported using Basic Local Alignment Search Tool (BLAST). The conserved regions did not match with any previously reported microorganisms or viruses, which confirmed the novelty of SARS-CoV-2. Currently there is no approved drug for the prevention and treatment of COVID-19, but researchers globally are attempting to come up with one or more soon. Therapeutic strategies need to be addressed urgently to combat COVID-19. Successful drug repurposing is a tool that uses old and safe drugs, is time effective and requires lower development costs, and was thus considered for the study. Molecular docking was used for repurposing drugs from our own comprehensive database of approximately 300 highly characterized, existing drugs with known safety profile, to identify compounds that will inhibit the chosen molecular targets - SARS-CoV-2, ACE2, and TMPRSS2. The study has identified and proposed twenty seven candidates for further in vitro and in vivo studies for the treatment of SARS-CoV-2 infection.
Keywords: SARS-CoV-2, COVID-19, Drug repurposing, Molecular modeling, ACE2, TMPRSS2, Lopinavir, Remdesivir, Pacritinib
SARS-CoV-2; COVID-19; Drug repurposing; Molecular modeling; ACE2; TMPRSS2; Lopinavir; Remdesivir; Pacritinib
1. Introduction
The coronavirus disease (COVID-19) pandemic has escalated into an unprecedented global crisis in just a few weeks. On the 30th of January 2020, World Health Organization (WHO) officially declared the COVID-19 epidemic as a public health emergency of international concern. The confirmed cases were reported to exceed 105,856,046 globally, with the death toll of above 2,311,048, according to the dashboard from Johns Hopkins University on the 7th of February, 2021, though the actual figures may be much higher. Data had initially shown a difference in the severity of corona virus outbreaks between countries in the global map that reacted quickly and decisively with social distancing and preventive measures such as Taiwan, Vietnam, etc. compared with those that acted late such as Iran, Italy, UK, etc. However, some countries had a larger outbreak after that. Scientists all over the world are working hard to explain why the virus has been behaving in such an extreme and unpredictable way. However, the structure of the virus does explain its rapid transmission [1].
Coronaviruses (CoVs) are enveloped viruses having a positive RNA genome. They belong to the Coronaviridae family of the order Nidovirales, which are of four genera (α, β, γ and δ). CoVs have been reported to contain at least four structural proteins - spike (S) protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein [2]. It has been further reported that the S protein is involved in the host attachment and virus-cell membrane fusion when infected with the virus. The types of the Severe Acute Respiratory Syndrome Coronavirus - SARS-CoV and SARS-CoV-2 - interface with the renin–angiotensin–aldosterone system (RAAS) through the angiotensin-converting enzyme 2 (ACE2), an enzyme that physiologically counters RAAS activation but also functions as a receptor for both the SARS viruses. This RAAS is a cascade of vasoactive peptides that are responsible for key processes in the human body. The interaction between the SARS viruses and ACE2 has been proposed as a potential factor in the way they affect the human body [2].
The SARS-CoV-2 that is responsible for the COVID-19 belongs to the β genus. It has been suggested that coronaviruses rely on the binding of the viral spike proteins to cellular receptors and on spike protein priming by host cell proteases to enter a cell. SARS-CoV-2 binds to the ACE2 receptor for entry and the serine protease TMPRSS2 for spike protein priming. Both of the key factors that mediate SARS-CoV-2 pathogenicity are highly expressed in urogenital organs, suggesting that these organs could also be susceptible to damage by this virus [3]. SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming. A drug already approved for clinical use, if can block the entry of the virus by acting as a TMPRSS2 inhibitor, could be considered a treatment option [4]. However, there are more proteins that will need to be considered for the holistic treatment of the disease caused by this deadly virus.
1.1. SARS-CoV-2 and ACE2
Coronaviruses are RNA viruses enveloped in a lipid bilayer, with each virus particle being a small set of genes, enclosed by a sphere of fatty lipid molecules resembling a spiky ball under the microscope. The spike protein S has two regions, S1 and S2, where S1 is for host cell receptor binding and S2 is for membrane fusion. Furthermore, the S1 region includes an N-terminal domain (NTD) and three C-terminal domains (CTD1, CTD2, and CTD3). The coronavirus attaches to the human host cells through the binding of its receptor binding domain (RBD) protein to the angiotensin-converting enzyme II (ACE2) found in the heart, lung, vessels, gut, kidney, testis and the brain [5, 6], resulting in the fusion of the viral membrane and the host cell membrane during infection. This is a known mechanism of entry of the coronavirus into the human body. Given the high homology between SARS-CoV and SARS-CoV-2, it is expected that SARS-CoV-2 would also use the ACE2 molecule as the receptor to enter human cells. However, what is new for the SARS-CoV-2 is that it has undergone mutations that allow the virus to bind more strongly with ACE2. Therefore, its ability to infect people has become much higher than the classical SARS-CoV [7].
The first viral protein created inside the infected cell is a chain of 16 proteins joined together. Two of these proteins act like scissors, snipping the links between the different proteins and freeing them to do their jobs. The interaction between the RBD of this virus, located in the CTD1 of the S1 region of the S protein, and ACE2 of the human host is a prerequisite for the human infection with SARS-CoV. Although it has been clear that SARS-CoV-2 infects human cells through the binding of its RBD domain with the human ACE2 receptor, the molecular mechanism of the binding between the RBD protein and the ACE2 receptor is still unknown [8]. The brain has been reported to express ACE2 receptors, which makes them a potential target of COVID-19. Patients with acute SARS-CoV-2 illness have also demonstrated the presence of the virus in the cerebrospinal fluid [9].
1.2. SARS-CoV-2 and TMPRSS2
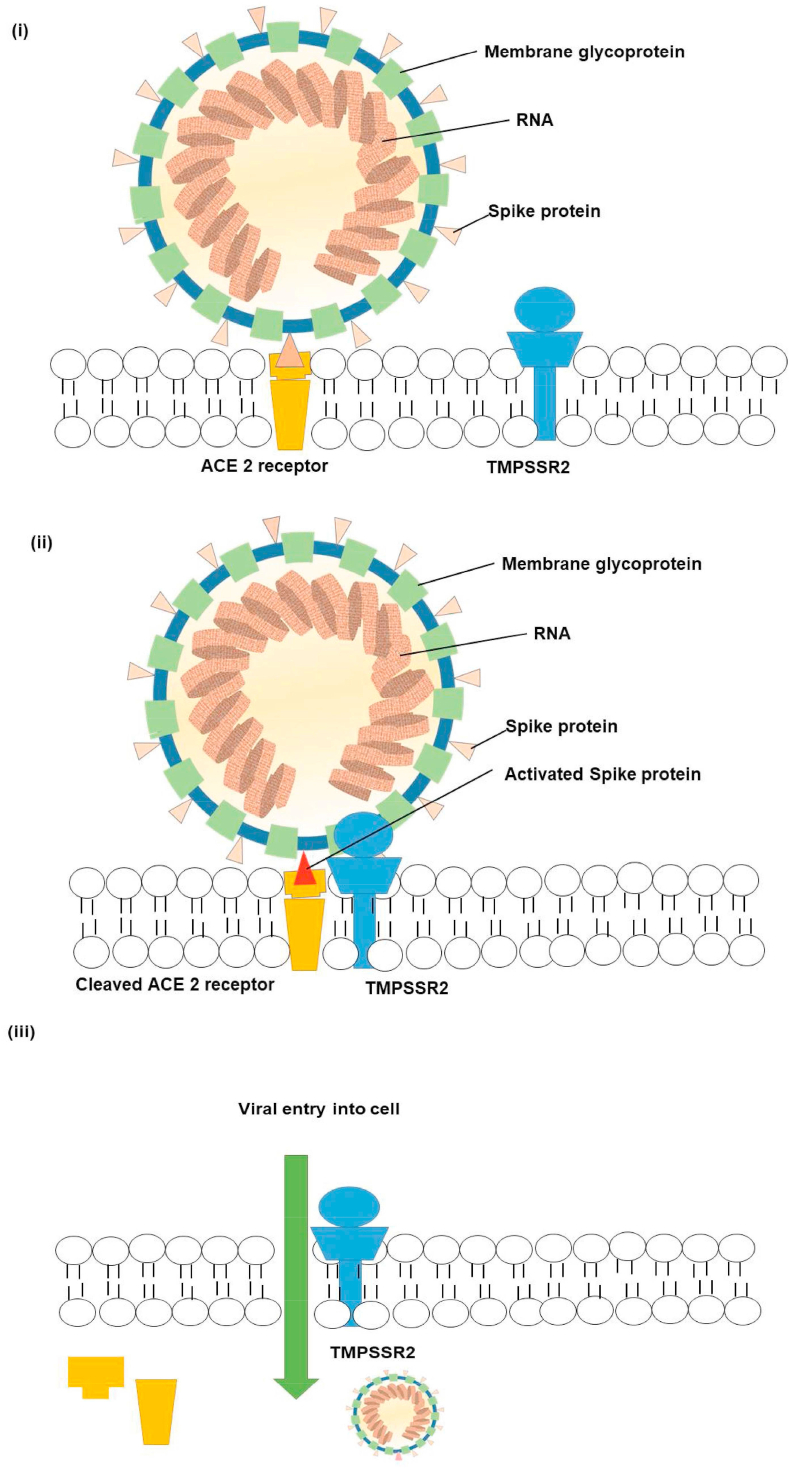
As already mentioned, the first step of the process of the viral entry is the binding of the N-terminal portion of the viral protein unit S1 to the ACE2 receptor. The second step, which is significant for viral entry, is the protein cleavage between the S1 and S2 units. The receptor transmembrane protease serine 2 (TMPRSS2), a member of the Hepsin/TMPRSS subfamily, is responsible for the second step [5, 10], as shown in Figure 1.
Figure 1.
Mechanism of viral entry.
The strategy assumed in the current study is that if either or both of these two factors can be blocked by a clinically proven inhibitor, it might help to propose options for prevention and treatment of the disease caused by this novel virus [11] since the cell entry of coronaviruses depends on the binding of the viral spike (S) proteins to cellular receptors, followed by the S protein priming by host cell proteases. The current study considers the assumption that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming [11]. These two receptors have been thus considered for the current in silico study.
1.3. Phylogenetic tree
A phylogenetic tree was constructed for the virus in South Asia to find evolutionary relationships among the novel virus and these phylogenetic trees are used to guide the sampling of taxa for comparative research. There are approximately 1.7 million identified species, which is just a fraction of the total number of species that exist. Approximately 80,000 of these species have been analyzed for evolutionary relationships and assigned into a hierarchy called the phylogenetic tree [13]. Evolutionary biologists use phylogenetic trees to conceptualize, visualize, and analyze the relationships among the biological lineages [14]. Ideally, all the species recognized have a place in the phylogenetic tree. The study of these tree data structures and representations, essential in biological research, display evolutionary relationships between different species and how they have evolved over time. These so-called tree of life show the hierarchical organizations of biological data and concepts, where some of the most promising efforts for hierarchical representations are the Gene Ontology (GO) [15]. GO describes the functional annotation of genes by a hierarchically organized set of terms and phrases, and UMLS (Unified Medical Language System) that has a biomedical focus. These representations are also useful in the classification and the clustering visualization of biological data [16].
1.4. Drug repurposing
Drug repurposing aims to identify multiple targets of already existing drugs. It is considered an important branch in drug discovery as it helps to bypass optimization issues related to drug discovery and development and preclinical development. Repositioning of already approved drugs can be cheaper and faster, since the pharmacological and toxicological data of the existing drugs are available. It not only makes use of previous investments but also minimizes clinical activities, thus reducing expenses, time efforts, and failures that are typically related to the drug discovery process [17, 18, 19]. Furthermore, it provides an understanding of the drug-target interaction matrix. In silico methods have the advantage of using high-performance computers for a safe virtual screening. Virtual screening can be vital in large-scale production of drugs, working with large number of data sets. In silico molecular docking, part of computational biology, is used for drug repurposing. Docking predicts the preferred orientation of one molecule to a second one when binding with each other to form a stable complex [20]. This method also helps find the gaps in the drug-target interaction matrix, providing safety and efficacy data [21, 22]. Research into repositioning known drugs to treat diseases other than the originally intended disease continues to grow and develop, encouraged in part, by several recent success stories. Drugs could have many off-target effects and can be intelligently repurposed if the off-target effects can be employed for therapeutic purposes, in times of crisis such as the current COVID pandemic.
1.5. Potential candidates for drug repurposing
SARS-CoV-2 infection depends on the host cell factors ACE2 and TMPRSS2 and may be blocked by a clinically proven protease inhibitor. These findings were used to help establish options for the treatment and prevention of the current COVID-19 pandemic [11]. Currently there is no approved vaccine or drug for the prevention and treatment of COVID-19, but researchers globally are attempting to come up with one or more by the end of the year. Antiviral drugs used in influenza such as remdesivir and a combination of anti-HIV drugs lopinavir-ritonavir are being reported to be promising drug candidates in the treatment of COVID-19 [22]. Corticosteroid (anti-inflammatory drug) treatment is not routinely recommended to treat SARS-CoV-related pneumonia. However, according to new pathological findings of pulmonary edema and hyaline membrane formation, timely and appropriate use of corticosteroids together with ventilator support may be considered for the severe COVID-19 patients to prevent acute respiratory distress syndrome (ARDS) development, which is thought to be majorly associated with COVID-19 related mortality.
The development of vaccines is also being done at an accelerated rate. There is a pressing urgency to accelerate therapeutics for COVID-19 patients worldwide since the world is facing an unprecedented and growing challenge with increasing numbers of cases and of deaths due to the COVID-19 pandemic. To navigate the immediate impacts of this unprecedented crisis, short-term and immediate solutions are also imperative. In such situations, drug repurposing is a rapid and economic approach of leveraging existing drugs that can be used in a pandemic such as the COVID-19 pandemic. The reference drugs chosen for the SARS-CoV-2 protein were remdesivir and lopinavir [22], for ACE2 protein benazepril and lisinopril [23] and for TMPRSS2 protein camostat mesylate and gabexate mesylate. Camostat mesylate and gabexate mesylate were taken since they are known inhibitors of the protein TMPRSS2 [24]. For this study, an exhaustive list of commonly used anticoagulants, antiplatelets, anti-viral drugs, anti-inflammatory drugs, antifungals, antibiotics, antihypertensives, antidiabetic drugs, statins, janus kinase inhibitors, natural molecules and glycogen synthase kinase inhibitors were taken from our drug database and systematically screened. Most of these drugs are currently in the market, as well as undergoing clinical trials for SARS-CoV-2. The aim of the current study was to propose candidates for further in vitro and in vivo studies as strategies to treat this epidemic SARS-CoV-2 infection that may be effective in treating people exposed to COVID-19.
2. Materials and method
2.1. Sequence retrieval from NCBI
Coding sequences from 635 genomes of SARS-CoV-2 from different regions, including the reference sequence from the first novel corona virus (NC_045512.2) were retrieved from NCBI Virus [25] (Table 1). Nucleocapsid phosphoprotein and surface glycoprotein sequences were manually extracted and curated from the retrieved data [26].
Table 1.
Conserved sequences in South Asian Virus Sequences (Obtained from NCBI).
| Conserved Region | Sequences |
|---|---|
| Conserved region 1 | ATTAAAGGTTTATACCTTCCCAGGTAACAAACCAACCAACTT TCGATCTCTTGTAGATCTGTTCTCTAAACGAACTTTAAAATCT GTGTGGCTGTCACTCGGCTGCATGCTTAGTGCACTCACGCAG TATAATTAATAACTAATTACTGTCGTTGACAGGACACGAGTA ACTCGTCTATCTTCTGCAGGCTGCTTACGGTTTCGTCCGTGTT GCAGCCGATCATCAGCACATCTAGGTTT |
| Conserved region 2 | GTCCGGGTGTGACCGAAAGGTAAGATGGAGAGCCTTGTCCC TGGTTTCAACGAGAAAACACACGTCCAACTCAGTTTGCCTGT TTTACAGGTTCGCGACGTGCTCGTACGTGGCTTTGGAGACTC CGTGGAGGAGGTCTTATCAGAGGCACGTCAACATCTTAAAG ATGGCACTTGTGGCTTAGTAGAAGTTGAAAAAGGCGTTTTGC CTCAACTTGAACAGCCCTATGTGTTCATCAAACGTTCGGATG CTCGAACTGCACCTCATGGTCATGTTATGGTTGAGCTGGTAG CAGAACTCGAAGGCATTCAGTACGGTCGTAGTGGTGAGACA CTTGGTGTCCTTGTCCCTCATGTGGGCGAAATACCAGTGGCT TACCGCAAGGTTCTTCTTCGTAAGAACGGTAATAAAGGAGCT GGTGGCCATAGTTACGGCGCCGATCTAAAGTCATTTGACTTA GGCGACGAGCTTGGCACTGATCCTTATGAAGATTTTCAAGAA AACTGGAACACTAAACATAGCAGTGGTGTTACCCGTGAACTC ATGCGTGAGCTTAACGGAGGGGCATACACTCGCTATGTCGAT AACAACTTCTGTGGCCCTGATGGCTACCCTCTTGAGTGCATT AAAGACCTTCTAGCACGTGCTGGTAAAGCTTCATGCACTTTG TCCGAACAACTGGACTTTATTGACACTAAGAGGGGTGTATAC TGCTGCCGTGAACATGAGCATGAAATTGCTTGGTACACGGAA CGTTCTGAAAAGAGCTATGAATTGCAGACACCTTTTGAAATT AAATTGGCAAAGAAATTTGACACCTTCAATGGGGAATGTCCA AATTTTGTATTTCCCTTAAATTCCATAATCAAGACTATTCAAC CAAGGGTTGAAAAGAAAAAGCTTGATGGCTTTATGGGTAGAA TTCGATCTGTCTATCCAGTTGCGTCACCAAATGAATGCAACC AAATGTGCCTTTCAACTCTCATGAAGTGTGATCATTGTGGTGA AACTTCATGGCAGACGGGCGATTTTGTTAAAGCCACTTGCGAA TTTTGTGGCACTGAGAATTTGACTAAAGAAGGTGCCACTACTT GTGGTTACTTACCCCAAAATGCTGTTGTTAAAATTTATTGTCCA GCATGTCACAATTCAGAAGTAGGACCTGAGCATAGTCTTGCCGA ATACCATAATGAATCTGGCTTGAAAACCATTCTTCGTAAGGGTGG TCGCACTATTGCCTTTGGAGGCTGTGTGTTCTCTTATGTTGGTTGC CATAACAAGTGTGCCTATTGGGTTCCACGTGCTAGCGCTAACATA GGTTGTAACCATACAGGTGTTGTTGGAGAAGGTTCCGAAGGTCTTA ATGACAACCTTCTTGAAATACTCCAAAAAGAGAAAGTCAACATCA ATATTGTTGGTGACTTTAAACTTAATGAAGAGATCGCCATT |
| Conserved region 3 | TTTTGGCATCTTTTTCTGCTTCCACAAGTGCTTTTGTGGAAAC TGTGAAAGGTTTGGATTATAAAGCATTCAAACAAATTGTTGA ATCCTGTGGTAATTTTAAAGTTACAAAAGGAAAAGCTAAAAA AGGTGCCTGGAATATTGGTGAACAGAAATCAATACTGAGTCC TCTTTATGCATTTGCATCAGAGGCTGCTCGTGTTGTACGATC AATTTTCTCCCGCACTCTTGAAACTGCTCAAAATTCTGTGCG TGTTTTACAGAAGGCCGCTATAACAATACTAGATGGAATTTC ACAGTATTCACTGAGACTCATTGATGCTATGATGTTCACATC TGATTTGGCTACTAACAATCTAGTTGTAATGGCCTACATTAC AGGTGGTGTTGTTCAGTTGACTTCGCAGTGGCTAACTAACAT CTTTGGCACTGTTTATGAAAAACTCAAACCCGTCCTTGATTG GCTTGAAGAGAAGTTTAAGGAAGGTGTAGAGTTTCTTAGAGA CGGTTGGGAAATTGTTAAATTTATCTCAACCTGTGCTTGTGA AATTGTCGGTGGACAAATTGTCACCTGTGCAAAGGAAATTAA GGAGAGTGTTCAGACATTCTTTAAGCTTGTAAATAAATTTTT GGCTTTGTGTGCTGACTCTATCATTATTGGTGGAGCTAAACT TAAAGCCTTGAATTTAGGTGAAACATTTGTCACGCACTCAAA GGGATTGTACAGAAAGTGTGTTAAATCCAGAGAAGAAACTGG CCTACTCATGCCTCTAAAAGCCCCAAAAGAAATTATCTTCTT AGAGGGAGAAACACTTCCCACAGAAGTGTTAACAGAGGAAGT TGTCTTGAAAACTGGTGATTTACAACCATTAGAACAACCTAC TAGTGAAGCTGTTGAAGCTCCATTGGTTGGTACACCAGTTTG TATTAACGGGCTTATGTTGCTCGAAATCAAAGACACAGAAAA GTACTGTGCCCTTGCACCTAATATGATGGTAACAAACAATAC CTTCACACTCAAAGGCGGTGCACCAACAAAGGTTACTTTTGG TGATGACACTGTGATAGAAGTGCAAGGTTACAAGAGTGTGAA TATCACTTTTGAACTTGATGAAAGGATTGATAAAGTACTTAA TGAGAAGTGCTCTGCCTATACAGTTGAACTCGGTACAGAAGT AAATGAGTTCGCCTGTGTTGTGGCAGATGCTGTCATAAAAAC TTTGCAACCAGTATCTGAATTACTTACACCACTGGGCATTGA TTTAGATGAGTGGAGTATGGCTACATACTACTTATTTGATGA GTCTGGTGAGTTTAAATTGGCTTCACATATGTATTGTTCTTT CTACCCTCCAGATGAGGATGAAGAAGAAGGTGATTGTGAAGA AGAAGAGTTTGAGCCATCAACTCAATATGAGTATGGTACTGA AGATGATTACCAAGGTAAACCTTTGGAATTTGGTGCCACTTC TGCTGCTCTTCAACCTGAAGAAGAGCAAGAAGAAGATTGGTT AGATGATGATAGTCAACAAACTGTTGGTCAACAAGACGGCAG TGAGGACAATCAGACAACTACTATTCAAACAATTGTTGAGGT TCAACCTCAATTAGAGATGGAACTTACACCAGTTGTTCAGAC TATTGAAGTGAATAGTTTTAGTGGTTATTTAAAACTTACTGA CAATGTATACATTAAAAATGCAGACATTGTGGAAGAAGCTAA AAAGGTAAAACCAACAGTGGTTGTTAATGCAGCCAATGTTTA CCTTAAACATGGAGGAGGTGTTGCAGGAGCCTTAAATAAGGC TACTAACAATGCCATGCAAGTTGAATCTGATGATTACATAGC TACTAATGGACCACTTAAAGTGGGTGGTAGTTGTGTTTTAAG CGGACACAATCTTGCTAAACACTGTCTTCATGTTGTCGGCCC AAATGTTAACAAAGGTGAAGACATTCAACTTCTTAAGAGTGC TTATGAAAATTTTAATCAGCACGAAGTTCTACTTGCACCATT ATTATCAGCTGGTATTTTTGGTGCTGACCCTATACATTCTTT AAGAGTTTGTGTAGATACTGTTCGCACAAATGTCTACTTAGC TGTCTTTGATAAAAATCTCTATGACAAACTTGTTTCAAGCTT TTTGGAAATGAAGAGTGAAAAGCAAGTTGAACAAAAGATCGC TGAGATTCCTAAAGAGGAAGTTAAGCCATTTATAACTGAAAG TAAACCTTCAGTTGAACAGAGAAAACAAGATGATAAGAAAAT CAAAGCTTGTGTTGAAGAAGTTACAACAACTCTGGAAGAAAC TAAGTTCCTCACAGAAAACTTGTTACTTTATATTGACATTAA TGGCAATCTTCATCCAGATTCTGCCACTCTTGTTAGTGACAT TGACATCACTTTCTTAAAGAAAGATGCTCCATATATAGTGGG TGATGTTGTTCAAGAGGGTGTTTTAACTGCTGTGGTTATACC TACTAAAAAGGCTGGTGGCACTACTGAAATGCTAGCGAAAGC TTTGAGAAAAGTGCCAACAGACAATTATATAACCACTTACCC GGGTCAGGGTTTAAATGGTTACACTGTAGAGGAGGCAAAGAC AGTGCTTAAAAAGTGTAAAAGTGCCTTTTACATTCTACCATC TATTATCTCTAATGAGAAGCAAGAAATTCTTGGAACTGTTTC TTGGAATTTGCGAGAAATGCTTGCACATGCAGAAGAAACACG CAAATTAATGCCTGTCTGTGTGGAAACTAAAGCCATAGTTTC AACTATACAGCGTAAATATAAGGGTATTAAAATACAAGAGGG TGTGGTTGATTATGGTGCTAGATTTTACTTTTACACCAGTAA AACAACTGTAGCGTCACTTATCAACACACTTAACGATCTAAA TGAAACTCTTGTTACAATGCCACTTGGCTATGTAACACATGG CTTAAATTTGGAAGAAGCTGCTCGGTATATGAGATCTCTCAA AGTGCCAGCTACAGTTTCTGTTTCTTCACCTGATGCTGTTAC AGCGTATAATGGTTATCTTACTTCTTCTTCTAAAACACCTGA AGAACATTTTATTGAAACCATCTCACTTGCTGGTTCCTATAA AGATTGGTCCTATTCTGGACAATCTACACAACTAGGTATAGA ATTTCTTAAGAGAGGTGATAAAAGTGTATATTACACTAGTAA TCCTACCACATTCCACCTAGATGGTGAAGTTATCACCTTTGA CAATCTTAAGACACTTCTTTCTTTGAGAGAAGTGAGGACTAT TAAGGTGTTTACAACAGTAGACAACATTAACCTCCACACGCA AGTTGTGGACATGTCAATGACATATGGACAACAGTTTGGTCC AACTTATTTGGATGGAGCTGATGTTACTAAAATAAAACCTCA TAATTCACATGAAGGTAAAACATTTTATGTTTTACCTAATGA TGACACTCTACGTGTTGAGGCTTTTGAGTACTACCACACAAC TGATCCTAGTTTTCTGGGTAGGTACATGTCAGCATTAAATCA CACTAAAAAGTGGAAATACCCACAAGTTAATGGTTTAACTTC TATTAAATGGGCAGATAACAACTGTTATCTTGCCACTGCATT GTTAACACTCCAACAAATAGAGTTGAAGTTTAATCCACCTGC TCTACAAGATGCTTATTACAGAGCAAGGGCTGGTGAAGCTGC TAACTTTTGTGCACTTATCTTAGCCTACTGTAATAAGACAGT AGGTGAGTTAGGTGATGTTAGAGAAACAATGAGTTACTTGTT TCAACATGCCAATTTAGATTCTTGCAAAAGAGTCTTGAACGT GGTGTGTAAAACTTGTGGACAACAGCAGACAACCCTTAAGGG TGTAGAAGCTGTTATGTACATGGGCACACTTTCTTATGAACA ATTTAAGAAAGGTGTTCAGATACCTTGTACGTGTGGTAAACA AGCTACAAAATATCTAGTACAACAGGAGTCACCTTTTGTTAT GATGTCAGCACCACCTGCTCAGTATGAACTTAAGCATGGTAC ATTTACTTGTGCTAGTGAGTACACTGGTAATTACCAGTGTGG TCACTATAAACATATAACTTCTAAAGAAACTTTGTATTGCAT AGACGGTGCTTTACTTACAAAGTCCTCAGAATACAAAGGTCC TATTACGGATGTTTTCTACAAAGAAAACAGTTACACAACAAC CATAAAACCAGTTACTTATAAATTGGATGGTGTTGTTTGTAC AGAAATTGACCCTAAGTTGGACAATTATTATAAGAAAGACAA TTCTTATTTCACAGAGCAACCAATTGATCTTGTACCAAACCA ACCATATCCAAACGCAAGCTTCGATAATTTTAAGTTTGTATG TGATAATATCAAATTTGCTGATGATTTAAACCAGTTAACTGG TTATAAGAAACCTGCTTCAAGAGAGCTTAAAGTTACATTTTT CCCTGACTTAAATGGTGATGTGGTGGCTATTGATTATAAACA CTACACACCCTCTTTTAAGAAAGGAGCTAAATTGTTACATAA ACCTATTGTTTGGCATGTTAACAATGCAACTAATAAAGCCAC GTATAAACCAAATACCTGGTGTATACGTTGTCTTTGGAGCAC AAAACCAGTTGAAACATCAAATTCGTTTGATGTACTGAAGTC AGAGGACGCGCAGGGAATGGATAATCTTGCCTGCGAAGATCT AAAACCAGTCTCTGAAGAAGTAGTGGAAAATCCTACCATACA GAAAGACGTTCTTGAGTGTAATGTGAA |
| Conserved region 4 | ACTACCGAAGTTGTAGGAGACATTATACTTAAAC |
| Conserved region 5 | AGCAAATAATAGTTTAAAAATTACAGAAGAGGTTGGCCACAC AGATCTAATGGCTGCTTATGTAGACAATTCTAGTCTTACTAT TAAGAAACCTAATGAATTATCTAGAGTATTAGGTTTGAAAAC CCTTGCTACTCATGGTTTAGCTGCTGTTAATAGTGTCCCTTG GGATACTATAGCTAATTATGCTAAGCCTTTTCTTAACAAAGT TGTTAGTACAACTACTAACATAGTTACACGGTGTTTAAACCG TGTTTGTACTAATTATATGCCTTATTTCTTTACTTTATTGCT ACAATTGTGTACTTTTACTAGAAGTACAAATTCTAGAATTAA AGCATCTATGCCGACTACTATAGCAAAGAATACTGTTAAGAG TGTCGGTAAATTTTGTCTAGAGGCTTCATTTAATTATTTGAA GTCACCTAATTTTTCTAAACTGATAAATATTATAATTTGGTT TTTACTATTAAGTGTTTGCCTAGGTTCTTTAATCTACTCAAC CGCTGCTTTAGGTGTTTTAATGTCTAATTTAGGCATGCCTTC TTACTGTACTGGTTACAGAGAAGGCTATTTGAACTCTACTAA TGTCACTATTGCAACCTACTGTACTGGTTCTATACCTTGTAG TGTTTGTCTTAGTGGTTTAGATTCTTTAGACACCTATCCTTC TTTAGAAACTATACAAATTACCATTTCATCTTTTAAATGGGA TTTAACTGCTTTTGGCTTAGTTGCAGAGTGGTTTTTGGCATA TATTCTTTTCACTAGGTTTTTCTATGTACTTGGATTGGCTGC AATCATGCAATTGTTTTTCAGCTATTTTGCAGTACATTTTAT TAGTAATTCTTGGCTTATGTGGTTAATAATTAATCTTGTACA AATGGCCCCGATTTCAGCTATGGTTAGAATGTACATCTTCTT TGCATCATTTTATTATGTATGGAAAAGTTATGTGCATGTTGT AGACGGTTGTAATTCATCAACTTGTATGATGTGTTACAAACG TAATAGAGCAACAAGAGTCGAATGTACAACTATTGTTAATGG TGTTAGAAGGTCCTTTTATGTCTATGCTAATGGAGGTAAAGG CTTTTGCAAACTACACAATTGGAATTGTGTTAATTGTGATAC ATTCTGTGCTGGTAGTACATTTATTAGTGATGAAGTTGCGAG AGACTTGTCACTACAGTTTAAAAGACCAATAAATCCTACTGA CCAGTCTTCTTACATCGTTGATAGTGTTACAGTGAAGAATGG TTCCATCCATCTTTACTTTGATAAAGCTGGTCAAAAGACTTA TGAAAGACATTCTCTCTCTCATTTTGTTAACTTAGACAACCT GAGAGCTAATAACACTAAAGGTTCATTGCCTATTAATGTTAT AGTTTTTGATGGTAAATCAAAATGTGAAGAATCATCTGCAAA ATCAGCGTCTGTTTACTACAGTCAGCTTATGTGTCAACCTAT ACTGTTACTAGATCAGGCATTAGTGTCTGATGTTGGTGATAG TGCGGAAGTTGCAGTTAAAATGTTTGATGCTTACGTTAATAC GTTTTCATCAACTTTTAACGTACCAATGGAAAAACTCAAAAC ACTAGTTGCAACTGCAGAAGCTGAACTTGCAAAGAATGTGTC CTTAGACAATGTCTTATCTACTTTTATTTCAGCAGCTCGGCA AGGGTTTGTTGATTCAGATGTAGAAACTAAAGATGTTGTTGA ATGTCTTAAATTGTCACATCAATCTGACATAGAAGTTACTGG CGATAGTTGTAATAACTATATGCTCACCTATAACAAAGTTGA AAACATGACACCCCGTGACCTTGGTGCTTGTATTGACTGTAG TGCGCGTCATATTAATGCGCAGGTAGCAAAAAGTCACAACAT TGCTTTGATATGGAACGTTAAAGATTTCATGTCATTGTCTGA ACAACTACGAAAACAAATACGTAGTGCTGCTAAAAAGAATAA CTTACCTTTTAAGTTGACATGTGCAACTACTAGACAAGTTGT TAATGTTGTAACAACAAAGATAGCACTTAAGGGTGGTAAAAT TGTTAATAATTGGTTGAAGCAGTTAATTAAAGTTACACTTGT GTTCCTTTTTGTTGCTGCTATTTTCTATTTAATAACACCTGT TCATGTCATGTCTAAACATACTGACTTTTCAAGTGAAATCAT AGGATACAAGGCTATTGATGGTGGTGTCACTCGTGACATAGC ATCTACAGATACTTGTTTTGCTAACAAACATGCTGATTTTGA CACATGGTTTAG |
2.2. Multiple sequence alignment & phylogenetic tree construction
ClustalW [27] algorithm was employed to perform multiple sequence alignment. Maximum likelihood phylogenetic trees were constructed with a bootstrap value of 500. Tamura Nei model of evolution was selected while constructing the phylogenetic tree. Molecular Evolutionary Genetics Analysis (MEGAX) program was used for alignment formation and phylogenetic tree construction. The complete sequences of 635 SARS-CoV-2 from different regions, including the reference sequence from the first coronavirus in Wuhan, were retrieved from NCBI. Multiple Sequence Alignment (MSA) was then carried out with the different sequences using MEGAX [28], and the conserved regions identified through MSA using ClustalW. Using the MEGA file, a phylogenetic tree was then constructed, where South Asia was targeted to find a conserved region within the virus. The full structures of the viruses that were reported in India, Pakistan and Nepal were taken, and accordingly the sequences from South Asia were used to find the conserved regions within the virus. A phylogenetic tree was thus constructed for the virus in South Asia to find evolutionary relationships among the novel virus. The search for off target similarities with other microorganisms has been reported using Basic Local Alignment Search Tool (BLAST). The aim was to find microorganisms with 99%–100% similarity, and if the conserved regions matched with any previously reported microorganisms or viruses, treatment options could easily be suggested for the novel corona virus.
2.3. Off target similarity search using BLAST
BLAST search was performed against human genome and transcriptome using the standalone blast package [29] to identify the possible off target matches. The e-value was set to reduce the stringency of the search condition thereby increasing the chances of random matches.
2.4. Selection of proteins and drugs for computational studies
Based on literature review from articles in authentic journals, a number of reference drugs for treating the coronavirus were chosen. In this study, both the virus proteins as well as two human proteins were targeted to propose drug candidates that may be helpful in the treatment of COVID-19 patients. For SARS-CoV-2 protein, the reference drugs taken were remdesivir and lopinavir. These drugs were suggested to inhibit the protein, and thus useful as starting points and for the validation of the results obtained. They also assisted in predicting whether the proposed candidates worked in a similar mechanism.
The study was limited to a number of commonly used drugs that are either approved or of natural origin, belonging to different classes of drugs such as antidiabetic, antiviral, anti-inflammatory, corticosteroids, opioid, antihistamine, statins, cholesterol lowering drugs, janus kinase inhibitors, anticoagulants, anti-platelets, component of ginger, garlic, flavonoid, antibiotics, corticosteroids, glycogen synthase kinase 3 inhibitor, natural compounds such as shikonin, component of black cumin, plant sterol, antifungal. However, only a few were finally selected after considering their binding affinity with the three proteins (SARS-CoV-2, ACE-2 and TMPRSS2), followed by superimposition of reference and candidate drugs with the proteins, common amino acids while binding with the reference drugs, and finally evaluation of the pharmacokinetic properties.
The study also aimed to inhibit the proteins, ACE2 and TMPRSS2, with the assumption to prevent viral entry. For ACE2 receptor, benazepril and lisinopril were taken as the reference drugs to repurpose the drugs in the treatment of COVID-19 patients. The ACE2 receptor protein was selected as the spike proteins on the surface of the virus bind with the ACE2 receptors on the surface of the target cell. In response, the type II transmembrane serine protease (TMPRSS2) binds and cleaves the ACE2 receptor. In the process, the spike protein is activated, and the cleaved ACE2 and activated spike protein facilitate entry of the coronavirus [12]. The reference drugs chosen for inhibiting TMPRSS2 were camostat mesylate and gabexate mesylate.
The different classes of drugs used were retrieved from PubChem to carry out the in silico study. The main objective was to determine whether they had the potential to inhibit the viral protein and the two human proteins.
2.5. Software and online tools
Several software were used for the computational methods during this in silico study, as shown in Table 2. It is also worth mentioning that a small number of databases, such as RCSB-PDB (Protein Data Bank), PubChem Project, EMBL-EBI (European Molecular Biology Laboratory-European Bioinformatics Institute), were used to accumulate data which enhanced the reliability and validation of this study.
Table 2.
Software and online tools.
2.6. In silico study
The SARS-CoV-2 protein was retrieved from RCSB PDB (PDB ID: 6M17). The resolution of the protein was 2.9Å and the expression system was Homo sapiens. The protein was curated using PyMOL by removing the native ligands and heteroatoms. Chain E of the protein, which consisted of the SARS-CoV-2 Receptor Binding Domain, was kept intact.
The ACE2 receptor protein was also obtained from RCSB PDB (PDB ID: 6CS2). Chain D, which consisted of the ACE2 enzyme, was selected for further curation.
The structure of the protein, TMPRSS2 was not available in the protein data banks, and thus the FASTA sequence from NCBI was retrieved and modeled using Swiss-Model Interactive Space. Three structures were generated, but only the one that passed the validation tests using ERRAT [36] and Verify 3D [37, 38] was taken for further docking studies.
The different classes of drugs were then obtained from PubChem in their 3D sdf format and converted to pdb format using Open Babel. The 3D structure of the drugs not available in PubChem were converted to 3D using Avogadro and optimized.
Molecular docking was next carried out using AutoDock Vina and PyRx. The drugs and natural molecules that demonstrated strong binding affinities were selected for further studies. A more negative value (or a lower value) indicated a stronger affinity of the drug or the natural molecule towards the protein, and the next step was to check whether these drugs superimposed with the reference drugs. This was done using PyMOL, visualization software, and the superimposed drugs further screened to determine their antagonistic effect.
Discovery Studio was used to provide a representation of the actual distance between the ligands with their bonding types and the amino acids of the protein (non-bonding interactions). Interactions of amino acids were investigated to find out the amino acids involved in the binding of the drugs with the protein of interest, that is, the protein-ligand interactions were investigated. These were then compared with the reference drugs to check for the amino acid similarities. The ADME properties of the potential inhibitors were then evaluated using QikProp [35].
The drug molecules and natural compounds that passed all the above-mentioned steps were shortlisted and considered, while the rest were not taken.
3. Results
3.1. Conserved regions
From the Multiple Sequence Alignment using MEGAX, fourteen conserved regions were found within the viral protein from South Asia (Table 1). The conserved regions were searched for off-target similarities with other microorganisms previously reported using BLAST).
In Figure 2, each sequence ID referred to the SARS-CoV-2 species and each South Asian country denoted different geometrical shapes and colors (from top to bottom). It is also clear that the branches denoting SARS-CoV-2 from Wuhan, China and Rajkot, India are closely related with each other and have the same branch point which indicated the same ancestor. The top branch consisting of SARS-CoV-2 from Wuhan, China and Rajkot, India is also closely related with SARS-CoV-2 from Italy. The bottom branch of SARS-CoV-2 from Italy and Kerala, India having the same branch point indicated that the virus may have migrated from Italy to India [12]. On the other hand, the tree also shows that the SARS-CoV-2 from Pakistan and Nepal are closely related.
Figure 2.
Phylogenetic tree of SARS-CoV-2 in South Asia [39].
3.2. In Silico Binding Affinity Values and Non-Bonded Protein Ligand Interactions with the SARS-CoV-2 Protein
Molecular docking was carried out with the viral protein. The reference drugs were chosen based on the stages of clinical trials the drugs were in at the time. These included remdesivir, lopinavir, favipiravir, danoprevir, chloroquine, hydroxychloroquine and methyl prednisolone. Based on the binding affinity, remdesivir and lopinavir were selected as the reference drugs.
As shown in Table 3, the binding affinity of the reference drug remdesivir was found to be -7.8 kcal/mol and binding affinity of the proposed drug, naloxegol was found to be -7.5 kcal/mol. The binding affinity of the reference drug lopinavir was also found to be -8.8 kcal/mol and the binding affinity of the corresponding proposed drug, gemigliptin was found to be -7.7 kcal/mol.
Table 3.
Binding affinity values of reference drugs and selected drugs with the viral protein.
| Drugs | Binding Affinities (kcal/mol) |
|---|---|
| Remdesivir (Reference drug) | -7.8 |
| Naloxegol (Opioid) | -7.5 |
| Lopinavir (Reference drug) | -8.8 |
| Gemigliptin (Antidiabetic drug) | -7.7 |
For the viral protein, the drugs with the highest binding affinities were superimposed with the chosen reference drug, remdesivir. Naloxegol was found to superimpose with remdesivir. On the other hand, gemigliptin superimposed with lopinavir. The protein-ligand interactions were visualized using Discovery Studio, and the results are shown in Table 4. Naloxegol showed to have two common amino acids (ARG355, LYS462) with remdesivir, and gemigliptin had three common amino acids (LEU517, LEU390, VAL382) with lopinavir.
Table 4.
Common amino acids involved in the viral protein-ligand interaction.
| Drugs | Amino acids |
|---|---|
| Remdesivir (Reference drug) | ARG355, PHE464, PRO426, PHE429, LYS462 |
| Naloxegol (Opioid) | ARG355, LYS462 |
| Lopinavir (Reference drug) | LEU517, CYS391, LEU390,VAL382 |
| Gemigliptin (Antidiabetic drug) | LEU517, LEU390,VAL382 |
3.3. In Silico Binding Affinity Values and Non-Bonded Protein Ligand Interactions with the ACE2 receptor
Benazepril, trandolapril and lisinopril were initially taken as reference drugs for docking. However, benazepril and lisinopril were only later taken based on strong protein-ligand interactions. The binding affinities of the two reference drugs (ACE inhibitors), benazepril and lisinopril, as well as the binding affinities of the different drug molecules and natural molecules with them, are shown in Table 5.
Table 5.
Binding affinity values of reference drugs and selected drugs, binding with the ACE2 receptor.
| Drugs | Binding Affinities (kcal/mol) |
|---|---|
| Benazepril (Reference drug) | -9.3 |
| Glimepiride (Antidiabetic drug) | -9.4 |
| Empaflozin (Antidiabetic drug) | -8.7 |
| Rosiglitazone (Antidiabetic drug) | -7.8 |
| Dapagliflozin (Antidiabetic drug) | -9.3 |
| Englitazone (Antidiabetic drug) | -8.2 |
| Glyburide (Antidiabetic drug) | -8.7 |
| Fluvastatin (Statin) | -8.9 |
| Mevastatin (Statin) | -8.2 |
| Pravastatin (Statin) | -9.6 |
| Ezetimibe (Cholesterol lowering drug) | -9.0 |
| Remdesivir (Antiviral drug) | -9.4 |
| Pacritinib (Janus kinase inhibitor) | -10 |
| Enoxaparin (Anticoagulant) | -13.8 |
| Fondaparinux (Anticoagulant) | -14.4 |
| Edoxaban (Anticoagulant) | -8.5 |
| Prasugrel (Antiplatelet) | -8.9 |
| Eptifibatide (Antiplatelet) | -10 |
| Ticagrelor (Antiplatelet) | -9.8 |
| Dipyridamol/aspirin (Antiplatelet) | -8.7 |
| 10-gingerdione (Component of ginger) | -8.2 |
| Epigallocatechin (Flavonoid) | -9.3 |
| Lisinopril (Reference drug) | -9.8 |
| Empaflozin (Antidiabetic drug) | -8.7 |
| Voglibose (Antidiabetic drug) | -7.5 |
| Glyburide (Antidiabetic drug) | -8.7 |
| Rosiglitazone (Antidiabetic drug) | -7.8 |
| Fluvastatin (Statin) | -8.9 |
| Mevastatin (Statin) | -8.2 |
| Pravastatin (Statin) | -9.6 |
| Ezetimibe (Cholesterol lowering drug) | -9.0 |
| Doxycycline (Antibiotic) | -9.8 |
| Erythromycin (Antibiotic) | -10.6 |
| Danoprevir (Antiviral drug) | -10.6 |
| Remdesivir (Antiviral drug) | -9.4 |
| Pacritinib (Janus kinase inhibitor) | -10 |
| Edoxaban (Anticoagulant) | -8.5 |
| Eptifibatide (Antiplatelet) | -10 |
| Ticagrelor (Antiplatelet) | -9.8 |
| Dipyridamol/aspirin (Antiplatelet) | -8.7 |
| Rutin (Component of garlic) | -10.6 |
| Epigallocatechingallate (Flavonoid) | -8.4 |
| Epigallocatechin (Flavonoid) | -9.3 |
| 10-gingerdione (Component of ginger) | -8.2 |
Using PyMOL, these drugs and natural molecules were also found to superimpose with the reference drugs, benazepril and lisinopril, respectively.
Table 6 gives the common amino acids involved in the ACE2 receptor-ligand interaction. Remdesivir was found to have five amino acids in common with the reference drug, benazepril. Glyburide and ticagrelor showed four amino acid similarities with the reference drug, indicating strong protein-ligand interactions. On the other hand, glimepiride, ezetimibe, pravastatin, pacritinib, enoxaparin, fondaparinux, edoxaban, eptifibatide, 10-gingerdione and epigallocatechin had three amino acids in common with the reference drug and therefore had significant protein-ligand interactions. Dipyridamol/aspirin, prasugrel, empaflozin, fluvastatin, dapagliflozin, mevastatin, rosiglitazone, englitazone had two amino acids in common with the reference drug. Thus, it can be predicted that they had moderate protein-ligand interaction. On the other hand, ticagrelor demonstrated six amino acid similarities with the reference drug, lisinopril, indicating the strongest protein-ligand interaction among all the candidate drugs. Eptifibatide was found to have five amino acid similarities; remdesivir, edoxaban and 10-gingerdione demonstrated four amino acid similarities; empaflozin, danoprevir, ezetimibe, pravastatin, glyburide, dipyridamol/aspirin, had three amino acids; and voglibose, fluvastatin, mevastatin, rosiglitazone, doxycycline, erythromycin, pacritinib, epigallocatechin gallate and epicatechin had two common amino acids with lisinopril, indicating moderate protein-ligand interactions.
Table 6.
Common amino acids involved in the ACE2 receptor-ligand interaction.
| Drugs | Amino acids |
|---|---|
| Benazepril (Reference drug) | ALA348, ASN394, PHE40, PHE390, HIS401 |
| Glimepiride (Antidiabetic drug) | PHE390, HIS401, PHE40 |
| Empaflozin (Antidiabetic drug)) | ASN394, HIS401 |
| Rosiglitazone (Antidiabetic drug)) | ASN394, PHE40 |
| Dapagliflozin (Antidiabetic drug)) | ALA348, PHE40 |
| Englitazone (Antidiabetic drug)) | PHE40, PHE390 |
| Glyburide (Antidiabetic drug)) | ASN394, PHE40, PHE390, HIS401 |
| Fluvastatin (Statin) | ALA348, HIS401 |
| Mevastatin (Statin) | ALA348, PHE40 |
| Pravastatin (Statin) | ALA348, PHE40, HIS401 |
| Ezetimibe (Cholesterol lowering drug) | ALA348, ASN394, PHE40 |
| Remdesivir (Antiviral drug) | ASN394, ALA348, HIS401, PHE40, PHE390 |
| Pacritinib (Janus kinase inhibitor) | ASN394, PHE40, PHE390 |
| Enoxaparin (Anticoagulant) | ALA348, ASN394, PHE40, |
| Fondaparinux (Anticoagulant) | HIS378, ASN394, HIS401 |
| Edoxaban (Anticoagulant) | ALA348, PHE40, PHE390 |
| Prasugrel (Antiplatelet) | ASN394, PHE40 |
| Eptifibatide (Antiplatelet) | ALA348, HIS401, ASN394 |
| Ticagrelor (Antiplatelet) | ALA348, ASN394, PHE40, HIS401 |
| Dipyridamol/aspirin (Antiplatelet) | ALA348, HIS401 |
| 10-gingerdione (Component of ginger) | ALA348, PHE390, HIS401 |
| Epigallocatechin (Flavonoid) | ASN394, PHE40, HIS401 |
| Lisinopril (Reference drug) | HIS378, HIS401, ASP382, ASN394, PHE40, ALA348 |
| Empaflozin (Antidiabetic drug) | ASN394, HIS401, ASP382 |
| Voglibose (Antidiabetic drug) | ALA348, HIS378 |
| Glyburide (Antidiabetic drug) | ASN394, PHE40, HIS401 |
| Rosiglitazone (Antidiabetic drug) | ASN394, PHE40 |
| Fluvastatin (Statin) | ALA348,HIS401 |
| Mevastatin (Statin) | ALA348,PHE40 |
| Pravastatin (Statin) | ALA348,PHE40, HIS401 |
| Ezetimibe (Cholesterol lowering drug) | ALA348, ASN394, PHE40 |
| Doxycycline (Antibiotic) | HIS401, ALA348 |
| Erythromycin (Antibiotic) | HIS401, ASP382 |
| Danoprevir (Antiviral drug) | ASN394,HIS401, ASP382 |
| Remdesivir (Antiviral drug) | ASN394, ALA348, HIS401, PHE40 |
| Pacritinib (Janus kinase inhibitor) | ASN394, PHE40 |
| Edoxaban (Anticoagulant) | ALA348, PHE40, PHE390, HIS378 |
| Eptifibatide (Antiplatelet) | HIS378, ALA348, HIS401, ASN394, ASP382 |
| Ticagrelor (Antiplatelet drug) | ALA348, ASN394, PHE40, HIS401, ASP382, HIS378 |
| Dipyridamol/aspirin (Antiplatelet) | ALA348, HIS401, HIS378 |
| Rutin (Component of garlic) | PHE40, ASP382, HIS378 |
| Epigallocatechin gallate (Flavonoid) | HIS378, HIS401 |
| Epigallocatechin (Flavonoid) | ASN394, ASP382 |
| 10-gingerdione (Component of ginger) | ALA348, HIS401, HIS378, ASP382 |
3.4. In Silico Binding Affinity Values and Non-Bonded Protein Ligand Interactions with TMPRSS2
Camostat mesylate and gabexate mesylate are known inhibitors of the protein, TMPRSS2 and were thus taken as reference drugs for inhibiting the protein [11]. Their binding affinities are shown in Table 7.
Table 7.
Binding affinity values of reference drugs and selected drugs, binding with the TMPSSR2 protein.
| Drugs | Binding Affinities (kcal/mol) |
|---|---|
| Camostat mesylate (Reference drug) | -8.2 |
| Rosiglitazone (Antidiabetic drug) | -7.8 |
| Mevastatin (Statin) | -7.6 |
| Pitavastatin (Statin) | -8.7 |
| Ezetimibe (Cholesterol lowering drug) | -8.2 |
| Methyl prednisolone (Corticosteroid) | -9.5 |
| Danoprevir (Antiviral drug) | -8.9 |
| Warfarin (Anticoagulant) | -7.8 |
| Dipyridamole/aspirin (Antiplatelet) | -7.8 |
| Cilostazol (Antiplatelet) | -8.5 |
| Pacritinib (Janus kinase inhibitor) | -8.6 |
| Tideglusib (Glycogen synthase kinase 3 inhibitor) | -8.6 |
| Shikonin (Natural compound) | -7.7 |
| Epigallocatechin gallate (Flavonoid) | -7.2 |
| Fucosterol (Component of black cumin) | -8.9 |
| Dithymoquinone (Component of black cumin) | -7.6 |
| Campesterol (Plant sterol) | -8.9 |
| Gabexate mesylate (Reference Drug) | -8.5 |
| Danoprevir (Antiviral drug) | -8.9 |
| Pacritinib (Janus kinase inhibitor) | -8.6 |
| Tideglusib (Glycogen synthase kinase 3 inhibitor) | -8.6 |
| Shikonin (Natural compound) | -7.7 |
| Dithymoquinone (Component of black cumin) | -7.6 |
For TMPRSS2, the drugs with the highest binding affinities were chosen and were superimposed with the two reference drugs, camostat mesylate and gabexate mesylate, respectively (Table 8).
Table 8.
Common amino acids involved in the TMPSSR 2-ligand interaction.
| Drugs | Amino acids |
|---|---|
| Camostat mesylate (Reference drug) | ASP491, GLU289, ASN192, PHE357, MET488, PHE194, ILE242, PRO288 |
| Rosiglitazone (Antidiabetic drug) | PHE194, ILE242 |
| Mevastatin (Statin) | ILE242, PRO288, |
| Pitavastatin (Statin) | PHE357, PHE194, PRO288, ILE242 |
| Ezetimibe (Cholesterol lowering drug) | ASN192, GLU289, PHE194, ILE242 |
| Methyl prednisolone (Corticosteroid) | GLU289, ILE242, PHE194, PRO288, PHE357 |
| Danoprevir (Antiviral drug) | PHE357, PRO288 |
| Warfarin (Anticoagulant) | ASN192,PHE194,ILE242 |
| Dipyridamole/aspirin (Antiplatelet) | PHE357,ILE242,PRO288,PHE194, |
| Cilostazol (Antiplatelet) | GLU289, PHE194, ILE242, PRO288 |
| Pacritinib (Janus kinase inhibitor) | PHE357, PHE194, ILE242 |
| Tideglusib (Glycogen synthase kinase 3 inhibitor) | PHE357, PRO288 |
| Shikonin (Natural compound) | PHE357, PHE194, ILE242 |
| Epigallocatechin gallate (Flavonoid) | GLU289, ASN192, ASP491 |
| Fucosterol (Component of black cumin) | ILE242, PRO288, PHE194, PHE357 |
| Dithymoquinone (Component of black cumin) | ILE242, PRO288, PHE194, PHE357 |
| Campesterol (Plant sterol) | ILE242, PRO288, PHE357 |
| Gabexate mesylate (Reference drug) | PHE357, ALA243 |
| Danoprevir (Antiviral drug) | PHE357, ALA243 |
| Pacritinib (Janus kinase inhibitor) | PHE357, ALA243 |
| Tideglusib (Glycogen synthase kinase 3 inhibitor) | PHE357, ALA243 |
| Shikonin (Natural compound) | PHE357, ALA243 |
| Dithymoquinone (Component of black cumin) | PHE357, ALA243 |
Methyl prednisolone had five amino acids in common with the reference drug, camostat mesylate while interacting with the protein, TMPRSS2. Ezetimibe, pitavastatin, dipyridamole/aspirin, cilostazol, dithymoquinone and fucosterol had four common amino acids indicating they interacted well with the protein. Shikonin, warfarin, campesterol and epicatechin gallate had three amino acids in common with the reference drug. On the other hand, danoprevir, mevastatin and rosiglitazone demonstrated two common amino acids with the reference drug, camostat mesylate. Danoprevir, shikonin, pacritinib, tideglusib and dithymoquinone had two common amino acids with the reference drug, gabexate mesylate, indicating they bind to the same pocket as the reference drug.
3.5. Pharmacokinetic evaluation of the reference and candidate drugs
The target organ of the virus causing COVID-19 infection is the lungs, affecting the pneumocytes and macrophages, where both the ACE2 receptor and the TMPRSS2 protein, that facilitate the entry of the virus, are present. Cases of encephalitis have been reported in patients with COVID-19, associated with either negative or positive detection of SARS-CoV-2 in the CSF. Studies on the CNS of SARS patients at autopsy have suggested that the coronavirus enters the CNS and causes neurological effects [40]. A possible access route thus includes the blood brain barrier to penetrate the CNS, facilitated by the expression of the SARS-CoV-2 receptor ACE2 in the brain, where it would act as a cell surface peptidase present on the surface of endothelial cells and neurons. Thus, in this uncontrollable pandemic, the well-tolerated brain penetrating drugs to minimize any neurological consequences of SARS-CoV-2 infection could be considered [40, 41]. For this reason, we have chosen in our study both BBB+ and BBB- drugs (drugs that penetrate the CNS and drugs that do not penetrate the CNS).
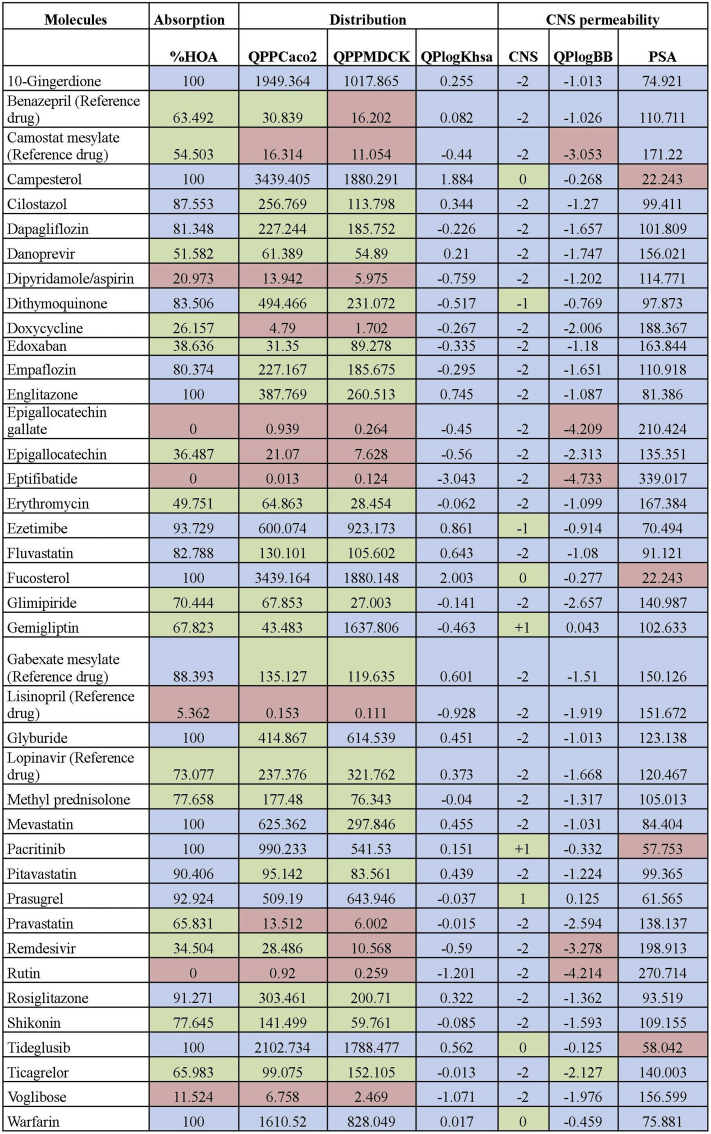
The ADME properties of the drugs were evaluated using QikProp (Figure 3), where 10-gingerdione, campesterol, englitazone, fucosterol, glyburide, mevastatin, pacritinib, tideglusib and warfarin showed 100% oral absorption. However, except for dipyridamol/aspirin, epigallocatechin gallate, eptifibatide, lisinopril and rutin, all the other molecules showed good values for oral absorption. The selected molecules were also found to show high permeability through the intestinal cells (QPPCaco2), except camostat mesylate, dipyridamol/aspirin, doxycycline, epigallocatechin gallate, epigallocatechin, eptifibatide, lisinopril, rutin and voglibose. Most of the drugs showed permeability for renal cells (QPPMDCK), except benazepril, camostat mesylate, dipyridamol/aspirin, doxycycline, epigallocatechin gallate, epigallocatechin, eptifibatide, lisinopril, remdesivir, rutin and voglibose. For the binding to serum albumin, all the molecules were within the range with the exception of campesterol and eptifibatide. Cell distribution was predicted considering median binding to plasma proteins, median to high %HOA, and from median to high for Caco2 as well as MDCK cells.
Figure 3.
Pharmacokinetic properties of the reference and candidate drugs. %HOA: Percentage of Human Oral Absorption; QPPCaco2: intestinal permeability in nm/s; QPPMDCK: renal permeability in nm/s; QPlogKHSA: Binding to human serum albumin; CNS: Central Nervous System activity; QPlogBB: brain/blood partition coefficient; PSA: Van der Waals surface area of polar nitrogen and oxygen atoms.
Except for camostat mesylate, epigallocatechin gallate, eptifibatide, remdesivir and rutin, all the molecules evaluated had a predicted brain/blood partition coefficient within the acceptable range (–3.0 to 1.2). Most of the drugs also showed no permeability to the CNS except campesterol, dithymoquinone, erythromycin, fucosterol, gemigliptin, pacritinib, prasugrel, tideglusib and warfarin. Finally, the PSA value indicated campesterol, fucosterol, pacritinib and tideglusib cross the blood brain barrier.
Considering the ADME parameters, specifically absorption, distribution and CNS permeability based on the QikProp analysis, we chose the following twenty-seven molecules as potential candidates to be further explored in the treatment of COVID-19 - 10-gingerdione, campesterol, cilastazol, dapagliflozin, danoprevir, dithymoquinone, doxycycline, edoxaban, empaflozin, englitazone, erythromycin, ezetimibe, fluvastatin, fucosterol, glimepiride, gemigliptin, glyburide, methyl prednisolone, mevastatin, pacritinib, pitavastatin, prasugrel, rosiglitazone, shikonin, tideglusib, ticagrelor and warfarin.
4. Discussion
Multiple sequence alignment of the viral structure in South Asia showed fourteen conserved regions. The analysis of the phylogenetic tree obtained in this study suggests that the same SARS-CoV-2 virus of Wuhan origin may have migrated from Italy to India.
After analyzing the docking results, superimposition, non-bonded protein-ligand interaction and ADME properties, both remdesivir and lopinavir were found to be potential inhibitors of the SARS-CoV-2 protein, and thus suggested as the potential reference drugs targeting SARS-CoV-2. Based on the binding affinity using AutoDock Vina or PyRx, non-bonded protein-ligand interactions by Discovery Studio, and superimposition by PyMOL, the strongest drug candidate, gemigliptin (antidiabetic drug) was then chosen. QikProp results indicated gemigliptin had higher renal permeability compared to lopinavir. It can thus be suggested that gemigliptingemigliptinmay be used in treating COVID-19, and thus prevent the cases of stroke that have been reported in COVID-19 patients. However, doses of gemigliptin should be adjusted before prescribing this drug to patients taking other drugs [42].
On the other hand, the ACE2 receptor is significant in combating the COVID-19 disease as the virus spike protein needs to bind to the ACE2 receptor in order to get into the host cell's cytosol. If the ACE2 receptors are made unavailable in this way, it could potentially block the entry of the SARS-CoV-2 into the human body. In case of ACE2 receptors, several drugs from different classes were screened to find the potential inhibitors of the receptor. The reference drugs that were chosen for inhibiting ACE2 were benazepril and lisinopril. They were also found to have a good binding affinity with SARS-CoV-2 and suggested in the treatment of COVID-19 [43]. Although other ACE2 inhibitors showed strong binding affinity, these were selected as they showed strong protein-ligand interactions it. The drugs with the strongest binding affinities were shortlisted and were superimposed individually with the two reference drugs. Accordingly, the repurposed drugs selected were voglibose and glyburide (antidiabetic drug), doxycycline and erythromycin (antibiotic), remdesivir (antiviral drug) and danoprevir (antiviral drug), They had strong binding affinities, strong protein-ligand interactions as well as safe drug profile, suggesting that they could be potential drug candidates against COVID-19. The results indicate that they may follow the same pathway as the reference drugs to inhibit the ACE2 receptor. Dapagliflozin, edoxaban, empaflozin, englitazone, ezetimibe, glimepiride, glyburide, mevastatin, pacritinib, prasugrel, rosiglitazone and ticagrelor had a higher renal permeability than the reference drug, Benazepril. All the molecules showed a higher oral absorption compared to the reference drug, except rutin. The intestinal permeability of all the molecules were higher except eptifibatide and the renal permeability of all the molecules were higher. Ezetimibe and pacritinib showed high renal permeability. Most of the selected molecules showed no CNS permeability, except pacritinib and ezetimibe. The PSA results suggested pacritinib crosses the blood brain barrier.
The reference drugs chosen for inhibiting TMPSSR2 were camostat mesylate and gabexate mesylate, and the drugs that could be repurposed as potential inhibitors of TMPSSR2 included danoprevir (antiviral), ezetimibe (cholesterol lowering drug), methyl prednisolone (corticosteroid), mevastatin (statin), pitavastatin (statin), rosiglitazone (antidiabetic), shikonin (natural compound), tideglusib (glycogen synthase kinase 3 inhibitor), pacritinib (janus kinase inhibitor), warfarin (anticoagulant), dipyridamole/aspirin (antiplatelet), cilostazol (antiplatelet) and natural molecules dithymoquinone, fucosterol, campesterol. These sixteen drugs can be suggested to prevent viral entry and thus help in the prevention of the fatal conditions of COVID-19.
The approach considered for the study was totally based on in silico methods, using molecular docking software for repurposing existing drugs from our own comprehensive database of approximately 300 highly characterized, existing drugs and natural molecules with known safety profile, to identify compounds that may inhibit the SARS-CoV-2, ACE2, and TMPRSS2. Detecting similar drug binding sites in more than one protein has a wide range of applications in drug repurposing [44]. We thus investigated the similar binding sites of drug with the proteins - SARS-CoV-2, ACE2 and TMPSSR2. RRosiglitazone, mevastatin, ezetimibe, danoprevir, pacritinib and the combination of dipyridamole/aspirin inhibited ACE2 and TMPSSR2. After analysis of the pharmacokinetic properties, it was found that all the molecules had better pharmacokinetic properties than the reference drug, Camostat mesylate except dipyridamole/aspirin. The combination had lower oral absorption, intestinal and renal permeability. With gabexate mesylate as the reference drug, most of the selected candidates showed better pharmacokinetic properties. Pacritinib and tideglusib crossed the blood brain barrier as per the predicted PSA results.
After the following twenty-seven drugs were finally chosen after evaluating the pharmacokinetic properties of the chosen drugs based on human oral absorption, intestinal and renal cell permeability, binding to human serum albumin, CNS permeability, predicted brain/blood partition coefficient and Van der Waals surface area of polar nitrogen and oxygen atoms, we finally chose the following twenty-seven drugs: 10-gingerdione, campesterol, cilastazol, dapagliflozin, danoprevir, dithymoquinone, doxycycline, edoxaban, empaflozin, englitazone, erythromycin, ezetimibe, fluvastatin, fucosterol, glimepiride, gemigliptin, glyburide, methyl prednisolone, mevastatin, pacritinib, pitavastatin, prasugrel, rosiglitazone, shikonin, tideglusib, ticagrelor and warfarin.
These twenty seven candidates may be proposed to inhibit the entry of the virus by blocking the activity of any one of the three proteins, shown either earlier in step (i) or (ii) of Figure 1. However, the aim was to publish these results immediately so that the next steps of in vivo and in vitro evaluations could be done as soon as possible, and the crisis of facing unprecedented difficulties in dealing with the current COVID-19 crisis throughout the world could be addressed with specific drug therapies successfully. Lung tissue distribution of the drugs proposed must also be considered, since there are high viral loads in the lung tissue of COVID-19 patients, and thus high volume of the lung distribution of drugs used in the treatment is required to stop or block the replication of the coronavirus [45]. This is also required in determining an appropriate dosing and route of administration in humans. Overall pharmacodynamic markers must also be considered in such drug repurposing studies, particularly in determining whether the proposed drugs are truly effective.
Thus, after screening and repurposing almost 300 drugs, the current study proposes twenty twenty-seven candidates as potential candidates for further in vitro and in vivo studies for the treatment of the SARS-CoV-2 infection. Future research could also examine the inhibition of the other proteases such as furin, cathepsin B and L, etc. responsible for SARS-CoV-2 entry into the host with the drugs proposed in the study for a holistic treatment option for COVID-19 patients.
Declarations
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fehr A.R., Perlman S. Chapter 1 coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282 doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang S., Zhou X., Zhang T., Wang Z. The need for urogenital tract monitoring in COVID-19. Nat. Rev. Urol. 2020:1–2. doi: 10.1038/s41585-020-0319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdecchia P., Cavallini C., Spanevello A., Angeli F. 2020. Since January 2020 Elsevier Has Created a COVID-19 Resource centre with Free Information in English and Mandarin on the Novel Coronavirus COVID- 19 . The COVID-19 Resource centre Is Hosted on Elsevier Connect, The company’s Public News and Information. [Google Scholar]
- 6.Xu X., Chen P., Wang J., Feng J., Zhou H., Li X., Zhong W., Hao P. Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its spike protein for risk of human transmission. Sci. China Life Sci. 2020;63:457–460. doi: 10.1007/s11427-020-1637-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on Decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He J., Tao H., Yan Y., Huang S.-Y., Xiao Y. Molecular mechanism of evolution and human infection with the novel coronavirus (2019-nCoV) BioRxiv. 2020 doi: 10.3390/v12040428. 2020.02.17.952903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mannan Baig A., Khaleeq A., Ali U., Syeda H. 2020. Evidence of the COVID-19 Virus Targeting the CNS: Tissue Distribution, Host−Virus Interaction, and Proposed Neurotropic Mechanisms. [DOI] [PubMed] [Google Scholar]
- 10.Maier H.J., Bickerton E., Britton P. 2015. Coronaviruses: Methods and Protocols, Coronaviruses Methods Protoc; pp. 1–282. [DOI] [PubMed] [Google Scholar]
- 11.Hoffmann M., Kleine-Weber H., Schroeder S., Mü M.A., Drosten C., Pö S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabi F.A., Al Zoubi M.S., Kasasbeh G.A., Salameh D.M., Al-Nasser A.D. SARS-CoV-2 and coronavirus disease 2019: what we know so far. Pathogens. 2020;9:231. doi: 10.3390/pathogens9030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hebert A., States U. Modernizing the tree of life. Science (80-. ) 2003;300:1692–1697. doi: 10.1126/science.300.5626.1692. [DOI] [PubMed] [Google Scholar]
- 14.Smith S.A., Brown J.W., Hinchliff C.E. Analyzing and synthesizing phylogenies using tree alignment graphs. PLoS Comput. Biol. 2013;9 doi: 10.1371/journal.pcbi.1003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., Harris M.A., Hill D.P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J.C., Richardson J.E., Ringwald M., Rubin G., Sherlock Gerald M. The gene Ontology consortium, Michael Ashburner1, catherine A. Ball3, Judith A. Blake4, David Botstein3, Heather Butler1, J. Michael Cherry3, allan P. Davis4, Kara Dolinski3, Selina S. Dwight3, janan T. Eppig4, Midori A. Harris3, David P. Hill4, Laurie is. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pavlopoulos G.A., Soldatos T.G., Barbosa-Silva A., Schneider R. A reference guide for tree analysis and visualization. BioData Min. 2010;3:1–16. doi: 10.1186/1756-0381-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berdigaliyev N., Aljofan M. An overview of drug discovery and development. Future Med. Chem. 2020;12:939–947. doi: 10.4155/fmc-2019-0307. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Y., Wang F., Tang J., Nussinov R., Cheng F. Artificial intelligence in COVID-19 drug repurposing. Lancet Digit. Heal. 2020;2:e667–e676. doi: 10.1016/S2589-7500(20)30192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gore Mohini, Jagtap B. Umesh. Springer; New York, NY: 2018. Computational Drug Discovery and Design. [Google Scholar]
- 20.Kabir E.R., Siam M.K.S., Mustafa N., Kabir S.M. Proc. 9th Int. Conf. Comput. Syst. Bioinforma. - CSBio 2018. ACM Press; New York, USA: 2018. Molecular docking reveals pitavastatin and related molecules antagonize 1DHF and its pseudogene DHFR2 in cancer treatment; pp. 1–9. [Google Scholar]
- 21.Cichonska A., Ravikumar B., Parri E., Timonen S., Pahikkala T., Airola A., Wennerberg K., Rousu J., Aittokallio T. Computational-experimental approach to drug-target interaction mapping: a case study on kinase inhibitors. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kupferschmidt K. WHO launches global megatrial of the four most promising coronavirus treatments. Science (80-. ) 2020 [Google Scholar]
- 23.Common Generic and Brand Names for ACE Inhibitors and ARBs. 2010. [Google Scholar]
- 24.Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of nafamostat as a potent inhibitor of middle east respiratory syndrome Coronavirus s protein-mediated membrane fusion using the split-protein-based cell-cell fusion assay. Antimicrob. Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Severe Acute Respiratory Syndrome Coronavirus 2 Isolate Wuhan-Hu-1. co - Nucleotide - NCBI; 2020. https://www.ncbi.nlm.nih.gov/nuccore/NC_045512 [Google Scholar]
- 26.SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) Sequences. 2021. https://www.ncbi.nlm.nih.gov/genbank/sars-cov-2-seqs/ [Google Scholar]
- 27.Thompson J.D., Higgins D.G., Gibson T.J., CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stecher G., Tamura K., Kumar S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2021:1237–1239. doi: 10.1093/molbev/msz312. PMID: 31904846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 30.Dallakyan S., Olson A.J. Small-molecule library screening by docking with PyRx. Methods Mol. Biol. 2015:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
- 31.Huey R., Morris G.M., Olson A.J., Goodsell D.S. Software news and update a semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 32.DeLano W.L. Pymol: an open-source molecular graphics tool. Newsl. Protein Crystallogr. 2002;40:1–8. https://www.ccp4.ac.uk/newsletters/newsletter40/11_pymol.pdf [Google Scholar]
- 33.Gao Y.D., Huang J.F. An extension strategy of Discovery Studio 2.0 for non-bonded interaction energy automatic calculation at the residue level. Dongwuxue. Yanjiu. 2011;32:262–266. doi: 10.3724/SP.J.1141.2011.03262. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen W.L. The many roles of computation in drug discovery. Science (80-. ) 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 35.User Q. 2015. Manual, Schrödinger Press QikProp 4.4 User Manual. [Google Scholar]
- 36.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowie J.U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. http://www.ncbi.nlm.nih.gov/pubmed/1853201 [DOI] [PubMed] [Google Scholar]
- 38.Lüthy R., Bowie J.U., Eisenberg D. Assessment of protein models with three-dimensional profiles. Nature. 1992;356:83–85. doi: 10.1038/356083a0. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S., Stecher G., Li M., Knyaz C., Tamura K., Mega X. Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson P.J., Ottaviani S., Prelle A., Stebbing J., Casalini G., Corbellino M. CNS penetration of potential anti-COVID-19 drugs. J. Neurol. 2020;267:1880–1882. doi: 10.1007/s00415-020-09866-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mao L., Jin H., Wang M., Hu Y., Chen S., He Q., Chang J., Hong C., Zhou Y., Wang D., Miao X., Li Y., Hu B. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol. 2020 doi: 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Noh Y.H., Lim H.S., Jin S.J., Kim M.J., Kim Y.H., Sung H.R., Choi H.Y., Bae K.S. Effects of ketoconazole and rifampicin on the pharmacokinetics of gemigliptin, a dipeptidyl peptidase-IV inhibitor: a crossover drug-drug interaction study in healthy male Korean volunteers. Clin. Ther. 2012;34:1182–1194. doi: 10.1016/j.clinthera.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 43.Rismanbaf A. Potential treatments for COVID-19; a narrative literature review. Arch. Acad. Emerg. Med. 2020;8:e29. [PMC free article] [PubMed] [Google Scholar]
- 44.Govindaraj R.G., Brylinski M. Comparative assessment of strategies to identify similar ligand-binding pockets in proteins. BMC Bioinf. 2018;19:91. doi: 10.1186/s12859-018-2109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Chen L. Lung tissue distribution of drugs as a key factor for COVID-19 treatment. Br. J. Pharmacol. 2020;177:4995–4996. doi: 10.1111/bph.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.