Abstract
The crab Cancer borealis nervous system is an important model for understanding neural circuit dynamics and its modulation, but the identity of neuromodulatory substances and their influence on circuit dynamics in this system remains incomplete, particularly with respect to behavioral state-dependent modulation. Therefore, we used a multifaceted mass spectrometry (MS) method to identify neuropeptides that differentiate the unfed and fed states. Duplex stable isotope labeling revealed that the abundance of 80 of 278 identified neuropeptides was distinct in ganglia and/or neurohemal tissue from fed vs. unfed animals. MS imaging revealed that an additional 7 and 11 neuropeptides exhibited altered spatial distributions in the brain and the neuroendocrine pericardial organs (POs), respectively, during these two feeding states. Furthermore, de novo sequencing yielded 69 newly identified putative neuropeptides that may influence feeding state-related neuromodulation. Two of these latter neuropeptides were determined to be upregulated in PO tissue from fed crabs and one of these two peptides influenced heartbeat in ex vivo preparations. Overall, the results presented here identify a cohort of neuropeptides that are poised to influence feeding-related behaviors, providing valuable opportunities for future functional studies.
Keywords: Mass spectrometry, neuropeptides, imaging, stable isotope labeling, stomatogastric nervous system, cardiac neuromodulation
Graphical Abstract
Introduction:
Feeding, like all behaviors, involves a complex coordination of numerous component movements (e.g. chewing and swallowing), with each component often being driven by a separate neural circuit. Obtaining a comprehensive understanding of how neural circuits generate behaviors is a long-standing goal of neuroscience research that continues to be pursued in many model systems using in vivo and ex vivo approaches.1–9 One current challenge is to understand how the well-characterized neural circuits that have been studied at the cellular level in isolated nervous systems operate in vivo during actual behavior. This challenge is being met using cutting-edge tools, such as molecular genetics and optogenetics, to manipulate neural circuits in vivo. An alternative approach is to take advantage of the unparalleled access to neural circuits afforded by studying them ex vivo and introducing events that normally only occur in vivo.
Studying neural circuit operation in the isolated nervous system, however, excludes the presence of the neuroendocrine system, which is a pivotal source for circulating hormones that modify neural circuit activity.10, 11 Identifying the full complement of these hormones in a behavioral state-dependent manner would enable a detailed examination of their collective impact on circuit activity in the isolated system. One model system that displays the complementary values of having well-defined neural circuits and providing the opportunity to identify the full complement of circulating hormones in behavioral states related to these circuits is the stomatogastric nervous system (STNS) of the crab Cancer borealis.12
The STNS has been studied for over 50 years in several decapod crustacean species as a model for understanding how neuronal circuits generate rhythmic motor patterns that underlie rhythmic movements (e.g. chewing).9, 12–14 This numerically small yet elegant extension of the central nervous system (CNS) has enabled investigators to gain extensive knowledge about neural circuit dynamics, including how individual neurons contribute to circuit output.9, 12–14
The STNS is composed of four linked ganglia, including the stomatogastric ganglion (STG), oesophageal ganglion (OG), and paired commissural ganglia (CoG).12, 14, 15 In addition to the neuronal interactions between these ganglia, the STNS is influenced by other key neural tissues, such as the brain (i.e. supraoesophageal ganglion), and by circulating peptide and small molecule hormones released from the neuroendocrine system, prominently including the sinus glands (SGs) in the eye stalks, and the paired pericardial organs (POs), which release hormones into the cardiac sinus.
In the crab Cancer borealis, the STG contains only 26 neurons, but overlapping sets of these neurons are sufficient to comprise the gastric mill (chewing) and pyloric (pumping/filtering of chewed food) motor circuits. Due to their small number, their large soma size (30–120 μm), the presence of most neuron types as single copies, and the ability of both circuits to continue operating in the isolated STNS, the associated connectomes and how they operate under particular conditions are well-defined.14 The ability of applied- and neurally-released modulators to influence these circuits has also received considerable attention,14, 16, 17 but the complete repertoire of modulators in this system and how they act collectively in a behavioral state-dependent manner to influence the STG circuits remains to be determined.
Neuropeptides comprise the most diverse class of neurotransmitters, and the full array of mechanisms by which they affect the STG circuits remains elusive. The primary structure of these neuropeptides can vary widely, with many of them being present as numerous isoforms. These isoforms can differ by as little as a single amino acid, yet in most cases it remains to be determined whether they have conserved or distinct functions.18, 19 This uncertainty of function among isoforms is illustrated by three neuropeptides with conserved sequence homology (AST-C I, II, III), which have distinct effects and spatial distributions within the cardiac neuromuscular system of the lobster Homarus americanus.20 Furthermore, the same neuropeptide can have different functions in different neurons or at different concentrations, such as PevPK2, which affects the contraction frequency, duration, and fall time of the lobster heart in a concentration-dependent manner.21 In addition to the high degree of complexity of neuropeptide function, these molecules are challenging to study due to their low abundance in vivo and propensity for being rapidly degraded.22–24 For example, application of peptidase inhibitors alters the response of the STG circuits to neurally-released peptides.25–27
To obtain a more complete understanding of what neuropeptides influence the STNS, large-scale, untargeted profiling has been successfully employed using mass spectrometry (MS). MS is a highly suitable tool for identifying the key neuropeptides responsible for behavioral outputs because it is fast, sensitive, and can theoretically detect all analytes above its limit of detection (i.e., several attomoles) in a sample, and can do so in a single analysis without prior knowledge about the specific analytes. As a result, MS has been used to profile the neuropeptides present in key tissues29–31 and to identify those involved in numerous biological processes such as adaptation to temperature-,32 salinity-,33 and pH stress.34 The role of neuropeptides in feeding behavior has also been explored previously, both in various tissues and in hemolymph.35–37
In this study, a multifaceted MS method using advanced techniques was employed to gain deeper insight into the neuropeptides likely to be involved in feeding behavior in the crab Cancer borealis.12 Quantitative analysis, using duplex formaldehyde chemical labeling, was performed to compare the relative abundance of neuropeptides in five key neural tissues, the STG, CoGs, brain, SGs, and POs, when crabs were in both unfed and fed states. Additionally, MS imaging was used to spatially localize neuropeptides within the brain and POs, with the goal of understanding how neuropeptide organization changed within these tissues after feeding. Finally, de novo sequencing was utilized to identify novel putative neuropeptides whose abundance and/or localization were changed during the feeding process, thereby suggesting that they act as modulators of feeding-related circuits in C. borealis.
The results of this study revealed a cohort of neuropeptides that exhibited changes in abundance and/or localization in various tissues and thus may alter the outputs of feeding-related circuits within the STNS. Two of these novel neuropeptides were identified and their sequences confirmed with custom-synthesized standards. One of these two peptides consistently influenced the cardiac neuromuscular system, which is known to be influenced by the feeding process.38, 39
Results and Discussion
Feeding regulation is an important biological process that is controlled by multiple neural circuits.40 To identify the individual neuropeptides likely to influence this process, we analyzed neuropeptide abundance levels in unfed and recently fed (i.e. 30 minutes post-feeding, see Methods) crabs in several neuronal (brain, STG, CoGs) and neuroendocrine (PO, SG) tissues. These tissues were chosen for their key roles in neuropeptide secretion or transmission during feeding.12 The time point of 30 minutes was chosen because it is early in the feeding process and is likely to reveal changes triggered by the initiation of food intake. In parallel, we spatially mapped neuropeptide localization within the brain and POs in the unfed and recently fed conditions, using MALDI-MSI (matrix-assisted laser desorption/ionization-mass spectrometry imaging). While the results presented here highlight neuropeptides that may be implicated in feeding using a multifaceted method, there may be additional peptides involved that were not detected with these methods. Furthermore, it should be noted that a lack of change in abundance, particularly in a ganglion, does not preclude a role in feeding.
Changes in Tissue Abundance
Neuropeptide samples acquired from the five aforementioned tissues in fed and unfed crabs were differentially labeled using isotopic reductive methylation. For complementary coverage, two different methods, MALDI-MS and LC-ESI-MS/MS (liquid chromatography-electrospray ionization-tandem mass spectrometry), were used to study the tissues. For each approach, 8 biological replicates were collected from all tissues. Across all 5 tissues, 278 neuropeptides from 35 peptide families were detected, as listed in Supporting Information II. In some cases, it is possible that an identified peptide in fact represents a degradation product of a parent peptide (e.g. APSGFLGM and APSGFLGMRamide). If so, it nevertheless remains possible that such a degradation product retains biological activity25. As presented below, neuropeptide abundance post-feeding either increased, decreased, or did not change relative to that in unfed animals. Increased abundance indicates that these peptides accumulated in the studied tissue(s) after feeding, perhaps because their release was suppressed due to inhibition of the peptide-containing neuron(s) and/or in preparation for subsequent release. Measured increases could also be due to increased production of the neuropeptide, though this change may not yield measurable results in the short time tested. In contrast, decreased abundance suggests that these peptides exhibited increased release during/after feeding, presumably due to increased excitatory drive to the peptide-releasing neurons. Decreases in abundance could also result from enzyme-triggered degradation within the tissue. Those that did not show changes may not have a role in feeding processes at this early time point, but may still affect the feeding process at later time points or may do so without exhibiting an appreciable abundance change within the tissue.
The stomatogastric nervous system.
Despite the small size of the STG, 44 neuropeptides were identified in both unfed and fed tissue (see Supporting Information II). Six of these forty-four peptides were up-regulated by feeding, and none were found to be down-regulated. The up-regulated peptides included two pyrokinins (PKs), a tachykinin-related peptide (TRP), a N-terminally extended RFamide, an A-type allatostatin (AST-A), and a diuretic hormone 44 precursor-related peptide (DH 44-PRP) (Fig. 1a and Table 1). Previous studies have shown that PKs, TRP, RFamide and AST peptides all modulate the gastric mill and/or pyloric rhythms in the STG.41–46 The two PK peptides that exhibited an increased abundance, TNFAFAPRPamide and ANFAFAPRPamide, are novel identifications for C. borealis. However, ANFAFAPRPamide was originally predicted using in silico mining of the C. borealis transcriptome,47 while TNFAFAPRPamide was recently identified in C. irroratus.48 While other, previously identified PK peptides were not detected in the STG in this study, they were identified in other tissues. This suggests that these peptides were not present in quantifiable amounts in the STG throughout the early stages of the feeding process.
Figure 1.
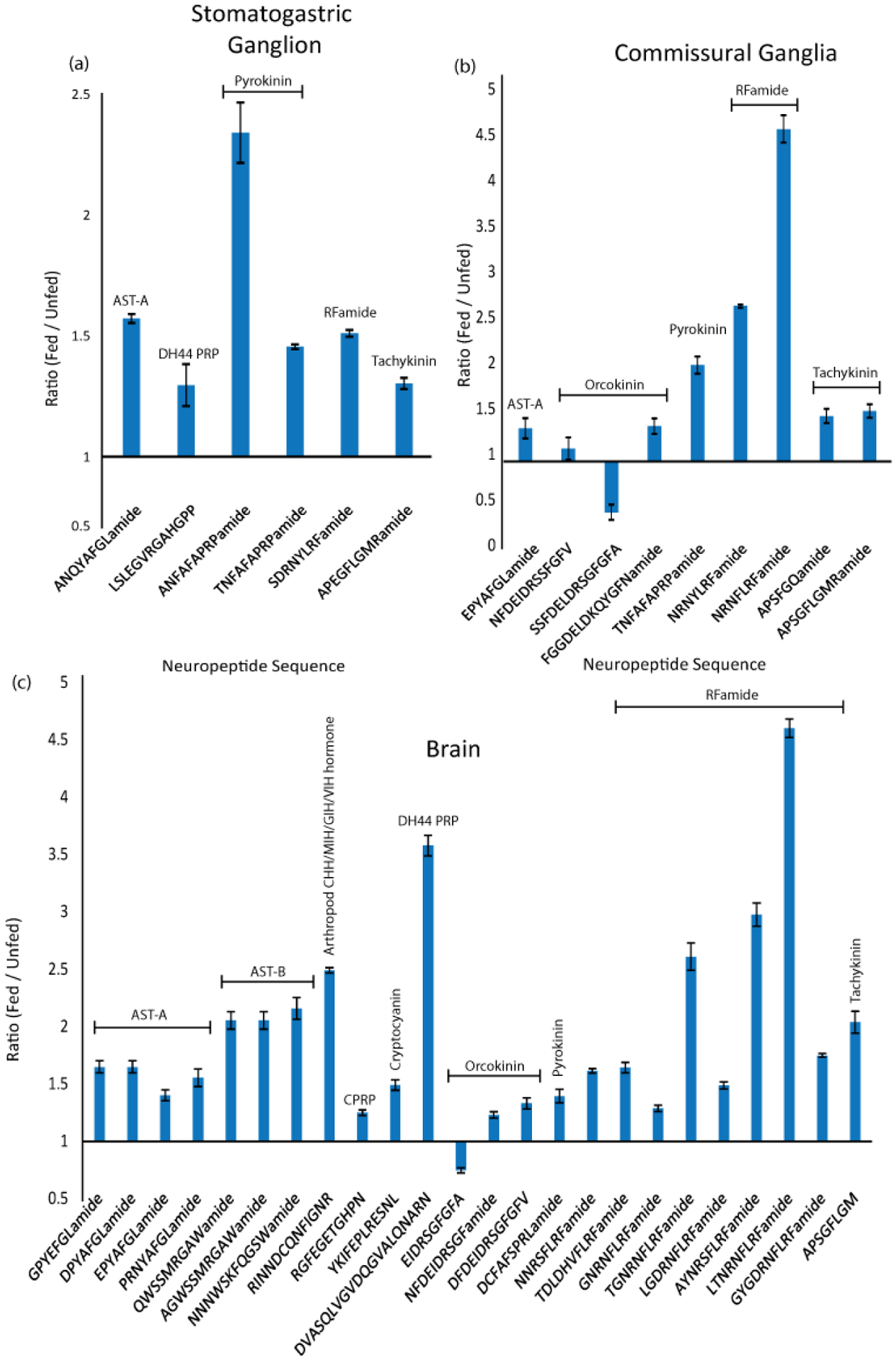
Neuropeptides exhibiting changes in abundance after feeding, in the stomatogastric ganglion, commissural ganglion and brain of C. borealis. Bar graphs show neuropeptides displaying significantly increased or decreased abundance in 30 minutes post-feeding tissues compared to unfed tissue. The y-axis represents the ratio of fed- to unfed abundance. Significance based on p < 0.05, Unpaired, Two-Tailed Student’s t-Test, n=5. Error bars represent SEM. Neuropeptide families are indicated, allatostatin (AST), crustacean hyperglycemic hormone precursor related peptide (CPRP), precursor related peptide (PRP), diuretic hormone (DH).
Table 1.
Neuropeptides exhibiting changes in abundance after feeding in the brain, commissural ganglion (CoG) and stomatogastric ganglion (STG) of C. borealis, including each neuropeptide’s family, sequence, and ratio (fed/unfed) and p-value for each tissue. Significance is based on p < 0.05, Unpaired, Two-Tailed Student’s t-Test, n=5. Neuropeptide families are indicated, allatostatin (AST), crustacean hyperglycemic hormone precursor related peptide (CPRP), precursor related peptide (PRP), diuretic hormone (DH).
| Family | Sequence | Brain | CoG | STG | |||
|---|---|---|---|---|---|---|---|
| Ratio | p-value | Ratio | p-value | Ratio | p-value | ||
| AST A-type | GPYEFGLamide | 1.78 | 0.0064 | ||||
| AST A-type | DPYAFGLamide | 1.78 | 0.0064 | ||||
| AST A-type | EPYAFGLamide | 1.46 | 0.0467 | 1.46 | 0.024 | ||
| AST A-type | ANQYAFGLamide | 1.51 | 0.001 | ||||
| AST A-type | PRNYAFGLamide | 2.13 | 0.0188 | ||||
| AST B-type | QWSSMRGAWamide | 2.74 | 0.0205 | ||||
| AST B-type | AGWSSMRGAWamide | 2.74 | 0.0205 | ||||
| AST B-type | NNNWSKFQGSWamide | 6.34 | 0.0086 | ||||
| MIH hormone | RINNDCQNFIGNR | 2.53 | 0.0001 | ||||
| CPRP | RGFEGETGHPN | 1.31 | 0.0012 | ||||
| Cryptocyanin | YKIFEPLRESNL | 2.58 | 0.0242 | ||||
| DH44 PRP | LSLEGVRGAHGPP | 1.28 | 0.025 | ||||
| DH44 PRP | DVASQLVGVDQGVALQNARN | 4.79 | 0.0070 | ||||
| Orcokinin | NFDEIDRSSFGFV | 3.22 | 0.037 | ||||
| Orcokinin | SSFDELDRSGFGFA | 0.49 | 0.021 | ||||
| Orcokinin | FGGDELDKQYGFNamide | 1.47 | 0.028 | ||||
| Orcokinin | EIDRSGFGFA | 0.79 | 0.0070 | ||||
| Orcokinin | NFDEIDRSGFamide | 1.28 | 0.0183 | ||||
| Orcokinin | DFDEIDRSGFGFV | 18.58 | 0.0308 | ||||
| Pyrokinin | ANFAFAPRPamide | 3.09 | 0.049 | ||||
| Pyrokinin | TNFAFAPRPamide | 2.69 | 0.010 | 1.41 | 1.05E-04 | ||
| Pyrokinin | DCFAFSPRLamide | 1.72 | 0.0201 | ||||
| RFamide | NNRSFLRFamide | 1.85 | 0.0007 | ||||
| RFamide | NRNYLRFamide | 2.77 | 3.25E-06 | ||||
| RFamide | TDLDHVFLRFamide | 1.76 | 0.0015 | ||||
| RFamide | NRNFLRFamide | 5.65 | 4.59E-04 | ||||
| RFamide | GNRNFLRFamide | 1.31 | 0.0215 | ||||
| RFamide | SDRNYLRFamide | 1.45 | 2.52E-04 | ||||
| RFamide | TGNRNFLRFamide | 10.30 | 0.0385 | ||||
| RFamide | LGDRNFLRFamide | 1.51 | 0.0072 | ||||
| RFamide | AYNRSFLRFamide | 5.23 | 0.0153 | ||||
| RFamide | LTNRNFLRFamide | 12.94 | 0.0005 | ||||
| RFamide | GYGDRNFLRFamide | 1.89 | 0.0002 | ||||
| Tachykinin | APSFGQamide | 1.76 | 0.010 | ||||
| Tachykinin | APSGFLGM | 3.29 | 0.0210 | ||||
| Tachykinin | APSGFLGMRamide | 2.53 | 0.012 | ||||
| Tachykinin | APEGFLGMRamide | 1.27 | 0.002 | ||||
The ~400 neurons in each CoG include ~20 projection neurons that innervate the STG.49 In this tissue, 58 neuropeptides belonging to 20 different families were detected. Of these neuropeptides, 8 were up-regulated and one was down-regulated in fed tissue (Fig. 1b and Table 1). Among these were three members of the orcokinin family, which activate hindgut contractions in several species and modulate the STNS output.50–53 Interestingly, one of these peptides (FGGDELDKQYGFN) exhibited a large increase in abundance in fed tissues, while another one (SSFDELDRSGFGFA) exhibited a 2-fold decrease in abundance. These differences suggest that, despite their sequence similarity, these peptides have different feeding-related functions. Two TRPs, APSFGQamide and APSGFLGMRamide (aka CabTRP Ia),54 also showed increased abundance in fed CoG tissue samples. TRPs play at least two roles in feeding-related behaviors in C. borealis. Specifically, these peptides are present in the anterior commissural organ (ACO), a neuroendocrine structure embedded within the CoG neuropil. The ACO originates from the CoG-projecting post-oesophageal commissure (POC) neurons,55, 56 whose activation triggers a particular version of the gastric mill rhythm both in vitro and in vivo.55, 57 CabTRP Ia is also present in MCN1, an identified CoG projection neuron that innervates the STG and modulates the gastric mill and pyloric rhythms.54, 58, 59 MCN1 is the sole source of CabTRP Ia in the C. borealis STG.60
The brain.
The OG and CoGs are directly connected with the brain, and one pair of projection neurons (IVN) in the brain innervates both the CoGs and the STG.61, 62 Therefore, the brain potentially plays a pivotal role in modulating feeding behavior. In this study, 138 neuropeptides were detected in fed and unfed brain tissue, with 24 exhibiting changes in abundance in fed tissue (Fig. 1c and Table 1). Only one of these peptides, orcokinin EIDRSGFGFA, exhibited a decreased abundance post-feeding, with the other 23 peptides displaying increased abundance. Among this latter cohort were 4 A-type and 4 B-type ASTs. Bath application of A-type ASTs inhibits various activities, including the pyloric rhythm.45, 63, 64 Although the effects of neurally-released AST-A are not known, it is present in the gastro-pyloric receptor (GPR) neurons, a set of muscle stretch-sensitive sensory neurons that influence the STG circuits both directly and via actions on CoG projection neurons.65 B-type ASTs, while less well-documented, also elicit decreases in pyloric rhythm activity when bath-applied.66 As suggested above, the accumulation of these neuropeptides in the tissue may indicate their preparation for release to terminate feeding behavior or the inhibition of their release to allow feeding to take place. Eight members of the RFamide family also showed increased abundance in the brain after feeding. Members of this family are broadly distributed throughout the crustacean CNS and neuroendocrine system, and some of their functions are known. For example, bath-applying different family members modulates the gastric mill and pyloric rhythms.46, 67, 68 Two orcokinin peptides (DFDEIDRSGFGFV, NFDEIDRSGF), and one TRP (APSGFLGM) also increased in abundance in fed tissue. As described above, these families have been implicated in STNS activity and their observed feeding-related changes in the brain indicate there may be additional feeding-related functions of these specific isoforms.
Neuroendocrine system.
In addition to acting on the STG through direct neural projections, neuropeptides gain access to the STG after being released as circulating hormones from neuroendocrine tissue such as the paired POs and the paired SGs. Neuropeptides released from these tissues are transported via the hemolymph, which is pumped from the pericardial sinus. The STG is located within the dorsal ophthalmic artery, which exits directly from the heart, and so it continuously receives hormonal influences.12, 69
The PO is a paired neurohemal site located in the lateral interior walls of the pericardial cavity. It contains the terminals of thoracic ganglion neurons that release neuropeptides directly into the pericardial sinus. After their release, they are transported via the hemolymph throughout the animal. Many (137) neuropeptides were detected in both fed and unfed POs; 21 of them differed in abundance in fed and unfed tissue (Fig. 2a and Table 2). Five of these twenty-two peptides exhibited reduced levels after feeding, including two B-type ASTs (AGWSSLKAWamide, NNNWTKFQGSWamide), an arthropod CHH/MIH/GIH/VIH hormone (RVINDDCPNLIGNR), a RFamide (DGGRNFLRFamide), and a RYamide (QGFYSQRYamide). The effect of RYamide and CHH/MIH/GIH/VIH peptides on the STG circuits is unknown. However, their decreased abundance after feeding suggests that they have a feeding-related function in the STG and/or elsewhere. While some B-type AST- and RFamide family members showed decreased abundance in fed tissue, other isoforms from these families exhibited increased abundance in fed tissue, indicating again the likely diversity of function even between neuropeptide isoforms within the same family.
Figure 2.
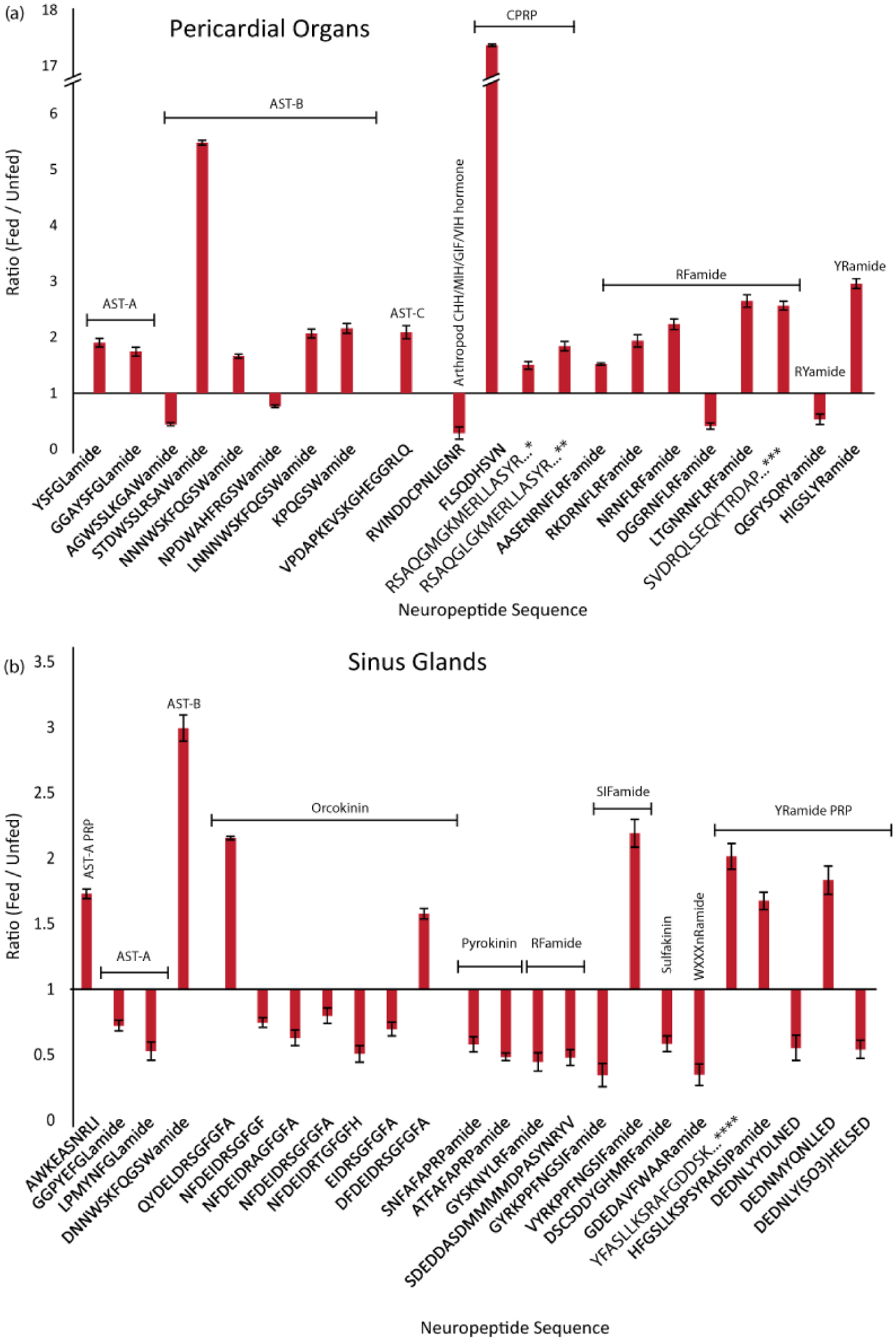
Feeding state-related changes in neuropeptide abundance in the POs and SGs, two regions of the C. borealis neuroendocrine system. Bar graphs show neuropeptides that were significantly increased or decreased in fed- compared to unfed tissue. The y-axis represents the ratio of fed- to unfed abundance. Significance based on p < 0.05, Unpaired Two-Tailed Student’s t-Test, n=5. Error bars represent SEM. Neuropeptide families are indicated, allatostatin (AST), crustacean hyperglycemic hormone precursor related peptide (CPRP), precursor related peptide (PRP), diuretic hormone (DH), pigment dispersing hormone (PDH). Full sequences: *RSAQGLGKMERLLASYRGALEPNTPLGDLSGSLGHPVE, **RSAQGLGKMEHLLASYRGALEPNTPLGDLSGSLGHPVE, ***SVDRQLSEQKTRDAPVAPTATIHSPAKTQESHRS, ****YFASLLKSRAFGDDSKLIPHNAAGDSEPHLQ
Table 2.
Neuropeptides exhibiting changes in abundance after feeding in the pericardial organ (PO) of C. borealis, including each neuropeptide’s family, sequence, and ratio (fed/unfed) and p-value for each tissue. Significance is based on p < 0.05, Unpaired, Two-Tailed Student’s t-Test, n=5. Neuropeptide families are indicated, allatostatin (AST), crustacean hyperglycemic hormone precursor related peptide (CPRP), precursor related peptide (PRP).
| Family | Sequence | Ratio | p-value |
|---|---|---|---|
| AST A-type | YSFGLamide | 2.19 | 0.012 |
| AST A-type | GGAYSFGLamide | 2.60 | 0.049 |
| AST B-Type | AGWSSLKGAWamide | 0.97 | 3.53E-04 |
| AST B-type | STDWSSLRSAWamide | 6.44 | 1.84E-04 |
| AST B-type | NNNWSKFQGSWamide | 8.49 | 9.52E-05 |
| AST B-type | NPDWAHFRGSWamide | 0.77 | 0.006 |
| AST B-type | LNNNWSKFQGSWamide | 11.41 | 0.002 |
| AST B-type | KPQGSWamide | 2.51 | 0.029 |
| AST C-type PRP | VPDAPKEVSKGHEGGRLQ | 42.76 | 0.022 |
| MIH hormone | RVINDDCPNLIGNR | 0.32 | 0.013 |
| CPRP | FLSQDHSVN | 20.44 | 9.37E-07 |
| CPRP | RSAQGLGKMEHLLASYRGALEPNTPLGDLSGSLGHPVE | 4.31 | 0.031 |
| CPRP | RSAQGLGKMERLLASYRGALEPNTPLGDLSGSLGHPVE | 5.45 | 0.013 |
| RFamide | AASENRNFLRFamide | 1.56 | 0.001 |
| RFamide | RKDRNFLRFamide | 5.97 | 0.045 |
| RFamide | NRNFLRFamide | 2.97 | 0.013 |
| RFamide | DGGRNFLRFamide | 0.59 | 0.003 |
| RFamide | LTGNRNFLRFamide | 12.38 | 0.007 |
| RFamide PRP | SVDRQLSEQKTRDAPVAPTATIHSPAKTQESHRS | 4.52 | 0.011 |
| RYamide | QGFYSQRYamide | 0.65 | 0.032 |
| YRamide | HIGSLYRamide | 3.78 | 0.010 |
The SGs, located in the crab eyestalks, are another major source of circulating hormones. In this tissue, 171 neuropeptides were detected, of which 25 displayed distinct abundance between the unfed and fed states (Fig. 2b and Table 3). Unlike the other tissues, where most abundance changes were increases when examined 30 minutes post-feeding, 16 of the aforementioned 25 SG peptides (64%) decreased in abundance. These peptides belong to the AST-A, orcokinin, YRamide precursor-related peptide, pyrokinin (PK), sulfakinin (SK), and WXXXnRamide families. While these neuropeptides were also identified in other tissues, only one of them exhibited an abundance change in another tissue. Specifically, the orcokinin peptide family member EIDRSGFGFA (m/z 1097.503) was lower in abundance after feeding in brain as well as in SG tissue. SG peptides that exhibited an increased abundance belonged to the A-type and B-type ASTs, orcokinin, YRamide precursor-related peptide, and SIFamide families. While the various isoforms of SIFamide have been widely characterized,70–73 functional studies have only recently been conducted in crab species.73, 74 This decreased abundance suggests that these SG peptides were released into the hemolymph to act long-range on neuronal circuits and possibly non-neural tissues within the animal. As above, the reason for the increased abundance of other SG peptides remains speculative. It may indicate inhibition of neuronal release leading to an accumulation in preparation for later release to modulate subsequent feeding-related behavior, or perhaps to enable initiation of later aspects of the feeding process. Overall, these changes appeared to generally be tissue-specific.
Table 3.
Neuropeptides exhibiting changes in abundance after feeding in sinus gland (SG) of C. borealis, including each neuropeptide’s family, sequence, and ratio (fed/unfed) and p-value for each tissue. Significance is based on p < 0.05, Unpaired, Two-Tailed Student’s t-Test, n=5. Neuropeptide families are indicated, allatostatin (AST), precursor related peptide (PRP).
| Family | Sequence | Ratio | p-value |
|---|---|---|---|
| AST A-type PRP | AWKEASNRLI | 1.78 | 0.001 |
| AST A-type | GGPYEFGLamide | 0.76 | 0.015 |
| AST A-type | LPMYNFGLamide | 0.56 | 0.022 |
| AST B-type | DNNWSKFQGSWamide | 4.78 | 0.015 |
| Ocrokinin | QYDELDRSGFGFA | 2.19 | 2.01E-05 |
| Orcokinin | NFDEIDRSGFGF | 0.79 | 0.007 |
| Orcokinin | NFDEIDRAGFGFA | 0.65 | 0.039 |
| Orcokinin | NFDEIDRSGFGFA | 4.82 | 0.037 |
| Orcokinin | NFDEIDRTGFGFH | 0.53 | 0.013 |
| Orcokinin | EIDRSGFGFA | 0.75 | 0.016 |
| Orcokinin | DFDEIDRSGFGFA | 4.29 | 4.55E-04 |
| Pyrokinin | SNFAFAPRPamide | 0.60 | 0.007 |
| Pyrokinin | ATFAFAPRPamide | 0.49 | 0.001 |
| RFamide | GYSKNYLRFamide | 1.04 | 0.003 |
| RFamide | SDEDDASDMMMMDPASYNRYV | 0.88 | 0.001 |
| SIFamide | GYRKPPFNGSIFamide | 0.91 | 0.001 |
| SIFamide | VYRKPPFNGSIFamide | 3.43 | 0.022 |
| Sulfakinin | DSCSDDYGHMRFamide | 0.70 | 0.024 |
| WXXXnRamide | GDEDAVFWAARamide | 0.38 | 0.002 |
| Yramide PRP | YFASLLKSRAFGDDSKLIPHNAAGDSEPHLQ | 5.46 | 0.011 |
| YRamide PRP | HFGSLLKSPSYRAISIPamide | 8.44 | 0.009 |
| YRamide PRP | DEDNLYYDLNED | 0.74 | 0.043 |
| YRamide PRP | DEDNMYQNLLED | 6.26 | 0.041 |
| YRamide PRP | DEDNLY(SO3)HELSED | 40.97 | 0.005 |
Changes in tissue localization
In addition to measuring changes in peptide abundance between the fed and unfed states, we visualized neuropeptide distribution within brain tissue and POs in a feeding state-dependent manner, to provide additional clues regarding the function of these peptides. Specifically, we implemented MALDI-MS imaging to spatially localize neuropeptides in these two regions. Twenty-nine neuropeptides were reproducibly detected in brain sections across biological replicates, and 59 neuropeptides were consistently detected in PO biological replicates. Of these, a subset in each tissue exhibited visualized localization differences between unfed and fed animals. All images were normalized to the total ion current (TIC). As the abundance of peptides throughout each tissue cannot be absolutely quantified, the results here are provided as complementary data to the quantitative data obtained from isotopically labeled tissue extracts in the previous section.
In the brain tissue sections, 7 neuropeptides exhibited spatial distributions that may be differential between animals in unfed and fed groups (Fig. 4). These distributions were conserved across 3 fed and 3 unfed biological replicates (Fig. S1). The optical images of two representative brain sections are shown in Figure 4a. The localization of two of the detected neuropeptides, the tachykinin APSGFLGMRamide (Fig. 4b) and the YRamide HL/IGSL/IYRamide (Fig. 4d), was limited primarily towards the outer regions of the brain in unfed crabs. These peptides were also observed in fed crabs with a more broadly distributed signal. Another peptide, KPKTEKK (Fig. 4c), showed a different pattern, where the peptide in the unfed section was localized toward the perimeter of the brain whereas peptide in the fed tissue showed a more centralized distribution pattern. The 4 other neuropeptides, all members of the RFamide superfamily including GRPNFLRFamide, YGAHVFLRFamide, GNSFLRFamide, and RARPRFamide, showed a broader distribution pattern throughout the brain in tissue sections in fed compared to unfed crabs. While some of these neuropeptides appear to also show differences in abundance in the quantitative analysis of tissue extracts (previous section), there were no statistically significant quantitative differences in abundance between the fed and unfed states for any of these peptides based on the tissue extract analysis. These changes in localization patterns suggest several possible functions, such as a relatively rapid release or accumulation of these peptides during or after feeding, perhaps to influence neuronal systems that regulate some aspect of the feeding process, such as the STNS-generated rhythms. The observed shift in localization could also represent two separate changes in abundance in different neurons, with one decreasing and the other increasing. While these differences may indicate a feeding-related function, the exact role(s) cannot be determined until downstream physiological analyses are performed.
Figure 4.
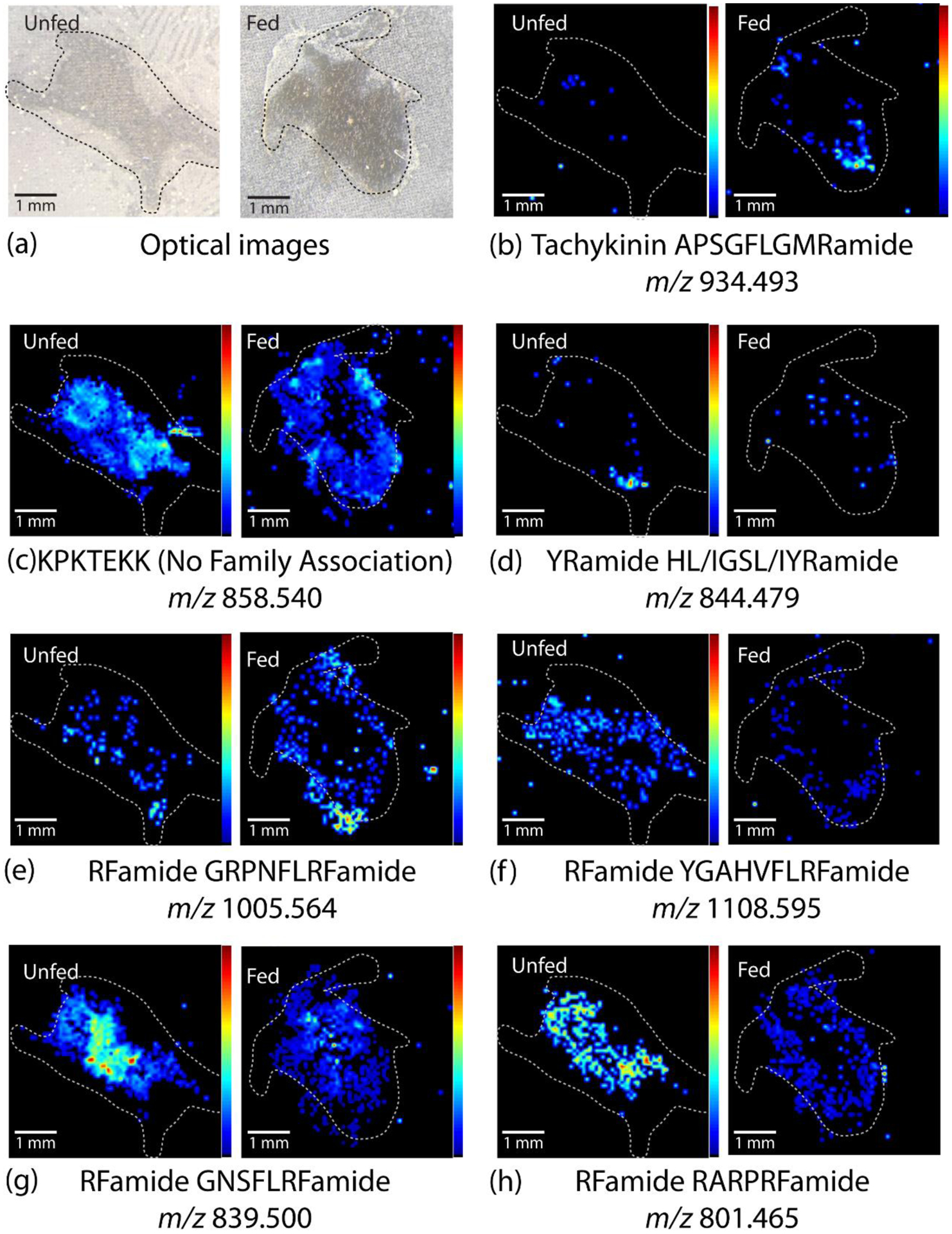
Representative MALDI-MS images of unfed and fed brain sections, including (a) the optical images of the brain section and (b-h) heatmap images of intensity distribution of specific neuropeptides (NPs) within each tissue. All images are normalized to the total ion current (TIC) and oriented such that the anterior side is top right and posterior side is bottom left. The images show differences in neuropeptide distribution between unfed and fed brain tissue for most neuropeptides. White dotted lines indicate approximate outline of tissue. Images from all replicates are shown in Fig S1.
Eleven detected neuropeptides showed localization patterns in fed and unfed POs that may be distinct between the two groups. Figure 5 shows representative MALDI-MSI results for these peptides; the results for both pairs of all 3 biological replicates are shown in Figure S2. The optical images of 2 representative POs are shown in Figure 5a. Three AST A-type neuropeptides, GHYNFGLamide, PKTYSFGLamide, and PRAYSFGLamide (Fig. 5b–d), localized to the side of the PO in unfed tissue, but showed an increased abundance at the posterior and anterior junctions in the fed tissue. Neuropeptides from the RFamide family (Fig. 5e–h) exhibited a similar trend in that the abundance appeared to decrease on the sides of the tissue and increase at the posterior and especially the anterior junction. These differences suggest a possible role for AST-A and RFamides that is region-specific within the POs, though the presence of region-specific release sites or a region-specific function for neuropeptides in the PO is not currently known. Four additional neuropeptides in the PO revealed by MALDI-MSI did not show any localization patterns that were conserved within their families but exhibited distinct patterns in the fed and unfed animals, such as a change in localization from the anterior to the posterior side, as occurred for the tachykinin SGFLGMRamide (Fig. 5i), KPKTEKK (Fig. 5j), and the RYamide FVGGSRYamide (Fig. 5k). Proctolin (RYLPT), a neuropeptide well characterized in the STNS and cardiac neuromuscular system, also displayed increased abundance at the posterior and anterior junctions in fed- compared to unfed animals, with less abundance in the central part of the PO (data not shown). This observation suggests that proctolin plays a modulatory role as a circulating hormone related to the feeding process in addition to being released from projection neurons directly into the crab STG neuropil to modulate the pyloric rhythm.75–79 These peptides exhibiting changes in localization were not found to have significant abundance changes in the extract analysis, indicating that the changes shown are based on relative changes in distribution only. Overall, these observed changes in localization of specific neuropeptides in the brain and PO complement the quantitative data from the previous section assessing total peptide content to provide more detail as to which neuropeptides may contribute to the behavioral outputs involved in the feeding process.
Figure 5.

Representative MALDI-MS images of unfed and fed pericardial organs, including (a) optical images of the PO and (b-l) heatmap images of intensity distribution of specific neuropeptides within each tissue. PO images were selected from 1 of 3 replicates for each m/z. All images are normalized to the total ion current (TIC) and oriented such that the anterior side is on top and posterior side is on the bottom. The images show differences between unfed and fed tissue for various neuropeptides, such as distinct differences in presence within the tissue or relative abundance in various areas of the tissue. White dotted lines indicate approximate outline of tissue. Images from all replicates are shown in Fig S1.
Putative novel neuropeptides identified with de novo sequencing
In addition to observing changes in abundance and localization of known neuropeptides in various tissues, we searched the data for putative neuropeptides not currently present in the database. To this end, with the use of PEAKS de novo sequencing, peptide sequences of MS/MS spectra were obtained. These sequences were mined for peptides that either possessed the sequence motifs characteristic of common crustacean neuropeptide families or exhibited a large degree of sequence similarity to those already present in the database. After filtering through neuropeptide sequences, each MS/MS spectrum was manually examined to ensure that it had a score ≥75% average local confidence (ALC), representing an average of the confidence of each amino acid within the sequence, and cleavage between every amino acid to ensure proper sequence assignment. We discovered 69 peptides that adhered to these specifications, with motifs similar to those belonging to the A-type AST, orcomyotropin, RFamide, and RYamide peptide families. An “other” category was also included for neuropeptides not matching a sequence motif, but having high sequence similarity to existing neuropeptides in the database. The sequences are listed in Table S1 (see Supporting Information), along with their molecular weight, tissue(s) in which they were detected, and ALC. The MS/MS spectra for the de novo sequences are shown in Figures S3–71.
Figure 6a shows the number of putative novel neuropeptides identified in each tissue. The largest numbers of neuropeptides were identified in the SGs, POs, and brain, with 32, 29, and 26 neuropeptides identified in each tissue, respectively. These tissues play key neuromodulatory functions, either by releasing circulating hormones or through direct neural projections onto other tissues. A small number of putative novel neuropeptides were also detected in the CoG. Notably, the tissues with the smallest number of neurons, the STG and OG, also contained putative neuropeptides not present in the database. Two RFamides, SPANFLRFamide and EHNFLRFamide, were identified in the STG. In the OG, one peptide, DLPKVDVSLK, was identified that has a sequence similar to several common neuropeptides already present in the database. None of these neuropeptides appear to have been previously identified in a crustacean species. However, one neuropeptide identified in the SGs, neuropeptide PF1 (SDPNFLRFamide), was previously identified in several other invertebrate species, including two species of roundworm, Ascaridia galli80 and Ascaris suum81, a land snail, Helix aspersa82, and three species of nematode, Caenorhabiditis briggsae83, Caenorhabditis elegans84, and Panagrellus redivivus85. Previous studies of this neuropeptide revealed a role in inhibiting muscle contractions in A. galli and A. suum, but it is not known if the peptide possesses the same function in C. borealis80, 86–88.
Figure 6.
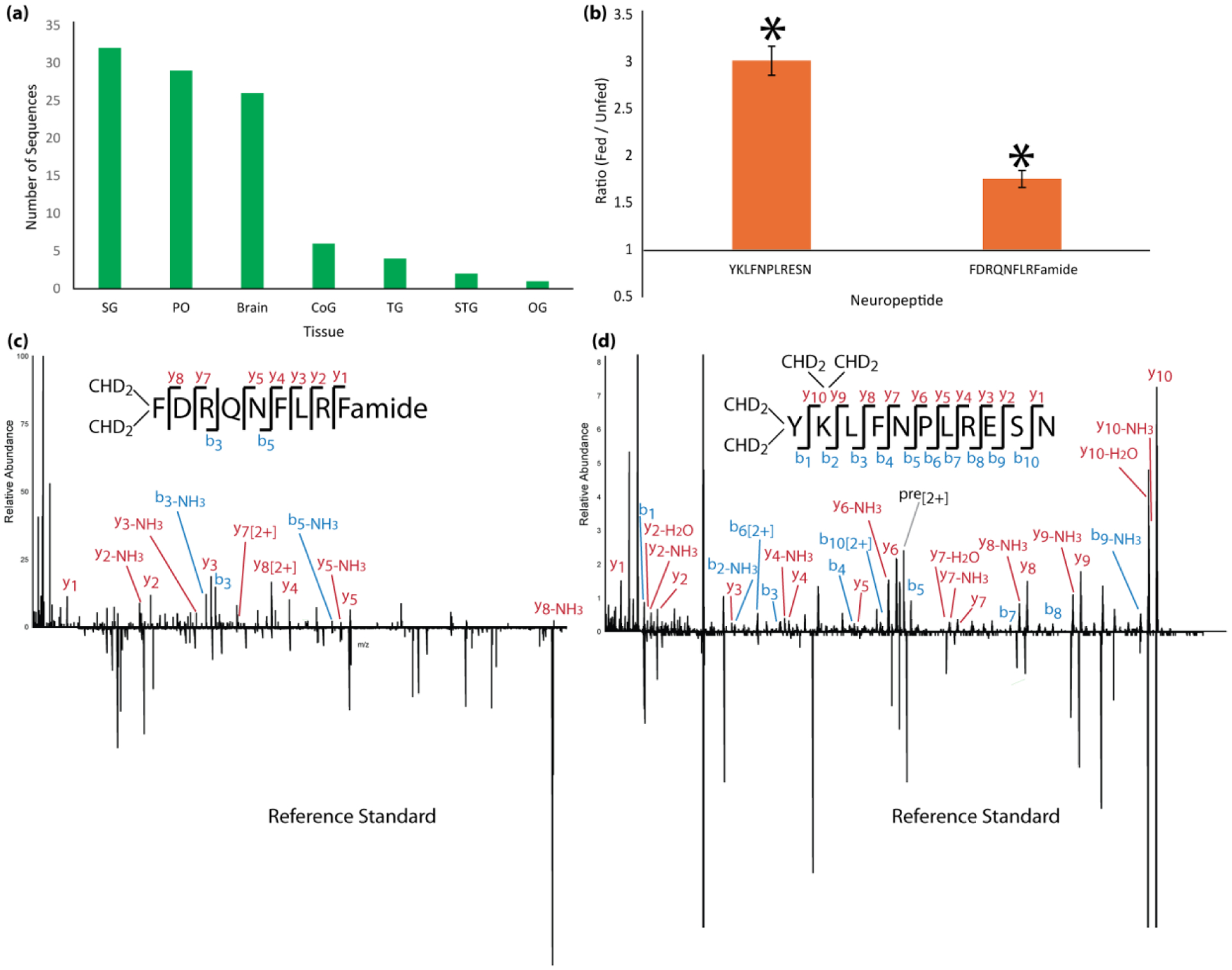
Summary of results of putative novel neuropeptide sequences identified, including (a) the number of novel neuropeptides identified in each tissue and (b) the relative ratio of two of these neuropeptides in fed and unfed POs that were significantly different (p < 0.05, n=5). Error bars represent standard error of the mean. No other novel sequences showed changes that were statistically significant. (c, d) MS/MS spectra mirror plots of the two novel neuropeptides identified with de novo sequencing, including (c) RFamide peptide FDRQNFLRFamide and (d) Cryptocyanin-like peptide YKLFNPLRESN. The top spectrum shows the experimentally-obtained spectrum of each from tissue extract and the bottom mirrored spectrum shows the spectrum obtained from a commercially-synthesized standard. Most key fragment ions are present in both spectra. The difference in intensity of some of the ions is likely due to ion suppression from interfering artifacts in the sample.
It was unclear whether any of these newly identified neuropeptides are biologically active, but two that we studied (YKLFNPLRESN and FDRQNFLRFamide) exhibited increased abundance in the POs of fed compared to unfed animals (p<0.05; Fig. 6b). The MS/MS spectra of these two peptides are shown in Figures 6c and 6d. As can be observed, both spectra appear to be good quality with complete backbone fragmentation and sufficient ion intensity. MS/MS spectra of all other neuropeptides are shown in Figures S3–71 (see Supporting Information). No other novel neuropeptides exhibited statistically significant changes in abundance between fed and unfed animals.
Two novel neuropeptides influence the cardiac neuromuscular system
To determine whether the two aforementioned, putative novel neuropeptides that are up-regulated in the PO of fed crabs are biologically active, we custom synthesized standards of each peptide for use in functional studies. Figures 6c and 6d show a mirror plot of the MS/MS spectrum obtained for each peptide in PO extract compared to the MS/MS spectrum of the respective standard, confirming the identity of each peptide. The peptides were each perfused through the cardiac sinus within the intact crab heart at a concentration of 10−6 M. In all preparations, perfusion of FDRQNFLRFamide elicited a reversible increase in both amplitude and frequency of heart contractions (p<0.05, n=10, Paired Student’s t-Test) (Fig. 7a,b). YKLFNPLRESN elicited a reversible decrease in contraction frequency in five of six heart preparations (Fig. 7c,d). In the sixth preparation, this peptide instead elicited a decreased contraction frequency. While this difference could indicate inter-animal variability in response to this peptide, the observed trend was that perfusing this peptide decreases contraction frequency, though it did not reach statistical significance (p>0.05, n=6). Unlike the change in contraction frequency, applying YKLFNPLRESN did not change the contraction amplitude in any preparations (p>0.05, n=6). These changes in heartbeat are summarized in Figure 7e, with the individual values shown in Figure S72 (See Supporting Information), and they indicate that at least one of these novel neuropeptides can modulate heartbeat, thus demonstrating their biological activity. However, as the concentrations of peptides tested in this study are relatively high, a threshold concentration would need to be determined to assess whether these effects are likely to be behaviorally relevant. Moreover, although many peptides do modulate both the STNS and the heart in crustaceans, additional experiments are needed to determine whether these peptides modulate the feeding circuitry.
Figure 7.
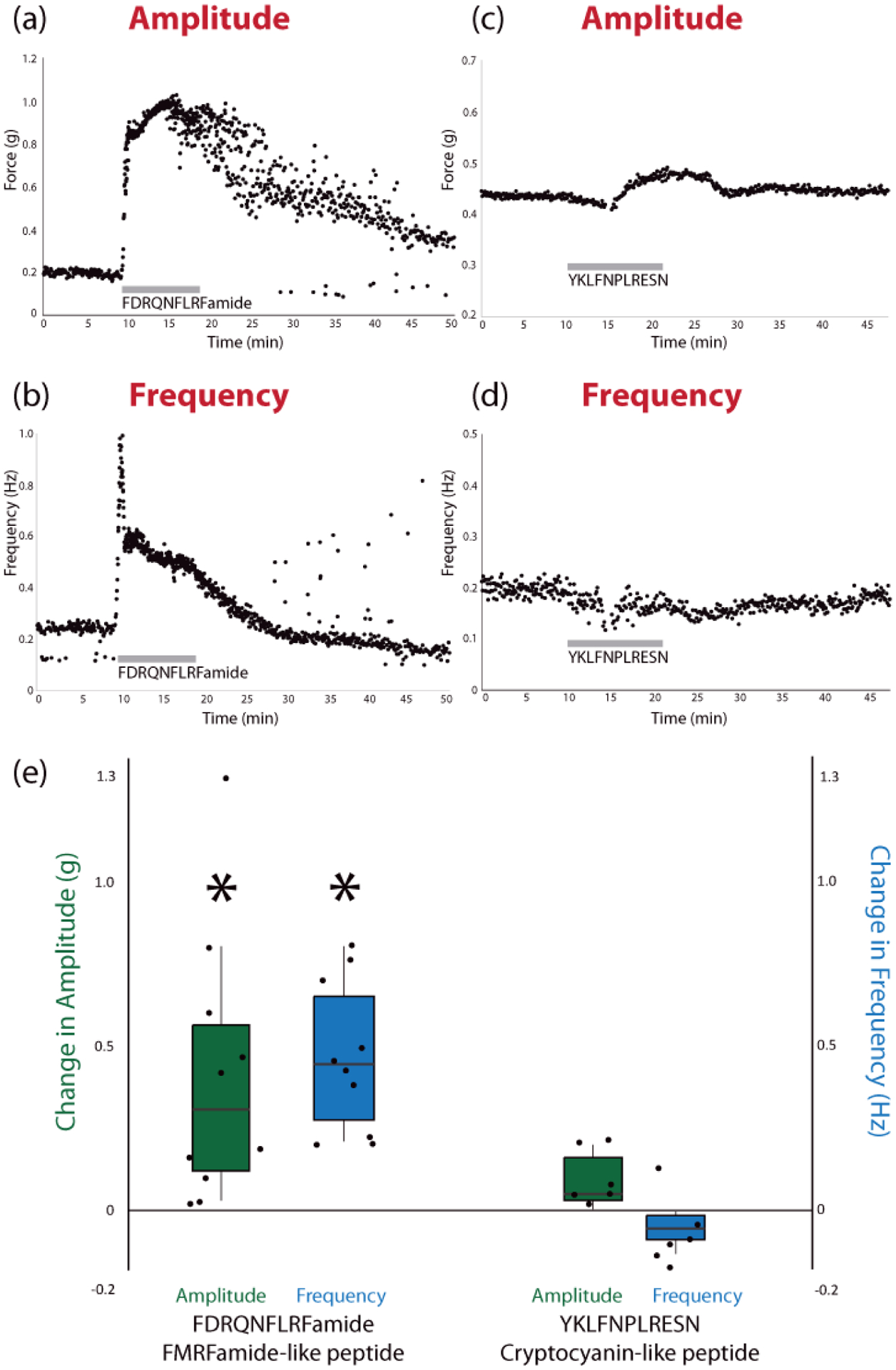
Results from functional assessment of synthesized peptides on ex vivo whole heart preparations, including examples of the change in amplitude and frequency during perfusion of each peptide (a-d) and the average change in amplitude and frequency for each peptide (e), relative to controls, across all biological replicates (*p < 0.05, Student’s Paired Two-Tailed t-Test, n = 10 for FDRQNFLRFamide, n = 6 for YKLFNPLRESN). Individual values for control and experimental conditions are shown in Fig S72. The average control amplitude was 0.17 g and the average control frequency was 0.20 Hz across all replicates (n = 15). In the boxplots in (e), the center line indicates the median value, the top and bottom of the box represent the 75% quartile and 25% quartile, respectively, and the upper and lower whiskers indicate the largest and smallest values, respectively, not exceeding 1.5 times the inter-quartile range.
Conclusions
This study examined neuropeptide abundances and distributions in key neural and neuroendocrine tissues of C. borealis in a feeding state-dependent manner. By employing a multifaceted mass spectrometric approach using complementary ionization- and acquisition methods, we detected 278 neuropeptides, based on our in-house crustacean neuropeptide database, which was compiled from various sources.47, 89, 90 Through the use of duplex formaldehyde labeling, we determined that a subset of these peptides exhibited either increased or decreased abundance in the analyzed tissues of fed compared to unfed animals, suggesting functional roles in the feeding and digestion process (though other peptides known to be present may also have functional effects that are not captured in this study). The results show that neuropeptides in the ganglia mostly exhibited increases in abundance, while neuropeptides in the neurohemal tissues exhibited deceases as well as increases. Additionally, using MALDI-MSI, we found changes in the spatial distribution of some peptides during these two feeding states. By combining the information gained from these two methodologies, we have obtained a list of neuropeptides that are positioned to influence one or more aspects of feeding behavior. Follow-up functional studies (e.g. electrophysiological recordings) should shed light on the feeding-related role of each peptide. We also identified 69 new C. borealis neuropeptides as part of this study. While the biological activity of these neuropeptides is unknown, at least two of them were up-regulated in the POs of fed crabs compared to unfed crabs and one of them consistently modulated cardiac activity when applied to the isolated crab heart. Overall, this study provides deeper insight into the complexity of neuropeptide modulation in the STNS as well as providing a target cohort of feeding-related peptides that can be further investigated with subsequent electrophysiological experiments. The success of these experimental approaches in the crustacean nervous system offers promise for their successful use in studying neuropeptides in the larger and more complex vertebrate nervous system, thereby providing groundwork for comparable studies of more complex circuits driving rhythmic movements.
Methods:
Chemicals and Materials
Formaldehyde (FH2), deuterated formaldehyde (FD2), and ACS-grade formic acid (FA) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Gelatin was purchased from Becton Dickinson (Franklin Lakes, NJ, USA). All other chemicals and solvents were purchased from Fisher Scientific (Pittsburgh, PA, USA). Amicon Ultra 10 kDa 0.5 mL molecular weight cutoff (MWCO) devices and 10 μL C18 Ziptips were purchased from Merck Millipore (Billerica, MA USA), and 100 μL C18 Ziptips were purchased from Agilent Technologies (Santa Clara, CA, USA). Acidified methanol was prepared using 90/9/1 water/methanol/acetic acid. ACS-grade solvents and Milli-Q water (Merck Millipore, Billerica, MA, USA) were used for sample preparation, and Optima-grade solvents were used for MS analysis. Peptide standards were custom synthesized by GenScript Corporation (Piscataway, NJ, USA).
Animals and Feeding Experiments
C. borealis were purchased from the Fresh Lobster Company, LLC (Gloucester, MA USA). Animals were adjusted to their environment for two weeks prior to experiments. Animals were housed in artificial seawater tanks (12° – 13° C) with an alternating 12-hour light/dark cycle. All animals were kept in an unfed state for 2 weeks prior to feeding experiments.
For tissue experiments, 3 control (unfed) animals and 3 experimental (fed) animals were used per biological replicate. Experimental animals were fed an excess of tilapia for 30 minutes and control animals were left in an unfed state. Animals were then cold-anesthetized on packed ice for 30 minutes, and dissected in chilled physiological saline (440 mM NaCl, 11 mM KCl, 26 mM MgCl2, 13 mM CaCl2, 11 mM Trizma base, 5 mM maleic acid, adjusted to pH 7.45 with NaOH) as described elsewhere.91 The SGs, POs, brain, commissural ganglia, and STG were collected separately and placed in chilled acidified methanol, with 5 biological replicates total. For imaging of the brain and POs, the experiment was conducted as described above except that the tissue was quickly dipped in water to desalt after dissection. The brain was embedded in gelatin and flash-frozen in dry ice. The POs were placed directly on a glass microscope slide and placed in a desiccator box. The tissue was then stored at −80°C. A total of 3 control and 3 experimental biological replicates (using distinct animals) were performed for MS imaging of each tissue.
Extraction and Desalting
Tissues were extracted in chilled acidified methanol. For each tissue type, samples from the three experimental animals were pooled, as were those from the control animals. The sets of tissue were manually homogenized with 150 μL acidified methanol (50 μL per tissue), sonicated using a bath sonicator, and then centrifuged for 10 minutes at 16.1 rcf. The supernatant was transferred to a new tube, and 50 μL of acidified methanol was added to the pellet. The pellet was then homogenized, sonicated, and centrifuged as previously. The resulting supernatant was again kept, and the process was repeated a second time. The combined supernatant from the three extractions was evaporated in a SpeedVac on medium heat and stored at −80° C.
Tissue samples were desalted with 10 μL C18 ziptips using the manufacturer’s instructions, except that a three-step elution was used, consisting of 25/75 acetonitrile/water with 0.1% FA, followed by 50/50 acetonitrile/water with 0.1% FA, and finally 75/25 acetonitrile/water with 0.1% FA. The three elutions were then combined and evaporated in the SpeedVac on medium heat.
In-Solution Formaldehyde Labeling
Control and experimental samples were differentially labeled using 2-plex isotopic formaldehyde. Samples were suspended in 10 μL of water, after which 10 μL of 1% formaldehyde solution (FH2 or FD2) was added to each, followed by 10 μL borane pyridine. The samples were then incubated in a 37° C water bath for 15 minutes, after which 10 μL of 100 mM ammonium bicarbonate was added to quench the labeling reaction. Light and heavy-labeled samples were then combined and evaporated in the SpeedVac on medium heat. The labels used for control and experimental samples were alternated for different biological replicates to account for possible labeling bias.
Samples were reconstituted in 15 μL of 0.1% formic acid (FA) in water. A 2.5 μL-aliquot was removed from each sample and combined with 2.5 μL of 150 mg/mL 2,5-dihydroxy benzoic acid (DHB) matrix. The mixture was then spotted on a stainless steel MALDI plate, 1 μL per spot, for MALDI-MS analysis. The remaining 12.5 μL of sample was desalted using 10 μL C18 zip tips with the three-step gradient described above and evaporated in the SpeedVac on medium heat. The sample was then dissolved in 15 μL of 0.1% FA in water for LC-ESI-MS/MS analysis.
Brain Sectioning and Matrix Application for Imaging
Brain tissues that were embedded in gelatin were sectioned on a cryostat at −23° C. Sections of 12 μm thickness were thaw-mounted on a glass microscope slide. An automated matrix deposition sprayer (TM sprayer, HTX Technologies LLC, Chapel Hill, NC, USA) was used to apply 12 layers of 40 mg/mL DHB matrix to brain tissue sections and POs, with 30 sec drying time between each coating.
Mass Spectrometry and MALDI Imaging
MALDI-MS analysis was performed using a Thermo Scientific MALDI-LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany) equipped with a 337.1 nm, 60 Hz nitrogen laser for analysis of spots of tissue and hemolymph extract. Spots and tissue sections were analyzed in positive-ion mode with an m/z range of 500 to 2000 and 20 kJ laser energy. Crystal positioning system (CPS) plate motion was used to acquire a total of 50 scans (2 microscans/scan) for each spot of extract (tissue and hemolymph). Three spots were analyzed from each sample, generating 3 technical replicates per sample. For brain tissue imaging, optical images were acquired with an HP scanner and imported into the LTQ tune page. A raster plate motion was used to obtain spectra over the tissue section area with a step size of 75 μm.
For acquisition of MS/MS spectra, a Thermo Scientific Q Exactive instrument (Thermo Scientific, Bremen, Germany) coupled to a nano-ESI source was used with a Waters nanoAcquity LC system (Waters Corp, Milford, MA, USA). A 15 cm self-packed C18 column (75 μm internal diameter, 1.7 μm particle size) was used for LC separation. Mobile phases consisted of water with 0.1% FA (solution A) and acetonitrile with 0.1% FA (solution B), and the flow rate was set to either 0.300 or 0.350 μL/min (to ensure a pressure of approximately 7000 psi at 100% solution A). An injection volume of 2 μL of each sample was loaded onto the column and separated over a 108 minute gradient as follows: 0–0.5 min 0–4% B; 0.5–70 min 4–35% B; 70–80 min 35–75% B; 80–81 min 75% B; 81–82 min 75–95% B; 82–92 min 95% B; followed by column re-equilibration at 92–93 min 95–0% B; 93–108 min 0% B. MS acquisition took place in positive ESI mode with a top 10 data dependent acquisition (DDA), in which the top 10 most abundant precursor ions were selected for HCD fragmentation with an isolation window of 2 m/z and a collision energy of 30 eV. The MS scan range was 300 – 1800 m/z at 35,000 resolution, and the MS/MS scan range was adjusted depending on the parent mass, with a fixed first mass of 120 m/z at 17,500 resolution.
Whole Heart Functional Assessment of Neuropeptides on Cardiac Neuromuscular System
The function of two newly identified neuropeptides was tested on the isolated whole heart of C. borealis, as described in previous literature20, 21, 92. Briefly, crabs were anesthetized on ice for 30 minutes, after which the heart and carapace above the heart were removed and placed in a dissecting dish filled with chilled artificial saline. The heart remained attached to the carapace such that the amount of stretch placed on the heart was identical to in vivo conditions. Artificial saline at 12° C was perfused both through the heart via the posterior artery and over the heart to maintain temperature, at a flow rate of 2.5 mL/min using a Peltier temperature control system (CL100 bipolar temperature controller and SC-20 solution heater/cooler; Warner Instruments, Hamden, CT, USA). The anterior arteries were tied with size 6–0 suture silk to a Grass FT03 force-displacement transducer (AstroNova, Inc., West Warwick, RI, USA) at a 45° angle and 1 g baseline force for heart contraction measurements. Heart preparations were left to stabilize for at least 1 hr, and then peptide standards at 1×10−6 M in saline were perfused into the heart. The force displacement transducer was connected to an ETH-250 Bridge amplifier (CB Sciences, Dover, NH, USA). Data were acquired on a computer using a Micro 1401 data acquisition unit with Spike2 version 9 software (Cambridge Electronic Design Limited, Cambridge, UK).
Data Analysis
For analysis of MALDI-MS data, neuropeptides were identified by accurate-mass matching to an in-house crustacean neuropeptide database, with a 5 ppm mass tolerance. The database included neuropeptides previously identified in our lab, those predicted by the C. borealis transcriptome47, and crustacean neuropeptides reported in NeuroPep90. For LC-ESI-MS/MS data analysis, raw data acquired from the Q Exactive instrument was processed using PEAKS de novo software (Bioinformatics Solutions Inc., Waterloo, ON, CAN)93. PEAKS parameters were set as follows: no enzyme cleavage specified, instrument orbi-orbi, HCD fragmentation, and precursor correction. For de novo sequencing and database searching, modifications were set to light and heavy dimethylation, amidation, pyroglutamate, and oxidation. All other parameters were set to the default. For quantitative analysis, neuropeptides were included only if both the light and heavy-labeled peaks were detected. Ratios were calculated by dividing the experimental intensity by the control intensity. For putative neuropeptide identifications, results from PEAKS were filtered using PepExplorer94 for sequence similarity to our crustacean neuropeptide database, and peptides were selected that had an average local confidence (ALC) score of 75% or greater. MS/MS spectra were manually inspected for fragment ions indicative of cleavage at every amino acid to ensure the appropriate sequence was assigned. Each peptide sequence was searched in the NeuroPep database to determine if it had been previously identified in other species90.
MALDI-MS images of brain and pericardial organ tissue were generated and exported as imzml files using Thermo Scientific ImageQuest software. Data were then loaded into MSiReader to generate images of neuropeptides in the crustacean neuropeptide database with a 5 ppm mass error95. All images were normalized to the TIC and 5th order linear smoothing was applied.
Statistical Information
For quantitative analysis, ratios (fed / unfed) were averaged across 8 biological replicates for MALDI-MS and 5 biological replicates for ESI-LC-MS/MS, each using the average of 3 technical replicates as its value, and the standard error of the mean (SEM) was calculated. Averaging was done in the same manner for peptides detected with both ionization sources. Statistical analysis was done using a Student’s Unpaired Two-Tailed t-Test, with p<0.05 indicating statistical significance. For analysis of whole-heart preparations, a Student’s Paired Two-Tailed t-Test was used to compare the average heart contraction amplitude and frequency between baseline and peptide conditions. For the peptide FDRQNFLRFamide, 10 biological replicates were used. For YKLFNPLRESN, 6 biological replicates were used for determining significance.
Supplementary Material
Figure 3.
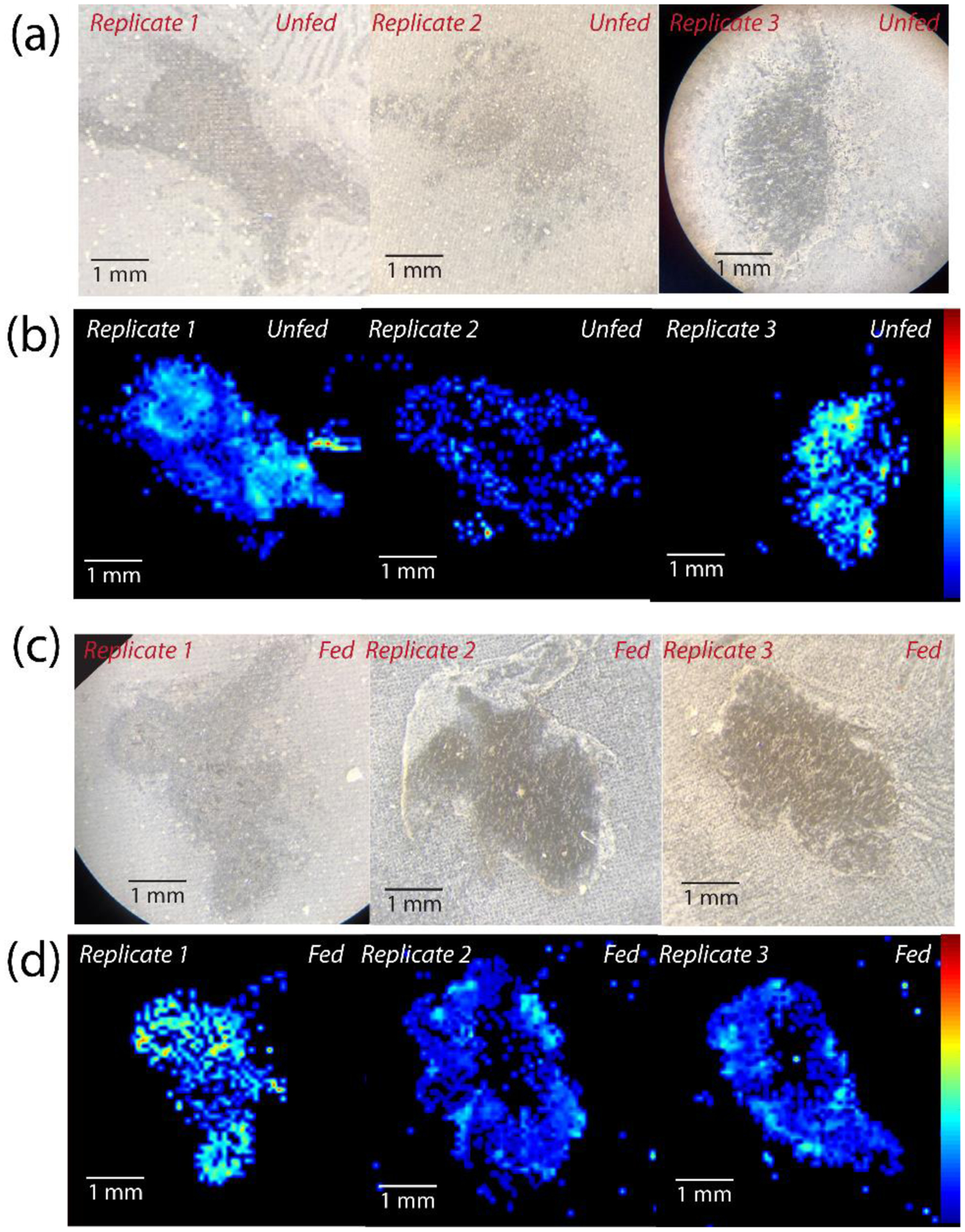
Optical and MALDI-MS images of fed and unfed brain sections for all 3 biological replicates of each condition for one representative peptide, KPKTEKK (m/z 858.540), showing the biological reproducibility between sections originating from distinct animals, including (a) optical images of 3 unfed brain sections, (b) MALDI-MS images of unfed brain sections, (c) optical images of fed brain sections, and (d) MALDI-MS images of fed brain sections. Brain sections are oriented such that the anterior is on the right and posterior is on the left.
Acknowledgment
This work was supported by the National Institutes of Health through grants R01DK071801 (LL), R01NS029436 (MPN, LL), and P20GM0103423 (PSD). Partial support of this research was also provided by National Science Foundation (CHE-1710140 LL; IOS-1354567 PSD). The Orbitrap instruments were purchased through the support of an NIH shared instrument grant (NIH-NCRR S10RR029531) and Office of the Vice Chancellor for Research and Graduate Education at the University of Wisconsin-Madison. KD acknowledges a predoctoral fellowship supported by the National Institutes of Health, under Ruth L. Kirschstein National Research Service Award from the National Institutes of Health-General Medical Sciences (1F31GM126870-01A1) and a predoctoral traineeship from the National Heart Lung and Blood Institute to the University of Wisconsin-Madison Cardiovascular Research Center (T32 HL 007936). LL acknowledges a Vilas Distinguished Achievement Professorship and Charles Melbourne Johnson Professorship with funding provided by the Wisconsin Alumni Research Foundation and University of Wisconsin-Madison School of Pharmacy.
Footnotes
Supporting Information
The following supporting information is available free of charge at ACS website http://pubs.acs.org
- MALDI-MS images of all biological replicates of brain and POs, table of putative neuropeptides detected with de novo sequencing, MS/MS spectra of all de novo sequences, Amplitude and frequency measurements for heart preparations when perfused with saline as a control (C) and when perfused with selected peptides (P) with each data point indicating a unique biological replicate from distinct animals.
- Spreadsheet of all detected neuropeptides across all tissues.
- Spreadsheet of putative novel neuropeptides identified with de novo sequencing.
Data Availability
The data used in this study are available from the corresponding author upon reasonable request.
Competing Interests
The authors declare no competing interests.
References
- 1.Zimmerman CA, Knight ZA Layers of signals that regulate appetite. Curr Opin Neurobiol 2020, 64, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu A, Yu B, Komiyama T Plasticity in olfactory bulb circuits. Curr Opin Neurobiol 2020, 64, 17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hulse BK, Jayaraman V Mechanisms underlying the neural computation of head direction. Annu Rev Neurosci 2019, 43, 31–54. [DOI] [PubMed] [Google Scholar]
- 4.Currier TA, Nagel KI Multisensory control of navigation in the fruit fly. Curr Opin Neurobiol 2019, 64, 10–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barkan CL, Zornik E Feedback to the future: motor neuron contributions to central pattern generator function. J Exp Biol 2019, 222 (Pt 16), 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira-Pinto MJ, Ruder L, Capelli P, Arber S Connecting circuits for supraspinal control of locomotion. Neuron 2018, 100 (2), 361–374. [DOI] [PubMed] [Google Scholar]
- 7.Cropper EC, Jing J, Vilim FS, Weiss KR Peptide cotransmitters as dynamic, intrinsic modulators of network activity. Front Neural Circuits 2018, 12, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emmons SW Neural circuits of sexual behavior in Caenorhabditis elegans. Annu Rev Neurosci 2018, 41, 349–369. [DOI] [PubMed] [Google Scholar]
- 9.Daur N, Nadim F, Bucher D The complexity of small circuits: the stomatogastric nervous system. Curr Opin Neurobiol 2016, 41, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeLong ND, Nusbaum MP Hormonal modulation of sensorimotor integration. J Neurosci, 2010, Vol. 30, pp 2418–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mykles DL, Adams ME, Gäde G, Lange AB, Marco HG, Orchard I Neuropeptide action in insects and crustaceans. Physiol Biochem Zool 2010, 83 (5), 836–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marder E, Bucher D Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol 2007, 69, 291–316. [DOI] [PubMed] [Google Scholar]
- 13.Marder E, O’Leary T, Shruti S Neuromodulation of circuits with variable parameters: single neurons and small circuits reveal principles of state-dependent and robust neuromodulation. Annu Rev Neurosci 2014, 37, 329–346. [DOI] [PubMed] [Google Scholar]
- 14.Nusbaum MP, Blitz DM, Marder E Functional consequences of neuropeptide and small-molecule co-transmission. Nat Rev Neurosci 2017, 18 (7), 389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Follmann R, Goldsmith CJ, Stein W Spatial distribution of intermingling pools of projection neurons with distinct targets: A 3D analysis of the commissural ganglia in Cancer borealis. J Comp Neurol 2017, 525 (8), 1827–1843. [DOI] [PubMed] [Google Scholar]
- 16.Stein W Modulation of stomatogastric rhythms. J Comp Physiol A 2009, 195 (11), 989–1009. [DOI] [PubMed] [Google Scholar]
- 17.Marder E Neuromodulation of neuronal circuits: back to the future. Neuron 2012, 76 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nässel DR Neuropeptides in the nervous system of Drosophila and other insects: multiple roles as neuromodulators and neurohormones. Prog Neurobiol 2002, 68 (1), 1–84. [DOI] [PubMed] [Google Scholar]
- 19.Nusbaum MP, Blitz DM Neuropeptide modulation of microcircuits. Curr Opin Neurobiol 2012, 22 (4), 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dickinson PS, Armstrong MK, Dickinson ES, Fernandez R, Miller A, Pong S, Powers BW, Pupo-Wiss A, Stanhope ME, Walsh PJ, et al. Three members of a peptide family are differentially distributed and elicit differential state-dependent responses in a pattern generator-effector system. J Neurophysiol 2018, 119 (5), 1767–1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson PS, Sreekrishnan A, Kwiatkowski MA, Christie AE Distinct or shared actions of peptide family isoforms: I. Peptide-specific actions of pyrokinins in the lobster cardiac neuromuscular system. In J Exp Biol, 2015, Vol. 218, pp 2892–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeLaney K, Buchberger AR, Atkinson L, Grunder S, Mousley A, Li L New techniques, applications and perspectives in neuropeptide research. J Exp Biol 2018, 221 (Pt 3), 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Y, Wong JM, Mabrouk OS, Kennedy RT Reducing multifunctional adsorption to improve recovery and in vivo detection of neuropeptides by microdialysis with LC-MS. Anal Chem 2015, 87 (19), 9802–9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stemmler EA, Barton EE, Esonu OK, Polasky DA, Onderko LL, Bergeron AB, Christie AE, Dickinson PS C-terminal methylation of truncated neuropeptides: an enzyme-assisted extraction artifact involving methanol. Peptides 2013, 46, 108–125. [DOI] [PubMed] [Google Scholar]
- 25.Coleman MJ, Nusbaum MP Functional consequences of compartmentalization of synaptic input. J Neurosci 1994, 14, 6544–6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood DE, Stein W, Nusbaum MP Projection neurons with shared cotransmitters elicit different motor patterns from the same neural circuit. J Neurosci 2000, 20, 8943–8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood DE, Nusbaum MP, Extracellular peptidase activity tunes motor pattern modulation. J Neurosci 2002, 22 (10), 4185–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Bucher D, Nadim F Distinct co-modulation rules of synapses and voltage-gated currents coordinate interactions of multiple neuromodulators. J Neurosci 2018, 38 (40), 8549–8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behrens HL, Chen RB, Li LJ Combining microdialysis, nanoLC-MS, and MALDI-TOF/TOF to detect neuropeptides secreted in the crab, Cancer borealis. Anal Chem 2008, 80 (18), 6949–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma M, Wang J, Chen R, Li L Expanding the crustacean neuropeptidome using a multifaceted mass spectrometric approach. J Proteome Res 2009, 8 (5), 2426–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLaney K, Li L Data independent acquisition mass spectrometry method for improved neuropeptidomic coverage in crustacean neural tissue extracts. Anal Chem 2019, 91 (8), 5150–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen RB, Xiao MM, Buchberger A, Li LJ Quantitative neuropeptidomic study of the effects of temperature change in the crab Cancer borealis. J Proteome Res 2014, 13 (12), 5767–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Buchberger A, Muthuvel G, Li L Expression and distribution of neuropeptides in the nervous system of the crab Carcinus maenas and their roles in environmental stress. Proteomics 2015, 15(23–24), 3969–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y, Buchberger AR, DeLaney K, Li Z, Li L Multifaceted mass spectrometric investigation of neuropeptide changes in atlantic blue crab, Callinectes sapidus, in response to low pH stress. J Proteome Res 2019, 18 (7), 2759–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmerberg CM, Liang ZD, Li LJ Data-independent MS/MS quantification of neuropeptides for determination of putative feeding-related neurohormones in microdialysate. ACS Chem Neurosci 2015, 6 (1), 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, DeLaney K, Hui L, Wang J, Sturm RM, Li L A multifaceted mass spectrometric method to probe feeding related neuropeptide changes in Callinectes sapidus and Carcinus maenas. J Am Soc Mass Spectrom 2018, 29 (5), 948–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen RB, Hui LM, Cape SS, Wang JH, Li LJ Comparative neuropeptidomic analysis of food intake via a multifaceted mass spectrometric approach. ACS Chem Neurosci 2010, 1 (3), 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGaw IJ, Reiber CL Integrated physiological responses to feeding in the blue crab Callinectes sapidus. J Exp Biol 2000, 203 (Pt 2), 359–368. [DOI] [PubMed] [Google Scholar]
- 39.McGaw IJ, Curtis DL A review of gastric processing in decapod crustaceans. J Comp Physiol B 2013, 183 (4), 443–465. [DOI] [PubMed] [Google Scholar]
- 40.Sohn JW, Elmquist JK Williams KW, Neuronal circuits that regulate feeding behavior and metabolism. Trends Neurosci 2013, 36 (9), 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saideman SR, Ma M, Kutz-Naber KK, Cook A, Torfs P, Schoofs L, Li L, Nusbaum MP Modulation of rhythmic motor activity by pyrokinin peptides. J Neurophysiol 2007, 97 (1), 579–595. [DOI] [PubMed] [Google Scholar]
- 42.Nassel DR Tachykinin-related peptides in invertebrates: a review. Peptides 1999, 20 (1), 141–158. [DOI] [PubMed] [Google Scholar]
- 43.Nassel DR Functional roles of neuropeptides in the insect central nervous system. Naturwissenschaften 2000, 87 (10), 439–449. [DOI] [PubMed] [Google Scholar]
- 44.Severini C, Improta G, Falconieri-Erspamer G, Salvadori S, Erspamer V The tachykinin peptide family. Pharmacol Rev 2002, 54 (2), 285–322. [DOI] [PubMed] [Google Scholar]
- 45.Jorge-Rivera J, Marder E Allatostatin decreases stomatogastric neuromuscular transmission in the crab Cancer borealis. J Exp Biol 1997, 200 (Pt 23), 2937–2946. [DOI] [PubMed] [Google Scholar]
- 46.Bechtold DA, Luckman SM The role of RFamide peptides in feeding. J Endocrinol 2007, 192 (1), 3–15. [DOI] [PubMed] [Google Scholar]
- 47.Christie AE, Pascual MG Peptidergic signaling in the crab Cancer borealis: Tapping the power of transcriptomics for neuropeptidome expansion. Gen Comp Endocrinol 2016, 237, 53–67. [DOI] [PubMed] [Google Scholar]
- 48.OuYang C Neuropeptides in Crustaceans: Expression, Quantitation and Distribution Probed by Multi-Dimensional Mass Spectrometric Approaches. PhD Dissertation, University of Wisconsin-Madison, Madison, WI, USA, 2016. [Google Scholar]
- 49.Coleman MJ, Nusbaum MP, Cournil I, Claiborne BJ Distribution of modulatory inputs to the stomatogastric ganglion of the crab, Cancer borealis. J Comp Neurol 1992, 325 (4), 581–594. [DOI] [PubMed] [Google Scholar]
- 50.Stangier J, Hilbich C, Burdzik S, Keller R Orcokinin – a novel myotropic peptide from the nervous-system of the crayfish, Orconectes limosus. Peptides 1992, 13 (5), 859–864. [DOI] [PubMed] [Google Scholar]
- 51.Li L, Pulver SR, Kelley WP, Thirumalai V, Sweedler JV, Marder E Orcokinin peptides in developing and adult crustacean stomatogastric nervous systems and pericardial organs. J Comp Neurol 2002, 444 (3), 227–244. [DOI] [PubMed] [Google Scholar]
- 52.Skiebe P, Dreger M, Meseke M, Evers JF, Hucho F Identification of orcokinins in single neurons in the stomatogastric nervous system of the crayfish, Cherax destructor. J Comp Neurol 2002, 444 (3), 245–259. [DOI] [PubMed] [Google Scholar]
- 53.Dickinson PS, Stemmler EA, Barton EE, Cashman CR, Gardner NP, Rus S, Brennan HR, McClintock TS, Christie AE Molecular, mass spectral, and physiological analyses of orcokinins and orcokinin precursor-related peptides in the lobster Homarus americanus and the crayfish Procambarus clarkii. Peptides 2009, 30 (2), 297–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christie AE, Lundquist CT, Nassel DR, Nusbaum MP Two novel tachykinin-related peptides from the nervous system of the crab Cancer borealis. J Exp Biol 1997, 200 (17), 2279–2294. [DOI] [PubMed] [Google Scholar]
- 55.Blitz DM, White RS, Saideman SR, Cook A, Christie AE, Nadim F, Nusbaum MP A newly identified extrinsic input triggers a distinct gastric mill rhythm via activation of modulatory projection neurons. J Exp Biol 2008, 211 (Pt 6), 1000–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Messinger DI, Kutz KK, Le T, Verley DR, Hsu YW, Ngo CT, Cain SD, Birmingham JT, Li L, Christie AE Identification and characterization of a tachykinin-containing neuroendocrine organ in the commissural ganglion of the crab Cancer productus. J Exp Biol 2005, 208 (Pt 17), 3303–3319. [DOI] [PubMed] [Google Scholar]
- 57.Diehl F, White RS, Stein W, Nusbaum MP Motor circuit-specific burst patterns drive different muscle and behavior patterns. J Neurosci 2013, 33 (29), 12013–12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Christie AE, Kutz-Naber KK, Stemmler EA, Klein A, Messinger DI, Goiney CC, Conterato AJ, Bruns EA, Hsu YWA, Li L et al. Midgut epithelial endocrine cells are a rich source of the neuropeptides APSGFLGMRamide (Cancer borealis tachykinin-related peptide Ia) and GYRKPPFNGSIFamide (Gly(1)-SIFamide) in the crabs Cancer borealis, Cancer magister and Cancer productus. J Exp. Biol 2007, 210 (4), 699–714. [DOI] [PubMed] [Google Scholar]
- 59.DeLong ND, Kirby MS, Blitz DM, Nusbaum MP Parallel regulation of a modulator-activated current via distinct dynamics underlies co-modulation of motor circuit output. J Neurosci 2009, 29 (39), 12355–12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blitz DM, Nusbaum MP Distinct functions for cotransmitters mediating motor pattern selection. J Neurosci 1999, 19 (16), 6774–6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Christie AE, Stein W, Quinlan JE, Beenhakker MP, Marder E, Nusbaum MP Actions of a histaminergic/peptidergic projection neuron on rhythmic motor patterns in the stomatogastric nervous system of the crab Cancer borealis. J Comp Neurol 2004, 469 (2), 153–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kirby MS, Nusbaum MP Central nervous system projections to and from the commissural ganglion of the crab Cancer borealis. Cell Tissue Res 2007, 328 (3), 625–637. [DOI] [PubMed] [Google Scholar]
- 63.Skiebe P, Schneider H Allatostatin peptides in the crab stomatogastric nervous system: inhibition of the pyloric motor pattern and distribution of allatostatin-like immunoreactivity. J Exper Biol 1994, 194, 195–208. [DOI] [PubMed] [Google Scholar]
- 64.Dircksen H, Skiebe P, Abel B, Agricola H, Buchner K, Muren JE, Nassel DR Structure, distribution, and biological activity of novel members of the allatostatin family in the crayfish Orconectes limosus. Peptides 1999, 20 (6), 695–712. [DOI] [PubMed] [Google Scholar]
- 65.Blitz D, Nusbaum MP Neural circuit flexibility in a small sensorimotor system. Curr Opin Neurobiol 2011, 21 (4), 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Szabo TM, Chen RB, Goeritz ML, Maloney RT, Tang LS, Li LJ, Marder E Distribution and physiological effects of B-type allatostatins (myoinhibitory peptides, MIPs) in the stomatogastric nervous system of the crab Cancer borealis. J Comp Neurol 2011, 519 (13), 2658–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dockray GJ The expanding family of -RFamide peptides and their effects on feeding behaviour. Exp Physiol 2004, 89 (3), 229–235. [DOI] [PubMed] [Google Scholar]
- 68.Heinzel HG, Weimann JM, Marder E The behavioral repertoire of the gastric mill in the crab, Cancer pagurus: an in situ endoscopic and electrophysiological examination. J Neurosci 1993, 13 (4), 1793–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kirby MS, Nusbaum MP Peptide hormone modulation of a neuronally modulated motor circuit. J Neurophysiol 2007, 98 (6), 3206–3220. [DOI] [PubMed] [Google Scholar]
- 70.Christie AE, Stemmler EA, Peguero B, Messinger DI, Provencher HL, Scheerlinck P, Hsu Y-WA, Guiney ME, de la Iglesia HO, Dickinson PS Identification, physiological actions, and distribution of VYRKPPFNGSIFamide (Val1-SIFamide) in the stomatogastric nervous system of the American lobster Homarus americanus. J. Comp. Neurol 2006, 496 (3), 406–421. [DOI] [PubMed] [Google Scholar]
- 71.Dickinson PS, Stemmler EA, Cashman CR, Brennan HR, Dennison B, Huber KE, Peguero B, Rabacal W, Goiney CC, Smith CM, et al. SIFamide peptides in clawed lobsters and freshwater crayfish (Crustacea, Decapoda, Astacidea): a combined molecular, mass spectrometric and electrophysiological investigation. Gen Comp Endocrinol 2008, 156 (2), 347–360. [DOI] [PubMed] [Google Scholar]
- 72.Vazquez-Acevedo N, Rivera NM, Torres-Gonzalez AM, Rullan-Matheu Y, Ruiz-Rodriguez EA, Sosa MA GYRKPPFNGSIFamide (Gly-SIFamide) Modulates Aggression in the Freshwater Prawn Macrobrachium rosenbergii. Biol Bull US 2009, 217 (3), 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dickinson PS, Samuel HM, Stemmler EA, Christie AE SIFamide peptides modulate cardiac activity differently in two species of Cancer crab. Gen Comp Endocrinol 2019, 282, 113204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blitz DM, Christie AE, Cook AP, Dickinson PS, Nusbaum MP Similarities and differences in circuit responses to applied Gly1-SIFamide and peptidergic (Gly1-SIFamide) neuron stimulation. J Neurophysiol 2019, 121 (3), 950–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dickinson PS, Stemmler EA, Christie AE The pyloric neural circuit of the herbivorous crab Pugettia producta shows limited sensitivity to several neuromodulators that elicit robust effects in more opportunistically feeding decapods. J Exp Biol 2008, 211 (Pt 9), 1434–1447. [DOI] [PubMed] [Google Scholar]
- 76.Jorge-Rivera JC, Sen K, Birmingham JT, Abbott LF, Marder E Temporal dynamics of convergent modulation at a crustacean neuromuscular junction. J Neurophysiol 1998, 80 (5), 2559–2570. [DOI] [PubMed] [Google Scholar]
- 77.Marder E, Hooper SL, Siwicki KK Modulatory action and distribution of the neuropeptide proctolin in the crustacean stomatogastric nervous system. J Comp Neurol 1986, 243 (4), 454–467. [DOI] [PubMed] [Google Scholar]
- 78.Dickinson PS, Marder E Peptidergic modulation of a multioscillator system in the lobster. I. Activation of the cardiac sac motor pattern by the neuropeptides proctolin and red pigment-concentrating hormone. J Neurophysiol 1989, 61 (4), 833–844. [DOI] [PubMed] [Google Scholar]
- 79.Blitz DM, Christie AE, Coleman MJ, Norris BJ, Marder E, Nusbaum MP Different proctolin neurons elicit distinct motor patterns from a multifunctional neuronal network. J Neurosci 1999, 19 (13), 5449–5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Franks CJ, Walker RJ, Holden-Dye L A structure-activity study of the neuropeptide PF1, SDPNFLRFamide, using the dorsal body wall muscle of the chicken nematode, Ascaridia galli. Acta Biol Hung 2004, 55 (1–4), 343–351. [DOI] [PubMed] [Google Scholar]
- 81.Thompson DP, Davis JP, Larsen MJ, Coscarelli EM, Zinser EW, Bowman JW, Alexander-Bowman SJ, Marks NJ, Geary TG Effects of KHEYLRFamide and KNEFIRFamide on cyclic adenosine monophosphate levels in Ascaris suum somatic muscle. Int J Parasitol 2003, 33 (2), 199–208. [DOI] [PubMed] [Google Scholar]
- 82.Pedder SM, Walker RJ The actions of FxRFamide related neuropeptides on identified neurones from the snail, Helix aspersa. Acta Biol Hung 1999, 50 (1–3), 185–198. [PubMed] [Google Scholar]
- 83.Husson SJ, Landuyt B, Nys T, Baggerman G, Boonen K, Clynen E, Lindemans M, Janssen T, Schoofs L Comparative peptidomics of Caenorhabditis elegans versus C. briggsae by LC-MALDI-TOF MS. Peptides 2009, 30 (3), 449–457. [DOI] [PubMed] [Google Scholar]
- 84.Rosoff ML, Doble KE, Price DA, Li C The flp-1 propeptide is processed into multiple, highly similar FMRFamide-like peptides in Caenorhabditis elegans. Peptides 1993, 14 (2), 331–338. [DOI] [PubMed] [Google Scholar]
- 85.Geary TG, Price DA, Bowman JW, Winterrowd CA, Mackenzie CD, Garrison RD, Williams JF, Friedman AR Two FMRFamide-like peptides from the free-living nematode Panagrellus redivivus. Peptides 1992, 13 (2), 209–214. [DOI] [PubMed] [Google Scholar]
- 86.Franks CJ, Holden-Dye L, Williams RG, Pang FY, Walker RJ A nematode FMRFamide-like peptide, SDPNFLRFamide (PF1), relaxes the dorsal muscle strip preparation of Ascaris suum. Parasitology 1994, 108 (Pt 2), 229–236. [DOI] [PubMed] [Google Scholar]
- 87.Bowman JW, Winterrowd CA, Friedman AR, Thompson DP, Klein RD, Davis JP, Maule AG, Blair KL, Geary TG Nitric oxide mediates the inhibitory effects of SDPNFLRFamide, a nematode FMRFamide-related neuropeptide, in Ascaris suum. J Neurophysiol 1995, 74 (5), 1880–1888. [DOI] [PubMed] [Google Scholar]
- 88.Holden-Dye L, Franks CJ, Williams RG, Walker RJ The effect of the nematode peptides SDPNFLRFamide (PF1) and SADPNFLRFamide (PF2) on synaptic transmission in the parasitic nematode Ascaris suum. Parasitology 1995, 110 (Pt 4), 449–455. [DOI] [PubMed] [Google Scholar]
- 89.Christie AE, Stemmler EA, Dickinson PS Crustacean neuropeptides. Cell Mol Life Sci 2010, 67 (24), 4135–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang Y, Wang M, Yin S, Jang R, Wang J, Xue Z, Xu T NeuroPep: a comprehensive resource of neuropeptides. Database (Oxford) 2015, 2015, bav038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Grashow RG, Gutierrez GJ Cancer borealis stomatogastric nervous system dissection. Grashow RG, Ed. JOVE, 2009, 25, 1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dickinson PS, Kurland SC, Zu X, Parker BO, Sreekrishnan A, Kwiatkowski MA, Williams AH, Ysasi AB, Christie AE Distinct or shared actions of peptide family isoforms: II. Multiple pyrokinins exert similar effects in the lobster stomatogastric nervous system. J Exp Biol, 2015, Vol. 218, pp 2905–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ma B, Zhang KZ, Hendrie C, Liang CZ, Li M, Doherty-Kirby A, Lajoie G PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectr 2003, 17 (20), 2337–2342. [DOI] [PubMed] [Google Scholar]
- 94.Leprevost FV, Valente RH, Lima DB, Perales J, Melani R, Yates JR 3rd, Barbosa VC, Junqueira M, Carvalho PC PepExplorer: a similarity-driven tool for analyzing de novo sequencing results. Mol Cell Proteomics 2014, 13 (9), 2480–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robichaud G, Garrard KP, Barry JA, Muddiman DC MSiReader: An open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J Am Soc Mass Spectr 2013, 24 (5), 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


