Abstract
The stability of the HIV-1 core in the cytoplasm is crucial for productive HIV-1 infection. Mutations that stabilize or destabilize the core showed defects in HIV-1 reverse transcription and infection. We developed a novel and simple assay to measure stability of in vitro-assembled HIV-1 CA-NC complexes. This assay allowed us to demonstrate that cytosolic extracts strongly stabilize the HIV-1 core (Fricke et al., J Virol 87:10587–10597, 2013). By using our novel assay, one can measure the ability of different drugs to modulate the stability of in vitro-assembled HIV-1 CA-NC complexes, such as PF74, CAP-1, IXN-053, cyclosporine A, Bi2, and the peptide CAI. We also found that purified CPSF6 (1–321) protein stabilizes in vitro-assembled HIV-1 CA-NC complexes (Fricke et al., J Virol 87:10587–10597, 2013). Here we describe in detail the use of this capsid stability assay. We believe that our assay can be a powerful tool to assess HIV-1 capsid stability in vitro.
Keywords: HIV-1, Capsid, Stability, Core, Uncoating
1. Introduction
In its simplest definition, uncoating is the shedding of monomeric capsid proteins from the retroviral core or ribonucleoprotein complex. Because only ~40 % of the total capsid in the virion comprises the retroviral core [1–3], a simple model is that the monomeric capsid is in dynamic equilibrium with the assembled capsid (viral core). This implies that the core might exist in a meta-stable state only when the soluble capsid is in high concentration, keeping the equilibrium shifted toward the core formation by mass action. The fact that complexes containing capsid have been detected in the cytoplasm of cells early during infection implies that cellular factors might be involved in stabilization of the core [4, 5].
The capsid protein is required for the successful completion of several early steps of HIV-1 replication: (1) successful infection requires that capsid uncoating occurs during or after reverse transcription [6–9]; (2) elegant experiments have shown that the capsid sequence is the genetic determinant for the ability of lentiviruses to infect nondividing cells [10–13]; and (3) the stability provided by the capsid protein assembled into the viral core is important for the occurrence of reverse transcription and productive infection [14–16].
Over the years, sensitive assays to biochemically measure core stability have been developed, such as the “fate of the capsid” assay that measures core stability during infection of cells over time [6, 7, 17]. Even though this assay is widely used [7, 8, 14, 17–23], it is intensive and laborious and a more rapid assay to measure core stability is desirable.
Here we present a rapid and simple assay to measure capsid stability in vitro using in vitro-assembled HIV-1 CA-NC complexes as a surrogate for the HIV-1 core [24]. This assay will assist the evaluation of drugs or proteins that change the stability of capsid. Furthermore, this assay could be used to identify novel cellular factors that bind to the HIV-1 core.
2. Materials
Prepare all solutions using ultrapure water and analytical grade reagents. Prepare and store all reagents at room temperature (unless indicated otherwise). Diligently follow all waste disposal regulations when disposing waste materials.
2.1. Capsid Assembly
1 M Tris–HCl, pH 8. 0.
5 M NaCl.
10 mg/ml oligo (TG) 25 (store at −20 °C).
HIV-1 CA-NC protein purified as described previously [25].
2.2. Stability Assay
Destabilization buffer (DB) 5× : 50 mM Tris–HCl (pH 8.0), 300 mM NaCl, 2 mM MgCl 2, 10 % (v/v) glycerol, 0.5 % (v/v) (NP-40). Make fresh every time.
Stabilization buffer (SB): 10 mM Tris–HCl (pH 8.0), 10 mM KCl, 2 mM MgCl 2, 0.5 mM DTT. Store at 4 °C for up to 1 week.
PBS 1×.
Human 293 T cells.
Refrigerated tabletop centrifuge.
CAI peptide (amino acid sequence, ITFEDLLDYYGP) and the CAIctrl peptide (amino acid sequence, IYDPTLYGLEFD) (95 % purity) [26]. Prepare stock solutions at 10 mM in dimethyl sulfoxide (DMSO). Store at −20 °C.
PF74 (PF-3450074), stock solution 100 mM in DMSO (store at −20 °C) [27].
CPIPB (4-{2-[3-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid dihydrochloride) stock solution of 20 mM in DMSO [28–30]. Store at −20 °C.
CPSF6 (1–321) purified as described previously [31].
2.3. Differential Ultracentrifugation
70 % (w/v) sucrose (ACS reagent > 99%purity) in 1× PBS.
Ultracentrifuge.
SW55 Rotor.
SW55 centrifuge tubes.
2.4. PAGE and Western Blot
1× SDS sample buffer: 60 m M Tris–HCl (pH 6.8), 2 % (w/v) SDS, 5 % (v/v) β-mercaptoethanol, 0.02 % (w/v) bromophenol blue, 9 % (v/v) glycerol.
5× SDS sample buffer: 0.3 M Tris–HCl (pH 6.8), 10 % (w/v) SDS, 25 % (v/v) β-mercaptoethanol, 0.1 % (w/v) bromophenol blue, 45 % (v/v) glycerol. Store aliquots at −20 °C.
10 % (w/v) polyacrylamide NuPAGE gel.
3. Methods
To measure HIV-1 core stability, we developed an assay that measures the stability of HIV-1 CA-NC complexes in vitro (Fig. 1). The assay consists of determining the stability of in vitro-assembled HIV-1 CA-NC complexes in the presence of different agents such as proteins or small-molecule inhibitors to subsequently measure the remaining amount of assembled HIV-1 CA-NC complexes using a sucrose cushion. To measure whether an agent stabilizes the capsid, in vitro-assembled HIV-1 CA-NC complexes are incubated with the agent in question in destabilization buffer (Fig. 1). The total amount of stabilized capsid complexes is measured using a sucrose cushion, and the amount of stabilized complexes is compared to complexes obtained in the absence of the agent in question. To measure whether an agent destabilizes the capsid, in vitro-assembled HIV-1 CA-NC complexes are incubated with the agent in stabilization buffer (Fig. 1). The amount of destabilized capsid complexes is measured by subtracting the amount of complexes obtained in the presence of the agent from the amount of complexes obtained in the absence of the agent.
Fig. 1.
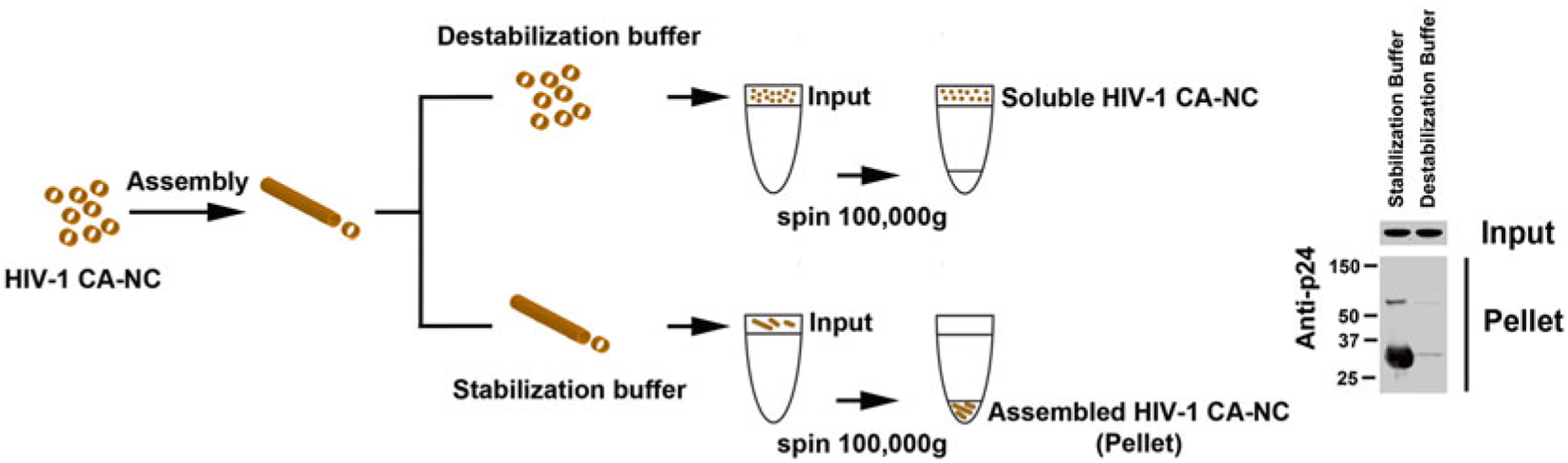
Diagram of the stabilization assay. In vitro-assembled HIV-1 CA-NC complexes, shown here as tubular structures, are formed by monomeric recombinant HIV-1 CA-NC fusion proteins under highly ionic conditions and recapitulate the surface of the HIV-1 core. When HIV-1 CA-NC complexes are incubated in destabilization buffer, they disassemble spontaneously. Disassembled capsids are layered on top of a 70 % sucrose cushion; however, the disassembled capsid does not cross the cushion after spinning at 100,000 × g for 1 h. In contrast, incubation of HIV-1 CA-NC in stabilization buffer preserves the assembled structures, which pellet when layered onto a 70 % sucrose cushion with spinning at 100,000 × g for 1 h. Input, a fraction of the sample layered onto the sucrose cushion before the centrifugation step. Pellet, a fraction of the capsid pelleted after the sample has been centrifuged at 100,000 × g for 1 h. Input and pellet samples were analyzed by Western blotting using anti-p24 antibodies.
3.1. Capsid Assembly
Add 2.5 μl 1 M Tris–HCl (pH 8.0), 5 μl 5 M NaCl, and 10 μl oligo(TG) 25 to 25 μl of 10–20 mg/ml purified CA-NC protein (see Note 1).
Add 7.5 μl water to the suspension. The mixture is incubated for 5 min at room temperature. Successful assembly is noted by the appearance of white complexes in solution. The size of the complexes can be determined by dynamic light scattering (see Note 2).
3.2. Stabilization Assay
Prepare up to six Eppendorf test tubes with 400 μl stabilization buffer. Add additional 100 μl stabilization buffer in the first test tube. To the other test tubes, add 100 μl of 5× destabilization buffer. Final volume is 500 μl.
Add the indicated agents to test tubes 3–6 (Fig. 2a). Incubate 3 μl of in vitro-assembled HIV-1 CA-NC complexes in the presence of PF74, CAI peptide, CAIctrl peptide, or purified CPSF6 (1–321) (Fig. 2a). As a second example, the ability of CPIPB to change the stability of in vitro-assembled HIV-1 CA-NC complexes is determined. For this purpose, incubate 3 μl of in vitro-assembled HIV-1 CA-NC complexes in the presence of different concentrations of CPIPB (Fig. 3) (see Note 3).
Incubate 3 μl of in vitro-assembled HIV-1 CA-NC complexes with the different agents at room temperature for 1 h (see Notes 4 and 5).
Store an aliquot of this mixture at −20 °C for further analysis, henceforth referred to as “input.” For this purpose, add 5× SDS sample buffer. Proceed with ultracentrifugation below.
Fig. 2.
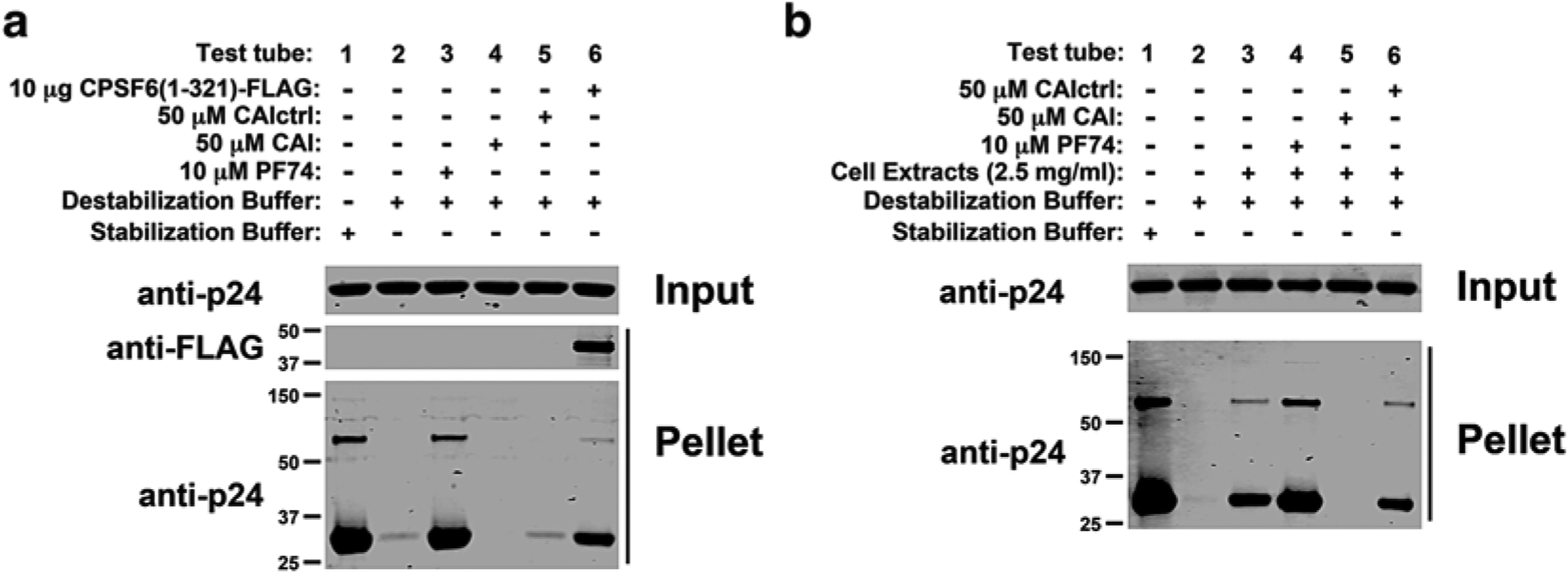
PF74 and CPSF6 (1–321) increase the stability of in vitro-assembled HIV-1 CA-NC complexes, whereas the CAI peptide decreases the stability. In contrast, the CAI peptide with a randomized sequence (CAIctrl) did not show an effect on the stability of HIV-1 CA-NC complexes. The stability of HIV-1 CA-NC complexes in destabilization buffer (stabilization assay) (a) or destabilization buffer in cell extracts (destabilization assay) (b) supplemented with 10 μM PF74, 50 μM CAI, or CAIctrl peptide was measured. The assay without cellular extract was performed additionally using 10 μg of the protein CPSF6 (amino acids 1–321).
Fig. 3.
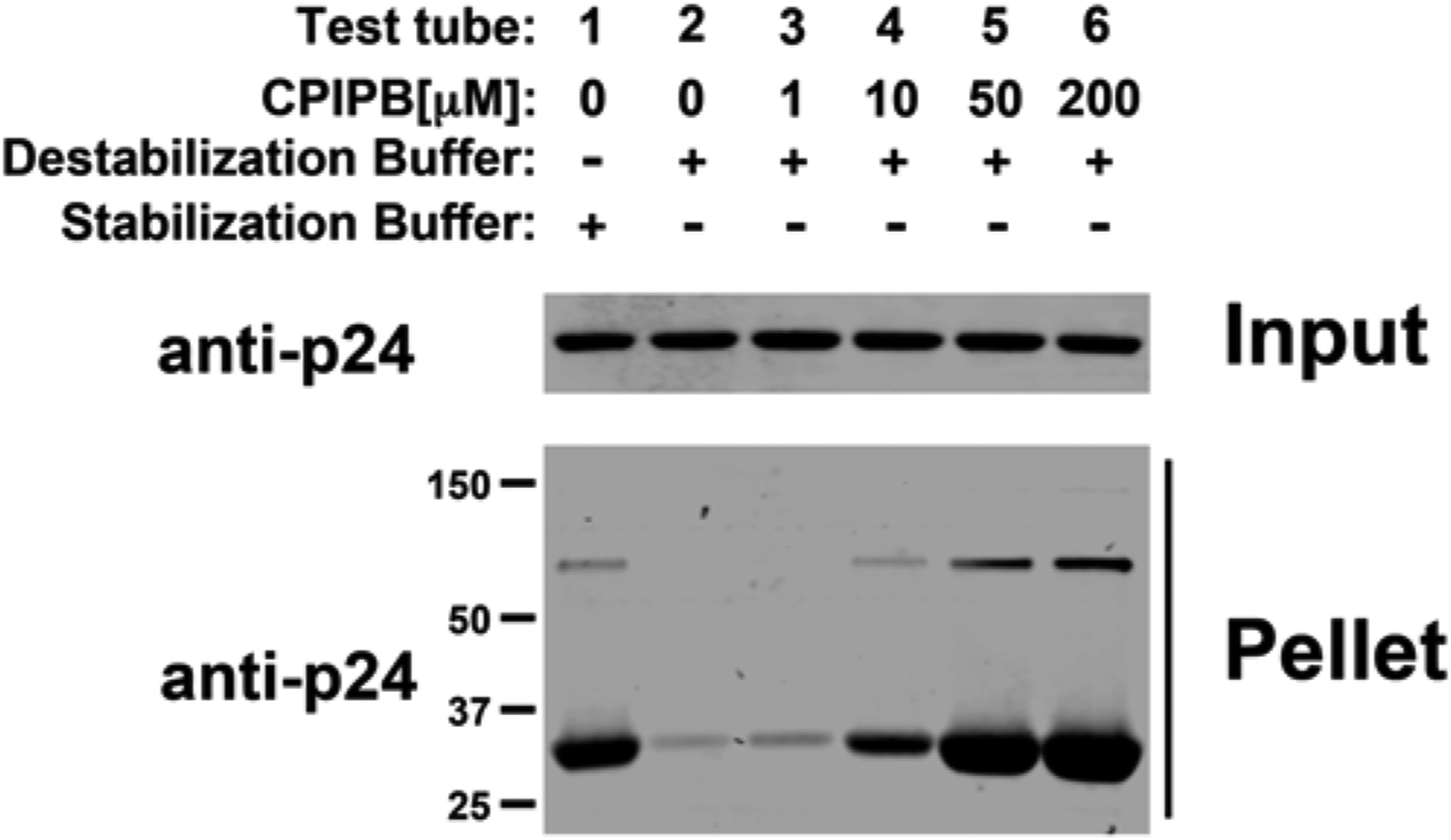
The capsid drug CPIPB (4-{2-[3-(3-chlorophenyl)-1H-pyrazol-4-yl]-1-[3-(1H-imidazol-1-yl)propyl]-1H-benzimidazol-5-yl}benzoic acid dihydrochloride) stabilizes in vitro-assembled HIV-1 CA-NC complexes. The stability of HIV-1 CA-NC complexes in destabilization buffer using increasing concentrations of the drug CPIPB was measured. CPIPB has been shown previously to strongly increase HIV-1 capsid crystallization and competes with the novel restriction factor M×B for binding to the HIV-1 capsid [29, 30].
3.3. Destabilization Assay
-
Culture 293 T cells in DMEM containing 10 % FBS and antibiotics (penicillin and streptomycin) in an incubator calibrated at 37 °C and 5 % CO 2. Cells are detached from nearly confluent dishes with trypsin-EDTA. Five million cells are seeded in 10 ml medium in 100 mm plastic culture dishes and cultured for 1 day prior to the assay. The next day, cultures will have grown to approximately 100 % confluence.
Discard the media by decanting and add 400 μl of stabilization buffer (see Note 6).
Scrape the cells on the 100 mm plate using a cell scraper and transfer cells into 1.5 ml tube.
Incubate sample for 15 min on ice.
Centrifuge with maximum speed using a 4 °C refrigerated tabletop centrifuge.
Carefully take the tubes from the centrifuge and transfer the supernatant (cell extract) into a fresh tube.
Prepare up to six test tubes and fill the first two with 400 μl stabilization buffer. Add 400 μl of cell extracts resuspended in stabilization buffer into test tubes 3–6. Add additional 100 μl of stabilization buffer in the first test tube. To the test tubes 2–6, add 100 μl of 5× destabilization buffer.
Add the agents/drugs you would like to assay to test tubes 4–6. In this example, the well-known HIV-1 capsid drug PF74 and the peptides CAI and CAIctrl are tested (Fig. 2b) (see Note 3).
Add 3 μl of in vitro-assembled HIV-1 CA-NC complexes to all test tubes, and incubate the mixture at room temperature for 1 h (see Notes 4 and 5).
An aliquot of this mixture, henceforth referred to as “input,” was stored at −20 °C for further analysis. For this purpose, add 5× SDS sample buffer.
3.4. Ultracentrifugation
Prepare six centrifuge tubes containing 3 ml 70 % sucrose at 4 °C (see Note 7). Check the sucrose levels and if necessary adjust using a 200 μl pipette (keep the tubes in the cold room until needed).
Load the sample from Subheading 3.2, step 4 above carefully on top of the sucrose cushion (see Note 8).
Centrifuge at 100,000 × (30,000 rpm using a SW55 rotor) for 1 h at 4 °C.
Aspirate the supernatant carefully and resuspend pellet in 1× SDS loading buffer (see Note 9).
3.5. PAGE and Western Blot
Load input and pellet with a molecular size marker on a 10 % acrylamide NuPAGE gel.
Analyze gel by standard western blot using anti-p24 antibodies against HIV-1 capsid and anti-FLAG antibodies to detect CPSF6 (1–321).
4. Notes
The protein should assemble within 5 min; the solution turns into a white suspension. These complexes could be used for at least 5 days.
The capsid should be assembled at least one night before the experiment and stored at 4 °C.
If the drug is soluble in an organic solvent, then add the same amount of solvent to the control in test tubes 1 and 2.
Use the 10 μl pipette about halfway into the solution and slowly add the capsid.
Do not shake the tube during the incubation time.
Remove remaining media using vacuum aspirator.
Use a serological 5 ml pipette and load 3.0 ml of the 70 % sucrose solution into the SW55 tube. Consider that the 5 ml serological pipette retains at least 0.5 ml of the sucrose.
Mix the sample carefully before applying to the sucrose cushion by pipetting once up and down.
First remove the supernatant carefully, change the tip and remove the sucrose layer, and allow time for the sucrose to move down from the tube walls.
Acknowledgments
Projects AI087390, AI10282401, R56AI108432, and AI104476 to F.D.-G supported this work. We are grateful to the NIH HIV-1/ AIDS repository for providing reagents such as antibodies and small-molecule inhibitors that were crucial for this work.
References
- 1.Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC (2004) The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11:672–675 [DOI] [PubMed] [Google Scholar]
- 2.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD (2003) Structural organization of authentic, mature HIV-1 virions and cores. EMBO J 22:1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lanman J, Lam TT, Emmett MR, Marshall AG, Sakalian M, Prevelige PE Jr (2004) Key interactions in HIV-1 maturation identified by hydrogen-deuterium exchange. Nat Struct Mol Biol 11:676–677 [DOI] [PubMed] [Google Scholar]
- 4.McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ (2002) Visualization of the intracellular behavior of HIV in living cells. J Cell Biol 159:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fassati A, Goff SP (2001) Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J Virol 75:3626–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J (2006) Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 103:5514–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diaz-Griffero F, Kar A, Lee M, Stremlau M, Poeschla E, Sodroski J (2007) Comparative requirements for the restriction of retrovirus infection by TRIM5alpha and TRIMCyp. Virology 369:400–410, Epub 2007 Oct 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roa A, Hayashi F, Yang Y, Lienlaf M, Zhou J, Shi J, Watanabe S, Kigawa T, Yokoyama S, Aiken C, Diaz-Griffero F (2012) RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J Virol 86:1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arfi V, Lienard J, Nguyen XN, Berger G, Rigal D, Darlix JL, Cimarelli A (2009) Characterization of the behavior of functional viral genomes during the early steps of human immunodeficiency virus type 1 infection. J Virol 83:7524–7535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamashita M, Emerman M (2004) Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol 78:5670–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita M, Perez O, Hope TJ, Emerman M (2007) Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog 3:1502–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamashita M, Emerman M (2006) Retroviral infection of non-dividing cells: old and new perspectives. Virology 344:88–93 [DOI] [PubMed] [Google Scholar]
- 13.Diaz-Griffero F (2012) The role of TNPO3 in HIV-1 replication. Mol Biol Int 2012:868597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Fricke T, Diaz-Griffero F (2013) Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J Virol 87:683–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forshey BM, von Schwedler U, Sundquist WI, Aiken C (2002) Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 76:5667–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ohagen A, Gabuzda D (2000) Role of Vif in stability of the human immunodeficiency virus type 1 core. J Virol 74:11055–11066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Griffero F, Kar A, Perron M, Xiang SH, Javanbakht H, Li X, Sodroski J (2007) Modulation of retroviral restriction and proteasome inhibitor-resistant turnover by changes in the TRIM5alpha B-box 2 domain. J Virol 81:10362–10378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi J, Zhou J, Shah VB, Aiken C, Whitby K (2011) Small-molecule inhibition of human immunodeficiency virus type 1 infection by virus capsid destabilization. J Virol 85:542–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Iaco A, Santoni F, Vannier A, Guipponi M, Antonarakis S, Luban J (2013) TNPO3 protects HIV-1 replication from CPSF6-mediated capsid stabilization in the host cell cytoplasm. Retrovirology 10:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J (2007) The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol 81:2138–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Griffero F, Perron M, McGee-Estrada K, Hanna R, Maillard PV, Trono D, Sodroski J (2008) A human TRIM5alpha B30.2/SPRY domain mutant gains the ability to restrict and prematurely uncoat B-tropic murine leukemia virus. Virology 378:233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohkura S, Goldstone DC, Yap MW, Holden-Dye K, Taylor IA, Stoye JP (2011) Novel escape mutants suggest an extensive TRIM5alpha binding site spanning the entire outer surface of the murine leukemia virus capsid protein. PLoS Pathog 7, e1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berube J, Bouchard A, Berthoux L (2007) Both TRIM5alpha and TRIMCyp have only weak antiviral activity in canine D17 cells. Retrovirology 4:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fricke T, Brandariz-Nunez A, Wang X, Smith AB 3rd, Diaz-Griffero F (2013) Human cytosolic extracts stabilize the HIV-1 core. J Virol 87:10587–10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI (1999) Assembly and analysis of conical models for the HIV-1 core. Science 283:80–83 [DOI] [PubMed] [Google Scholar]
- 26.Sticht J, Humbert M, Findlow S, Bodem J, Muller B, Dietrich U, Werner J, Krausslich HG (2005) A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol 12: 671–677 [DOI] [PubMed] [Google Scholar]
- 27.Blair WS, Pickford C, Irving SL, Brown DG, Anderson M, Bazin R, Cao J, Ciaramella G, Isaacson J, Jackson L, Hunt R, Kjerrstrom A, Nieman JA, Patick AK, Perros M, Scott AD, Whitby K, Wu H, Butler SL (2010) HIV capsid is a tractable target for small molecule therapeutic intervention. PLoS Pathog 6, e1001220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goudreau N, Lemke CT, Faucher AM, Grand-Maitre C, Goulet S, Lacoste JE, Rancourt J, Malenfant E, Mercier JF, Titolo S, Mason SW (2013) Novel inhibitor binding site discovery on HIV-1 capsid N-terminal domain by NMR and X-ray crystallography. ACS Chem Biol 8:1074–1082 [DOI] [PubMed] [Google Scholar]
- 29.Lemke CT, Titolo S, Goudreau N, Faucher AM, Mason SW, Bonneau P (2013) A novel inhibitor-binding site on the HIV-1 capsid N-terminal domain leads to improved crystallization via compound-mediated dimerization. Acta Crystallogr D Biol Crystallogr 69:1115–1123 [DOI] [PubMed] [Google Scholar]
- 30.Fricke T, White TE, Schulte B, de Souza Aranha Vieira DA, Dharan, Campbell, Brandariz-Nunez, Diaz-Griffero F (2014) MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fricke T, Valle-Casuso JC, White TE, Brandariz-Nunez A, Bosche WJ, Reszka N, Gorelick R, Diaz-Griffero F (2013) The ability of TNPO3-depleted cells to inhibit HIV-1 infection requires CPSF6. Retrovirology 10:46. [DOI] [PMC free article] [PubMed] [Google Scholar]


