Abstract
The ciliate Tetrahymena thermophila has been a powerful model system for molecular and cellular biology. However, some investigations have been limited due to the incomplete closure and sequencing of the macronuclear genome assembly, which for many years has been stalled at 1,158 scaffolds, with large sections of unknown sequences (available in Tetrahymena Genome Database, TGD, http://ciliate.org/). Here we completed the first chromosome-level Tetrahymena macronuclear genome assembly, with approximately 300× long Single Molecule, Real-Time reads of the wild-type SB210 cells—the reference strain for the initial macronuclear genome sequencing project. All 181 chromosomes were capped with two telomeres and gaps were entirely closed. The completed genome shows significant improvements over the current assembly (TGD 2014) in both chromosome structure and sequence integrity. The majority of previously identified gene models shown in TGD were retained, with the addition of 36 new genes and 883 genes with modified gene models. The new genome and annotation were incorporated into TGD. This new genome allows for pursuit in some underexplored areas that were far more challenging previously; two of them, genome scrambling and chromosomal copy number, were investigated in this study. We expect that the completed macronuclear genome will facilitate many studies in Tetrahymena biology, as well as multiple lines of research in other eukaryotes.
Keywords: Tetrahymena thermophila, macronuclear genome, 181 chromosomes, DNA scrambling, copy number
INTRODUCTION
As a unicellular eukaryote model organism, Tetrahymena thermophila is the most well-studied of all protozoa and has contributed to fundamental biological discoveries in multiple aspects (Cervantes et al., 2013; Feng et al., 2017; Gao et al., 2013; Mochizuki et al., 2002; Mochizuki and Gorovsky, 2004a, 2005; Orias et al., 2017; Wang et al., 2017b; Wang et al., 2019b; Xiong et al., 2016; Xu et al., 2019; Zhao et al., 2019). Like other ciliates, Tetrahymena possess two types of nuclei in a single cell, distinct in their appearance and function (Cheng et al., 2019; Collins and Gorovsky, 2005; Karrer, 2012; Wang et al., 2017a; Yan et al., 2019). The smaller, diploid, germline-like micronucleus (MIC) directly contributes DNA to the sexual progeny (Ray Jr, 1956; Karrer, 1999) and consists of five pairs of chromosomes (Karrer, 2012). The larger, polyploid, soma-like macronucleus (MAC) supports all the vegetative functions of the cell and contains 181 chromosomes (Coyne et al., 2012). This unique nuclear dimorphism has fascinated researchers in many fields of biology.
The macronuclear genome of Tetrahymena was among the first few to be sequenced in the dawn of the genomic era. Its first version was reported in 2006 (Eisen et al., 2006), with reads produced by first-generation shotgun sequencing. Since then, the assembly was continuously improved in sequence accuracy, genome assembly, and gene model annotation, with efforts from the whole Tetrahymena research community (Coyne et al., 2008; Hamilton et al., 2006; Stover et al., 2006; Xiong et al., 2012). The current community-annotated assembly (TGD 2014) contains a 103.01 Mb genome with highly accurate sequences and gene model annotation, providing invaluable resources for the Tetrahymena community and other fields as well. However, as the TGD genome was assembled from reads with limited length (~2,000 bp), its 181 chromosomes are split into 1,158 scaffolds (129 with two telomeres, collectively ~58.9 Mb; 29 with only one telomere), with 0.06% unknown sequences (Ns) (Fraser et al., 2002). An updated genome assembly with complete chromosomes is demanded for future research.
For example, the occurrence of genome-wide DNA scrambling/unscrambling in Tetrahymena has not been systematically tested. This phenomenon is exaggerated in some ciliates (Chen et al., 2019; Zhang et al., 2018), such as Oxytricha, wherein massive DNA segments are reshuffled to functional genes in the MAC from their interrupted and scrambled germline precursors (Chen et al., 2014; Fang et al., 2012; Nowacki et al., 2008), after the fragmentation of MIC chromosomes (Klobutcher et al., 1988; Prescott, 2000) and the removal of internally eliminated sequences (IESs) (Prescott, 1994). In Tetrahymena, however, the remaining macronuclear-destined sequences (MDSs) were generally thought to be linear between MAC and MIC (Mochizuki and Gorovsky, 2004b; Ruehle et al., 2016; Stover et al., 2012). It was proposed that shuffling of discontinuous MIC segments also occurs during the new MAC development (Hamilton et al., 2016), but the parallel comparison between MAC and MIC was challenging, given that MAC chromosomes are not complete and a considerable part of the current MAC genome assembly is comprised of interscaffold gaps.
Chromosomal copy number is another area yet to be fully explored. The T. thermophila MAC genome consists of 181 chromosomes, which was identified by physical and genetic mapping (Coyne et al., 2012). Of these, the 21 kb ribosomal DNA (rDNA) minichromosome is an inverted repeat with ~9,000 copies (Gall, 1974; Mohammad et al., 2007; Yao and Yao, 1989). The remaining non-rDNA chromosomes were inferred to be maintained at an average of ~45 copies per cell, based on phenotype assortment rates from a handful of loci (Doerder et al., 1992). The uniformity was validated for limited chromosomes in the initial MAC assembly (Eisen et al., 2006), but its generality for all chromosomes remain inconclusive. Less is known about how or do Tetrahymena chromosomes maintain a stable copy number (~45C), as the MAC divides amitotically and its chromosomes are distributed unequally during each cell division (Orias and Flacks, 1975). The complete sequence of all 181 chromosomes is a prerequisite to decipher their copy number control mechanism.
The long-read sequencing technology such as Single Molecule, Real-Time (SMRT) sequencing permitted span of repeats and missing bases, thereby closing gaps and completing chromosomes. Indeed, SMRT was employed in a hypotrich ciliate Oxytricha trifallax with highly fragmented genome, showing the ability to capture tiny nanochromosomes in single reads (Lindblad et al., 2019). It was also used for the high-quality and near-complete macronuclear genome assembly of another ciliate Paramecium bursaria (He et al., 2019).
We here report the complete closure of the MAC genome of T. thermophila. Using SMRT sequencing data with an ultra-high depth (~300×), we completed the first chromosome-level MAC genome assembly of T. thermophila and updated the gene model annotation. With the help of the completed genome, we tested two underexplored topics in Tetrahymena, genome scrambling and chromosomal copy number. We conclude that the completed MAC genome will greatly facilitate many investigations of Tetrahymena biology.
RESULTS AND DISCUSSION
Completion of the Tetrahymena thermophila macronuclear genome
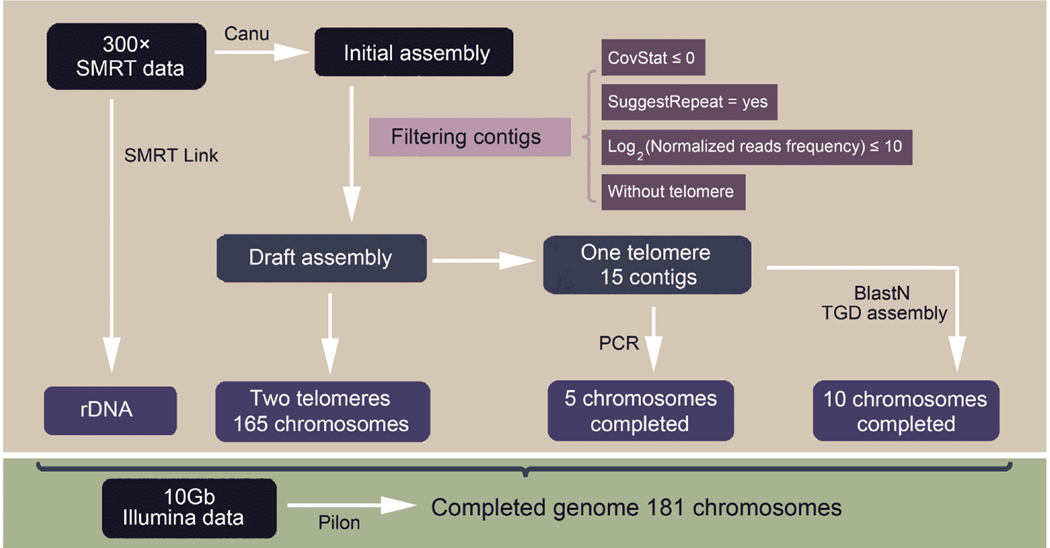
The whole-genome shotgun sequence of the T. thermophila MAC genome presents a unique challenge, with more than 1,000 scaffolds and about 650 intrascaffold gaps (average length of 271 bp) (Eisen et al., 2006). As scaffolds could not be assembled directly into superscaffolds due to a large amount of intra- and inter-scaffold gaps (Eisen et al., 2006; Hamilton et al., 2016), we employed SMRT sequencing in this study to generate long reads, ideal for resolving long tandem repeats and closing gaps (English et al., 2012; Rasko et al., 2011; Roberts et al., 2013). In total, SMRT reads (average sub-read length of 11.2 kb) with an ultra-high sequencing depth (300×) of T. thermophila wild-type (WT) SB210 strain were generated and assembled into a draft assembly composed of 346 contigs using Canu (Koren et al., 2017). After filtering erroneous contigs (repetitive contigs and contigs with low mapped reads or with no telomeres), 180 contigs were retained. Of these, 165 contigs were capped with telomeric repeats at both ends, and 15 contigs were telomere-capped at one end. The gap closures of 10 uncompleted contigs were finished by BLAST to the TGD 2014 assembly, and the remaining five contigs were completed by polymerase chain reaction (PCR) amplification and sequencing (Figure S1A in Supporting Information). The 21 kb rDNA minichromosome was separately assembled using SMRT link v5.10 (Pacific Biosciences). 10 Gb Illumina reads were used to error correct the PacBio assembly using Pilon (Walker et al., 2014). In total, we obtained a 103.3 Mb T. thermophila MAC genome assembly consisting of 181 complete chromosomes (Figure 1), named from 1 to 181 by their order along the 5 MIC chromosomes (chr181 for rDNA minichromosome) (Hamilton et al., 2016).
Figure 1.
The assembly flow of the completed genome.
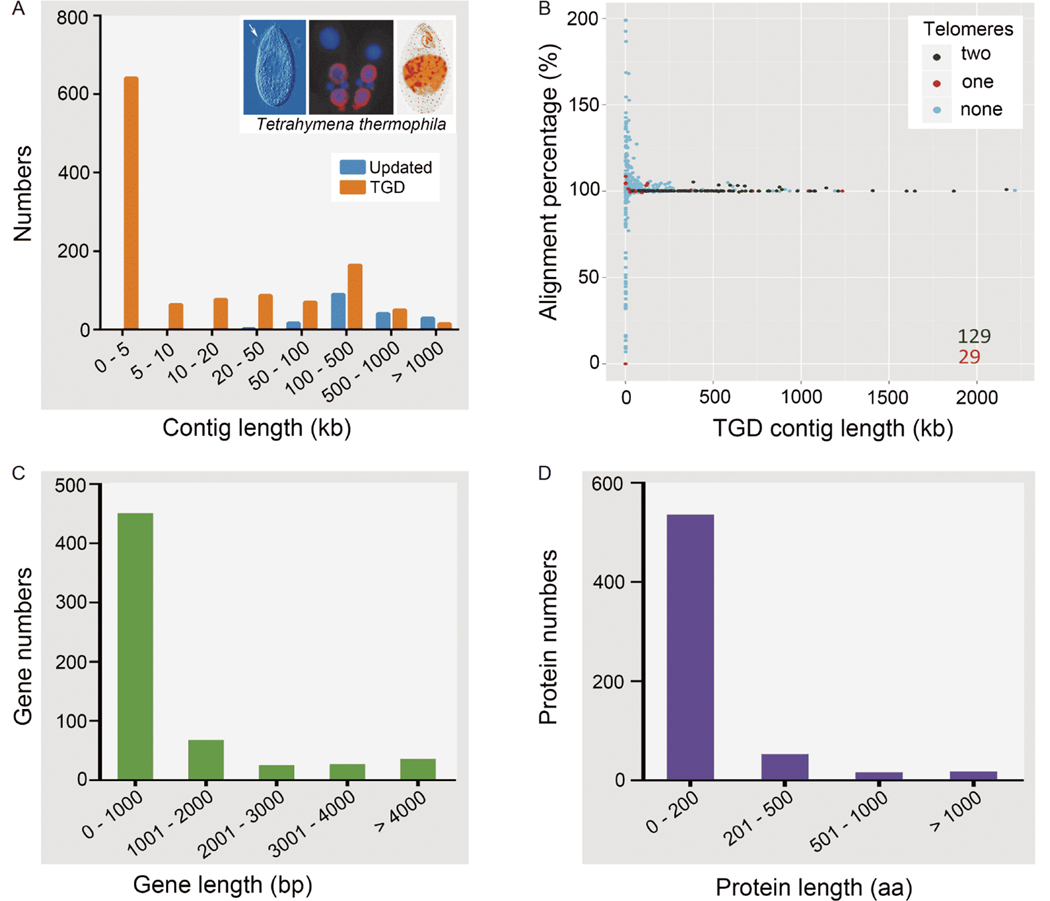
The genome size and GC content of the completed genome are nearly identical to the TGD 2014 assembly (Table 1). In the TGD 2014 assembly, a large portion (54.5%) of scaffolds is shorter than 5 kb (Figure 2A). In the updated assembly, the N50 length was increased about two-fold, from 521 to 930 kb, with the longest being 3.3 Mb (Table 1, Figure 2A). Six hundred and twenty intrascaffold gaps, representing 0.06% sequences of the genome, were entirely closed. In particular, 432 gaps were located in genic regions, closure of which resulted in an amino acid sequence change for 266 corresponding genes.
Table 1.
The comparison between the completed genome and the current assembly
| Completed genome | TGD 2014 genome | |
|---|---|---|
| Genome size | 103.34 M | 103.01 M |
| Gene models | 26,258 | 26,996 |
| Total contigs | 181 | 1,158 |
| N’s per 100 kb | 0.00 | 61.83 |
| N50 | 929,705 | 520,943 |
| Longest contig | 3,329,751 | 2,216,158 |
| Contigs with two telomeres | 181 | 129 |
| Contigs with one telomere | 0 | 29 |
| GC (%) | 22.30 | 22.32 |
Figure 2.
Comparisons between the completed genome and the TGD 2014 assembly. A, The length distribution of chromosomes in the completed genome and TGD 2014. B, The completed genome shows high sequence similarity to TGD 2014. Green dots and red dots represent the scaffolds with two telomeres and one telomere in TGD 2014 respectively. The x-axis represents the length of TGD 2014 scaffolds, and the y-axis represents the alignment percentage (alignment length in the new genome/TGD scaffold length). C, The length distribution of 607 TGD genes which had no significant match (E>1.0×e−5) among the updated set. D, The length distribution of 623 TGD proteins which had no significant match (E>1.0×e−5) among the updated set.
To estimate the sequence accuracy and integrity, the completed genome was compared with the TGD 2014 assembly (Stover et al., 2012). The total alignment percentage (alignment length in the completed genome/length of TGD scaffolds) is 100.55%, because the completed genome increases slightly in size after closing gaps. The alignment percentage of each TGD scaffold was shown in Figure 2B. Of these, most long scaffolds, including 129 completed chromosomes and 28 one telomere capped scaffolds in TGD 2014, showed high concordance to corresponding chromosomes in the completed genome, presented as dots with alignment percentage approaching 100%. Dots with alignment percentage under 100% indicated that these scaffolds were merged in the completed genome, while above 100% indicated that these scaffolds belonged to part of repetitive sequences in the completed genome. The only exception (red dot, left bottom in Figure 2B) was a misassembled scaffold (scf_8255776, 251 bp), completely made up of telomeric repeats.
To remove MIC DNA contamination in the MAC genome assembly, we searched for the presence of all 7,544 IES sequences (Hamilton et al., 2016) in the completed genome using BLASTN (E-value<1.0×10−5, identity>95%, alignment length>90%). Only one effective hit (IES-05521-r13) was detected, which however was incorrectly predicted and should be reassigned as an MDS (Figure S1B, Table S1 in Supporting Information).
Together, these results demonstrated that the sequences in the completed genome had high accuracy and integrity.
Optimized gene model annotation of the completed MAC genome
Considering that the TGD gene model annotation has been constantly improved and widely accepted, gene and CDS sequences from TGD were used to make hintsfile for gene prediction conducted by Augustus (Stanke et al., 2006). In total, 26,258 protein-coding genes were predicted. 25,339 of them matched the TGD models completely, so their “TTHERM” identifier number and functional annotation were inherited. One hundred and sixty-seven genes were merged from 393 genes in TGD with numerically adjacent GenBank IDs; they were split in TGD due to unconnected scaffolds and unclosed gaps. Another 716 adjacent genes were split from TGD genes, due to newly predicted stop codons after the completion of the genome. Intriguingly, we found 36 new genes that are not present in TGD, coding for proteins homologous to extracellular matrix protein FRAS1, proteasome subunit beta 2, and so on.
A BLASTN search was performed between ours and the TGD 2014 gene models (Table S2 in Supporting Information). Six hundred and seven older gene models had no significant match (E>1.0×e−5) to the updated gene sequences. Most of these genes were too short to code for functional proteins (74%<1,000 bp, median length =1,072 bp) (Figure 2C), and 34 of them contained unknown sequences (N≥10). The comparison was also performed using BLASTP for peptides (Table S3 in Supporting Information). Six hundred and twenty-three previous TGD proteins had no significant match (E>1.0×e−5) with the updated protein sets, and their median length was 162 amino acids (Figure 2D), including 92 with gaps and 471 “hypothetical proteins”. These results collectively confirmed the accuracy of the updated gene model annotation.
Scrambled regions detected in the completed MAC genome
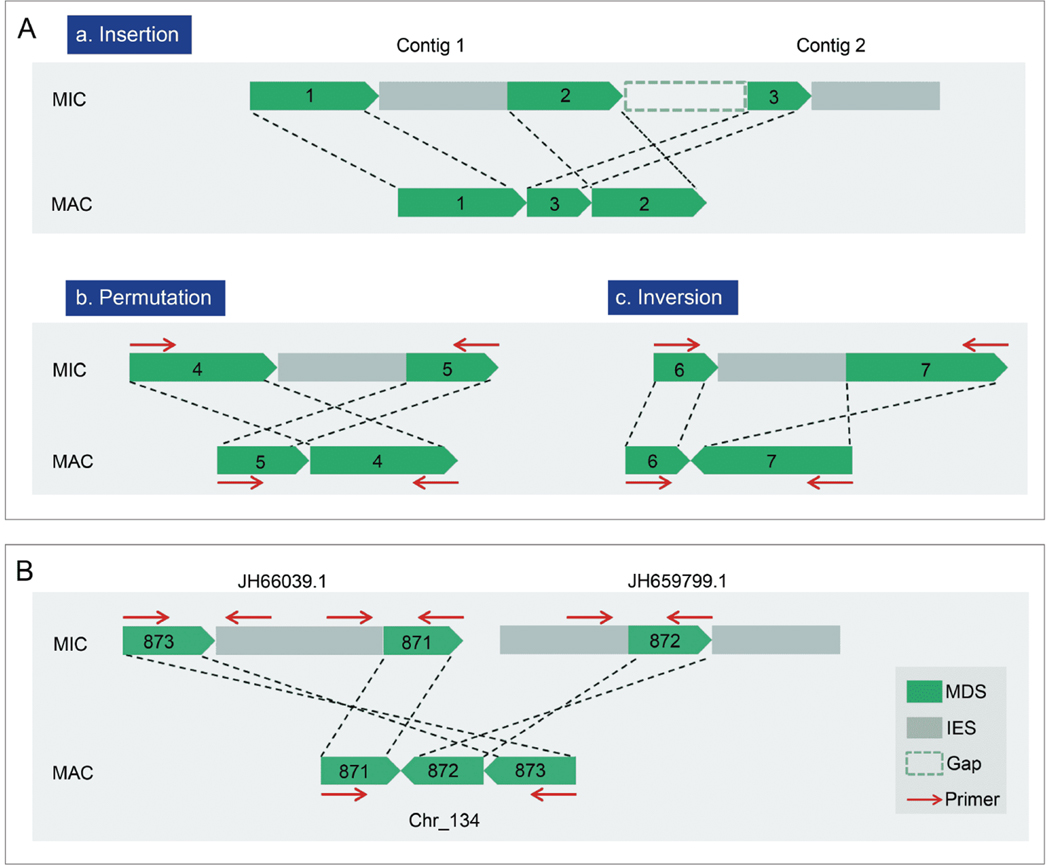
To detect potential DNA scrambling in the MAC, MDSs identified by MIDAS (http://knot.math.usf.edu/midas/index.html) were aligned to the MIC genome. 2,711 scrambled regions were detected and categorized into three types: insertion (1,622), permutation (1,033), and inversion (56) (Figure 3A). Four hundred and seventy-four of the 1,622 insertions could possibly be re-categorized as a variant of permutation, considering that the MIC genome sequence used in this study has not been fully completed, and the five micronuclear chromosomes remain split into numerous contigs.
Figure 3.
DNA scrambling in the macronuclear genome. A, Three types of scrambled regions, categorized by possible ways for the MDSs to be unscrambled. B, A typical scrambled region in T. thermophila confirmed by PCR amplification and sequencing.
In particular, we confirmed a typical scrambled region by PCR amplification and sequencing, in which three MDSs from two MIC contigs were reversed (inversion), inserted (insertion) and reordered (permutation) in the MAC (Figure 3B). PCR testing validated three other scrambled regions, two for permutation and one for inversion (Figure S1C in Supporting Information). Descrambling of these three regions is required to assemble functional genes, coding for two transmembrane proteins (TTHERM_00229940 and TTHERM_000229949) and one kinase (TTHERM_00171610) respectively, suggesting that scrambling is a biologically relevant event in Tetrahymena. A better assembled MIC genome will allow us to further explore the scale, function and molecular mechanism of scrambling in Tetrahymena.
Analysis of the chromosomal copy number
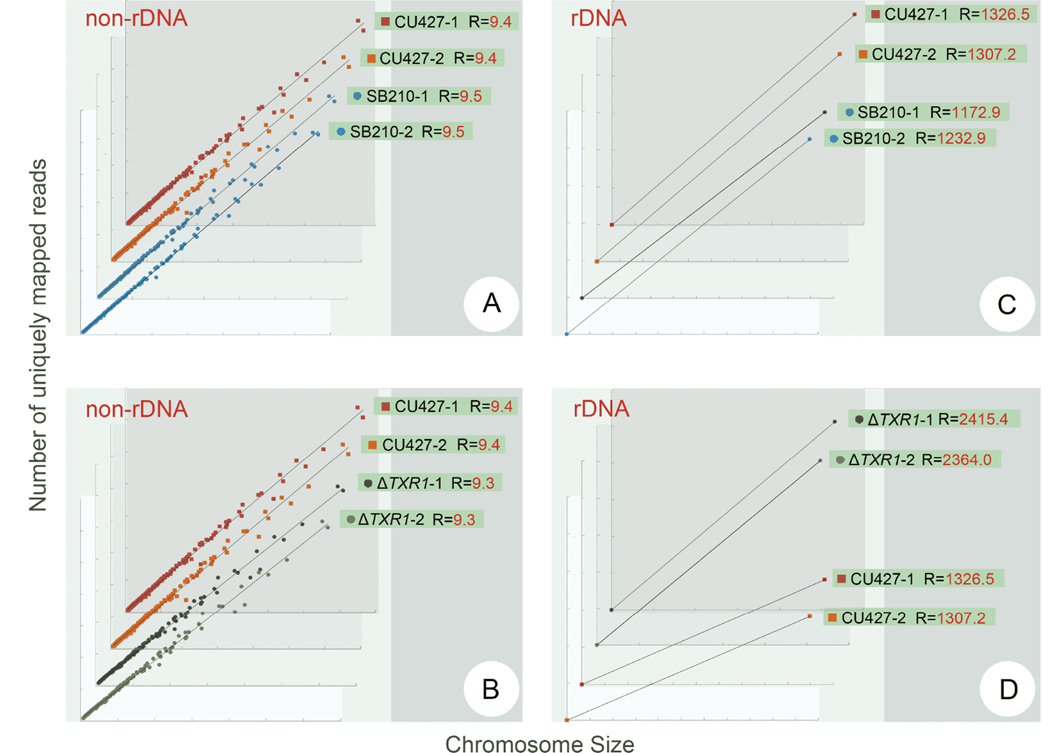
The number of reads mapped to the reference chromosome should have a direct proportionality with chromosomal length and copy number, given that whole-genome sequencing (WGS) generates reads with highly uniform coverage of the genome (Illumina sequencing official document) (Xu et al., 2017). Therefore, we used the ratio between unique mapped reads and chromosomal length to reflect the relative copy number (R) of chromosomes (R=unique mapped reads/chromosomal length). In this study, WT (SB210 and CU427) and replication-deficient (ΔTXR1) cells were sequenced and analyzed (Wang et al., 2019a).
The 180 non-rDNA chromosomes were maintained at the same level in all tested strains, represented as dots along the same trend line (Figure 4A and B). Moreover, strain background (SB210 vs. CU427) had no influence on copy number in both rDNA minichromosome and non-rDNA chromosomes, showed as trend lines with similar slopes (Figure 4A and C). The ratio of rDNA minichromosome to non-rDNA chromosomes (1,200/9≈130) was slightly different from expected (9,000/45=200), possibly because the repetitive sequences in the palindromic rDNA reduced the number of uniquely mapped reads.
Figure 4.
The copy number variation between two WT strains (SB210, CU427) and replication-deficient ΔTXR1 in non-rDNA chromosomes and rDNA minichromosome. The y-axis represents the length of chromosomes and the x-axis represents the number of unique reads (normalized by total reads number of each sample) mapped to the corresponding chromosome.
In the replication-deficient strain ΔTXR1 (Gao et al., 2013; Zhao et al., 2017), the copy number of non-rDNA chromosomes is identical to that of WT (Figure 4B). However, the copy number of the rDNA minichromosome increased significantly (Figure 4D). This was verified by quantitative polymerase chain reaction (qPCR) showing that the amount of rDNA minichromosome was much higher in ΔTXR1 cells (Figure S2 in Supporting Information), consistent with a previous finding revealed by plug gel electrophoresis (Gao et al., 2013). These results suggested that the copy number of rDNA minichromosome and non-rDNA chromosomes was regulated by different mechanisms (Larson et al., 1986), although the nature and role of the regulatory elements are largely unknown (Larson et al., 1986; Larson et al., 1991).
MATERIALS AND METHODS
Cell growth, DNA isolation, and library construction
WT strains for T. thermophila (SB210, CU427) were obtained from the Tetrahymena Stock Center (http://tetrahymena.vet.cornell.edu). Replication-deficient ΔTXR1 was a homozygous homokaryon strain generated by genetic manipulation (Gao et al., 2013; Zhao et al., 2017).
Genomic DNA was collected from vegetative log-phase cells (~2×105 cells mL–1) using Wizard® Genomic DNA Purification Kit (Promega, A1120). SMRT sequencing libraries of SB210 and Illumina sequencing libraries of SB210, CU427 and ΔTXR1 were constructed according to manufacturer-recommended protocols and sequenced by Novogene Co. Ltd (Beijing, China).
Genome assembly
SMRT sub-reads generated in this study and previously (Wang et al., 2017b; Wang et al., 2019b) were assembled into a draft assembly using Canu (Koren et al., 2017) software (correctedErrorRate=0.040, corMaxEvidenceErate=0.15, genomeSize=100 m), including error correction, read trimming and sequence assembly. Subsequently, erroneous contigs (CovStat≼0, contigs more likely to be repetitive; SuggestRepeat=yes, contigs detected as a repeat based on graph topology or read overlaps to other sequences; log2 (Normalized reads frequency)≼10, contigs with low mapped reads; without telomeres) were filtered. Reads frequency was defined as number of reads mapped to each contig, normalized by contig length. The remaining 180 contigs were separated into two groups: 165 contigs with telomeres on both sides and 15 contigs with telomeres on one side. Telomeres were identified by searching contigs for exact matches to a 12-mer encompassing two telomeric repeats (GGGGTTGGGGTT or CCCCAACCCCAA) (Eisen et al., 2006). Among these 15 contigs, 10 contigs were completed by BLAST to the TGD 2014 assembly. PCR primers (Table S1 in Supporting Information) were designed for another five contigs to amplify the uncompleted sequences (Figure S1A in Supporting Information) and were sequenced by Sanger sequencing. The 21 kb rDNA minichromosome was assembled with the same sub-reads using the assembly protocol (HGAP4, default parameters) in the SMRT link v5.10 (Pacific Biosciences). Finally, PE150 reads from short insert-size (200 bp) libraries were imported to Pilon (–genome genome.fasta, –bam input.bam) to correct the draft genome (Walker et al., 2014). The assembly flow is shown in Figure 1. The comparison between the completed genome and the TGD 2014 assembly was conducted by MUMmer (Delcher et al., 2003) with parameters (-i 90 -q).
Gene prediction and annotation
To identify protein-coding genes, gene and CDS sequences from TGD were aligned to the completed genome using BLAST (E-value≤1.0×10−5, identity>98%, alignment length>95%) for making hintsfile for gene prediction conducted by Augustus (Stanke et al., 2006) (–species=tetrahymena). Of these genes, 241 mispredicted genes (sequences length<40, encompassing telomeric repeats, false start codon or stop codon) were discarded. The Reciprocal Best Hits (RBH) approach was used in BLAST to generate similar genes between the remaining 26,258 predicted genes and the 26,996 genes from TGD 2014, with only effective hits retained (E-value≤1.0×10−20, identity≥93%) (Table S3 in Supporting Information). Approximately 96.5% (25,339) predicted genes matched corresponding TGD 2014 genes in the RBH list; in these cases, the “TTHERM” number and functional annotation of these genes were also inherited by the gene models in this study. Molecular function, biological process, and cellular component predictions for new genes found in this study were annotated using the Gene Ontology (GO). All genes were first aligned by BLASTP to sequences in the NCBI non-redundant database, and Blast2GO (Conesa et al., 2005) was used subsequently to annotate the sequences with GO terms.
Nuclei purification, unscrambling illustration
Purification of MACs and MICs was carried out following established protocols (Chen et al., 2016). The MDSs were identified by MIDAS (http://knot.math.usf.edu/midas/index.html) and aligned to the MIC assembly from TGD. Scrambled regions were detected and classified by customized scripts in Perl. To further confirm this phenomenon, corresponding PCR primers were designed (Table S1 in Supporting Information).
Copy number analysis
A total of six DNA samples of T. thermophila were sequenced, two replicates each for WT (SB210 and CU427) and ΔTXR1 respectively. After trimming sequencing adapters and filtering low quality reads with Trimmomatic (Bolger et al., 2014) (TruSeq3-PE.fa: 2:30:10, leading: 3, trailing: 3, sliding window: 4:15, minlen: 80), reads were mapped to the updated Tetrahymena genome by TopHat2 (Kim et al., 2013). For non-rDNA chromosomes, uniquely mapped reads were defined as reads mapped only once to the reference genome. For the palindromic rDNA minichromosome, uniquely mapped reads were defined as reads mapped twice, once to each palindromic half. The number of unique reads mapped to each chromosome was determined using customized Perl scripts. qPCR validation of rDNA copy number disparity between CU427 and ΔTXR1 was performed with genomic DNA as template and with serial rDNA-specific primers covering the 11 kb arm of rDNA (Table S1 in Supporting Information). JMJ1 (TTHERM_00185640) was used for loading control and normalization.
Data availability
The new genome and gene model annotation data were incorporated into Tetrahymena Genome Database (TGD, http://ciliate.org/). All sequencing data are accessible from NCBI under BioProject numbers PRJNA 611686.
Supplementary Material
Figure S1 Gel electrophoresis results in this study.
Figure S2 qPCR analysis confirmed that the copy number of the rDNA minichromosome increases in replication-deficient ΔTXR1 cells.
Table S1 Primers used in this study
Table S2 TGD genes BLAST to the predicted genes
Table S3 TGD proteins BLAST to the predicted proteins
Acknowledgements
This work was supported by the Natural Science Foundation of Shandong Province (JQ201706), The Marine S&T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (2018SDKJ0406-2), Fundamental Research Funds for the Central Universities (201841005), the Blue Life Breakthrough Program of LMBB of Qingdao National Laboratory for Marine Science and Technology (MS2018NO04), and National Institutes of Health (2P40OD010964). The authors would like to thank the following people for assistance with this study: Dr. Weibo Zheng (Ocean University of China) for providing advice during data analysis; Mr. Bo Pan (OUC) for helping revise the manuscript; and Dr. Mingjian Liu (OUC) for taking photos of T. thermophila. Our special thanks are given to Dr. Weibo Song (OUC) and Dr. Yifan Liu (University of Michigan) for their helpful suggestions during drafting the manuscript. We also acknowledge the computing resources provided on IEMB-1, a high-performance computing cluster operated by the Institute of Evolution and Marine Biodiversity.
Footnotes
Compliance and ethics The author(s) declare that they have no conflict of interest.
SUPPORTING INFORMATION
The supporting information is available online at http://life.scichina.com and https://link.springer.com. The supporting materials are published as submitted, without typesetting or editing. The responsibility for scientific accuracy and content remains entirely with the authors.
References
- Bolger AM, Lohse M, and Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes MD, Hamilton EP, Xiong J, Lawson MJ, Yuan D, Hadjithomas M, Miao W, and Orias E. (2013). Selecting one of several mating types through gene segment joining and deletion in Tetrahymena thermophila. PLoS Biol 11, e1001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, et al. (2014). The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 158, 1187–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Gao S, Liu Y, Wang Y, Wang Y, and Song W. (2016). Enzymatic and chemical mapping of nucleosome distribution in purified micro- and macronuclei of the ciliated model organism, Tetrahymena thermophila. Sci China Life Sci 59, 909–919. [DOI] [PubMed] [Google Scholar]
- Chen X, Jiang Y, Gao F, Zheng W, Krock TJ, Stover NA, Lu C, Katz LA, and Song W. (2019). Genome analyses of the new model protist Euplotes vannus focusing on genome rearrangement and resistance to environmental stressors. Mol Ecol Resour 19, 1292–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T, Wang Y, Huang J, Chen X, Zhao X, Gao S, and Song W. (2019). Our recent progress in epigenetic research using the model ciliate, Tetrahymena thermophila. Mar Life Sci Technol 1, 4–14. [Google Scholar]
- Collins K, and Gorovsky MA (2005). Tetrahymena thermophila. Curr Biol 15, R317–R318. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, and Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Stover NA, and Miao W. (2012). Whole genome studies of Tetrahymena. In Methods in Cell Biology (Oxford: Academic Press; ), pp. 53–81. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Thiagarajan M, Jones KM, Wortman JR, Tallon LJ, Haas BJ, Cassidy-Hanley DM, Wiley EA, Smith JJ, Collins K, et al. (2008). Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genomics 9, 562–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Salzberg SL, and Phillippy AM (2003). Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinf 00, 10.3.1–10.3.18. [DOI] [PubMed] [Google Scholar]
- Doerder FP, Deak JC, and Lief JH (1992). Rate of phenotypic assortment in Tetrahymena thermophila. Dev Genet 13, 126–132. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. (2006). Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4, e286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. (2012). Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 7, e47768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht JR, Nowacki M, and Landweber LF (2012). Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151, 1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Wang G, Hamilton EP, Xiong J, Yan G, Chen K, Chen X, Dui W, Plemens A, Khadr L, et al. (2017). A germline-limited piggyBac transposase gene is required for precise excision in Tetrahymena genome rearrangement. Nucleic Acids Res 45, 9481–9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Eisen JA, Nelson KE, Paulsen IT, and Salzberg SL (2002). The value of complete microbial genome sequencing (you get what you pay for). J Bacteriol 184, 6403–6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG (1974). Free ribosomal RNA genes in the macronucleus of Tetrahymena. Proc Natl Acad Sci USA 71, 3078–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Xiong J, Zhang C, Berquist BR, Yang R, Zhao M, Molascon AJ, Kwiatkowski SY, Yuan D, Qin Z, et al. (2013). Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev 27, 1662–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Dear PH, Rowland T, Saks K, Eisen JA, and Orias E. (2006). Use of HAPPY mapping for the higher order assembly of the Tetrahymena genome. Genomics 88, 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton EP, Kapusta A, Huvos PE, Bidwell SL, Zafar N, Tang H, Hadjithomas M, Krishnakumar V, Badger JH, Caler EV, et al. (2016). Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. eLife 5, e19090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M, Wang J, Fan X, Liu X, Shi W, Huang N, Zhao F, and Miao M. (2019). Genetic basis for the establishment of endosymbiosis in Paramecium. ISME J 13, 1360–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrer KM (1999). Tetrahymena genetics: two nuclei are better than one. In Methods in Cell Biology (Oxford: Academic Press; ), pp. 127–186. [DOI] [PubMed] [Google Scholar]
- Karrer KM (2012). Nuclear dualism. In Methods in Cell Biology (Oxford: Academic Press; ), pp. 29–52. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, and Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Huff ME, and Gonye GE (1988). Alternative use of chromosome fragmentation sites in the ciliated protozoan Oxytricha nova. Nucl Acids Res 16, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, and Phillippy AM (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 27, 722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DD, Blackburn EH, Yaeger PC, and Orias E. (1986). Control of rDNA replication in Tetrahymena involves a cis-acting upstream repeat of a promoter element. Cell 47, 229–240. [DOI] [PubMed] [Google Scholar]
- Larson DD, Umthun AR, and Shaiu WL (1991). Copy number control in the Tetrahymena macronuclear genome. J Protozool 38, 258–263. [DOI] [PubMed] [Google Scholar]
- Lindblad KA, Pathmanathan JS, Moreira S, Bracht JR, Sebra RP, Hutton ER, and Landweber LF (2019). Capture of complete ciliate chromosomes in single sequencing reads reveals widespread chromosome isoforms. BMC Genomics 20, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, and Gorovsky MA (2002). Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell 110, 689–699. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, and Gorovsky MA (2004a). Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev 18, 2068–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, and Gorovsky MA (2004b). Small RNAs in genome rearrangement in Tetrahymena. Curr Opin Genet Dev 14, 181–187. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, and Gorovsky MA (2005). A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev 19, 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MM, Donti TR, Sebastian Yakisich J, Smith AG, and Kapler GM (2007). Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J 26, 5048–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, and Landweber LF (2008). RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, and Flacks M. (1975). Macronuclear genetics of Tetrahymena I. Random distribution of macronuclear gene copies in T. pyriformis, syngen 1. Genetics 79, 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orias E, Singh DP, and Meyer E. (2017). Genetics and epigenetics of mating type determination in Paramecium and Tetrahymena. Annu Rev Microbiol 71, 133–156. [DOI] [PubMed] [Google Scholar]
- Prescott DM (1994). The DNA of ciliated protozoa. Microbiol Mol Biol Rev 58, 233–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM (2000). Genome gymnastics: unique modes of DNA evolution and processing in ciliates. Nat Rev Genet 1, 191–198. [DOI] [PubMed] [Google Scholar]
- Rasko DA, Webster DR, Sahl JW, Bashir A, Boisen N, Scheutz F, Paxinos EE, Sebra R, Chin CS, Iliopoulos D, et al. (2011). Origins of the E. coli strain causing an outbreak of Hemolytic-Uremic syndrome in Germany. N Engl J Med 365, 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray C Jr. (1956). Meiosis and nuclear behavior in Tetrahymena pyriformis. J Protozool 3, 88–96. [Google Scholar]
- Roberts RJ, Carneiro MO, and Schatz MC (2013). The advantages of SMRT sequencing. Genome Biol 14, 405–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruehle MD, Orias E, and Pearson CG (2016). Tetrahymena as a unicellular model eukaryote: genetic and genomic tools. Genetics 203, 649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, and Morgenstern B. (2006). AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34, W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover NA, Krieger CJ, Binkley G, Dong Q, Fisk DG, Nash R, Sethuraman A, Weng S, and Cherry JM (2006). Tetrahymena Genome Database (TGD): a new genomic resource for Tetrahymena thermophila research. Nucleic Acids Res 34, D500–D503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover NA, Punia RS, Bowen MS, Dolins SB, and Clark TG (2012). Tetrahymena Genome Database Wiki: a community-maintained model organism database. Database 2012, bas007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wang C, Jiang Y, Katz LA, Gao F, and Yan Y. (2019a). Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny. Sci China Life Sci 62, 203–214. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang Y, Sheng Y, Huang J, Chen X, Al-Rasheid KAS, and Gao S. (2017a). A comparative study of genome organization and epigenetic mechanisms in model ciliates, with an emphasis on Tetrahymena, Paramecium and Oxytricha. Eur J Protistol 61, 376–387. [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen X, Sheng Y, Liu Y, and Gao S. (2017b). N6-adenine DNA methylation is associated with the linker DNA of H2A.Zcontaining well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acids Res 45, 11594–11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng Y, Liu Y, Zhang W, Cheng T, Duan L, Pan B, Qiao Y, Liu Y, and Gao S. (2019b). A distinct class of eukaryotic MT-A70 methyltransferases maintain symmetric DNA N6-adenine methylation at the ApT dinucleotides as an epigenetic mark associated with transcription. Nucleic Acids Res 47, 11771–11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Gao S, Dui W, Yang W, Chen X, Taverna SD, Pearlman RE, Ashlock W, Miao W, and Liu Y. (2016). Dissecting relative contributions of cis- and trans-determinants to nucleosome distribution by comparing Tetrahymena macronuclear and micronuclear chromatin. Nucleic Acids Res 44, 10091–10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong J, Lu X, Zhou Z, Chang Y, Yuan D, Tian M, Zhou Z, Wang L, Fu C, Orias E, et al. (2012). Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS ONE 7, e30630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Li H, Perry JM, Singh VP, Unruh J, Yu Z, Zakari M, McDowell W, Li L, and Gerton JL (2017). Ribosomal DNA copy number loss and sequence variation in cancer. PLoS Genet 13, e1006771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Li X, Song W, Wang W, and Gao S. (2019). Cyclin Cyc2p is required for micronuclear bouquet formation in Tetrahymena thermophila. Sci China Life Sci 62, 668–680. [DOI] [PubMed] [Google Scholar]
- Yan Y, Maurer-Alcalá XX, Knight R, Kosakovsky Pond SL, and Katz LA (2019). Single-cell transcriptomics reveal a correlation between genome architecture and gene family evolution in ciliates. mBio 10, 10.1128/mBio.02524-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao MC, and Yao CH (1989). Accurate processing and amplification of cloned germ line copies of ribosomal DNA injected into developing nuclei of Tetrahymena thermophila. Mol Cell Biol 9, 1092–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Wang C, Katz LA, and Gao F. (2018). A paradox: rapid evolution rates of germline-limited sequences are associated with conserved patterns of rearrangements in cryptic species of Chilodonella uncinata (Protista, Ciliophora). Sci China Life Sci 61, 1071–1078. [DOI] [PubMed] [Google Scholar]
- Zhao X, Wang Y, Wang Y, Liu Y, and Gao S. (2017). Histone methyltransferase TXR1 is required for both H3 and H3.3 lysine 27 methylation in the well-known ciliated protist Tetrahymena thermophila. Sci China Life Sci 60, 264–270. [DOI] [PubMed] [Google Scholar]
- Zhao X, Xiong J, Mao F, Sheng Y, Chen X, Feng L, Dui W, Yang W, Kapusta A, Feschotte C, et al. (2019). RNAi-dependent Polycomb repression controls transposable elements in Tetrahymena. Genes Dev 33, 348–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Gel electrophoresis results in this study.
Figure S2 qPCR analysis confirmed that the copy number of the rDNA minichromosome increases in replication-deficient ΔTXR1 cells.
Table S1 Primers used in this study
Table S2 TGD genes BLAST to the predicted genes
Table S3 TGD proteins BLAST to the predicted proteins
Data Availability Statement
The new genome and gene model annotation data were incorporated into Tetrahymena Genome Database (TGD, http://ciliate.org/). All sequencing data are accessible from NCBI under BioProject numbers PRJNA 611686.