Abstract
Native capillary zone electrophoresis-mass spectrometry (CZE-MS) has attracted attentions for the characterization of monoclonal antibodies (mAbs) due to the potential of CZE for highly efficient separations of mAbs under native conditions as well as its compatibility with native electrospray ionization (ESI)-MS. However, the low sample loading capacity and limited separation resolution of native CZE for large proteins and protein complexes (e.g. mAbs) impede the widespread adoption of native CZE-MS. Here, we present a novel native capillary isoelectric focusing (cIEF)-assisted CZE-MS method for the characterization of mAbs with much larger sample loading capacity and significantly better separation resolution than native CZE-MS alone. The native cIEF-assisted CZE-MS employed separation capillaries with a new carbohydrate-based neutral coating, a commercilized electrokinetically pumped sheathflow CE-MS interface, and a high-end quadrupole-time-of-flight (Q-TOF) mass spectrometer. Using the method, we documented the separations of different proteoforms of the SigmaMAb and the detection of its various glyco-proteoforms and homodimer. The native cIEF-assisted CZE-MS separated the NIST mAb into three peaks with a submicroliter sample loading volume, corresponding to its different proteoforms. We observed that both the NIST mAb and its homodimer had eight glyco-proteoforms, four of which had low abundance. The results demonstrate the potential of our native cIEF-assisted CZE-MS method for advancing the characterization of large proteins and protein complexes under native conditions.
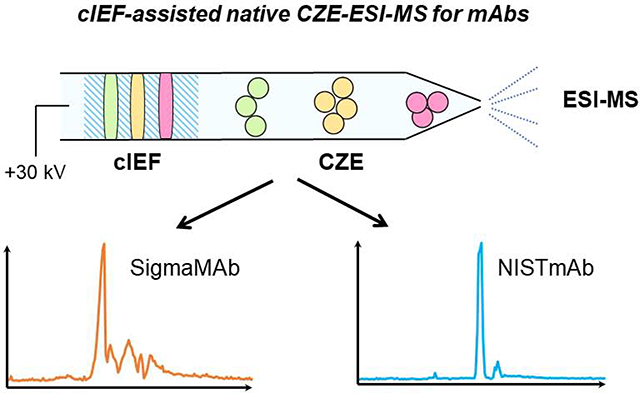
Graphical Abstract
Introduction
Monoclonal antibodies (mAbs) have become a dominant class of therapeutics for the treatment of cancer and autoimmune diseases because of their specificity and affinity to diverse targets [1,2]. Since first commercialized in 1985, over 80 therapeutic mAbs have been approved by FDA [3,4]. However, the complex production process of mAbs usually introduces various post-translational modifications (e.g., glycosylation, oxidation, deamidation, etc.) and structure changes (e.g., misfolding, denaturation, aggregation, etc.), leading to heterogeneities in the final products, which affect the potency, stability and efficacy of the therapeutics [5–7]. Thus, critical quality attributes of mAbs need to be closely monitored to ensure desired product quality.
Capillary zone electrophoresis (CZE) has been widely used for the quality control of mAbs in biopharmaceutical fields due to its high separation efficiency and straightforward operation [8]. Compared to other electrophoresis techniques, one advantage of CZE is that it has better compatibility with electrospray ionization-mass spectrometry (ESI-MS). A large number of reports illustrate CZE-ESI-MS for the characterization of mAbs from peptide mapping to intact protein analysis [9–19]. Although the peptide-level analysis can reveal detailed information on primary structure and PTMs of mAbs, intact mAb analysis better defines accurate mass and heterogeneity of mAb proteoforms. Han et al. performed an intact mass analysis of reduced and deglycosylated IgG1 by CZE-ESI-MS implemented by an electrokinetically pumped sheath-flow nanospray interface [16]. Redman et al. developed a microfluidic CZE-ESI device with online MS detection for separation and characterization of charge variants of intact Infliximab [14]. We note that most of the intact mAb analyses by CZE-MS are performed under denaturing conditions, which most likely lead to the information loss of mAb’s structure changes.
Recently, native CZE-ESI-MS has emerged as a promising technology for the characterization of mAb variants and aggregates as well as complex proteomes under native conditions [20,21]. The Ivanov group published the pioneering works on the analysis of mAbs by native CZE-MS, revealing major proteoforms due to glycosylations as well as low-abundance truncated species and mAb aggregates [22,23]. A sheathless CZE-MS interface [24], a linear polyacrylamide (LPA)-coated capillary, and an Orbitrap EMR mass spectrometer [25] were employed in the study. Le-Minh et al. investigated the conformational changes of Infliximab under stressed conditions using native CZE-MS [26]. A co-axial sheath liquid interface [27], a separation capillary with cationic coatings, and a Q-TOF mass spectrometer were utilized. Some challenges still exsit in native CZE-MS for the characterization of mAbs. First, the CZE separation of different mAb variants or conformations under native conditions needs to be improved. Second, the sample loading capacity of CZE is low under native conditions, which impedes the detection of low-abundance proteoforms of mAbs. Our group has demonstrated the capability of dynamic pH junction sample stacking method [28] for substantially boosting the sample loading capacity of CZE for large-scale top-down proteomics under denaturing conditions [29]. However, the dynamic pH junction method is hard to deploy for native CZE due to the fact that it employs the drastic difference in pH between sample buffer and background electrolyte (BGE) (i.e., pH 8–11 vs. 3). Alternative sample stacking methods are required for native CZE-MS.
In this work, we present the successful coupling of CZE with a high-end Q-TOF mass spectrometer using the electrokinetically pumped sheath-flow CE-MS interface [30,31] for the characterization of mAbs under native conditions. We first optimized the Q-TOF instrument parameters and CZE separation conditions, together with employing a new capillary coating to fulfill the maximum signal and mass resolution for native mAb detection. An online sample stacking method based on capillary isoelectric focusing (cIEF) in a narrow pH range was developed to expand the loading capacity as well as improve the CZE separation in the native condition for the first time.
Material and methods
1. Materials and reagents
The SILu Lite SigmaMAb universal antibody standard human (MSQC4), Pharmalyte 3–10, ammonium persulfate, ammonium acetate, ammonium formate and ammonium bicarbonate were purchased from Millipore Sigma Inc. (St Louis, MO, USA). Micro Bio-Spin™ P-6 gel columns were purchased from Bio-Rad Laboratories (Hercules, CA, USA). Hydrofluoric acid (HF) and acrylamide were purchased from Acros Organics (NJ, USA). The fused silica capillary (50 μm i.d., 360 μm o.d.) was purchased from Polymicro Technologies (Phoenix, AZ).
2. Antibody purification
SigmaMAb lyophilized powder was dissolved in water (10 mg/mL). NISTmAb was received in solution (10 mg/mL). Antibody samples were purified and buffer exchanged with 10 mM ammonium acetate (pH 6.8) by Bio-Spin™ P-6 gel columns according to the instruction. Briefly, after the removal of remaining buffer in the column, the column was washed with 500 μL 10 mM ammonium acetate for four times. Then 20 μL of the stock sample was loaded in the column, and the protein sample was collected in the flowthrough solution after centrifugation. The samples were diluted to desired concentrations for native CZE-MS analysis.
3. Preparation of separation capillaries
Two 70-cm-long capillaries (50 μm i.d., 360 μm o.d.) coated with a new linear carbohydrate polymer (LCP) coating from a synthesized sugar monomer (for details about the monomer synthesis and characterizations, see the Scheme S1 and Figures S1 and S2 in the Supporting Information) and a linear polyacrylamide (LPA) coating from a commercially available acrylamide monomer (Figure S3A) were used for CZE separation. The newly designed sugar monomer (Figure S3B) was used to form the LCP coating. The LPA coating was prepared on the inner wall of the capillary based on the literature [30,31]. The new coating procedure is similar to the LPA coating. Briefly, a bare fused silica capillary was successively flushed with 1 M hydrochloric acid, water, 1 M sodium hydroxide, water, and methanol, followed by treatment with 3-(trimethoxysilyl) propyl methacrylate to introduce carbon-carbon double bonds on the inner wall of the capillary. Then the pretreated capillary was filled with degassed sugar monomer (3-O-acryloyl-α/β-D-glucopyranose) solution (0.5 mg/mL) containing ammonium persulfate, followed by incubation at 35 °C water bath for 25–30 min with both ends sealed by silica rubber. After that, the capillary was flushed with water to remove the unreacted reagents. Both capillaries were etched with HF to reduce the outer diameter of one ends of the capillaries to ~ 70 μm [32].
4. Native CZE-ESI-MS analysis
A 7100 CE System from Agilent Technologies (Santa Clara, CA) was used for automated operation of CZE. The commercialized electrokinetically pumped sheath-flow nanospray interface (EMASS-II CE-MS Ion Source, CMP Scientific, Brooklyn, NY) [33,34] was used to couple CZE to a 6545XT AdvanceBio LC/Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA). The ESI emitters of the CE-MS interface were pulled from borosilicate glass capillaries (1.0 mm o.d., 0.75 mm i.d., 10 cm length) with a Sutter P-1000 flaming/brown micropipet puller. The opening size of the ESI emitters was 30–40 μm. Voltage for ESI ranged from +2.2 to +2.5 kV.
High voltage (+30 kV) and 50 mbar assisting pressure were applied for CZE separation unless specified. Samples were injected into the capillary by applying 100–950 mbar air pressure and the injection volume was calculated based on Poiseuille’s law. The background electrolyte (BGE) was 25 mM ammonium acetate (pH 6.8), and the sheath liquid (SL) was 10 mM ammonium acetate (pH 6.8) for CZE separation unless specified otherwise. For cIEF, SigmaMAb was dissolved in 10 mM ammonium acetate (pH 6.8) with 0.25% Pharmalyte 3–10. Before sample injection, 160 nL (50 mM) ammonium acetate (AA, pH 9.0) was injected as the catholyte. After sample injection, 12 nL BGE was injected to make sure the sample would not move back into the stock BGE solution. At the beginning of the separation, the sample was first focused without assisting pressure for 5–20 min depending on the injection volume. Then assisting pressure of 50 mbar was applied to the sample injection end of the separation capillary.
6545XT AdvanceBio LC/Q-TOF mass spectrometers (Agilent Technologies, Santa Clara, CA) with and without an electromagnetostatic ExD cell (e-MSion, Corvallis, OR) were used for the experiments. The instrument without the ExD cell was used for optimizations of MS parameters, CZE background electrolytes (BGEs) and sheath liquid (SL). The instrument with the ExD cell was used for other experiments. The ExD cell was set for positive transmission without ECD fragmentation (ECD off). Collision energy of 10 V was required for the maximum transmission efficiency of mAbs in the system with the ExD cell. A regular ESI spray shield and a nanoESI spray shield were used in the experiments. The gas temperature and flow rate of nitrogen drying gas was 365 °C and 1 L/min. The voltage applied on the ion transfer capillary was 0 V. The mass range option was set as High (10000 m/z). The slicer mode was High Resolution. The mass range of detection was 3000–10000 m/z, and the scan rate was 0.25 spectrum/sec. Fragmentor voltage, skimmer voltage and collision energy were set as specified.
5. Data analysis
Native CZE-ESI-MS runs were analyzed with Agilent MassHunter Qualitative Navigator B.08.00. Mass spectra were averaged across the electropherographic peaks. Average charge state (Zavg) of the mAb was calculated based on the equation [35]:
where q is the net charge of the given charge state, I is the intensity of the given charge state.
Deconvolution was performed using Agilent MassHunter BioConfirm 10.0 using Maximum Entropy algorithm. The mass step was 0.05 Da. Other parameters for deconvolution were set as default.
Results and Discussion
1. Optimizations of mass spectrometric parameters
Different from the detection of denaturing proteins, native MS requires mild MS parameters to prevent protein complex dissociation, denaturation and fragmentation in the system. To achieve high-quality spectra as well as maintain the native condition of native mAbs, we investigated three MS parameters (fragmentor voltage, skimmer voltage, collision energy) of the Q-TOF instrument through direct infusion MS of SigmaMAb (3 mg/mL in 10 mM NH4Ac, pH 6.8) with CZE system.
Fragmentor voltage is designed to promote ion transmission and perform in-source fragmentation. For large molecules like mAbs, the high fragmentor voltage could improve transmission and sensitivity. Moreover, high salt concentration is usually used for native conditions and caused salt adduction on proteins. The high fragmentor voltage could help decluster salts and water molecules complexing with the proteins. Therefore, we chose the fragmentor voltage of 380 V for the later experiments, which was the largest value we could set for the instrument.
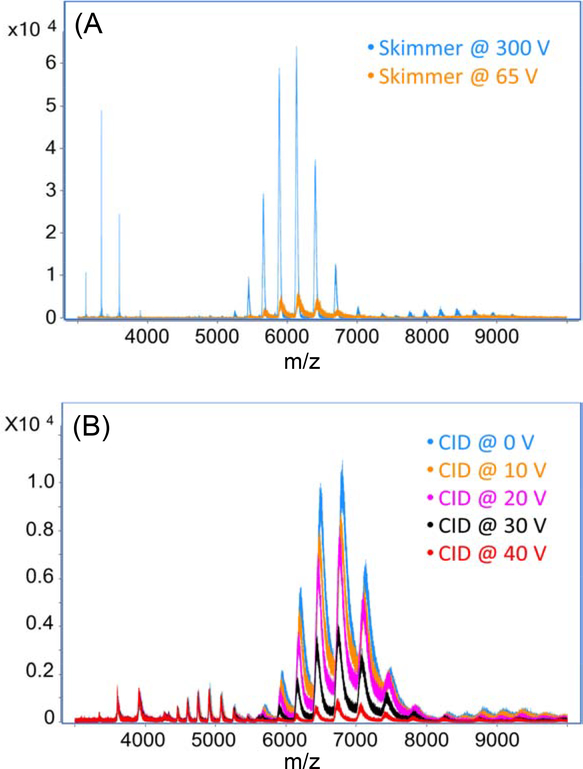
Skimmer voltage and collision energy also had significant impacts on the mass resolution and signal intensity of the mAb. Skimmer is used to sample the analytes into the high vacuum compartment. It can also focalize the ions and reduce ion beam broadening. When the skimmer voltage was raised from 65 to 300 V, we observed a three-fold improvement in mass resolution and a ten-fold increase in mAb intensity, Figure 1A. The higher skimmer energy could add more internal energy on the analytes and further remove the salt and solvent adducts on the mAbs. As a result, the heterogeneity of the mAb was reduced and higher mass resolution and signal intensity were achieved.
Figure 1.
Mass spectra of the SigmaMAb through direct infusion MS with the CZE system and the nanospray sheathflow CE interface. (A) Skimmer and (B) Collision energy were investigated.
Applying collision voltage is also commonly used to help remove salt and neutral adducts from proteins during native MS experiments. We investigated five different collision energy voltages (0 V to 40 V) in the CID cell. As we increased the collision voltage, the intensity of both monomer and homodimer of SigmaMAb decreased, Figure 1B. One possible reason is that we already applied enough energy on the proteins for declustering, and the high collision energy could cause fragmentation of the proteins and lead to the reduction of protein intensity. Therefore, collision energy was not necessary for native MS of the SigmaMAb. We used 10 V for later experiments because we have an ExD cell in our instrument and we needed the addition of collision energy to increase the transmission efficiency.
Another setting that affects the acquired spectra is the spray shield on the inlet of the Q-TOF mass spectrometer. We tested two kinds of the spray shield: a regular ESI spray shield and a nanoESI spray shield. With the regular ESI spray shield that has a larger orifice, more ions could be transmitted into the instrument and higher signal intensity was obtained. With nanoESI spray shield that has a smaller orifice and allows fewer ions to pass into the instrument, the signal intensity dropped about 10 times. However, the mass resolution was greatly improved and the peak broadening effect was reduced. As a result, we were able to see clear signals of mAb proteoforms due to glycosylations in the spectra with the nanoESI spray shield.
2. Optimizations of the CZE conditions for mAb
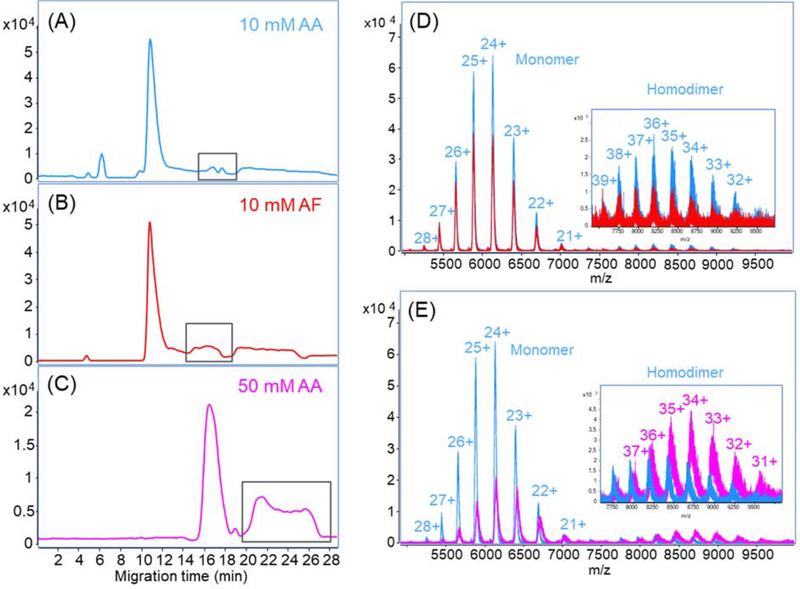
Volatile salt solutions around neutral pH are usually used to preserve the higher order structure of proteins in native liquid-phase separations. We first investigated two kinds of salt, ammonium acetate (AA) and ammonium formate (AF), for BGE and SL in native CZE-ESI-MS. An LPA-coated capillary was used for CZE separation and SigmaMAb (3 mg/mL) was still used as a standard to test the system. For each CZE run, 60 nL mAb sample was injected. Here, we used the regular ESI spray shield. Both BGE and SL were prepared with 10 mM concentration of the salt buffers. AA and AF presented almost identical separation profiles, Figures 2A and 2B, but AA had slightly better separation performance in the labeled part where two minor peaks were separated. The mass spectra of the major peaks in both electropherograms have a charge state distribution from 21+ to 28+ in the range of 5000–7200 m/z, Figure 2D. Besides the monomer, we also observed the homodimer of SigmaMAb with a charge state distribution of 32+ to 39+, demonstrating two salt conditions were gentle enough to preserve the noncovalent interaction. However, when comparing these two spectra, AA presented higher intensity for both monomer and homodimer of the SigmaMAb. We further calculated the average charge state of the mAb in two conditions. The mAb in AA has a marginally lower average charge state than that in AF (24.4 vs. 24.5). The result indicates that mAb in AF might be a little more denatured, which is consistent with previous reports that AF has a destabilizing effect and can cause structure unfolding of proteins [35,36].
Figure 2.
Investigation of native CZE separation conditions for the SigmaMAb. (A-C) Base peak electropherograms of native CZE-MS for SigmaMab with 10 mM AA, 10 mM AF and 50 mM AA as the BGE and SL. The peaks labeled with black boxes in the electropherograms represent the same mAb species. The spectra of the main peak in (D) 10 mM AA vs. 10 mM AF and (E) 10 mM AA vs. 50 mM AA are overlapped for comparisons. The insets are the zoom-in spectra of 7500–9500 m/z range. A LPA-coated capillary was used.
We also tested 50 mM AA for BGE and SL and the electropherogram is shown in Figure 2C. The CZE separation in 50 mM AA has a similar separation profile, but longer migration time and wider peak width compared to that in 10 mM AA, probably due to the increased viscosity of BGE as the salt concentration increased. From the mass spectra, we observed the reduction of mAb monomer intensity as well as the mass resolution with increased concentration of AA, Figure 2E, which could be explained by the higher salt concentration interfered with the ionization and caused ion suppression. Nonetheless, it should be noted that the intensity of mAb dimer is higher in 50 mM AA, Figure 2E. Also, the average charge state of mAb in 50 mM AA is lower than that in 10 mM AA (23.8 vs. 24.4), because the higher salt concentration could better maintain protein higher order structure. After an overall consideration of signal intensity and native conformation, we finally decided to use 10 mM AA as SL and 25 mM AA as BGE for the following native CZE-ESI-MS experiments.
The assisting pressure used in CZE separation was also studied. In native condition, the mAb is folded and carries much fewer charges compared to that in denaturing conditions. Consequently, the electrophoretic mobility of the mAb in native CZE is lower than regular CZE, and its migration time can be very long. We applied assisting pressure in native CZE separation of the mAb to shorten the migration time and increase the throughput of experiments. Figure S4 shows the electropherograms of SigmaMAb with 0–50 mbar assisting pressure during the native CZE separation. The injection amount is 60 nL. One thing we need to additionally indicate is that from here we changed the regular ESI spray shield to a nanoESI spray shield. Thus, the peak intensity was decreased significantly compared to the experiments above. As the assisting pressure decreased, the migration time of the mAb turned longer. At the same time, the peak width became wider and peak shape became worse. When applying 50 mbar assisting pressure, two peaks were observed in the electropherogram. However, only one peak was observed with lower assisting pressure. When no assisting pressure was applied, we could not even find the mAb signal. The longer migration time gave the analytes more chance to diffuse during the separation and eventually led to the peak broadening. Considering both the throughput and separation performance, assisting pressure of 50 mbar was used for the following experiments.
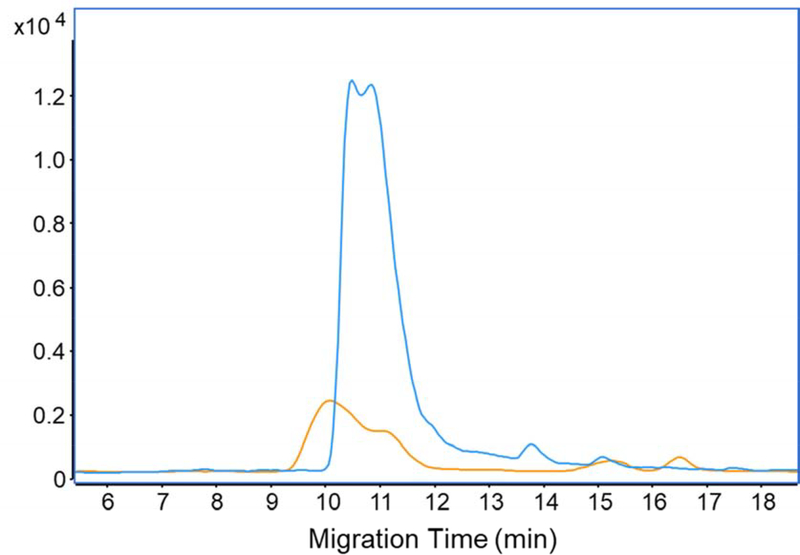
Capillary coating is another key factor in CZE separation. The LPA coating has been widely used in both peptide and intact protein analysis to eliminate electroosmotic flow in the capillary and improve CZE separation performance. However, we noticed from the data above that the LPA-coated capillary still had protein adsorption on the inner wall as evidenced by wide peaks of the mAb. It has been demonstrated that the polymers having the nitrogen element lead to significant protein adsorption [37] and carbohydrates-based polymers have excellent resistance to protein non-specific adsorption [38, 39]. Recently, we developed a new linear carbohydrate polymer (LCP)-based neutral coating, which is based on a glucose monomer (for details, see ESI) and applied this new coating for mAb studies. With the same MS settings and CZE conditions, the new carbohydrate coating showed a 6-fold increment in mAb intensity, Figure 3. The result suggests that the new LCP coating produces less interaction with the mAb during separation, boosting the sensitivity of native CZE-MS for the mAb significantly. We employed capillaries with the LCP coating for the rest of the experiments.
Figure 3.
Base peak electropherograms of native CZE-MS for SigmaMAb with the LCP coated-capillary (blue) and the LPA coated-capillary (orange). Same MS settings and CZE conditions were applied for both CZE runs.
3. Evaluating native capillary isoelectric focusing (cIEF)-assisted CZE-MS for mAbs
One drawback of CZE is the low sample loading capacity. Less than 1% of the total capillary volume is typically filled with the sample to obtain high separation efficiency, which limits the detection of low-abundance species in the sample. Using online sample stacking methods could help solve this problem. In addition, it could reduce peak width and lead to higher separation resolution. Many sample stacking methods have been evaluated for denaturing CZE-MS characterization of proteins, e.g., dynamic pH junction and field-amplified sample stacking (FASS) [29]. However, it is difficult to apply them in native conditions efficiently. An efficient sample stacking method under native conditions is urgently needed for native CZE-MS.
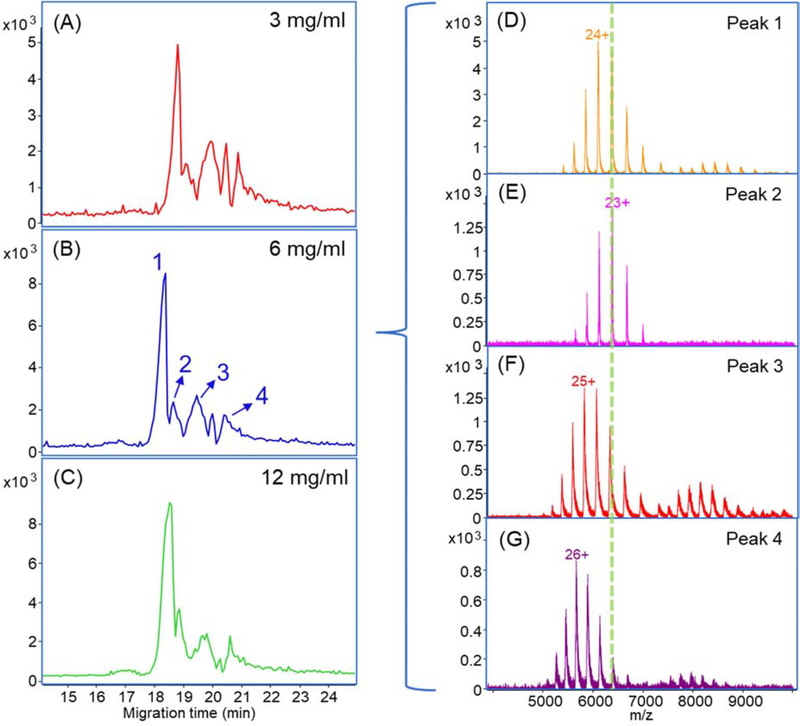
Here we investigated the possibility of employing cIEF for online sample concentration in native CZE-MS. cIEF separates analytes based on their isoelectric points (pIs). Although cIEF is mostly used for protein and peptide separation under denaturing conditions, several studies proved its feasibility for protein complex separation under native environments without destroying the native conformation and noncovalent interactions [40–44]. Its feature of focusing and ability of operation in native conditions provide us the possibility to utilize cIEF in a narrow pH range (i.e., pH 6–9) as a sample stacking method in the native CZE separation. First, a short plug of 50 mM AA (pH 9.0, about 160 nL) was injected as the catholyte for cIEF. Then, we injected a 30-nL SigmaMAb sample (3 mg/mL) dissolving in 0.25% Pharmalyte and 10 mM AA into the capillary. After that, 12-nL BGE (25 mM ammonium acetate, pH 6.8) was injected. After a high voltage was applied across the capillary, the SigmaMAb was first focused by native cIEF in the sample plug. After the focusing was completed, the mAb was further separated by native CZE. The native cIEF stacking had an obvious contribution to the CZE separation, Figure 4A. Four major peaks of SigmaMAb were separated, which was not observed by regular native CZE separations in Figure 2.
Figure 4.
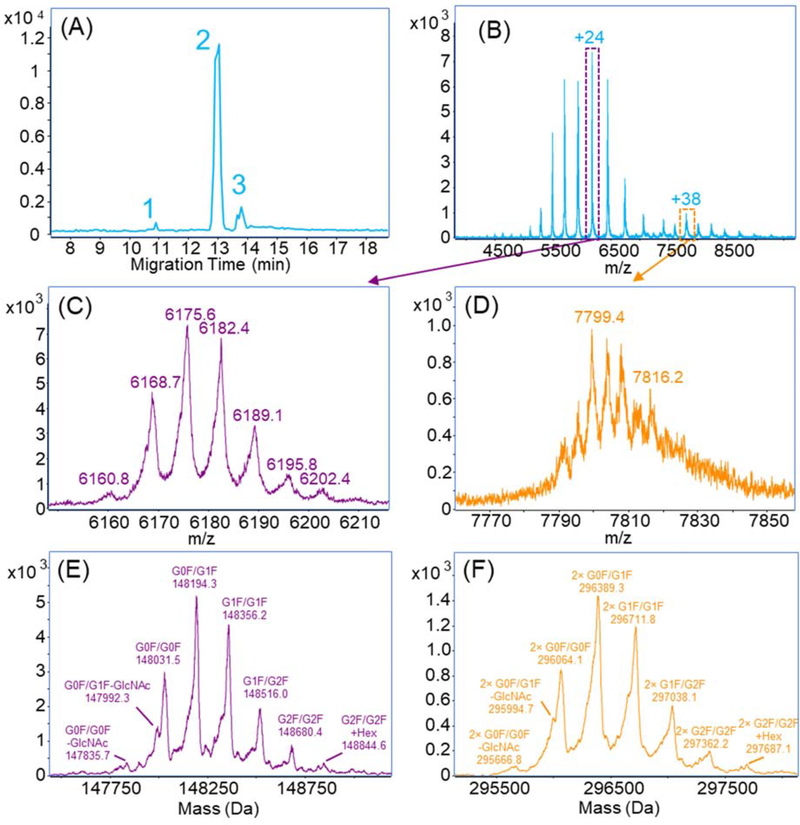
(A-C) Base peak electropherograms of native cIEF-assisted CZE-MS for 3, 6 and 12 mg/mL SigmaMAb; (D-G) Averaged mass spectra of the four major peaks separated in (B). Herein the LCP-coated capillary was used.
We increased the sample concentration for higher intensity as shown in Figures 4B and 4C. The electropherograms of the three concentrations are reproducible in terms of migration time and separation profile. The highest intensity was achieved with 6-mg/mL SigmaMAb. The higher protein concentration (12 mg/mL) did not further improve the signal intensity but widened the peak instead, which indicated that the system was saturated. We observed four major peaks of SigmaMAb with the 6 mg/mL sample (Figures 4D-G). The mass spectra show charge state distributions (CSDs) of mAb monomer from 20+ to 28+ and homodimer from 32+ to 39+. The zoom-in mass spectrum of the 23+ charge state from Figure 4E is shown in Figure S5A. The proteoforms due to different glycosylations could be resolved in the spectrum. The deconvoluted mass spectrum of Figure 4E shows five known glycosylated proteoforms of SigmaMAb (Figure S5B, Table S1), which were not achieved in the previous work with native CZE separation and Q-TOF instrument [26]. Interestingly, the four major peaks of SigmaMAb have minor differences in mass after deconvolution (within 300 Da, ~0.2% of the mAb mass) but are significantly different in CSD. As shown in Figures 4D–G, ions in peaks 1 and 2 carried significantly fewer charges than peaks 3 and 4 (e.g., +23 in peak 2 vs. +26 in peak 4). We need to note that the mass resolution in Figures 4D, F, and G is much lower than that in Figure 4E evidenced by much wider peaks, which certainly contributed to the mass differences across the four peaks to some extent. Considering the situations, we speculated that the four peaks could correspond to different groups of proteoforms of SigmaMAb due to variations in the protein sequence, PTM, or conformation.
We further tested different sample injection volumes using the native cIEF-assisted CZE-MS for the mAb. With the help of cIEF sample stacking, we were able to inject a large sample volume without losing separation resolution significantly. The concentration of Pharmalyte was decreased from 0.25% to 0.1% to reduce the interference of ampholytes to the mass spectrometer. Four different sample injection volumes from 30 nL to 800 nL were evaluated in two aspects: peak intensity and peak width, Figure S6. The SigmaMAb sample we used was 3 mg/mL. When the injection volume increased from 30 nL to 200 nL, the peak intensity was boosted about 6.5-folds and the peak width was doubled. When we increased the injection volume from 200 nL to 800 nL, the peak intensity was increased slightly, but the peak width was increased by nearly 100%. Although we adopted cIEF to stack the sample in the capillary, the stacking ability of cIEF was limited in native conditions. When too much mAb sample was injected, it would cause peak broadening and even protein precipitation in the capillary, and finally could not provide the expected increment of intensity. Furthermore, excess injection volume also resulted in excess ampholytes in the capillary, which interfered with the ionization of the mAb, enlarged the background noise and decreased the S/N ratio. Therefore, we selected 200 nL as the injection volume for later experiments.
4. Native cIEF-assisted CZE-MS for the NISTmAb
We further applied the native cIEF-assisted CZE-MS in the characterization of another recombinant humanized monoclonal IgG1 antibody, NISTmAb. The NISTmAb was dissolved in a buffer containing 0.1% Pharmalyte and 10 mM AA with a concentration of 1 mg/mL. Because NISTmAb has an isoelectric point as 9.18, we increased the catholyte pH to 9.5 to ensure the mAb could be focused by cIEF. For each run, 200 nL sample was injected into the capillary. Figure 5A shows the separation of the NIST mAb with three peaks. The main peak (peak 2) reveals the presence of both monomer and homodimer of the mAb, Figure 5B. Zoom-in mass spectra of the monomer at 24+ and dimer at 38+ are shown in Figures 5C and 5D. The deconvolution of the monomer signal, Figure 5E, identified four major and four minor biantennary glyco-proteoforms of the NISTmAb. The minor species included G2F/G2F, the addition of one hexose and the loss of N-acetylglucosamine (GlcNAc). These glyco-proteoforms have been previously reported for the NISTmAb [45]. The peaks of major glyco-proteoforms in the spectrum had fronting shapes, which were caused by the C-terminal lysine variants that could not be resolved due to the limited mass resolution. We also observed eight corresponding glyco-proteoforms of the homodimeric NIST mAb, Figures 5D and 5F. The assignments of all proteoforms for monomer and homodimers are listed in Table 1. The mass spectra and deconvoluted spectra of peak 1 and peak 3 are shown in Figure S7. Both monomer and dimer signals were detected in the averaged spectrum across peak 1, Figure S7A. We could not get a clear deconvolution result for glyco-proteoforms from the spectrum due to the low intensity, Figure S7C. However, it is clear that the deconvolution mass of the mAb proteoform in peak 1 is roughly 1500 Da smaller than that in the major peak 2. The mass shift is close to the mass of one glycan, thus, peak 1 probably represents the hemi-glycosylated mAb. The mass spectrum of peak 3 (Figure S7B) shows a shift of CSD of monomer to lower charge states compared to the major proteoforms of the NISTmAb, and the dimer is not observed. Several major glyco-proteoforms can be identified by deconvolution, Figure S7D. The most abundant charge state (23+) in peak 3 is one less than that in peak 2 (24+). One possible explanation is the deamidation of the mAb, which is previously reported as a common post-translational modification for NISTmAb [46]. Unfortunately, we cannot accurately distinguish this 1-Da mass difference in our experimental condition. Nonetheless, the cIEF-assisted CZE shows great potential for separation of different variants of mAbs in native conditions with large sample loading capacity.
Figure 5.
(A) Base peak electropherogram of native cIEF-assisted CZE-MS for the NISTmAb. (B) Mass spectrum averaged across the peak 2 in (A). (C, D) Zoom-in mass spectra of +24 and +38 charge states and (E, F) Deconvolution of NISTmAb proteoforms in the main peak (peak 2). Herein the LCP-coated capillary was used.
Table 1.
Theoretical and observed masses of the glyco-proteoforms of NISTmAb monomer and homodimer detected in the main peak (peak 2) with the native cIEF-assisted CZE-MS.
| Structure | Glyco-proteoform | Theoretical Mass (Da)a | Observed Mass (Da) | Mass Error (Da) | Mass Error (ppm) |
|---|---|---|---|---|---|
| Monomer | G0F/G0F – GlcNAc | 147834.0 | 147835.7 | 1.7 | 11.5 |
| G0F/G1F – GlcNAc | 147996.1 | 147992.3 | 3.8 | 25.7 | |
| G0F/G0F | 148037.2 | 148031.5 | 5.7 | 38.5 | |
| G0F/G1F | 148199.3 | 148194.3 | 5.0 | 33.7 | |
| G1F/G1F | 148361.4 | 148356.2 | 5.2 | 35.1 | |
| G1F/G2F | 148523.6 | 148516.0 | 7.6 | 51.2 | |
| G2F/G2F | 148685.7 | 148680.4 | 5.3 | 35.6 | |
| G2F/G2F + Hex | 148847.7 | 148844.6 | 3.1 | 20.8 | |
| Homodimer | 2× G0F/G0F – GlcNAc | 295668.0 | 295666.8 | 1.2 | 4.1 |
| 2× G0F/G1F – GlcNAc | 295992.2 | 295994.7 | 2.5 | 8.4 | |
| 2× G0F/G0F | 296074.4 | 296064.1 | 10.3 | 34.8 | |
| 2× G0F/G1F | 296398.6 | 296389.3 | 9.3 | 31.4 | |
| 2× G1F/G1F | 296722.8 | 296711.8 | 11.0 | 37.1 | |
| 2× G1F/G2F | 297047.2 | 297038.1 | 9.1 | 30.6 | |
| 2× G2F/G2F | 297371.4 | 297362.2 | 9.2 | 31 | |
| 2× G2F/G2F + Hex | 297695.4 | 297687.1 | 8.3 | 28 |
The theoretical masses are from reference 45.
Conclusions
We developed a novel cIEF-assisted CZE-MS platform for the analysis of mAbs under native conditions with large sample loading capacity. The optimizations of the Q-TOF parameters and CZE conditions provide a reference guide to the community for the characterization of native mAbs with a Q-TOF mass spectrometer and CZE separation. With the new capillary coating and the online cIEF sample stacking, this platform achieved high-quality characterization of glyco-proteoforms, variants and aggregates of two mAbs. Using cIEF in a narrow pH range for sample stacking is the first attempt and a proof of concept to preconcentrate the analytes and increase the loading capacity in native CZE conditions. We expect our novel platform could be a useful analytical tool for the characterization of various mAbs and large protein complexes.
Although we used Pharmalyte 3–10 in the experiments, we believe the ampholytes in a narrow pI range (e.g., 6–9) would improve the stacking performance in the native conditions further and will be investigated in future studies. Another direction of improvement is to integrate gas-phase fragmentation in the platform (i.e. electron capture dissociation), which could offer more precise information of PTMs on mAbs and help us to understand the formation of different variants and aggregates during production processes. We also need to note that the separation performance of our native CZE-MS system for mAb charge variants needs to be boosted further via investigating different additives to the separation buffer of CZE and evaluating different sugar monomers for the LCP coatings.
Supplementary Material
A native CZE-MS method with large sample loading capacity was developed.
A new carbohydrate-based neutral capillary coating was employed.
Native cIEF-assisted CZE-MS enabled the characterization of mAbs.
Delineation of different proteoforms and homodimers of mAbs were achieved.
Acknowledgements
We thank Agilent and CMP Scientific for their help for this project. In particular, we thank John Sausen (Director of Strategic Initiatives-Mass Spectrometry), Dr. David Wong, Dr. Caroline S. Chu, Dr. Christian Klein, Dr. Christopher Colangelo at Agilent and Dr. James Xia at CMP Scientific for their useful discussions about the data. We thank the support from the National Institute of General Medical Sciences (NIGMS) through Grant R01GM125991 and the National Science Foundation through Grant DBI1846913 (CAREER Award).
Footnotes
Conflict of Interest
The authors are collaborating with Agilent and CMP Scientific.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Carter PJ, Lazar GA, Next generation antibody drugs: pursuit of the “high-hanging fruit,” Nat. Rev. Drug Discov 17 (2017) 197–223. doi: 10.1038/nrd.2017.227. [DOI] [PubMed] [Google Scholar]
- [2].Chames P, Van Regenmortel M, Weiss E, Baty D, Therapeutic antibodies: successes, limitations and hopes for the future, Br. J. Pharmacol 157 (2009) 220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Goulet DR, Atkins WM, Considerations for the Design of Antibody-Based Therapeutics, J. Pharm. Sci 109 (2020) 74–103. doi: 10.1016/j.xphs.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kaplon H, Reichert JM, Antibodies to watch in 2019, mAbs. 11 (2019) 219–238. doi: 10.1080/19420862.2018.1556465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Groves K, Cryar A, Cowen S, Ashcroft AE, Quaglia M, Mass Spectrometry Characterization of Higher Order Structural Changes Associated with the Fc-glycan Structure of the NISTmAb Reference Material, RM 8761, J. Am. Soc. Mass Spectrom 31 (2020) 553–564. doi: 10.1021/jasms.9b00022. [DOI] [PubMed] [Google Scholar]
- [6].Srzentić K, Fornelli L, Tsybin YO, Loo JA, Seckler H, Agar JN, Anderson LC, Bai DL, Beck A, Brodbelt JS, van der Burgt YEM, Chamot-Rooke J, Chatterjee S, Chen Y, Clarke DJ, Danis PO, Diedrich JK, D’Ippolito RA, Dupré M, Gasilova N, Ge Y, Goo YA, Goodlett DR, Greer S, Haselmann KF, He L, Hendrickson CL, Hinkle JD, Holt MV, Hughes S, Hunt DF, Kelleher NL, Kozhinov AN, Lin Z, Malosse C, Marshall AG, Menin L, Millikin RJ, Nagornov KO, Nicolardi S, Paša-Tolić L, Pengelley S, Quebbemann NR, Resemann A, Sandoval W, Sarin R, Schmitt ND, Shabanowitz J, Shaw JB, Shortreed MR, Smith LM, Sobott F, Suckau D, Toby T, Weisbrod CR, Wildburger NC, Yates JR, Yoon SH, Young NL, Zhou M, Interlaboratory Study for Characterizing Monoclonal Antibodies by Top-Down and Middle-Down Mass Spectrometry, J. Am. Soc. Mass Spectrom 31 (2020) 1783–1802. doi: 10.1021/jasms.0c00036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Beck A, Wagner-Rousset E, Ayoub D, Van Dorsselaer A, Sanglier-Cianférani S, Characterization of Therapeutic Antibodies and Related Products, Anal. Chem. 85 (2012) 715–736. doi: 10.1021/ac3032355. [DOI] [PubMed] [Google Scholar]
- [8].Gahoual R, Beck A, Leize-Wagner E, François Y-N, Cutting-edge capillary electrophoresis characterization of monoclonal antibodies and related products, J. Chromatogr. B. 1032 (2016) 61–78. doi: 10.1016/j.jchromb.2016.05.028. [DOI] [PubMed] [Google Scholar]
- [9].Whitmore CD, Gennaro LA, Capillary electrophoresis-mass spectrometry methods for tryptic peptide mapping of therapeutic antibodies, Electrophoresis. 33 (2012) 1550–1556. doi: 10.1002/elps.201200066. [DOI] [PubMed] [Google Scholar]
- [10].Gahoual R, Burr A, Busnel J-M, Kuhn L, Hammann P, Beck A, François Y-N, Leize-Wagner E, Rapid and multi-level characterization of trastuzumab using sheathless capillary electrophoresis-tandem mass spectrometry, mAbs. 5 (2013) 479–490. doi: 10.4161/mabs.23995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dada OO, Zhao Y, Jaya N, Salas-Solano O, High-Resolution Capillary Zone Electrophoresis with Mass Spectrometry Peptide Mapping of Therapeutic Proteins: Peptide Recovery and Post-translational Modification Analysis in Monoclonal Antibodies and Antibody-Drug Conjugates. Anal Chem. 89 (2017) 11236–11242. doi: 10.1021/acs.analchem.7b03643. [DOI] [PubMed] [Google Scholar]
- [12].Lew C, Gallegos-Perez J-L, Fonslow B, Lies M, Guttman A, Rapid Level-3 Characterization of Therapeutic Antibodies by Capillary Electrophoresis Electrospray Ionization Mass Spectrometry, J. Chromatogr. Sci. 53 (2015) 443–449. doi: 10.1093/chromsci/bmu229. [DOI] [PubMed] [Google Scholar]
- [13].Chevreux G, Tilly N, Bihoreau N, Fast analysis of recombinant monoclonal antibodies using IdeS proteolytic digestion and electrospray mass spectrometry, Anal. Biochem. 415 (2011) 212–214. doi: 10.1016/j.ab.2011.04.030. [DOI] [PubMed] [Google Scholar]
- [14].Redman EA, Batz NG, Mellors JS, Ramsey JM, Integrated Microfluidic Capillary Electrophoresis-Electrospray Ionization Devices with Online MS Detection for the Separation and Characterization of Intact Monoclonal Antibody Variants, Anal. Chem. 87 (2015) 2264–2272. doi: 10.1021/ac503964j. [DOI] [PubMed] [Google Scholar]
- [15].Zhao Y, Sun L, Knierman MD, Dovichi NJ, Fast separation and analysis of reduced monoclonal antibodies with capillary zone electrophoresis coupled to mass spectrometry, Talanta. 148 (2016) 529–533. doi: 10.1016/j.talanta.2015.11.020. [DOI] [PubMed] [Google Scholar]
- [16].Han M, Rock BM, Pearson JT, Rock DA, Intact mass analysis of monoclonal antibodies by capillary electrophoresis—Mass spectrometry, J. Chromatogr. B 1011 (2016) 24–32. doi: 10.1016/j.jchromb.2015.12.045. [DOI] [PubMed] [Google Scholar]
- [17].Jooß K, Hühner J, Kiessig S, Moritz B, Neusüß C, Two-dimensional capillary zone electrophoresis–mass spectrometry for the characterization of intact monoclonal antibody charge variants, including deamidation products, Anal. Bioanal. Chem 409 (2017) 6057–6067. doi: 10.1007/s00216-017-0542-0. [DOI] [PubMed] [Google Scholar]
- [18].Belov AM, Zang L, Sebastiano R, Santos MR, Bush DR, Karger BL, Ivanov AR, Complementary middle-down and intact monoclonal antibody proteoform characterization by capillary zone electrophoresis - mass spectrometry, Electrophoresis. 39 (2018) 2069–2082. doi: 10.1002/elps.201800067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Cheng J, Wang L, Rive CM, Holt RA, Morin GB, Chen DDY, Complementary Methods for de Novo Monoclonal Antibody Sequencing to Achieve Complete Sequence Coverage. J Proteome Res. 19 (2020) 2700–2707. doi: 10.1021/acs.jproteome.0c00223. [DOI] [PubMed] [Google Scholar]
- [20].Shen X, Kou Q, Guo R, Yang Z, Chen D, Liu X, Hong H, Sun L, Native Proteomics in Discovery Mode Using Size-Exclusion Chromatography-Capillary Zone Electrophoresis-Tandem Mass Spectrometry. Anal Chem. 90 (2018) 10095–10099. doi: 10.1021/acs.analchem.8b02725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Shen X, Yang Z, McCool EN, Lubeckyj RA, Chen D, Sun L, Capillary zone electrophoresis-mass spectrometry for top-down proteomics, Trends Anal. Chem. 120 (2019) 115644. doi: 10.1016/j.trac.2019.115644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Belov AM, Viner R, Santos MR, Horn DM, Bern M, Karger BL, Ivanov AR, Analysis of Proteins, Protein Complexes, and Organellar Proteomes Using Sheathless Capillary Zone Electrophoresis - Native Mass Spectrometry, J. Am. Soc. Mass Spectrom. 28 (2017) 2614–2634. doi: 10.1007/s13361-017-1781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Belov AM, Zang L, Sebastiano R, Santos MR, Bush DR, Karger BL, Ivanov AR, Complementary middle-down and intact monoclonal antibody proteoform characterization by capillary zone electrophoresis - mass spectrometry, Electrophoresis. 39 (2018) 2069–2082. doi: 10.1002/elps.201800067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Moini M, Simplifying CE-MS operation. 2. Interfacing low-flow separation techniques to mass spectrometry using a porous tip, Anal. Chem. 79 (2007), 4241–4246. 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- [25].Maple HJ, Scheibner O, Baumert M, Allen M, Taylor RJ, Garlish RA, Bromirski M, Burnley RJ, Application of the Exactive Plus EMR for automated protein-ligand screening by non-covalent mass spectrometry. Rapid Commun Mass Spectrom. 28 (2014) 1561–8. doi: 10.1002/rcm.6925. [DOI] [PubMed] [Google Scholar]
- [26].Le-Minh V, Tran NT, Makky A, Rosilio V, Taverna M, Smadja C, Capillary zone electrophoresis-native mass spectrometry for the quality control of intact therapeutic monoclonal antibodies, J. Chromatogr. A. 1601 (2019) 375–384. doi: 10.1016/j.chroma.2019.05.050. [DOI] [PubMed] [Google Scholar]
- [27].Smith RD, Barinaga CJ, Udseth HR, Improved electrospray ionization interface for capillary zone electrophoresis-mass spectrometry, Anal. Chem. 60 (1988), 1948–1952. 10.1021/ac00169a022. [DOI] [Google Scholar]
- [28].Britz-McKibbin P, Chen DDY, Selective Focusing of Catecholamines and Weakly Acidic Compounds by Capillary Electrophoresis Using a Dynamic pH Junction, Anal. Chem. 72 (2000), 1242–1252. 10.1021/ac990898e. [DOI] [PubMed] [Google Scholar]
- [29].Lubeckyj RA, McCool EN, Shen X, Kou Q, Liu X, Sun L, Single-Shot Top-Down Proteomics with Capillary Zone Electrophoresis-Electrospray Ionization-Tandem Mass Spectrometry for Identification of Nearly 600 Escherichia coli Proteoforms, Anal. Chem. 89 (2017), 12059–12067. 10.1021/acs.analchem.7b02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhu G, Sun L, Dovichi NJ, Thermally-initiated free radical polymerization for reproducible production of stable linear polyacrylamide coated capillaries, and their application to proteomic analysis using capillary zone electrophoresis–mass spectrometry, Talanta. 146 (2016) 839–843. doi: 10.1016/j.talanta.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].McCool EN, Lubeckyj R, Shen X, Kou Q, Liu X, Sun L, Large-scale Top-down Proteomics Using Capillary Zone Electrophoresis Tandem Mass Spectrometry, J. Vis (2018) e58644. doi: 10.3791/58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun L, Zhu G, Zhao Y, Yan X, Mou S, Dovichi NJ, Ultrasensitive and Fast Bottom-up Analysis of Femtogram Amounts of Complex Proteome Digests, Angew. Chem. Int. 52 (2013) 13661–13664. doi: 10.1002/anie.201308139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wojcik R, Dada OO, Sadilek M, Dovichi NJ, Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis, Rapid Commun. Mass Spectrom. 24 (2010), 2554–2560. 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- [34].Sun L, Zhu G, Zhang Z, Mou S, Dovichi NJ, Third-generation electrokinetically pumped sheath-flow nanospray interface with improved stability and sensitivity for automated capillary zone electrophoresis-mass spectrometry analysis of complex proteome digests, J Proteome Res 14 (2015), 2312–2321. 10.1021/acs.jproteome.5b00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Ventouri IK, Malheiro DBA, Voeten RLC, Kok S, Honing M, Somsen GW, Haselberg R, Probing Protein Denaturation during Size-Exclusion Chromatography Using Native Mass Spectrometry, Anal. Chem 92 (2020) 4292–4300. doi: 10.1021/acs.analchem.9b04961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Konermann L, Addressing a Common Misconception: Ammonium Acetate as Neutral pH “Buffer” for Native Electrospray Mass Spectrometry, J. Am. Soc. Mass Spectrom 28 (2017) 1827–1835. doi: 10.1007/s13361-017-1739-3. [DOI] [PubMed] [Google Scholar]
- [37].Metzke M, Guan Z, Structure-property studies on carbohydrate-derived polymers for use as protein-resistant biomaterials, Biomacromolecules. 9 (2008) 208–15. doi: 10.1021/bm701013y. [DOI] [PubMed] [Google Scholar]
- [38].Metzke M, Bai JZ, Guan Z, A novel carbohydrate-derived side-chain polyether with excellent protein resistance, J Am Chem Soc. 125 (2003) 7760–1. doi: 10.1021/ja0349507. [DOI] [PubMed] [Google Scholar]
- [39].Maiti S, Manna S, Shen J, Esser-Kahn AP, Du W, Mitigation of Hydrophobicity-Induced Immunotoxicity by Sugar Poly(orthoesters), J Am Chem Soc 141 (2019) 4510–4514. doi: 10.1021/jacs.8b12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Martinović S, Paša-Tolić L, Masselon C, Jensen PK, Stone CL, Smith RD, Characterization of human alcohol dehydrogenase isoenzymes by capillary isoelectric focusing — mass spectrometry, Electrophoresis. 21 (2000) 2368–2375. doi:. [DOI] [PubMed] [Google Scholar]
- [41].Przybylski C, Mokaddem M, Prull-Janssen M, Saesen E, Lortat-Jacob H, Gonnet F, Varenne A, Daniel R, On-line capillary isoelectric focusing hyphenated to native electrospray ionization mass spectrometry for the characterization of interferon-γ and variants, Analyst. 140 (2015) 543–550. doi: 10.1039/c4an01305k. [DOI] [PubMed] [Google Scholar]
- [42].Wu X-Z, Asai S, Yamaguchi Y, Study of protein-protein binding reaction by whole-column fluorescence-imaged CIEF, Electrophoresis. 30 (2009) 1552–1557. doi: 10.1002/elps.200800506. [DOI] [PubMed] [Google Scholar]
- [43].Cunliffe JM, Liu Z, Pawliszyn J, Kennedy RT, Use of a native affinity ligand for the detection of G proteins by capillary isoelectric focusing with laser-induced fluorescence detection, Electrophoresis. 25 (2004) 2319–2325. doi: 10.1002/elps.200405953. [DOI] [PubMed] [Google Scholar]
- [44].Okun VM, Affinity mode of capillary isoelectric focusing for the characterization of the biotin-binding protein actinavidin, Electrophoresis. 19 (1998) 427–432. doi: 10.1002/elps.1150190311. [DOI] [PubMed] [Google Scholar]
- [45].Formolo T, Ly M, Levy M, Kilpatrick L, Lute S, Phinney K, Marzilli L, Brorson K, Boyne M, Davis D, Schiel J, Determination of the NISTmAb Primary Structure, American Chemical Society, 2015. doi: 10.1021/bk-2015-1201.ch001. [DOI] [Google Scholar]
- [46].Dong Q, Liang Y, Yan X, Markey SP, Mirokhin YA, Tchekhovskoi DV, Bukhari TH, Stein SE, The NISTmAb tryptic peptide spectral library for monoclonal antibody characterization, mAbs. 10 (2018) 354–369. doi: 10.1080/19420862.2018.1436921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.