Abstract
Pulsed-electromagnetic-field (PEMF) treatment was found to enhance cellular differentiation of the mouse preosteoblast, MC3T3-E1, to a more osteoblastic phenotype. Differentiation genes such as Alp, BSPI, cFos, Ibsp, Osteocalcin, Pthr1 and Runx2 showed increased expression in response to PEMF stimulation. Detailed molecular mechanisms linking PEMF to the activation of these genes are limited. Two adenosine receptors known to be modulated in response to PEMF, Adora2A and Adora3, were functionally impaired by CRISPR-Cas9-mediated gene disruption, and the consequences of which were studied in the context of PEMF-mediated osteoblastic differentiation. Disruption of Adora2A resulted in a delay of Alp mRNA expression, but not alkaline phosphatase protein expression, which was similar to that found in wild type cells. However, Adora3 disruption resulted in significantly reduced responses at both the alkaline phosphatase mRNA and protein levels throughout the PEMF stimulation period. Defects observed in response to PEMF were mirrored using a chemically defined growth and differentiation-inducing media (DM). Moreover, in cells with Adora2A disruption, gene expression profiles showed a blunted response in cFos and Pthr1 to PEMF treatment; whereas cells with Adora3 disruption had mostly blunted responses in AlpI, BSPI, Ibsp, Osteocalcin and Sp7 gene activation. To demonstrate specificity for Adora3 function, the Adora3 open reading frame was inserted into the ROSA26 locus in Adora3 disrupted cells culminating in rescued PEMF responsiveness and thereby eliminating the possibility of off-target effects. These results lead us to propose that there are complementary and parallel positive roles for adenosine receptor A2A and A3 in PEMF-mediated osteoblast differentiation.
Introduction
Bone fracture is the most common musculoskeletal injury, and healing of such injury involves complex cellular and molecular processes [1,2]. Fracture healing is a multistep biological process, involving temporally defined stages including hematoma, inflammation, fibro-vasculature, bone formation and remodeling. Intermittent pulsed-electromagnetic-field (PEMF) stimulation has long been reported to heal bone fractures, especially delayed and non-unions [3–9]. PEMF, a noninvasive and safe procedure, has received extensive attention in clinical applications over last two decades. However, the biological mechanisms behind its success remain mostly enigmatic. Proliferation of different cell types, including neural stem cells [10], bone marrow mesenchymal stem cells [11], intervertebral disc cells [12], tendon cells [13], myoblasts [14], and finally osteoblasts [15] have been reported to be enhanced by PEMF stimulation. Reinforcement of intracellular calcium transients [15] and activation of the mammalian target of rapamycin (mTOR) signaling pathway leading to proliferation [16] of osteoblast cells have been described as possible effector pathways activated through intermittent treatment with electromagnetic pulses. PEMF has been reported to also modulate cAMP levels [17] and thus to induce an anti-inflammatory response in a variety of cell types [18–21].
Adenosine receptors (ADRs) have been shown to play a crucial role in both the anti-inflammatory response and in bone formation. ADR A2A has been reported to regulate anti-inflammatory effects, bone metabolism and hemostasis, increase cAMP levels through activation of adenylcyclase and Akt-mediated nuclear localization of the transcription factor β-catenin [22–25]. ADR A3 has been shown to regulate inflammatory responses and to decrease adenylcyclase activity leading to lower cAMP levels [26]. However, the role of ADR A3 in bone formation is largely unknown although it is expressed on osteoblasts and osteoblast precursors [27] and A3 selective agonists have been shown to stimulate cell proliferation while A3 inhibitors blocked this effect [28]. Interestingly, PEMF has been known to increase plasma-membrane densities of A2A and A3 [22,29], the physiological implications of which relative to pre-osteoblast to osteoblast differentiation is unclear and is a driving force for our current study.
Though the enhanced healing of non-union bone fractures by PEMF-mediated osteoblast proliferation and differentiation has been documented for decades, leading to an FDA approval of PEMF devices [30], the cellular and molecular mechanisms leading to such biological effects have not been well-defined in terms of their role in PEMF-mediated fracture healing. Here, we propose that the Adenosine Receptors A2A and A3 play positive roles of PEMF-mediated osteoblast differentiation and provide evidence of the operative molecular mechanisms each contributes to.
Materials and methods
Plasmids and stable lines
Homology Directed Repair (HDR) mediated CRISPR-Cas9 gene disruption technique (Origene Technologies, Inc.) was used to gene knock-out Adora A2A and A3, according to the manufacturer’s protocol. Briefly, in this two-plasmid system, cells were transfected with a plasmid harboring Cas9 and gRNA specific for early in the first exon of the target genes and one with a cassette expressing antibiotic marker genes (blasticidin for A2A and puromycin for A3) flanked by homology arms for the target genes. After seven passages, cells were treated with antibiotics and individual clones were isolated, expanded and PCR screened for integration.
For inducible gene knock-in, the cumate inducible expression system was used (System Biosciences, LLC) with modification. Briefly, to make the donor plasmid, the A3 open reading frame (ORF) was subcloned in the SparQ-T2A plasmid downstream of the cumate promoter, and left and right homology arms for mouse ROSA26 locus were synthesized from GenScript and were subcloned upstream of cumate promoter and downstream of SV40 polyA sequence, respectively. pX330-Cas9-ROSA26gRNA was made by subcloning ROSA26gRNA sequence in pX330-Cas9 (Addgene #42230). This donor plasmid together with Cas9-Rosa26gRNA plasmid were co-transfected into A3 disrupted cells, and stable cloned were isolated and screened, and the best expressing clone under cumate treatment was used for further experiments.
Cell culture
Mouse preosteoblast cell MC3T3-E1 was bought from ATCC and grown in the growth medium (GM), made of the MEM-α without ascorbic acid supplemented with 10% fetal bovine serum and penicillin-streptomycin. The GM additionally supplemented with 50 μg/ml ascorbic acid and 10 mM β-glycerophosphate was used as a chemically defined differentiation medium (DM) [31].
PEMF treatment
The PEMF signal used in this study had similar waveform characteristics to the clinically approved PhysioStim® (Orthofix Medical Inc., Lewisville TX) signal used to heal long-bone non-unions. The signal has a pulse frequency of 3.85 kHz and slew-rate of 10 T/s (30). Cells were cultured in GM, and exposed to daily 4 hours PEMF treatment for up to 16 days.
Quantitative RT-PCR
Osteoblast differentiation was measured by activation of genes, Alp, cFos, Ibsp (Bone Sialoprotein II, BSPII), Osteocalcin (BGLP), Osteopontin (Bone Sialoprotein I, BSPI, SPPI), Pthr1, Runx2 and Sp7, with 4 hours daily PEMF treatment, followed by total RNA extraction using TRIzol reagent (Ambion/Life Technologies) following the manufacturer’s protocol. First-stand cDNAs were synthesized from mRNAs by reverse transcription using Superscript IV (Invitrogen) as per manufacturer’s protocol and expression levels were assayed by quantitative RT-PCR with Taqman Gene Expression (Applied Biosystems) primers-probes (AlpI: Mm00475834_m1; BSPI/SPP1/Osteopontin: Mm00436767_m1; Ibsp/BSPII: Mm0049255_m1; cFos: Mm00487425_m1; Osteocalcin/Bglap: Mm03413826_mH; Pthr1: Mm00441046_m1; Runx2: Mm00501584_m1; Sp7/Osterix: Mm04209856_m1) in a StepOne Plus (Applied Biosystems) real-time PCR instrument. Each data set was normalized by untreated cells grown for 8 days, and GAPDH (Mm99999915_g1) was used as housekeeping gene. Each experiment was performed in biological triplicates and qRT-PCRs were performed for each in duplicates.
Alkaline phosphatase assays and Alizarin Red staining
The differentiation process was also monitored by induction of alkaline phosphatase enzymatic activity in cell lysates and by cell staining every 4 days up to 16 days. For alkaline phosphatase assay, cells were lysed in homogenizer (Fisherbrand Pellet Pestle Microtubes), aliquots were incubated with assay buffer containing pNPP at room temperature for 30 mins, and OD405 was read [31,32]. For alkaline phosphatase staining, cells were fixed for 30 sec with 4% paraformaldehyde, washed and incubated with alkaline-dye mixture containing 0.01% naphthol AS-MX phosphate and 0.24 mg/ml Fast Blue RR at room temperature for 30 min, washed and photographed.
For mineralization assay, Alizarin Red staining was performed at similar time points as above [31]. Briefly, cells were fixed with 4% paraformaldehyde, washed and incubated with 2% Alizarin red for 5 minutes at room temperature, washed and photographed.
Western Blot
To examine at the effect on protein expression, cells were treated with PEMF (4 hours daily) or DM for 12 days, whole-cell proteins were extracted using lysis buffer containing 1% NP-40 and 0.5% TX-100, and protein concentration determined with DC Protein Assay (Bio-Rad). Total cell proteins (50 μg/lane) were fractionated on denaturing, reducing 10% or 16% SDS-PAGE and transferred onto PVDF membranes. The membranes were blocked with 4% Blotting-grade Blocker (Bio-Rad) in PBS and probed for one hour at room temperature with 1:1000 dilutions (in PBS) of antibodies against the following proteins: BSPI (PA5-79423, Invitrogen), IBSP (5468S, Cell Signaling) and Osteocalcin (ab93876, Abcam) with GAPDH (MA5-15738, Invitrogen) serving as loading control. Blots were washed three times 5 minutes each with PBS containing 0.05% Tween-20 and then incubated with 1:10000 dilution of secondary antibodies (HRP-conjugated) anti-mouse (7076S, Cell Signaling) or ant-rabbit (7074S, Cell Signaling), for 1 hour at room temperature. Finally, the blots were washed 3 times by 5 minutes each, before being developed with ECL Prime Western Blotting Detection reagent (RPN2232, GE Healthcare) per manufacturer’s direction.
Images and statistical analysis
All experiments were performed at least in biological triplicate. The Student’s two-tailed T-test was used to analyze experimental vs control cell types or treatment conditions, and a σ value of p<0.05 denoted with asterisks to emphasis significance.
Results
PEMF stimulates osteoblast differentiation
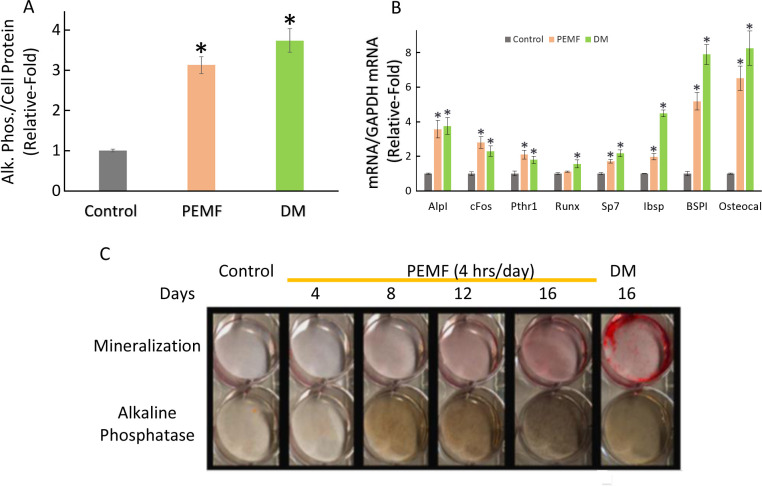
We examined the effect that PEMF elicited on the expression of select genes known to be associated with the early stages of pre-osteoblast to osteoblast differentiation. Pre-osteoblast MC3T3-E1 cells were treated with PEMF with 4 hours daily cycles, or differentiation media (DM) for up to 16 days. Cell proteins were extracted and assayed for enhanced alkaline phosphate enzymatic activity normalized to cell protein, a typical hallmark of osteoblast differentiation, and significant activation was detectable due to both treatments (Fig 1A). Apart from Alp, the other osteoblast differentiation genes, viz. BSPI, cFos, Ibsp, Osteocalcin, Pthr1, and Sp7, were also found to have significantly increased expression in response to PEMF treatment as measured by qRT-PCR from total RNA normalized to glyceraldehyde dehydrogenase (GAPDH) (Fig 1B). Alp, BSPI, Ibsp and Osteocalcin are the top four most prominent responding genes both to PEMF and DM treatments under these experimental conditions. PEMF was also found to increase alkaline phosphate cell staining, and Alizarin Red staining for mineralization in a dose-dependent manner (Fig 1C). These results further confirmed that PEMF stimulates osteoblastic differentiation of MC3T3-E1 under the experimental conditions employed here.
Fig 1. PEMF induces osteoblast differentiation.
A. Relative alkaline phosphate enzymatic activity normalized to cell protein after 4 hours daily treatment with PEMF for 12 days or differentiation medium (DM) for 12 days. * indicates p<0.05, Student’s t-test compared to untreated control. B. Relative expression of osteoblast differentiation genes as indicated, normalized to GAPDH after 4 hours/day treatment with PEMF for 12 days or DM for 12 days. * indicates p<0.05, Student’s t-test compared to untreated control of same gene. C. Time series of Alizarin red staining for mineralization and alkaline phosphate activity staining after PEMF or DM treatment.
Adora3 disruption decreases alkaline phosphatase activation by PEMF in osteoblast differentiation
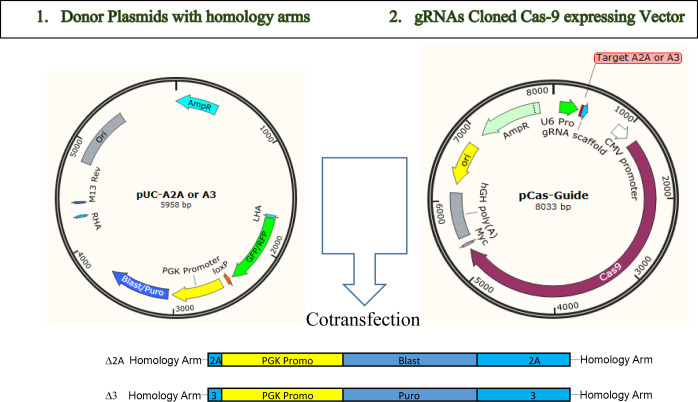
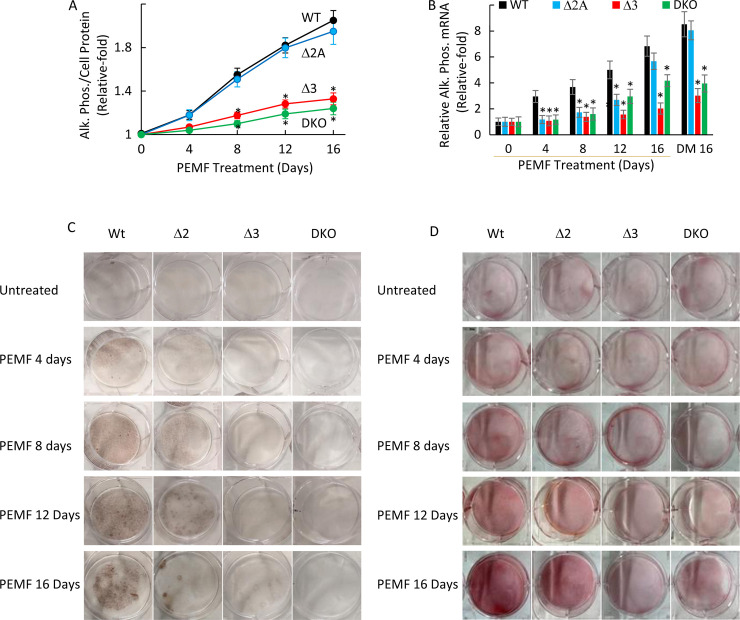
To investigate the role of Adenosine receptors A2A and A3, we began by disrupting the expression of these two genes individually or in combination in MC3T3-E1 cells employing a homology directed recombination (HDR)-based CRISPR-Cas9 methodology strategy (see Fig 2) and then exposing the wild-type (WT), single disrupted and doubly disrupted cellular clones to PEMF (4 hr/day) for the indicated time periods. The PCR screening results confirmed that disrupted clones were homologously disrupted for the targeted receptors. Disruption of Adora2A resulted in almost no significant difference in its enzymatic activity and only a delayed induction of alkaline phosphatase overexpression upon PEMF stimulation compared to the WT cells up to 16 days as measured by enzymatic assay, qRT-PCR and cell staining (Fig 3A–3C). On the other hand, disruption of Adora3 resulted in a reduced response compared to the WT cells as measured by both Alp in protein and mRNA levels throughout the stimulation periods. Incidentally, treatment with DM for 16 days showed a similar pattern of more prominent loss of alkaline phosphatase activation in Adora3 or doubly disrupted (A2A/A3) cells, but not in Adora2A disrupted in MC3T3-E1 (Fig 3B). For PEMF-mediated activation of Alp expression, a similar dependence was observed for Adora 3, but not Adora 2A, as measured by alkaline phosphatase staining (Fig 3C), and by Alizarine Red staining (Fig 3D).
Fig 2. Adora 2A and 3 disruption by CRISPR-Cas9.
MC3T3-E1 cells were co-transfected with a pUC donor plasmid with RFP-PGK-BSD or GFP-PGK-Puro flanked by homolog arms (1) and pCas-Guide plasmid containing Cas9 and gRNA corresponding to cleavage site in the first exon in each gene (2). Expected insertions of the cassette after homology-dependent recombination (HDR) are shown at the bottom.
Fig 3. Adora3 disruption affects PEMF activation of alkaline phosphatase.
A. Relative alkaline phosphate enzymatic activity normalized to cell protein after 4 hours daily treatment with PEMF for 4, 8, 12 and 16 days. * indicates p<0.05, Student’s t-test compared to WT control for each time-point. B. Relative expression of alkaline phosphatase gene normalized to GAPDH after 4 hours daily treatment with PEMF up to 16 days or DM for 16 days. * indicates p<0.05, Student’s t-test compared to WT for each time-point. C. Alkaline phosphatase staining of the MC3T3-E1 cells without and with PEMF treatment. D. Alizarin Red staining of the MC3T3-E1 cells without and with PEMF treatment.
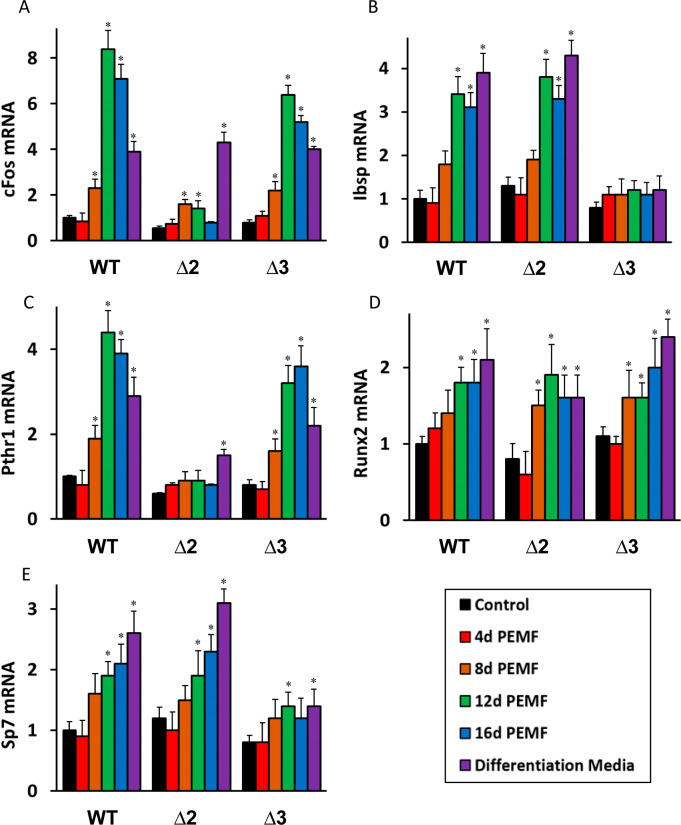
PEMF-induced cFos and Pthr1 expression is blunted in Adora2A disruption
Next we went on to investigate the roles of the A2A and A3 receptors in PEMF-mediated activation of other osteoblast differentiation genes, viz. cFos, Ibsp, Pthr1, Runx2 and Sp7. In the wild-type MC3T3-E1 cells, all these genes showed increased expression up to 12–16 days of PEMF stimulation (Fig 4A–4E). DM-mediated increases in Ibsp, Runx2 and Sp7 were also observed, however for cFos and Pthr1 the activation was very modest or non-existent. After Adora2A disruption, a general tendency of dramatically blunted response to PEMF stimulation was observed for both cFos and Pthr1 gene expression, whereas in the case of Adora3 disruption, the effect on expression of those genes was less remarkable. On the other hand, PEMF stimulation was more blunted in Ibsp and Sp7 due to Adora3 disruption, but not Adora2 disruption; while Runx2 activation seemed to be independent of either.
Fig 4.
Adora2 and Adora3 disruption affects PEMF activation of cFos, Ibsp, Pthr1, Runx2 and Sp7 (A-E). Relative fold-induction of each gene expression as measured by qRT-PCR with Gapdh as standard, with PEMF treatment up to 16 days (4 hours daily) or DM for 16 days. * indicates p<0.05, Student’s t-test compared to untreated control at day 8 of same gene of same cell-type.
PEMF-induced expression of differentiation genes and the effect of Adora disruption on their message levels are also seen at the protein level
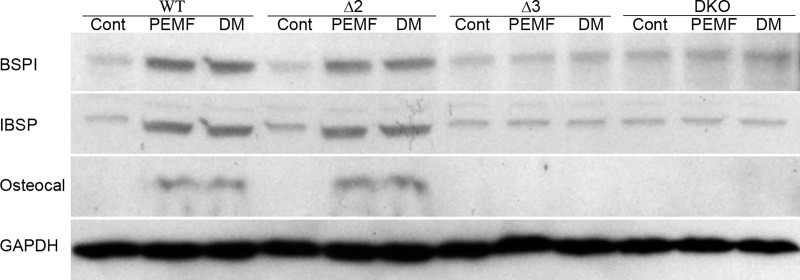
To correlate the molecular data at the messenger RNA level with protein expression of osteoblast markers, effects of PEMF on the mature osteoblast markers BSP, IBSP, and osteocalcin in wild-type and Adora-disrupted MC3T3-E1 cells was evaluated by Western Blot (Fig 5). Normalized to GAPDH expression serving as a loading control, both PEMF and DM exposure induced increased expression of all the above proteins compared to control cells in the wild-type and Adora2 disrupted cells, but not in Adora3 or doubly disrupted cells.
Fig 5. Adora3 disruption affects PEMF mediated overexpression of BSP, IBSP and Osteocalcin.
Wild-type (WT), Adora2 (Δ2), Adora3 (Δ3) and Double Knock-Out (DKO) MC3T3-E1 cells were treated with PEMF (4 hours daily) or differentiation medium (DM) for 12 days and Western Blots for fractionated whole-cell proteins were probed with each of the indicated antibodies.
Adora3 reconstitution in Adora3 disrupted cells restores alkaline phosphatase activation by PEMF
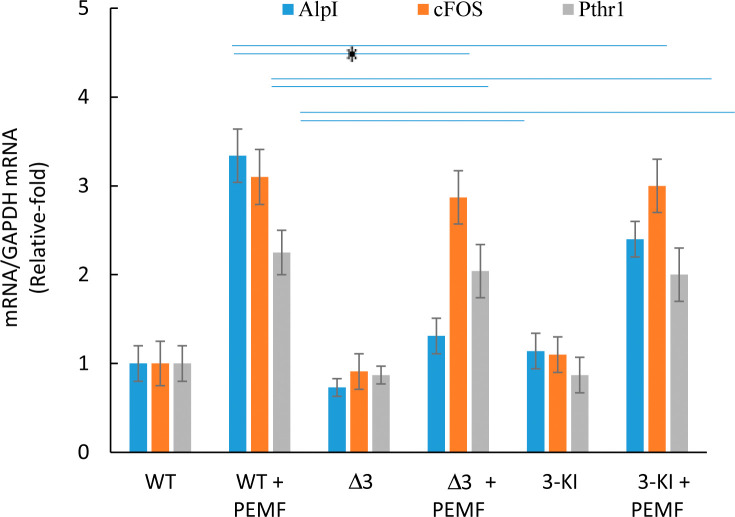
To address the specificity of gene disruption, and to eliminate the possibility of off-target effects masquerading as an A3-specific result, we took the Adora3 disrupted MC3T3-E1 cells and “knocked-in” the Adora3 ORF into the ROSA26 locus under the control of a cumate-inducible promoter. When Adora3 function was thus reconstituted in Adora3 disrupted cells, PEMF response as determined through Alp gene activation was found to be restored close to the typical response level observed in the wild-type cells (Fig 6). Expression of cFos and Pthr1 mRNA remained unaltered either as a result of Adora3 disruption or Adora3 reconstitution.
Fig 6. Adora3 knock-in in Adora3 disrupted cells restores PEMF-mediated induction of alkaline phosphatase in PEMF-treated cells.
Expressions of alkaline phosphate, cFos and Pthr1 genes were measured by qRT-PCR with Gapdh as standard with PEMF treatment for 8 days. * indicates p<0.05, Student’s t-test compared to PEMF treatment in WT cells.
Discussion
Though PEMF has been known to stimulate non-union fracture healing for more than four decades [3,4,7], its effect on the initial step of wound healing, inflammation, and oxidative stress has been investigated only recently in terms modulation of plasma membrane Ca2+, ATP/ATPase or heat shock responses [18–21]. Multiple studies examining the pre-osteoblast to osteoblast transition and differentiation have focused on a core set of genes that include alkaline phosphatase (Alp), cellular proto-oncogene derived from the Finkel–Biskis–Jinkins murine osteogenic sarcoma virus (cFos), integrin-binding sialoprotein (Ibsp), parathyroid hormone 1 receptor (Pthr1), Runt-related transcription factor 2 (Runx2) and transcription factor Sp7, also called Osterix (Sp7/Osterix) [33–36]. PEMF has been shown to activate ALP, insulin-like growth factor 1, collagen type 1, osteocalcin and Runx2 expression in a Ca2+-dependent fashion [15,37]; however, the upstream signaling pathways leading to these activations are poorly understood. Prior studies mentioned above and others have shown that the only adenosine receptors that are clearly modulated by PEMF were the A2A and A3 ADRs [22,29,38–42]. The present study explores a more detailed profiling of PEMF-responsive genes, and focuses on the role of a subset of adenosine receptors in PEMF-mediated osteoblast differentiation.
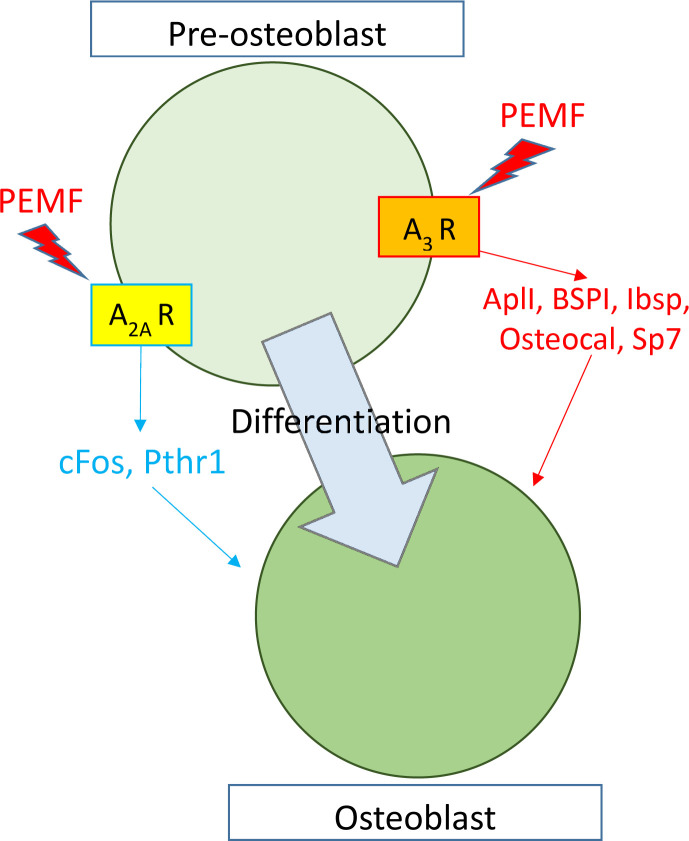
Previous studies showed a connection between PEMF-mediated effects on proliferation and inflammation in multiple cell types through adenosine receptors, where specifically A2A and A3 expression were amplified and proteins migrated to the plasma membrane [22,29,42]. Furthermore, in separate contexts, these receptors, A2A and A3 were shown to be involved in osteoblast differentiation under non-PEMF physiological conditions [24,43]. These two separate but complementary observations in turn led us to test the hypothesis whether these receptors are modulators of osteoblast differentiation under PEMF stimulation conditions as well. Using CRISPR-Cas9-based gene editing techniques to disrupt these two genes, individually or in combination, we were able to show functional implications of Adora2A and Adora3 in PEMF-mediated osteoblast differentiation. We kept our study focused mainly on mRNA expression of differentiation genes because it has been reported in several previous studies that their protein expressions correlated mRNA expression [44–46], which also matched our observation in the case of AlpI enzymatic assay (Fig 3). Our results indicate that the A2A and A3 receptors are required for at least two parallel and complementary signaling pathways during the differentiation process: A2A seems to be important in PEMF-mediated stimulation of cFos and Pthr1 pathways while A3 appears to modulate Alp, BSPI, Ibsp, Osteocalcin and Sp7 pathways (summarized in Fig 7). The messenger RNA data determined by qRT-PCR was confirmed at the protein level for the key osteoblast differentiation protein markers. Curiously, a similar pattern of gene modulation was observed under differentiation using a chemically defined medium (DM), which indicates a commonality of molecular pathways being activated by these two disparate stimuli. One plausible scenario is that PEMF, like DM, activates release of certain ligands that modulate the activity of these receptors. This possibility is currently under investigation.
Fig 7. Schematic diagram role of Adenosine Receptors in PEMF-mediated MC3T3-E1 differentiation.
PEMF stimulated osteoblastic differentiation depends on Adenosine Receptor A2A for cFos and Pthr1 overexpression, and A3 for AlpI activation.
To exclude the possibility that the loss of function observed in Adora3 gene disrupted cells was due to off-target effects, “knock-in” experiments were performed to reintroduce the Adora3 ORF into the open chromatin ROSA26 locus under control of the inducible cumate promoter in Adora3-disrupted MC3T3-E1 cells. These reconstituted A3 cells showed a PEMF response pattern similar to that observed for the wild-type pre-osteoblast MC3T3-E1 cells, which confirmed the specificity of the A3 receptor’s role in Alp pathway activation by both PEMF and DM.
This study utilized a targeted approach focusing on a subset of adenosine receptors that were shown to be modulated by PEMF during osteoblast differentiation, and to behave as facilitators of PEMF action in the initial phases of osteoblast differentiation. This modulation was observed to operate through at least two parallel paths. We do not envision these receptors as direct PEMF-sensing receptors; however they do appear to respond to upstream PEMF-initiated signals. In order to identify PEMF receptors, key PEMF-signaling node genes and downstream PEMF-responsive genes, an untargeted approach can be envisioned where genome-wide gene silencing and screening can be carried out using a knock-out library and reporter system followed by informatics analysis to identify the most upstream PEMF sensing genes. This strategy is currently being explored.
Supporting information
(PDF)
(PDF)
Abbreviations
- DM
Differentiation Medium
- PEMF
Pulsed-electromagnetic-field
- qRT-PCR
Quantitative Reverse Transcription-Polymerase Chain Reaction
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
JAD was funded by a sponsored research grant from Orthofix (https://www.orthofix.com/). NSK also received salary support from this grant. Orthofix also provided support for this study in the form of salaries for NZ, EIW, and JTR. The specific roles of these authors are articulated in the ‘author contributions’ section. No additional external funding was received for this study.
References
- 1.Loi F, Cordova LA, Pajarinen J, Lin TH, Yao Z, Goodman SB. Inflammation, fracture and bone repair. Bone. 2016;86:119–30. Epub 2016/03/08. 10.1016/j.bone.2016.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahney CS, Zondervan RL, Allison P, Theologis A, Ashley JW, Ahn J, et al. Cellular biology of fracture healing. J Orthop Res. 2019;37(1):35–50. Epub 2018/10/30. 10.1002/jor.24170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sedel L, Christel P, Duriez J, Duriez R, Evrard J, Ficat C, et al. Results of non unions treatment by pulsed electromagnetic field stimulation. Acta Orthop Scand Suppl. 1982;196:81–91. Epub 1982/01/01. 10.3109/17453678209158551 . [DOI] [PubMed] [Google Scholar]
- 4.Muhsin AU, Islam KM, Ahmed AM, Islam MS, Rabbani KS, Rahman SM, et al. Effect of pulsed electromagnetic field on healing of experimental nonunion in rat tibiae. Bangladesh Med Res Counc Bull. 1991;17(1):1–10. Epub 1991/06/01. . [PubMed] [Google Scholar]
- 5.Nelson FR, Brighton CT, Ryaby J, Simon BJ, Nielson JH, Lorich DG, et al. Use of physical forces in bone healing. J Am Acad Orthop Surg. 2003;11(5):344–54. Epub 2003/10/21. 10.5435/00124635-200309000-00007 . [DOI] [PubMed] [Google Scholar]
- 6.Assiotis A, Sachinis NP, Chalidis BE. Pulsed electromagnetic fields for the treatment of tibial delayed unions and nonunions. A prospective clinical study and review of the literature. J Orthop Surg Res. 2012;7:24. Epub 2012/06/12. 10.1186/1749-799X-7-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray HB, Pethica BA. A follow-up study of the in-practice results of pulsed electromagnetic field therapy in the management of nonunion fractures. Orthop Res Rev. 2016;8:67–72. Epub 2016/12/01. 10.2147/ORR.S113756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohajerani H, Tabeie F, Vossoughi F, Jafari E, Assadi M. Effect of pulsed electromagnetic field on mandibular fracture healing: A randomized control trial, (RCT). J Stomatol Oral Maxillofac Surg. 2019. Epub 2019/03/06. 10.1016/j.jormas.2019.02.022 . [DOI] [PubMed] [Google Scholar]
- 9.Stewart SK. Fracture Non-Union: A Review of Clinical Challenges and Future Research Needs. Malays Orthop J. 2019;13(2):1–10. Epub 2019/08/31. 10.5704/MOJ.1907.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng D, Xu T, Guo F, Yin W, Peng T. The effects of high-intensity pulsed electromagnetic field on proliferation and differentiation of neural stem cells of neonatal rats in vitro. J Huazhong Univ Sci Technolog Med Sci. 2009;29(6):732–6. Epub 2009/12/29. 10.1007/s11596-009-0612-4 . [DOI] [PubMed] [Google Scholar]
- 11.Sun LY, Hsieh DK, Yu TC, Chiu HT, Lu SF, Luo GH, et al. Effect of pulsed electromagnetic field on the proliferation and differentiation potential of human bone marrow mesenchymal stem cells. Bioelectromagnetics. 2009;30(4):251–60. Epub 2009/02/11. 10.1002/bem.20472 . [DOI] [PubMed] [Google Scholar]
- 12.Lee HM, Kwon UH, Kim H, Kim HJ, Kim B, Park JO, et al. Pulsed electromagnetic field stimulates cellular proliferation in human intervertebral disc cells. Yonsei Med J. 2010;51(6):954–9. Epub 2010/09/30. 10.3349/ymj.2010.51.6.954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Girolamo L, Stanco D, Galliera E, Vigano M, Colombini A, Setti S, et al. Low frequency pulsed electromagnetic field affects proliferation, tissue-specific gene expression, and cytokines release of human tendon cells. Cell Biochem Biophys. 2013;66(3):697–708. Epub 2013/01/25. 10.1007/s12013-013-9514-y . [DOI] [PubMed] [Google Scholar]
- 14.Xu H, Zhang J, Lei Y, Han Z, Rong D, Yu Q, et al. Low frequency pulsed electromagnetic field promotes C2C12 myoblasts proliferation via activation of MAPK/ERK pathway. Biochem Biophys Res Commun. 2016;479(1):97–102. Epub 2016/09/16. 10.1016/j.bbrc.2016.09.044 . [DOI] [PubMed] [Google Scholar]
- 15.Tong J, Sun L, Zhu B, Fan Y, Ma X, Yu L, et al. Pulsed electromagnetic fields promote the proliferation and differentiation of osteoblasts by reinforcing intracellular calcium transients. Bioelectromagnetics. 2017;38(7):541–9. Epub 2017/08/24. 10.1002/bem.22076 . [DOI] [PubMed] [Google Scholar]
- 16.Miyamoto H, Sawaji Y, Iwaki T, Masaoka T, Fukada E, Date M, et al. Intermittent pulsed electromagnetic field stimulation activates the mTOR pathway and stimulates the proliferation of osteoblast-like cells. Bioelectromagnetics. 2019;40(6):412–21. Epub 2019/07/25. 10.1002/bem.22207 . [DOI] [PubMed] [Google Scholar]
- 17.Fang QQ, Li ZZ, Zhou J, Shi WG, Yan JL, Xie YF, et al. [Low-frequency pulsed electromagnetic fields promotes rat osteoblast differentiation in vitro through cAMP/PKA signal pathway]. Nan Fang Yi Ke Da Xue Xue Bao. 2016;36(11):1508–13. Epub 2016/11/25. . [PubMed] [Google Scholar]
- 18.Vincenzi F, Ravani A, Pasquini S, Merighi S, Gessi S, Setti S, et al. Pulsed Electromagnetic Field Exposure Reduces Hypoxia and Inflammation Damage in Neuron-Like and Microglial Cells. J Cell Physiol. 2017;232(5):1200–8. Epub 2016/09/18. 10.1002/jcp.25606 . [DOI] [PubMed] [Google Scholar]
- 19.Merighi S, Gessi S, Bencivenni S, Battistello E, Vincenzi F, Setti S, et al. Signaling pathways involved in anti-inflammatory effects of Pulsed Electromagnetic Field in microglial cells. Cytokine. 2019;125:154777. Epub 2019/08/11. 10.1016/j.cyto.2019.154777 . [DOI] [PubMed] [Google Scholar]
- 20.Selvam R, Ganesan K, Narayana Raju KV, Gangadharan AC, Manohar BM, Puvanakrishnan R. Low frequency and low intensity pulsed electromagnetic field exerts its antiinflammatory effect through restoration of plasma membrane calcium ATPase activity. Life Sci. 2007;80(26):2403–10. Epub 2007/06/01. 10.1016/j.lfs.2007.03.019 . [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Liu Y, Wang Y, Wei Z, Suo D, Ning G, et al. Lowfrequency pulsed electromagnetic field promotes functional recovery, reduces inflammation and oxidative stress, and enhances HSP70 expression following spinal cord injury. Mol Med Rep. 2019;19(3):1687–93. Epub 2019/01/11. 10.3892/mmr.2019.9820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vincenzi F, Targa M, Corciulo C, Gessi S, Merighi S, Setti S, et al. Pulsed electromagnetic fields increased the anti-inflammatory effect of A(2)A and A(3) adenosine receptors in human T/C-28a2 chondrocytes and hFOB 1.19 osteoblasts. PLoS One. 2013;8(5):e65561. Epub 2013/06/07. 10.1371/journal.pone.0065561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He W, Mazumder A, Wilder T, Cronstein BN. Adenosine regulates bone metabolism via A1, A2A, and A2B receptors in bone marrow cells from normal humans and patients with multiple myeloma. FASEB J. 2013;27(9):3446–54. Epub 2013/05/18. 10.1096/fj.13-231233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mediero A, Wilder T, Shah L, Cronstein BN. Adenosine A2A receptor (A2AR) stimulation modulates expression of semaphorins 4D and 3A, regulators of bone homeostasis. FASEB J. 2018;32(7):3487–501. Epub 2018/02/03. 10.1096/fj.201700217R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borhani S, Corciulo C, Larranaga-Vera A, Cronstein BN. Adenosine A2A receptor (A2AR) activation triggers Akt signaling and enhances nuclear localization of beta-catenin in osteoblasts. FASEB J. 2019;33(6):7555–62. Epub 2019/03/15. 10.1096/fj.201900014R . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salvatore CA, Tilley SL, Latour AM, Fletcher DS, Koller BH, Jacobson MA. Disruption of the A(3) adenosine receptor gene in mice and its effect on stimulated inflammatory cells. J Biol Chem. 2000;275(6):4429–34. Epub 2000/02/08. 10.1074/jbc.275.6.4429 . [DOI] [PubMed] [Google Scholar]
- 27.Evans BA, Elford C, Pexa A, Francis K, Hughes AC, Deussen A, et al. Human osteoblast precursors produce extracellular adenosine, which modulates their secretion of IL-6 and osteoprotegerin. J Bone Miner Res. 2006;21(2):228–36. 10.1359/JBMR.051021 . [DOI] [PubMed] [Google Scholar]
- 28.Costa MA, Barbosa A, Neto E, Sa-e-Sousa A, Freitas R, Neves JM, et al. On the role of subtype selective adenosine receptor agonists during proliferation and osteogenic differentiation of human primary bone marrow stromal cells. J Cell Physiol. 2011;226(5):1353–66. 10.1002/jcp.22458 . [DOI] [PubMed] [Google Scholar]
- 29.Varani K, Vincenzi F, Targa M, Corciulo C, Fini M, Setti S, et al. Effect of pulsed electromagnetic field exposure on adenosine receptors in rat brain. Bioelectromagnetics. 2012;33(4):279–87. Epub 2011/10/21. 10.1002/bem.20704 . [DOI] [PubMed] [Google Scholar]
- 30.Waldorff EI, Zhang N, Ryaby JT. Pulsed electromagnetic field applications: A corporate perspective. J Orthop Translat. 2017;9:60–8. Epub 2018/04/18. 10.1016/j.jot.2017.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balint E, Szabo P, Marshall CF, Sprague SM. Glucose-induced inhibition of in vitro bone mineralization. Bone. 2001;28(1):21–8. Epub 2001/02/13. 10.1016/s8756-3282(00)00426-9 . [DOI] [PubMed] [Google Scholar]
- 32.Sabokbar A, Millett PJ, Myer B, Rushton N. A rapid, quantitative assay for measuring alkaline phosphatase activity in osteoblastic cells in vitro. Bone Miner. 1994;27(1):57–67. Epub 1994/10/01. 10.1016/s0169-6009(08)80187-0 . [DOI] [PubMed] [Google Scholar]
- 33.Ferroni L, Gardin C, Dolkart O, Salai M, Barak S, Piattelli A, et al. Pulsed electromagnetic fields increase osteogenetic commitment of MSCs via the mTOR pathway in TNF-alpha mediated inflammatory conditions: an in-vitro study. Sci Rep. 2018;8(1):5108. Epub 2018/03/25. 10.1038/s41598-018-23499-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvamurugan N, He Z, Rifkin D, Dabovic B, Partridge NC. Pulsed Electromagnetic Field Regulates MicroRNA 21 Expression to Activate TGF-beta Signaling in Human Bone Marrow Stromal Cells to Enhance Osteoblast Differentiation. Stem Cells Int. 2017;2017:2450327. Epub 2017/05/18. 10.1155/2017/2450327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selvamurugan N, Kwok S, Vasilov A, Jefcoat SC, Partridge NC. Effects of BMP-2 and pulsed electromagnetic field (PEMF) on rat primary osteoblastic cell proliferation and gene expression. J Orthop Res. 2007;25(9):1213–20. Epub 2007/05/16. 10.1002/jor.20409 . [DOI] [PubMed] [Google Scholar]
- 36.Bagheri L, Pellati A, Rizzo P, Aquila G, Massari L, De Mattei M, et al. Notch pathway is active during osteogenic differentiation of human bone marrow mesenchymal stem cells induced by pulsed electromagnetic fields. J Tissue Eng Regen Med. 2018;12(2):304–15. 10.1002/term.2455 . [DOI] [PubMed] [Google Scholar]
- 37.Zhai M, Jing D, Tong S, Wu Y, Wang P, Zeng Z, et al. Pulsed electromagnetic fields promote in vitro osteoblastogenesis through a Wnt/beta-catenin signaling-associated mechanism. Bioelectromagnetics. 2016;37(3):152–62. Epub 2016/02/19. 10.1002/bem.21961 . [DOI] [PubMed] [Google Scholar]
- 38.Ongaro A, Varani K, Masieri FF, Pellati A, Massari L, Cadossi R, et al. Electromagnetic fields (EMFs) and adenosine receptors modulate prostaglandin E(2) and cytokine release in human osteoarthritic synovial fibroblasts. J Cell Physiol. 2012;227(6):2461–9. 10.1002/jcp.22981 . [DOI] [PubMed] [Google Scholar]
- 39.Varani K, De Mattei M, Vincenzi F, Gessi S, Merighi S, Pellati A, et al. Characterization of adenosine receptors in bovine chondrocytes and fibroblast-like synoviocytes exposed to low frequency low energy pulsed electromagnetic fields. Osteoarthritis Cartilage. 2008;16(3):292–304. Epub 2007/08/19. 10.1016/j.joca.2007.07.004 . [DOI] [PubMed] [Google Scholar]
- 40.Varani K, Gessi S, Merighi S, Iannotta V, Cattabriga E, Pancaldi C, et al. Alteration of A(3) adenosine receptors in human neutrophils and low frequency electromagnetic fields. Biochem Pharmacol. 2003;66(10):1897–906. 10.1016/s0006-2952(03)00454-4 . [DOI] [PubMed] [Google Scholar]
- 41.Varani K, Gessi S, Merighi S, Iannotta V, Cattabriga E, Spisani S, et al. Effect of low frequency electromagnetic fields on A2A adenosine receptors in human neutrophils. Br J Pharmacol. 2002;136(1):57–66. 10.1038/sj.bjp.0704695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vincenzi F, Targa M, Corciulo C, Gessi S, Merighi S, Setti S, et al. The anti-tumor effect of A3 adenosine receptors is potentiated by pulsed electromagnetic fields in cultured neural cancer cells. PLoS One. 2012;7(6):e39317. Epub 2012/07/05. 10.1371/journal.pone.0039317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mediero A, Cronstein BN. Adenosine and bone metabolism. Trends Endocrinol Metab. 2013;24(6):290–300. Epub 2013/03/19. 10.1016/j.tem.2013.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian DY, Yan GB, Bai B, Chen Y, Zhang SJ, Yao YC, et al. Differential circRNA expression profiles during the BMP2-induced osteogenic differentiation of MC3T3-E1 cells. Biomed Pharmacother. 2017;90:492–9. Epub 2017/04/11. 10.1016/j.biopha.2017.03.051 . [DOI] [PubMed] [Google Scholar]
- 45.Hou Q, Huang Y, Liu Y, Luo Y, Wang B, Deng R, et al. Profiling the miRNA-mRNA-lncRNA interaction network in MSC osteoblast differentiation induced by (+)-cholesten-3-one. BMC Genomics. 2018;19(1):783. Epub 2018/10/31. 10.1186/s12864-018-5155-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin N, Zhu L, Ding L, Yuan J, Du L, Pan M, et al. MiR-135-5p promotes osteoblast differentiation by targeting HIF1AN in MC3T3-E1 cells. Cell Mol Biol Lett. 2019;24:51. Epub 2019/08/15. 10.1186/s11658-019-0177-6 [DOI] [PMC free article] [PubMed] [Google Scholar]