Abstract
Genome-wide association studies (GWASs) have identified multiple susceptibility loci for Alzheimer’s disease (AD), which is characterized by early and progressive damage to the hippocampus. However, the association of hippocampal gene expression with AD and the underlying neurobiological pathways remain largely unknown. Based on the genomic and transcriptomic data of 111 hippocampal samples and the summary data of two large-scale meta-analyses of GWASs, a transcriptome-wide association study (TWAS) was performed to identify genes with significant associations between hippocampal expression and AD. We identified 54 significantly associated genes using an AD-GWAS meta-analysis of 455,258 individuals; 36 of the genes were confirmed in another AD-GWAS meta-analysis of 63,926 individuals. Fine-mapping models further prioritized 24 AD-related genes whose effects on AD were mediated by hippocampal expression, including APOE and two novel genes (PTPN9 and PCDHA4). These genes are functionally related to amyloid-beta formation, phosphorylation/dephosphorylation, neuronal apoptosis, neurogenesis and telomerase-related processes. By integrating the predicted hippocampal expression and neuroimaging data, we found that the hippocampal expression of QPCTL and ERCC2 showed significant difference between AD patients and cognitively normal elderly individuals as well as correlated with hippocampal volume. Mediation analysis further demonstrated that hippocampal volume mediated the effect of hippocampal gene expression (QPCTL and ERCC2) on AD. This study identifies two novel genes associated with AD by integrating hippocampal gene expression and genome-wide association data and reveals candidate hippocampus-mediated neurobiological pathways from gene expression to AD.
Author summary
The hippocampus is a potential neuroimaging endophenotype for Alzheimer’s disease (AD). This study identifies genes whose expression in hippocampal tissue is associated with AD and establishes the pathways from hippocampal gene expression to hippocampal volume to AD. We demonstrate that 24 genes are associated with AD in hippocampal tissue, and these genes are enriched for AD-related biological processes of amyloid-beta formation, phosphorylation/dephosphorylation, neuronal apoptosis, neurogenesis and telomerase-related processes. We further show that hippocampal volume mediates the effects of the hippocampal gene expression of QPCTL and ERCC2 on AD. These findings improve our understanding of the genetic and neural mechanisms of AD.
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disorder clinically characterized by progressive dementia and pathologically featured by senile plaques composed of amyloid beta peptide (Aβ) and intracellular neurofibrillary tangles (NFTs), which themselves are composed of hyperphosphorylated tau [1,2]. AD is a highly heritable disease with an estimated heritability of 58%-79% [3], emphasizing the importance of exploring the genetic mechanisms of AD. Despite rapid progress in utilizing genome-wide association studies (GWASs) and meta-analyses to identify AD-related genetic variants [4–11], the pathogenic mechanisms of the identified genetic loci in AD remain largely unknown.
Expression quantitative trait loci (eQTLs) are considered links between GWAS loci and disease susceptibility [12,13]. By integrating the large-scale gene expression data of a given tissue and disease-related GWAS data, transcriptome-wide association study (TWAS) has been proposed as a powerful approach to identify genes with significant associations between gene expression in certain tissues and the disease of interest [14–16]. By incorporating transcriptomic data of available human tissues and GWAS data of AD, several TWAS studies have confirmed multiple AD-related genes identified by GWASs and found novel genes that have not been previously reported [17–21]. Although the inclusion of all available tissues in these TWAS studies could improve the power, they provide little tissue-specific information, which is important for exploring pathogenic mechanisms of AD because tissues show different eQTLs [22] and TWAS is more reliable for trait-related tissues than for trait-unrelated tissues [23].
In neuroimaging studies, hippocampal atrophy is the most prominent imaging feature of AD [24–26]. Most of the neuropathological hallmarks (neurofibrillary tangles, neuronal loss, synaptic loss, amyloid plaques, and glial responses) of AD can be observed in the hippocampus, and neurofibrillary tangles, neuronal and synaptic loss are present in the hippocampus at an early stage of AD, which are closely associated with the progression of AD [27]. Moreover, eQTLs of hippocampal tissue are significantly enriched for AD-GWAS-identified associations [28]. These findings indicate that hippocampal tissue is an ideal candidate for AD-TWAS and that hippocampal volume is a potential neuroimaging marker to investigate the mediation effect of the hippocampus on the association between hippocampal gene expression and AD.
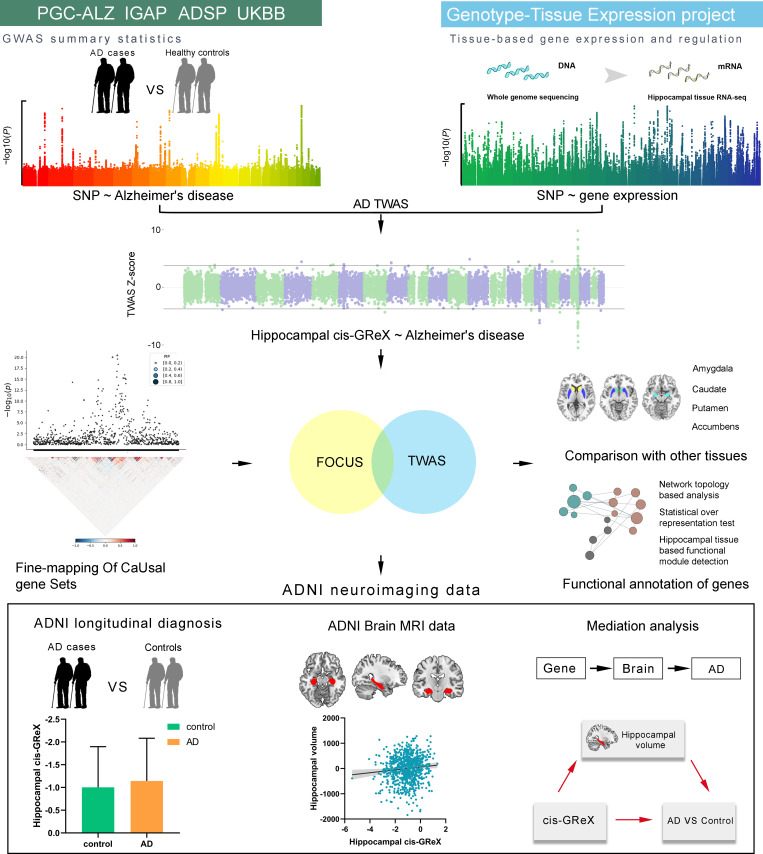
In this study, we first determined the relationship between each single nucleotide polymorphism (SNP) and hippocampal gene expression using whole genome sequencing (WGS) data and hippocampal tissue RNA-seq data provided by GTEx [22]. Second, based on the obtained SNP-expression associations and SNP-AD associations identified by AD-GWAS, TWAS was performed to identify hippocampus- and AD-related genes, which were defined as genes whose cis-genetically regulated expression (cis-GReX) in hippocampal tissue was associated with AD. Third, fine-mapping analysis was used to prioritize these genes, and associations of the expression of these genes in four other subcortical tissues (amygdala, caudate, nucleus accumbens and putamen) with AD were also studied. The identified genes were functionally annotated by network topology-based analysis, statistical over-representation test and hippocampal-based functional module detection. Finally, we further validated the identified genes in Alzheimer’s Disease Neuroimaging Initiative (ADNI) neuroimaging data and established the pathway from hippocampal gene expression to hippocampal volume to AD diagnosis. A schematic overview of the study design is presented in Fig 1.
Fig 1. A schematic overview of the study design.
AD = Alzheimer’s disease; ADNI = Alzheimer’s disease Neuroimaging Initiative; ADSP = Alzheimer’s disease Sequencing Project; cis-GReX = cis-genetically regulated expression; FOCUS = Fine-mapping of causal gene sets; GWAS = Genome-wide association study; IGAP = International Genomics of Alzheimer’s Project; PGC-ALZ = Alzheimer’s disease working group of the Psychiatric Genomics Consortium; TWAS = Transcriptome-wide association study; UKBB = UK Biobank.
Results
Prediction models for hippocampal gene expression
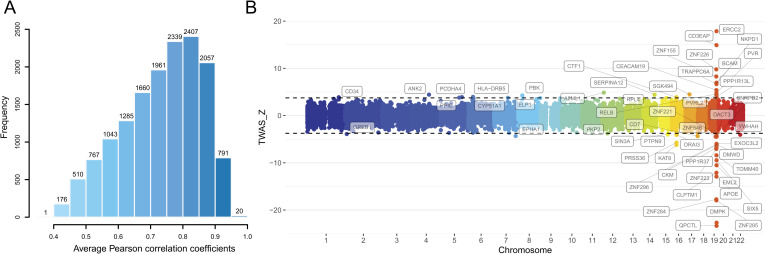
We established correspondences between SNPs and the expression of each gene in hippocampal tissue and calculated the weighted value of each SNP in predicting the expression of the gene using the WGS and RNA-seq data of 111 hippocampal samples from GTEx. For each gene, QTLtools (https://qtltools.github.io/qtltools/) [29] was used to perform conditional analysis to identify cis-eQTLs with independent effects on gene expression. The GTEx V7 pipeline (https://github.com/hakyimlab/PredictDB_Pipeline_GTEx_v7) was applied to train prediction models for hippocampal expression of 15,831 protein-coding genes with the nested cross validated elastic-net [14]. A prediction model was significant if the average Pearson correlation coefficient between predicted and measured gene expression during nested cross validation was greater than 0.1 and the estimated p-value for this statistic passed the multiple testing correction threshold of familywise error (FWE) (pc < 0.05/15,831 = 3.16 × 10−6). Among the 15,831 protein-coding genes, we built significant hippocampal gene expression prediction models for 15,017 genes (pc < 0.05, FWE corrected), indicating a rather high success rate (94.9%). The average Pearson correlation coefficients between predicted and observed expression levels during nested cross validation in hippocampal tissue for these genes (mean ± SD = 0.73 ± 0.12, range 0.40–0.96) are shown in Fig 2A, which were relatively high, suggesting good performance of these prediction models. For each gene, the prediction model generated the weighted value for each SNP’s relative contribution to the gene’s expression level in hippocampal tissue.
Fig 2.
Gene expression prediction models of hippocampal tissue (A) and TWAS results of AD (B). (A) Average Pearson correlation coefficients of 15,017 significant hippocampal gene expression prediction models (pc < 0.05, FWE corrected). (B) Manhattan plot of all TWAS associations of AD in the discovery stage. Each point represents a single gene, with physical position in chromosome plotted on the x-axis and z-score of the association statistics between gene and AD plotted on the y-axis. Transcriptome-wide significant threshold (qc < 0.05, FDR corrected) is highlighted as black dotted lines and the significant associations are labeled with gene names. AD, Alzheimer’s disease; FDR, false discovery rate; FWE, familywise error; TWAS, transcriptome-wide association study.
Identification and validation of AD-related genes using TWAS
TWAS can integrate the gene expression of certain tissues and GWAS data to test correlations between cis-GReX and disease/complex traits [14–16]. In our study, summary-PrediXcan (S-PrediXcan) [15] was used to perform TWAS to identify significant associations between AD and gene expression in hippocampal tissue. Two sets of GWAS summary statistics of AD were used: the meta-analysis (n = 455,258 including 71,880 AD or AD-by-proxy and 383,378 controls) [4] collected from the Alzheimer’s disease working group of the Psychiatric Genomics Consortium (PGC-ALZ), the International Genomics of Alzheimer’s Project (IGAP), the Alzheimer’s Disease Sequencing Project (ADSP) and the UK Biobank (UKBB) was used as the discovery sample. The updated GWAS meta-analysis of IGAP (n = 63,926 including 21,982 AD and 41,944 controls) [11] was used as the replication sample. Based on SNP-AD associations obtained from the GWAS summary statistics of 455,258 individuals (the discovery sample) and SNP-expression associations obtained from the hippocampal gene expression prediction models, we found 54 genes whose cis-GReX values in hippocampal tissue were significantly associated with AD at the 5% false discovery rate (FDR) threshold (qc < 0.05) (Fig 2B and S1 Table). Based on the GWAS summary statistics of 63,926 individuals from IGAP (the replication sample) and the hippocampal gene expression prediction models, we successfully replicated 36 of 54 genes at a nominal threshold of p < 0.05 with consistent direction of z-scores between discovery and validation stage, among which 23 genes passed the FDR correction for multiple testing (qc < 0.05) in the replication samples (S1 Fig and S1 Table).
Fine-mapping prioritizes AD-related genes
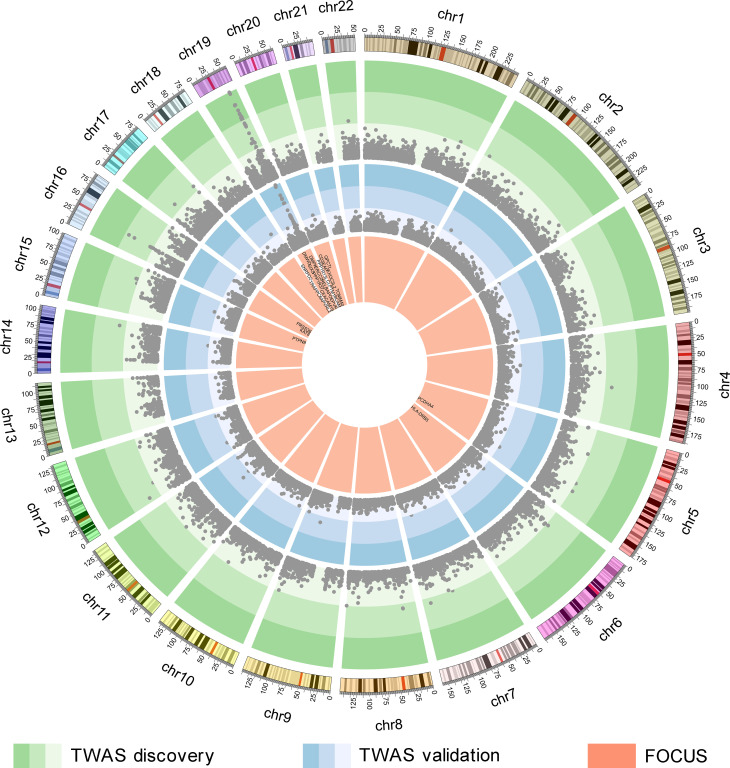
The reliability of TWAS-identified AD-related genes was challenged by linkage disequilibrium (LD) among the SNPs and coregulation of gene models [23]. Here, FOCUS (fine-mapping of causal gene sets) [30] was further used to prioritize the 36 genes for AD by assigning a probability for each gene based on prediction modules, recommended LD reference data, and AD-GWAS summary statistics (n = 455,258 including 71,880 AD or AD-by-proxy and 383,378 controls). FOCUS inferred whether each of the 36 TWAS-identified genes was included in credible set at the nominal confidence level (90%). Among the 36 TWAS-identified AD-related genes, FOCUS prioritized 24, which were in credible sets (Fig 3 and S1 Table).
Fig 3. Genes identified by TWAS and FOCUS.
The green circle shows the genes identified by the AD-TWAS in hippocampal tissue, each point represents a single gene, with physical position in human genome plotted on the x-axis and -log10(p) of association between cis-GReX in hippocampal tissue and AD plotted on the y-axis. The color gradients represent significant levels and points located in the green and darker green regions indicate significant associations with AD at the 5% FDR threshold. The blue circle shows the AD-TWAS results of validation stage. The orange circle shows the results of FOCUS, the 24 genes showed in the figure are included in 90% credible gene sets. AD, Alzheimer’s disease; cis-GReX, cis-genetically regulated expression; FDR, false discovery rate; FOCUS, fine-mapping of causal gene sets; TWAS, transcriptome-wide association study.
Specificity for hippocampal tissue
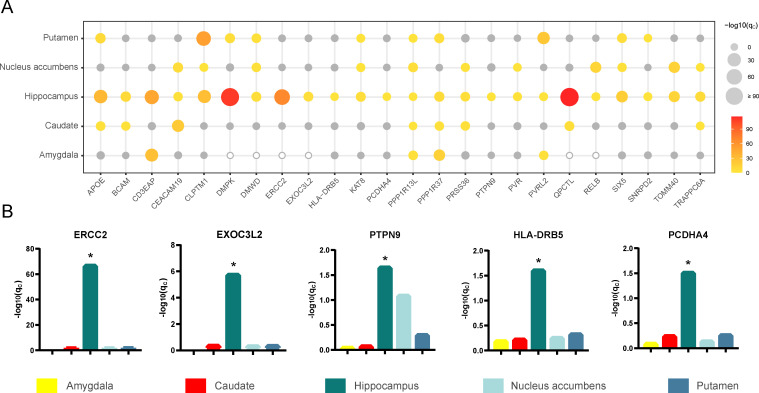
To determine whether the 24 identified AD-related genes specifically affect the hippocampus, we also investigated the associations of the expression of these genes in four other subcortical tissues with AD. These subcortical tissues included the amygdala, caudate, nucleus accumbens and putamen, and the volume loss of the latter two nuclei appears earlier than that of the hippocampus in AD patients [31]. The same pipeline used for the hippocampus was applied to train prediction models and to perform TWAS for the other four tissues. The average Pearson correlation coefficients between predicted and observed expression levels for the amygdala, caudate, nucleus accumbens and putamen are shown in S2 Fig. We used the prediction models of the four tissues and the AD-GWAS summary statistics (n = 455,258 including 71,880 AD or AD-by-proxy and 383,378 controls) to perform TWAS. Manhattan plots of the TWAS results are shown in S3–S6 Figs. We compared the TWAS results of the 24 prioritized AD-related genes between hippocampal tissue and other tissues (Fig 4A). The gene expression prediction models of six genes were not established successfully in the amygdala, which meant that the SNPs could not predict the gene expression in this tissue. The gene expression of ERCC2, EXOC3L2, PTPN9, HLA-DRB5 and PCDHA4 was associated with AD only in hippocampal tissue at the 5% FDR threshold (Fig 4B). More genes showed shared genetic contributions to AD in at least two tissues. For example, AD was affected by the gene expression of CD3EAP in the hippocampus and amygdala; TOMM40, PVR and RELB in the hippocampus and nucleus accumbens; DMPK and SNRPD2 in the hippocampus and putamen; and QPCTL and BCAM in the hippocampus and caudate. In addition, some genes showed extensive cross-tissue effects on AD, such as AD was associated with the expression of APOE, CEACAM19, CLPTM1, DMWD, KAT8, PRSS36, PVRL2, SIX5, TRAPPC6A, PPP1R13L and PPP1R37 in at least three tissues.
Fig 4. TWAS results of different subcortical tissues.
(A) The bubble plot shows TWAS results of different subcortical tissues. The x-axis shows the 24 prioritized AD-related genes, the y-axis shows the five types of brain tissues. The size and the color of the bubbles demonstrate the significance of each gene in TWAS of a given tissue. The gray bubbles represent non-significant associations in TWAS, the hollow bubbles reflect the genes whose prediction models are not established successfully. (B) The bar plots show the FDR corrected p-values in TWAS for the five genes associated with AD only in hippocampal tissue (qc < 0.05, FDR corrected). AD, Alzheimer’s disease; FDR, false discovery rate; TWAS, transcriptome-wide association study. *qc < 0.05.
Functional annotation of AD-related genes
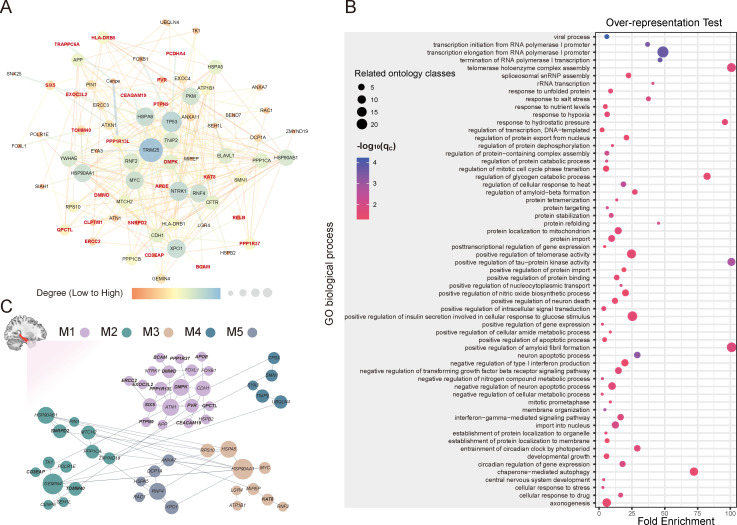
To identify the functional relationship and the involved biological processes of the 24 prioritized AD-related genes, we first constructed a protein-protein interaction (PPI) network by network topology-based analysis embedded in the WEB-based gene set analysis toolkit (Webgestalt, https://www.webgestalt.org) [32]. The PPI network contained 22 seed genes (prioritized AD-related genes) and 50 top-ranking neighbors based on network proximity (Fig 5A and S2 Table), PVRL2 and PRSS36 were not included in the network due to the lack of connectivity. APP, a known susceptibility protein of AD [33], was a hub node of the network. Notably, the prioritized gene QPCTL was directly connected with APP, suggesting their molecular interaction and AD relevance. In addition, the AD-related genes we identified were included in the common network with APP, which means that they may have coherent biological functions. Second, the statistical over-representation test of PANTHER [34] was used to identify enriched gene ontology (GO) terms of biological process for the generated network. The 72 genes in the network were enriched in 260 GO biological process terms (qc < 0.05, Benjamini-Hochberg FDR (BH-FDR) corrected) (S3 Table). These GO terms were divided into different ontologies according to hierarchical relations. Specifically, the 72 genes were mainly over-represented in a GO subclass of biological processes for neuron-related functions, such as neuron apoptotic process (fold enrichment = 28.56, qc = 0.000776), positive regulation of neuron death (fold enrichment = 11.54, qc = 0.0324), negative regulation of neuron apoptotic process (fold enrichment = 9.27, qc = 0.0215), axonogenesis (fold enrichment = 5.07, qc = 0.0344), and central nervous system development (fold enrichment = 3.34, qc = 0.0215). The genes were also correlated with amyloid-beta and tau phosphorylation related processes, including positive regulation of amyloid fibril formation (fold enrichment > 100, qc = 0.0181), regulation of amyloid-beta formation (fold enrichment = 26.78, qc = 0.022), positive regulation of tau-protein kinase activity (fold enrichment > 100, qc = 0.00178), and regulation of protein dephosphorylation (fold enrichment = 9.99, qc = 0.0174), which are well-known neuropathology of AD. In addition, they were associated with telomerase-related processes, such as telomerase holoenzyme complex assembly (fold enrichment > 100, qc = 0.018) and positive regulation of telomerase activity (fold enrichment = 23.8, qc = 0.0267) (Fig 5B). These results demonstrated that the 22 prioritized AD-related genes were interconnected in a common PPI network and contributed to the neuropathological process of AD.
Fig 5. Functional annotation of AD-related genes.
(A) The PPI network containing 22 seed proteins (marked in bold and dark red) and 50 top-ranking neighbors. (B) The bubble plot shows the enriched GO terms of biological process (the most specific subclass of each ontology is shown). The x-axis shows the fold enrichment of statistical over-representation test for each term (y-axis). The size of the bubbles reflects the number of related enriched terms of biological process. The color of the bubbles demonstrates the significance of each term based on the statistical over-representation test. (C) Hippocampal-based functional modules formed by AD-TWAS genes (marked in bold) and tightly connected genes. AD, Alzheimer’s disease; FDR, false discovery rate; GO, gene ontology; M, module; PPI, protein-protein interaction; TWAS, transcriptome-wide association study.
Modulization analysis of AD-related genes in the hippocampal network
Functional modules were built using the HumanBase online tool [35] (https://hb.flatironinstitute.org/) in the context of hippocampal tissue networks. The 72 genes in the constructed PPI network were clustered into five cohesive functional modules in hippocampal tissue. Module 1 (M1) included 13/24 prioritized AD-related genes (APOE, BCAM, CEACAM19, DMPK, DMWD, ERCC2, EXOC3L2, PPP1R13L, PPP1R37, PTPN9, PVR, QPCTL and SIX5), module 2 (M2) contained 3/24 prioritized AD-related genes (CD3EAP, TOMM40 and SNRPD2) and module 3 (M3) contained 1/24 prioritized AD-related genes (KAT8) (Fig 5C). In the enrichment analysis (qc < 0.05, BH-FDR corrected), M1 genes were enriched for neurogenesis-, neuron differentiation-, amyloid-beta formation- and dephosphorylation-related processes, suggesting that a large proportion of detected AD-related genes aggregated in the neuron-relevant functional module and critical processes for AD in the hippocampus. M2 genes were enriched for ribonucleoprotein complex- and protein localization to mitochondrion-related processes. M3 genes were enriched for autophagy-, immune system development- and histone modification-related processes (S4 Table), indicating that several detected AD-related genes could modulate common cellular functions in the hippocampus.
Hippocampal gene expression differences in ADNI data
In the TWAS and FOCUS analyses, we prioritized 24 genes with significant differences in the predicted cis-GReX in the hippocampus between the AD and control groups. We further validated this finding in ADNI imaging genetics dataset (http://www.loni.usc.edu/). We used the genotyping data, structural brain MRI data and demographic information from ADNI1, ADNIGO and ADNI2. After quality control and preprocessing of genetic and hippocampal volume data from brain MRI (see Materials and methods), 1410 ADNI subjects were finally included. At baseline, the 1410 ADNI subjects were diagnosed as cognitively normal (CN, n = 415), mild cognitive impairment (MCI, n = 720) and AD (n = 275). After up to 13 years of follow-up, the diagnoses were 317 CN, 416 MCI and 599 AD, and 78 individuals were excluded due to uncertain diagnoses. The baseline MCI patients (n = 567) with a follow-up period of more than 2 years were further divided into the conversion (MCI-C, n = 300) group and the stable (MCI-S, n = 267) group. The demographic information of the 1332 subjects with definite diagnoses is shown in Table 1.
Table 1. Demographics and MRI data of the included sample.
| Demographic variables | CN | MCI | AD |
|---|---|---|---|
| Number | 317 | 416 | 599 |
| Age (years) | 74.18 ± 5.63 | 73.75 ± 7.40 | 74.64 ± 7.35 |
| Sex (Male/Female) | 157/160 | 256/160 | 354/245 |
| Education (years) | 16.50 ± 2.68 | 16.01 ± 2.90 | 15.53 ± 2.88 |
| Hippocampal volume (ml) | 3738.88 ± 445.46 | 3474.60 ± 529.06 | 3002.31 ± 538.27 |
Data are shown as mean ± standard deviation.
CN = cognitively normal, MCI = mild cognitive impairment, AD = Alzheimer’s disease.
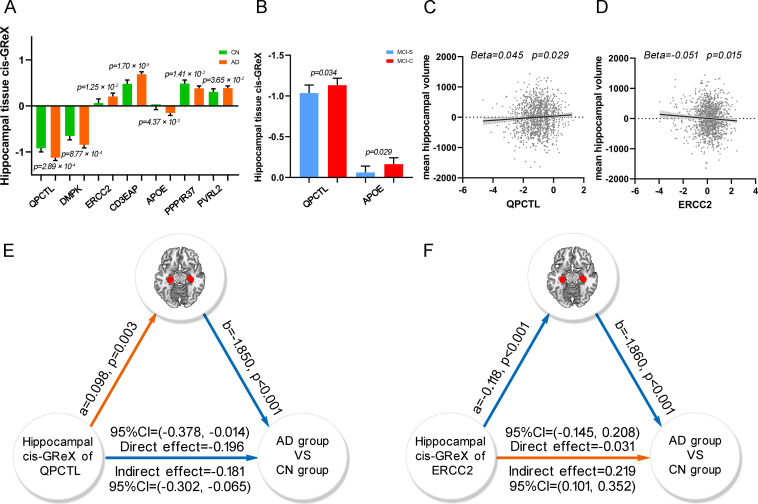
For each gene, Predixcan [14] was used to predict the cis-GReX of the gene in hippocampal tissue for each ADNI subject by integrating genotypic data of the subject with the weighted value of each SNP derived from the prediction models. For each of the 24 AD-related genes, binary logistic regression was performed to test the difference in gene expression in hippocampal tissue (predicted cis-GReX) between the AD (n = 599) and CN (n = 317) groups, controlling for age, sex, education and the first 4 components of multidimensional scaling (MDS). The hippocampal expression of QPCTL, DMPK, ERCC2, CD3EAP, APOE, PPP1R37 and PVRL2 was significantly different between the two groups (Table 2 and Fig 6A). The hippocampal expression of QPCTL and APOE was also significantly different between the MCI-C and MCI-S groups (Fig 6B).
Table 2. Seven TWAS significant genes were validated in the ADNI neuroimaging data.
| Gene | AD TWAS | ADNI validation | ||||
|---|---|---|---|---|---|---|
| TWAS_Z | TWAS_P | OR | 95%CI | SE | P | |
| QPCTL | -23.332 | 2.10 × 10−120 | 0.760 | 0.656 to 0.882 | 0.076 | 2.89 × 10−4 |
| DMPK | -22.672 | 8.54 × 10−114 | 0.774 | 0.666 to 0.900 | 0.077 | 8.77 × 10−4 |
| ERCC2 | 17.870 | 2.01× 10−71 | 1.199 | 1.040 to 1.382 | 0.073 | 1.25 × 10−2 |
| CD3EAP | 14.916 | 2.60× 10−50 | 1.396 | 1.199 to 1.625 | 0.078 | 1.70 × 10−5 |
| APOE | -12.901 | 4.43× 10−38 | 0.814 | 0.707 to 0.938 | 0.072 | 4.37 × 10−3 |
| PPP1R37 | -6.710 | 1.95× 10−11 | 0.834 | 0.722 to 0.964 | 0.074 | 1.41 × 10−2 |
| PVRL2 | 4.458 | 8.26× 10−6 | 1.166 | 1.010 to 1.346 | 0.073 | 3.65 × 10−2 |
OR-values, 95% CI, SE-values and P-values are from binary logistic regression.
AD = Alzheimer’s disease, ADNI = Alzheimer’s Disease Neuroimaging Initiative
TWAS = transcriptome-wide association study, OR = odds ratio, CI = confidence interval, SE = standard error.
The abbreviation of genes is referred to at https://www.ncbi.nlm.nih.gov/gen.
Fig 6. ADNI neuroimaging data analysis.
(A) The bar plot shows the significant difference of hippocampal tissue cis-GReX of QPCTL, DMPK, ERCC2, CD3EAP, APOE, PPP1R37 and PVRL2 between AD and CN groups. The p-values are calculated by binary logistic regression between the hippocampal tissue cis-GReX of the seven genes and diagnoses. (B) The bar plot shows the significant difference of hippocampal tissue cis-GReX of QPCTL and APOE between MCI-C and MCI-S groups. (C and D) The scatter plots show correlations between the hippocampal tissue cis-GReX of QPCTL (C), ERCC2 (D) and mean hippocampal volume using linear regression. The y-axis shows the residual of mean hippocampal volume after regressed age, sex, education, MR field strength, first 4 components of MDS and clinical diagnoses. (E and F) The mediation analysis shows that hippocampal volume mediates the effect of the hippocampal tissue cis-GReX of QPCTL (E) and ERCC2 (F) on the diagnosis of AD. The colors of the lines demonstrate the positive correlation (orange color) and the negative correlation (blue color) in the analysis. AD, Alzheimer’s disease; cis-GReX, cis-genetically regulated expression; CN, cognitively normal; MCI-C, mild cognitive impairment conversion; MCI-S, mild cognitive impairment stable; MDS, multidimensional scaling.
Hippocampal gene expression and hippocampal volume
Since the identified AD-related genes showed abnormal expression in the hippocampus and AD is characterized by hippocampal atrophy, we further wanted to identify which of these genes are associated with hippocampal volume. Seven genes were validated in ADNI whose expression in hippocampal tissue was associated with AD. We performed linear regression between the expression of each validated AD-related gene in the hippocampus and the mean hippocampal volume in the 1332 ADNI subjects while controlling for age, sex, education, MR field strength, clinical diagnosis and the first 4 components of MDS. We found that the mean hippocampal volume was nominally correlated with the expression of QPCTL (Beta = 0.045, p = 0.029) and ERCC2 (Beta = -0.051, p = 0.015) in the hippocampus (Fig 6C and 6D). In addition, we performed multiple linear regression to identify the total effect of the 24 genes on the mean hippocampal volume of AD (n = 599), controlling for all confounders. The 24 genes impacted the hippocampal volume of AD (p = 0.039, R2 = 0.062). R2 is the proportion of variance of the dependent variable (hippocampal volume) that can be explained by the independent variables (predicted cis-GReX in hippocampal tissue of the 24 genes).
Given that the expression of QPCTL and ERCC2 in hippocampal tissue was associated with hippocampal volume, we further explored whether the expression of these two genes in other subcortical tissues was also associated with the volumes of the corresponding structures. We used the same procedure as that of the hippocampus, wherein the predicted cis-GReX for each gene was calculated by integrating genotypic data of the ADNI subjects and the prediction models of the amygdala, caudate, nucleus accumbens and putamen, respectively. For the 1332 subjects we included, two individuals were excluded due to the failure of the segmentation of the four brain structures. We explored the correlations between the predicted cis-GReX of the two genes (QPCTL, ERCC2) in each of the three brain tissues (caudate, nucleus accumbens and putamen) and the corresponding volumes (n = 1330). There was no significant correlation between the cis-GReX of the two genes in these tissues and corresponding brain structure volumes (S5 Table). In contrast to the significant joint effect of the 24 prioritized AD-related genes on the hippocampal volume of AD, these genes showed no joint effect on the volumes of the amygdala, caudate, nucleus accumbens and putamen in AD patients (n = 599) (S7 Fig). Taken together, these results suggest that these genes show greater impacts on hippocampal volume than volumes of other subcortical nuclei.
Hippocampal volume mediates the effect of hippocampal gene expression on AD
As an important intermediate phenotype of AD, hippocampal volume may mediate the association between gene expression (QPCTL and ERCC2) in hippocampal tissue and AD. To identify the hippocampus-mediated pathway from gene expression to AD, we performed mediation analysis, in which the predicted cis-GReX of QPCTL or ERCC2 in hippocampal tissue was set as an independent variable, mean hippocampal volume as a mediator variable, and disease state (AD versus CN) as a dependent variable. The significance of the indirect effect was tested by calculating bias-corrected 95% bootstrap confidence interval with 5000 resampling, and the statistical significance of other effects was set at p < 0.05. For QPCTL, we found a significant indirect effect (effect = -0.181, 95% CI = -0.302 to -0.065) from cis-GReX to the disease state, indicating that the expression of QPCTL in hippocampal tissue could affect AD via modulating hippocampal volume. In addition, the direct effect (effect = -0.196, p = 0.034, 95% CI = -0.378 to -0.014) from the cis-GReX of QPCTL in hippocampus to the disease state was also significant, which represented a portion of the effect of the gene expression on AD being not mediated by hippocampal volume (Fig 6E). For ERCC2, the hippocampal volume could mediate the effect of the cis-GReX on the disease state with a significant indirect effect (effect = 0.219, 95% CI = 0.101 to 0.352), but the direct effect from the cis-GReX of ERCC2 on the disease state was not significant (p = 0.727, 95% CI = -0.145 to 0.208) (Fig 6F). These findings indicate that the expression of ERCC2 in hippocampal tissue could affect AD mainly by modulating hippocampal volume.
Discussion
In this study, we jointly used TWAS and fine mapping approaches to identify genes whose expression in hippocampal tissue was associated with AD and screened 24 AD- and hippocampus-related genes involved in crucial biological processes of AD and functional modules in hippocampal tissue. We further validated the associations of QPCTL, DMPK, ERCC2, CD3EAP, APOE, PPP1R37 and PVRL2 with AD in ADNI data, and found relations of QPCTL and APOE with the conversion from MCI to AD. We also found that hippocampal volume mediated the associations of hippocampal tissue cis-GReX of ERCC2 and QPCTL with AD. These findings provide candidate genes linked to AD by regulating gene expression in hippocampal tissue and underline the importance of the hippocampus in explaining the genetic risks of AD.
This study extends AD-related genetic loci identified by prior AD-GWAS studies [4–6,8–11] by providing evidence that some loci (APOE, TOMM40, PVRL2, EXOC3L2, KAT8 and HLA-DRB5) may lead to AD by affecting the gene expression levels in the hippocampus. Among the 24 AD- and hippocampus-related genes identified in this study, previous studies only provide clues for the associations of hippocampal expression of PRSS36, KAT8, HLA-DRB5 [4], TOMM40 [20], CEACAM19 and PVRL2 [21] with AD. However, some indirect evidence may support other associations. For example, we found that the expression of APOE in hippocampal tissue was associated with AD and the conversion from MCI to AD, which is consistent with the concept that APOE is an important genetic risk gene for AD [36] and with the reduced APOE protein level in the hippocampus in patients with AD [37]. In addition, we identified 18/24 genes that have not been found in GWAS, which may be due to differences in methodology or the lack of statistical power in GWAS. However, our findings are highly consistent (19/24) with previous TWAS results [17–21].
Comparing with previous TWAS and GWAS studies, we found two novel genes, PTPN9 and PCDHA4, affecting AD through hippocampal expression. Given the two loci are non-significant in AD GWASs, our analysis leveraged the hippocampal gene expression data and combined the effects of SNPs on each gene by TWAS to increase statistical power for discovery. Our prediction models successfully established the relationship between SNPs and gene expression for PTPN9 and PCDHA4. In addition, compared to the other four brain tissues, the expression of the two genes was associated with AD only in hippocampal tissue, suggesting that mechanistically related tissue and high-performance prediction models of TWAS are important for identifying context-specific disease genes. The functional annotation revealed that PTPN9 participated in neurogenesis (GO:0022008) and dephosphorylation (GO:0016311), and both PTPN9 and PCDHA4 were involved in nervous system development (GO:0007399). Modulization analysis based on the extended AD-related gene set and hippocampal-based network revealed that PTPN9 was a member of the M1 functional module affecting neuron-related biological processes. PTPN9 belongs to the protein tyrosine phosphatase family, which is involved in numerous important biological processes [38], and PTPN9 knockout mice show severe neurodevelopmental disorders [39]. In addition, PCDHA4 is a member of the protocadherin alpha gene family, and neural cadherin-like cell adhesion proteins encoded by these genes play a critical role in establishing complex brain networks of neuronal connections [40]. Knockdown of mouse protocadherin alpha proteins results in abnormalities in learning and memory [41]. Together, both functional annotation and previous studies provided evidence that PTPN9 and PCDHA4 may affect hippocampus-dependent AD development.
By combining network topology-based analysis, statistical over-representation test and hippocampal-based functional module detection, we can better understand the function of the identified AD- and hippocampus-related genes. These genes were interconnected in the PPI network and interacted with the causal proteins of AD, such as a key PPI network member, APP, which could generate neurotoxic Aβ peptide and play a crucial role in the development of AD [33,42]. The component genes of the constructed PPI network were related to many important processes for AD, such as amyloid-beta formation- and phosphorylation/dephosphorylation-related biological processes. In the AD brain, phosphorylation/dephosphorylation imbalance is an important mechanism for hyperphosphorylation of tau [43]. In addition, neuronal apoptosis- and neurogenesis-related processes have been identified, and in the AD brain, adult hippocampal neurogenesis is impaired with immature differentiation of neurons [44]. Telomerase is expressed in mature human hippocampal neurons [45], and telomerase-deficient mice with short telomeres exhibit loss of neurons in the hippocampus [46]. Neuronal telomeres are shorter in hippocampal neurons of AD [47]. Therefore, telomere-related processes may participate in AD pathogenesis. Moreover, 17/24 prioritized AD-related genes were involved in hippocampal-based functional modules and enriched in key pathways of AD, further supporting their pathogenicity in the etiology of AD.
In the present study, we also investigated the relationship of the predicted gene expression in hippocampal tissue with hippocampal volume in ADNI data. We found that QPCTL and ERCC2 were associated with hippocampal volume and that hippocampal volume mediated the effect of the two genes on AD. The 24 AD- and hippocampus-related genes had combined effects on the hippocampal volume of AD. These associations were only found in the hippocampus (compared with the amygdala, caudate, nucleus accumbens and putamen). Thus, hippocampal volume, which is an important endophenotype of AD, could fill gaps between gene expression in hippocampal tissue and AD. In our analysis, QPCTL interacted with APP, and QPCTL and ERCC2 were involved in M1, which is related to many important pathways for AD. Thus, the two genes may affect hippocampal volume by modulating neurogenesis, neuron differentiation, amyloid-beta formation and dephosphorylation related processes and further increase the risk of AD.
There are limitations in our study. First, although probabilistic fine-mapping was used in this study, it only yields credible sets of genes that contain potential causal genes by estimating the probability of causality, so it could be used to prioritize genes rather than to identify true causal genes. Further biological validation of the discovered genes needs to be performed in future studies. Second, the discovery and replication samples are partially overlapped (11.9% of the discovery sample and 84.7% of the replication sample). The discovery patients contain the AD-by-proxy phenotype, and the replication patients have defined diagnosis. We used the GWAS summary data with the largest sample size as the discovery sample to increase the statistical power. Due to the sample overlap, the replication sample could exclude the influence of AD-by-proxy phenotype, but not replicate the results in an independent dataset, therefore, the reproducibility of the identified genes in different dataset is challenged. To eliminate the effect of sample overlap on the reliability of the TWAS results, we used two independent databases to validate our results and successfully replicated 20/36 TWAS genes (S1 Text and S6 Table). In addition, we used the multiple trait analysis of GWAS (MTAG) [48] approach to perform meta-analysis using discovery and replication GWAS summary statistics while accounting for potential sample overlap, and 25/36 genes were successfully replicated (qc < 0.05) (S1 Text and S7 Table). Taken together, we could replicate 31/36 of our identified genes (S8–S10 Figs); however, a completely independent large-scale GWAS data of AD will be needed to fully validate our discovery.
Materials and methods
Ethics statement
All the data used in this study were obtained from public data repositories and got approval by their medical ethics review committees. Details about informed consent of GTEx can be found in the original paper [22] (dbGaP accession number phs000424.v7. p2). For ADNI, written informed consent was provided for all participants, and the study protocol was approved by each participating sites’ institutional review board (http://www.loni.usc.edu/). For the GWAS summary data of AD, all cohorts obtained written informed consent and each study protocol was approved by the institutional review boards. Full details can be found in the original paper [4,11]. We followed the instructions of accessing summary data on the websites (https://ctg.cncr.nl/software/summary_statistics, https://www.niagads.org/datasets/ng00075).
Data resources
WGS and RNA-seq data of hippocampal tissue
WGS and RNA-seq data of 111 hippocampal samples from GTEx Version 7 were used to build prediction models for gene expression in hippocampal tissue based on genomic variants. The pipelines for processing WGS and RNA-seq data were available at the GTEx portal (https://gtexportal.org/home/). For WGS data, the reads were annotated according to the human reference genome (hg19/GRCh37). The sample-level quality control (QC) included genotyping call rate per individual (> 98%), sex concordance check and identity check. The SNP-level QC included SNP call rate (> 85%), Hardy-Weinberg equilibrium (HWE) (p > 1 × 10−6), minor allele frequency (MAF) (> 1%), and with non-ambiguous strand (no A/T or C/G SNPs). The obtained SNPs were pre-phased by SHAPEIT2 [49] and imputed by IMPUTE2 [50] with the 1000 Genomes Phase 3 reference panel. A total of 7,920,040 SNPs were finally selected from the imputed SNPs based on the criteria of biallelic and single-character allele codes only, non-ambiguous stranded SNPs, SNP call rate = 100%, HWE p > 1 × 10−6, MAF > 0.01 and IMPUTE2 info quality score > 0.8. GTEx standard quantification and QC procedures were conducted for hippocampal tissue RNA-seq data by GTEx consortium. All reads were aligned to the human reference genome (hg19/GRCh37) based on GENCODE v19 reference annotations [51] (https://www.gencodegenes.org/human/release_19.html). The same pipeline used for the hippocampus was applied to process WGS and RNA-seq data of the amygdala, caudate, putamen and nucleus accumbens (S8 Table).
GWAS summary data of AD
In the discovery stage, GWAS summary statistics of AD was derived from a meta-analysis collected from the PGC-ALZ (n = 17,477), IGAP (n = 54,162), ADSP (n = 7,506) and UKBB (n = 376,113), including 455,258 (71,880 AD or AD-by-proxy and 383,378 controls) unrelated individuals of European ancestry. Details about genotyping, quality control and genome-wide meta-analysis can be found in the original paper [4]. In UKBB, each proxy case had a clear family history of AD. In the validation stage, we replicated our findings in a GWAS meta-analysis of diagnosed AD from the updated IGAP (n = 63,926 including 21,982 AD and 41,944 controls) [11]. 54,162 participants from IGAP were overlapped between the discovery and validation samples.
Neuroimaging and genotyping data of ADNI
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a comprehensive imaging genetics dataset containing genetic, neuroimaging, biochemical and clinical data. The genotyping data, structural brain MRI data and demographic information used in this study were downloaded from the ADNI data repository (http://www.loni.usc.edu/). ADNI was designed as an ongoing, longitudinal project. Initially, ADNI1 enrolled participants of CN, MCI and AD. Subsequent studies, including ADNIGO and ADNI2, further extended the study with additional cohorts and followed up with rollovers. The 757 participants from ADNI1 were genotyped by Illumina Human610-Quad BeadChip, and the 793 participants from ADNIGO/2 were genotyped by Illumina HumanOmniExpress BeadChip (http://www.illumina.com). The intensity data was processed with GenomeStudio v2009.1. Detailed information for the QC and processing procedures is shown in the S1 Text. After QC and imputation, 1423 individuals and 8,035,650 autosomal SNPs were retained for subsequent analysis.
The ADNI repository provides hippocampal volume data and volumetric data of the amygdala, caudate, nucleus accumbens and putamen calculated by FreeSurfer (http://surfer.nmr.mgh.harvard.edu/) with the pipeline for cross-sectional comparisons; for details, please see the manual (http://www.loni.usc.edu/). First, QC was performed for hippocampal volumetric data. One individual without hippocampal volume data, eleven individuals with failed segmentation and one individual without clinical data were excluded. In the remaining 1410 individuals, the hippocampal volume data of 1377 individuals were extracted from the baseline data, and those of 33 individuals were extracted from the nearest time point data to the baseline. Second, QC was performed for the volumetric data of the amygdala, caudate, nucleus accumbens and putamen in the 1410 individuals who had qualified hippocampal volumetric data, and two individuals were excluded due to the failure of brain tissue segmentation of these structures.
Predicting hippocampal gene expression by SNPs
For each gene, conditional analysis in QTLtools (https://qtltools.github.io/qtltools/) [29] was used to identify cis-eQTLs with independent effects on gene expression in a cis-window of ± 1 Mb from the transcription start site (TSS). In this analysis, forward variable selection was used to decide the number of independent signals per gene expression at a moderate threshold (p < 0.01), and backward elimination was used to assign nearby variants to the independent signals. For each candidate SNP, the genotype of each sample was encoded as 0, 1 and 2 based on the counts of the effect allele. For the 111 hippocampal samples, we used the candidate SNPs to predict gene expression in hippocampal tissue with an additive genetic model. After cis-eQTLs mapping, the prediction models were only built for protein-coding genes (n = 15,831). Prediction models were built using the nested cross validated elastic-net procedure following the GTEx V7 pipeline (https://github.com/hakyimlab/PredictDB_Pipeline_GTEx_v7) [14]. First, the 10-fold cross-validated elastic-net was performed 5 times to estimate the significance of the models. The 111 hippocampal samples were split into 5 folds randomly, one-fold was removed at a time, the remaining samples (four folds) were used to train the prediction models by elastic-net with 10-fold cross-validation to tune the lambda parameter, and then the prediction models were applied to the samples of the removed fold to evaluate the correlations between the predicted and measured expression levels of the hippocampal samples. The performance of each prediction model was assessed by the average Pearson correlation coefficient between predicted and measured expression across subjects, which was the averaged value of the 5 times 10-fold nested cross validation tests. A prediction model was significant if the estimated p-value for the average Pearson correlation coefficient passed the multiple testing correction threshold of FWE (pc < 0.05/15,831 = 3.16 × 10−6). In addition, the threshold value of the average Pearson correlation coefficient was greater than 0.1 to avoid the negative correlation according to the suggestion of the pipeline. Second, for each significant prediction model, a new elastic-net model was trained using 10-fold cross validation to tune the lambda parameter based on all hippocampal samples to calculate weights. The pipeline could avoid the bias caused by using the same data to tune the parameter and assess the performance. The same procedure as that used for hippocampal tissue was applied to establish prediction models for the tissues of amygdala (15,827 protein-coding genes), caudate (15,926 protein-coding genes), putamen (15,629 protein-coding genes) and nucleus accumbens (15,937 protein-coding genes). The sex, 15 expression residuals, top 3 genetic principal components and sequencing platforms were controlled during both eQTLs mapping and prediction model construction. The prediction models generated the weighted value of each candidate SNP’s relative contribution to the gene’s expression level in the corresponding tissue.
Identifying AD-related genes by TWAS
In this study, TWAS was used to identify AD-related genes by testing correlations between cis-GReX and AD diagnosis with S-PrediXcan [15], which was embedded in the MetaXcan framework (https://github.com/hakyimlab/MetaXcan). In TWAS, SNP-AD associations were derived from GWAS summary data of AD, SNP-expression associations were assessed by the weighted value of each SNP to corresponding gene expression in the hippocampal and other tissues, and LD reference set was created by the prediction models. Multiple testing was corrected by FDR method (qc < 0.05).
Fine-mapping TWAS-identified AD-related genes
To exclude the possibility that TWAS-identified AD-related genes resulted from the genomic architecture of LD or co-regulation of gene models, FOCUS [30] was used to estimate the posterior inclusion probabilities for causality while accounting for the correlation structures of LD and co-regulation of gene expression prediction models. As a recommended strategy to improve the power, FOCUS also included SNP weights for gene expression in nonhippocampal tissues (PrediXcan weights of the GTEx v7 data (http://predictdb.org/) [14] and FUSION (functional summary-based imputation) weights of the METSIM, NTR, YFS, CMC data (http://gusevlab.org/projects/fusion/) [16]. FOCUS estimated credible gene sets based on the posterior inclusion probabilities at the 90% confidence level.
Network topology-based analysis
We conducted network topology-based analysis using Webgestalt [32] with reference to the human PPI of the Biological General Repository for Interaction Datasets (BIOGRID) (Build 3.5.167) [52]. For the seed genes we mapped to the PPI network, random walk analysis was performed to expand the network by ranking all genes based on their network proximity with the seed genes. The resulting PPI network was constructed with the input seeds and the 50 top-ranking neighbors.
Statistical over-representation test
The generated network was investigated by performing a statistical over-representation test using PANTHER classification system (v.14.0) [34] based on GO biological processes. For the genes in the PPI network, a statistical over-representation test was applied to detect statistical over representation of input genes compared to the human genome reference gene list. We used Fisher’s exact test to calculate the p-value based on the comparison between the number of input genes in a certain term and the number of reference genes in the same term. Fold enrichment represents the ratio of the value of observed gene number over that of expected. We used the BH-FDR correction for multiple testing (qc < 0.05).
Finding functional modules composed of the identified AD-related genes
Gene network analysis was used to test whether the identified AD-related genes were involved in certain cohesive gene clusters in hippocampal tissue by the HumanBase online tool [35]. Functional enrichment was performed for the resulting functional modules using GO terms. The statistical significance of each GO term was tested by one-sided Fisher’s exact test, and multiple testing was corrected by BH-FDR (qc < 0.05).
Mediation analysis
The PROCESS macro for SPSS (v3.4) was used for mediation analysis [53]. Only genes with both intergroup expression differences between the AD and CN groups (p < 0.05) and correlations with hippocampal volume (p < 0.05) were selected for the mediation analysis. In this model, the hippocampal cis-GReX for each gene was defined as an independent variable, the mean hippocampal volume as a mediator variable, the disease states (AD versus CN) as a binary dependent variable, hippocampal volume was adjusted by a linear regression with MR field strength, and the covariates included age, sex and education. For the dichotomous outcome in our analysis, PROCESS generated the direct effects, indirect effects, and paths from the mediator variables to the binary dependent variables by logistic regression. The coefficients between independent variables and mediator variables were estimated by ordinary least squares (OLS) regression.
Detailed protocols of the methods used above are available in https://dx.doi.org/10.17504/protocols.io.bp4amqse.
Supporting information
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Each point represents a single gene, with physical position in chromosome plotted on the x-axis and z-score of association statistics between gene and AD plotted on the y-axis. Significant associations (p < 0.05, FDR corrected) are labeled with gene names.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
The x-axis shows the five subcortical tissues, the y-axis shows the R2 obtained from multiple linear regression, which represents the proportion of variance of the dependent variable that can be explained by the independent variables. R2, coefficient of determination.
(PDF)
The blue circle represents the 36 genes identified in the discovery stage of TWAS (qc < 0.05, FDR corrected) and validated at nominal threshold of p < 0.05 with consistent direction of z-scores between discovery and validation stage; The grey circle represents the 25 genes identified by using two independent data sets of GWAS summary statistics of AD; The yellow circle represents the 74 genes identified by using the GWAS summary statistics accounting for sample overlap.
(PDF)
(PDF)
(PDF)
Extended validation of TWAS results.
(DOCX)
Acknowledgments
We are grateful to the GTEx, ADNI for providing data for our analysis. We would like to thank Hae Kyung Im and Bogdan Pasaniuc for their outstanding work and sharing the source code; IGAP and Danielle Posthuma team for providing GWAS summary statistics.
A complete list of Alzheimer’s disease Neuroimaging Initiative investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Data Availability
All relevant data are within the manuscript and its Supporting Information files. The protected WGS data from the GTEx project is available from dbGaP (accession number phs000424.v7. p2). GWAS summary statistics of the AD meta-analysis collected from the PGC-ALZ (n = 17,477), IGAP (n = 54,162), ADSP (n = 7,506) and UKBB (n = 376,113), including 455,258 (71,880 AD or AD-by-proxy and 383,378 controls) participants is available online (https://ctg.cncr.nl/software/summary_statistics). The GWAS meta-analysis from the updated IGAP (21,982 AD and 41,944 controls) has been deposited in the National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS) (https://www.niagads.org/datasets/ng00075). The genotyping data, structural brain MRI data and demographic information of ADNI are available in the ADNI data repository (http://adni.loni.usc.edu/). The data are available to researchers via application to ADNI. Application is required to ensure proper protection of confidentiality of the participants. The summary data of the prediction models we trained (S9–S13 Tables) and the TWAS summary statistics (S14–S16 Tables) are available in the supporting information.
Funding Statement
This work was supported by the National Key Research and Development Program of China (2018YFC1314300) to CY, the National Natural Science Foundation of China to CY (82030053, 81425013), the National Natural Science Foundation of China to MJL (31871327), the National Natural Science Foundation of China to JX (82001797), the National Natural Science Foundation of China to HL (81701668) and the Tianjin Key Technology R&D Program (17ZXMFSY00090) to CY. The authors report no biomedical financial interests or potential conflicts of interest. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Spires-Jones TL, Hyman BT. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron. 2014;82(4):756–71. 10.1016/j.neuron.2014.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyman BT, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Carrillo MC, et al. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease. Alzheimers Dement. 2012;8(1):1–13. 10.1016/j.jalz.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, et al. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63(2):168–74. 10.1001/archpsyc.63.2.168 [DOI] [PubMed] [Google Scholar]
- 4.Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51(3):404–13. 10.1038/s41588-018-0311-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–93. 10.1038/ng.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45(12):1452–8. 10.1038/ng.2802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1094–9. 10.1038/ng.439 [DOI] [PubMed] [Google Scholar]
- 8.Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. 2010;303(18):1832–40. 10.1001/jama.2010.574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43(5):429–35. 10.1038/ng.803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43(5):436–41. 10.1038/ng.801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. 2019;51(3):414–30. 10.1038/s41588-019-0358-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10(3):184–94. 10.1038/nrg2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albert FW, Kruglyak L. The role of regulatory variation in complex traits and disease. Nat Rev Genet. 2015;16(4):197–212. 10.1038/nrg3891 [DOI] [PubMed] [Google Scholar]
- 14.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–8. 10.1038/ng.3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9(1):1825. 10.1038/s41467-018-03621-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BW, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nature genetics. 2016;48(3):245–52. 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj T, Li YI, Wong G, Humphrey J, Wang M, Ramdhani S, et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat Genet. 2018;50(11):1584–92. 10.1038/s41588-018-0238-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y, Li M, Lu Q, Weng H, Wang J, Zekavat SM, et al. A statistical framework for cross-tissue transcriptome-wide association analysis. Nat Genet. 2019;51(3):568–76. 10.1038/s41588-019-0345-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hao S, Wang R, Zhang Y, Zhan H. Prediction of Alzheimer’s Disease-Associated Genes by Integration of GWAS Summary Data and Expression Data. Front Genet. 2018;9:653. 10.3389/fgene.2018.00653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu W, Li M, Zhang W, Zhou G, Wu X, Wang J, et al. Leveraging functional annotation to identify genes associated with complex diseases. bioRxiv. 2020. 10.1371/journal.pcbi.1008315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerring ZF, Lupton MK, Edey D, Gamazon ER, Derks EM. An analysis of genetically regulated gene expression across multiple tissues implicates novel gene candidates in Alzheimer’s disease. Alzheimers Res Ther. 2020;12(1):43. 10.1186/s13195-020-00611-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–13. 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51(4):592–9. 10.1038/s41588-019-0385-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ridha BH, Barnes J, Bartlett JW, Godbolt A, Pepple T, Rossor MN, et al. Tracking atrophy progression in familial Alzheimer’s disease: a serial MRI study. The Lancet Neurology. 2006;5(10):828–34. 10.1016/S1474-4422(06)70550-6 [DOI] [PubMed] [Google Scholar]
- 25.Fox NC, Warrington EK, Freeborough PA, Hartikainen P, Kennedy AM, Stevens JM, et al. Presymptomatic hippocampal atrophy in Alzheimer’s disease. A longitudinal MRI study. Brain. 1996;119 (Pt 6):2001–7. 10.1093/brain/119.6.2001 [DOI] [PubMed] [Google Scholar]
- 26.Henneman WJ, Sluimer JD, Barnes J, van der Flier WM, Sluimer IC, Fox NC, et al. Hippocampal atrophy rates in Alzheimer disease: added value over whole brain volume measures. Neurology. 2009;72(11):999–1007. 10.1212/01.wnl.0000344568.09360.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serrano-Pozo A, Frosch MP, Masliah E, Hyman BT. Neuropathological alterations in Alzheimer disease. Cold Spring Harb Perspect Med. 2011;1(1):a006189. 10.1101/cshperspect.a006189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gamazon ER, Segre AV, van de Bunt M, Wen X, Xi HS, Hormozdiari F, et al. Using an atlas of gene regulation across 44 human tissues to inform complex disease- and trait-associated variation. Nat Genet. 2018;50(7):956–67. 10.1038/s41588-018-0154-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delaneau O, Ongen H, Brown AA, Fort A, Panousis NI, Dermitzakis ET. A complete tool set for molecular QTL discovery and analysis. Nat Commun. 2017;8:15452. 10.1038/ncomms15452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mancuso N, Freund MK, Johnson R, Shi H, Kichaev G, Gusev A, et al. Probabilistic fine-mapping of transcriptome-wide association studies. Nat Genet. 2019;51(4):675–82. 10.1038/s41588-019-0367-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oxtoby NP, Young AL, Cash DM, Benzinger TLS, Fagan AM, Morris JC, et al. Data-driven models of dominantly-inherited Alzheimer’s disease progression. Brain. 2018;141(5):1529–44. 10.1093/brain/awy050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199–w205. 10.1093/nar/gkz401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Brien RJ, Wong PC. Amyloid precursor protein processing and Alzheimer’s disease. Annu Rev Neurosci. 2011;34:185–204. 10.1146/annurev-neuro-061010-113613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi H, Muruganujan A, Huang X, Ebert D, Mills C, Guo X, et al. Protocol Update for large-scale genome and gene function analysis with the PANTHER classification system (v.14.0). Nat Protoc. 2019;14(3):703–21. 10.1038/s41596-019-0128-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47(6):569–76. 10.1038/ng.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belloy ME, Napolioni V, Greicius MD. A Quarter Century of APOE and Alzheimer’s Disease: Progress to Date and the Path Forward. Neuron. 2019;101(5):820–38. 10.1016/j.neuron.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beffert U, Cohn JS, Petit-Turcotte C, Tremblay M, Aumont N, Ramassamy C, et al. Apolipoprotein E and beta-amyloid levels in the hippocampus and frontal cortex of Alzheimer’s disease subjects are disease-related and apolipoprotein E genotype dependent. Brain Res. 1999;843(1–2):87–94. 10.1016/s0006-8993(99)01894-6 [DOI] [PubMed] [Google Scholar]
- 38.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. 10.1016/j.cell.2004.05.018 [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Vachon E, Zhang J, Cherepanov V, Kruger J, Li J, et al. Tyrosine phosphatase MEG2 modulates murine development and platelet and lymphocyte activation through secretory vesicle function. J Exp Med. 2005;202(11):1587–97. 10.1084/jem.20051108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97(6):779–90. 10.1016/s0092-8674(00)80789-8 [DOI] [PubMed] [Google Scholar]
- 41.Fukuda E, Hamada S, Hasegawa S, Katori S, Sanbo M, Miyakawa T, et al. Down-regulation of protocadherin-alpha A isoforms in mice changes contextual fear conditioning and spatial working memory. The European journal of neuroscience. 2008;28(7):1362–76. 10.1111/j.1460-9568.2008.06428.x [DOI] [PubMed] [Google Scholar]
- 42.Zhang YW, Thompson R, Zhang H, Xu H. APP processing in Alzheimer’s disease. Molecular brain. 2011;4:3. 10.1186/1756-6606-4-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iqbal K, Liu F, Gong CX, Grundke-Iqbal I. Tau in Alzheimer disease and related tauopathies. Curr Alzheimer Res. 2010;7(8):656–64. 10.2174/156720510793611592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moreno-Jimenez EP, Flor-Garcia M, Terreros-Roncal J, Rabano A, Cafini F, Pallas-Bazarra N, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med. 2019;25(4):554–60. 10.1038/s41591-019-0375-9 [DOI] [PubMed] [Google Scholar]
- 45.Spilsbury A, Miwa S, Attems J, Saretzki G. The role of telomerase protein TERT in Alzheimer’s disease and in tau-related pathology in vitro. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2015;35(4):1659–74. 10.1523/JNEUROSCI.2925-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rolyan H, Scheffold A, Heinrich A, Begus-Nahrmann Y, Langkopf BH, Hölter SM, et al. Telomere shortening reduces Alzheimer’s disease amyloid pathology in mice. Brain. 2011;134(Pt 7):2044–56. 10.1093/brain/awr133 [DOI] [PubMed] [Google Scholar]
- 47.Franco S, Blasco MA, Siedlak SL, Harris PL, Moreira PI, Perry G, et al. Telomeres and telomerase in Alzheimer’s disease: epiphenomena or a new focus for therapeutic strategy? Alzheimers Dement. 2006;2(3):164–8. 10.1016/j.jalz.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 48.Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229–37. 10.1038/s41588-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5–6. 10.1038/nmeth.2307 [DOI] [PubMed] [Google Scholar]
- 50.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–D73. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oughtred R, Stark C, Breitkreutz B-J, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Research. 2019;47(D1):D529–D41. 10.1093/nar/gky1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayes AF. Introduction to mediation, moderation, and conditional process analysis: A regression-based approach: Guilford publications; 2017. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Each point represents a single gene, with physical position in chromosome plotted on the x-axis and z-score of association statistics between gene and AD plotted on the y-axis. Significant associations (p < 0.05, FDR corrected) are labeled with gene names.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
The x-axis shows the five subcortical tissues, the y-axis shows the R2 obtained from multiple linear regression, which represents the proportion of variance of the dependent variable that can be explained by the independent variables. R2, coefficient of determination.
(PDF)
The blue circle represents the 36 genes identified in the discovery stage of TWAS (qc < 0.05, FDR corrected) and validated at nominal threshold of p < 0.05 with consistent direction of z-scores between discovery and validation stage; The grey circle represents the 25 genes identified by using two independent data sets of GWAS summary statistics of AD; The yellow circle represents the 74 genes identified by using the GWAS summary statistics accounting for sample overlap.
(PDF)
(PDF)
(PDF)
Extended validation of TWAS results.
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. The protected WGS data from the GTEx project is available from dbGaP (accession number phs000424.v7. p2). GWAS summary statistics of the AD meta-analysis collected from the PGC-ALZ (n = 17,477), IGAP (n = 54,162), ADSP (n = 7,506) and UKBB (n = 376,113), including 455,258 (71,880 AD or AD-by-proxy and 383,378 controls) participants is available online (https://ctg.cncr.nl/software/summary_statistics). The GWAS meta-analysis from the updated IGAP (21,982 AD and 41,944 controls) has been deposited in the National Institute on Aging Genetics of Alzheimer's Disease Data Storage Site (NIAGADS) (https://www.niagads.org/datasets/ng00075). The genotyping data, structural brain MRI data and demographic information of ADNI are available in the ADNI data repository (http://adni.loni.usc.edu/). The data are available to researchers via application to ADNI. Application is required to ensure proper protection of confidentiality of the participants. The summary data of the prediction models we trained (S9–S13 Tables) and the TWAS summary statistics (S14–S16 Tables) are available in the supporting information.