Abstract
Laboratory strains of mice are typically housed in specific pathogen free facilities to minimize exposure to microbes. This method encourages uniformity in responses to experimentally induced parameters and prevents loss of animals, allowing for the survival and study of immunodeficient mice. However, these restrictions also limit physiologic relevance to humans who are exposed to numerous microbes from birth. Recent evidence from several groups has demonstrated that exposure of laboratory mice to commensal and pathogenic microbes normally found in wild or pet store mice, can dramatically impact the cellular makeup and function of the immune system. This article outlines procedures for physiologically exposing laboratory strains of mice to the diverse array of microbes typically found in pet store mice. Suggested methods for characterization of the immune system following this exposure are also described.
Basic Protocol 1:
Cohousing Laboratory Strains of Mice with Pet Store Mice
Basic Protocol 2:
Exposure of Laboratory Strains of Mice to Fomite Bedding
Keywords: Mouse model, Microbial Diversity, Microbiome, T cells, Serology
INTRODUCTION:
Research involving murine models typically begins by purchasing mice from vendors that certify the animals are free of common mouse pathogens. Once purchased and transported to contemporary facilities, laboratory mice continue to be housed under ‘specific pathogen free’ (SPF) conditions. Although this term can be ambiguous, it refers to animals that are confirmed to be free of specific pathogens by diagnostic testing (LANE-PETTER, 1962). The exact pathogens and the method of testing are left up to institutional discretion. Common agents include microorganisms associated with clinical signs or pathogenic lesions as well as those that can induce phenotypic changes, affecting the interpretation of research results(Mähler Convenor et al., 2014). The perceived research benefits of maintaining animals under SPF conditions include a reduction in the potential loss of animals – especially immunocompromised strains - to infectious diseases as well as mitigation of confounding variables that may affect the interpretation of results(Franklin, 2006).
In stark contrast to this approach, several studies have recently highlighted that both wild mice and laboratory mice exposed to a varied array of microbes offer important insights into the immune system (Beura et al., 2016; Japp et al., 2017; Reese et al., 2016; Rosshart et al., 2019; Rosshart et al., 2017). This is particularly relevant given that humans experience many encounters with acute and chronic viruses, bacteria, and parasites throughout life. Additional influences by commensal organisms that make up the microbiome are also now well appreciated to impact the immune system, and the makeup of such communities is dynamic and influenced by many factors including the housing/environmental circumstances of the host(Berg et al., 2020).
Given the limitations imposed by maintaining mice as SPF, the push to develop models which may more faithfully represent the human experience have now been pursued. One such method of introducing robust exposure to a diverse set of microbes is through cohousing research mice with pet store mice (Beura et al., 2016; Huggins et al., 2019). A second method which can be employed involves the transfer of fomite bedding from pet store mouse cages into cages of research mice. Over several weeks, these two techniques cause quite dramatic changes to the cellular makeup of the immune system, which can be documented in the blood by using flow cytometry (Beura et al., 2016; Huggins et al., 2019). Following these housing procedures, animals can be utilized as ‘dirty mice’ in the particular model of research interest pursued by the investigator.
BASIC PROTOCOL 1
COHOUSING LABORATORY STRAINS OF MICE WITH PET STORE MICE
Diverse exposure to microbes commonly carried by mice can be achieved by cohousing research strains of mice with mice obtained from pet stores. This protocol can only be performed with female mice, as male mice will undergo territorial fighting. Success is dependent on the number of microbes introduced by the pet store mouse at the time of cohousing. Importantly, the housing of pet store mice and transmission to other animals must be performed in facilities well-separated from SPF colonies. Housing is discussed in more detail under Critical Parameters.
Materials:
6–12 weeks old female research mouse strain of interest (C57BL/6, BALB/c, etc.) purchased from vendor (e.g. The Jackson Laboratory, Charles River, Taconic)
6–20 weeks old female pet store mice of any coat color (1 per cohousing cage) purchased from commercial vendors
Autoclavable mouse cage bottoms: standard size (Ancare, N10) or large size (10 ½” x 19” x 6 1/8”, Ancare, N40) with matching wire lid and microfilter top
Water bottles (Ancare, reduced height bottle, 16 oz.) with Drinking Tube (Ancare, OT-199) and rubber stopper (Ancare, #8.5R)
Irradiated corncob bedding (Teklad, 7902)
Mouse chow (non-autoclaved) (Teklad, 2018)
Obtain all mice from appropriate vendors.
Combine 1 pet store mouse with 4 research strain mice (1:4) in a standard mouse cage (5 mice total) or (1:8) in a large mouse cage (9 mice total). Maintain an equivalent number of research strain mice in SPF housing as age-matched controls.
Note that only female mice can be used in this protocol due to fighting amongst males that are not of the same litter.
The ratio of pet store mice to laboratory mice can be increased (2:3 or 2:7). This is likely to intensify the number of microbes transmitted, but will also risk increased lethality.
3. Provide chow and water ad libitum. Both chow and water are not autoclaved or irradiated.
4. Perform bedding changes weekly.
During the first two weeks, mice may show signs of mild illness (hunched posture, ruffled fur) but most immunocompetent mice (~80–90%) will not succumb. The survival of immunodeficient or genetically altered strains will need to be determined on a case by case basis.
5. After 30 days, mice should be assessed to confirm the presence of natural microbes. This can be done either by surveying for serological antibodies against common microbes carried by mice using serum or by direct detection of common microbes using PCR testing.
This may be accomplished by using assays that can be developed within the laboratory or by outsourcing to a vendor such as Charles River using their blood/serum based Assessment Plus test (which detects antibodies against 23 microbes) and/or their direct PCR based Surveillance Plus test (which measures for the presence of 44 microbes). Results from these assays reveal the presence or absence of each antibody or microbe (but not the quantity). Additional evidence of microbial exposure can be collected using flow cytometry to assess immune cell alterations (see Support Protocol 1). Which assays are chosen depends on available funds (serology is much cheaper than PCR) and whether the experiment requires knowing about active infection (PCR) vs. exposure (serology). Most experimental goals are likely to be satisfied by a serological assessment of microbial exposure.
If at this stage poor microbial transmission or impact on the immune system is observed, a fresh pet store mouse can be added to the cage.
6. After 60 days, mice should be surveyed again by serological and immune cell characterization (as in step 5) and then used in experimental protocols as ‘dirty mice’. Comparisons should always be made to an age-matched SPF control group to reduce variability and highlight small but critical differences.
SUPPORT PROTOCOL 1
ANTIBODY STAINING OF CIRCULATING IMMUNE CELLS AND ANALYSIS BY FLOW CYTOMETRY
The staining of immune cells in the blood with fluorescent antibodies followed by flow cytometric analysis allows the researcher to rapidly obtain evidence that the immune system is responding to a newly introduced microbial milieu. Additionally, the blood can be analyzed from the same mice at multiple time points during Basic Protocol 1 or Basic Protocol 2 to provide longitudinal information.
Materials:
Fresh blood collected from SPF control and ‘dirty mice’ (from basic protocol steps 5 and 6)
Heparin Sodium (Sagent, NDC Code 25021-400-10)
18 Gauge Hypodermic Needle (Becton-Dickinson, 305196) 5ml Polystyrene Round Bottom Tube (BD Falcon, 352008) RBC lysis buffer (ThermoFisher, cat. no. 00-4333-57)
Staining buffer: 1X PBS (ThermoFisher, cat. no. 10010023) + 1%FCS Flourescent Antibodies (See Table 1)
Table 1.
Antibodies Used for Analysis of Immune Cells in ‘Dirty Mice’
| Marker | Clone | Manufacturer | Reference No. | Dilution |
|---|---|---|---|---|
| CD11c | N418 | Biolegend | 117311 | 1:100 |
| CD115 | AFS98 | Tonbo Biosciences | 50–1152-U100 | 1:100 |
| KLRG1 | 2F1 | eBiosciences | 25–5893-82 | 1:100 |
| CD8a | 53–6.7 | Invitrogen | 45–0081-87 | 1:100 |
| Ly6C | HK1.4 | Biolegend | 128015 | 1:400 |
| Nkp46 | 29A1.4 | Biolegend | 137612 | 1:50 |
| MHCII (I:A/I:E) | M5/114.15.2 | Biolegend | 107635 | 1:200 |
| CD62L | MEL-14 | Biolegend | 104438 | 1:100 |
| CD4 | RM4–5 | Biolegend | 100546 | 1:100 |
| CD45 | 30-F11 | BD Biosciences | 563709 | 1:200 |
| CD44 | IM7 | Biolegend | 103041 | 1:200 |
| Ly6G | 1A8 | Biolegend | 127622 | 1:100 |
| CD19 | 1D3 | BD Biosciences | 563557 | 1:100 |
| CD11b | M1/70 | BD Biosciences | 564443 | 1:100 |
| L/D | N/A | Tonbo Biosciences | 13–0865-T100 | 1:400 |
Fixative- 2% paraformaldehyde (Fisher Scientific, cat. no. 50-980-487) or equivalent 96-well round bottom microtest plate (Sarstedt, 82.1582)
96-well Polypropylene Cluster Tubes (Corning, 4401)
Refrigerated centrifuge (Beckman Coulter Allegra X-14R) or equivalent LSR II flow cytometer (BD Biosciences) or equivalent
Prepare a 5mL collection tube for each mouse by adding 80 usp units of heparin to each tube.
Collect 4–5 drops of blood [or 40–50 microliters of blood] from one mouse by cheek stick using an 18g needle, or similar IACUC-approved blood draw procedure.
Add 2 mL of RBC lysis buffer. Mix and incubate for 2 minutes at room temperature.
Add 2 mL of staining buffer. Centrifuge at 1600 RPM for 5 minutes and discard supernatant.
Repeat steps 3 and 4.
Resuspend cell pellet in 100 microliters of staining buffer and transfer to a 96 well plate.
Centrifuge at 2000 RPM for 2 minutes and discard supernatant.
Resuspend cell pellet in 50 microliters of staining buffer containing fluorescent antibody cocktail (see Table 1). Incubate for 30 minutes at 40°C.
Add 150ul of staining buffer and centrifuge at 2000 RPM for 2 minutes then discard supernatant.
Wash sample with 200 microliters staining buffer and centrifuge at 2000 RPM for 2 minutes. Repeat.
Fix sample by resuspending the cell pellet in 100 microliters of 2% paraformaldehyde or equivalent fixative.
When ready to read on the flow cytometer, wash sample with 200 microliters staining buffer and centrifuge. Resuspend cell pellet in 250 microliters of staining buffer, transfer to a cluster tube and run the sample. Analyze data using appropriate software (FlowJo or equivalent).
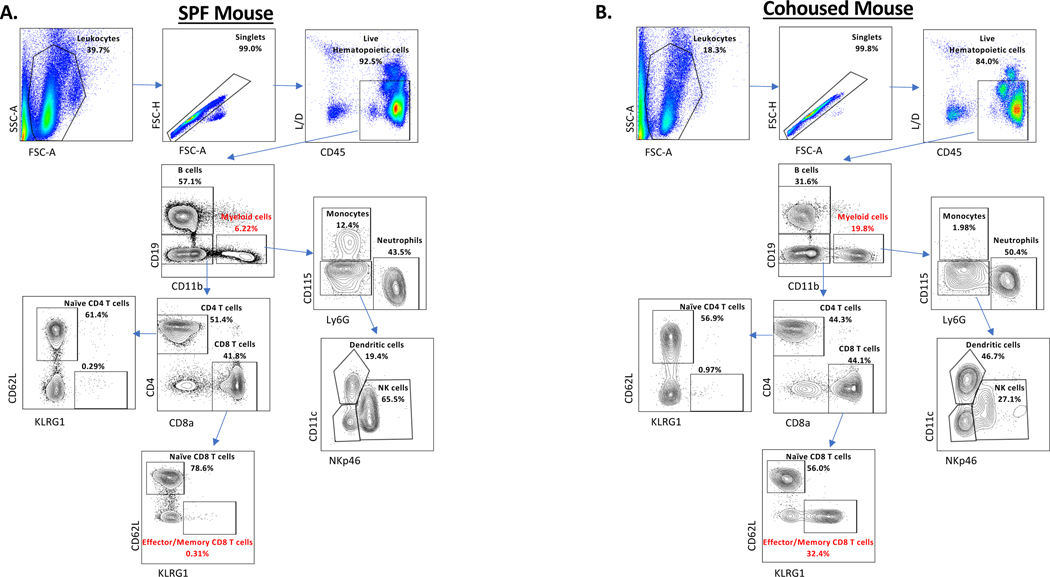
Results of antibody staining on mice exposed to diverse natural pathogens are highly variable due to the method of exposure, although they do follow a loose pattern when compared to the results of antibody staining on SPF mice. The most prominent difference is the drastic increase in CD8 T cells most notably CD44Hi KLRG1+ cells as well as an increase in CD62L negative CD8 T cells. Additional variations are seen in most cell populations but are not as consistent as the T-cell compartment and require age-matched SPF controls to fully characterize differences. Example data is shown in Figure 1 and Table 2.
Figure 1. Flow cytometry gating strategy for assessing immune cells after cohousing.
Blood was collected from either (A) a female C57BL6 SPF mouse or (B) a female C57BL6 mouse cohoused for 30 days with a pet store mouse. Blood was processed and stained with fluorescent antibodies as described in Support Protocol 1. Frequencies listed represent percentages of the parent population. Populations in red identify the largest consistently altered populations between SPF and cohoused mice. These and other expected differences in the frequency of antibody stained cells are listed in Table 2.
Table 2.
Relative Frequency of Antibody Staining Cells
| SPF | Cohoused | Fomite | Pet Store | |
|---|---|---|---|---|
| CD8 | + | ++ | ++ | + |
| CD4 | ++ | + | +++ | +++ |
| CD44 | + | ++++ | ++++ | +++ |
| CD62L | ++++ | +++ | ++ | ++ |
| KLRG1 | + | ++++ | ++++ | +++ |
| Ly6C Hi | + | + | ++ | + |
| CD11b | + | +++ | ++++ | +++ |
| Ly6G | + | ++ | +++ | ++ |
| CD115 | ++ | + | +++ | + |
| NKp46 | + | + | + | ++ |
BASIC PROTOCOL 2
EXPOSURE OF LABORATORY STRAINS OF MICE TO FOMITE BEDDING
Fomite bedding collected from the cages in which pet store mouse are housed can also be used to introduce a mixture of microbes to laboratory SPF mice. This method has the advantage that both male and female cohorts of mice can be used. Additionally, fewer pet store mice are required, as a cage of 4–5 mice can produce useful fomite bedding for 10–12 cages of research mice (50–60 mice in total) for several weeks. Another benefit is that a homogeneous mixture of bedding from several pet store mice can be spread among multiple cages of experimental mice. However, fomite exposure reliably transmits a reduced number of microbes as compared to cohousing and thus can lead to less impressive immune system alterations(Hamilton et al., 2020). It is often useful to use this protocol in conjunction with the cohousing method when possible, so that the investigator can determine the best choice for their specific experimental goals.
Materials:
6–12 week old female or male research mouse strain of interest (C57BL/6, BALB/c, etc.) purchased from vendor (e.g. The Jackson Laboratory, Charles River, Taconic)
Female or male pet store mice of any coat color or age purchased from commercial vendors
Autoclavable mouse cage bottoms: standard size (Ancare, N10) or large size (10 ½” x 19” x 6 1/8”, Ancare, N40) with matching wire lid and microfilter top
Water bottles (Ancare, reduced height bottle, 16 oz.) with Drinking Tube (Ancare, OT-199) and rubber stopper (Ancare, #8.5R)
Irradiated corncob bedding (Teklad, 7902)
Mouse chow (non-autoclaved) (Teklad, 2018)
Polypropylene Beaker (Nalgene, 76038)
Obtain all mice from appropriate vendors.
House 4 male or 5 female pet store mice in a standard mouse cage (or 8 male or 9 female pet store mice in a large cage) for a minimum of 5–7 days to generate fomite bedding.
Using a plastic beaker, transfer 100mls/100cm3 of the soiled bedding from the pet store mouse cage into a standard cage or 200mls/200cm3 into a large cage containing research mice. Maintain an equivalent number of research mice in SPF housing as age- matched controls.
Fresh fomite bedding should be added weekly to further encourage microbial exposure.
4. Provide food and water ad libitum.
5. Perform bedding changes weekly or as standard for the housing facility.
During the first two weeks, mice may show signs of mild illness (hunched posture, ruffled fur) but most immunocompetent mice will not succumb to fomite exposure. The survival of immunodeficient or genetically altered strains will need to be determined on a case-by-case basis.
6. After 30 days, mice can be surveyed for serological antibodies against common microbes carried by mice. This may be accomplished by outsourcing to a vendor such as Charles River (Mouse PRIA panels, FELASA Complete) or assays can be developed within the laboratory. Additional evidence of microbial exposure can be collected using flow cytometry to assess immune cell alterations (see Support Protocol 1).
If at this stage poor microbial transmission or impact on the immune system is observed, fresh pet store mice should be obtained to produce a new source of fomite bedding.
7. After 60 days, mice can be surveyed again by serological and immune cell assays (as in step 5) and then used in experimental protocols as ‘dirty mice’. Comparisons should always be made to an age-matched SPF control group to reduce variability and highlight small but critical differences (see Table 2).
COMMENTARY
BACKGROUND INFORMATION
The observation that microbial exposure has long term effects on the immune system is not new. However, the pressure to limit unknown variables and generate data that could be easily reproduced pushed the development of increasingly sterile environments in which to house laboratory mice. This led to the concept of ‘specific pathogen free’ mice that has been strictly adhered to by most research institutions for decades. However, growing evidence in mice and humans also continued to support that both chronic pathogen infection or commensal organisms making up the microbiome, could substantially influence the host response to new insults (injury, infection, vaccination, etc.). Since 2016, several methods to generate mice with diverse microbial experience (generally known as ‘dirty mice’) have been established. These approaches include: sequential infections with multiple pathogens(Reese et al., 2016), introduction of the microbiome from wild mice(Rosshart et al., 2019; Rosshart et al., 2017; Wilmore et al., 2018), housing in outdoor enclosures (rewilding)(Leung et al., 2018; Lin et al., 2020; Yeung et al., 2020), and cohousing with pet store mice or exposure to their fomite bedding as described here (Beura et al., 2016; Huggins et al., 2019). Each of these techniques offers a different approach to introduce a diverse milieu of commensals and/or pathogens to laboratory mice, and all publications using these methods have noted alterations to the cellular makeup and function of the immune system following exposure. Some have concluded that the dirty mouse immune system is more akin to the human one(Beura et al., 2016), and provided evidence that these mouse models may be a powerful tool to increase the successful translation of pre-clinical studies(Rosshart et al., 2019). The method chosen by an investigator will depend on the particular questions being addressed and the type of animal housing permissible at their institution.
CRITICAL PARAMETERS
Pet Store Mouse Microbial Burden
The number of microbes carried by the pet store mouse is the most critical factor related to the success of transmission to laboratory mice, and different vendors may sell mice with varying degrees of microbial burden. Thus, multiple sources of mice may need to be pursued. Pre- screening of pet shop mice for either serological antibodies against common mouse pathogens or the presence of microbes by PCR can assist in identifying microbes that are present and likely to be transferred to other mice. However, empirical experience suggests delaying cohousing with research mice to complete testing decreases the efficiency of microbe transfer: we find that pet store mice are most effective at transferring microbes within 4 days of purchase from the vendor. As most pet store vendors house mice until sold, it is ideal to purchase as close to the date of delivery to the vendor as possible. The issue of timing is likely related to when pet store mice are initially exposed to the microbial milieu themselves, and the kinetics of active infection. Some organisms are characterized as chronically shed from mice (e.g. mouse hepatitis virus, mouse cytomegalovirus), but others will undergo more acute kinetics followed by clearance from the host.
Housing of Pet Store Mice
In addition to the regular health monitoring and surveillance that laboratory mice undergo, SPF colonies are further maintained and protected by other biocontainment measures. Although these specific practices are dependent on the institution, the goal of excluding pathogens permeates most policies and procedures in SPF vivaria. The type of housing and sanitation, personal protective equipment and aseptic techniques, importation and movement of animals, and even the screening of biologics are dictated in part by the goal of protecting the animal colony from unwanted microorganisms(White, Anderson, Geistfeld, & Martin, 1998).
Only after recognizing the great lengths that institutions take to maintain SPF conditions in their vivaria, can the challenges of housing dirty mice be understood. The main concern of dirty mice is the introduction of undesirable pathogens. The pathogen burden of the dirty mice will depend on the source, but based on published serologic and PCR profiles from various pet store mice, infection with microorganisms that are often excluded in SPF colonies are prevalent (e.g. mouse hepatitis virus, Mycoplasma pulmonis, pinworms)(Beura et al., 2016; Hamilton et al., 2020; Huggins et al., 2019). The desired presence of these pathogens is incompatible with the goal of SPF facilities. Therefore, the housing and husbandry of pet store mice present a logistical challenge for vivaria where SPF colonies are maintained. Considerations in determining appropriate housing of pet store mice include the facility and equipment, sanitation and disposal, airflow, personnel traffic, and risk to the rest of the animal colony. Our institution has overcome this hurdle by housing pet store mice in a biosafety level-3 (BSL-3) containment facility (Beura et al., 2016; Huggins et al., 2019). Physically separating SPF colonies from dirty mice, combined with the use of dedicated staff and equipment may also be sufficient to prevent colony cross- contamination. Housing and care of pet store mice ultimately depend on the institution and the resources and facilities that it has at its disposal. Success depends on collaborating with institutional laboratory animal management personnel to determine the best option given the risks and the resources available.
TROUBLESHOOTING
Pet store mice which are free of most common mouse microbes will not be useful to generate ‘dirty mice’. If low microbial transmission (lack of serological response) or lack of immune cell changes are observed, an alternative source of pet store mice should be pursued. In contrast, if laboratory mouse strains are found to overwhelmingly succumb to cohousing, fomite bedding can be used as an alternative method. Fomite exposure rarely induces death in immunocompetent mice and transfers a lower microbial burden(Hamilton et al., 2020). The replacement of this bedding (or the amount of bedding) can also be modified to reduce (or increase) potential exposure as desired.
ANTICIPATED RESULTS
Immunocompetent mice cohoused with pet store mice or housed on fomite bedding, will generate antibodies to multiple microbes that can be detected in the serum(Hamilton et al., 2020). Typically, testing is performed at least 1 month post-exposure to pet store mice or fomite bedding, but antibodies may be detected earlier. The most commonly transmitted microbes detected by serological analysis are: Mouse hepatitis virus, Mouse norovirus, Theiler’s Murine Encephalomyelitis Virus, and Mycoplasma pulmonis (Beura et al., 2016; Huggins et al., 2019). Mycoplasma pulmonis is only transmitted via the cohousing method in our experience (Hamilton et al., 2020). The most dramatic immune cell change observed with diverse microbial exposure is the increased frequency of activated/memory CD8+ T cells in both the blood and tissues (Figure 1) (Beura et al., 2016; Beura et al., 2018; Huggins et al., 2019). This will start to be apparent in the blood after 5–7 days of cohousing, and will be maintained long term at an elevated frequency as compared to SPF mice (Beura et al., 2016; Huggins et al., 2019).
Additional common observations include an increased frequency of neutrophils and/or monocytes in the blood and decreases in NK cells (Huggins et al., 2019). Increases in CD4+ T cell subsets and switched B cells are also common in lymphoid tissues(Beura et al., 2016).
Serological testing provides basic characterization of the microbial experience of dirty mice, but is dependent on the robust generation of an antibody response in the infected animal. Additional evidence can be obtained by pursuing other assays such as: PCR analysis, tissue histology, microbiome analysis(Huggins et al., 2019), or RNA sequencing. The major limitation for pursuing these assays is cost, but if that can be overcome, a wealth of information about the microbes inhabiting dirty mice can be obtained.
With both of the protocols outlined, the investigator should be prepared to observe increased variability compared to expectations based on working with SPF mice. Microbial burden and the resulting immune system changes will vary from cage to cage and even between mice within the same cage. The number and diversity of microbes transmitted, the route and kinetics of exposure, etc. are all uncontrolled variables. However, reproducible biological differences and statistical significance can still be achieved without the need for excessive numbers of animals (typically 5–10 dirty mice/group).
TIME CONSIDERATIONS
Once appropriate institutional approvals are obtained for housing ‘dirty mouse’ and all mice have been purchased, cohousing can begin immediately. A cage of pet store mice will produce useful fomite bedding after 5–7 days. Microbial transmission to research mice occurs over the ensuing days-weeks, with mice typically considered ‘dirty’ and exhibiting a stable immune phenotype after two months of cohousing or fomite bedding exposure.
SIGNIFICANCE STATEMENT:
Diverse microbial burden can be physiologically transferred from pet store mice to research mice through either cohousing or fomite bedding exposure, inducing long term changes to the immune system.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (AI116678 to SEH). We gratefully acknowledge the pioneering work of Drs. David Masopust, Stephen Jameson, and Lalit Beura in the initial generation and characterization of ‘dirty mice’ that led to this protocol. We also thank the essential support provided by research animal resources and BSL-3 facility staff at the University of Minnesota.
LITERATURE CITED
- Berg G, Rybakova D, Fischer D, Cernava T, Vergès MC, Charles T, . . . Schloter M. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome, 8(1), 103. doi: 10.1186/s40168-020-00875-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Hamilton SE, Bi K, Schenkel JM, Odumade OA, Casey KA, . . . Masopust D. (2016). Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature, 532(7600), 512–516. doi: 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura LK, Wijeyesinghe S, Thompson EA, Macchietto MG, Rosato PC, Pierson MJ, . . . Masopust D. (2018). T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity, 48(2), 327–338.e325. doi: 10.1016/j.immuni.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin CL (2006). Microbial considerations in genetically engineered mouse research. ILAR J, 47(2), 141–155. doi: 10.1093/ilar.47.2.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton SE, Badovinac VP, Beura LK, Pierson M, Jameson SC, Masopust D, & Griffith TS (2020). New Insights into the Immune System Using Dirty Mice. J Immunol, 205(1), 3–11. doi: 10.4049/jimmunol.2000171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins MA, Sjaastad FV, Pierson M, Kucaba TA, Swanson W, Staley C, . . . Hamilton SE (2019). Microbial Exposure Enhances Immunity to Pathogens Recognized by TLR2 but Increases Susceptibility to Cytokine Storm through TLR4 Sensitization. Cell Rep, 28(7), 1729–1743 e1725. doi: 10.1016/j.celrep.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Japp AS, Hoffmann K, Schlickeiser S, Glauben R, Nikolaou C, Maecker HT, . . . Thiel A. (2017). Wild immunology assessed by multidimensional mass cytometry. Cytometry A, 91(1), 85–95. doi: 10.1002/cyto.a.22906 [DOI] [PubMed] [Google Scholar]
- LANE-PETTER W. (1962). Provision of pathogen-free animals. Proc R Soc Med, 55, 253–256. [PubMed] [Google Scholar]
- Leung JM, Budischak SA, Chung The H, Hansen C, Bowcutt R, Neill R, . . . Graham AL. (2018). Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol, 16(3), e2004108. doi: 10.1371/journal.pbio.2004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JD, Devlin JC, Yeung F, McCauley C, Leung JM, Chen YH, . . . Loke P. (2020). Rewilding Nod2 and Atg16l1 Mutant Mice Uncovers Genetic and Environmental Contributions to Microbial Responses and Immune Cell Composition. Cell Host Microbe, 27(5), 830–840.e834. doi: 10.1016/j.chom.2020.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mähler Convenor M, Berard M, Feinstein R, Gallagher A, Illgen-Wilcke B, Pritchett-Corning K, . . . rabbits F. w. g. o. r. o. g. f. h. m. o. r. a. (2014). FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab Anim, 48(3), 178–192. doi: 10.1177/0023677213516312 [DOI] [PubMed] [Google Scholar]
- Reese TA, Bi K, Kambal A, Filali-Mouhim A, Beura LK, Bürger MC, . . . Virgin HW. (2016). Sequential Infection with Common Pathogens Promotes Human-like Immune Gene Expression and Altered Vaccine Response. Cell Host Microbe, 19(5), 713–719. doi: 10.1016/j.chom.2016.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, . . . Rehermann B. (2019). Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science, 365(6452). doi: 10.1126/science.aaw4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Vassallo BG, Angeletti D, Hutchinson DS, Morgan AP, Takeda K, . . . Rehermann B. (2017). Wild Mouse Gut Microbiota Promotes Host Fitness and Improves Disease Resistance. Cell, 171(5), 1015–1028.e1013. doi: 10.1016/j.cell.2017.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White WJ, Anderson LC, Geistfeld J, & Martin DG (1998). Current Strategies for Controlling/Eliminating Opportunistic Microorganisms. ILAR J, 39(4), 291–305. doi: 10.1093/ilar.39.4.291 [DOI] [PubMed] [Google Scholar]
- Wilmore JR, Gaudette BT, Gomez Atria D, Hashemi T, Jones DD, Gardner CA, . . . Allman D. (2018). Commensal Microbes Induce Serum IgA Responses that Protect against Polymicrobial Sepsis. Cell Host Microbe, 23(3), 302–311.e303. doi: 10.1016/j.chom.2018.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F, Chen YH, Lin JD, Leung JM, McCauley C, Devlin JC, . . . Cadwell K. (2020). Altered Immunity of Laboratory Mice in the Natural Environment Is Associated with Fungal Colonization. Cell Host Microbe, 27(5), 809–822.e806. doi: 10.1016/j.chom.2020.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]