Abstract
The ongoing COVID-19 pandemic has highlighted the need to incorporate pathogen genomics for enhanced disease surveillance and outbreak management in Africa. The genomics of SARS-CoV-2 has been instrumental to the timely development of diagnostics and vaccines and in elucidating transmission dynamics. Global disease control programmes, including those for tuberculosis, malaria, HIV, foodborne pathogens, and antimicrobial resistance, also recommend genomics-based surveillance as an integral strategy towards control and elimination of these diseases. Despite the potential benefits, capacity remains low for many public health programmes in Africa. The COVID-19 pandemic presents an opportunity to reassess and strengthen surveillance systems and potentially integrate emerging technologies for preparedness of future epidemics and control of endemic diseases. We discuss opportunities and challenges for integrating pathogen genomics into public health surveillance systems in Africa. Improving accessibility through the creation of functional continent-wide networks, building multipathogen sequencing cores, training a critical mass of local experts, development of standards and policies to facilitate best practices for data sharing, and establishing a community of practice of genomics experts are all needed to use genomics for improved disease surveillance in Africa. Coordination and leadership are also crucial, which the Africa Centres for Disease Control and Prevention seeks to provide through its institute for pathogen genomics.
Introduction
The ongoing COVID-19 pandemic continues to highlight the crucial need to strengthen systems for epidemic preparedness and surveillance in Africa, including the need to build genomic and digital surveillance capacity,1 biobanks,2 and local diagnostics and therapeutics manufacturing capacity.3 Over the past decade, Africa has grappled with two Ebola virus epidemics, with substantial mortality and economic losses,4, 5 and continues to be greatly impacted by the COVID-19 pandemic. Overall, an estimated 140 disease outbreaks are reported annually within the continent.6 These outbreaks are in addition to endemic infectious disease threats, which altogether account for at least 35% of the 10 million deaths reported on the continent annually and the loss of more than 227 million years of healthy life.7 Antimicrobial resistance is also a serious challenge that is projected to result in millions of deaths and hard-to-treat infections, and an increased burden on health-care systems.8 Prevention, control, and elimination of emerging, re-emerging, and endemic infections including antimicrobial resistance is thus a crucial goal of national disease control programmes in Africa.9 However, attainment of this goal is a daunting task given the weak infrastructure and restricted capacity and resources to support surveillance, preparedness, control, and prevention of infectious diseases.10
The rapid innovation in sequencing technologies has led to the development of robust next-generation sequencing (NGS) equipment with the ability for high pathogen resolution at increasingly affordable prices. This development subsequently led to the incorporation of pathogen genomics in disease surveillance systems in high-income countries, allowing for timely and in-depth pathogen characterisation leading to targeted and effective control of disease threats.11, 12, 13 In the COVID-19 pandemic, for example, genomics has been used for timely development of diagnostics,14 guiding vaccine development, monitoring for viral evolution that affects diagnosis,15 transmissibility,16 and virulence,17 elucidating transmission dynamics,18, 19 supporting timely control of nosocomial outbreaks,18, 19 and the overall assessment of the effectiveness of infection prevention and control measures.19 More recently, genomics-based surveillance has been cited as an important tool to identify and track the spread of new concerning variants of SARS-CoV-2, such as B.1.1.7 (N501Y) and B.1.351 (N501Y.V2), which have high transmission rates and the potential to affect COVID-19 medical countermeasures.20
However, NGS use in Africa is sparse, despite the greater need to control the high burden of infectious diseases. In this Personal View, we discuss the potential applications, opportunities, and challenges of integrating pathogen genomics into existing public health surveillance systems in Africa.
Genomics use cases for improving public health surveillance in Africa
Pathogen genomics has the potential to transform public health surveillance by improving outbreak detection and investigation, tracking transmission routes and networks, monitoring genetic changes that impact pathogenicity, diagnostics, therapeutics, and vaccines, and assessing the effectiveness of policies and interventions.13
Recommended and well established genomics use cases
WHO guidance for global surveillance of HIV drug resistance,21 tuberculosis drug resistance,22 malaria,23 antimicrobial resistance,24 vaccine-preventable diseases,25 and foodborne pathogens26 already recommend or encourage the use of NGS as an additional surveillance tool (table 1 ).
Table 1.
WHO-recommended and other pathogen genomics use cases for disease control programmes
| Disease control programme | Purpose | Pending concerns | |
|---|---|---|---|
| HIV drug resistance | HIV | Guiding treatment policy to ensure sustained effectiveness of restricted regimens and sustained progress towards achieving epidemic control by 2030; potential for use in clinical management, especially in heavily treated patients with few therapy options | Methods depend mainly on Sanger-based sequencing; standardisation of NGS methods still ongoing, including establishment of clinically significant thresholds for minority variants and standardisation of bioinformatics pipelines and NGS-based EQA panels or programmes |
| Tuberculosis drug resistance | Tuberculosis | Guiding treatment policy; forecasting for second-line drugs; overall prevention efforts for minimising emergence and spread of drug resistance; individualised resistance test is essential for guiding clinical management, especially for people with multidrug-resistant or extensively drug-resistant tuberculosis | Optimisation and standardisation of laboratory methods and bioinformatics tools ongoing; need for more sensitive methods to allow for non-culture WGS-based approaches; need for optimisation of the sensitivity of resistance detection for certain drugs |
| Malaria drug resistance | Malaria | Ensuring sustained effectiveness of the few available regimens | Need to identify clinically significant resistant markers, and establish sampling strategies, standardisation of methods, and guidelines on use of genomic data for policy changes |
| Resistance to diagnostics (target sites: Pfhrp2/3 deletions) | Malaria | Guiding policy and choice of rapid diagnostic tests, ensuring their continuous effectiveness, and minimising misdiagnoses | Need to establish sampling strategies and standardisation of methods |
| Vector or insecticide resistance | Malaria | Guiding policy on vector control and ensuring continuous effectiveness of insecticides | Need to identify resistant markers and reference genome for the vectors; need to develop appropriate spatial sampling strategy and standardisation of methods |
| Transmission dynamics and elimination surveillance | Malaria | Quantification of malaria importation risk useful in elimination surveillance, determining transmission chains in elimination surveillance for targeted interventions, and characterising changing transmission intensity potentially guiding control measures | Need to standardise methods; more local, regional, and global data are needed for adequate resolution of imported vs indigenous cases and the characterisation of changes in parasite population structure |
| Vector dynamics | Malaria | Monitoring changes in vector species and their effect on transmission and monitoring effectiveness of mosquito gene-drive interventions | Need for a database mapping local vector species, entomology capacity building, development of appropriate spatial sampling strategy, and standardisation of methods |
| Antimicrobial resistance | Antimicrobial resistance | Monitoring antimicrobial resistance emergence, prevention, and transmission dynamics; identifying clusters and mechanism of resistance; reconstruction of transmission; outbreak management; vital for ensuring continuous effectiveness of antimicrobial drugs | Need to develop a more comprehensive database or set of algorithms for genotypic-to-phenotypic profiling of more organisms; need to identify resistance mechanisms for more organisms |
| Outbreak management and surveillance | Foodborne pathogens | Outbreak management; tracing transmission routes and networks; identifying contamination sources; surveillance and effective outbreak management ensuring food safety | Need to standardise methods; quality assurance or quality control needs to be done, including EQA |
| Vaccine efficacy monitoring, transmission dynamics, and elimination surveillance | Vaccine-preventable diseases (eg, polio, rubella, measles, and influenza) | Monitoring the effectiveness and impact of vaccines; outbreak management; identifying new strains, including imported cases, vaccine escape variants, and vaccine-derived variants; inform vaccine development for pathogens such as influenza | Need to standardise methods; quality assurance or quality control needs to be done, including EQA |
EQA=external quality assessment. NGS=next-generation sequencing. Pfhrp2=Plasmodium falciparum histidine-rich protein 2. WGS=whole genome sequencing.
The WHO Drug Resistance Report 2019 showed that one in ten adults with HIV globally harbour a drug-resistant strain before treatment initiation, as do one in two children infected with HIV in Africa.27 Equally, tuberculosis drug resistance is a major public health problem in Africa, which hinders effective tuberculosis control and prevention by national programmes. In 2018, 7·3% of the 1·06 million cases of tuberculosis reported in Africa had resistance to rifampicin or were cases of multidrug-resistant tuberculosis (resistant to rifampicin and isoniazid).28 Managing drug-resistant tuberculosis is challenging, leading to a high mortality rate of about 18%.28 Overall, the emergence of drug-resistant strains is challenging in Africa, which has comparatively high levels of antimicrobial resistance despite having fewer affordable antimicrobial drugs relative to other regions. With few drug options, WHO recommends genomic surveillance for antimicrobial resistance as part of the global strategy for infectious disease control.22, 23, 24, 29, 30
Sub-Saharan Africa accounts for 93% of the 228 million malaria cases and 94% of the 405 000 annual malaria deaths worldwide.31 To support control and elimination efforts, WHO's global malaria programme recommends the following genomics use cases: (1) monitoring changes in the frequency of molecular markers for drug resistance over time and space, (2) monitoring the effectiveness of rapid diagnostic tests, (3) assessing transmission dynamics to improve classification of cases as either indigenous or imported, (4) monitoring the effectiveness of insecticide vector-control measures, and (5) assessing vector dynamics including the emergence of new species.23
Africa also bears disproportionately high incidence and mortality from foodborne diseases, leading to nearly 1200 disability-adjusted life years (DALYs) per 100 000 people.32 WHO recommends the use of NGS as a crucial surveillance tool for detection of foodborne pathogen outbreaks, and for case definition, ascertainment, and source attribution.26
With 30 million yearly infections among children in Africa younger than 5 years, vaccine-preventable diseases, such as pneumonia and measles, pose a considerable threat to the continent, with the potential to cause nearly half a million deaths annually.33 Genomic surveillance is considered an essential tool in elimination strategies. This kind of surveillance can monitor the effectiveness and impact of vaccines by assessing expansion, decline, or extinction of specific lineages and identify gaps in the control efforts, including potential reservoirs, outbreak investigation, and new strains (eg, imported cases, vaccine escape variants, vaccine-derived variants), as well as inform on vaccine development for pathogens such as influenza. Currently, WHO's global surveillance of vaccine-preventable diseases encourages the use of genomic surveillance for such diseases as polio, measles, rubella, influenza, and invasive bacterial disease.25
Genomics use cases for disease surveillance and control in Africa
Besides the aforementioned use cases, incorporating genomics into surveillance of emerging and re-emerging infectious diseases is also important, especially in Africa. In the past decade, the continent has faced two Ebola epidemics that have caused around 13 000 direct deaths and more than 20 000 indirect deaths.33 During the COVID-19 pandemic, a total of 3·52 million cases have been reported in Africa, including around 89 000 deaths as of Jan 29, 2021, and a substantial number of indirect deaths are projected.34, 35 More than ever, surveillance systems for prevention, preparedness, and control of emerging and re-emerging infections need to be strengthened. Adding pathogen genomics tools to the existing strategies will be beneficial for early detection and prevention of zoonotic diseases before they jump to humans, improve outbreak management by rapidly identifying the causative agent, and facilitate the designing of diagnostics and preventive, therapeutic, and other countermeasures, while monitoring their effectiveness. Pathogen genomics can also help identify gaps in infection prevention and control measures, improve assessment of transmission dynamics, identify networks to aid in tracing potential contacts, assess the spatial and temporal distribution of the epidemic, and identify the reproduction rate (R0) and missing transmission chains, which are especially important early in the epidemic.13, 18, 36, 37, 38, 39
In Africa, genomic surveillance is increasingly being adopted for prevention and control of emerging infections.18, 36, 38, 39 Notable instances include the 2018 outbreak of Lassa fever virus in Nigeria39 and the Ebola virus outbreaks in the Democratic Republic of the Congo (2018–20) and west Africa (2014–16), in which genomic data were used in combination with other data to elucidate transmission dynamics, including the role played by survivors in sustaining the epidemic by acting as carriers, and assess the spatiotemporal aspects of the epidemic and the effectiveness of diagnostic tests and vaccines.36, 38 More recently, genomic data are being used to track the evolution of SARS-CoV-2 and were notably used to identify and track nosocomial infection spread in South Africa,18 sources of introduction and lineages,40 as well as the emergence and spread of the more transmissible B.1.351 variant and other variants of interest with potential to affect disease severity and countermeasures such as vaccines.20, 41
Genomic surveillance might also have a role in elimination surveillance of both endemic and neglected tropical diseases (table 2 ). For example, genomics has already been incorporated into routine surveillance strategies for tuberculosis and HIV control in some high-income countries.22, 42 For HIV, genomic data have the potential to guide targeted prevention efforts through characterising transmission dynamics and identifying unrecognised clusters and untreated individuals who are probable drivers of HIV transmission.42, 43 The Phylogenetics and Networks for Generalized Epidemics in Africa (PANGEA) consortium is already implementing this strategy on the continent.43 Equally, real-time molecular surveillance of tuberculosis enables increased accuracy in the identification of resistance hotspots and transmission clusters, which overall maximises prevention efforts.22 Adoption of routine molecular-based surveillance, as is the practice in high-income countries, might potentially improve case detection and targeted interventions for these endemic diseases.42
Table 2.
Potential pathogen genomics use cases for disease control programmes in Africa
| Disease control programme | Purpose | Pending concerns | |
|---|---|---|---|
| Outbreak management and surveillance | Emerging and re-emerging infections | Rapid identification of causative agent; design and effectiveness monitoring of diagnostics, therapeutics, preventive, and other countermeasures; determination of reproductive rate and transmission dynamics (especially early in the epidemic); prevention, surveillance, and management of outbreaks | Need for guidelines on standardised sampling strategies and sequencing and analysis methods, including EQA programmes; need for development of and more widespread access to more simple-to-use tools for advanced analysis as well as a comprehensive database that includes regional data for effective identification and assessment of transmission dynamics |
| Molecular network surveillance | HIV | Elucidating transmission dynamics in high-risk groups for targeted prevention efforts; identifying undetected clusters for targeted testing, treatment, improvement in viral suppression, and overall prevention efforts | Need for guidelines to support routine use of molecular network surveillance and standardisation of methods, including EQA programmes; need for development of and more widespread access to simple-to-use tools for advanced analysis |
| Molecular network surveillance | Tuberculosis | Identifying clusters and potential hotspots for tuberculosis and drug-resistant tuberculosis, thus enabling targeted prevention efforts | Need for guidelines to support routine use of molecular network surveillance and standardisation of methods, including EQA programmes; need for development of and more widespread access to simple-to-use tools for advanced analysis |
| Outbreak management and surveillance, and elimination surveillance | Neglected tropical diseases | Outbreak management; tracking eradication and emergence of new strains including imported cases; identifying evolution history and transmission patterns; supporting vaccine development | Need for guidelines on standardised sampling strategies and sequencing and analysis methods, including EQA programmes; need for development of and more widespread access to simple-to-use tools for advanced analysis as well as a comprehensive database that includes regional data for effective identification and assessment of transmission dynamics |
EQA=external quality assessment.
Africa has nearly 40% of the 1·5 billion people globally who are most at risk for neglected tropical diseases.44 Genomic surveillance can potentially also be used for elimination surveillance, outbreak management, and for informing the development of vaccines and diagnostics.
Challenges and solutions to genomic disease surveillance in Africa
We and others have previously reviewed the strengths and challenges of the different genomic assays—ie, Sanger-based assays, point mutation assays, and NGS methods.45, 46, 47, 48 Most countries in Africa rely on Sanger-based assays,49 but NGS is frequently being adopted for various use cases. However, because of the comparative advantages in terms of cost, potential wide application, and depth of information obtainable with NGS-based methods,45, 46, 47, 48 we have limited the discussions in this Personal View to NGS.
Infrastructure and human capacity building
Sequencing, data computation, and storage infrastructure
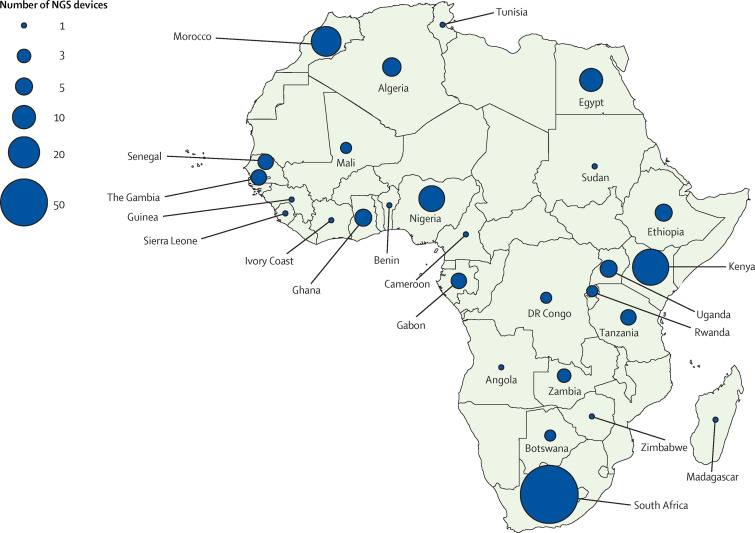
A landscape assessment of sequencing capacity within Africa reveals that capacity is sparse and concentrated in a few countries (figure 1 ). Up to 71% of next-generation sequencers are concentrated in five countries: South Africa (n=79; 38%), Kenya (n=28; 14%), Nigeria (n=13; 6%), Morocco (n=18; 9%), and Egypt (n=10; 5%). However, most of the capacity (144 [70%] of 206) is found outside of national public health institutes (appendix). This distribution highlights the need to increase capacity in national public health institutes and create functional networks with academic and research facilities and between countries.
Figure 1.
Next-generation sequencing capacity in Africa
The circles show the number and distribution of NGS devices on the continent. Data was obtained primarily from manufacturers of the most used NGS equipment—ie, Illumina (Illumina, CA, USA), Oxford Nanopore (Oxford Nanopore Technologies, Oxford, UK), and Ion Torrent (Thermo Fisher Scientifics, MA, USA). However, these data might not be conclusive as they might not include equipment donated or bought outside Africa. Overall, we identified a total of 206 NGS devices in Africa, which includes South Africa (79), Kenya (28), Morocco (18), Nigeria (13), Egypt (10), Algeria (6), Ethiopia (5), Ghana (5), Uganda (5), Gabon (4), The Gambia (4), Senegal (4), Tanzania (4), Zambia (3), Botswana (2), Mali (2), the Democratic Republic of the Congo (2), Rwanda (2), and one each for Angola, Benin, Cameroon, Cote d'Ivoire, Guinea, Madagascar, Sierra Leone, Sudan, Tunisia, and Zimbabwe. NGS=next-generation sequencing.
Guidance for setting up sequencing infrastructure has been discussed elsewhere.22, 26, 46, 50 In brief, the initial capital for establishing sequencing capacity is estimated at US$100 000–700 000, depending on the sequencing platform.26 In addition, substantial investments are needed for ancillary equipment for library preparation and quality control. The choice of platform is dependent on the type of application it is used for, taking into account read depth (throughput), read length, and error rates, and involves trade-offs in accuracy, efficiency, time, capital, cost of reagents, and computational requirements.
Guidance on computational infrastructure and analytical tools has also been reviewed elsewhere.22, 26, 46, 50 It is generally recommended for facilities with minimal bioinformatics expertise to consider automated, standardised, and validated end-to-end tools that could either be commercial or cloud-based. User-friendly automated platforms that rapidly process and analyse genomic data with simplified interpretation into actionable information are preferred.51, 52, 53 The choice of platform for a data storage and computing system (local, offsite, or cloud-based) will need to be evaluated against cost, privacy risk, internet reliability, ease of accessibility or retrieval, and other security measures. This evaluation should be done in agreement with national data ethics requirements. Due to restricted internet access in some settings, a hybrid approach of cloud-based and onsite computing infrastructure would be desirable. Overall, a comprehensive sequencing, data analysis, and archiving framework for Africa might be helpful, but countries could also use WHO and other international guidelines.22, 26, 46, 50
Skilled workforce
A wide cadre of experts is needed to support genomic surveillance, including molecular biologists for wet laboratory analysis, service bioinformaticians and molecular epidemiologists for data analysis and interpretation, field epidemiologists, disease specialists for interpretation of the generated data, and public health specialists for adoption of the findings into policy. Furthermore, there is a need to develop local expertise for equipment installation and maintenance to prevent long delays and additional costs accrued from overseas outsourcing.
Several programmes in Africa can be taken advantage of, including the Human Heredity and Health in Africa (H3Africa) bioinformatics network training and education programmes44 and their associated networks, African Genomic Medicine Training Initiative (AGMT),54 the Wellcome Trust-led Developing Excellence in Leadership and Genetics Training for Malaria Elimination in Sub-Saharan Africa (DELGEME),55 graduate programmes and short courses offered under the South African National Bioinformatics Institute (SANBI) and other universities,56, 57 training programmes by genomics institutions such as Kwazulu-Natal Research Innovation and Sequencing Platform (KRISP), the African Center of Excellence for Genomics of Infectious Disease (ACEGID),58 and Medical Research Council-The Gambia,59 among others.
Overall, these initiatives should be supplemented by mentorship programmes involving hands-on training, especially for wet laboratory analysis, to enhance competency. In the long run, it would be more desirable to integrate pathogen genomics training into existing curricula for molecular biologists, public health scientists, and biomedical engineers at college or university. Such programmes might use or borrow from the experience of other successful and comprehensive competency-based training programmes, such as the Field Epidemiology and Laboratory Training Program.60 However, sustaining the skilled workforce might be a challenge. Innovative incentives, such as opportunities for further training and involvement in scientific research, might be needed to retain the diverse genomics workforce in addition to continuous training strategies for both biomedical undergraduates and people at different career stages.
Integrating genomics into existing laboratory surveillance systems
Building multipathogen laboratories
We expect that only a few multipathogen core laboratories would be sufficient to support the different national disease control programmes. Compared with siloed pathogen-specific sequencing laboratories, multipathogen core sequencing units are advantageous in: (1) maximising the available capacity for both infrastructure (sequencing and computation) and human resources; (2) enhancing cost reductions in equipment maintenance costs; (3) potentially increasing multiplexing and subsequent sequencing cost reductions; (4) facilitating bulk procurement; (5) strengthening the surveillance of neglected diseases and pathogens; and (6) standardisation and streamlining of workflows across the different pathogens. A core facility offers the opportunity to pool resources for setting up and sustaining the genomics centres across national disease control programmes, which might in turn result in increased cohesion and integration of these programmes. However, integrated models could be too slow if the workforce is small, workload is high, or machines break down. A hybrid-integrated approach might be preferable, in which downstream processes are done in disease-specific laboratories and only the upstream steps, such as quality checks, sequencing, and analysis, are done at the core sequencing and bioinformatics facilities.
Laboratory networks and quality assurance systems
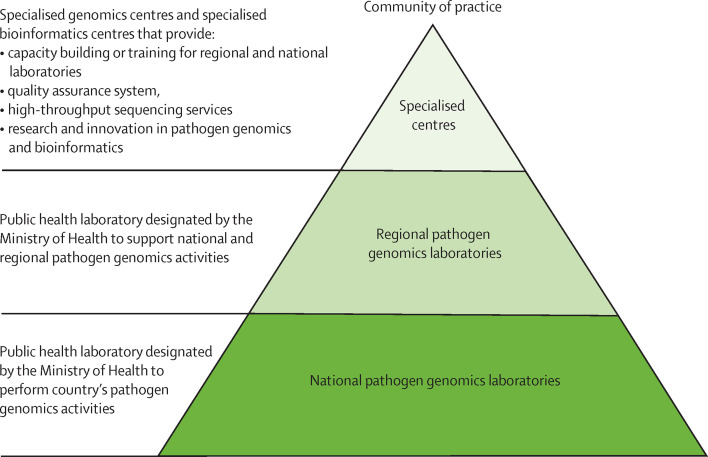
A network approach across Africa would help use the few resources available, especially for national public health institutes and disease control programmes that do not have in-country capacity. A potential strategy might involve a three-tiered pyramidal structure (figure 2 ), synonymous to that used in other successful WHO disease networks.25 At the base are national-level pathogen genomic laboratories, whose primary role is to support timely and accurate genomic surveillance. These laboratories are linked to regional laboratories that, in addition to serving as national laboratories, could also serve as regional reference centres to support national public health institutes and disease control programmes in countries with no sequencing capacity. In addition, these regional laboratories could serve to train and support the national referral laboratories for pathogen genomics. Finally, the specialised laboratories and bioinformatics centres could provide training, technical assistance, and quality assurance support to the national and regional laboratories, research innovative and more affordable methods, and serve as referral testing sites. Overall, the network could use and work jointly with other disease-specific or use-case-specific networks, such as the Malaria Genomics Epidemiology Network and the different WHO laboratory networks, particularly for the implementation of disease-specific standardised tools and the enhancement of quality assurance systems. In addition, the Africa Centres for Disease Control and Prevention (CDC) Regional Integrated Surveillance and Laboratory Network (RISLNET)61 could offer a platform to integrate the different networks, laboratories, and experts in a region for efficient services.
Figure 2.
Proposed pathogen genomics laboratory and bioinformatics network structure
Establishment of quality standards and continuous monitoring is essential to generate accurate data for policy making. In most cases, these services are provided by the disease-specific networks. However, there is still a need to develop or strengthen quality standards, external quality assessment (EQA) programmes, and frameworks for monitoring quality aspects for pathogens or applications not covered within the existing disease-specific networks. Quality assurance standards for NGS including EQA have been discussed elsewhere, with recommendations to incorporate both wet laboratory analysis for genomics and dry laboratory analysis for bioinformatics aspects.62, 63 Organisations such as the African Society of Laboratory Medicine (ASLM) and the African Society for Bioinformatics and Computational Biology (ASBCB) can work jointly with other stakeholders to establish standards for quality testing and analysis.
Enabling mechanisms
Policies and regulatory frameworks
Integrating genomic surveillance into public health will require enabling policies and regulatory mechanisms. These mechanisms can facilitate good practice in the collection, storage, and use of specimens, specimen transfer between countries, genomic archiving and biorepository storage and sharing, analysis and use of pathogen genomic data, among others. Although the benefits of rapid data sharing, especially for effective cross-border disease control and pandemic prevention, have long been recognised, gaps in this domain exist in Africa.64 A harmonised legal framework for the continent, developed through a consultative approach, that identifies and addresses local barriers is needed. This framework should preferably use existing international frameworks, such as that developed by the WHO Research and Development Blueprint meeting on pathogen genetic sequence data65 and the Nagoya Protocol on fair and equitable sharing of benefits arising from the sharing of genetic resources,66 and can be facilitated by continental bodies (eg, Africa CDC) with engagement from other groups, such as H3Africa, the Public Health Alliance for Genomic Epidemiology (PHA4GE), and the African Academy of Sciences, many of which have already been working on these aspects.
Cost-reduction mechanisms
The per-sample cost of NGS is generally high and prohibitive for routine use by national public health institutes or disease control programmes, but this cost can be greatly reduced through multiplexing (in which many samples that have been individually barcoded with a primer are pooled and sequenced together),67 which is favourable mainly with high-throughput facilities. However, the cost of library preparation remains high even with multiplexing,45 and pricing needs to be negotiated with manufacturers to make sequencing more affordable. In addition, negotiated pricing is also needed to address the disparity in equipment, maintenance, and reagent costs, which are substantially higher in Africa than in other areas, partly due to profit margins set by local companies and distributors, in addition to shipment and customs costs. Other approaches could include bulk procurement through regional or continental systems.
Community of practice
A community of experts in pathogen genomics, including public health and animal health experts, epidemiologists, genomics specialists, ethicists, biostatisticians, and bioinformaticians, needs to be established. The community will facilitate sharing of best practices, affordable wet laboratory genomic standards, statistical and bioinformatics tools, training materials, and innovations throughout Africa. Moreover, a community of practice will help foster collaborative efforts for control or eradication of common diseases by building mutual trust for data sharing, strengthening technical capacities, developing joint strategies, harmonising data reporting systems, and informal routine reporting of potential disease threats. Various networks, including the HIV PANGEA, H3Africa, and associated networks (eg, Pan-African Bioinformatics Network, ASBCB, African Society of Human Genetics, and ASLM) can be used in establishing this community.
Leadership and coordination
Establishing a functional pathogen genomics network that relies on shared resources and joint efforts for cross-border disease control and elimination within the continent will require an effective coordination mechanism. The Africa CDC was established as a continental body to support and work with all African countries to improve surveillance, outbreak management, and prevention of infectious diseases.68 Subsequently, the Africa CDC, through its pathogen genomics institute, has been working jointly with continental peers to provide leadership for integrating pathogen genomics into public health systems, as part of its mandate to improve disease surveillance within the continent. This collaboration includes mobilising resources for capacity building and facilitating SARS-CoV-2 sequencing, which is expected to be useful for targeted and timely control of epidemic resurgence in case of a second wave but also for monitoring the effectiveness of diagnostics, therapeutics, and vaccines. It is expected that the institute will help develop policies and standards, facilitate bulk purchasing and negotiated pricing with manufacturers, support training of a skilled workforce, establish and coordinate a functional pan-African laboratory network with sample shipment and quality-assured testing, and coordinate and ensure the effective functioning of the community of practice, among other contributions.
Impact assessment and sustainability
Monitoring and evaluation
Regular assessments of the efficiency and effectiveness of incorporating genomic data in routine public health surveillance systems will be crucial.69 This evaluation should include assessing completeness and timeliness of data collection, reporting on and using the data for policy making, identifying gaps and areas for improvement, and assessing costs and benefits of the genomic data compared with other surveillance approaches.
Funding and sustainability
Countries should also develop a sustainability framework for genomic surveillance. Genomic surveillance will need to be considered as part of the broader public health goods and as part of national disease control programmes, emergency responses, surveillance of antimicrobial resistance, and other surveillance programmes—and thus commit sufficient resources to it. Equally, mainstream funding bodies, such as global funds and other partners, should continue supporting the implementation of recommended genomics use cases, which are necessary for the success of disease control programmes. Countries can also use other groups that support various genomics use cases in Africa.31, 70 Overall, national public health institutes and national disease control programmes should jointly develop with global partners definite financial commitment and sustainability plans.
Conclusion and future direction
Pathogen genomics has the potential to improve disease surveillance, and outbreak detection and management, as well as accelerate control and eradication of endemic diseases in Africa. Despite its potential, considerable challenges exist. Incorporating pathogen genomics into public health will require substantial investments in NGS and computing infrastructure, but these investments can be minimised by using functional networks of multipathogen genomics facilities. The cost of sequencing can also be considerably reduced through high-level multiplexing, centralised bulk purchasing, and price negotiations with manufacturers, whereas capacity building can take advantage of established African institutions. Furthermore, harmonised policies and regulatory frameworks for the whole continent would be needed to guide best policies around materials and data sharing and protection, which will further support cross-border and regional joint disease control efforts.
Establishing systems for timely reporting of genomic data in a policy-digestible language is also essential. Monitoring, evaluation, and sustainability frameworks will be needed to assess the added value of NGS over other surveillance tools, identify gaps and areas of improvement, and ensure availability of sufficient resources for its operations. Finally, integration of pathogen genomics for surveillance in Africa will require leadership and coordination, which could be provided by a mandated technical institution such as the Africa CDC in consultation with experts.
Acknowledgments
Acknowledgments
We thank Alan Christoffels and Benjamin Djoudalbaye for their constructive comments on the draft of the manuscript. We are also grateful to participants of the consultative workshop on the integration of pathogen genomics into public health practice in Africa (Jan 15–17, 2020) for their contributions and deliberations towards this topic.
Editorial note: the Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Contributors
SCI, SKT, YK, AEOO, and JNK conceptualised the paper. SCI drafted the manuscript, with assistance from SKT and JNK. SCI and SKT accessed and verified the data on sequencing capacity. All authors reviewed and contributed to subsequent drafts for important intellectual content and approved the final manuscript. The authors alone are responsible for the views expressed in this publication and they do not necessarily represent the decisions or policies of the Africa CDC.
Declaration of interests
All authors are employed by the Africa CDC.
Supplementary Material
References
- 1.European Investment Bank Africa's digital solutions to tackle COVID-19. July, 2020. https://www.eib.org/attachments/country/africa_s_digital_solutions_to_tackle_covid_19_en.pdf
- 2.Peeling RW, Boeras D, Wilder-Smith A, Sall A, Nkengasong J. Need for sustainable biobanking networks for COVID-19 and other diseases of epidemic potential. Lancet Infect Dis. 2020;20:e268–e273. doi: 10.1016/S1473-3099(20)30461-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nkengasong J. Let Africa into the market for COVID-19 diagnostics. Nature. 2020;580:565. doi: 10.1038/d41586-020-01265-0. [DOI] [PubMed] [Google Scholar]
- 4.Delamou A, Delvaux T, El Ayadi AM, et al. Public health impact of the 2014–2015 Ebola outbreak in West Africa: seizing opportunities for the future. BMJ Glob Heal. 2017;2 doi: 10.1136/bmjgh-2016-000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mbala-Kingebeni P, Pratt CB, Wiley MR, et al. 2018 Ebola virus disease outbreak in Équateur Province, Democratic Republic of the Congo: a retrospective genomic characterisation. Lancet Infect Dis. 2019;19:641–647. doi: 10.1016/S1473-3099(19)30124-0. [DOI] [PubMed] [Google Scholar]
- 6.WHO Emergency operations: annual report. Saving lives and reducing suffering: WHO's work in emergency response operations in the WHO African Region in 2018. https://reliefweb.int/sites/reliefweb.int/files/resources/WHO-AF-WHE-EMO-01-2020.pdf
- 7.Roser M, Ritchie H. Burden of disease. 2018. OurWorldInData.orghttps://ourworldindata.org/burden-of-disease
- 8.WHO Antimicrobial resistance: global report on surveillance. 2014. https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf?sequence=1
- 9.Kandel N, Chungong S, Omaar A, Xing J. Health security capacities in the context of COVID-19 outbreak: an analysis of International Health Regulations annual report data from 182 countries. Lancet. 2020;395:1047–1053. doi: 10.1016/S0140-6736(20)30553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert M, Pullano G, Pinotti F, et al. Preparedness and vulnerability of African countries against importations of COVID-19: a modelling study. Lancet. 2020;395:871–877. doi: 10.1016/S0140-6736(20)30411-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant K, Jenkins C, Arnold C, Green J, Zambon M. Public Health England; 2018. Implementing pathogen genomics: a case study.https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/731057/implementing_pathogen_genomics_a_case_study.pdf [Google Scholar]
- 12.European Centre for Disease Prevention and Control ECDC strategic framework for the integration of molecular and genomic typing into European surveillance and multi-country outbreak investigations—2019–2021. March, 2019. https://www.ecdc.europa.eu/sites/default/files/documents/framework-for-genomic-surveillance.pdf
- 13.Armstrong GL, MacCannell DR, Taylor J, et al. Pathogen genomics in public health. N Engl J Med. 2019;381:2569–2580. doi: 10.1056/NEJMsr1813907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vogels CBF, Brito AF, Wyllie AL, et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT-qPCR primer-probe sets. Nat Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Young BE, Fong S-W, Chan Y-H, et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giandhari J, Pillay S, Wilkinson E, et al. Early transmission of SARS-CoV-2 in South Africa: an epidemiological and phylogenetic report. Int J Infect Dis. 2020;103:234–241. doi: 10.1016/j.ijid.2020.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meredith LW, Hamilton WL, Warne B, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect Dis. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WHO SARS-CoV-2 variants. Dec 31, 2020. https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/
- 21.Parkin NT, Avila-Rios S, Bibby DF, et al. Multi-laboratory comparison of next-generation to Sanger-based sequencing for HIV-1 drug resistance genotyping. Viruses. 2020;12:e694. doi: 10.3390/v12070694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.WHO . World Health Organization; Geneva: 2018. The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide.https://apps.who.int/iris/bitstream/handle/10665/274443/WHO-CDS-TB-2018.19-eng.pdf?ua=1 [Google Scholar]
- 23.WHO Technical consultation on the role of parasite and anopheline genetics in malaria surveillance. June, 2019. https://www.who.int/malaria/mpac/mpac-october2019-session7-report-consultation-on-genomics.pdf?ua=1
- 24.WHO . World Health Organization; Geneva: 2019. Molecular methods for antimicrobial resistance (AMR) diagnostics to enhance the Global Antimicrobial Resistance Surveillance System.https://apps.who.int/iris/bitstream/handle/10665/310993/WHO-WSI-AMR-2019.1-eng.pdf?ua=1 [Google Scholar]
- 25.Mulders MN, Serhan F, Goodson JL, Icenogle J, Johnson BW, Rota PA. Expansion of surveillance for vaccine-preventable diseases: building on the Global Polio Laboratory Network and the Global Measles and Rubella Laboratory Network platforms. J Infect Dis. 2017;216(suppl 1):S324–S330. doi: 10.1093/infdis/jix077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.WHO . World Health Organization; Geneva: 2018. Whole genome sequencing for foodborne disease surveillance: landscape paper.https://apps.who.int/iris/bitstream/handle/10665/272430/9789241513869-eng.pdf?ua=1 [Google Scholar]
- 27.WHO . World Health Organization; Geneva, Switzerland: 2019. HIV Drug Resistance Report 2019.https://www.who.int/hiv/pub/drugresistance/hivdr-report-2019/en/ [Google Scholar]
- 28.WHO . World Health Organization; Geneva: 2019. Global tuberculosis report 2019.https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf [Google Scholar]
- 29.WHO . World Health Organization; Geneva: 2017. Global action plan on HIV drug resistance 2017–2021.https://www.who.int/hiv/pub/drugresistance/hivdr-action-plan-2017-2021/en/ [Google Scholar]
- 30.WHO HIV drug resistance surveillance guidance: 2015 update. December, 2015. http://apps.who.int/iris/bitstream/10665/204471/1/9789241510097_eng.pdf?ua=1
- 31.Foundation for Innovative Diagnostics SEQ&TREAT. https://www.finddx.org/tb/seq-treat/
- 32.WHO . World Health Organization; Geneva: 2015. WHO estimates of the global burden of foodborne diseases.https://apps.who.int/iris/bitstream/handle/10665/199350/9789241565165_eng.pdf?sequence=1 [Google Scholar]
- 33.WHO . WHO Regional Office for Africa; Brazzaville: 2019. Investment case for vaccine-preventable diseases surveillance in the African Region, 2020–2030.https://www.afro.who.int/sites/default/files/2019-11/VPD_Surv_Brochure_Final_20190918_WEB.pdf [Google Scholar]
- 34.Hogan AB, Jewell BL, Sherrard-Smith E, et al. Potential impact of the COVID-19 pandemic on HIV, tuberculosis, and malaria in low-income and middle-income countries: a modelling study. Lancet Glob Health. 2020;8:e1132–e1141. doi: 10.1016/S2214-109X(20)30288-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberton T, Carter ED, Chou VB, et al. Early estimates of the indirect effects of the COVID-19 pandemic on maternal and child mortality in low-income and middle-income countries: a modelling study. Lancet Glob Health. 2020;8:e901–e908. doi: 10.1016/S2214-109X(20)30229-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dudas G, Carvalho LM, Bedford T, et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature. 2017;544:309–315. doi: 10.1038/nature22040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mbala-Kingebeni P, Aziza A, Di Paola N, et al. Medical countermeasures during the 2018 Ebola virus disease outbreak in the North Kivu and Ituri Provinces of the Democratic Republic of the Congo: a rapid genomic assessment. Lancet Infect Dis. 2019;19:648–657. doi: 10.1016/S1473-3099(19)30118-5. [DOI] [PubMed] [Google Scholar]
- 39.Siddle KJ, Eromon P, Barnes KG, et al. Genomic analysis of Lassa virus during an increase in cases in Nigeria in 2018. N Engl J Med. 2018;379:1745–1753. doi: 10.1056/NEJMoa1804498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tessema SK, Inzaule SC, Christoffels A, et al. Accelerating genomics-based surveillance for COVID-19 response in Africa. Lancet Microbe. 2020;1:e227–e228. doi: 10.1016/S2666-5247(20)30117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyoshima Y, Nemoto K, Matsumoto S, Nakamura Y, Kiyotani K. SARS-CoV-2 genomic variations associated with mortality rate of COVID-19. J Hum Genet. 2020;65:1075–1082. doi: 10.1038/s10038-020-0808-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.US Centers for Disease Control and Prevention Detecting and responding to HIV transmission clusters: a guide for health departments. June, 2018. https://www.cdc.gov/hiv/pdf/funding/announcements/ps18-1802/CDC-HIV-PS18-1802-AttachmentE-Detecting-Investigating-and-Responding-to-HIV-Transmission-Clusters.pdf
- 43.Abeler-Dörner L, Grabowski MK, Rambaut A, Pillay D, Fraser C, on behalf of the PANGEA consortium PANGEA-HIV 2: phylogenetics and networks for generalised epidemics in Africa. Curr Opin HIV AIDS. 2019;14:173–180. doi: 10.1097/COH.0000000000000542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.WHO . World Health Organization; Geneva: 2017. Integrating neglected tropical diseases into global health and development: fourth WHO report on neglected tropical diseases.https://apps.who.int/iris/bitstream/handle/10665/255011/9789241565448-eng.pdf?sequence=1 [Google Scholar]
- 45.Inzaule SC, Hamers RL, Paredes R, Yang C, Schuurman R, Rinke de Wit TF. The evolving landscape of HIV drug resistance diagnostics for expanding testing in resource-limited settings. AIDS Rev. 2017;19:219–230. [PubMed] [Google Scholar]
- 46.Maljkovic Berry I, Melendrez MC, Bishop-Lilly KA, et al. Next generation sequencing and bioinformatics methodologies for infectious disease research and public health: approaches, applications, and considerations for development of laboratory capacity. J Infect Dis. 2020;221(suppl 3):S292–S307. doi: 10.1093/infdis/jiz286. [DOI] [PubMed] [Google Scholar]
- 47.Tessema SK, Raman J, Duffy CW, Ishengoma DS, Amambua-Ngwa A, Greenhouse B. Applying next-generation sequencing to track falciparum malaria in sub-Saharan Africa. Malar J. 2019;18:268. doi: 10.1186/s12936-019-2880-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Poelvoorde LAE, Saelens X, Thomas I, Roosens NH. Next-generation sequencing: an eye-opener for the surveillance of antiviral resistance in influenza. Trends Biotechnol. 2020;38:360–367. doi: 10.1016/j.tibtech.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 49.Inzaule SC, Ondoa P, Peter T, et al. Affordable HIV drug-resistance testing for monitoring of antiretroviral therapy in sub-Saharan Africa. Lancet Infect Dis. 2016;16:e267–e275. doi: 10.1016/S1473-3099(16)30118-9. [DOI] [PubMed] [Google Scholar]
- 50.Association of Public Health Laboratories Next generation sequencing implementation guide. October, 2016. https://www.aphl.org/aboutAPHL/publications/Documents/ID-NGS-Implementation-Guide102016.pdf
- 51.Neher RA, Bedford T. Real-time analysis and visualization of pathogen sequence data. J Clin Microbiol. 2018;56:e00480–e00518. doi: 10.1128/JCM.00480-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hadfield J, Megill C, Bell SM, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Argimón S, Abudahab K, Goater RJE, et al. Microreact: visualizing and sharing data for genomic epidemiology and phylogeography. Microb Genomics. 2016;2 doi: 10.1099/mgen.0.000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nembaware V, Mulder N, Mulder N. The African Genomic Medicine Training Initiative (AGMT): showcasing a community and framework driven genomic medicine training for nurses in Africa. Front Genet. 2019;10 doi: 10.3389/fgene.2019.01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.The Wellcome Trust Grants awarded: Developing Excellence in Leadership, Training and Science Initiative. https://wellcome.ac.uk/what-we-do/directories/developing-excellence-leadership-training-and-science-initiative
- 56.Shaffer JG, Mather FJ, Wele M, et al. Expanding research capacity in sub-Saharan Africa through informatics, bioinformatics, and data science training programs in Mali. Front Genet. 2019;10:331. doi: 10.3389/fgene.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.H3Africa Nurturing genomics and bioinformatics research capacity in Africa. https://h3africa.org/index.php/consortium/nurturing-genomics-and-bioinformatics-research-capacity-in-africa-breca/
- 58.Medical Research Council Unit The Gambia Genomics sequencing in The Gambia. Https://www.mrc.gm/genomics-sequencing-in-the-gambia/
- 59.Medical Research Council Unit The Gambia Research training and career development. https://www.mrc.gm/research-training-career-development/
- 60.Nsubuga P, Johnson K, Tetteh C, et al. Field epidemiology and laboratory training programs in sub-Saharan Africa from 2004 to 2010: need, the process, and prospects. Pan Afr Med J. 2011;10:24. doi: 10.4314/pamj.v10i0.72235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amukele T. Africa CDC: establishing integrated surveillance and laboratory networks for rapid disease detection and response, control, prevention, and clinical care in Africa. Afr J Lab Med. 2017;6:638. doi: 10.4102/ajlm.v6i1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gargis AS, Kalman L, Lubin IM. Assuring the quality of next-generation sequencing in clinical microbiology and public health laboratories. J Clin Microbiol. 2016;54:2857–2865. doi: 10.1128/JCM.00949-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee ER, Gao F, Sandstrom P, Ji H. External quality assessment for next-generation sequencing-based HIV drug resistance testing: unique requirements and challenges. Viruses. 2020;12:550. doi: 10.3390/v12050550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Vries J, Munung SN, Matimba A, et al. Regulation of genomic and biobanking research in Africa: a content analysis of ethics guidelines, policies and procedures from 22 African countries. BMC Med Ethics. 2017;18:8. doi: 10.1186/s12910-016-0165-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Miller CM, Ketlhapile M, Rybasack-Smith H, Rosen S. Why are antiretroviral treatment patients lost to follow-up? A qualitative study from South Africa. Trop Med Int Health. 2010;15(suppl 1):48–54. doi: 10.1111/j.1365-3156.2010.02514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Secretariat of the Convention on Biological Diversity . Secretariat of the Convention on Biological Diversity; Montreal: 2011. Nagoya protocol on access to genetic resources and the fair and equitable sharing of benefits arising from their utilization.https://www.cbd.int/abs/ [Google Scholar]
- 67.Ekici H, Rao SD, Sönnerborg A, Ramprasad VL, Gupta R, Neogi U. Cost-efficient HIV-1 drug resistance surveillance using multiplexed high-throughput amplicon sequencing: implications for use in low- and middle-income countries. J Antimicrob Chemother. 2014;69:3349–3355. doi: 10.1093/jac/dku278. [DOI] [PubMed] [Google Scholar]
- 68.Nkengasong JN, Maiyegun O, Moeti M. Establishing the Africa Centres for Disease Control and Prevention: responding to Africa's health threats. Lancet Glob Health. 2017;5:e246–e247. doi: 10.1016/S2214-109X(17)30025-6. [DOI] [PubMed] [Google Scholar]
- 69.WHO . World Health Organization; Geneva: 2018. International Health Regulations (2005) IHR Monitoring and Evaluation Framework.https://apps.who.int/iris/bitstream/handle/10665/276651/WHO-WHE-CPI-2018.51-eng.pdf?sequence=1 [Google Scholar]
- 70.Fleming Fund The petri dish. March, 2020. https://www.flemingfund.org/wp-content/uploads/7d0b543c8f054814d0e0bb3247d8b77f.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.