Abstract
Background
The clinical predictors and biological mechanisms for localized prostate cancer (PCa) outcomes remain mostly unknown. We aim to evaluate the role of serum immune-checkpoint-related (ICK) proteins and genetic variations in predicting outcomes of localized PCa.
Methods
We profiled the serum levels of 14 ICK-related proteins (BTLA, GITR, HVEM, IDO, LAG-3, PD-1, PD-L1, PD-L2, Tim-3, CD28, CD80, 4-1BB, CD27, and CTLA-4) in 190 patients with localized PCa. The genotypes of 97 single nucleotide polymorphisms (SNPs) from 19 ICK-related genes were analyzed in an extended population (N = 1762). Meta-data from ArrayExpress and TCGA was employed to validate and to probe functional data. Patients were enrolled and tumor aggressiveness, biochemical recurrence (BCR), and progression information were obtained. Statistical analyses were performed analyzing associations between serum biomarkers, genotypes, mRNA and outcomes.
Results
We showed that serum (s)BTLA and sTIM3 levels were associated with PCa aggressiveness (P < 0.05). sCD28, sCD80, sCTLA4, sGITR, sHVEM and sIDO correlated with both BCR and progression risks (all P < 0.05). We further identified ICK variants were significantly associated with aggressiveness, BCR and progression. Among them, 4 SNPs located in CD80 (rs7628626, rs12695388, rs491407, rs6804441) were not only associated with BCR and progression risk, but also correlated with sCD80 level (P < 0.01). rs491407 was further validated in an independent cohort. The CD80 mRNA expression was associated with BCR (HR, 1.85, 95% CI 1.06–3.22, P = 0.03) in meta-analysis of validation cohorts.
Conclusion
We highlight the prognostic value of serum ICK-related proteins for predicting aggressiveness, BCR and progression of PCa. The genetic variations and mRNA expression in CD80 could be predictors and potential targets of localized PCa.
Electronic supplementary material
The online version of this article (10.1007/s00262-020-02718-1) contains supplementary material, which is available to authorized users.
Keywords: Serum immune checkpoint, Genetic variations, Localized prostate cancer, Aggressiveness, Biochemical recurrence, Progression free survival
Introduction
Treatment of localized prostate cancer (PCa) is largely dependent on risk stratification based on aggressiveness [1]. Pathological Gleason score (GS) and serum prostate-specific antigen (PSA) are mainly applied in outcome prediction for PCa [2] but remain imperfect in predicting properties of the tumor and biochemical recurrence (BCR), which may lead to over-treatment in patients with indolent cancer [3].
Immunotherapy has shown promise in the battle against PCa. Though Ipilimumab failed in the treatment of metastatic castration-resistant PCa (mCRPC) in a recent clinical trial [4], combinatorial immunotherapy with Immune checkpoint blockade (ICB) and Myeloid-derived suppressor cells (MDSCs) targeted therapy showed efficacy in mCRPC [5]. Overexpression of immune checkpoint (ICK) genes like HAVCR2 (TIM-3) in T cells could cause dysfunction of PSA-specific CD8+ T cells in PCa [6]. PD-L1 expression is independently associated with BCR in aggressive PCa [7]. These studies implicate the involvement of ICK-related genes during PCa development.
Soluble T cell regulatory proteins (mostly ICK-related proteins) released from immune and tumor cells may affect the efficacy of treatment and outcomes [8]. High soluble (s)PD-L1 is associated with impaired immunity and poor outcomes in aggressive renal cell cancer [9]. However, the association of these markers with PCa outcomes and potential mechanisms is not clear, which requires more studies.
Therefore, we implemented a three-stage study to determine whether soluble ICK-related proteins were associated with PCa outcomes and functionally explored the potential mechanism. First, we evaluated serum levels of 14 ICK-related proteins and their associations with outcomes in 190 patients with localized PCa in the MD Anderson Cancer Center PCa (MDACC-PCa) cohort. Second, the genotype data of 97 single nucleotide polymorphisms (SNPs) from 19 ICK-related genes were obtained in an extended cohort of patients with PCa (N = 1762), and the correlation between genotypes and outcomes were further analyzed. Further, we investigated the genotype–phenotype correlation between SNPs and serum ICK-related proteins and used data from ArrayExpress and The Cancer Genome Altas (TCGA) to validate our findings and to derive functional relevance to support our findings.
Materials and methods
Study population and data collection
The study was approved by the MDACC Institutional Review Board. Each subject consented to having their clinical data obtained and providing blood samples for research purposes.
This study used data from a previously described cohort of patients with PCa enrolled at MDACC [10, 11]. In brief, the cohort involved non-Hispanic white men with previously untreated PCa. The MDACC-PCa cohort recruited patients who underwent treatment for localized PCa from 2003 to 2013. Clinical data, including diagnosis date, PSA level at diagnosis, biopsy-proven GS, clinical tumor stage, treatment details, tissue pathology, and follow up information were collected from medical records. BCR after local therapy was defined as a single measure of PSA ≥ 0.2 ng/mL after radical prostatectomy [12], or a PSA rise of 2 ng/mL or more above the nadir PSA in patients who received radiotherapy [13]. Progression was defined by either BCR or clinical/radiographic progression of the disease [1, 14]. All pathologic slides from outside institutions were reviewed by a single genitourinary pathologist at MDACC. In all cases, the GS assessed at MDACC was used.
Another independent validation cohort derived from ArrayExpress, which included 132 patients with PCa from Tumour Identity Card Program (CIT), France [15]. All patients were post-prostatectomy, and the criteria for aggressiveness and BCR were the same as for the MDACC-PCa cohort (Table S1).
Serum ICK-related proteins detection
Samples were tested in duplicate using ProcartaPlex Human Immuno-Oncology Checkpoint Panel (Thermo Fisher, USA) in 96-well plate format to quantify 14 human immune checkpoint markers. Assay was conducted according to protocols provided by the manufacturer using Luminex 200™ instrument and xPONENT® software (Luminex Corp, USA). All inter-assay and intra-assay coefficients of variation (CV) were below 15%.
Genes of interest in ICK pathway
Based on previous literature, a total of 19 ICK-related genes, which includes most immune checkpoint receptors and ligands, were selected [16]. The chromosome positions of transcriptional start and end points of each gene were obtained from the USCS Genome Browser (https://genome.ucsc.edu/, build version: GRCh37/hg19).
Genotyping and quality control
All DNA samples were extracted from peripheral whole blood using the QIAamp DNA extraction Kit (QIAGEN). Custom Infinium OncoArray-500 K Beadchip was used to genotype both populations. Assays were run on the iScan system (Illumina, USA). Genotyping data were analyzed and exported using the Genome Studio software (Illumina, USA). All subjects had a call rate > 95%. Genotyping data of 97 SNPs found within 10 kb of upstream and downstream flanking regions for each gene of interest were extracted from the OncoArray dataset and included in the study.
Functional characterization and immune phenotype association
SNP array (Illumina HumanOmniExpress-12 v1.0) and mRNA expression array (Affymetrix Human Gene 2.0 ST Array) data of 132 patients with PCa from CIT cohort were downloaded from ArrayExpress portal (https://www.ebi.ac.uk/arrayexpress). Bioinformatic analysis was applied to investigate tumor (N = 494) and normal tissue (N = 52) data of patients with PCa from TCGA databases. Gene level expression based on RNA-seq data (normalized, RSEM level 3) and clinical data for each sample were downloaded directly from the TCGA data and analyzed using Firebrowse API (https://firebrowse.org/). CD80 mRNA was dichotomized at the median level. Positive Tumor infiltrate lymphocyte (TIL) was defined as TIL percent greater than 0%.
Statistical analysis
Levels of serum proteins and gene expressions were dichotomized using a logistic regression spline model [17]. All potential biomarkers including SNPs were separately evaluated for individual association with aggressive disease (D’Amico high-risk vs. low-risk) using unconditional multivariable logistic regression adjusted by age at diagnosis (continuous) and PSA levels at diagnosis (categorical). All biomarkers and SNPs of interest were examined for association with time to BCR or progression using multivariate Cox proportional hazard models adjusted for age, GS, T stage and baseline PSA level. Kaplan–Meier analyses and log-rank tests were used to calculate survival differences. To reduce the likelihood of false discovery, q-value for multiple testing was applied in both soluble ICK-related protein and ICK-related genetic variation analysis [18]. The association between genotypes and soluble ICK-related protein levels was analyzed with Spearman correlation. Meta-analysis was performed with the ‘meta’ package in R.
All data were analyzed and visualized with Excel (Microsoft office 365), R software (v3.4.1), PLINK (v1.07), and STATA 14.2 (STATA Corp). All P values were two-sided, with values less than 0.05 considered statistically significant.
Results
Patient characteristics
We included 1762 individuals with localized PCa in this study. All patients were genotyped, and their clinicopathological features were listed in Table 1. In general, there were 1109 (62.9%) clinical T1, 575 (32.6%) clinical T2, 69 (3.9%) T3–4 PCa. Approximately two thirds (63.0%) of all patients had PSA levels ranging from 4 to 9.9 ng/mL, whereas there were 8.2% of patients at 10–19.9 ng/mL and 3.7% of patients over 20 ng/mL. One third (37.3%) of patients had GS value as 6, whereas 50.3% of patients had GS value as 7 and 7.0% of patients had GS value higher than 7. Among all patients, 100 (5.7%) showed BCR and 39 (2.2%) died.
Table 1.
Host characteristics of MDACC-Pca, CIT and TCGA cohorts
| Variables | MDACC-PCa cohort (n %) | Subset of MDACC-Pca cohort | CIT cohort (n %) | TCGA cohort (n %) | |
|---|---|---|---|---|---|
| All | High-risk groupc | Low-risk groupc | All | All | |
| Age, mean (SD) | 61.60 (7.90) | 64.06 (7.60) | 64.01 (7.50) | 63.21 (6.26) | 61.56 (6.78) |
| Smoking | |||||
| Current | 149 (8.5) | 31 (32.6) | 39 (41.1) | NA | NA |
| Former | 770 (43.7) | 45 (47.4) | 47 (49.5) | NA | NA |
| Never | 826 (46.9) | 19 (20.0) | 9 (9.5) | NA | NA |
| Clinical T stage | |||||
| T1 | 1109 (62.9) | 34 (35.8) | 95 (100.0) | 0 | 0 |
| T2 | 575 (32.6) | 12 (12.6) | 0 (0.0) | 91 (68.9) | 187 (37.9) |
| T3–T4 | 69 (3.9) | 47 (49.5) | 0 (0.0) | 40 (30.3) | 300 (60.7) |
| PSA level at diagnosis, ng/mL | |||||
| < 4 | 442 (25.1) | 14 (14.7) | 33 (34.7) | 8 (6.1)) | 53 (10.7) |
| 4–9.9 | 1108 (63.0) | 54 (56.8) | 62 (65.3) | 97 (73.4) | 275 (55.7) |
| 10–19.9 | 145 (8.2) | 7 (7.4) | 0 (0.0) | 21 (15.9) | 98 (19.8) |
| ≥ 20 | 65 (3.7) | 20 (21.1) | 0 (0.0) | 3 (2.3) | 53 (10.7) |
| Biopsy-proven GS | |||||
| 6 | 657 (37.3) | 8 (8.4) | 95 (100.0) | 66 (50) | 45 (9.1) |
| 7 | 887 (50.3) | 27 (28.4) | 0 (0.0) | 45 (34.1) | 246 (49.8) |
| 8 | 123 (7.0) | 35 (36.8) | 0 (0.0) | 12 (9.1) | 64 (13.0) |
| 9 | 90 (5.1) | 23 (24.2) | 0 (0.0) | 8 (6.1) | 135 (27.3) |
| 10 | 5 (0.3) | 2 (2.1) | 0 (0.0) | 0 | 4 (0.8) |
| Dead | |||||
| No | 1352 (76.7) | 89 (93.7) | 95 (100.0) | NA | 486 (98.4) |
| Yes | 39 (2.2) | 6 (6.3) | 0 (0.0) | NA | 8 (1.6) |
| Treatment | |||||
| Radical prostatectomy | 918 (52.10) | 53 (55.8) | 31 (32.6) | 132 | NA |
| Radiotherapy | 378 (21.45) | 27 (28.4) | 14 (14.7) | 0 | NA |
| Surveillance/unknown | 429 (24.35) | 13 (13.7) | 47 (49.5) | ||
| Otherb | 37 (2.10) | 2 ( 2.1) | 3 (3.2) | 0 | NA |
| Biochemical recurrence (BCR)a | |||||
| No | 1291 (73.3) | 79 (83.2) | 95 (100.0) | 95 (72.0) | 369 (74.7) |
| Yes | 100 (5.7) | 16 (16.8) | 0 (0.0) | 15 (11.4) | 57 (11.5) |
| Total | 1762 | 95 | 95 | 132 | 494 |
GS Gleason Score
aBiochemical recurrence (BCR) after local therapy was defined as a single measure of PSA ≥ 0.2 ng/mL after radical prostatectomy, and a PSA rise of 2 ng/mL or more above the nadir PSA in patients who received radiotherapy
bOthers indicate cryoablation, high-intensity focused ultrasound, transurethral resection
cBased on D’Amico criteria for aggressiveness of prostate cancer
A subset of 190 patients was retrieved from the MDACC-PCa cohort, whose serum samples were evaluated for serum ICK-related protein levels. According to D’Amico risk classification, there were 95 patients in the low-risk group and another 95 patients in the high-risk group. There were 34 clinical T1 (35.8%), 12 T2 (12.6%) and 47 T3–4 (49.6%) in the high-risk group, whereas all the patients in the low-risk group had T1 disease. About two thirds (65.3%) of the patients in the low-risk group presented with PSA levels at 4–9.9 ng/mL and in the high-risk group, there were 21.1% of patients over 20 ng/mL, 7.4% at 10–19.9 ng/mL, 56.8% at 4–9.9 ng/mL and 14.7% lower than 4 ng/mL. All patients in the low-risk group had GS = 6, while most patients in the high-risk group had GS > 6 (91.6%). In addition, 84 patients received radical prostatectomy, 41 patients received radiotherapy, 60 patients were under surveillance and 5 patients underwent treatment such as cryoablation.
Demographic information from CIT cohort and TCGA cohort are listed in Table 1 and a schematic diagram is depicted in Figure S1.
Association of serum ICK-related proteins with PCa aggressiveness, BCR and tumor progression
We evaluated the association between soluble ICK-related proteins and PCa aggressiveness by dichotomizing patients by spline method into high- and low-level groups (Table 2). Among all biomarkers, sTIM3 and sBTLA were associated with D’Amico high-risk disease, and sBTLA had q-value lower than 0.15 in multiple comparisons (OR = 2.7, 95% CI 1.3–5.6, P = 0.01, q-value = 0.14).
Table 2.
Soluble immune checkpoint factors associated with PCa outcomes
| Soluble markers | D'Amico high vs low | BCR | Progression | Cut-off value (pg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| high vs. low levelsa | Adjusted ORb (95% CI) | P value | q value | Adjusted HRc (95% CI) | P value | q value | Adjusted HRc (95% CI) | P value | q value | |
| sBTLA | 2.7 (1.3–5.6) | 0.010 | 0.140 | 4.1 (0.9–17.4) | 0.060 | 0.084 | 6.5 (1.9–22.8) | 0.003 | 0.028 | 506.56 |
| sCD27 | 1.4 (0.8–2.5) | 0.250 | 0.404 | 1.3 (0.2–6.4) | 0.780 | 0.780 | 0.4 (0.04–3.6) | 0.390 | 0.455 | 1846.83 |
| sCD28 | 0.6 (0.3–1.5) | 0.260 | 0.404 | 12.8 (1.6–101.9) | 0.020 | 0.047 | 7.1 (1.2–42.6) | 0.030 | 0.047 | 2896.50 |
| sCD80 | 1.1 (0.4–2.6) | 0.870 | 0.870 | 5.1 (1.4–19.3) | 0.020 | 0.047 | 4.2 (1.1–15.9) | 0.030 | 0.047 | 51.24 |
| sCD137 | 0.8 (0.3–1.8) | 0.550 | 0.592 | 1.3 (0.2–6.4) | 0.780 | 0.780 | 2.2 (0.4–13.0) | 0.380 | 0.455 | 70.62 |
| sCTLA4 | 5.3 (0.6–46.6) | 0.130 | 0.327 | 8.9 (1.3–59.2) | 0.020 | 0.047 | 8.8 (1.6–48.5) | 0.010 | 0.028 | 89.28 |
| sGITR | 0.3 (0.03–3.1) | 0.330 | 0.462 | 8.9 (2.1–38.3) | 0.003 | 0.047 | 7.3 (1.7–30.9) | 0.007 | 0.028 | 124.82 |
| sHVEM | 1.4 (0.6–3.4) | 0.500 | 0.583 | 7.0 (1.7–29.5) | 0.008 | 0.047 | 7.0 (1.7–28.9) | 0.007 | 0.028 | 29.00 |
| sIDO | 1.9 (0.9–3.7) | 0.070 | 0.245 | 6.7 (1.3–35.1) | 0.020 | 0.047 | 7.9 (1.5–41.9) | 0.020 | 0.047 | 10.51 |
| sLAG3 | 0.4 (0.1–1.0) | 0.060 | 0.245 | 1.3 (0.3–5.4) | 0.720 | 0.780 | 1.1 (0.2–4.7) | 0.930 | 0.930 | 57.86 |
| sPDCD1 | 0.3 (0.02–2.3) | 0.220 | 0.404 | 5.7 (1.2–27.5) | 0.030 | 0.053 | 6.4 (1.5–27.2) | 0.010 | 0.028 | 296.17 |
| sPD–L1 | 1.7 (0.4–7.6) | 0.460 | 0.583 | 4.8 (0.9–25.7) | 0.060 | 0.084 | 4.0 (0.8–20.9) | 0.100 | 0.140 | 17.22 |
| sPD–L2 | 1.6 (0.9–2.9) | 0.140 | 0.327 | 5.5 (1.2–26.2) | 0.030 | 0.053 | 4.6 (1.1–18.5) | 0.030 | 0.047 | 1420.71 |
| sTIM3 | 2.0 (1.0–3.7) | 0.040 | 0.245 | 1.3 (0.4–4.7) | 0.650 | 0.780 | 1.5 (0.5–4.5) | 0.520 | 0.560 | 1413.69 |
PCa prostate cancer, BCR biochemical recurrence, OR odds ratio, HR hazard ratio, CI confidence interval
aHigh and low levels were determined after dichotomization using the spline model (Rosenberg et al., 2003). Significant values in bold font
bAdjusted for age and baseline PSA level
cAdjusted for age, GS, T stage and baseline PSA level
dBenjaminiand Hochberg correction method was applied
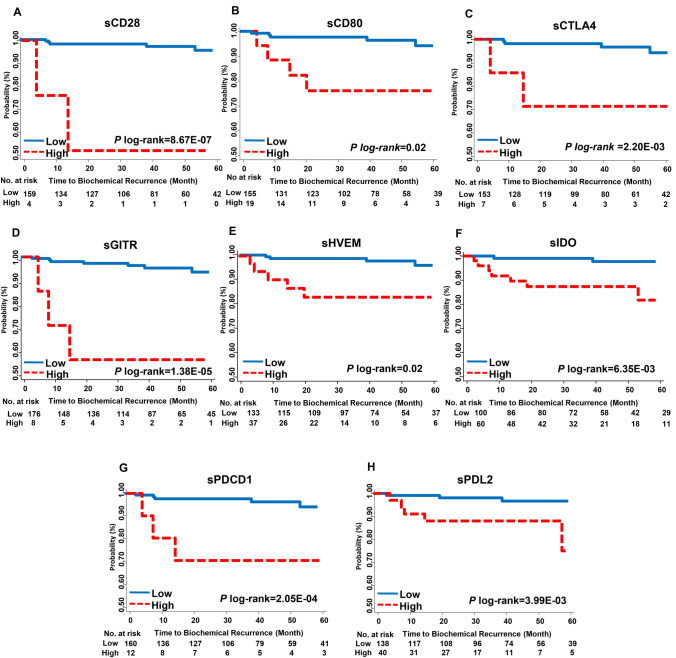
We also analyzed the association between 14 ICK-related proteins and BCR in patients with PCa. sCD28, sCTLA4, sHVEM, sIDO, sGITR, sCD80 were significantly correlated with BCR after multiple comparison (Table 2). sGITR demonstrated the most significant association with BCR. The high-sGITR group demonstrated approximately ninefold increased risk of BCR than that of the low-sGITR group (HR, 8.9, 95% CI 2.1–38.3, P = 3.4E−03, q-value = 0.047). High sGITR was a predictor of poor BCR-free survival in Kaplan–Meier analysis (log-rank-P = 1.38E−05; Fig. 1). Similarly, sCD28, sCD80, sCTLA4, sHVEM, sIDO, sPDCD1 and sPD-L2 were also significantly associated with BCR-free survival (log-rank-P < 0.05; Fig. 1).
Fig. 1.
Kaplan Meier analyses of biochemical recurrence (BCR)-free survival by levels of soluble immune checkpoint proteins in localized prostate cancer (PCa) patients. BCR-free survival was estimated according to the levels of a sCD28, b sCD80, c sCTLA4, d sHVEM, e sIDO, f sGITR, g sPDCD1, and h sPDL2. Patients were dichotomized into high- and low-level groups based on the spline model
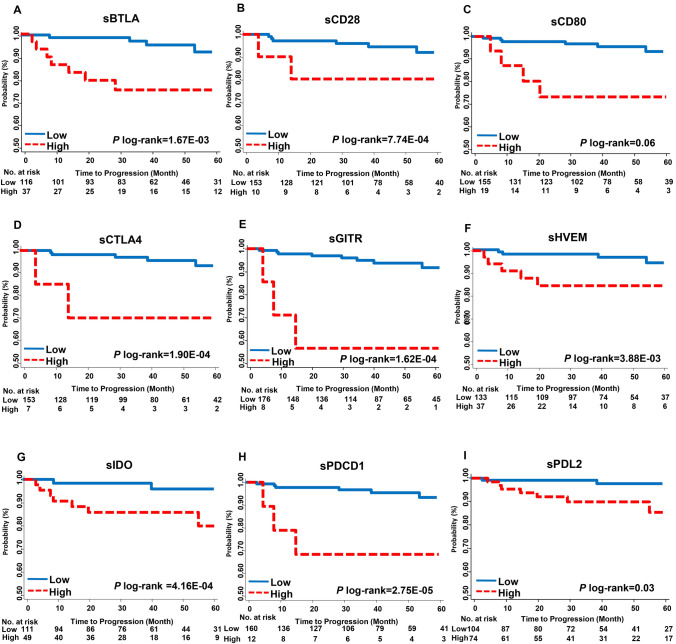
Further, the associations between serum ICK-related proteins and cancer progression were analyzed. sBTLA, sCD28, sCD80, sCTLA4, sGITR, sHVEM, sIDO, sPDCD1 and sPD-L2 were identified as predictors of PCa progression after multiple comparison (P < 0.05). sBTLA is the most significant ICK factor associated with progression (HR, 6.5, 95% CI 1.9–22.8, P = 3.3E−03, q-value = 0.028). Kaplan–Meier analysis showed the serum factor as a significant predictor of progression-free survival (PFS) (log-rank-P = 1.67E−03, Table 2, Fig. 2). sCD28, sCTLA4, sGITR, sHVEM, sIDO, sPDCD1 and sPD-L2 also significantly correlated with PFS (Fig. 2).
Fig. 2.
Kaplan Meier analyses of progression-free survival (PFS) by levels of soluble immune checkpoint proteins in patients with localized PCa. Progression-free survival was estimated according to the levels of a sBTLA, b sCD28, c sCD80, d sCTLA4, e sHVEM, f sIDO, g sGITR, h sPDCD1, and i sPDL2. Patients were dichotomized into high- and low-level groups based on the spline model
Associations of ICK-related genetic variants with PCa outcomes and functional validation
To investigate the association between ICK-related genetic variations and PCa outcomes, we compared the allele frequencies of 97 SNPs from 19 ICK-related genes for risks of aggressiveness, BCR, and progression. Nine SNPs were associated with risk of aggressive disease. Among those, LAG3:rs1997510 conferred the strongest association (OR, 0.34, 95% CI 0.17–0.67, P = 0.002, q-value = 0.11, Table S1). In addition D86:rs17203439 was identified as the most significant SNP correlated with BCR risk among 18 SNPs (HR, 49.2, 95% CI 5.83–414.0, P = 3.42E-04, q-value = 0.03, Table S2), whereas CD274:rs822335 showed the strongest association with PCa progression among 22 SNPs (HR, 1.73, 95% CI 1.31–2.29, P = 9.53E−05, q-value = 0.009, Table S3). Among all identified SNPs, we observed 8 SNPs significantly associated with both risk of BCR and risk of progression (P < 0.05, Table 3).
Table 3.
Genetic variants of immune checkpoint genes associated with PCa aggressiveness, BCR and progression
| Gene | SNP | Location | Model | D'Amico high vs. low | BCR | Progression | |||
|---|---|---|---|---|---|---|---|---|---|
| ORa (95% CI) | P value | HRb (95% CI) | P value | HRb (95% CI) | P value | ||||
| BTLA | |||||||||
| rs2633562 | Intron | DOM | 0.61 (0.30–1.24) | 0.169 | 2.56 (1.40–4.69) | 0.002 | 2.29 (1.35–3.87) | 0.002c | |
| CD80 | |||||||||
| rs6804441d | Intron | REC | – | – | 4.00 (1.72–9.31) | 0.001 | 3.82 (1.84–7.93) | 3.24E-04c | |
| rs12695388d | Intron | DOM | 0.95 (0.66–1.35) | 0.763 | 0.61 (0.38–0.98) | 0.043 | 0.52 (0.36–0.77) | 0.001c | |
| rs7628626d | 3 ‘UTR | REC | – | – | 3.27 (1.47–7.31) | 0.004 | 2.35 (1.11–4.98) | 0.025 | |
| rs491407d | Intron | DOM | 2.01 (1.09–3.71) | 0.026 | 0.69 (0.45–1.04) | 0.075 | 0.78 (0.55–1.09) | 0.140 | |
| CD274 | |||||||||
| rs822335 | 5′ near gene | ADD | 1.07 (0.84–1.36) | 0.576 | 1.78 (1.28–2.47) | 0.001 | 1.73 (1.31–2.29) | 9.53E-05c | |
| rs3780395 | Intron | ADD | 1.12 (0.89–1.41) | 0.335 | 0.70 (0.50–0.99) | 0.044 | 0.63 (0.47–0.85) | 0.002c | |
| LAG3 | |||||||||
| rs12313899 | Intron | REC | 0.51 (0.31–0.84) | 0.008 | 0.50 (0.25–1.02) | 0.056 | 0.67 (0.39–1.14) | 0.136 | |
| rs12313899 | Intron | REC | 0.51 (0.31–0.84) | 0.008 | 0.50 (0.25–1.02) | 0.056 | 0.67 (0.39–1.14) | 0.136 | |
| PDCD1LG2 | |||||||||
| rs6476985 | Intron | DOM | 1.10 (0.75–1.60) | 0.632 | 1.74 (1.08–2.81) | 0.023 | 1.96 (1.32–2.92) | 0.001c | |
| TNFSF14 | |||||||||
| rs2277983 | Intron | REC | 0.96 (0.65–1.41) | 0.836 | 0.40 (0.18–0.86) | 0.020 | 0.54 (0.30–0.96) | 0.037 | |
Significant values in bold font
PCa prostate cancer, BCR biochemical recurrence, OR odds ratio, HR hazard ratio, CI confidence interval, DOM dominant, REC recessive, ADD additive
aAdjusted for age and baseline PSA level
bAdjusted for age, Gleason score, T stage and baseline PSA level
cSignificant after Benjaminiand Hochberg correction for multiple testing
dGenotype significant associated with soluble CD80 levels
Furthermore, we investigated the relationship between serum biomarkers and corresponding ICK-related genetic variations. Four SNPs located in CD80 (rs7628626, rs12695388, rs491407 and rs6804441) were significantly correlated with sCD80 level (P < 0.05). Among them, rs7628626 demonstrated the strongest association to the sCD80 level (Rho = 0.22, P = 0.004, Table S4).
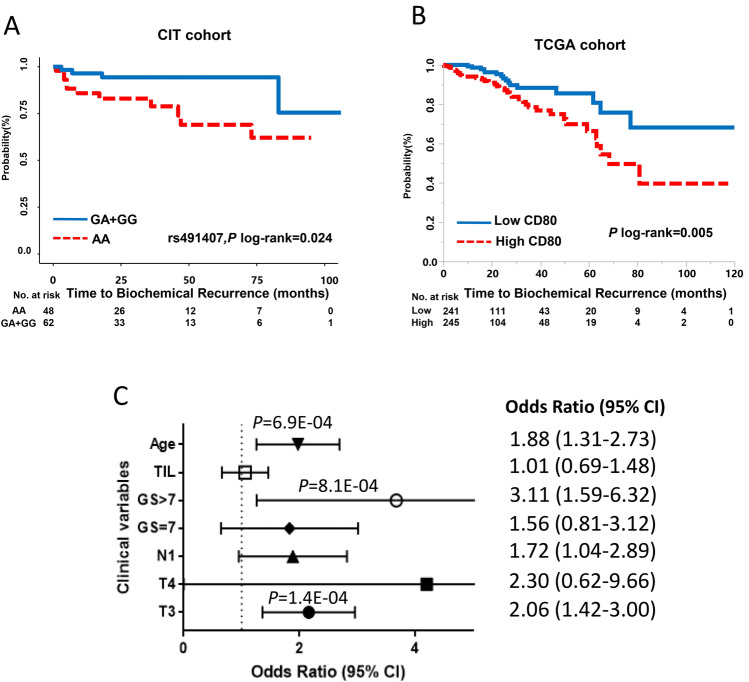
To validate our findings, we turned to the CIT and TCGA cohorts. rs491407 was significantly associated with BCR (HR, 0.23, 95% CI 0.06–0.73, log-rank-P = 0.024), which is consistent with MDACC-PCa cohort data (Fig. 3a, Table S5). To examine whether ICK-related genes were altered transcriptionally in PCa, we retrieved 19 ICK-related gene expression data from TCGA. The expression of CD28, CD274, CTLA4, LAG3, PDCD1LG2, TNFRSF14, TNFRSF18 and TNFSF18 demonstrated significant differential expression between tumor and normal tissues (P < 0.05, Figure S2). Among these genes, CD80 expression showed significant association with T3 stage, high-GS (> 7) and age. High CD80 expression correlated with poor BCR-free survival in patients with PCa (Log-rank P = 0.005) (Fig. 3b, c). In a meta-analysis of TCGA and CIT cohorts, CD80 was associated with BCR in multi-variate Cox model (HR, 1.85, 95% CI 1.06–3.22, P = 0.03) (Table S6).
Fig. 3.
Functional validation of the CD80 genetic variants in CIT cohort and TCGA cohort. a BCR-free survival was estimated according to the genotypes of rs491407, patients who carry G allele showed significant better BCR-free survival compare to AA genotype carriers in CIT cohort (log-rank P = 0.024). b High CD80 expression is significant associated with poor BCR-free survival in TCGA cohort (log-rank P = 0.005). c CD80 expression is closely associated with age, high GS (> 7), T3 in TCGA cohort
Discussion
In this study, we found that serum ICK-related proteins and genetic variations in ICK-related genes were associated with outcomes of localized PCa. Specifically, high levels of sBTLA and sTIM3 correlated with the risk of aggressive PCa. sCD28, sCD80, sCTLA4, sHVEM, sIDO, sGITR, sPDCD1, and sPDL2 were significantly correlated with both risks of BCR and PCa progression, whereas sBTLA was also identified as a predictor of PCa progression. We demonstrated that genetic variations of ICK-related genes were significantly associated with PCa outcomes. A total of 8 SNPs were associated with risks of BCR and progression. Interestingly, the genotypes of four CD80 variants were significantly correlated with sCD80 level. rs491407 was further validated in an independent cohort. CD80 expression was also identified as a prognostic biomarker for BCR-free survival in the combined analysis of CIT and TCGA cohort. To the best of our knowledge, this is the first study to demonstrate that serum proteins and SNPs of ICK-related genes are predictive of PCa outcomes, and the correlation between sCD80 level and CD80 genotypes may implicate a potential functional mechanism.
As an important co-signaling molecule expressed on the surface of antigen-presenting cells, CD80 binds to CD28 or CTLA4 to activate T cell co-stimulation or initiate T cell coinhibition, respectively [19]. One previous study has reported that B7 family members in tumors, specifically B7-H3 and B7x, are associated with disease spread and poor survival in PCa [20], but few studies have focused on B7-1 (CD80). Eugene et al. report that ectopic CD80 expression in a murine PCa cell line pTC1 transplanted to an in vivo mouse model could elicit immune elimination of tumor cells [21]. However, our study demonstrated consistent correlations between the cellular and circulating forms of CD80. It has been reported that human sCD80 is generated by alternative splicing of the gene and that a recombinant form inhibits T cell activation and proliferation in vitro [22]. Thus, sCD80 may bind to CTLA4 and initiate T cell co-inhibition, which could lead to immune escape of tumor cells and elevated risk of recurrence [23]. In addition, sCD80 may compete with membranous CD80 on APCs to modulate its costimulatory effect on T cells. To date, no evidence of sCD80′s therapeutic effect in human cell model has been reported, which requires more research.
We identified four SNPs located in CD80 loci that were associated with sCD80 level as well as cancer outcomes. rs491407 was further validated in another independent cohort as a predictor of BCR. rs7628626, located in the 3′UTR of CD80 gene, was associated with BCR and progression. This is consistent with a previous finding that genotype of rs7628626, targeted by miR-21-3p, is correlated with regional lymph node metastasis and tumor progression in colon cancer [24]. Interestingly, miR-21 could also promote tumor invasion and is associated with increased BCR in prostate cancer [25], which may explain our finding that CD80 expression predicts BCR-free survival in a combined analysis of CIT and TCGA cohort. Therefore, we propose that the mechanism between the genotype of rs7628626 and BCR and progression in localized PCa may involve miR-21 that can functionally regulate CD80 expression leading to differential serum and membranous CD80 levels.
BTLA is an inhibitory ICK interacting with HVEM and LIGHT in the surface of antigen-presenting cells [26]. In this study, we identified sBLTA as a predictor of aggressive disease and a high risk of progression in localized PCa for the first time. In addition, sHVEM was identified as a predictor of the high risk of BCR and progression. HVEM-BTLA binding resulted in an inhibitory effect on T cell activation and proliferation, which lead to impeded anti-tumor immunity [27], which may explain why both sHVEM and sBTLA predicting poor outcomes in PCa. Further, we identified rs2633562 and rs1982809 located in BTLA gene region, were associated with increased risk of BCR and progression. However, we failed to identify the correlation between sBTLA and the genotypes of this variant. Thus, our findings require further investigation to elucidate the underlying mechanism of association.
GITR is a co-stimulatory TNF receptor superfamily member, which affords the potential to expand CD8+ T cell population [28]. In our study, sGITR is associated with increased risk of BCR and progression, which is not consistent with its co-stimulatory role in T cell activation. The inconsistency may derive from the high GITR expression in Treg cells [29], which were highly infiltrated in prostate tissues [30]. Any immune suppression induced by Treg cell may contribute to BCR and progression. However, this will require further research to decipher the underlying mechanisms.
We also identified other serum ICK-related proteins associated with outcomes of PCa. For example, sPDL2 was correlated with increased risk of BCR, and sTIM3 was predictive of the high risk of aggressive PCa. Both PDL2 and Tim3 are reported as potential targets for cancer immunotherapy [31, 32].
Our study has distinct advantages in the multi-phase study design within a large and high-quality cohort, multiplex serum profiling of serum ICK-related proteins and further genotypic analysis of ICK-related genes, analysis of genotype–phenotype correlation, and validation which brings biological validity to some of our findings. However, we also acknowledge several limitations. First, our study was conducted in a single hospital-based cohort. Additional validation in another independent cohort is warranted. Second, cellular ICK gene expression in peripheral blood mononuclear cells (PBMCs) was not evaluated in this study due to the availability and viability of PBMC samples. The correlation between soluble and cellular ICK-related proteins in peripheral blood is unknown. Nevertheless, we analyzed the expression of selected ICK-related genes based on TCGA data to evaluate the ICK phenotypes in tumors. Third, we only assessed serum ICK protein levels in a small subset of patients with PCa from the cohort. The relatively small sample size and outcome events may restrict the study power. Fourth, as the functional implications in this study are conducted in silico, further validation in independent cohorts or in the laboratory is warranted.
In conclusion, we identified a panel of serum ICK-related proteins and genetic variations that were associated with outcomes of localized PCa. Significant genotype–phenotype associations were identified between sCD80 and genetic variants in CD80. Further validations indicated that rs491407 and CD80 expression were correlated with BCR. These findings point to the potential prognostic values of serum ICK-related proteins and genetic variants, which may also provide potential therapeutic targets for immunotherapy of localized PCa.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank the excellent work of the field team and laboratory staff for patient recruitment and for collection and/or processing of clinical/epidemiological information and biological specimens.
Author contributions
XW conceptualized and supervised the project. QW and XW were involved in the development of methodology. QW, HY, HT, DWC, DL, and XW participated in the acquisition of data or management of patients, and/or provided facilities. QW, SL, YY, DWC, and HT participated in the analysis and interpretation of data. XW participated in funding obtaining, QW, SL, MH and YY provided data management and analysis. All authors were involved in the writing, review, and/or revision of the manuscript.
Authors' information
XW has been one of the most funded researchers in MD Anderson Cancer Center and the entire US cancer epidemiology field. She is a highly productive cancer epidemiologist with over 545 publications, including Nature Genetics, Lancet, Lancet Oncology, JAMA, JNCI, JCO, Gut, NEJM, Science, etc. Her provocative studies showed that moderate exercise reduced mortality and extended life expectancy, which led to her proposed MPOWER measures to reduce the burden of physical inactivity. She identified novel cancer predisposition loci, discovered mutations with driver patterns and created a 20-gene panel to distinguish colorectal adenoma from adenocarcinoma. She constructed several powerful risk prediction models for cancer screening, early detection, prognosis, and toxicity. After her retirement from MD Anderson Cancer Center, she joined Zhejiang University and she was appointed Dean of the School of Public Health. This work was done in MD Anderson in 2017 by her research team.
QW is a promising young scholar in cancer research as well as an experienced surgeon in oncology. He achieved his M.D. and Ph.D in Zhejiang University, during his Ph.D, he was trained in City of Hope Comprehensive Cancer Center, which triggered his inspiration in cancer research. Therefore, after he finished his residential training in affiliated Sir Run Run Shaw Hospital, he joined Dr Wu Xifeng’s lab in MD Anderson and worked as a visiting scholar and assistant epidemiologist. This study is mainly finished by him under the direction of the XW.
HY is a post-doctoral fellow of Dr Wu Xifeng’s team in epidemiology department, MD Anderson Cancer Center.
SL is a post-doctoral fellow of Dr Wu Xifeng’s team in epidemiology department, MD Anderson Cancer Center.
HT is a post-doctoral fellow of Dr Wu Xifeng’s team in epidemiology department, MD Anderson Cancer Center.
YY is an associated professor from the school of public health, Zhejiang University. He used to be an associated professor in the department of epidemiology, The University of Texas MD Anderson Cancer Center.
DC is a senior scientist from the department of epidemiology, The University of Texas MD Anderson Cancer Center.
MH is a senior statistician from the department of epidemiology, The University of Texas MD Anderson Cancer Center.
Funding
This work was supported by National Cancer Institute (P50 CA140388), MD Anderson Cancer Center Prostate Cancer SPORE and Center for Translational and Public Health Genomics (CTPHG), Duncan Family Institute.
Availability of supporting data
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Ethics approval and consent to participate
The study was approved by MD Anderson's Institutional Review Board. Written informed consent to participate in the study was obtained from each participant before data and biospecimens were collected.
Consent for publication
Consent was achieved from all patients.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qinchuan Wang and Yuanqing Ye contributed equally to this work.
David W. Chang and Xifeng Wu equal co-last author.
References
- 1.Mohler JL, Antonarakis ES, Armstrong AJ, D'Amico AV, Davis BJ, Dorff T, et al. Prostate Cancer, Version 2.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2019;17(5):479–505. doi: 10.6004/jnccn.2019.0023. [DOI] [PubMed] [Google Scholar]
- 2.D'Amico AV, Whittington R, Malkowicz SB, Cote K, Loffredo M, Schultz D, et al. Biochemical outcome after radical prostatectomy or external beam radiation therapy for patients with clinically localized prostate carcinoma in the prostate specific antigen era. Cancer. 2002;95(2):281–286. doi: 10.1002/cncr.10657. [DOI] [PubMed] [Google Scholar]
- 3.Kader AK, Sun J, Isaacs SD, Wiley KE, Yan G, Kim ST, et al. Individual and cumulative effect of prostate cancer risk-associated variants on clinicopathologic variables in 5,895 prostate cancer patients. Prostate. 2009;69(11):1195–1205. doi: 10.1002/pros.20970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer TM, Kwon ED, Drake CG, Fizazi K, Logothetis C, Gravis G, et al. Randomized, double-blind, Phase III trial of ipilimumab versus placebo in asymptomatic or minimally symptomatic patients with metastatic chemotherapy-naive castration-resistant prostate cancer. J Clin Oncol. 2017;35(1):40–47. doi: 10.1200/JCO.2016.69.1584. [DOI] [PubMed] [Google Scholar]
- 5.Lu X, Horner JW, Paul E, Shang X, Troncoso P, Deng P, et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature. 2017;543(7647):728–732. doi: 10.1038/nature21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Japp AS, Kursunel MA, Meier S, Malzer JN, Li X, Rahman NA, et al. Dysfunction of PSA-specific CD8+ T cells in prostate cancer patients correlates with CD38 and Tim-3 expression. Cancer Immunol Immunother. 2015;64(11):1487–1494. doi: 10.1007/s00262-015-1752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gevensleben H, Dietrich D, Golletz C, Steiner S, Jung M, Thiesler T, et al. The immune checkpoint regulator PD-L1 Is highly expressed in aggressive primary prostate cancer. Clin Cancer Res. 2016;22(8):1969–1977. doi: 10.1158/1078-0432.CCR-15-2042. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Wang Q, Shi B, Xu P, Hu Z, Bai L, et al. Development of a sandwich ELISA for evaluating soluble PD-L1 (CD274) in human sera of different ages as well as supernatants of PD-L1+ cell lines. Cytokine. 2011;56(2):231–238. doi: 10.1016/j.cyto.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Frigola X, Inman BA, Lohse CM, Krco CJ, Cheville JC, Thompson RH, et al. Identification of a soluble form of B7–H1 that retains immunosuppressive activity and is associated with aggressive renal cell carcinoma. Clin Cancer Res. 2011;17(7):1915–1923. doi: 10.1158/1078-0432.CCR-10-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He Y, Gu J, Strom S, Logothetis CJ, Kim J, Wu X. The prostate cancer susceptibility variant rs2735839 near KLK3 gene is associated with aggressive prostate cancer and can stratify gleason score 7 patients. Clin Cancer Res. 2014;20(19):5133–5139. doi: 10.1158/1078-0432.CCR-14-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Q, Gregg JR, Gu J, Ye Y, Chang DW, Davis JW, et al. Genetic associations of T cell cancer immune response with tumor aggressiveness in localized prostate cancer patients and disease reclassification in an active surveillance cohort. Oncoimmunology. 2019;8(1):e1483303. doi: 10.1080/2162402X.2018.1483303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. Natural history of progression after PSA elevation following radical prostatectomy. JAMA. 1999;281(17):1591–1597. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 13.Bruce JY, Lang JM, McNeel DG, Liu G. Current controversies in the management of biochemical failure in prostate cancer. Clin Adv Hematol Oncol. 2012;10(11):716–722. [PubMed] [Google Scholar]
- 14.Wilt TJ, Jones KM, Barry MJ, Andriole GL, Culkin D, Wheeler T, et al. Follow-up of prostatectomy versus observation for early prostate cancer. N Engl J Med. 2017;377(2):132–142. doi: 10.1056/NEJMoa1615869. [DOI] [PubMed] [Google Scholar]
- 15.Kamoun A, Cancel-Tassin G, Fromont G, Elarouci N, Armenoult L, Ayadi M, et al. Comprehensive molecular classification of localized prostate adenocarcinoma reveals a tumour subtype predictive of non-aggressive disease. Ann Oncol. 2018;29(8):1814–1821. doi: 10.1093/annonc/mdy224. [DOI] [PubMed] [Google Scholar]
- 16.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discovery. 2015;14(8):561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg PS, Katki H, Swanson CA, Brown LM, Wacholder S, Hoover RN. Quantifying epidemiologic risk factors using non-parametric regression: model selection remains the greatest challenge. Stat Med. 2003;22(21):3369–3381. doi: 10.1002/sim.1638. [DOI] [PubMed] [Google Scholar]
- 18.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Flies DB. Molecular mechanisms of T cell co-stimulation and co-inhibition. Nat Rev Immunol. 2013;13(4):227–242. doi: 10.1038/nri3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zang X, Thompson RH, Al-Ahmadie HA, Serio AM, Reuter VE, Eastham JA, et al. B7–H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc Natl Acad Sci USA. 2007;104(49):19458–19463. doi: 10.1073/pnas.0709802104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon ED, Hurwitz AA, Foster BA, Madias C, Feldhaus AL, Greenberg NM, et al. Manipulation of T cell costimulatory and inhibitory signals for immunotherapy of prostate cancer. Proc Natl Acad Sci USA. 1997;94(15):8099–8103. doi: 10.1073/pnas.94.15.8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kakoulidou M, Giscombe R, Zhao X, Lefvert AK, Wang X. Human Soluble CD80 is generated by alternative splicing, and recombinant soluble CD80 binds to CD28 and CD152 influencing T-cell activation. Scand J Immunol. 2007;66(5):529–537. doi: 10.1111/j.1365-3083.2007.02009.x. [DOI] [PubMed] [Google Scholar]
- 23.Zang X, Allison JP. The B7 family and cancer therapy: costimulation and coinhibition. Clin Cancer Res. 2007;13(18 Pt 1):5271–5279. doi: 10.1158/1078-0432.CCR-07-1030. [DOI] [PubMed] [Google Scholar]
- 24.Wu D, Tang R, Qi Q, Zhou X, Zhou H, Mao Y, et al. Five functional polymorphisms of B7/CD28 co-signaling molecules alter susceptibility to colorectal cancer. Cell Immunol. 2015;293(1):41–48. doi: 10.1016/j.cellimm.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 25.Leite KR, Reis ST, Viana N, Morais DR, Moura CM, Silva IA, et al. Controlling RECK miR21 Promotes Tumor Cell invasion and is related to biochemical recurrence in prostate cancer. J Cancer. 2015;6(3):292–301. doi: 10.7150/jca.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pasero C, Truneh A, Olive D. Cosignaling molecules around LIGHT-HVEM-BTLA: from immune activation to therapeutic targeting. Curr Mol Med. 2009;9(7):911–927. doi: 10.2174/156652409789105589. [DOI] [PubMed] [Google Scholar]
- 27.Sedy JR, Ramezani-Rad P. HVEM network signaling in cancer. Adv Cancer Res. 2019;142:145–186. doi: 10.1016/bs.acr.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 28.Knee DA, Hewes B, Brogdon JL. Rationale for anti-GITR cancer immunotherapy. Eur J Cancer. 2016;67:1–10. doi: 10.1016/j.ejca.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Ronchetti S, Ricci E, Petrillo MG, Cari L, Migliorati G, Nocentini G, et al. Glucocorticoid-induced tumour necrosis factor receptor-related protein: a key marker of functional regulatory T cells. J Immunol Res. 2015;2015:171520. doi: 10.1155/2015/171520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sfanos KS, Bruno TC, Maris CH, Xu L, Thoburn CJ, DeMarzo AM, et al. Phenotypic analysis of prostate-infiltrating lymphocytes reveals TH17 and Treg skewing. Clin Cancer Res. 2008;14(11):3254–3261. doi: 10.1158/1078-0432.CCR-07-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin SP, Seo HH, Shin JH, Park HB, Lim DP, Eom HS, et al. Adenovirus expressing both thymidine kinase and soluble PD1 enhances antitumor immunity by strengthening CD8 T-cell response. Mol Ther. 2013;21(3):688–695. doi: 10.1038/mt.2012.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207(10):2187–2194. doi: 10.1084/jem.20100643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.