Abstract
The neuropeptide natalisin (NTL) has been determined to play essential roles in reproduction in two Diptera and one Coleoptera species. Whether NTL has similar or even different functions in Lepidoptera remains to be determined. Here, we cloned the NTL transcript in the common cutworm moth Spodoptera litura. This transcript encodes a 438-amino acid protein. Twelve putative Sl-NTL neuropeptides were defined by cleavage sites. These NTL peptides share a DDPFWxxRamide C-terminal motif. The expressions of Sl-NTL is low during the egg and larval stages, which increased to a higher level during the pupal stage, and then reached the maximum during the adult stage. Moreover, the expression pattern during the pupal stage is similar between sexes while during the adult stage, it is dimorphic. To explore the function of Sl-NTL and assess its potential as a target for pest control, we knocked down the expression of Sl-NTL in both sexes by using bacteria-mediated RNAi. This technique significantly down regulated (reduced up to 83%) the expression of Sl-NTL in both sexes. Knocking down Sl-NTL expression did not significantly affect its development, survival and morphology but significantly reduced adults’ reproductive behavior (including female calling, male courtship, mating and remating patterns and rates) and reproductive output (offspring gain reduced more than 70%).
Subject terms: Biological techniques, Molecular biology, Zoology
Introduction
Insects are currently the most speciose animal group, including nearly a million of species that occur almost everywhere on the earth1,2. The study of insect physiological and behavioral adaptations and diversity not only will facilitate our understanding of sexual selection mechanisms and evolutionary ecology of insects, but also will be helpful for the development of novel strategies to control pest species with a reduced or minimal harm to the environment and especially to non-target species3–5. Neuropeptides are one of the key players in information transfer, acting as important regulators of physiology and behavior in relation to development and reproduction in insects.
Tachykinin-related peptides (TRP)6 and Natalisin (NTL)7 are closely related neuropeptides found in insects. TRP and NTL share the common sequence motif FxxxRamide (C-terminally amidated), which is FxGxRamide in TRPs while in NTL is FxPxRamide for Diptera and FWxxRamide for Coleoptera and other insects. Similarly, the receptors of TRP and NTL are also closely related but unequivocally form separate clusters7,8. All known TKRP receptors are class A G-protein coupled receptors (GPCRs)6,9. One of these GPCRs, which was previously known as TRP receptor (TRPR)10,11, was identified as the NTL receptor (NTLR)7. Tachykinin-related peptides have multiple functions in insects, such as myotropic activity12, diuretic function13, control of lipid metabolism14 and modulation of olfactory neurons15,16. Comparatively, the function of NTL is exclusively linked to reproduction of insects. Knockdown of NTL by transgenic RNAi in Drosophila melanogaster showed lower mating rate and shorter inoculation duration than those of wild-type flies, whereas knockdown of NTL by dsRNA injection in Tribolium castaneum resulted in lower egg production than controls7. In the oriental fruit fly, Bactrocera dorsalis, RNA interference mediated by dsRNA injection in adults significantly reduced male and female mating frequencies8. Furthermore, a study showed that tachykinin controlled male-specific aggressive behaviour in D. melanogaster by acting on the NTLR17, suggesting that the NTL and tachykinin signalling systems might interact with each other.
So far, NTL orthologs have been found in almost twenty insect species from five orders, such as D. melanogaster7 and Glossina morsitans morsitans18 from Diptera, T. castaneum7 and Nicrophorus vespilloides19 from Coleoptera, Bombyx mori7 and S. exigua20 from Lepidoptera, and Locusta migratoria21 from Orthoptera. NTL orthologs have also been found in non-insect organisms, such as mites22,23 and crayfish24. However, the function of NTL has only been determined in three species from two insect orders, including T. castaneum from Coleoptera, B. dorsalis and D. melanogaster from Diptera7,8. Whether NTL has similar or even different functions in other insect or non-insect species remains to be determined. Interestingly, Jiang et al.22 found that the honey bees Apis mellifera lack both NTL and the NTL receptor in their genome sequences, while its ectoparasite Varroa destructor have these two genes, providing a foundation for the development of novel varroa-mite-specific control agents.
In the present study, we identified the NTL transcript in the common cutworm moth Spodoptera litura and characterized its structure and phylogenetic status among insect and non-insect species. We then tested the mRNA expression pattern of NTL in relation to different developmental stages and tissues of males and females. To shed some light on the function of NTL and explore its potential as a target for novel insecticides, we knockdown NTL expression in both sexes by using bacteria-mediated RNAi and tested whether and how silencing of NTL will affect the developmental and reproductive fitness in this insect. To our knowledge, this is the first study to investigate the function of NTL in Lepidoptera and for the first time using bacteria-mediated RNAi in S. litura.
RNA interference (RNAi) is a valuable tool for gene functional analysis and determination, and also is an emerging technology to provide novel approaches for pest control in agriculture and disease treatment in humans25. In insects, RNAi can be induced by injection of in vitro synthesized dsRNA or the oral route, either by feeding synthesized dsRNA directly or by feeding bacteria expressing the dsRNAs in vivo (bacteria-mediated RNAi)26,27. Bacterially expressed dsRNA is cheaper than producing dsRNA in vitro with a kit, particularly when used in large scale gene function analysis28. More importantly, bacteria-mediated RNAi is a promising technology to provide sustainable and environmentally sound approaches to control insect pests and plant pathogens25.
Spodoptera litura Fabricius (Lepidoptera: Noctuidae) is known as tobacco cutworm or cotton leafworm, and is one of the world’s key agricultural pests due to its strong pesticide resistance, alternating generations and omnivorous characteristics. The extensive and indiscriminate use of pesticides for the prevention and control of this pest have caused severe damage to the environment and even further increased its insecticide resistance29–31. Therefore, sustainable and environmentally sound control strategies such as bacteria-mediated RNAi based control techniques32–35 are imperative to control this pest.
Methods
Insects
Spodoptera litura larvae were reared on an artificial diet36 at 25 ± 1 °C and a relative humidity of 60–70% with a photoperiod of 14:10 h light:dark photoperiod regime. Newly eclosed male and female moths were maintained in separate cages to ensure virginity. Adult moths were maintained in the same environment and fed with 10% honey solution.
Molecular cloning
Total RNA was extracted from adult moths with Trizol (Takara, China) according to the manufacturer’s protocol, and the purity and concentration of RNA was measured by using a spectrophotometer (NanoDrop 2000, USA).
We obtained a partial mRNA fragment (3′ end missing) of NTL from S. litura in our previous RNA-seq analysis. Based on the partial sequence, gene specific primer (GSP) and nested gene specific primer (NGSP) were designed and synthesized for 3′-RACE (Table 1). The first round of 3′-RACE amplification was performed by using 3′-ready-cDNA with UPM and GSPf (SMARTer Kit, Clontech, USA); then the first round product was diluted at 1:100 and used as templates for nested PCR reactions with UPM and GSPf. All amplifications were performed with 50 μl reaction mixtures containing 1 μl of template, using the same program: 94 °C for 5 min, followed by 5 cycles of 94 °C for 30 s, 72 °C for 4 min, and 5 cycles of 94 °C for 30 s, 70 °C 30 s, 72 °C 4 min; then 25 cycles of 94 °C for 30 s, 68 °C 30 s, 72 °C for 4 min; and a final extension step of 72 °C for 10 min. The PCR products were then cloned into trans1-T1 cells using pEASY-T5 zero vector system (TransGen, China). The plasmids were isolated using TIANPrep (TransGen, China) and were sequenced by Beijing Huada Biological Company.
Table 1.
Primers for RACE, qPCR and RNAi.
| Primer names | Primer sequences (5ʹ–3ʹ) | Uses |
|---|---|---|
| NTL-GSPr | GAGGCAGGAGAGAAACAGACGACCCTTT | 3′-RACE |
| NTL-NGSPr | GGGTAACCGTGGAAGGCGGAAGAC | 3′-RACE |
| NTL-Qf | AGACAGGCGTGGTGCTATTG | qPCR |
| NTL-Qr | TTCCTCTTTGTGGAGTGTAGTTCC | qPCR |
| Actin-Qf | CATCTACGAAGGTTACGCCCT | qPCR |
| Actin-Qr | AGCGGTGGTGGTGAAAGAGTA | qPCR |
| dsNTL-F | AAGGAAAAAAGCGGCCGCGCGTGGTGCTATTGAAGA | RNAi |
| dsNTL-R | CCGCTCGAGTGTAATTGAGCTGCCAGTT | RNAi |
| dsEGFP-F | AAGGAAAAAAGCGGCCGCAAGCAGCACGACTTCTTC | RNAi |
| dsEGFP-R | CCGCTCGAGGCTCAGGTAGTGGTTGTC | RNAi |
Underlined were protecting nucleotides and bold ones were NotI and XhoI restriction sites.
Gene expression in relation to development, sex and tissues
The development duration of S. litura eggs, larvae and pupae is about 3, 18 and 11 days, respectively37. Adult moths can live up to 10 days but most matings and ovipositions occurred in the first three days after eclosion37. Therefore, in the present study, we collected samples from 1-d-old eggs (10 eggs per sample), 0- (1st instar), 9- (3rd instar) and 18-d-old (4th instar) larvae, 0-, 6- and 11-d-old male and female pupae, and 0-, 1- and 2-d-old adult male and female virgin moths (5 insects per sample), respectively. Heads, thoraxes and reproductive systems were also sampled from 0-d-old male and female adults (8 insects per sample). Sampled female reproductive systems included oviduct, vestibulum, bursa copulatrix, ductus seminalis, spermatheca and its duct and gland, while male reproductive systems contain vesicula seminalis, vas deferens, accessory glands and ductus ejaculatorius38. Total RNA was extracted from each of these samples using a RNA prep pure Tissue kit (TianGen, China). The purity and concentration of the RNA were assessed as above. Three replicates were used for each category.
First strand cDNA synthesis was performed using a PrimeScript RT reagent Kit (Perfect Real Time) (TaKaRa, China). Real-Time PCR was performed with gene specific primers for Sl-NTL (Table 1) using the TB Green Premix Ex Taq II (TaKaRa, China) in a volume of 25 μl. Actin was used as a reference gene39,40. Reactions were run in triplicate on the QuantStudio 7 Flex (Thermo Fisher Scientific, USA) using the following reaction condition: 95 °C for 5 min followed by 40 cycles of 95 °C for 30 s, 60 °C for 34 s. Analysis of the dissociation curves for the target and reference genes showed a single melt peak and the efficiencies of the target and reference genes were similar. The 2−ΔΔCT method41 was used to calculate the relative quantities of the target genes.
Vector construction and dsRNA preparation
A 255 bp fragment (nucleotide 1055–1309 of Sl-NTL transcript, GenBank ID: MK673156) was designed for RNAi target by using siDirect version 2.0. This fragment was amplified by RT-PCR using total RNA as a template and primers (Table 1) containing NotI and XhoI restriction sites. Amplification reactions comprised 30 cycles of 94 °C for 30 s, 60 °C for 30 s and 72 °C for 60 s, with a final extension step of 72 °C for 10 min. PCR products were confirmed by using a 1.5% agarose gel and purified by using a SanPrep DNA Gel Extraction Kit (Sangon Biotech, China). The PCR product was then cloned into the plasmid L4440 (obtained from Addgene)26,27 between the NotI and XhoI sites. The resulting recombinant vector L4440-SlNTL was introduced into competent HT115(DE3) cells. To produce dsRNA, single colonies of HT115(DE3) bacteria containing L4440-SlNTL or L4440 vector were grown for 14 h with shaking in LB with 100 μg/ml ampicillin and 12.5 μg/ml tetracycline at 37 °C. Synthesis of T7 polymerase was induced by addition of IPTG to 0.4 mM and the bacteria were incubated under shaking for an additional 4 h at 37 °C. The expressed NTL dsRNA was extracted from the bacteria by using RNAiso Plus (Takara, China) according to the manufacturer’s protocol. The length of the NTL dsRNA was confirmed by electrophoresis on 1% agarose gel. The quantity of NTL dsRNA was estimated by comparing the brightness of the NTL dsRNA band and a quantified RNA maker. The production of NTL dsRNA under above bacteria incubation system is about 7–9 μg dsRNA/1 ml bacteria culture.
A 372 bp fragment of the enhanced green fluorescence protein (EGFP) gene was selected as a control dsRNA and was amplified using designed specific primers (Table 1). The recombinant plasmid for EGFP dsRNA expression protocol was the same as that for L4440-SlNTL. The production of GFP dsRNA was also confirmed and quantified as above. The production of GFP dsRNA is also about 7–9 μg dsRNA/1 ml bacteria culture.
Bacterial-mediated RNAi and silencing efficiency
Above confirmed bacteria cultures were used for RNAi. To prepare bacterial cells for feeding, bacterial cells were collected from 30 ml IPTG-induced culture by centrifugation at 10,000 g for 2 min, resuspended in 1 ml sterile water. During our preliminary experiments, we have tried one-time ingestion test25 by feeding the third instar larvae with 20 μl per larva of the prepared bacteria solution, i.e., feed the solution only once in the lifetime. However, qPCR test of the expression of Sl-NTL in treated mature larvae and adults showed a low silencing efficiency (< 30%). We thus selected to use the continual ingestion method25 that may have better silencing efficiency in the present study. Briefly, the third instar larvae (9-d-old) were individually caged in 4 cm × 4 cm × 4 cm plastic cells and were fed with 20 μl of the prepared bacteria solution (dropped on the surface of the food) each day until larvae mature (18-d-old, stop feeding and preparing for pupation). S. litura larvae are gluttonous and thus starvation before bacteria feeding (often used in other insect species)25 is unneeded. To confirm the silencing efficiency, bacteria fed mature larvae (18-d-old) and 0-d-old males and 1-d-old females (maximum NTL expression during adult stage, Fig. 1) were collected and the transcription levels of Sl-NTL were quantified by qPCR as above. The heads, thoraxes and reproductive systems of adults were also dissected as above. Larvae feeding on the food added with 20 μl distilled water and food added with 20 μl solution containing the bacteria expressing EGFP dsRNA were set as controls. Three replicates were used for each treatment.
Figure 1.

Deduced amino acid sequences of NTL precursors (a) and prediction of mature peptides (b). The putative signal peptide at the N terminus is in italics and underlined. Canonical amidation with di-basic signals is marked by bold and underlined fonts. The putative mature peptides were marked by blue fonts. The calculated consensus logo is shown at the bottom.
Effect of NTL silencing on male and female reproductive fitness
The third instar larvae were individually fed with bacteria expressing Sl-NTL dsRNA continuously as above. Larvae feeding on the food added with distilled water and bacteria expressing EGFP dsRNA were also set as controls as above. After pupation, the male and female pupae were sexed according to the morphology of exterior paramera36 and maintained in separate cages to ensure virginity of adults after eclosion. Eclosed female and male moths were collected and reared in separate cages by feeding with 10% honey solution until they were used for the following experiments. Developmental duration, morphology and survival were also observed and recorded.
To determine the effect of NTL silencing on male reproductive behavior, L4440-SlNTL treated and control virgin males (n = 13) were paired with wild virgin females (one pair per box) at the beginning of the second scotophase after eclosion. Male reproductive behaviors were observed in the following two scotophases after pairing. Similarly, to determine the effect of NTL silencing on female reproductive behavior, L4440-SlNTL treated and control virgin females (n = 13) were paired with wild virgin males (one pair per box) at the beginning of the second scotophase after eclosion. Female reproductive behaviors were observed in the following two scotophases after pairing.
Following above treatments, behaviors were observed every 10 min by quickly scanning all pairs and recording the following: male courtship—the male jumping and fanning his wings over or around the female or if the male exposed his genitalia trying to engage the female’s genitalia; female calling—the area of the female ovipositor bearing the pheromone gland is extruded and exposed to the outside42; mating—the two insects engaged by the tip of the abdomen. The events and duration of female calling (a calling can be short, a few minutes, or long, a few hours, and usually can last up to tens of minutes) and mating behaviors (the mating duration of this species is about 45 min)43 were recorded. Male courtship is instantaneous (usually lasts a few seconds to tens of seconds) and thus is hard to record its duration. We thus recorded the number of courtships but did not record the duration of each courtship. The moths were provided with honey solution as food and paper strips as oviposition substrates. Illumination during observation was provided by a 15 W red light. Their lifetime mating patterns were recorded by observing all paired insect half-hourly during the scotophase as the mating duration of this species is about 45 min43. The first two days’ and lifetime fecundity (no. of eggs laid) and longevity were recorded. Matings were verified by dissecting dead females to count the number of spermatophores in their bursa copulatrix.
S. litura eggs are laid in clusters and are usually several layers thick (mostly 2–3 layers). Eggs adhered to each other and the paper tightly, and thus trying to separate them to count the numbers are very likely to damage the eggs. Therefore, we cut the egg clusters with the paper and then it was weighed (Wt1). The eggs were then incubated in boxes that contained food. After larvae emergence, the clean paper was weighed again (Wt2). The number of eggs was calculated by (Wt1–Wt2)/(weight of one egg). The weight of one egg was 0.046 mg, which was determined by weighing more than 10 thousand eggs and calculated previously. The number of emerged larvae were also recorded.
Statistics
Data on gene expression, number of eggs (the first two days after pairing and lifetime) and longevity were analysed using an ANOVA followed by Fisher’s LSD test for multiple comparisons. Data on reproductive behavior (calling, courtship and mating) and number of larvae were not normally distributed even after transformation and thus were analysed using the nonparametric Kruskal–Wallis test followed by Dunn’s procedure for multiple comparisons44. All analyses were conducted using SPSS 20.0. The rejection level was set at α < 0.05. All values were reported as mean ± SE.
Results
Molecular cloning and phylogenetic analysis
Based on RNA-seq and RACE, the full-length mRNA sequence of the putative NTL (Sl-NTL; 1593 bp; GenBank accession: MK673156) was cloned from S. litura. This mRNA contained a putative ORF of 1317 nucleotides encoding a 438-amino acid protein (Fig. 1a), flanked by a 5′-UTR of 95 bp and a 3′-UTR of 181 bp. BLASTP analysis showed that Sl-NTL has high identity to NTLs from other lepidopterans, such as S. exigua (AXY04276.1; 94.92% identity), but lower identity to NTLs from species of other insect orders. The first 22 amino acid residues of the precursor were predicted as a signal peptide for secretion by using SignalP 4.0. Twelve putative S. litura NTL peptides were defined by flanking dibasic cleavage sites (combinations of K and R) and the canonical amidation site (G) at the C-terminus. Alignment of these putative NTL peptides showed a consensus sequence DDPFWxxRamide (‘x’ indicates a variable residue and ‘amide’ represents the amidated C-terminus) at the C-terminus (Fig. 1b).
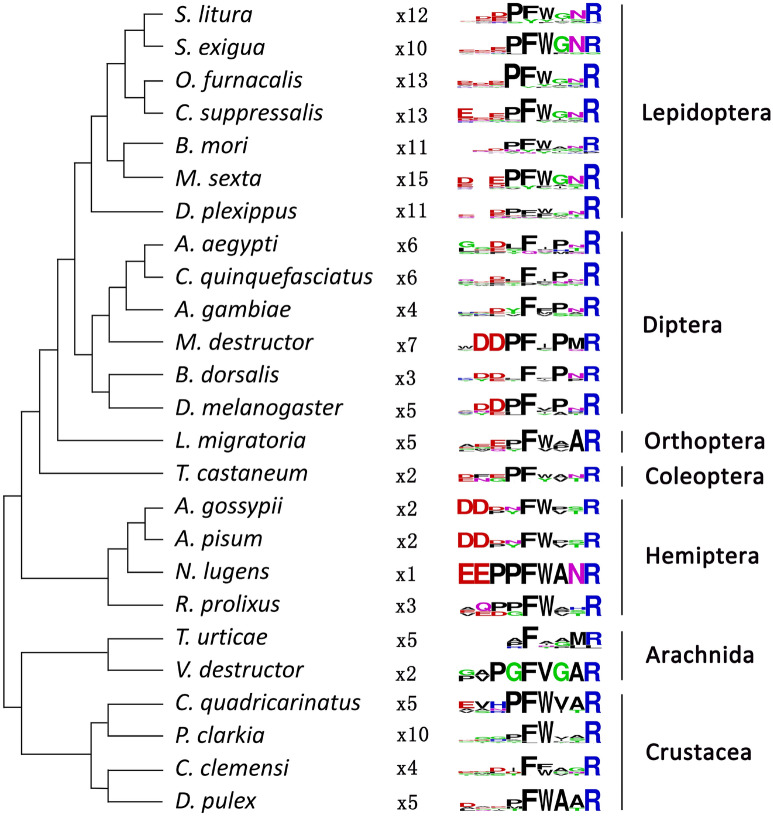
A hypothetical evolutionary tree and the C-terminal motifs of NTL from different organisms was drawn and presented in Fig. 2. The C-terminal motifs of NTL is FxPxRamide for Diptera while in other orders of Insecta, including Coleoptera, Lepidoptera, Orthoptera and Hemiptera, is FWxxRamide.
Figure 2.
The species tree and C-terminal motifs of NTL (drawn based on Jiang et al.7). The tree is based on the species tree. The calculated consensus logos of the C-terminal motifs of NTL from different species were shown behind the species names. The numbers of the paracopies carrying the motif are shown by x followed numbers. NTL sequences were pooled and provided in Dataset S1 (supplemental file).
Gene expression in relation to development, sex and tissues
Expression patterns of Sl-NTL in eggs and the whole body of insects from different developmental stages were examined using real time PCR and presented in Fig. 3a. Sl-NTL showed lower expression levels during the egg and larval stages, which increased to a higher level during the pupal stage, and then peaked during the adult stage. One-way ANOVA (F15,32 = 82.95, P < 0.0001) and post-hoc LSD test indicated that Sl-NTL showed the highest expression level in 1-d-old female adults (P < 0.05), followed by 2-d-old female adults (P < 0.05), then middle aged (6-d-old) male and female pupae (P < 0.05), and then 0-d-old male adults (P < 0.05). The expression in other stages were relative lower (P < 0.05).
Figure 3.
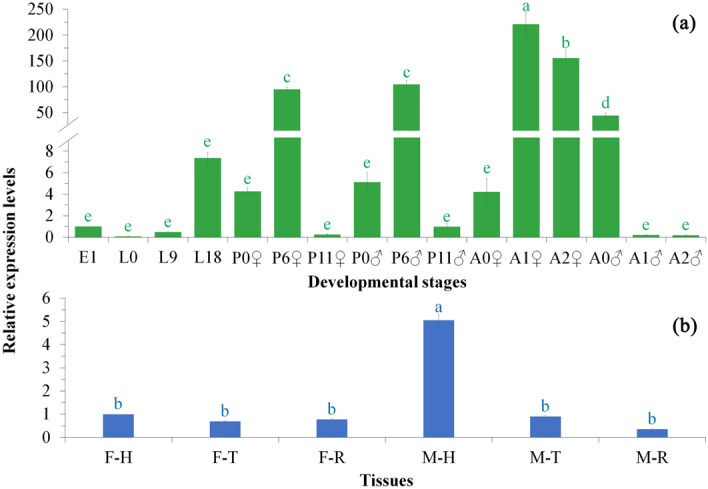
The expression of Sl-NTL in relation to development, sex and tissues. (a) expression patterns in the eggs and whole body of insects from different developmental stages. E1 refer to 1-d-old eggs; L0, L9 and L18 refer to 0-(1st instar), 9-(3rd instar) and 18-d-old (6th instar) larvae; P0♀, P6♀ and P11♀ refer to 0-, 6- and 11-d-old female pupae; P0♂, P6♂ and P11♂ refer to 0-, 6- and 11-d-old male pupae; A0♀, A1♀ and A2♀ refer to 0-, 1- and 2-d-old adult virgin females; and A0♂, A1♂ and A2♂ refer to 0-, 1- and 2-d-old adult virgin males. Expression in 1-d-old eggs was set as calibrator. (b) expression levels in 0-d-old male and female heads, thoraxes and reproductive systems. F–H, F–T and F–R refer to female heads, thoraxes and reproductive systems; M–H, M–T and M–R refer to male heads, thoraxes and reproductive systems; Expression in female heads was set as calibrator. For each parameter (Developmental stages or Tissues), bars with different letters are significantly different (P < 0.05); i.e., letters with the same color can be compared to derive the significance of difference between treatments.
Real-time fluorescence quantitative analysis also showed that Sl-NTL expressed in 0-d-old male and female heads, thoraxes and reproductive systems and showed significant variances (F15,32 = 54.45, P < 0.0001; Fig. 3b). The expression in male heads was significantly higher than that of thoraxes and reproductive systems (P < 0.05).
Bacterial-mediated RNAi and silencing efficiency
In this study, we constructed the dsRNA expression vector by inserting Sl-NTL into the plasmid L4440 within the NotI and XhoI sites, and then the recombinant plasmid was transformed into competent HT115 (DE3) cells for dsRNA expression. At the same time, we used a non-related gene, EGFP, as a control for dsRNA expression.
To knockdown the expression of Sl-NTL, the third instar larvae were individually caged in plastic cells and were fed with 20 μl of the prepared bacteria solution each day until larvae mature. Relative expression levels of Sl-NTL in NTL dsRNA males and females (feeding E. coli expressing NTL dsRNA) reduced (reduction percentages ranged from 45.88% to 82.70%) significantly in comparison with controls (feeding sterile water or E. coli expressing EGFP dsRNA) (Table 2).
Table 2.
Relative expression levels of the S. litura NTL in RNAi treated mature larvae and adults.
| Sample | NTL relative expression levels | |||||
|---|---|---|---|---|---|---|
| Water (control)a | GFP dsRNA (control) | NTL dsRNA | Reduction percentagesb | F-value (ANOVA) | P value | |
| Mature larvae (whole body) | 1.00a | 0.59 ± 0.28a | 0.21 ± 0.09a | 79.00; 64.41 | F2,6 = 1.90 | > 0.05 |
| Female adults (whole body) | 1.00a | 1.22 ± 0.03a | 0.37 ± 0.23b | 63.00; 69.67 | F2,6 = 36.16 | < 0.001 |
| Male adults (whole body) | 1.00a | 1.33 ± 0.69a | 0.23 ± 0.33b | 77.00; 82.70 | F2,6 = 19.47 | < 0.01 |
| Heads of female adults | 1.00a | 1.05 ± 0.13a | 0.36 ± 0.09b | 64.00; 65.71 | F2,6 = 17.02 | < 0.01 |
| Heads of male adults | 1.00a | 1.02 ± 0.20a | 0.32 ± 0.06b | 68.00; 68.03 | F2,6 = 10.85 | < 0.01 |
| Thoraxes of female adults | 1.00a | 0.85 ± 0.13a | 0.46 ± 0.12b | 54.00; 45.88 | F2,6 = 7.15 | < 0.05 |
| Thoraxes of male adults | 1.00a | 1.08 ± 0.09a | 0.25 ± 0.05b | 75.00; 76.85 | F2,6 = 58.44 | < 0.0001 |
| Reproductive systems of female adults | 1.00a | 1.02 ± 0.16a | 0.45 ± 0.14b | 55.00; 55.80 | F2,6 = 7.26 | < 0.05 |
| Reproductive systems of male adults | 1.00a | 1.02 ± 0.06a | 0.23 ± 0.090.b | 77.00; 77.45 | F2,6 = 48.79 | < 0.0001 |
aWater treated insects were used as calibrators. For each parameter (in each line), values with different letters are significantly different (P < 0.05).
bThe first value is the Reduction Percentage relative to Water, and the second value is the Reduction Percentage relative to GFP dsRNA.
Effects of NTL silencing on male and female reproductive fitness
Knock down NTL expression did not show significant effect on the development, survival and morphology in S. litura. The developmental duration and survival rate from the 3rd instar larvae (start to feed on bacteria) to adult emergence in treated and untreated insects were 19–21 days and 87–91%, respectively. No obvious morphological differences were found between treated and untreated insects.
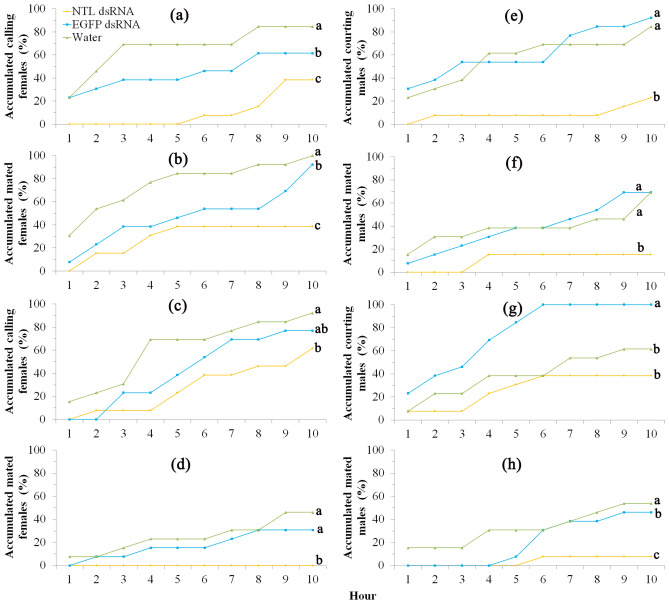
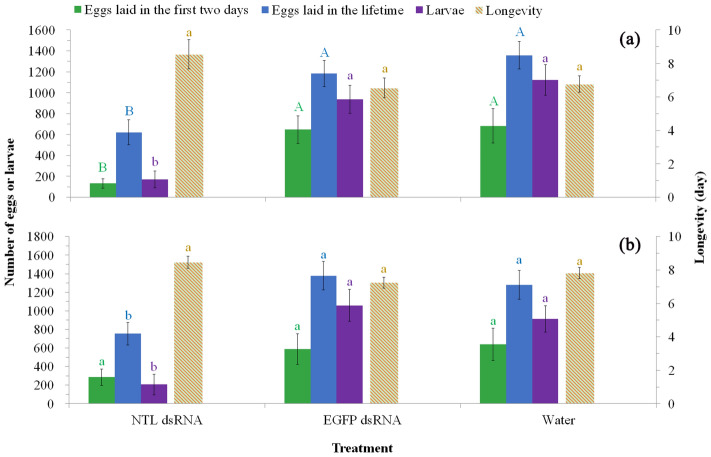
However, NTL dsRNA females (feeding on E. coli expressing NTL dsRNA) showed significant lower calling rate (χ2 = 20.29, P < 0.0001 for the first scotophase and χ2 = 6.68, P < 0.05 for the second scotophase; Fig. 4a,c) and shorter calling duration (Table 3), and lower mating rate with wild males (χ2 = 14.20, P < 0.001 for the first scotophase and χ2 = 19.43, P < 0.0001 for the second scotophase; Fig. 4b,d) in comparison with control females (feeding on sterile water or E. coli expressing EGFP dsRNA). The lifetime mating events in these females were also recorded and presented in Fig. 5a–c. Only 38% NTL dsRNA females mated in their lifetime and no remating occurred in these females (mean mating rate is 0.38 time per female) while almost all control females mated at least once (92% for EGFP dsRNA females and 100% for Water females) and some of them mated up to four times (mean mating rate is 1.54 and 1.77 times per female for EGFP dsRNA and Water females, respectively). Fecundity data showed NTL dsRNA females laid significant fewer eggs in the first two days after pairing (F2,36 = 6.13, P < 0.01) and lifetime (F2,36 = 9.41, P < 0.001), and have fewer offspring (larvae) (χ2 = 18.53, P < 0.0001) than those of control females (Fig. 6a). NTL dsRNA females showed relative higher longevity than controls but not significantly different (F2,36 = 2.66, P > 0.05).
Figure 4.
Effects of NTL RNAi on female (a-d; treated females paired with untreated males; thirteen pairs were used, i.e., n = 13) and male (e–h; treated males paired with untreated females; also thirteen pairs were used, i.e., n = 13) reproductive behavior in the first two days after pairing. Female calling behavior in the first (a) and second (c) scotophase. Female mating behavior in the first (b) and second (d) scotophase. Male courtship behavior in the first (e) and second (g) scotophase. Male mating behavior in the first (f) and second (h) scotophase. Data were not normally distributed even after transformation and thus were analysed using the nonparametric Kruskal–Wallis test followed by Dunn’s procedure for multiple comparisons. For each parameter, i.e., in each of the subgraphs, lines with different letters are significantly different (P < 0.05).
Table 3.
Effects of NTL RNAi on female calling duration, number of male courtships and mating duration in S. litura.
| Pairing patterns | Parameters measured | Water (control) | GFP dsRNA (control) | NTL dsRNA | χ2-value | P value |
|---|---|---|---|---|---|---|
| Treated female paired with untreated male | Female calling duration in the 1st scotophase (min) | 68.5 ± 24.4a | 55.4 ± 23.1a | 7.69 ± 4.55b | 3.53 | 0.039 |
| Female calling duration in the 2nd scotophase (min) | 79.2 ± 27.4a | 61.5 ± 24.3a | 53.1 ± 20.9a | 0.27 | 0.767 | |
| Female mating duration (min) | 43.3 ± 1.3a | 43.5 ± 1.2a | 42.0 ± 2.0a | 0.20 | 0.823 | |
| Treated male paired with untreated female | Number of male courtships in the 1st scotophase | 1.46 ± 0.33a | 1.23 ± 0.23a | 0.46 ± 0.27b | 3.90 | 0.029 |
| Number of male courtships in the 2nd scotophase | 1.46 ± 0.43ab | 2.62 ± 0.56a | 0.77 ± 0.34b | 4.67 | 0.015 | |
| Male mating duration (min) | 44.7 ± 1.7a | 44.0 ± 1.6a | 43.3 ± 3.3a | 0.07 | 0.930 |
Thirteen pairs were used for each Pairing pattern. Female calling durations or the numbers of male courtships were the means of all tested females or males in the 1st or 2nd scotophase. Female or male mating durations were the means of male or female matings occurred in the first two scotophases. Data were not normally distributed even after transformation and thus were analysed using the nonparametric Kruskal–Wallis test followed by Dunn’s procedure for multiple comparisons. For each parameter (in each line), values with different letters are significantly different (P < 0.05).
Figure 5.
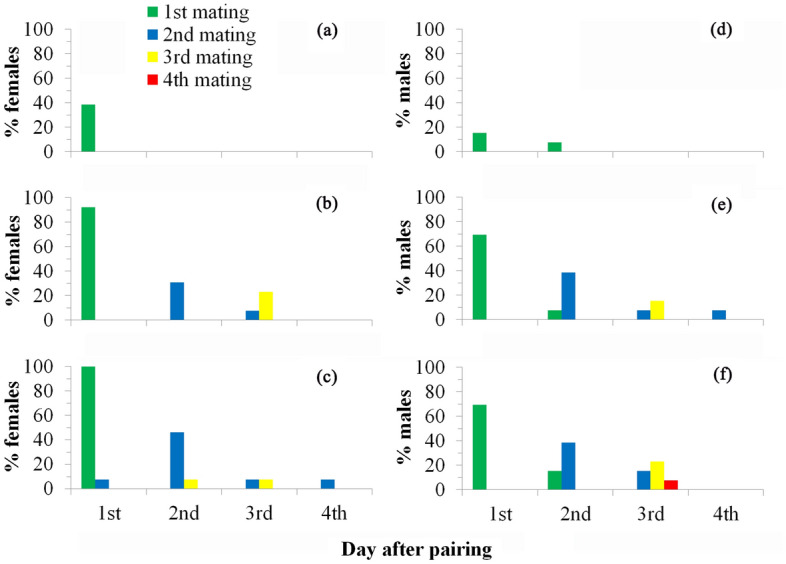
Effects of NTL RNAi on male and female lifetime mating and remating patterns. (a) mating pattern of NTL dsRNA females; (b) mating pattern of EGFP dsRNA females; (c) mating pattern of Water females; (d) mating pattern of NTL dsRNA males; (e) mating pattern of EGFP dsRNA males; (f) mating pattern of Water males.
Figure 6.
Effects of NTL RNAi on female (a) and male (b) reproductive output and longevity (mean number of days the adults live, i.e., the duration from moth eclosion to death). For each parameter (Eggs laid in the first two days, Eggs laid in the lifetime, Larvae or Longevity) of RNAi on female (a) or RNAi on male (b), bars with different letters are significantly different (different uppercase letters indicate P < 0.01 and different lowercase letters indicate P < 0.05); i.e., letters with the same color can be compared to derive the significance of difference between treatments in the same subgraph, (a) or (b).
Similarly, NTL dsRNA males showed significant lower courting rate (χ2 = 19.57, P < 0.0001 for the first scotophase and χ2 = 12.55, P < 0.01 for the second scotophase; Fig. 4e,g) and fewer courtships (Table 3), and lower mating rate with wild females (χ2 = 14.92, P < 0.001 for the first scotophase and χ2 = 12.24, P < 0.01 for the second scotophase; Fig. 4f,h) in comparison with control males. The lifetime mating events in these males were also recorded and presented in Fig. 5d–f. NTL dsRNA males showed lower mating rate in their lifetime (23% mated) and no remating occurred in these males (mean mating rate is 0.23 time per female) while most control males mated at least once (77% for EGFP dsRNA males and 85% for Water males) and some of them mated up to three times (mean mating rate is 1.54 and 1.62 times per male for EGFP dsRNA and Water males, respectively). Fecundity test showed that wild females mated with dsNTL males laid fewer eggs in the first two days after pairing (but not significant: F2,36 = 1.70, P > 0.05) and lifetime (significant: F2,36 = 5.42, P < 0.01), and have significant fewer offspring (larvae) (F2,36 = 12.59, P < 0.01) than those of wild females mated with control males (Fig. 6b). NTL dsRNA males showed relative higher longevity than controls but not significant (F2,36 = 3.21, P > 0.05).
Discussion
To date, NTL orthologs have been found in more than twenty arthropod species (Fig. 2). Most of these orthologs belong to Insecta species (Fig. 2) and more importantly, NTL has been proven to play an important function in the reproduction in three insect species, D. melanogaster, T. castaneum and B. dorsalis7,8. This evidence suggests that NTL is pervasive and may play essential roles in insect reproduction process.
The common cutworm moth S. litura is one of the key agricultural pests and is also notorious for developing insecticide resistance30,31. Developing environmental friendly control methods such as RNAi mediated management technique is imperative to control this pest. In the present study, we for the first time cloned and sequenced the NTL transcript in S. litura. This transcript contains a 1317 nucleotides ORF encoding a 438-amino acid protein (Fig. 1a). Twelve putative Sl-NTL neuropeptides were defined by flanking dibasic cleavage sites (combinations of K and R). Moreover, an amidated C-terminus was predicted for each mature peptide by a canonical amidation site (G). A complex enzymatic process is involved during the release of neuropeptides from their precursor proteins45,46. The C-terminal motifs of Sl-NTL is FWxxRamide, which is consistent to other species from Lepidoptera, Coleoptera, Orthoptera, Hemiptera and other non-insect species from Crustacea (Fig. 2). However, the C-terminal motifs of NTL is FxPxRamide in Diptera and in mites might be FxxxRamide. In addition, as showed in S. litura (Fig. 1) and other insects (Dataset S1), not all predicted peptides have the “Ramide” C-terminal structure, which can varied in the same or different species7,8. These variances on the C-terminal structure may reflect an evolutionary process based on the ligand–receptor activities7,47.
Real-time fluorescence quantitative analysis also showed that Sl-NTL expressed in both male and female adults’ heads, thoraxes and reproductive systems and showed variances. Tissue specific expression levels of NTL have been shown in other insects and the highest transcript levels of NTL were found in the CNS in T. castaneum and B. dorsalis7,8. The expression pattern of Sl-NTL in different developmental stages from larvae to adults (Fig. 3a) is quite similar to that of B. dorsalis8, i.e., lower expression levels during the larval stage, which increased to a higher level during the pupal stage, and then peaked at the beginning of the adult stage. S. litura adults eclose at dusk and no mating occurs in the eclosion scotophase (0-d-old). Maximum mating (approximately 70%) occurs during the subsequent scotophase after eclosion (1-d-old)48. The remaining unmated moths mate during the third scotophase (2-d-old). The correlation between NTL expression and adult sexual maturation in this species suggests this peptide has an important function in reproduction. However, the expression of NTL in S. litura is much lower during the egg stage while in B. dorsalis it is much higher (even higher than adult stage)8. This may suggest that NTL might have different functions during different developmental stage in different species.
To shed some light on the function of NTL and explore its potential as a target for novel insecticides, we knockdown NTL expression in both sexes by using bacteria-mediated RNAi and tested whether and how silencing of NTL will affect the developmental and reproductive fitness in this pest. Relative expression levels of Sl-NTL in NTL dsRNA males and females reduced (reduction percentage ranged from 45.88 to 82.70%) significantly in comparison with controls (Table 1). Both RNAi49 and bacteria intaking50 are likely to induce insect immune responses and thus may bring side effects on gene function study using bacteria-mediated RNAi. In the present study, knock down of NTL expression did not significantly affect the development, survival and morphology in S. litura. Moreover, our previous study also showed that feeding bacteria did not significantly affect the reproductive behavior and fecundity in S. litura (J. Xu, unpl. data). These results suggest that bacteria-mediated RNAi can be used for reproduction-related gene function analysis in this insect. Other studies25,51 also suggested that the bacteria-mediated RNAi can be a reliable technique for gene function study as these bacteria are non-pathogenic and the same and similar bacteria species are widely found in the gut of insects53. In the present study, NTL knock down significantly reduced adults’ reproductive behavior, including male courtship and female calling behavior in the first night (Fig. 4a,f), mating (reduced more than 50%) and remating pattern and rate (remating rate reduced more than 75%; no remating occurred in NTL knock down individuals while wild ones can mate up to 4 times) in both sexes in their lifetime (Figs. 4, 5). In D. melanogaster7 and B. dorsalis8, knocking down the expression of NTL also significantly reduced mating rate by about 40–75%. Similar to T. castaneum7, fecundity test also demonstrated that NTL knock down significantly reduced male and female productive output in S. litura (offspring gain reduced more than 70%; Fig. 6).
Based on the above results and similar results on NTL in other insect species7,8, we suggest that NTL may also play important roles in the process of reproduction in S. litura. However, the exact mechanism of NTL for influencing mating behaviors is still unclear. Previous studies have demonstrated that NTLs may regulate reproductive behaviors via activating their receptors (NTLRs; G protein-coupled receptors, GPCR) in insect based on ligand-receptor interaction analysis and NTLR-RNAi7,8,47. NTLR was expressed dominantly in the CNS in comparison with other tissues and NTLR mRNA was also confirmed in the digestive (midgut and hindgut) and reproductive systems7,47. These evidences suggested that NTL was released into the periphery, where it may bind to the receptor and implement various functions through GPCR signal systems. However, more investigation is required to reveal the signal cascades of NTLR in regulation reproductive behavior. Moreover, two NTLRs (Bm A32 and A33) were found in B. mori, which is different to other insect species that have only one copy of the NTLR7. And interestingly, Bm A32 was specific to BmNTL1, 3, and 5, which have the C-terminal FxxxRa consensus sequence, whereas BmNTLR A33 was specific to BmNTL10 and BmNTL11, which have the YxxxRa consensus sequence7. These results have suggested that the mechanism of NTL for influencing mating behaviors may be more complicated and varied in different insect taxa. Studies on the Sex Peptide (SP) and its receptor (SPR; also a GPCR) for the function on reproductive behavior in Drosophila have shown that the regulation mechanism is complicated, which is related to neural, physiological and molecular processes52–54. Therefore, future studies on NTL regulation mechanisms by taking account these (neural, physiological and molecular) processes are likely to achieve better progress in this field. The bacterial-mediated RNAi targeting NTL developed in this study have built a foundation for a better understanding on how the neuropeptide NTL is involved in reproductive physiology and may be a potential technique for pest control.
Supplementary information
Acknowledgements
We thank Dr. Su Ping Ong from Forest Research Institute Malaysia (FRIM) for English improvements, and the editor and four anonymous reviewers for their constructive comments. Research reported here was supported by projects from the National Natural Science Foundation Program of P.R. China (31560606; 32060357; 31760635), Science and Technology Planning Project in Key Areas of Yunnan Province (202001BB050002) and Joint Special Project of Yunnan Province for Agricultural Basic Research (2018FG001-002).
Author contributions
X.F.W., Z.C. and X.B.W. performed the experiments. J.X., P.C. and H.Y. designed the experiments. J.X. and X.F.W. wrote the paper.
Data availability
All data generated or analysed during this study are included in this article and its Supplementary Information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xia-Fei Wang and Zhe Chen.
Contributor Information
Jin Xu, Email: xujin2798@126.com.
Peng Chen, Email: pengchenn@126.com.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-84104-0.
References
- 1.Tomioka K, Matsumoto A. Circadian molecular clockworks in non-model insects. Curr. Opin. Insect Sci. 2015;7:58–64. doi: 10.1016/j.cois.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Derst C, et al. Evolution of neuropeptides in non-pterygote hexapods. BMC Evol. Biol. 2016;16:51. doi: 10.1186/s12862-016-0621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wenninger EJ, Averill AL. Effects of delayed mating on reproductive output of female oriental beetle Anomala orientalis (Coleoptera: Scarabaeidae) Agric. For. Entomol. 2006;8:221–231. doi: 10.1111/j.1461-9563.2006.00300.x. [DOI] [Google Scholar]
- 4.Michereff MFF, Vilela EF, Michereff Filho M, Nery DMS, Thierbaut JT. Effects of delayed mating and male mating history on the reproductive potential of Leucoptera coffeella (Lepidoptera: Lyonetiidae) Agric. For. Entomol. 2004;6:241–247. doi: 10.1111/j.1461-9555.2004.00227.x. [DOI] [Google Scholar]
- 5.Polajnar J, et al. Manipulating behaviour with substrate-borne vibrations: potential for insect pest control. Pest Manag. Sci. 2015;71:15–23. doi: 10.1002/ps.3848. [DOI] [PubMed] [Google Scholar]
- 6.Van Loy T, et al. Tachykinin-related peptides and their receptors in invertebrates: a current view. Peptides. 2010;31:520–524. doi: 10.1016/j.peptides.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Jiang HB, et al. Natalisin, a tachykinin-like signaling system, regulates sexual activity and fecundity in insects. Proc. Natl. Acad. Sci. USA. 2013;110:E3526–E3534. doi: 10.1073/pnas.1310676110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gui SH, Jiang HB, Liu XQ, Xu L, Wang JJ. Molecular characterizations of natalisin and its roles in modulating mating in the oriental fruit fly, Bactrocera dorsalis (Hendel) Insect Mol. Biol. 2017;26:103–112. doi: 10.1111/imb.12274. [DOI] [PubMed] [Google Scholar]
- 9.Satake, H. in Handbook of Hormones (eds Takei, Y., Ando, H. & Tsutsui, K.) 364–365 (Academic Press, London, 2016).
- 10.Monnier D, et al. NKD, a developmentally regulated tachykinin receptor in Drosophila. J. Biol. Chem. 1992;267:1298–1302. doi: 10.1016/S0021-9258(18)48429-3. [DOI] [PubMed] [Google Scholar]
- 11.Poels J, et al. Characterization and distribution of NKD, a receptor for Drosophila tachykinin-related peptide 6. Peptides (New York) 2009;30:545–556. doi: 10.1016/j.peptides.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 12.Nassel DR. Insect myotropic peptides: differential distribution of locustatachykinin- and leucokinin-like immunoreactive neurons in the locust brain. Cell Tissue Res. 1993;274:27–40. doi: 10.1007/BF00327982. [DOI] [PubMed] [Google Scholar]
- 13.Johard HAD, Coast GM, Mordue W, Nassel DR. Diuretic action of the peptide locustatachykinin I: cellular localisation and effects on fluid secretion in Malpighian tubules of locusts. Peptides (New York) 2003;24:1571–1579. doi: 10.1016/j.peptides.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Song W, Veenstra JA, Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung JW, et al. Neuromodulation of olfactory sensitivity in the peripheral olfactory organs of the American cockroach Periplaneta americana. PLoS ONE. 2013;8:e81361. doi: 10.1371/journal.pone.0081361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gui S-H, et al. Role of a tachykinin-related peptide and its receptor in modulating the olfactory sensitivity in the oriental fruit fly, Bactrocera dorsalis (Hendel) Insect Biochem. Mol. 2017;80:71–78. doi: 10.1016/j.ibmb.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Asahina K, et al. Tachykinin-expressing neurons control male-specific aggressive arousal in Drosophila. Cell. 2014;156:221–235. doi: 10.1016/j.cell.2013.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Caers J, et al. Peptidomics of neuropeptidergic tissues of the tsetse fly Glossina morsitans morsitans. J. Am. Soc. Mass Spectr. 2015;26:2024–2038. doi: 10.1007/s13361-015-1248-1. [DOI] [PubMed] [Google Scholar]
- 19.Benowitz KM, McKinney EC, Cunningham CB, Moore AJ. Predictable gene expression related to behavioral variation in parenting. Behav. Ecol. 2019;30:402–407. doi: 10.1093/beheco/ary179. [DOI] [Google Scholar]
- 20.Llopis-Gimenez A, Han Y, Kim Y, Ros VID, Herrero S. Identification and expression analysis of the Spodoptera exigua neuropeptidome under different physiological conditions. Insect Mol. Biol. 2019;28:161–175. doi: 10.1111/imb.12535. [DOI] [PubMed] [Google Scholar]
- 21.Hou L, Jiang F, Yang P, Wang X, Kang L. Molecular characterization and expression profiles of neuropeptide precursors in the migratory locust. Insect Biochem. Mol. 2015;63:63–71. doi: 10.1016/j.ibmb.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, et al. Ligand selectivity in tachykinin and natalisin neuropeptidergic systems of the honey bee parasitic mite Varroa destructor. Sci Rep. 2016;6:19547. doi: 10.1038/srep19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gui S-H, Jiang H-B, Smagghe G, Wang J-J. The neuropeptides and protein hormones of the agricultural pest fruit fly Bactrocera dorsalis: what do we learn from the genome sequencing and tissue-specific transcriptomes? Peptides (New York) 2017;98:29–34. doi: 10.1016/j.peptides.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Veenstra JA. The power of next-generation sequencing as illustrated by the neuropeptidome of the crayfish Procambarus clarkii. Gen. Comp. Endocr. 2015;224:84–95. doi: 10.1016/j.ygcen.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 25.Xu J, et al. RNA interference in moths: mechanisms, applications, and progress. Genes. 2016;7:88. doi: 10.3390/genes7100088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/S0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 27.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854–854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 28.Fraser AG, et al. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- 29.Zhou XM, Huang BQ. Insecticide resistance of the common cutworm (Spodoptera litura) and its control strategies. Entomol. Knowl. 2002;39:98–102. [Google Scholar]
- 30.Rehan A, Saleem MA, Freed S. Baseline susceptibility and stability of insecticide resistance of Spodoptera litura (F.) (Lepidoptera: Noctuidae) in the absence of selection pressure. Pak. J. Zool. 2011;43:973–978. [Google Scholar]
- 31.Xue L, Noll M. Drosophila female sexual behavior induced by sterile males showing copulation complementation. Proc. Natl. Acad. Sci. USA. 2000;97:3272–3275. doi: 10.1073/pnas.97.7.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang X, Liu X, Ma J, Zhao J. Silencing of cytochrome P450 CYP6B6 gene of cotton bollworm (Helicoverpa armigera) by RNAi. Bull. Entomol. Res. 2013;103:584–591. doi: 10.1017/S0007485313000151. [DOI] [PubMed] [Google Scholar]
- 33.Qi XL, et al. The effect of silencing arginine kinase by RNAi on the larval development of Helicoverpa armigera. Bull. Entomol. Res. 2015;105:555–565. doi: 10.1017/S0007485315000450. [DOI] [PubMed] [Google Scholar]
- 34.Tian H, et al. Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing dsRNA of a non-midgut gene. PLoS ONE. 2009;4:e6225. doi: 10.1371/journal.pone.0006225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo YQ, et al. Comparative analysis of cytochrome P450-like genes from Locusta migratoria manilensis: expression profiling and response to insecticide exposure. Insect Sci. 2012;19:75–85. doi: 10.1111/j.1744-7917.2011.01450.x. [DOI] [Google Scholar]
- 36.Li W, Zou WJ, Wang LH. The bionomics and control of Prodenia litura in Kunming. Southwest China J. Agric. Sci. 2006;19:85–89. [Google Scholar]
- 37.Li, C. Molecular Characterization and Functional Analysis of the Sex-Peptide Receptor in the Tobacco Cutworm Spodoptera litura. Master thesis, Southwest Forestry University (2014).
- 38.Ahmed A, Etman M, Hooper HS. Developmental and reproductive biology of Spodoptera litura (F.) (Lepidoptera: Noctuidae) Aust. J. Entomol. 1980;18:363–372. doi: 10.1111/j.1440-6055.1979.tb00868.x. [DOI] [Google Scholar]
- 39.Li C, et al. Molecular characterization and functional analysis of a putative sex-peptide receptor in the tobacco cutworm Spodoptera litura (Fabricius, 1775) (Lepidoptera: Noctuidae) Aust. Entomol. 2014;53:424–431. doi: 10.1111/aen.12088. [DOI] [Google Scholar]
- 40.Lu Q, et al. Identification and RNA interference of the pheromone biosynthesis activating neuropeptide (PBAN) in the common cutworm moth Spodoptera litura. J. Econ. Entomol. 2015;108:1344–1353. doi: 10.1093/jee/tov108. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Rainaa AK, Werginb WP, Murphyb CA, Erbe EF. Structural organization of the sex pheromone gland in Helicoverpa zea in relation to pheromone production and release. Arthropod Struct. Dev. 2000;29:343–353. doi: 10.1016/S1467-8039(01)00014-7. [DOI] [PubMed] [Google Scholar]
- 43.Li C, Yu J-F, Xu J, Liu J-H, Ye H. Reproductive rhythms of the tobacco cutworm, Spodoptera litura (Lepidoptera: Noctuidae) GSTF J. BioSci. 2012;2:25–29. doi: 10.5176/2251-3140_2.1.18. [DOI] [Google Scholar]
- 44.Zar JH. Biostatistical analysis. Upper Saddle River: Prentice Hall; 1999. [Google Scholar]
- 45.Veenstra JA. Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect Biochem. 2000;43:49–63. doi: 10.1002/(SICI)1520-6327(200002)43:2<49::AID-ARCH1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 46.Southey BR, Sweedler JV, Rodriguez-Zas SL. Prediction of neuropeptide cleavage sites in insects. Bioinformatics. 2008;24:815–825. doi: 10.1093/bioinformatics/btn044. [DOI] [PubMed] [Google Scholar]
- 47.Gui S-H, et al. Function of the natalisin receptor in mating of the oriental fruit fly, Bactrocera dorsalis (Hendel) and testing of peptidomimetics. PLoS ONE. 2018;13:e0193058. doi: 10.1371/journal.pone.0193058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloch G, Hazan E, Rafaeli A. Circadian rhythms and endocrine functions in adult insects. J. Insect Physiol. 2013;59:56–69. doi: 10.1016/j.jinsphys.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Fritz JH, Girardin SE, Philpott DJ. Innate immune defense through RNA interference. Sci. STKE. 2006;2006:27. doi: 10.1126/stke.3392006pe27. [DOI] [PubMed] [Google Scholar]
- 50.Eigenbrod T, Dalpke AH. Bacterial RNA: an underestimated stimulus for innate immune responses. J. Immunol. 2015;195:411–418. doi: 10.4049/jimmunol.1500530. [DOI] [PubMed] [Google Scholar]
- 51.Xiang S, et al. In vitro and in vivo gene silencing by TransKingdom RNAi (tkRNAi) Methods Mol. Biol. 2009;487:147–160. doi: 10.1007/978-1-60327-547-7_7. [DOI] [PubMed] [Google Scholar]
- 52.McGeary MK, Findlay GD. Molecular evolution of the sex peptide network in Drosophila. J. Evol. Biol. 2020;33:629–641. doi: 10.1111/jeb.13597. [DOI] [PubMed] [Google Scholar]
- 53.Misra S, Wolfner MF. Drosophila seminal sex peptide associates with rival as well as own sperm, providing SP function in polyandrous females. eLife. 2020;9:e58322. doi: 10.7554/eLife.58322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sturm S, Dowle A, Audsley N, Isaac RE. The structure of the Drosophila melanogaster sex peptide: identification of hydroxylated isoleucine and a strain variation in the pattern of amino acid hydroxylation. Insect Biochem. Mol. 2020;124:103414. doi: 10.1016/j.ibmb.2020.103414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this article and its Supplementary Information files.