Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is an important pathogen associated with a wide variety of infections in humans. The ability of MRSA to infect companion animals has gained increasing attention in the scientific literature. In this study, 334 dogs were screened for MRSA in two cities located in Rio de Janeiro State. The prevalence of MRSA in dogs was 2.7%. Genotyping revealed isolates from sequence types (ST) 1, 5, 30, and 239 either colonizing or infecting dogs. The genome of the canine ST5 MRSA (strain SA112) was compared with ST5 MRSA from humans—the main lineage found in Rio de Janeiro hospitals—to gain insights in the origin of this dog isolate. Phylogenetic analysis situated the canine genome and human strain CR14-035 in the same clade. Comparative genomics revealed similar virulence profiles for SA112 and CR14-035. Both genomes carry S. aureus genomic islands νSAα, νSAβ, and νSAγ. The virulence potential of the canine and human strains was similar in a Caenorhabditis elegans model. Together, these results suggest a potential of canine MRSA to infect humans and vice versa. The circulation in community settings of a MRSA lineage commonly found in hospitals is an additional challenge for public health surveillance authorities.
Subject terms: Antimicrobials, Bacteriology, Bacterial genetics, Clinical microbiology, Phylogeny
Introduction
Staphylococcus aureus is a common human pathogen involved in a wide variety of diseases. Infections caused by this agent range from skin and soft tissue infections to life-threatening bacteremia, endocarditis, osteomyelitis, and necrotizing pneumonia1,2. The high incidence of human infections caused by antimicrobial-resistant bacteria, especially methicillin-resistant S. aureus (MRSA), has led to an increased awareness related to the presence of resistant bacteria in domestic animals. Although S. aureus is not the most common staphylococcal species isolated from dogs3, the incidence of MRSA compared with that of methicillin-susceptible S. aureus (MSSA) is increasing in this host4,5. Consequently, from the one health perspective, domestic animals may serve as MRSA reservoirs in the community.
To date, both hospital-associated MRSA (HA-MRSA) and community-associated MRSA (CA-MRSA) lineages have been identified in companion animals6–8. Additionally, MRSA has been isolated from livestock, a finding with clear economic and public health implications9,10. The risk of zoonotic transmission of MRSA between humans and companion animals has been described in households, community, and healthcare settings11,12. However, to our knowledge, there is no study in the scientific literature addressing phylogenetic analysis and comparative genomics between MRSA strains from human and dog origins. In the present study we detected and molecularly characterized MRSA isolates obtained from asymptomatic and infected dogs in two cities in Rio de Janeiro State, Brazil: Campos dos Goytacazes and Rio de Janeiro. The antimicrobial resistance profile and the presence of lukSF-PV genes (encoding Panton-Valentine leukocidin) were also assessed. Additionally, because the ST5 lineage is emerging as the predominant MRSA in Rio de Janeiro hospitals13, we sequenced the whole genome of human and canine isolates from the ST5 lineage to gain some insights into the origin of the canine ST5 MRSA.
Results
Genotyping and resistance profile of MRSA from canine origins
The overall percentage of MRSA in the dogs analyzed was 2.7%; this corresponds to five isolates from Rio de Janeiro (5/210; 2.4%) and four isolates from Campos dos Goytacazes (4/124; 3.3%). A high percentage of MRSA (56.2%; n = 9; p < 0.0001) was detected amongst the 16 S. aureus isolates collected from 334 dogs (16/334; 4.8%). The three MRSA isolates (PA68, PA69, and PA73) from nasal carriers (3/88; 3.4%) were susceptible to most of the antimicrobials tested (Table 1). It is important to highlight that among the MRSA isolates cultured from infection cases (6/246; 2.4%) some displayed multiresistance profile (Table 1). MRSA strains SA07 and SA112 were resistant to six antibiotics (ciprofloxacin, clindamycin, chloramphenicol, erythromycin, gentamicin, and tetracycline) other than β-lactams (Table 1). The MRSA positive dogs were from different households; these dogs had no contact with each other.
Table 1.
Antimicrobial resistance and molecular characterization of the MRSA isolates collected from canines.
| Strain | Specimen | Antimicrobial resistance | ST | SCCmec | PFGE clone | lukSF |
|---|---|---|---|---|---|---|
| SA07* | Ear exudate | CP, CL, CH, ER, G, CF, TE | 5 | II | NY/J | − |
| SA112* | Ear exudate | CP, CL, CH, ER, G, CF, TE | 5 | II | NY/J | − |
| PA68* | Nasal swab | CL, ER, CF | 30 | IV | USA1100 | + |
| PA69* | Nasal swab | CF, TE | 30 | IV | USA1100 | + |
| PA73* | Nasal swab | CL, ER, CF | 30 | IV | USA1100 | + |
| NF62** | Ear exudate | CF, ER | 1 | IV | USA400 | + |
| NF191** | Exudate from soft tissue tumor | CF | 30 | IV | USA1100 | + |
| NF601** | Exudate from surgical site infection | CL, CH, ER, TE | 239 | III | BEC | − |
| NF661** | Skin exudate | CL, CH, ER, TE | 239 | III | BEC | − |
CP ciprofloxacin, CL clindamycin, CH chloramphenicol, ER erythromycin, G gentamicin, CF cefoxitin, TE tetracycline, ST MLST sequence typing, NY/J New York/Japan clone, BEC Brazilian epidemic clone, + presence, − absence, *Rio de Janeiro, **Campos de Goytacazes.
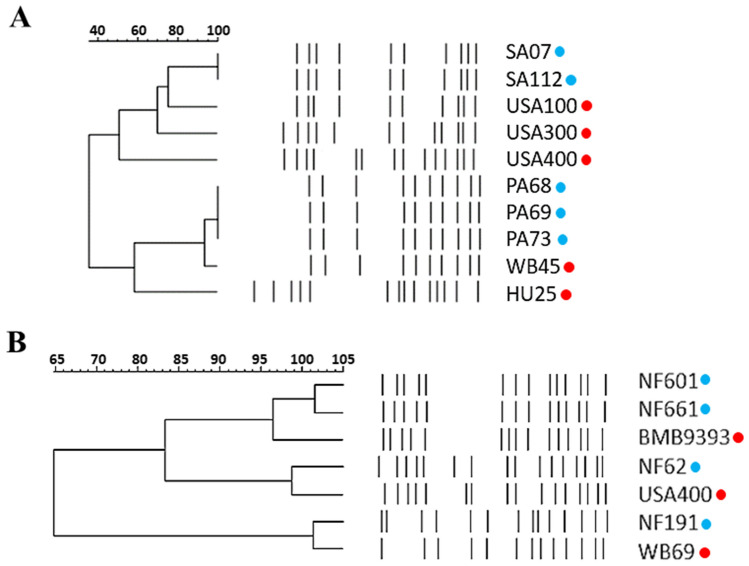
The isolates PA68, PA69, and PA73 (healthy dogs) and NF191 (infected soft tissue tumor) carried the SCCmec type IV, lukSF-PV genes, and displayed a PFGE pattern that resembled the strains WB45 (PA68, PA69, PA73) and WB69 (NF191), both of which were previously typed as PVL + and belong to the ST30-SCCmec IV linage, related to the Oceania Southwest Pacific Clone (OSPC/USA1100; Fig. 1). In addition, these USA1100-related isolates from dogs displayed the MLST allelic profile (2-2-2-2-6-3-2) corresponding to ST30. Two MRSA isolates (SA07 and SA112; both from ear exudates) carried SCCmec II. They did not harbor lukSF-PV genes and displayed a PFGE quite similar to that of HA-MRSA from the New York/Japan clone, also called USA100, which belongs to the ST5-SCCmec II lineage (Fig. 1A). In fact, these two canine SCCmec II isolates were typed as ST5. According to the dendrogram (Fig. 1B), the PFGE banding pattern of the MRSA isolate NF62 (collected from ear exudate) had 93.75% similarity with the band pattern of the strain USA400-0051, a representative of the USA400 clone. These data were further confirmed by MLST (allelic profile: 1-1-1-1-1-1-1), which allocated NF62 to ST1, and by SCCmec typing, which classified this isolate as SCCmec IV. In addition, the lukSF-PV genes were detected in NF62. Finally, the isolates NF601 (surgical site infection) and NF661 (pyoderma case) according to PFGE analysis were related to the strain BMB9393 (Fig. 1B); a representative of the Brazilian epidemic clone (BEC; ST239-SCCmec III). These canine isolates also carried SCCmec III and ST239 allelic profile (2-3-1-1-4-4-3), confirming the clonal association with BEC (Table 1).
Figure 1.
Pulsed-field gel electrophoresis (PFGE) of the SmaI-fragmented genomic DNA of MRSA isolates recovered from healthy and infected dogs in the state of Rio de Janeiro (red circles). (A) Dog isolates from Rio de Janeiro city and (B) dog isolates from Campos de Goytacazes city. USA100, USA300, USA400, WB45 and WB69 (USA1100), and BMB9393 and HU25 (BEC) were representatives of the MRSA international clones used for comparison purposes (blue circle). Dendrograms were generated from the PFGE patterns using GelCompar II software version 6.5.
Genomic comparison between canine and human CC5 MRSA
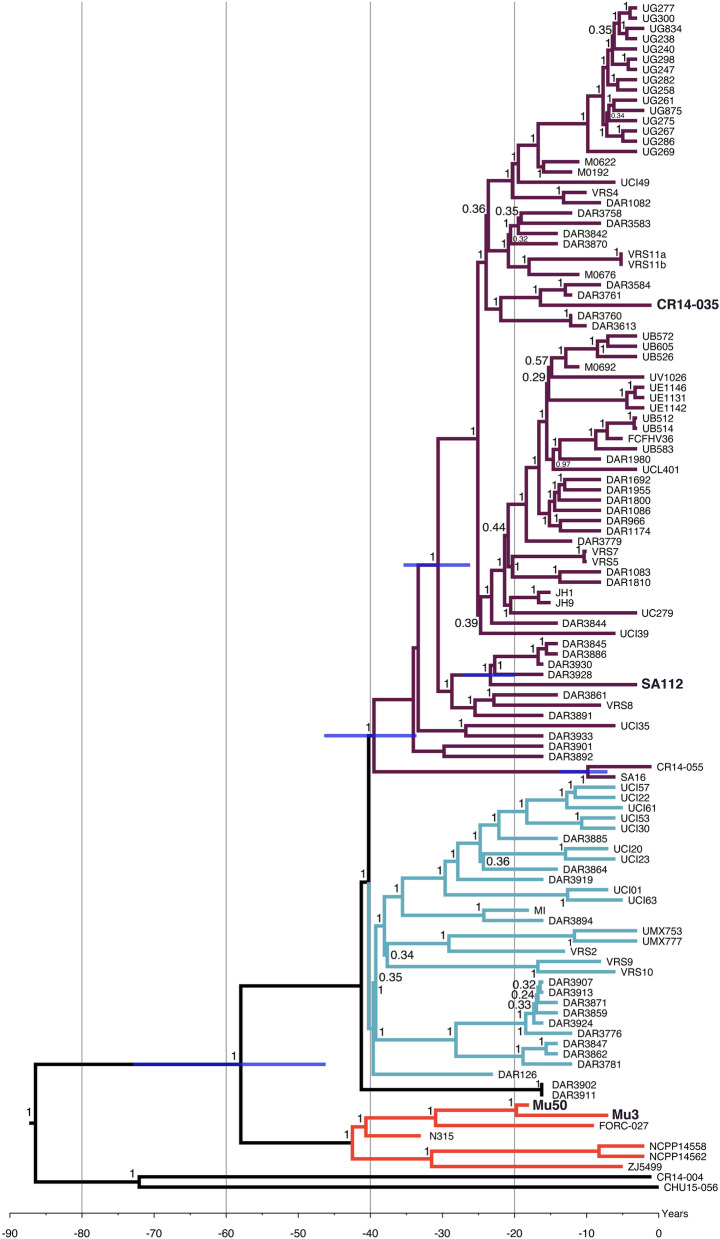
The general genomic characteristics of the MRSA of the ST5-SCCmec II lineage from a canine (strain SA112) and humans (strains CR14-035, CR14-004, CR14-055, and CHU15-056), used for whole genome sequencing, are presented in Table 2. The Bayesian phylogenetic inference using these and other CC5 genomes publicly available in GenBank revealed that the canine strain SA112 grouped in a clade (purple), which includes genomes of human-derived strains (Fig. 2). This clade grouped mostly the genomes of strains from the American continent (e.g.: USA, Brazil, Colombia, Chile, Mexico, Guatemala, among other countries) including CR14-035 and CR14-055 from hospitalized patients in Rio de Janeiro, and another Brazilian strain (FCFHV36) previously isolated from a case human osteomyelitis in the state of Santa Catarina, in the southern region of Brazil14. In addition, the purple clade clustered the genomes of the strains JH1 and JH9 collected from blood of a patient with congenital heart disease and endocarditis who was treated extensively with vancomycin without success in Baltimore, Maryland, USA (Fig. 2). The blue clade also grouped CC5 genomes from humans mostly from USA and the red clade from Asia; this last clade includes the genomes of Mu50 and Mu3 strains from Japan (Supplementary Table S2). It was also estimated that the canine SA112 strain shared a common ancestor with human MRSA strains from USA (e.g.: DAR3928, DAR3939, DAR3845, DAR3886) at 20.54 years before 2015 (19.97–21.17 95% HPD). The divergence of SA112 and CH14-035 (human strain from Rio de Janeiro) was estimated at 30.61 years before 2015 (26.17–35.4 95% HPD). However, SA112 and the human strains CH14-004 and CH15-056 from Rio de Janeiro might have diverged at more than 85 years before 2015. Given this older inferred date, the acquisition of mecA gene by these two human and animal strains seems more likely to have occurred as distinct genetic events in different MSSA strains. It is interesting that the archetype strain Mu50, a vancomycin-intermediate resistant S. aureus (VISA), isolated from a baby with a surgical site infection in Japan15, was grouped in an independent clade (red) with Mu316 and N31515, both MRSA strains isolated from patients in Japan. MU50 and Mu3 shared an ancestor at 20.04 years before 2015 (18.85–21.52 95% HPD). Therefore, in this prediction, Mu50 and Mu3 strains have emerged in Japan by 1996–1994 and the first report of these strains is dated 199616, in agreement with the calibration of the chronology calculated in our study. It was estimated that the genomes clustered in the red clade (Asia) diverged from those grouped in the blue and purple clades (America) approximately at 1957 (1967–1942%HPD) (Fig. 2).
Table 2.
Characteristics of the completely closed genomes from ST5 MRSA strains sequenced in this study.
| Isolate | Host | Specimen | Isolation year | GenBank accession number | Chromosome size (bp) | GC content (%) |
|---|---|---|---|---|---|---|
| SA112 | Dog | Ear exudate | 2012 | CP020553 | 2,982,283 | 32.9 |
| CR14-035 | Human | Nasal swab | 2014 | CP020544 | 2,989,067 | 32.9 |
| CR14-004 | Human | Tracheal secretion | 2014 | CP021105 | 2,745,711 | 32.9 |
| CR14-055 | Human | Blood | 2014 | CP021057 | 2,854,872 | 32.9 |
| CHU15-056 | Human | Blood | 2015 | CP021171 | 2,773,467 | 32.9 |
The strain SA112 is from canine origin and all others from humans.
Information about genome assembly and BioProject can be obtained by accessing the GenBank.
Figure 2.
A time-calibrated Bayesian phylogeny for MRSA strains of the lineage ST5-SCCmec II from canine (SA112) and human origins obtained from 116 core genome alignments. Values at nodes indicate the posterior probabilities for each node. The tree is drawn to a time-scale indicated in years. Blue bars indicate the 95% credibility intervals (CI) for some node ages. The 95% CI for all nodes can be seen at Supplementary Figure S1. The year of 2015 was used as a reference point for chronological estimates, which corresponds to the isolation year of the most recent strain used for the tree construction.
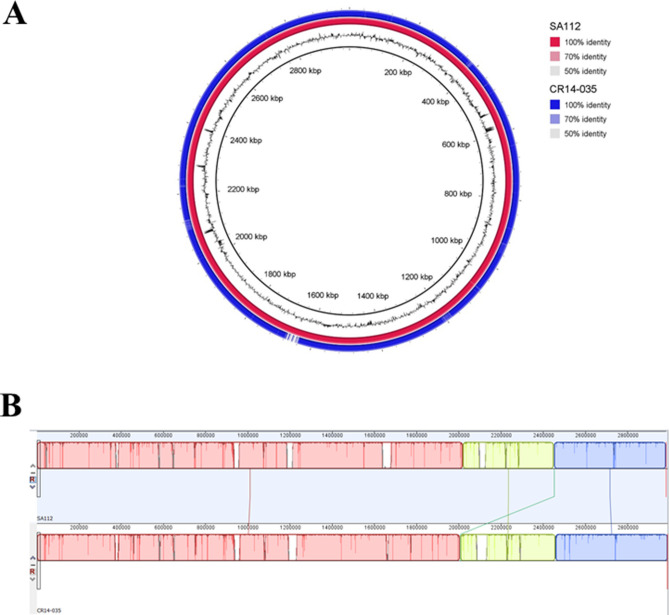
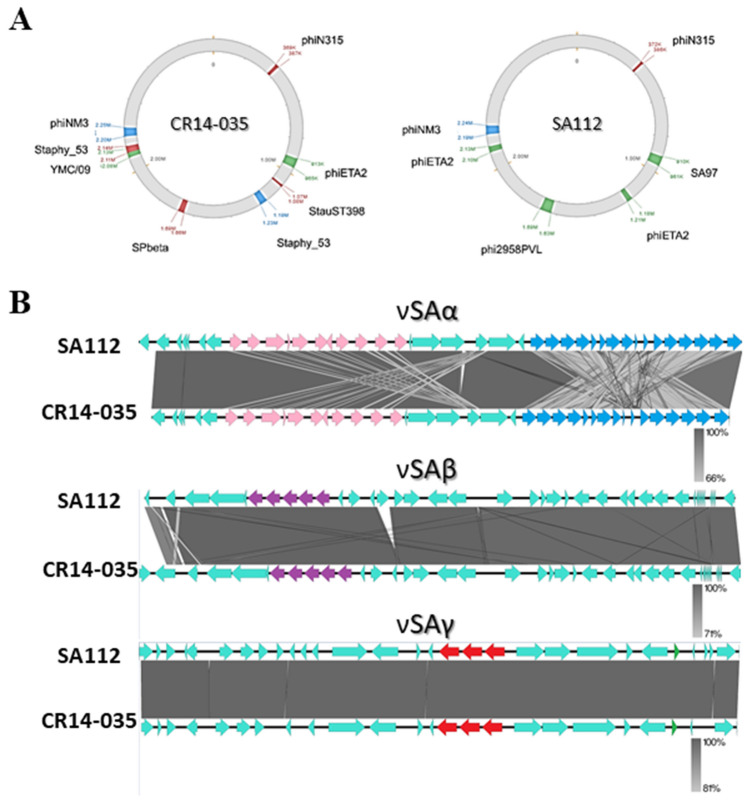
Based on the phylogenetic analysis, the completely closed genome of the human strain CR14-035 (ST5-SCCmecII) from Rio de Janeiro was chosen for the comparative genomic analysis with the genome of the canine strain SA112 (ST5-SCCmecII). Whole genome alignment revealed 95.36% of nucleotide identity between both genomes. The analysis of the circular genomes by BLAST Ring Image Generator (BRIG; Fig. 3A) and the chromosomal architecture using MauveProgressive alignment (Fig. 3B) confirmed the high nucleotide conservation. A total of four local co-linear blocks were obtained for these two genomes (Fig. 3B). The few synteny breaks observed were mostly associated with differences in phage profiles. A bacteriophage search revealed differences not only in the type and quantity of phages but also in the position of these phages on the bacterial chromosome, as shown in Fig. 4A. Only one bacteriophage, related to phiN315, was found in both genomes, in a similar position. Despite the differences between the human and canine MRSA genomes analyzed, all bacteriophages found in the SA112 genome have also been found in the genomes of other S. aureus strains of human origin as indicated in the BLAST searches.
Figure 3.
Genome alignments using blast ring generator (BRIG) and multiple alignment of conserved genomic sequence (MAUVE). (A) Overview of the completely closed genomes of the analyzed ST5 MRSA using BRIG. The color circles represent the genomes of each sequenced strain. The outer black circle represents the GC content and the inner the reference genome of the archetypal CA-MRSA strain MW2. Circular maps of ST5 genomes obtained from ST5 MRSA strains collected from canines (SA112—red circle) and humans (CR14-035—blue circle). The reference genome was the genome of the SA112. (B) Alignment between whole genomes of ST5 MRSA strains SA112 (from canine) and CR14-035 (from human) using MAUVE. Note that only four local collinear blocks were identified, with a translocation of the smallest block (represented by the green line).
Figure 4.
Regions of genomic plasticity corresponding to bacteriophages and important genomic islands. (A) Schematic representation of the position of bacteriophages in the ST5 genomes of the MRSA strains SA112 (from canine) and CR14-035 (from human) sequenced in this study. (B) Alignment of the genomic island carrying important virulence genes in the genome of the strains SA112 (from canine) and CR14-035 (from human). In νSAα the set genes are highlighted in pink and lpl genes are highlighted in blue. The splABCDF operon in νSAβ is highlighted in purple. In νSAγ, set genes are highlighted in red and psmβ is highlighted in green.
The isolate SA112 showed phenotypic resistance to ciprofloxacin, clindamycin, chloramphenicol, erythromycin, gentamycin, cefoxitin, and tetracycline (Table 1). Resfinder analysis detected genes encoding resistance to lincosamides (ermA), aminoglycosides (aadD) and beta-lactams (mecA) and found mutations on gyrA (S84L) and grlA (S80F) that leads to the phenotypic resistance to ciprofloxacin. No genes encoding resistance for chloramphenicol and tetracycline could be detected.
Both genomes carry the genomic islands νSAα, νSAβ, and νSAγ, for which the primary virulence associated genes are listed in Table 3 (Fig. 4B). Unlike other CC5 strains, such as Mu50, which carries staphylococcal pathogenicity island (SaPI), SA112 and CR14-035 genomes do not carry SaPI1, SaPI2, or SaPI3—which often harbor genes associated with toxic shock syndrome-TSST-1 (tst, seb, and sec genes).
Table 3.
Genomic islands detected in the completely closed genomes sequenced in this study obtained from ST5 MRSA strains collected from humans (CR14-035) and canine (SA112).
| Island | Reference | Main target | SA112 | CR14-035 |
|---|---|---|---|---|
| νSAα | Mu50 | set, lpl | + | + |
| νSAβ | Mu50 | splABCDF | + | + |
| νSAγ | Mu50 | set, psmβ | + | + |
| SaPI1 | COL | seb, tst, ear | − | − |
| SaPI2 | Mu50 | sel, sec, tst | − | − |
| SaPI3 | Mu50 | sel, sec, ear | − | − |
+ presence, − absence.
Additional analysis of the virulence gene repertoire between the canine and human genomes from Rio de Janeiro showed that many enterotoxins or enterotoxin-like encoding genes including seg, sei, sem, sen, and seo were detected. Some representative human genomes grouped in the purple, blue, and red clades (MU50, JH1, JH9, FCFHV36, CR14-035, CR14-055, and MI) were also included in this analysis. Not only were absent the genes sec, sel and tst in the MRSA genomes analyzed from humans but also they were equally missed in the CC5 genome from dog, SA112 (Supplementary Table S1). All genomes carried genes related to biofilm formation, such as the ica operon, the autolysin gene (atl), several exoproteins, and genes related to immune evasion (Supplementary Table S1). Notably, all virulence genes of the strain SA112 located in the island regions analyzed displayed high level of nucleotide identity (100%) with the correspondent genes carried by the MRSA strains of human origins; especially by those from Rio de Janeiro State (Supplementary Table S1). Exceptions were only observed for icaB, sak and scn for which all MRSA strains from Rio de Janeiro (dog and human origins) showed 99.89%, 99.39% and 99.72% of nucleotide identity, respectively (Supplementary Table S1).
Virulence assessment
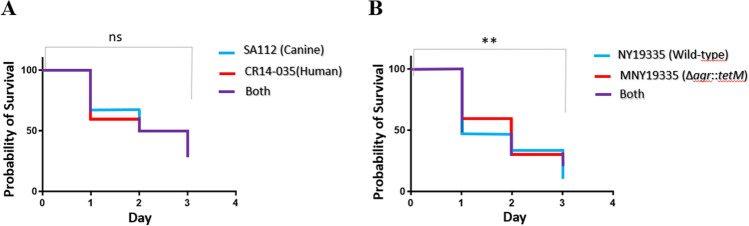
Caenorhabditis elegans model was used to compare the virulence potential of the canine-derived CC5 MRSA (SA112) with that of human MRSA (CR14-035). No difference was found between the killing curves of the CC5 strains with human and animal origins (31.4% and 29.1% of worm survival on the 3rd day of the experiment, respectively), as shown in Fig. 5A. Because agr-knockout mutants display significant reduction in their virulence potential17–19, we used a positive control Agr-functional S. aureus wild-type strain (NY19335) and its isogenic knockout mutant (MNY19335; Δagr::tetM) as a negative control, to validate this model for S. aureus. As expected, a difference was detected in the survival curves for the agr-functional strain and the isogenic knockout mutant (10.4% and 22.9% of worm survival on the 3rd day of the experiment, respectively), as shown in Fig. 5B.
Figure 5.
Comparison of survival curves of C. elegans to assess the virulence of the isolates. (A) The nematodes were separately infected with strain SA112, obtained from a dog and CR14-035, obtained from humans. (B) No significant difference was found. The set of strainsNY19335—an Agr-functional CC5 clinical isolate—and MNY19335∆agr::tetM—the isogenic agr knockout mutant—was used to control the experiments. Ns not significant; **p < 0.01.
Discussion
Methicillin resistance has been increasingly reported in staphylococcal isolates from canines in several countries20–24. The overall rate of MRSA among the 334 dogs in this study (2.7%) was similar to that observed elsewhere (varying from 1 to 6%) in locations such as Korea (0.6%), Japan (1%), Australia (4%) and the United States (5.7%)6,20–22. In our study, the prevalence of nasal colonization by MRSA in dogs (3.4%) was similar to that reported in this country for healthy human carriers (2.3%)25. However, it is of note that 56.2% of S. aureus isolates from the dogs were MRSA. Such high rates have also been observed in other countries—62.7% in Germany23 and 51.8% in the United States24. Although S. aureus is not the most common species isolated from canine infections, it is of concern that MRSA is becoming more prevalent than MSSA in dogs in some regions. The presence of MRSA in dogs has important implications for public health because of the high level of exposure between housed dogs and humans24. Studies have already suggested the possible transmission of S. aureus strains among dogs and their owners26,27. However, only a few studies have addressed this question using genomic strategies22,28,29.
The MRSA isolates from dogs detected in this study belonged to lineages commonly found infecting humans in Brazil and other countries2,30,31. The PVL-producer isolates from dogs were related to the CA-MRSA clones USA400 (NF62) or USA1100 (PA68, PA69, PA73). Of note, all USA400-related isolates previously detected from humans in Brazil have been associated with healthcare-associated infections and were PVL negative30. Although not as frequent as the USA300 clone, the USA400 clone is the second most common PVL-positive CA-MRSA associated with human community-acquired infections in North America1. The epidemiological data obtained relative to the canine isolate NF62 strongly suggest that the dog had acquired this PVL-positive USA400 isolate in the United States, a country where the animal had temporarily stayed prior to sample collection in Brazil, suggesting the potential that pets have for introducing new CA-MRSA clones in distant places.
The USA1100 CA-MRSA found in asymptomatic nasal carriers and infected dogs has previously been reported in Brazil in human infections in Rio de Janeiro29. Additionally, it was detected in the nasal cavity of a healthy cat in Rio de Janeiro7 and a healthy dog in Japan20. BEC-related isolates were the predominant MRSA in Rio de Janeiro hospitals from 1995 to 2009; they do not produce PVL and are typically human-associated hospital isolates32–34. As most BEC isolates from humans, the ST239-SCCmec III isolates recovered from diseased dogs (NF601 and NF661) are multidrug resistant.
The USA100-related MRSA recovered from the ear exudates of infected dogs is emerging as the predominant MRSA associated with human infections in Rio de Janeiro hospitals, replacing ST23913,35. Even though the overall incidence of MRSA in canines is still low in the cities analyzed—roughly 2–3 animals out of 100 will carry MRSA—some will be infected by these bacteria and may be untreatable with currently recommended drugs to treat companion animals. A research performed to access the risk factors for MRSA infections in dogs and cats reported as significant influences (1) the number of antimicrobial courses, (2) admission days to veterinary clinics, and (3) submission to surgical implants. In addition, the odds of contact with humans which had been hospitalized were higher in MRSA infected pets when compared with MSSA controls36.
Our Bayesian phylogenetic inference agrees with the previous dating done by Challagundla et al.37, although different clocks were used. Our purple clade, that grouped the animal strain SA112, and also our human strains CR14-035 and CR14-055, corresponded with the clade named CC5-II B by Challagundla et al. Also our blue clade corresponded with CC5-II-A and our red clade, that clustered the strains N315, Mu50 and Mu3 among others, matched the CC5-Basal clade of Challagundla et al.
It is conceivable that increased number of companion animals have intensified the exchange of bacteria between humans and their pets22,38,39. Worthing et al.22 analyzing MRSA isolates from Australia found that animal and veterinarian-derived MRSA were also intermingled on the phylogenetic tree. Likewise, Harrison et al.38 showed that ST22 MRSA isolates (EMRSA-15 clone) from cats and dogs in the United Kingdom were interspersed with human isolates throughout the epidemic EMRSA-15 pandemic clade. In many cases, human isolates were basal to those from companion animals, suggesting a human source for isolates infecting companion animals38,40.
It has been suggested that after transmission to a new host a pathogen may evolve and adapt by losing and/or acquiring new MGEs41,42. Loeffler et al.28 observed significant differences in MGE content between pets’ and their owners’ isolates. These authors concluded that the variation found amongst MGEs highlights the genetic adaptation of MRSA in different hosts28. In our study, the human and canine strains shared most MGEs and, despite the differences found in phage profiles, all phages detected in the animal strain had already been detected in human S. aureus isolates. Additionally, besides humans and canine genomes sharing a conserved set of virulence genes and comparable virulence potential in a C. elegans model, the canine genome was allocated between human genomes in the same phylogenetic clade. The C. elegans survival model used here was able to distinguish levels of virulence between the agr-null mutant and its isogenic wild-type validating this model for S. aureus.
Taken together, these results suggest that the canine ST5 strain may represent a human-to-animal transmission. The incidence of MRSA in the cities studied is still relatively low. In consequence, only one CC5 genome from a dog was analyzed. Therefore, our data should be analyzed considering such limitation. There have also been reports of isolation of USA100 (ST5-SCCmec II MRSA) from dogs and other domestic animals in the United States4,27,43,44. This finding is concerning because CC5 isolates can display high level of antibiotic multiresistance. Although still rare, VISA and VRSA resistances have mostly been described among CC5 strains45–47.
Conclusions
This work demonstrates that MRSA isolates were detected among infected and colonized dogs in the state of Rio de Janeiro. The isolates recovered from those animals are related to the CA-MRSA international clones USA1100/OSPC (ST30-SCCmec IV) and USA400 (ST1-SCCmec IV) and to the HA-MRSA clones USA100/NY/Japan (ST5-SCCmec II) and BEC ST239-SCCmec III) that are commonly involved in human infections in Brazil and in other countries. Because ST30 isolates are the primary CA-MRSA pathogens in Brazil, the presence of this PVL-producing MRSA in animals in two cities in Rio de Janeiro state is of great concern. Additionally, a high rate of methicillin resistance was found among the S. aureus detected, and two MRSA isolates were resistant to a wide variety of other antibiotics. We have demonstrated a high conservation of virulence traits between human and canine MRSA genomes. Indeed, the canine genome interspersed with human genomes of MRSA in the phylogenetic tree suggesting a possible human origin for the canine ST5 strain and raising the possibility that this strain can move easily between hosts without canine-specific adaptations. The circulation of pandemic lineages of MRSA, commonly found in hospitals and community settings is an additional challenge for public health authorities. Monitoring and surveillance of companion animals for MRSA would be beneficial to better define the circulation of these resistant strains and to assess the potential risk of animals as sources of resistant infections in humans.
Methods
Bacterial isolates
Three hundred and thirty-four dogs were screened for MRSA in a prospective study carried out between 2010 and 2013 in Rio de Janeiro (n = 210) and Campos dos Goytacazes (n = 124) cities, located in Rio de Janeiro state, 280 km apart. The dogs were male and female adults (age: 1–8 years) and either healthy (n = 88 nasal swabs) or infected animals (swabs of ear secretions and skin exudates; n = 246). The clinical material was collected from dogs brought to the Department of Small Animal Practice of the Veterinary Hospital at Universidade Estadual do Norte Fluminense (UENF), located in Campos dos Goytacazes, Rio de Janeiro, Brazil. Also, from dogs who visited a number of private veterinary clinics in Rio de Janeiro city. All of the samples were collected using sterile cotton swabs (Copan Diagnostic, Italy). Only one MRSA isolate from each dog was studied. This study was approved by the ethics committee of Federal Fluminense University (#218/10) and by the Ethics Committee for Animal Care and Use from UENF Darcy Ribeiro (#145/2011). The swabs were inoculated in tryptic soy agar (TSA; Difco, Franklin Lakes, NJ, USA) and mannitol salt agar (MSA; Merck, Darmstadt, Germany). After the samples were incubated at 37 °C/18 h, we examined cell morphology using the Gram-staining method. Gram-positive colonies were tested for catalase and free coagulase production. S. aureus isolates were identified using a polymerase chain reaction (PCR) nuc-based test described by Sasaki et al. (2010)48. The MRSA strains USA100-0022 (USA100), USA300-0114 (USA300), USA400-0051 (USA400), WB45 and WB49 (USA1100), BMB9393 and HU25 (BEC) were the representatives of MRSA international clones used to compare PFGE band differences.
Antimicrobial susceptibility
Resistance to methicillin was detected using 30-μg cefoxitin disks49. All of the MRSA isolates were also tested for susceptibility to ciprofloxacin (5 μg), clindamycin (2 μg), chloramphenicol (30 μg), erythromycin (15 μg), gentamicin (10 μg), rifampicin (5 μg), tetracycline (30 μg), and sulfamethoxazole + trimethoprim (23.75/1.25 μg) using a disk diffusion test, as recommended by the Clinical and Laboratory Standards Institute (CLSI)49. The disks were obtained from Cecon (São Paulo, SP, Brazil), and the S. aureus strain ATCC 25923 was used as a control.
Molecular characterization
The presence of the mecA gene in cefoxitin-resistant S. aureus was confirmed by PCR50. Methicillin-resistant S. aureus genotyping was performed using several methods including SCCmec typing51, restriction and modification (RM) testing52,53, pulsed-field gel electrophoresis (PFGE) of the SmaI-fragmented DNA54, and multilocus sequence typing (MLST)55. Dendrograms were generated from the PFGE patterns using GelCompar II software 6.5 (Applied Maths, Sint-Martens-Latem, Belgium). To assign the MLST sequence types, the allele sequences were trimmed and analyzed using the Public Database for Molecular Typing and Microbial Genome Diversity (http://pubmlst.org). Detection of lukSF-PV genes was performed using a PCR-based method described previously56.
Whole-genome sequencing
Because some MRSA isolates from dogs belonged to the ST5-SCCmec II lineage, which has recently emerged as the predominant lineage in Rio de Janeiro hospitals13, we aimed to investigate whether these canine ST5 isolates were from a human origin or were specific to animals. To achieve this goal, we performed whole genome sequencing of five ST5-SCCmec II isolates from humans and canines (Table 2). Genomic DNA was prepared from overnight cultures using the Wizard Genomic DNA Purification Kit (Promega Corporation, Madison, WI, USA). The bacterial cells were centrifuged, washed with TSM buffer (50 mM Tris, pH 7.5; 0.5 M sucrose; 10 mM MgCl2), lysed with 50 U/mL lysostaphin (Sigma-Aldrich, Merck, Darmstadt, Germany), and processed using the protocol suggested by the manufacturer. Genome libraries were prepared using the Nextera XT kit (Illumina, San Diego, CA, USA) and sequenced using the Illumina MiSeq instrument (paired end reads of 125 bp). Genome assembly was performed by combining de novo and map to reference assembly methods, through Velvet v7.0.457 and Geneious v11.0.558, respectively. The genomes were submitted to the NCBI Prokaryotic Genome Annotation Pipeline59.
Phylogenetic analysis
For this analysis, besides the five ST5-SCCmec II genomes sequenced in this work, we included 111 CC5-SCCmecII genomes publicly available in the GenBank (Supplementary Table S2). We ran core genome alignments with REALPHY60 and used ClonalFrameML61 to further verify the absence of recombination events that could possibly affect our molecular dating results. The phylogenetic tree under a Bayesian framework was obtained using the package BEAST v1.10.462, assuming a GTR + Γ nucleotide substitution model63,64. The tree was reconstructed using an uncorrelated log-normal relaxed molecular clock model65, with a random start tree and a coalescent constant size tree prior66. We used for the substitution rate prior a value of 1.5 × 10–6 as ucld.mean parameter (mean of the log-normal distribution), allowing the distribution to vary between 1.0 × 10–10 and 1.0 × 10–3. The final substitution rate inferred in this analysis was 2.00 × 10–6 substitutions per site year−1 (with 95% HPD interval of 1.62 × 10–6 and 2.40 × 10–6). This inferred rate is similar to that obtained by Duchêne et al.67. We calibrated the molecular clock using tip dates (sampling times) for all individuals. Nine independent MCMC chains for 200 million iterations each were run, combining them after discarding the first 10% as burn-in. We visualized the MCMC chains using Tracer v1.668 calculated the maximum clade credibility tree using TreeAnnotator v2.569 and edited the final tree using FigTree v1.4.370. The year of 2015 was used as a reference point for chronological estimates, which corresponds to the isolation year of the most recent strain used for tree constructions.
Genomic analysis
For most of the comparative genomics analysis, we selected the genome of the human strain CR14-035 from Rio de Janeiro city; the most closely related to that of the canine strain SA112 based on information in the phylogenetic tree. BLAST Ring Image Generator 0.9571 was used to display circular comparisons between the genomes, using the canine genome as reference. We analyzed chromosomal architecture and genome organization using the progressive Mauve genome alignment algorithm72, with default parameters. Bacteriophage analysis was conducted using Phage Search Tool Enhanced Release (PHASTER)73. Resfinder74,75 was used to identify the resistance genes in SA112 genome. The staphylococcal pathogenicity island (SaPI) and other genomic islands (GIs) were annotated with the support of Island Viewer76. We also relied on manual annotation using the scientific literature. Well known virulence genes from S. aureus were searched using Local BLAST77 not only in SA112 and CR14-035, but also in all other CC5 genomes included in the phylogenetic analysis. The genomes were considered positive for each gene applying a cut-off of 90% coverage and 96% identity using gene sequences from N315 (Genbank Accession number: BA000018).
Virulence potential assessment
The nematode Caenorhabditis elegans is increasingly recognized as relevant for the study of bacterial pathogenesis. This model is susceptible to different human pathogens that are able to infect the intestine of the nematode. Various genes related to mammalian immune response are not encoded in the nematode genome. However, C. elegans has an immune response that utilizes a number of evolutionarily conserved signaling pathways, including a highly conserved mitogen-activated protein kinase (MAPK) signaling pathways that activates the innate immune response to bacterial infections78. This system is easy to manipulate, has a good reproducibility and is not under subject of ethical consideration. Because of that, it is considered an interesting model to study virulence and other aspect of host-bacteria interactions. We used the C. elegans survival assay to compare the virulence potential of the strains CR14-035 and SA112 from humans and dogs, respectively, based on a model described previously in the literature79, with some modifications. Briefly, to produce the bacterial lawns, a single colony was inoculated in trypticase soy broth (TSB; BD) and the culture was incubated at 37 °C for 4 h with shaking (350 rpm). Then, a 10-μL aliquot of the culture was homogeneously spread to form a bacterial lawn on the wells of 96-well microtiter plates containing TSA supplemented with 5 μg/mL nalidixic acid and 5 μg/mL of cholesterol. For each assay, about 5–20 L4-stage nematodes were transferred to the wells containing lawns formed by each of the tested isolates. The plates were incubated at 25 °C and monitored every 24 h for live and dead worms over the course of 3 days. Each MRSA isolate was tested in three completely independent experiments with 12 replicates each and the data shown are an average of these experiments. Because agr-knockout mutants are less virulent than wild-type strains with functional agr systems in different in vivo and ex vivo models17–19, we carried out a control using the wild-type strain NY19335 (ST5-SCCmec II MRSA, isolated from bacteremia) and the isogenic agr knockout MNY19335 (∆agr::tetM)17 to assess the accuracy of this C. elegans model for distinguishing virulence potential in S. aureus human pathogen.
Statistical analysis
Nematode survival rates were calculated using the Kaplan–Meier method and tested for significance using the log rank test (Software, Inc., La Jolla, CA, USA). In addition, we used the chi-square test to calculate the significance of the increased rate of MRSA among the animals. p values less than 0.05 were considered to be statistically significant.
Guidelines statements
All methods were carried out in accordance with relevant guidelines and regulations. The study was carried out in compliance with the ARRIVE guidelines.
Supplementary information
Acknowledgements
This work was supported in part by Grants from the Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ # E-26/202.826/2018 to ATRV, # E-26/010.001764/2014 to AMSF and PJP, and E-26/201.147/2014 and E-26/202.803/2017 to AMSF), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq # 303170/2017-4 to ATRV and 303067/2015-2 to AMSF), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES # 864/15 to AMSF).
Author contributions
B.P. carried out strain isolation, identification, molecular characterization by P.F.G.E. and wrote the draft of the manuscript. M.B.S. carried out strain isolation, biochemical e molecular characterization of the isolates. A.E.R.S. and A.T.R.V. performed the phylogenetic analysis. M.S.R. and F.A.F. performed molecular typing by MLST and SCCmec typing. M.C.S.C., V.S.S. and R.F.R. carried out strain identification and molecular characterization by P.F.G.E. P.T.B. and V.S.S. performed virulence assessment using C. elegans model. P.J.P. was responsible for whole genome sequencing of the isolates of this study, some genomic analysis and revised the final version on the manuscript. O.V.M. carried out strain isolation, participated in the study designed and revised the final version of the manuscript. A.M.N.B. performed virulence assessment using C. elegans model, comparative genomics analysis and wrote the final version of the manuscript. A.M.S.F. designed the study and revised the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Bruno Penna and Marcella B. Silva.
Contributor Information
Ana M. N. Botelho, Email: abotelho@id.uff.br
Agnes M. S. Figueiredo, Email: agnes@micro.ufrj.br
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-83993-5.
References
- 1.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010;23:616–687. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardos de la Gandara M, et al. MRSA causing infections in hospitals in greater metropolitan New York: Major shift in the dominant clonal type between 1996 and 2014. PLoS One. 2016;11:e0156924. doi: 10.1371/journal.pone.0156924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Penna B, et al. Species distribution and antimicrobial susceptibility of staphylococci isolated from canine otitis externa. Vet. Dermatol. 2010;21:292–296. doi: 10.1111/j.1365-3164.2009.00842.x. [DOI] [PubMed] [Google Scholar]
- 4.Lin Y, et al. Evidence of multiple virulence subtypes in nosocomial and community-associated MRSA genotypes in companion animals from the upper midwestern and northeastern United States. Clin. Med. Res. 2011;9:7–16. doi: 10.3121/cmr.2010.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Balen J, et al. Presence, distribution, and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small animal teaching hospital: A year-long active surveillance targeting dogs and their environment. Vector Borne Zoonot. Dis. 2013;13:299–311. doi: 10.1089/vbz.2012.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoet AE, et al. Epidemiological profiling of methicillin-resistant Staphylococcus aureus-positive dogs arriving at a veterinary teaching hospital. Vector Borne Zoonot. Dis. 2013;13:385–393. doi: 10.1089/vbz.2012.1089. [DOI] [PubMed] [Google Scholar]
- 7.Quitoco IMZ, et al. First report in South America of companion animal colonization by the USA1100 clone of community-acquired meticillin-resistant Staphylococcus aureus (ST30) and by the European clone of methicillin-resistant Staphylococcus pseudintermedius (ST71) BMC Res. Notes. 2013;6:336. doi: 10.1186/1756-0500-6-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis JA, et al. Carriage of methicillin-resistant staphylococci by healthy companion animals in the US. Lett. Appl. Microbiol. 2014;59:1–8. doi: 10.1111/lam.12254. [DOI] [PubMed] [Google Scholar]
- 9.van Duijkeren E, et al. Methicillin-resistant Staphylococcus aureus in horses and horse personnel: An investigation of several outbreaks. Vet. Microbiol. 2010;141:96–102. doi: 10.1016/j.vetmic.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 10.Feingold BJ, et al. Livestock density as risk factor for livestock-associated methicillin-resistant Staphylococcus aureus, the Netherlands. Emerg. Infect. Dis. 2012;18:1841–1849. doi: 10.3201/eid1811.111850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manian FA. Asymptomatic nasal carriage of mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MRSA) in a pet dog associated with MRSA infection in household contacts. Clin. Infect. Dis. 2003;36:e26–e28. doi: 10.1086/344772. [DOI] [PubMed] [Google Scholar]
- 12.Nienhoff U, et al. Transmission of methicillin-resistant Staphylococcus aureus strains between humans and dogs: Two case reports. J. Antimicrob. Chemother. 2009;64:660–662. doi: 10.1093/jac/dkp243. [DOI] [PubMed] [Google Scholar]
- 13.Chamon RC, Ribeiro S, Da S, da Costa TM, Nouér SA, DosSantos KRN. Complete substitution of the Brazilian endemic clone by other methicillin-resistant Staphylococcus aureus lineages in two public hospitals in Rio de Janeiro, Brazil. Braz. J. Infect. Dis. 2017;21:185–189. doi: 10.1016/j.bjid.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCulloch JA, et al. Complete genome sequence of Staphylococcus aureus FCFHV36, a methicillin-resistant strain heterogeneously resistant to vancomycin. Genome Announc. 2015;3:20. doi: 10.1128/genomeA.00893-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuroda M, et al. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 16.Hiramatsu K, et al. Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. Lancet. 1997;350:1670–1673. doi: 10.1016/S0140-6736(97)07324-8. [DOI] [PubMed] [Google Scholar]
- 17.Coelho LR, et al. agr RNAIII divergently regulates glucose-induced biofilm formation in clinical isolates of Staphylococcus aureus. Microbiology. 2008;154:3480–3490. doi: 10.1099/mic.0.2007/016014-0. [DOI] [PubMed] [Google Scholar]
- 18.Gong J, et al. The accessory gene regulator (agr) controls Staphylococcus aureus virulence in a murine intracranial abscesses model. Braz. J. Infect. Dis. 2014;18:501–506. doi: 10.1016/j.bjid.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollitt EJG, West SA, Crusz SA, Burton-Chellew MN, Diggle SP. Cooperation, quorum sensing, and evolution of virulence in Staphylococcus aureus. Infect. Immun. 2014;82:1045–1051. doi: 10.1128/IAI.01216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishihara K, et al. Methicillin-resistant Staphylococcus aureus carriage among veterinary staff and dogs in private veterinary clinics in Hokkaido, Japan. Microbiol. Immunol. 2014;58:149–154. doi: 10.1111/1348-0421.12128. [DOI] [PubMed] [Google Scholar]
- 21.Jang Y, et al. Characterization of methicillin-resistant Staphylococcus spp. isolated from dogs in Korea. Jpn. J. Vet. Res. 2014;62:163–170. [PubMed] [Google Scholar]
- 22.Worthing KA, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from Australian animals and veterinarians. Microb. Drug Resist. 2017;100:mdr.2017.0032. doi: 10.1089/mdr.2017.0032. [DOI] [PubMed] [Google Scholar]
- 23.Vincze S, et al. Alarming proportions of methicillin-resistant Staphylococcus aureus (MRSA) in wound samples from companion animals, Germany 2010–2012. PLoS One. 2014;9:e85656. doi: 10.1371/journal.pone.0085656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iverson SA, et al. Anatomical patterns of colonization of pets with staphylococcal species in homes of people with methicillin-resistant Staphylococcus aureus (MRSA) skin or soft tissue infection (SSTI) Vet. Microbiol. 2015;176:202–208. doi: 10.1016/j.vetmic.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Bes TM, et al. Prevalence of methicillin-resistant Staphylococcus aureus colonization in individuals from the community in the city of Sao Paulo, Brazil. Rev. Inst. Med. Trop. São Paulo. 2018;60:e58. doi: 10.1590/s1678-9946201860058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferreira JP, et al. Transmission of MRSA between companion animals and infected human patients presenting to outpatient medical care facilities. PLoS One. 2011;6:e26978. doi: 10.1371/journal.pone.0026978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vincze S, et al. Risk factors for MRSA infection in companion animals: Results from a case–control study within Germany. Int. J. Med. Microbiol. 2014;304:787–793. doi: 10.1016/j.ijmm.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Loeffler A, et al. Whole-genome comparison of meticillin-resistant Staphylococcus aureus CC22 SCCmecIV from people and their in-contact pets. Vet. Dermatol. 2013;24:538–e128. doi: 10.1111/vde.12062. [DOI] [PubMed] [Google Scholar]
- 29.Davis MF, et al. Genome sequencing reveals strain dynamics of methicillin-resistant Staphylococcus aureus in the same household in the context of clinical disease in a person and a dog. Vet. Microbiol. 2015;180:304–307. doi: 10.1016/j.vetmic.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva-Carvalho MC, et al. Emergence of multiresistant variants of the community-acquired methicillin-resistant Staphylococcus aureus lineage ST1-SCCmecIV in 2 hospitals in Rio de Janeiro, Brazil. Diagn. Microbiol. Infect. Dis. 2009;65:300–305. doi: 10.1016/j.diagmicrobio.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 31.Martins A, Moraes Riboli DF, Cataneli Pereira V, de Lourdes Ribeiro de Souzada Cunha M. Molecular characterization of methicillin-resistant Staphylococcus aureus isolated from a Brazilian university hospital. Braz. J. Infect. Dis. 2014;18:331–335. doi: 10.1016/j.bjid.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Silva-Carvalho MC, et al. Comparison of different methods for detecting methicillin resistance in MRSA isolates belonging to international lineages commonly isolated in the American continent. Microbiol. Immunol. 2009;53:117–122. doi: 10.1111/j.1348-0421.2008.00096.x. [DOI] [PubMed] [Google Scholar]
- 33.de Sousa-Junior FC, et al. Genotyping of methicillin-resistant Staphylococcus aureus isolates obtained in the Northeast region of Brazil. Braz. J. Med. Biol. Res. 2009;42:877–881. doi: 10.1590/S0100-879X2009005000018. [DOI] [PubMed] [Google Scholar]
- 34.Costa MOC, et al. Complete genome sequence of a variant of the methicillin-resistant Staphylococcus aureus ST239 lineage, strain BMB9393, displaying superior ability to accumulate ica-independent biofilm. Genome Announc. 2013;1:1–2. doi: 10.1128/genomeA.00576-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teixeira MM, et al. Emergence of clonal complex 5 (CC5) methicillin-resistant Staphylococcus aureus (MRSA) isolates susceptible to trimethoprim-sulfamethoxazole in a Brazilian hospital. Braz. J. Med. Biol. Res. 2012;45:637–643. doi: 10.1590/S0100-879X2012007500065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soares Magalhães RJ, et al. Risk factors for methicillin-resistant Staphylococcus aureus (MRSA) infection in dogs and cats: A case–control study. Vet. Res. 2010;41:55. doi: 10.1051/vetres/2010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challagundla L, et al. Phylogenomic classification and the evolution of clonal complex 5 methicillin-resistant Staphylococcus aureus in the western hemisphere. Front Microbiol. 2018;9:1921. doi: 10.3389/fmicb.2018.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison E, Weinert L. A shared population of epidemic methicillin-resistant Staphylococcus aureus 15 circulates in humans and companion animals. MBio. 2014;5:1–10. doi: 10.1128/mBio.00985-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paterson GK, et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat. Commun. 2015;6:6560. doi: 10.1038/ncomms7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson EJ, et al. Gene exchange drives the ecological success of a multi-host bacterial pathogen. Nat. Ecol. Evol. 2018;2:1468–1478. doi: 10.1038/s41559-018-0617-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowder BV, et al. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guinane CM, et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2010;2:454–466. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbott Y, Leonard FC, Markey BK. Detection of three distinct genetic lineages in methicillin-resistant Staphylococcus aureus (MRSA) isolates from animals and veterinary personnel. Epidemiol. Infect. 2010;138:764–771. doi: 10.1017/S0950268809991580. [DOI] [PubMed] [Google Scholar]
- 44.Gómez-Sanz E, Torres C, Lozano C, Zarazaga M. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comp. Immunol. Microbiol. Infect. Dis. 2013;36:83–94. doi: 10.1016/j.cimid.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 45.Howe RA, Monk A, Wootton M, Walsh TR, Enright MC. Vancomycin susceptibility within methicillin-resistant Staphylococcus aureus lineages. Emerg. Infect. Dis. 2004;10:855–857. doi: 10.3201/eid1005.030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.da Costa TM, et al. Clinical and microbiological characteristics of heteroresistant and vancomycin-intermediate Staphylococcus aureus from bloodstream infections in a Brazilian teaching hospital. PLoS One. 2016;11:e0160506. doi: 10.1371/journal.pone.0160506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gardete S, Tomasz A. Mechanisms of vancomycin resistance in Staphylococcus aureus. J. Clin. Invest. 2014;124:2836–2840. doi: 10.1172/JCI68834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasaki T, et al. Multiplex-PCR method for species identification of coagulase-positive staphylococci. J. Clin. Microbiol. 2010;48:765–769. doi: 10.1128/JCM.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.The Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing CLSI supplement M100S. Wayne: Clinical and Laboratory Standards Institute; 2016. [Google Scholar]
- 50.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boye K, Bartels MD, Andersen IS, Møller JA, Westh H. A new multiplex PCR for easy screening of methicillin-resistant Staphylococcus aureus SCCmec types I-V. Clin. Microbiol. Infect. 2007;13:725–727. doi: 10.1111/j.1469-0691.2007.01720.x. [DOI] [PubMed] [Google Scholar]
- 52.Cockfield JD, Pathak S, Edgeworth JD, Lindsay JA. Rapid determination of hospital-acquired meticillin-resistant Staphylococcus aureus lineages. J. Med. Microbiol. 2007;56:614–619. doi: 10.1099/jmm.0.47074-0. [DOI] [PubMed] [Google Scholar]
- 53.Beltrame CO, et al. Restriction modification (RM) tests associated to additional molecular markers for screening prevalent MRSA clones in Brazil. Eur. J. Clin. Microbiol. Infect. Dis. 2012;31:2011–2016. doi: 10.1007/s10096-011-1534-1. [DOI] [PubMed] [Google Scholar]
- 54.Teixeira LA, et al. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J. Clin. Microbiol. 1995;33:2400–2404. doi: 10.1128/JCM.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 2000;38:1008–1015. doi: 10.1128/JCM.38.3.1008-1015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.von Eiff C, Friedrich AW, Peters G, Becker K. Prevalence of genes encoding for members of the staphylococcal leukotoxin family among clinical isolates of Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 2004;49:157–162. doi: 10.1016/j.diagmicrobio.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 57.Zerbino DR. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinform. 2010;11:20. doi: 10.1002/0471250953.bi1105s31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Angiuoli SV, et al. Toward an online repository of standard operating procedures (SOPs) for (meta)genomic annotation. OMICS. 2008;12:137–141. doi: 10.1089/omi.2008.0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 2014;31:1077–1088. doi: 10.1093/molbev/msu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Didelot X, Wilson DJ. ClonalFrameML: Efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suchard MA, et al. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:20. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tavaré. Some probabilistic and statistical problems in the analysis of DNA sequences. in Lectures on Mathematics in the Life Sciences. (ed. M., M. R.) 57–86 (1985).
- 64.Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 65.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kingman JFC. The coalescent. Stoch. Process. Appl. 1982;13:235–248. doi: 10.1016/0304-4149(82)90011-4. [DOI] [Google Scholar]
- 67.Duchêne S, et al. Genome-scale rates of evolutionary change in bacteria. Microb. Genom. 2016;2:20. doi: 10.1099/mgen.0.000094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rambaut, A. G. & Drummond, A. J. Tracer. http://beast.bio.ed.ac.uk/Tracer (2013).
- 69.Rambaut, A. & Drummond, A. J. TreeAnnotator. https://beast.community/treeannotator (2013).
- 70.Rambaut, A. FigTree. http://tree.bio.ed.ac.uk/software/figtree (2007).
- 71.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST ring image generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Darling AE, Mau B, Perna NT. ProgressiveMauve: Multiple genome alignm . PLoS One. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arndt D, et al. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zankari E, et al. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017;72:2764–2768. doi: 10.1093/jac/dkx217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bortolaia V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020;75:3421–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Langille MGI, Brinkman FSL. IslandViewer: An integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: Insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sifri CD, Begun J, Ausubel FM, Calderwood SB. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 2003;71:2208–2217. doi: 10.1128/IAI.71.4.2208-2217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.