Abstract
Tumor metastasis is a crucial impediment to the treatment of gastric cancer (GC), and the epithelial-to-mesenchymal transition (EMT) program plays a critical role for the initiation of GC metastasis. Thus, the aim of this study is to investigate the regulation of lnc-CTSLP4 in the EMT process during GC progression. We found that lnc-CTSLP4 was significantly downregulated in GC tumor tissues compared with adjacent non-tumor tissues, and its levels in GC tumor tissues were closely correlated with tumor local invasion, TNM stage, lymph node metastasis, and prognosis of GC patients. Loss- and gain-of-function assays indicated that lnc-CTSLP4 inhibited GC cell migration, invasion, and EMT in vitro, as well as peritoneal dissemination in vivo. Mechanistic analysis demonstrated that lnc-CTSLP4 could bind with Hsp90α/heterogeneous nuclear ribonucleoprotein AB (HNRNPAB) complex and recruit E3-ubiquitin ligase ZFP91 to induce the degradation of HNRNPAB, thus suppressing the transcriptional activation of Snail and ultimately reversing EMT of GC cells. Taken together, our results suggest that lnc-CTSLP4 is significantly downregulated in GC tumor tissues and inhibits metastatic potential of GC cells by attenuating HNRNPAB-dependent Snail transcription via interacting with Hsp90α and recruiting E3 ubiquitin ligase ZFP91, which shows that lnc-CTSLP4 could serve as a prognostic biomarker and therapeutic target for metastatic GC.
Keywords: lnc-CTSLP4, Hsp90α, HNRNPAB, ZFP91, Snail;LncRNA, EMT, Ubiquitin, Gastric Cancer, Metastasis
Graphical Abstract
lnc-CTSLP4 is significantly downregulated in GC tumor tissues and inhibits metastatic potential of GC cells by attenuating HNRNPAB-dependent Snail transcription via interacting with Hsp90α and recruiting E3-ubiquitin ligase ZFP91, which shows that lnc-CTSLP4 could serve as a prognostic biomarker and therapeutic target for metastatic GC.
Introduction
Gastric cancer (GC) is the fifth most common malignancy and the third leading cause of cancer-related death worldwide, with particularly high morbidity and mortality in China.1,2 Approximately 50% of GC patients present with metastases, and the 5-year overall survival (OS) rate of these patients is no more than 25%.3,4 Tumor metastasis is a crucial impediment to the successful treatment for GC,5 however, the mechanisms underlying GC metastasis have yet to be comprehensively elucidated.6 The epithelial-to-mesenchymal transition (EMT) program is a developmental process hijacked by cancer cells to facilitate their dissemination and metastatic cascade,7 during which epithelial cells progressively lose their cell identity and acquire a mesenchymal phenotype.8 Aberrant activation of EMT has been shown to play a key role in tumor metastatic cascade. Thus, understanding the molecular regulation of EMT is critical for elucidating the mechanism of GC metastasis and designing new therapeutic strategies.
Molecular networks involving the regulation of the EMT program are highly complicated, including various extracellular stimuli, classical growth factors, and other signaling pathways including Wnt, Notch, JAK-STAT, and nuclear factor κB (NF-κB).9 In response to these EMT-inducing pathways, a majority of EMT-inducing transcription factors (EMT-TFs), including SNAIL family (Slug, Snail, and Smuc), Twist, and ZEB family (ZEB1 and ZEB2), are activated to orchestrate the EMT program. Among these EMT-TFs, Snail is a zinc finger TF and is first described in Drosophila melanogaster.10 As a critical transcriptional repressor of E-cadherin expression, Snail plays an important role in the developmental and oncogenic EMT program.11,12 Recent studies have revealed that the expression of Snail is regulated by a complex signaling network at the transcriptional and post-translational level, and various signaling molecules within the tumor microenvironment, such as transforming growth factor beta (TGF-β), epidermal growth factor (EGF), tumor necrosis factor alpha (TNF-α), and fibroblast growth factor (FGF), have been shown to induce Snail expression in different cellular contexts. Snail protein stability, subcellular localization, and activity are regulated by various post-translational modifications, including phosphorylation, ubiquitination, and lysine oxidation.13 Notably, long non-coding RNAs (lncRNAs), eukaryotic cell genome-encoded transcripts of more than 200 nucleotides and generally lacking obvious open reading frames,14 have attracted more attention in the regulation of tumor progression, and a number of researchers have shown that lncRNAs can participate in the regulation of the EMT process.15, 16, 17 However, due to the large quantities and complex mechanisms, the vast majority of lncRNAs and their roles in the regulation of EMT and GC metastasis remain functionally uncharacterized.
In the present study, we identify that lnc-CTSLP4, a novel lncRNA, is significantly downregulated in GC tumor tissues and cell lines. lnc-CTSLP4 plays a role as a tumor suppressor by binding with Hsp90α and recruiting E3 ubiquitin ligase ZFP91, then decreases the stability and promotes the degradation of heterogeneous nuclear ribonucleoprotein AB (HNRNPAB), a TF of Snail, thus suppressing Snail transcription and ultimately reversing EMT.
Results
Lnc-CTSLP4 expression is downregulated in tumor tissues and inversely associated with the prognosis of GC patients
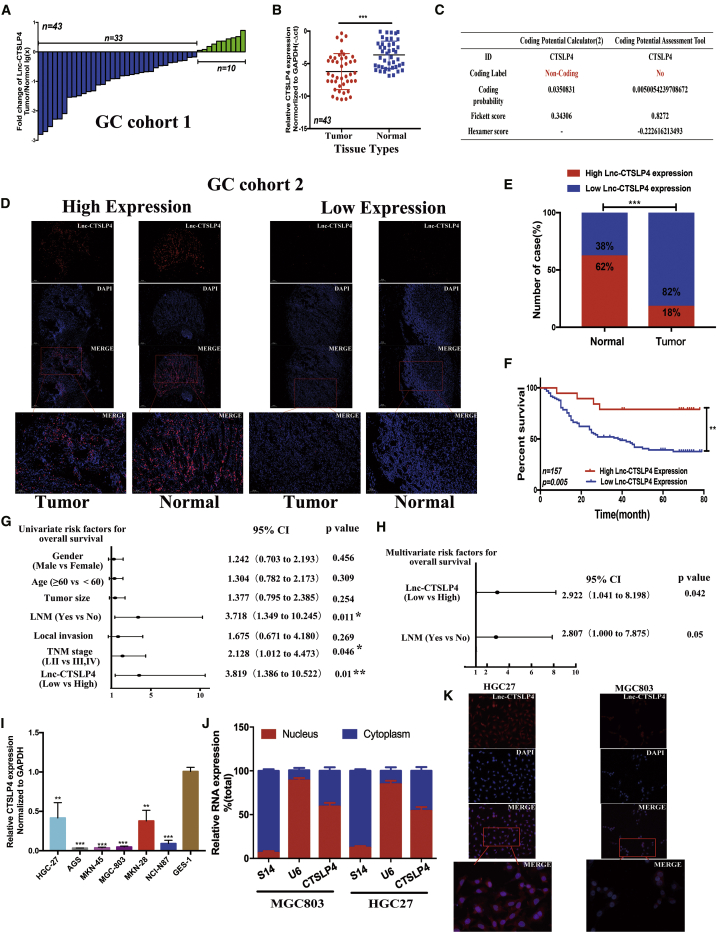
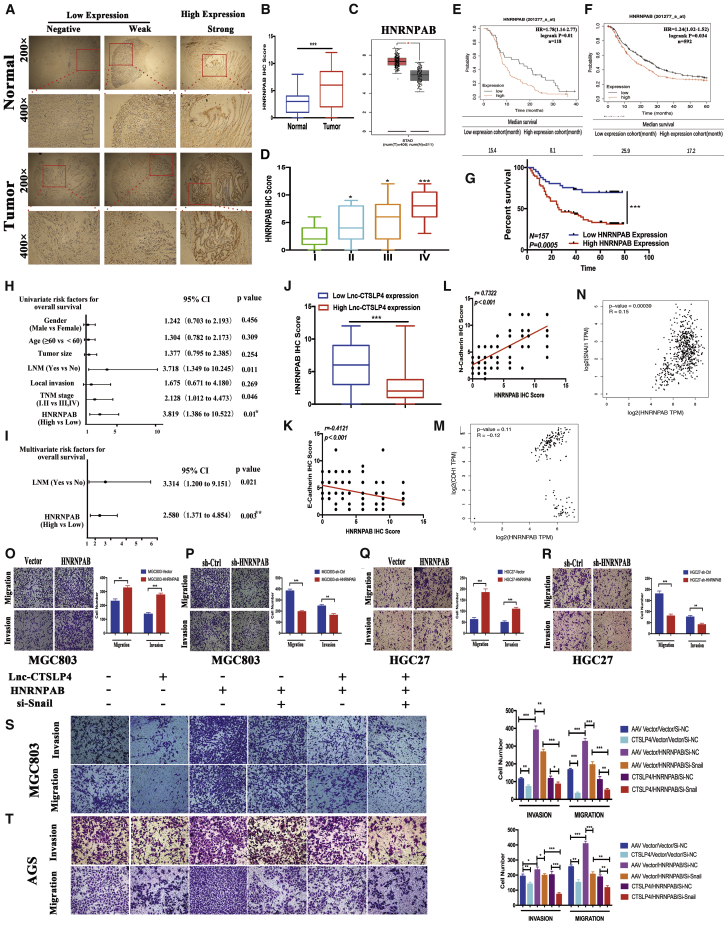
We have previously discovered that lnc-CTSLP4 is obviously dysregulated in GC.18 To identify the levels of lnc-CTSLP4 in GC, tumor tissues and adjacent non-tumor tissues from 43 GC patients (GC cohort 1) were subjected to qRT-PCR assays, and the results showed that the expression of lnc-CTSLP4 was decreased in 77% of GC tumor tissues (Figure 1A). Notably, lnc-CTSLP4 expression was significantly lower in GC tumor tissues than that in the adjacent non-tumor tissues (Figure 1B). We performed rapid amplification of cDNA ends (RACE) to identify the full sequence of lnc-CTSLP4 in GC cells (Figures S1A–S1F), and the protein-coding potential of lnc-CTSLP4 was predicted according to the CPC2 website (http://cpc2.gao-lab.org/) and CPAT website (http://lilab.research.bcm.edu/cpat/index.php; Figure 1C), which confirmed that lnc-CTSLP4 was affiliated to the non-coding RNA class. Furthermore, we checked lnc-CTSLP4 expression in 157 GC patients (GC cohort 2) by RNA-fluorescence in situ hybridization (RNA-FISH), and the results showed that lnc-CTSLP4 high expression rate was lower in GC tumor tissues (18.0%, 28 of 157) than that in adjacent non-tumor tissues (62%, 97 of 157; Figures 1D and 1E). In addition, we demonstrated that the expression of lnc-CTSLP4 was correlated with local invasion, lymph node metastasis, and TNM stage (Table 1; Figures S2A and S2B). Our data also revealed that low expression level of lnc-CTSLP4 in GC tumor tissues was positively correlated with shorter survival for patients with GC significantly (Figure 1F). Univariate analysis indicated that lnc-CTSLP4 expression in GC tumor tissues (hazard ratio (HR) = 3.819, 95% confidence interval [CI] [1.386–10.522], p = 0.01) was related to OS of GC patients (Figure 1G). Multivariate Cox analysis indicated that lnc-CTSLP4 expression in GC tumor tissues (HR = 2.922, 95% CI [1.041–8.198], p = 0.042) was an independent prognostic risk factor related to OS (Figure 1H).
Figure 1.
Lnc-CTSLP4 expression is downregulated in tumor tissues and inversely associated with prognosis of GC patients
(A) The expression of lnc-CTSLP4 in 43 GC tissues (GC cohort 1, the ratio of GC tumor tissues versus adjacent non-tumor tissues [−ΔCt]). (B) Expression levels of lnc-CTSLP4 in GC tumor tissues (GC cohort 1) and adjacent normal tissues (−ΔCt, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). (C) The coding ability of lnc-CTSLP4 calculated by CPC2 and CPAT. (D) RNA-FISH analysis of lnc-CTSLP4 expression in the paraffin-embedded GC tumor tissues and adjacent non-tumor tissues (red: lnc-CTSLP4; blue: nuclear; n = 157, scale bars: 100 μm and 200 μm). (E) lnc-CTSLP4 high and low expression rate in GC (GC cohort 2, RNA-FISH). (F) Kaplan-Meier analysis of the overall survival of GC (GC cohort 2, n = 157). The log-rank test revealed statistical significance between the low CTSLP4 expression group (n = 129) and the high CTSLP4 expression group (n = 28). (G and H) Univariate (G) and multivariate (H) analysis for prognostic features of GC patients (n = 157). (I) Expression of lnc-CTSLP4 in GC cell lines and a normal gastric epithelium cell line, GES-1. (J) The expression of lnc-CTSLP4 in the subcellular fractions of GC cells (MGC803 and HGC27, qRT-PCR; U6 and S14 were used as nuclear and cytoplasmic markers, respectively). (K) Representative RNA-FISH imaging of lnc-CTSLP4 (red) in GC cells (red: lnc-CTSLP4; blue: DAPI; magnification: 200× and 400×).
Table 1.
Correlation between the clinicopathological features and expression of lnc-CTSLP4
| Clinicopathologic parameters |
Number of cases |
lnc-CTSLP4 FISH |
p value |
|
|---|---|---|---|---|
| 157 | Low expression (n = 129) | High expression (n = 28) | ||
| Age (years) | ||||
| ≥60 | 89 | 75 | 14 | 0.431 |
| <60 | 68 | 54 | 14 | |
| Gender | ||||
| Male | 112 | 91 | 21 | 0.636 |
| Female | 45 | 38 | 7 | |
| Tumor size (cm) | ||||
| ≥5 | 102 | 89 | 13 | 0.023 |
| <5 | 55 | 40 | 15 | |
| Local invasion | ||||
| T1, T2 | 18 | 9 | 9 | <0.001 |
| T3, T4 | 139 | 120 | 19 | |
| Lymph node metastasis | ||||
| No | 27 | 15 | 12 | <0.001 |
| Yes | 130 | 114 | 16 | |
| TNM stage | ||||
| I, II | 34 | 17 | 17 | <0.001 |
| III, IV | 123 | 112 | 11 | |
Consistent with the downregulated expression of lnc-CTSLP4 expression in GC tumor tissues, GC cell lines also expressed relatively low levels of lnc-CTSLP4 compared with a normal gastric epithelium cell line, GES-1 (Figure 1I). To further define the cellular localization of lnc-CTSLP4 in GC cells, qRT-PCR and RNA-FISH were performed, and the results showed that lnc-CTSLP4 localized in both cytoplasm and nucleus of GC cells (HGC27 and MGC803, Figures 1J and 1K). Taken together, these results suggest that downregulated lnc-CTSLP4 is associated with the aggressive progression of GC, and lnc-CTSLP4 could be a potential prognostic indicator for patients with GC.
Lnc-CTSLP4 overexpression inhibits GC cell EMT, migration, and invasion in vitro
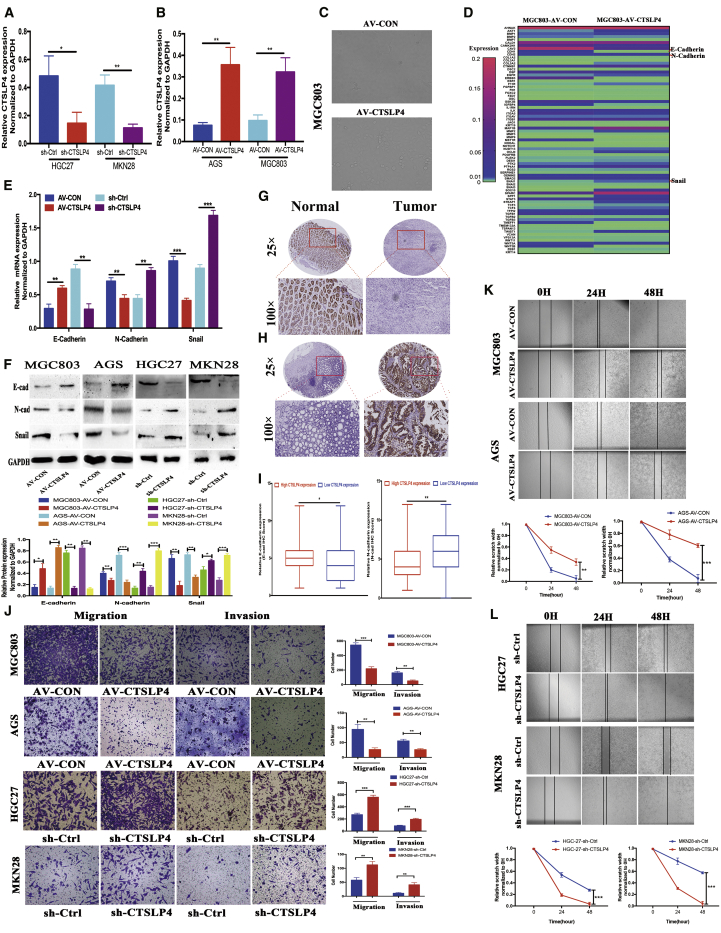
To determine the biological function of lnc-CTSLP4 in the GC cells, we constructed lnc-CTSLP4-overexpressing cells (MGC803-CTSLP4 and AGS-CTSLP4) and lnc-CTSL4-knockdown cells (HGC27-sh-CTSL4 and MKN28-sh-CTSL4) by transfecting GC cells with lnc-CTSLP4 AV-CTSLP4 or short hairpin RNA (shRNA), respectively (Figures 2A and 2B). EMT, a fundamental cell-biological process during embryonic development and organogenesis, is often hijacked by malignant cells to facilitate their dissemination to distant organs.9,19,20 We first determined whether lnc-CTSLP4 could affect GC cell morphology. GC cells expressing empty vector retained dispersed and mesenchymal properties. In contrast, GC cells overexpressing lnc-CTSLP4 exhibited a typical rounded (or amoeboid-like) morphology and reduction of lamellipodia formation, which indicated that lnc-CTSLP4 could potentially inhibit EMT of GC (Figure 2C). To further investigate the function of lnc-CTSLP4 in GC progression, we conducted PCR-array assays to detect the changed expression of EMT-associated genes in GC cells overexpressing lnc-CTSLP4. The results showed that the expression of Snail, the key EMT-inducing TF, and CDH2 (N-cadherin, the mesenchymal marker) was significantly decreased, and the expression of CDH1 (E-cadherin, the epithelial marker) was remarkably increased when lnc-CTSLP4 was overexpressed (Figure 2D). Consistent with these findings, the expression of E-cadherin, N-cadherin, and Snail in MGC803-AV-CTSLP4 cells and HGC27-sh-CTSLP4 cells was further confirmed by qRT-PCR, Western blot, and immunofluorescence (Figures 2E and 2F; Figures S3A and S3B). We validated the relationship between the lnc-CTSLP4 expression and EMT characters in 157 pairs of GC tumor tissues and adjacent non-tumor tissues, and the results showed that E-cadherin expression (based on immunohistochemistry [IHC] scores) in the low lnc-CTSLP4 expression group was significantly decreased when compared with that in the high lnc-CTSLP4 expression group; N-cadherin expression (based on IHC scores) in the low lnc-CTSLP4 expression group was significantly higher than that in the high lnc-CTSLP4 expression group (Figures 2G–2I). To further examine the effect of aberrant lnc-CTSLP4 expression on GC cell migration and invasion, we performed Transwell assays, and the results revealed that overexpression of lnc-CTSLP4 reduced the migratory and invasive capacity of MGC803-AV-CTSLP4 and AGS-AV-CTSLP4, while knockdown of lnc-CTSLP4 expression led to increased migratory and invasive capacity of HGC27-sh-CTSL4 and MKN28-sh-CTSL4 (Figure 2J). Moreover, wound healing assay confirmed that overexpression of lnc-CTSLP4 markedly reduced cell migration, whereas knockdown of lnc-CTSLP4 had the opposite effect (Figures 2K and 2L). Thus, these data demonstrate that lnc-CTSLP4 overexpression inhibits the metastatic capacity and the EMT program of GC cells.
Figure 2.
Lnc-CTSLP4 overexpression inhibits GC cell EMT, migration and invasion in vitro
(A and B) qRT-PCR analysis of lnc-CTSLP4 expression in GC cells after shRNA knockdown (A) or overexpression (B) of lnc-CTSLP4. ∗p < 0.05, ∗∗p < 0.01. (C) Morphological change of MGC803 cells after overexpression of CTSLP4 (100×). (D) PCR-array analysis of EMT-associated genes in MGC803-AV-CTSLP4 and MGC803-AV-CON cells. (E) qRT-PCR analysis of E-cadherin, N-cadherin, and Snail mRNA in GC cells after overexpression or shRNA knockdown of lnc-CTSLP4. ∗∗p < 0.01, ∗∗∗p < 0.001. (F) Western blot analysis of E-cadherin, N-cadherin, and Snail protein in GC cells (MGC803, AGS, HGC27, MKN28 cells) after knockdown of lnc-CTSLP4 (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). (G and H) Representative IHC imaging of E-cadherin and N-cadherin (magnification: 25× and 100×). (I) Correlation of lnc-CTSLP4 expression (RNA-FISH) and EMT makers (E-cadherin and N-cadherin, IHC) in GC (GC cohort 2, ∗p < 0.05, ∗∗p < 0.01). (J) The migration and invasion abilities of GC cells after overexpression or knockdown of lnc-CTSLP4 (Transwell assay, ∗∗p < 0.01, ∗∗∗p < 0.001, 200×). (K and L) Wound-healing assays of GC cells after transfection with AV-CTSLP4 or sh-CTSLP4 (∗∗p < 0.01, ∗∗∗p < 0.001, magnification: 200×).
Lnc-CTSLP4 binds to Hsp90α and promotes the degradation of HNRNPAB protein by recruiting ZFP91
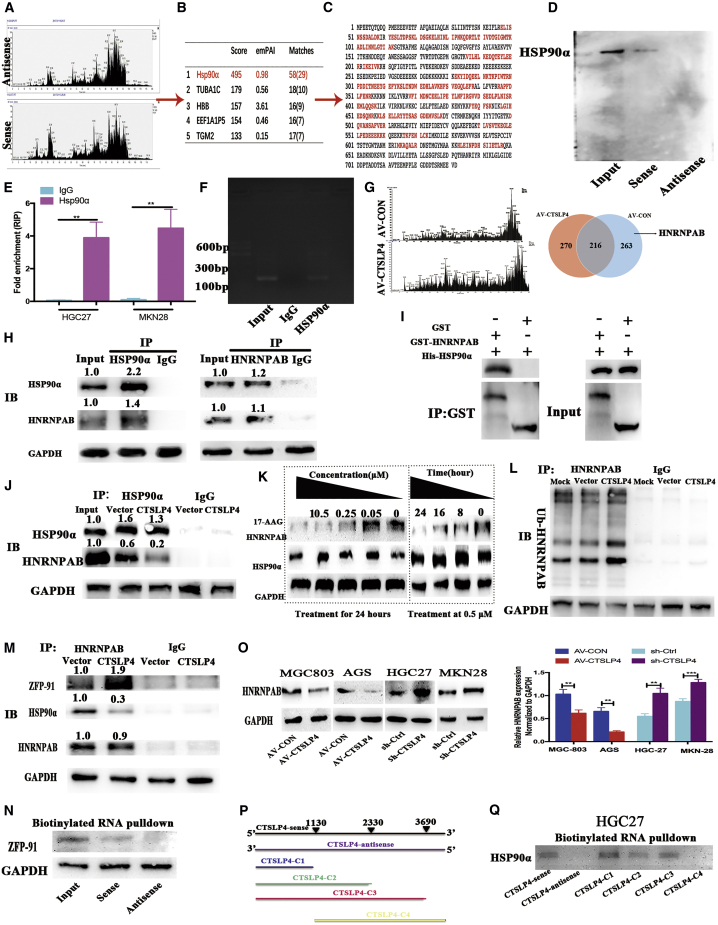
To explore the molecular mechanism and binding partners of lnc-CTSLP4 in GC cells, we performed RNA-pulldown assays with biotin-labeled lnc-CTSLP4 (lnc-CTSLP4 full-length sense and antisense), and the retrieved proteins were subjected to SDS-PAGE electrophoresis and mass spectrum analysis. Hsp90α was identified as a clear binding partner of lnc-CTSLP4 (Figures 3A–3C; Table S1). The specific interaction of lnc-CTSLP4 and Hsp90α was confirmed by Western blot using the proteins isolated from the RNA-pulldown assays (Figure 3D). RNA immunoprecipitation (RIP) assay was performed to confirm that Hsp90α could specifically bind to lnc-CTSLP4 using a specific Hsp90α antibody (Figures 3E and 3F). Hsp90α is known as a molecular chaperone that is essential for maintaining the proper folding and stability of specific target proteins (Hsp90 clients).21 We therefore performed co-immunoprecipitation (Co-IP) analysis with Hsp90α-specific antibody to evaluate binding affinity alterations of Hsp90α and its potential client proteins in MGC803-AV-CON cells and MGC803-AV-CTSLP4 cells, and the retrieved proteins were then subjected to SDS-PAGE electrophoresis and mass spectrum analysis. The results showed that Hsp90a could potentially bind 486 proteins in lnc-CTSLP4-overexpressing GC cells and 479 proteins in the AV-CON GC cells. 263 proteins were identified only in the AV-CON GC cells, and HNRNPAB, a member of ubiquitously expressed heterogeneous nuclear ribonucleoproteins (hnRNPs), was found to bind to Hsp90α in the absence of lnc-CTSLP4 (Figure 3G; Table S2).
Figure 3.
Lnc-CTSLP4 binds to Hsp90α and promotes the degradation of HNRNPAB protein by recruiting ZFP91
(A and B) RNA-pulldown assays and LC-MS/MS analysis of proteins that are specifically bound to lnc-CTSLP4 (anti-sense RNA as a negative control, A represents proteins that bound to sense or antisense of lnc-CTSLP4 and B represents top 5 proteins identified). (C) The peptide sequences of Hsp90α binding with lnc-CTSLP4 (red: matched peptide sequences). (D) Hsp90α was detected by Western blot after RNA-pulldown assays (A). (E and F) RIP and qRT-PCR analysis for the specific binding of Hsp90α to lnc-CTSLP4. ∗∗p < 0.01. (G) Co-IP followed by LC-MS/MS analysis showed that the Hsp90α could bind to HNRNPAB in the absence of lnc-CTSLP4. (H) The interaction of Hsp90α and HNRNPAB was verified by Co-IP in MGC803 cells. (I) GST-pulldown analysis of the binding of Hsp90α and HNRNPAB. (J) Analysis of proteins obtained in Co-IP in lnc-MGC803-AV-CTSLP4 and MGC803-AV-CON cells. (K) The protein levels of HNRNPAB and Hsp90α in MGC803 cells treated with 17-AAG (an Hsp90 inhibitor) for the indicated doses and times. (L) Ubiquitin modification of HNRNPAB in MGC803-AV-CTSLP4 and MGC803-AV-CON cells. (M) Co-IP and Western blot analysis of the binding of HNRNPAB to ZFP91 or Hsp90α in MGC803-AV-CTSLP4 and MGC803-AV-CON cells. (N) The specific binding of ZFP91 to lnc-CTSLP4 verified by RNA-pulldown assays (negative control: anti-sense). (O) Western blot analysis of HNRNPAB protein in GC cells (MGC803, AGS, HGC27, MKN28) after overexpression (AV-CTSLP4) or shRNA knockdown of lnc-CTSLP4 (sh-CTSLP4). (P) Truncated versions of lnc-CTSLP4, C1 (1-1130 bp), C2 (1-2330 bp), C3 (1-3690 bp), and C4 (1130-3690 bp) according to the predicted lnc-CTSLP4 structure. (Q) Western blot detection of Hsp90α after RNA-pulldown with biotinylated RNAs for different constructs of lnc-CTSLP4 or its antisense strand (negative control).
To determine whether HNRNPAB requires Hsp90α to maintain its structural stability, Co-IP was performed with Hsp90α- or HNRNPAB-specific antibodies (Figure 3H). The results confirmed that Hsp90α and HNRNPAB formed a complex in MGC803 cells. Glutathione S-transferase (GST)-pulldown experiments were then performed to detect the direct interactions of HNRNPAB and Hsp90α, and the results showed that GST-fused HNRNPAB, but not GST, could pulldown Hsp90α, which confirmed the direct interactions between HNRNPAB and Hsp90α (Figure 3I). Furthermore, the interaction between Hsp90α and HNRNPAB in MGC803 was markedly reduced in MGC803-CTSLP4 (Figure 3J). To confirm that HNRNPAB is a client of Hsp90, MGC803 cells were exposed to 17-AAG, an Hsp90 inhibitor, for a range of times and doses, and then the expression of HNRNPAB was analyzed. The results verified that upon HSP90α inhibition, the levels of HNRNPAB were reduced (Figure 3K; Figures S4A–S4D). Interestingly, the ubiquitin modification of HNRNPAB in MGC803-CTSLP4 cells was more enhanced than that in controls (Figure 3L). ZFP91 has been reported to regulate HNRNP F (a member of the hnRNP family) by its E3 ubiquitin ligase activity.22 We further performed Co-IP analysis to investigate the potential effect of lnc-CTSLP4 on the interaction of ZFP91 and HNRNPAB; we found that HNRNPAB could bind more ZFP91 and less Hsp90α in lnc-CTSLP4-overexpressing MGC803 cells compared with the control group, which indicated that the HNRNPAB-ZFP91 interaction was increased in GC cells after overexpression of lnc-CTSLP4, coincident with the decreased Hsp90α-HNRNPAB interaction (Figure 3M). RNA-pulldown assays verified that lnc-CTSLP4 could specifically bind to ZFP91 (Figure 3N). We determined the expression levels of HNRNPAB in GC cells and its potential correlation with lnc-CTSLP4 expression levels. The results showed that HNRNPAB protein expression was remarkably downregulated in MGC803-CTSLP4 and AGS-CTSLP4 cells, while it was upregulated in HGC27-sh-CTSLP4 and MKN28-sh-CTSLP4 cells (Figure 3O).
To further identify the site that mediates the interaction between lnc-CTSLP4 and Hsp90α, we predicted RNA-protein interactions and domains using CatRAPID (http://s.tartaglialab.com/page/catrapid_group), an online algorithm for estimating the protein-RNA binding propensity based on the secondary structure, hydrogen bonding, and molecular interatomic forces. Two regions (region 1 located around 469~626 bp; region 2 located around 3,065~3,222 bp) of lnc-CTSLP4 exhibited the greatest probability of binding Hsp90α (Figure S5A and S5B). Based on the predicted RNA secondary structure of lnc-CTSLP4 via the RNAfold website (http://rna.tbi.univie.ac.at//cgi-bin/RNAWebSuite/RNAfold.cgi; Figures S5C and S5D), we truncated lnc-CTSLP4 into four fragments (Figure 3P). RNA-pulldown assays demonstrated that the Hsp90α-binding activity mapped to 1~1,130 bp of lnc-CTSLP4 (Figure 3Q). Together, these data indicate that HNRNPAB is a new client of Hsp90α, and that lnc-CTSLP4 overexpression attenuates the interaction between HNRNPAB and Hsp90α, which promotes ZFP91-induced HNRNPAB ubiquitination and degradation.
Lnc-CTSLP4 inhibits EMT via suppressing HNRNPAB-mediated Snail transcriptional activation in GC cells
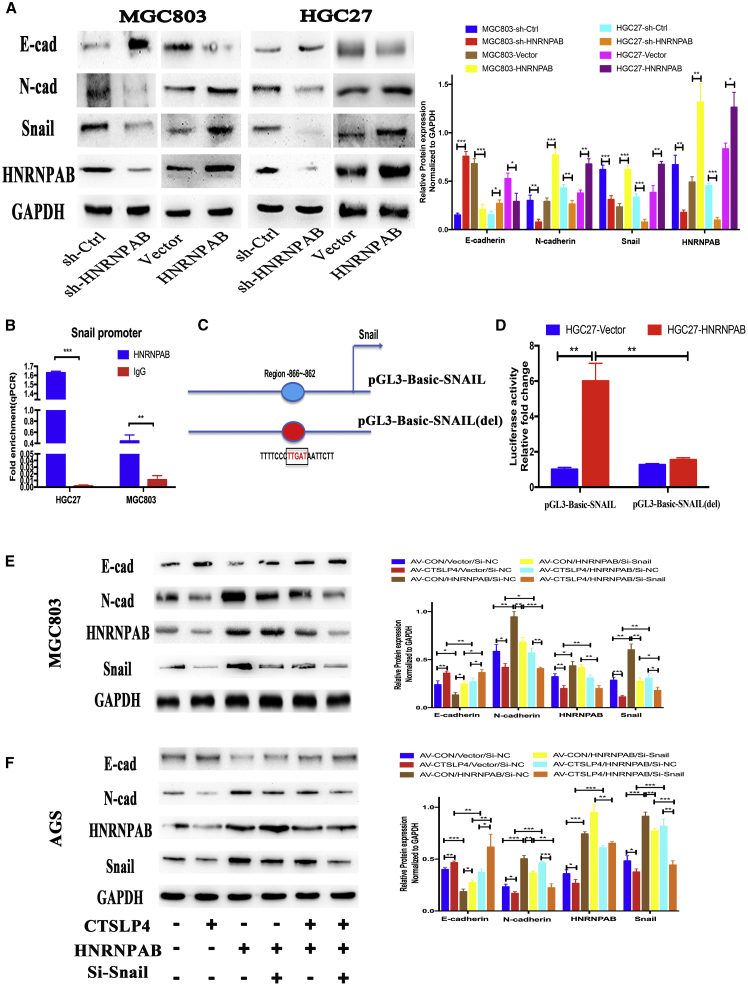
HNRNPAB is a member of hnRNPs and has been reported to act as a TF in activating Snail transcription.23 To better understand how lnc-CTSLP4 regulates GC cell metastatic potential, we considered the possibility that HNRNPAB-mediated Snail transcription and EMT were suppressed by lnc-CTSLP4 in GC cells. The expression of EMT markers was therefore assessed in both HNRNPAB-overexpressing and HNRNPAB-knockdown GC cells. Overexpression of HNRNPAB promoted the protein expression of Snail and N-cadherin and reduced the expression of E-cadherin, while knockdown of HNRNPAB had the reverse effect (Figure 4A). Moreover, chromatin IP (ChIP)-PCR and dual-luciferase reporter assays confirmed that HNRNPAB could bind the Snail promoter (Figure 4B) and induce Snail expression via its −866 to −862 bp promoter region (Figures 4C and 4D). To further ascertain whether lnc-CTSLP4 inhibits GC cell EMT by repressing the HNRNPAB/Snail pathway, we performed the rescue experiment. The results showed that the lnc-CTSLP4-mediated suppression of EMT was reversed by HNRNPAB overexpression, while inhibition of Snail expression blocked the HNRNPAB-induced EMT (Figures 4E and 4F). Taken together, these data indicate that lnc-CTSLP4 inhibits GC cell EMT via suppressing HNRNPAB-mediated Snail transcriptional activation.
Figure 4.
Lnc-CTSLP4 inhibits EMT via suppressing HNRNPAB-mediated Snail transcriptional activation in GC cells
(A) The expression level of E-cadherin, N-cadherin, HNRNPAB, and Snail in MGC803 and HGC27 cells after shRNA knockdown (sh-HNRNPAB) or overexpression (HNRNPAB) of HNRNPAB (∗∗p < 0.01, ∗∗∗p < 0.001). (B) The binding of HNRNPAB to the Snail promoter in GC cells (ChIP-PCR assays, ∗∗p < 0.01, ∗∗∗p < 0.001). (C and D) pGL3-Basic-SNAIL and pGL3-Basic-SNAIL-del (deletion at position −866 to −862 bp) constructs, along with the pRL-TK plasmid, were transfected into HGC27-HNRNPAB cells and HGC27-Vector cells, and then the relative luciferase activity was measured (∗∗p < 0.01, ∗∗∗p < 0.001). (E and F) Western blot (E) and densitometric analysis (F) of E-cadherin, N-cadherin, HNRNPAB, and Snail in MGC803 and AGS cells transfected with AV-CTSLP4, HNRNPAB-expressing vector, or Snail si-RNA (∗∗p < 0.01, ∗∗∗p < 0.001).
HNRNPAB is inversely correlated with lnc-CTSLP4 in human GC and rescues the suppressive effects of lnc-CTSLP4 on GC cells
To investigate the relationship between the expression of lnc-CTSLP4 and HNRNPAB in subjects with GC, we analyzed the public database and 157 pairs of human GC tumor tissues and adjacent non-tumor tissues from Ruijin Hospital, Shanghai Jiao Tong University School of Medicine. The results indicated that HNRNPAB was obviously upregulated in GC tumor tissues (Figures 5A and 5B), which was verified by GEPIA based on The Cancer Genome Atlas (TCGA) and GTEx database (http://gepia.cancer-pku.cn) (Figure 5C), and the expression of HNRNPAB in GC tumor tissues was positively correlative with tumor size, local invasion, lymph node metastasis, and TNM stage (Figure 5D; Figures S6A–S6C; Table 2). Based on Kaplan-Meier plotter analysis (http://kmplot.com/analysis) and our data, we found that HNRNPAB expression in GC tumor tissues was inversely proportional to the survival time of the patients with GC (Figures 5E–5G; Figures S6D–S6G). Univariate analysis showed that HNRNPAB expression in GC tumor tissues (HR = 2.580, 95% CI [1.371–4.854], p = 0.003) was related to OS in GC patients (Figure 5H). Multivariate analysis indicated that lnc-CTSLP4 expression in GC tumor tissues (hazard ratio (HR) = 2.580, 95% CI [1.371–4.854], p = 0.003) was an independent prognostic risk factor related to OS (Figure 5I). We also analyzed the association of the expression levels of HNRNPAB and lnc-CTSLP4 in GC tissues. We found that the HNRNPAB expression level was higher in the lnc-CTSLP4-low expression group compared with that in the lnc-CTSLP4-high expression group (Figure 5J). Pearson correlation analysis revealed that the expression level of HNRNPAB was correlated with E-cadherin (r = −0.4121, p < 0.001) and N-cadherin (r = 0.7322, p < 0.001) expression, which was verified by GEPIA (Figure 5K–5M). Meanwhile, bioinformatics analysis showed that the expression level of HNRNPAB was positively correlated with Snail expression based on GEPIA (Figure 5N). To further evaluate the biological function of HNRNPAB in GC progression, we performed Transwell assays, and the results revealed that upregulation of HNRNPAB promoted the migratory and invasive ability of GC cells, while knockdown of HNRNPAB expression inhibited GC cell (MGC803 and HGC27) migration and invasion (Figures 5O–5R). In addition, the inhibition of migratory capacity of GC cells induced by lnc-CTSLP4 was reversed by HNRNPAB, which was dependent on Snail expression (Figures 5S and 5T). Thus, these data demonstrate that HNRNPAB is inversely correlated with lnc-CTSLP4 in human GC and can rescue the suppressive effects of lnc-CTSLP4 on GC cells.
Figure 5.
HNRNPAB is inversely correlated with lnc-CTSLP4 in GC tissues and rescues the suppressive effects of lnc-CTSLP4 on GC cells
(A) Representative immunohistochemical staining of HNRNPAB in GC tumor tissues and adjacent non-tumor tissues (magnification: 200× and 400×). (B) The expression levels of HNRNPAB in 157 pairs of GC patients based on HNRNPAB IHC score (GC cohort 2, ∗∗∗p < 0.001). (C) Validation of HNRNPAB expression in GEPIA. (D) The expression levels of HNRNPAB in GC patients with different pathological stages based on HNRNPAB IHC score (∗∗p < 0.01, ∗∗∗p < 0.001). (E and F) The correlation of HNRNPAB expression levels and overall survival in the GSE14210 dataset (n = 118) and GSE14210, GSE15459, GSE22377, GSE29272, and GSE51105 dataset (n = 592) was analyzed by Kaplan-Meier plotter. (G) Kaplan-Meier survival analysis for OS of 157 patients with GC (GC cohort 2) reveals a statistically significant difference between the low HNRNPAB expression group (n = 129) and the high HNRNPAB group (n = 28). (H and I) Univariate (H) and multivariate (I) analysis of for prognostic features of GC patients (n = 157) showed that the expression of HNRNPAB was an independent prognostic factor for survival. (J) HNRNPAB is downregulated in the high lnc-CTSLP4 expression group compared to the low lnc-CTSLP4 expression group (GC cohort 2, ∗∗∗p < 0.001). (K and L) Correlation analysis of HNRNPAB with E-cadherin (K) or N-cadherin (L) in GC. (M and N) Validation of the relationship between HNRNPAB and E-cadherin (M) or Snail (N) based on GEPIA. (O–R) The metastatic abilities of GC cells (MGC803 and HGC27) after overexpression or shRNA knockdown of HNRNPAB (Transwell assays, magnification: 200×). (S and T) The metastatic abilities of GC cells (MGC803 and AGS cells) transfected with lnc-CTSLP4, HNRNPAB-expressing vector, or Snail si-RNA (Transwell assays, magnification: 200×, ∗∗∗p < 0.001).
Table 2.
Correlation between the clinicopathological features and expression of HNRNPAB
| Clinicopathologic parameters |
Number of cases |
HNRNPAB immunostaining |
p value |
|
|---|---|---|---|---|
| 157 | Weak positive (n = 57) | Strong positive (n = 100) | ||
| Age (years) | ||||
| ≥60 | 89 | 33 | 56 | 0.818 |
| <60 | 68 | 24 | 44 | |
| Gender | ||||
| Male | 112 | 42 | 70 | 0.623 |
| Female | 45 | 15 | 30 | |
| Tumor size (cm) | ||||
| ≥5 | 102 | 31 | 71 | 0.036 |
| <5 | 55 | 26 | 29 | |
| Local invasion | ||||
| T1, T2 | 18 | 8 | 10 | 0.445 |
| T3, T4 | 139 | 49 | 90 | |
| Lymph node metastasis | ||||
| No | 27 | 16 | 11 | 0.006 |
| Yes | 130 | 41 | 89 | |
| TNM stage | ||||
| I, II | 34 | 18 | 16 | 0.023 |
| III, IV | 123 | 39 | 84 | |
Lnc-CTSLP4 overexpression inhibits tumor growth, invasion, and metastasis in vivo
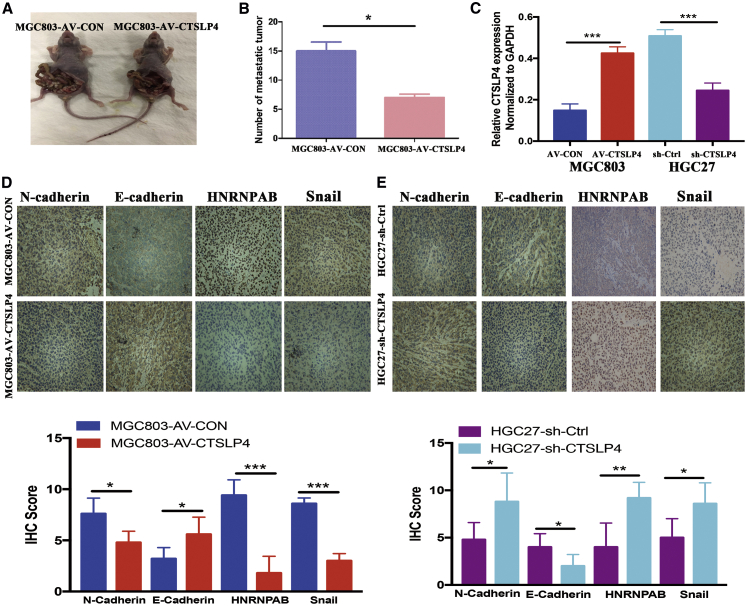
Next, we examined the effects of lnc-CTSLP4 on tumor growth and metastasis in vivo. MGC803-AV-CON, MGC803-AV-CTSLP4, HGC27-sh-Ctrl, and HGC27-sh-CTSLP4 cells were subcutaneously transplanted into nude mice and tumor growth was monitored. Overexpression of lnc-CTSLP4 resulted in dramatic inhibition of tumor growth, while genetic inhibition of lnc-CTSLP4 led to aggressive progression of tumor size. (Figures S7A–S7H). To directly verify that lnc-CTSLP4 overexpression attenuates the in vivo metastatic potential of GC cells, MGC803 cells transfected with AV-CTSLP4 or AV-CON were intraperitoneally inoculated into immunodeficient nude mice, and the occurrence of peritoneal metastases was assessed. GC cells transfected with AV-CTSLP4 developed fewer peritoneal metastatic nodules than those transfected with AV-CON (Figures 6A and 6B).
Figure 6.
Lnc-CTSLP4 overexpression inhibits GC cell invasion and metastasis in vivo
(A) The effect of CTSLP4 overexpression in MGC803 cell-derived peritoneal nodules in nude mice (n = 5 per group, ∗p < 0.05). (B) Average numbers of peritoneal nodules from nude mice. (C) The levels of lnc-CTSLP4 expression in tumor tissues formed from MGC803 cells (transfected with AV-CTSLP4 or AV-CON) and HGC27 cells (stably transfected with sh-CTSLP4 or sh-Ctrl lentivirus, ∗∗∗p < 0.001). (D) Representative immunohistochemical staining for E-cadherin, N-cadherin, HNRNPAB, and Snail in xenograft tumors derived from MGC803 cells transfected with AV-CTSLP4 or AV-CON (n = 5 per group, magnification: 200×). (E) Representative immunohistochemical staining for E-cadherin, N-cadherin, HNRNPAB, and Snail in xenograft tumors derived from HGC27 cells transfected with sh-CTSLP4 or sh-Ctrl lentivirus (n = 5 per group, magnification: 200×, ∗p < 0.05).
The expression of lnc-CTSLP4 was detected in the tumor tissue of the mice model by qRT-PCR. The results showed that lnc-CTSLP4 was significantly upregulated in the AV-CTSLP4 group and downregulated in the sh-CTSLP4 group in comparison with the control group, respectively (Figure 6C). IHC staining revealed that N-cadherin, HNRNPAB, and Snail expression were increased in the sh-CTSLP4 xenograft tumor tissues and reduced in the AV-CTSLP4 xenograft tumor tissues in comparison with the control group, respectively. However, E-cadherin expression was reduced in the sh-CTSLP4 xenograft tumor tissues and increased in the AV-CTSLP4 xenograft tumor tissues in comparison with the control group, respectively (Figures 6D and 6E). Together, these data demonstrate that lnc-CTSLP4 overexpression inhibits the proliferative and metastatic capacity of GC cells in vivo.
Discussion
Most GC patients diagnosed with advanced-stage disease present with cancer metastases, which leads to poor prognosis, less effective surgical treatment, and more resistant to drug therapy.24,25 Thus, there is an urgent need to explore more about the regulatory mechanisms underlying GC metastasis and to develop new therapeutic strategies. EMT describes a reversible phenotypic transformation from an epithelial form to a mesenchymal form and is a vital biological cellular process implicated in cancer metastasis, which could facilitate the adhesive and invasive ability of cells during the tumor development and progression.26,27 EMT could be regulated at multiple levels, including the epigenetic, transcriptional, posttranscriptional, translational, and posttranslational levels.27 LncRNAs have been demonstrated to affect gene regulation through various mechanisms and play important regulatory roles in tumorigenesis and cancer progression. In this study, we identify that lnc-CTSLP4 is a novel lncRNA in regulating GC cell EMT and metastasis and reveal that lnc-CTSLP4 is significantly downregulated in GC tissues, and low expression of lnc-CTSLP4 is positively correlated with advanced TNM stage, local invasion, lymph node metastasis, and poor prognosis. We further demonstrate that lnc-CTSLP4 significantly inhibits the metastatic potential of GC cells in vitro and in vivo by attenuating HNRNPAB-mediated Snail transcription. Our findings indicate that lnc-CTSLP4 is a novel EMT regulator in suppressing GC progression and could serve as an independent prognostic indicator and therapeutic target for metastatic GC.
LncRNAs mainly exert their functions upon formation of lncRNA-protein complexes. In this study we determine that lnc-CTSLP4 binds to Hsp90α. Hsp90α is an isoform of the Hsp90 family and has been shown to be upregulated in various human cancers.28, 29, 30 Recent studies have showed that Hsp90α, as a molecular chaperone, plays an important role in the post-translational conformational maturation, stabilization, and activation of numerous client proteins, many of which participate in signal transduction pathways that regulate proliferation, apoptosis, or metastasis of cancer cells.20,31,32 The Hsp90-client complexes vary in their forms, and increasing numbers of lncRNAs have been discovered to interact with Hsp90 and regulate their client proteins, leading to the term “Hsp90-associated lncRNAs”.33,34 In our study, integrative analysis including RNA-pulldown-mass spectrometry (MS) and RIP assays in GC cells identify lnc-CTSLP4 as an Hsp90α-associated lncRNA, and binding of lnc-CTSLP4 to Hsp90α blocks the interaction of Hsp90α with HNRNPAB, thus leading to the degradation of HNRNPAB via recruiting E3 ubiquitin ligase ZFP91. HNRNPAB belongs to the subfamily of hnRNPs, and we find that HNRNPAB is a TF of Snail in GC cells and promotes GC cell EMT by activating Snail transcription, indicating that lnc-CTSLP4 (an Hsp90-associated lncRNA) and HNRNPAB (an Hsp90-client protein) form an important pathway in regulating EMT during GC progression. Consistently, HNRNPAB is upregulated in GC tissues and predicts shorter OS of patients with GC. Moreover, HNRNPAB rescues the suppressive effects of lnc-CTSLP4 in GC cells. Thus, our findings reveal a novel mechanism of EMT regulation and provide a potential approach in preventing GC metastasis by targeting the lnc-CTSLP4-HNRNPAB pathway.
Epigenetic modulation of regulators in the Snail-mediated EMT program by ncRNAs has been studied previously,12,35 which showed that biotherapeutics based on lncRNA was a promising strategy for both vaccines and protein replacement therapy. Here, we find that lnc-CTSLP4 is a powerful suppressor of EMT, which may serve as an alternative target to treat metastatic GC. Although the detailed regulation of lnc-CTSLP4 in GC cells needs further investigation, our findings indicate that the activation of lnc-CTSLP4, such as via small activating RNA (saRNA) or via adeno-associated virus (AAV), could block the combination of Hsp90α and HNRNPAB to inhibit the transcription of Snail and EMT in vivo; thus, lnc-CTSLP4 may provide an alternative approach to treat metastatic GC. This is also the focus of our further research.
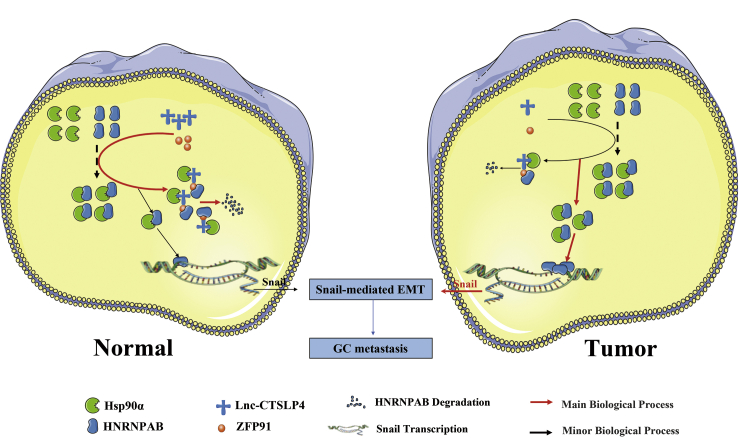
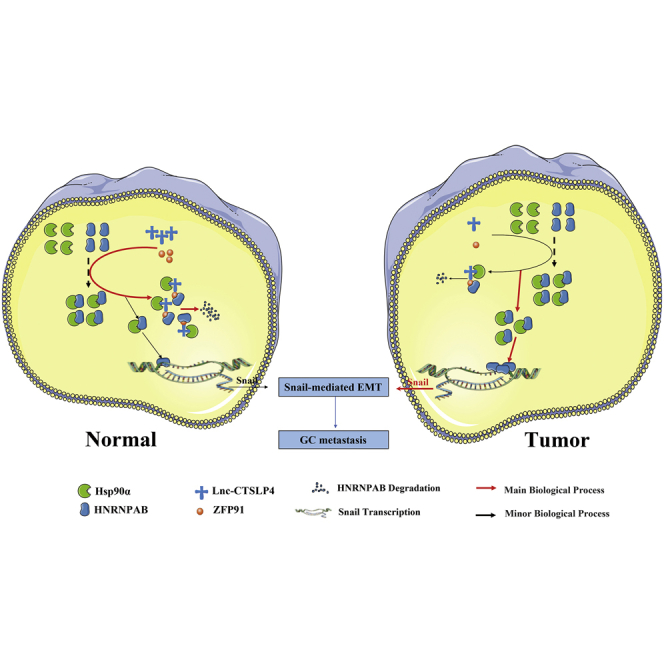
In summary, our findings support a model in which high levels of lnc-CTSLP4 recruit ZFP91 (E3 ubiquitin ligase) and bind Hsp90α/HNRNPAB complex in normal gastric epithelial cells, which prevents Hsp90α from binding HNRNPAB and promotes the degradation of HNRNPAB (a TF of Snail). While in GC cells, where lnc-CTSLP4 expression was downregulated, the Hsp90α-HNRNPAB interaction is increased, causing the stable expression of HNRNPAB and subsequent activating transcription of Snail, which leads to EMT and GC metastasis (Figure 7). Our findings suggest that lnc-CTSLP4 represents an additional layer in the regulation of GC progression and may serve as a novel prognostic biomarker and therapeutic target in GC.
Figure 7.
Schematic of the proposed mechanism of lnc-CTSLP4 in GC progression
Lnc-CTSLP4, as a regulator with decreased levels in GC, binds to Hsp90α and recruits the E3 ubiquitin ligase ZFP91, which can block the interaction of Hsp90α with HNRNPAB and decreases the stability of HNRNPAB, subsequently promotes its degradation, thus reducing the transcriptional activation of Snail and inhibiting Snail-mediated EMT.
Materials and methods
Cell culture and patient samples
GC cell lines HTB-103, AGS, and NCI-N87 used in our study were purchased from American Type Culture Collection (ATCC), and MGC803, MKN45, MKN28, HGC27, and GES-1 cell lines were purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences. All GC cell lines were authenticated by short tandem repeat analysis and had negative results for mycoplasma. HTB-103, AGS, NCI-N87, MKN45, MKN28, and HGC27 were cultured at 37°C in a humidified atmosphere of 5% CO2 with RPMI-1640 medium (Gibco, San Francisco, CA, USA) containing 10% fetal bovine serum (Gibco) with 100 U/mL penicillin and 100 U/mL streptomycin (Sangon Biotec, Shanghai, China). GES-1 and MGC803 were cultured at 37°C in a humidified atmosphere of 5% CO2 with DMEM medium (Gibco) containing 10% fetal bovine serum with 100 U/mL penicillin and 100 U/mL streptomycin.
A total of 200 patients in this study underwent D2 gastrectomy at Ruijin Hospital, Shanghai Jiao Tong University School of Medicine between 2011 and 2016. All the samples were obtained with the patients’ informed consent and were histologically confirmed. Forty-three pairs of GC tumor tissues and adjacent non-tumor tissues (GC cohort 1) were stored in RNA-latter reagent at −80°C for RNA extraction, and another 157 paired GC tumor tissues and adjacent non-tumor tissues (GC cohort 2) were fixed with formalin and made into tissue microarrays.
Plasmid construction, virus production, and infection
Full-length lnc-CTSLP4 sequence obtained by RACE analysis was constructed to a pAV-MCMV-EGFP-3FLAG vector using Ready-to-Use Seamless Cloning Kit (Sangon, Shanghai, China). Adenoviruses AV-CTSLP4 and AV-CON were generated in HEK293A cells and purified by cesium chloride (CsCl) density-gradient ultracentrifugation. Adenoviruses were added into the culture medium of HGC-27 and MGC-803 2 days before further experiments. shRNA sequences targeting lnc-CTSLP4 were designed by BLOCK-iT RNAi Designer (https://rnaidesigner.thermofisher.com/rnaiexpress/sort.do) and constructed to pLKO.1-puro plasmid. The lentiviruses were generated by co-transfecting shRNA plasmid with psPAX2 and pMD2.G plasmid to 293T cells. Lentivirus was precipitated by ultra-speed centrifugation. ShRNA lentivirus was added to the supernatant of HGC-27 and MGC-803 cells, and puromycin was used for screening stable clones.
RNA extraction, qRT-PCR, and RACE
Total RNA was extracted with Trizol reagent (Invitrogen, Carlsbad, CA, USA) and was reversely transcribed into cDNA using Reverse Transcription system (Toyobo, Kita-ku, Osaka, Japan). qRT-PCR was performed to quantify CTSLP4 mRNA level with the SYBER Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The primers for the PCR reaction are listed in Table S3. The 2-(ΔΔCt).method was used to calculate the relative abundance of RNA normalized to GAPDH. 5′-RACE and 3′-RACE analyses of CTSLP4 were performed using a GeneRacer Kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s manual. The primers for the qRT-PCR reaction and RACE are listed in Table S3.
Western blot
Cells were lysed using RIPA lysis buffer (Solarbio, Beijing, China) with proteinase inhibitor cocktail (Sigma, St. Louis, MO, USA) and phosphorylase inhibitor cocktail II and III (Sigma). The concentration of protein lysates was detected by BCA protein concentration detection kit (Thermo Fisher, San Francisco, CA, USA). A total of 30 μg protein is used at each Western blot. The protein was loaded to vertical electrophoresis and transferred to a 0.22 μm polyvinylidene fluoride (PVDF) membrane. The membrane was incubated with primary antibody and subsequent secondary antibody. The bands were detected by a Tanon ECL imaging system. ImageJ software was used to quantify the immunoblots. Area, mean gray value, and integrated density of immunoblots were measured during the process. For detailed description and the antibodies used this process, see Table S4.
IP and Co-IP
IP and Co-IP were performed using a magnetic Co-IP kit (Thermo Fisher, San Francisco, CA, USA) according to its user guide. Briefly, a total of 1 × 107 cells were seeded in a 10 cm plate, and 500 μL IP lysis buffer was used to obtain the cell lysate. After centrifugation, cell lysate was added with 5 μL magnetic beads pre-coupled with primary antibody and incubated at room temperature for 1 h. After washing with IP lysis buffer and followed by ultrapure water, the protein-protein complex was eluted from the magnetic beads with Elution buffer. After adding SDS loading buffer (Thermo Fisher), denaturation of the protein samples were performed at 100 degree for 10 minutes. The protein samples were then subjected to further analysis.
IHC staining and FISH
For IHC staining, the slides were incubated at 60°C for 60 min and immersed in xylene for 10 min. After that, the slides were soaked sequentially in anhydrous ethanol, 95% ethanol, and 75% ethanol for 5 min. Then the slides were incubated in 0.01 M sodium citrate solution for 15 min. After H2O2 incubation and sheep serum incubation, a primary antibody was added to the slides, and they were incubated at 4°C overnight. After incubation of horseradish peroxidase (HRP)-labeled secondary antibody, the slices were treated with DAB and hematoxylin. The resin was added and covered with a glass slide. Three pathologists evaluated the staining separately. The staining intensity and percentage were used to score the overall tissue sections. The intensity was graded as follows: 0 points (no staining), 1 point (light-brown staining), 2 points (brown staining), and 3 points (dark-brown staining). Percentage of positive cell number was divided into four grades: 0 points (<5%), 1 point (5%–30%), 2 points (31%–60%), and 3 points (61%–100%). Staining score was calculated as follows: overall staining score = intensity score × percentage score. A final score ≤3 was considered as negative staining and >3 as positive staining. The antibodies used are listed in the Supplemental information.
For FISH staining, the paraffin sections were taken out and sequentially put into xylene-xylene II-anhydrous alcohol-I-anhydrous alcohol II-85% alcohol-75% alcohol 5 min-diethyl pyrocarbonate (DEPC) water. The slides were boiled in the repair fluid and cooled naturally. They were digested for 25 min by adding protease K (20 μg/mL) at 37°C. After adding pre-hybrids, the lnc-CTSLP4 probe hybridization solution-containing probe was added to the slides. The slides were incubated at 37°C overnight. Then the slides were added sequentially with 2 × SSC for 37°C wash 10 min, 1 × SSC for 37°C wash 25 min, and 0.5 × SSC for room temperature wash 10 min. The slides were dripped with 2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) dye and incubated for 8 min. After washing, the anti-fluorescence quenching tablets were dripped. The images were observed and collected under a fluorescence microscope. The intensity was graded as follows: percentage of positive cell number was divided into four grades: 0 points (<5%), 1 point (5%–30%), 2 points (31%–50%), and 3 points (51%–100%). Percentage score ≤1was considered as negative staining or weakly positive staining (low lnc-CTSLP4 expression) and ≥2 as strongly positive staining (high lnc-CTSLP4 expression).
RNA pulldown and MS analysis
In vitro transcription of lnc-CTSLP4 full-length sense, antisense, and serial truncated sequences were performed using T7 or SP6 RNA Polymerase (New England Biolabs, Ipswich, Suffolk, England) according to the manufacturer’s instructions (T7 RNA polymerase was used to perform in vitro transcription of lnc-CTSLP4 full-length sense and SP6 RNA polymerase was used for lnc-CTSLP4 full-length antisense). RNA pulldown assays were performed with the Pierce Magnetic RNA-Protein Pull-Down Kit according to the manufacturer’s instructions (Thermo Fisher Scientific, Waltham, MA, USA). lncRNA-interacting proteins were obtained and subjected to MS analysis. LC-MS/MS detection was carried out on a hybrid quadrupole-TOF mass spectrometer (TripleTOF 5600+, SCIEX) equipped with a nanoLC system (nanoLC-Ultra 1D Plus, Eksigent).
RIP assays
RIP experiments were performed by using an RNA binding protein IP kit (Millipore). Generally, adherent cells were collected by RNase-free scraps and re-suspended in RIP lysis buffer. After freeze and thaw cycle to swell the cells, the RNA-protein complex was IP by magnetic beads pre-coupled with antibodies or immunoglobulin G (IgG) isotype. After washing with RIP wash buffer, the RNAs captured were extracted by phenol-chloroform-isoamylalcohol (125:25:1) extraction and were reverse transcribed to cDNA. RT-PCR was used for the detection of the protein-binding RNAs.
ChIP assays
ChIP assays were performed using a Simple ChIP Plus Enzymatic Chromatin IP Kit according to the manufacturer’s instructions (Cell Signaling Technology, Beverly, MA, USA). Briefly, cells were cross-linked by formalin and collected into an Eppendorf tube. After ultra-sonication, the chromosome was broken into small fragments. The lysates were added by primary antibody and stored at 4°C overnight. A total of 5 μg antibodies were used in 100 μL lysate originated from 1 × 106 cells in one experiment. Protein A/G beads were added into the lysate to capture antibody-protein-DNA complex. After centrifugation and washing, the DNA sequences were obtained by reverse-crosslinking and purification. Quantification of ChIP DNA was performed using qPCR. ChIP data were calculated as percentages relative to the input DNA using the equation: 2(Input Ct − Target Ct) × 100 (%). The primers used in QRT-PCR are listed in Table S3.
Dual-luciferase reporter assay
Full-length DNA sequences and mutated DNA sequence were synthesized and constructed to pGL3-basic plasmid via Ready-to-Use Seamless Cloning Kit (Sangon). pcDNA3.1-HNRNPAB plasmid was purchased from PPL (Nanjing, China). HGC27-vector and HGC27-HNRNPAB cells were obtained by transfecting with pcDNA3.1-HNRNPAB or pcDNA 3.1 plasmids to HGC-27 using Lipofectamine 3000 Transfection Reagent (Invitrogen). G418 was used to select stable clones. For dual-luciferase reporter assay, HGC27-vector and HGC27-HNRNPAB cells were seeded in 24-well plates and transfected with pGL3-Basic-SNAIL, pGL3-Basic-SNAIL(del), and pRL-TK plasmid using a Lipofectamine 3000 Transfection Reagent. Three days after transfection, dual-luciferase reporter assay was performed using Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
GST pulldown assay
6 × His tag sequences followed by CDS sequence of HSP90AA1 were synthesized and constructed to pET24a (+) plasmid. GST tag sequences followed by CDS of HNRNPAB were synthesized and constructed to pGEX-4T-1 plasmid. pET28a-HSP90AA1 or pGEX-4T-1-HNRNPAB plasmid was transformed to BL21(DE3) E. coli. IPTG was used to induce the expression of His-HSP90AA1 and GST-HNRNPAB. After incubation and collection, 400 μL Ni-IDA binding buffer was used to re-suspend the bacteria. Ultrasound sonication was used to release the intracellular proteins. The fusion proteins were purified by Ni-IDA-Sepharose CL-6B affinity chromatography column. After washing and elution, the proteins were subjected to further analysis.
The GST pulldown assays were performed using Pierce GST Protein Interaction Pulldown Kit (Thermo) as per its instructions. Briefly, 100 μg GST tag (Sangon, Shanghai, China) or GST-HNRNPAB was immobilized to 50 μL glutathione agarose (50% concentration). 800 μL pulldown-lysis buffer with 100 μg His-HSP90AA1 was added to immobilized GST or GST-HNRNPAB protein. After incubation at 4°C for 1 h, the uncaptured proteins were washed, and the captured proteins were eluted by glutathione elution buffer. The samples obtained were subjected to Western blot analysis. The materials used are listed in Table S4.
Cell migration and invasion assay
For the migration assay, cells were suspended in serum-free medium (1 × 105 cells/insert) and added to the upper chamber of the 24-well insert (membrane pore size, 8 μm; Corning Life Sciences, MA, USA). Medium containing 10% serum was added to the lower chamber. After incubation for 12 h, the cells that migrated to the bottom of the membranes were fixed and stained with 0.1% crystal violet for 30 min. For the invasion assay, chamber membranes were coated with diluted Matrigel (BD Bioscience, San Jose, CA, USA). After incubation for 24 h, the cells that invaded to the bottom of the membrane were fixed and stained with 0.1% crystal violet for 30 min. The stained cells were counted using a microscope and photographed. Ten fields were randomly selected to count, and the average number was presented.
In vivo tumorigenesis and peritoneal dissemination
Four-week-old male BALB/c nude mice were purchased from Institute of Zoology Chinese Academy of Science and housed at a specific pathogen-free environment in the Animal Laboratory Unit, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, China. MGC803 cells transduced by AV-CTSLP4 or AV-CON adenoviral particles (2 × 107) were harvested and resuspended in 500 μL of PBS and then were injected subcutaneously or peritoneally into the mice (4 × 106/100 μL PBS/mouse). HGC27 cells transduced by sh-CTSLP4 or sh-Ctrl lentivirus (2 × 107) were harvested and resuspended in 500 μL of PBS and then were injected subcutaneously into the mice (4 × 106/100 μL PBS/mouse). For tumorigenesis in vivo, all mice were sacrificed after 28 days, and the subcutaneous lump was removed, imaged, and placed into 4% formaldehyde for paraffin embedding. For the peritoneal dissemination model, all mice were sacrificed after 30 days and peritoneal metastasis nodules were counted.
Statistical analysis
All experiments were repeated at least three times and the results were summarized as means ± SD. The Student t test and one-way analysis of variance (ANOVA) were used to analyze the data, and chi-square tests were used to analyze categorical variables. All statistical tests were performed with SPSS 16.0 (SPSS, Chicago, IL, USA), and p values <0.05 were regarded as indicating statistically significant differences (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Availability of data and materials
All data and details can be obtained by contacting the corresponding author.
Ethics approval and consent to participate
The experimental protocol for animal studies was reviewed and approved by Ruijin Hospital Institutional Ethics Committee.
Acknowledgments
We thank LetPub (https://www.letpub.com/) for the linguistic assistance during the preparation of this manuscript. This work was supported by the National Natural Science Foundation of China (Nos. 81871902 to L.S., 81772509 to B.L., and 81772518 to Z. Yang); the Multicenter Clinical Trial of Shanghai Jiao Tong University School of Medicine (No. DLY201602 to Z.Z.); the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (No. 20152505 to L.S.); and the Eagle Program to Young Doctors of Shanghai Anticancer Association (No. SACA-CY19C10 to Z. Yu).
Author contributions
L.S. and B.L. made the original proposal and designed and supervised the project; T.P., Z. Y., and Z.J. performed all the experiments; T.P. and X.W. performed dada analysis; all participated in data interpretation; T.P. and Z.Y. prepared the first draft; L.S. and B.L. completed the final revisions. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2021.02.003.
Contributor Information
Bingya Liu, Email: liubingya@sjtu.edu.cn.
Liping Su, Email: suliping@shsmu.edu.cn.
Supplemental information
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Bernards N., Creemers G.J., Nieuwenhuijzen G.A., Bosscha K., Pruijt J.F., Lemmens V.E. No improvement in median survival for patients with metastatic gastric cancer despite increased use of chemotherapy. Ann. Oncol. 2013;24:3056–3060. doi: 10.1093/annonc/mdt401. [DOI] [PubMed] [Google Scholar]
- 4.Wagner A.D., Syn N.L., Moehler M., Grothe W., Yong W.P., Tai B.C., Ho J., Unverzagt S. Chemotherapy for advanced gastric cancer. Cochrane Database Syst. Rev. 2017;8:CD004064. doi: 10.1002/14651858.CD004064.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 6.Chambers A.F., Groom A.C., MacDonald I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 7.Yeung K.T., Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol. Oncol. 2017;11:28–39. doi: 10.1002/1878-0261.12017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nieto M.A. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 9.Diepenbruck M., Christofori G. Epithelial-mesenchymal transition (EMT) and metastasis: yes, no, maybe? Curr. Opin. Cell Biol. 2016;43:7–13. doi: 10.1016/j.ceb.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Grau Y., Carteret C., Simpson P. Mutations and Chromosomal Rearrangements Affecting the Expression of Snail, a Gene Involved in Embryonic Patterning in DROSOPHILA MELANOGASTER. Genetics. 1984;108:347–360. doi: 10.1093/genetics/108.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin Y., Dong C., Zhou B.P. Epigenetic regulation of EMT: the Snail story. Curr. Pharm. Des. 2014;20:1698–1705. doi: 10.2174/13816128113199990512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skrzypek K., Majka M. Interplay among SNAIL Transcription Factor, MicroRNAs, Long Non-Coding RNAs, and Circular RNAs in the Regulation of Tumor Growth and Metastasis. Cancers (Basel) 2020;12:209. doi: 10.3390/cancers12010209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Shi J., Chai K., Ying X., Zhou B.P. The Role of Snail in EMT and Tumorigenesis. Curr. Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopp F., Mendell J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell. 2018;172:393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C.Y., Li R.K., Qi Y., Li X.N., Yang Y., Liu D.L., Zhao J., Zhu D.Y., Wu K., Zhou X.D., Zhao S. Upregulation of long noncoding RNA SPRY4-IT1 promotes metastasis of esophageal squamous cell carcinoma via induction of epithelial-mesenchymal transition. Cell Biol. Toxicol. 2016;32:391–401. doi: 10.1007/s10565-016-9341-1. [DOI] [PubMed] [Google Scholar]
- 16.Sakai S., Ohhata T., Kitagawa K., Uchida C., Aoshima T., Niida H., Suzuki T., Inoue Y., Miyazawa K., Kitagawa M. Long Noncoding RNA ELIT-1 Acts as a Smad3 Cofactor to Facilitate TGFβ/Smad Signaling and Promote Epithelial-Mesenchymal Transition. Cancer Res. 2019;79:2821–2838. doi: 10.1158/0008-5472.CAN-18-3210. [DOI] [PubMed] [Google Scholar]
- 17.Xiong H.G., Li H., Xiao Y., Yang Q.C., Yang L.L., Chen L., Bu L.L., Zhang W.F., Zhang J.L., Sun Z.J. Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019;38:278. doi: 10.1186/s13046-019-1254-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Yu B., Li J., Su L., Yan M., Zhu Z., Liu B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5:2318–2329. doi: 10.18632/oncotarget.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dongre A., Weinberg R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019;20:69–84. doi: 10.1038/s41580-018-0080-4. [DOI] [PubMed] [Google Scholar]
- 21.Hoter A., El-Sabban M.E., Naim H.Y. The HSP90 Family: Structure, Regulation, Function, and Implications in Health and Disease. Int. J. Mol. Sci. 2018;19:2560. doi: 10.3390/ijms19092560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S.H., Zhu S., Wang Y., Huang J.Z., Chen M., Wu Q.X., He Y.T., Chen D., Yan G.R. ECD promotes gastric cancer metastasis by blocking E3 ligase ZFP91-mediated hnRNP F ubiquitination and degradation. Cell Death Dis. 2018;9:479. doi: 10.1038/s41419-018-0525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou Z.J., Dai Z., Zhou S.L., Hu Z.Q., Chen Q., Zhao Y.M., Shi Y.H., Gao Q., Wu W.Z., Qiu S.J. HNRNPAB induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by transcriptionally activating SNAIL. Cancer Res. 2014;74:2750–2762. doi: 10.1158/0008-5472.CAN-13-2509. [DOI] [PubMed] [Google Scholar]
- 24.Zhao J., Liu Y., Zhang W., Zhou Z., Wu J., Cui P., Zhang Y., Huang G. Long non-coding RNA Linc00152 is involved in cell cycle arrest, apoptosis, epithelial to mesenchymal transition, cell migration and invasion in gastric cancer. Cell Cycle. 2015;14:3112–3123. doi: 10.1080/15384101.2015.1078034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng Z., Wang C.X., Fang E.H., Wang G.B., Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J. Gastroenterol. 2014;20:5403–5410. doi: 10.3748/wjg.v20.i18.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bill R., Christofori G. The relevance of EMT in breast cancer metastasis: Correlation or causality? FEBS Lett. 2015;589:1577–1587. doi: 10.1016/j.febslet.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Cao Q.H., Liu F., Li C.Z., Liu N., Shu M., Lin Y., Ding L., Xue L. Testes-specific protease 50 (TSP50) promotes invasion and metastasis by inducing EMT in gastric cancer. BMC Cancer. 2018;18:94. doi: 10.1186/s12885-018-4000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elzakra N., Cui L., Liu T., Li H., Huang J., Hu S. Mass Spectrometric Analysis of SOX11-Binding Proteins in Head and Neck Cancer Cells Demonstrates the Interaction of SOX11 and HSP90α. J. Proteome Res. 2017;16:3961–3968. doi: 10.1021/acs.jproteome.7b00247. [DOI] [PubMed] [Google Scholar]
- 29.Lee Y.C., Chang W.W., Chen Y.Y., Tsai Y.H., Chou Y.H., Tseng H.C., Chen H.L., Wu C.C., Chang-Chien J., Lee H.T. Hsp90α Mediates BMI1 Expression in Breast Cancer Stem/Progenitor Cells through Facilitating Nuclear Translocation of c-Myc and EZH2. Int. J. Mol. Sci. 2017;18:1986. doi: 10.3390/ijms18091986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang H., Duan B., He C., Geng S., Shen X., Zhu H., Sheng H., Yang C., Gao H. Cytoplasmic HSP90α expression is associated with perineural invasion in pancreatic cancer. Int. J. Clin. Exp. Pathol. 2014;7:3305–3311. [PMC free article] [PubMed] [Google Scholar]
- 31.Cullinan S.B., Whitesell L. Heat shock protein 90: a unique chemotherapeutic target. Semin. Oncol. 2006;33:457–465. doi: 10.1053/j.seminoncol.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Trepel J., Mollapour M., Giaccone G., Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 2010;10:537–549. doi: 10.1038/nrc2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou C.C., Yang F., Yuan S.X., Ma J.Z., Liu F., Yuan J.H., Bi F.R., Lin K.Y., Yin J.H., Cao G.W. Systemic genome screening identifies the outcome associated focal loss of long noncoding RNA PRAL in hepatocellular carcinoma. Hepatology. 2016;63:850–863. doi: 10.1002/hep.28393. [DOI] [PubMed] [Google Scholar]
- 34.Guo H., Zhao L., Shi B., Bao J., Zheng D., Zhou B., Shi J. GALNT5 uaRNA promotes gastric cancer progression through its interaction with HSP90. Oncogene. 2018;37:4505–4517. doi: 10.1038/s41388-018-0266-4. [DOI] [PubMed] [Google Scholar]
- 35.Chen D.D., Cheng J.T., Chandoo A., Sun X.W., Zhang L., Lu M.D., Sun W.J., Huang Y.P. microRNA-33a prevents epithelial-mesenchymal transition, invasion, and metastasis of gastric cancer cells through the Snail/Slug pathway. Am. J. Physiol. Gastrointest. Liver Physiol. 2019;317:G147–G160. doi: 10.1152/ajpgi.00284.2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and details can be obtained by contacting the corresponding author.