Abstract
Congenital scoliosis (CS) is a congenital disease caused by malformations of vertebrae. Recent studies demonstrated that DNA modification could contribute to the pathogenesis of disease. This study aims to identify epigenetic perturbations that may contribute to the pathogenesis of CS. Four CS patients with hemivertebra were enrolled and underwent spine correction operations. DNA was extracted from the hemivertebrae and spinal process collected from the specimen during the hemivertebra resection. Genome-wide DNA methylation profiling was examined at base-pair resolution using whole-genome bisulfite sequencing (WGBS). We identified 343 genes with hyper-differentially methylated regions (DMRs) and 222 genes with hypo-DMRs, respectively. These genes were enriched in the mitogen-activated protein kinase (MAPK) signaling pathway, calcium signaling pathway, and axon guidance in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and were enriched in positive regulation of cell morphogenesis involved in differentiation, regulation of cell morphogenesis involved in differentiation, and regulation of neuron projection development in Biological Process of Gene Ontology (GO-BP) terms. Hyper-DMR-related genes, including IGHG1, IGHM, IGHG3, RNF213, and GSE1, and hypo DMR-related genes, including SORCS2, COL5A1, GRID1, RGS3, and ROBO2, may contribute to the pathogenesis of hemivertebra. The aberrant DNA methylation may be associated with the formation of hemivertebra and congenital scoliosis.
Keywords: Congenital Scoliosis (CS), DNA methylation, Whole Genome Bisulfite Sequencing (WGBS), Hemivertebra, Somitogenesis
Graphical abstract

Congenital scoliosis (CS) is a congenital disease caused by malformations of vertebra. Wu and his colleagues comprehensively described the whole-genome methylation difference in CS patients with hemivertebra using WGBS. The aberrant DNA methylation was identified to be associated with hemivertebra development, which can help elucidate the pathogenic mechanism of CS.
Introduction
Congenital scoliosis (CS) is a form of spinal deformity affecting 0.05%–0.1% of newborns.1,2 In the clinic, CS can be classified into three types based on the causes, including failure of formation, failure of segmentation, and mixed deformity.3 Hemivertebra is the most common consequence of failure of formation, which is characterized by the absence of one side of the vertebral body and one vertebral pedicle.
Genetic defects have been reported to be associated with congenital scoliosis.4, 5, 6, 7 Besides, environmental factors, including hypoxia2 and high altitude,8 could also increase the risk for CS. Although studies exploring the pathogenesis of CS have continuously boosted for decades, the disease mechanism of the majority of CS is still unclear.
DNA methylation is an epigenetic modification that usually gives rise to 5-methylcytosine (5-mC) by targeting the fifth carbon of the pyrimidine ring of cytosine. In mammals, transcriptional silencing is the most pivotal function of DNA methylation.9 It is postulated that the aberrant DNA methylation plays an important role in congenital diseases, such as congenital renal agenesis10 and congenital heart disease.11 Recent studies also indicated that differential methylation of key genes or the CpG site was related to scoliosis.12, 13, 14 However, there is no known methylation region associated with CS.
In this study, we enrolled four CS patients with hemivertebra. We compared their genome methylation difference between the hemivertebra body and spinal process, and explored the epigenetic perturbation to the pathogenesis of CS.
Results
Clinical information
Two of the CS patients were female, and the mean age of patients was 7.75 (from 2 to 14) years old (Table 1). Three of them were classified into type 1 CS, and one was classified into type 3 CS. The average angle of main Cobb angle was 66 degrees. All of the patients had hemivertebra (Figure 1).
Table 1.
Characteristics of the enrolled CS patients
| ID | Gender | Age (y) | CS classification | Main Cobb angle | Hemivertebra location |
|---|---|---|---|---|---|
| DISCO552 | F | 12 | 1 | 73 | T4 |
| DISCO644 | M | 14 | 3 | 75 | T7 |
| DISCO799 | F | 3 | 1 | 36 | L3 |
| DISCO831 | M | 2 | 1 | 80 | L1 |
CS, congenital scoliosis; F, female; M, male.
Figure 1.
Spine images of the four CS patients with hemivertebra
DNA methylation differences
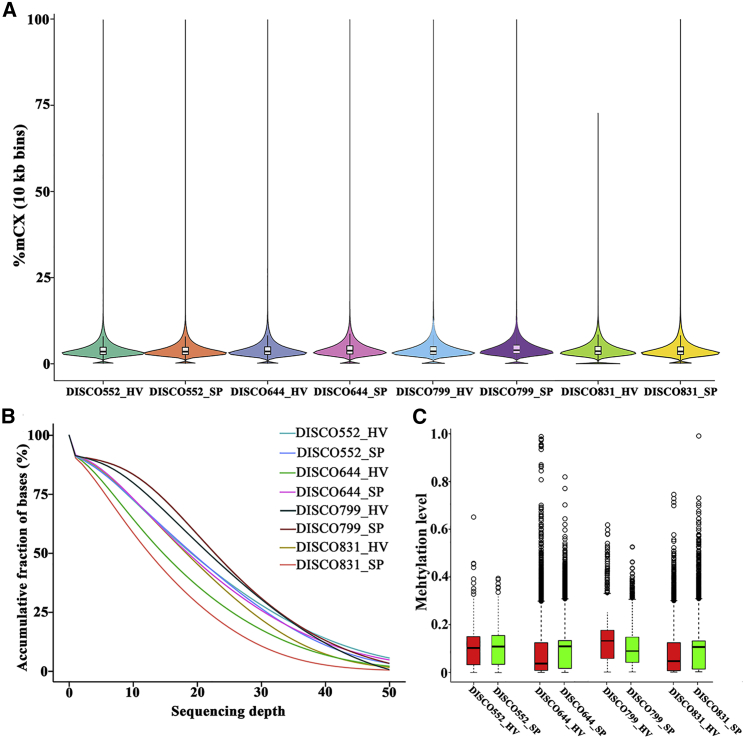
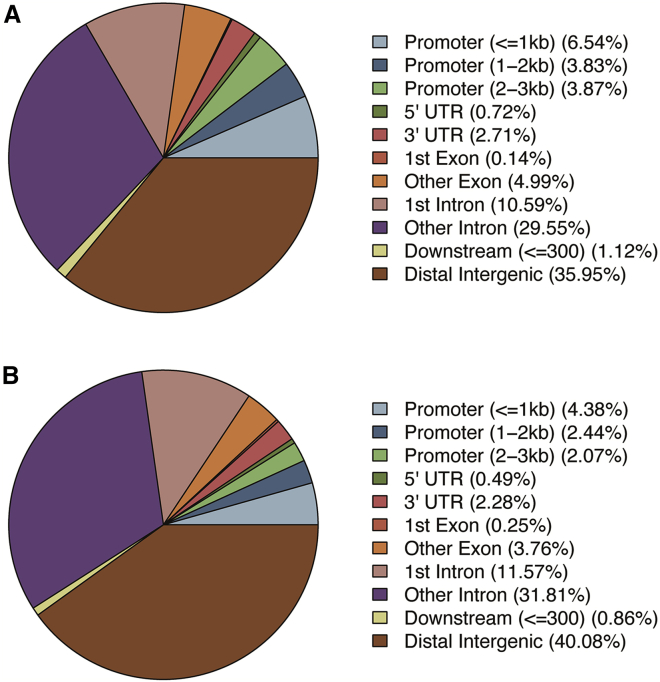
The distribution of global methylation level (Figure 2A) and sequencing depth (Figure 2B) between the hemivertebra and spinal process in the four CS patients were similar. We compared the methylation level distribution of the whole-genome DMRs between the hemivertebra body and the spinal process in each patient (Figure 2C). To characterize the distribution of DMRs, we assigned them to known genomic features, which include promoter, 5′ UTR, 3′ UTR, and intronic and intergenic regions. Among all of the hyper-DMRs, 36% were located in intergenic regions, whereas 14% were located in promoter regions. Similarly, for hypo-DMRs, 40% were located in intergenic regions, and a slightly lower percent (9%) was located in promoter regions (Figure 3). Finally, we identified 343 genes with hyper-differentially methylated regions (DMRs) and 222 genes with hypo-DMRs that are consistent among the four patients (Figure 4). We performed an unsupervised hierarchical clustering analysis using DNA methylation level of 343 hyper-DMR-related genes and 222 hypo-DMR-related genes to characterize the global methylation alterations between the hemivertebra group and spinal process group (Figure 5). We enriched these DMR-related genes and then subjected them to Kyoto Encyclopedia of Genes and Genomes (KEGG) and Biological Process of Gene Ontology (GO-BP) terms for pathway analysis. It revealed that DMR-related genes were enriched in several pathways. Three pathways with the most significant association were mitogen-activated protein kinase (MAPK) signaling pathway, calcium signaling pathway, and axon guidance in KEGG pathways. The top three pathways with the most significant association in GO-BP terms were positive regulation of cell morphogenesis involved in differentiation, regulation of cell morphogenesis involved in differentiation, and regulation of neuron projection development (Figure 6), indicating that these pathways could be involved in the development of hemivertebra in CS patients.
Figure 2.
DNA methylation differences in different samples
(A) Violin plot of the methylation level distribution in different samples. (B) Sequencing depth in different samples. (C) Boxplot of DMR methylation level.
Figure 3.
Characterization of identified DMRs between hemivertebra group and spinal process group
(A) Co-localization of hyper-DMRs with known genomic features. (B) Co-localization of hypo-DMRs with known genomic features.
Figure 4.
Matrix layout for visualization of all intersections of DMR-related genes between four patients, sorted by degree and size
Dark circles in the matrix indicate sets that are part of the intersection. (A) All intersections of hyper-DMR-related genes. (B) All intersections of hypo-DMR-related genes.
Figure 5.
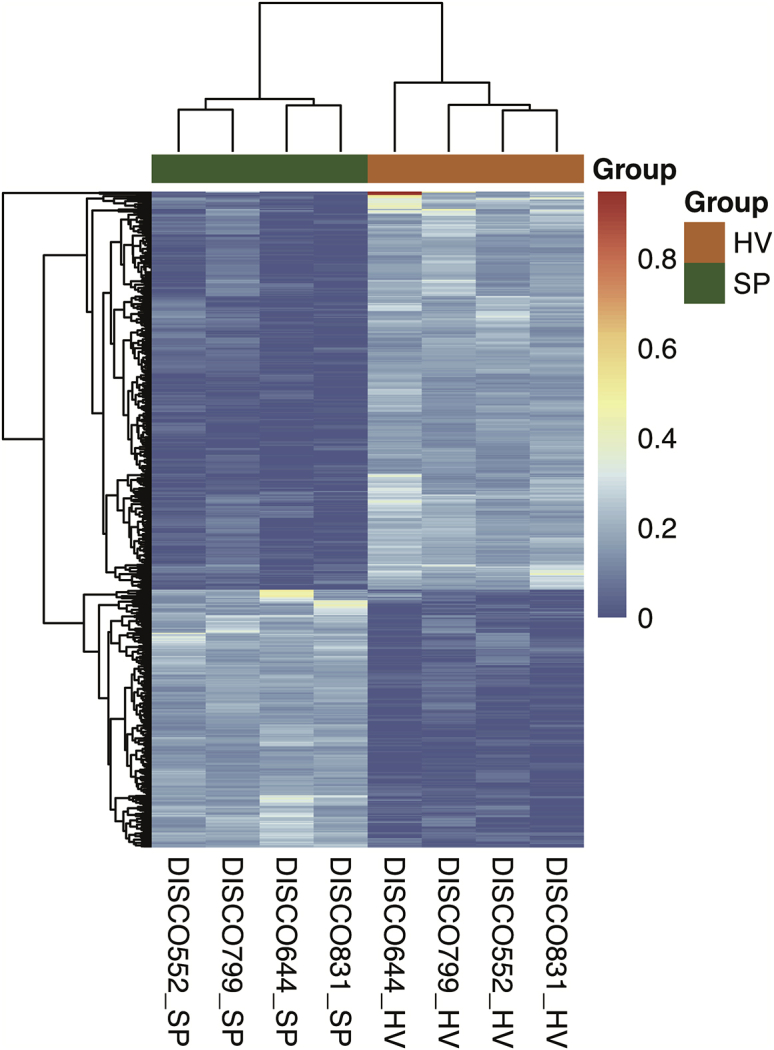
The unsupervised hierarchical clustering heatmap of hemivertebra and spinal process in four patients based on DNA methylation patterns of 343 hyper-DMR-related genes and 222 hypo-DMR-related genes
Figure 6.
Functional enrichment of DMR-related genes
(A) Bar chart of top 10 DMR-related genes enrichment with GO-BP. (B) Scatterplot of top 10 DMR-related genes enrichment with KEGG pathway.
Validation of candidate DMR-related genes
To identify the specific pathogenic epigenetic variants, we ranked the recurrent DMR-related genes according to the arithmetic mean value of the methylation difference between hemivertebra body and normal spinal process in each group (Table S1). The top five candidate DMR-related genes with greatest hyper-methylation differences were IGHG1, IGHM, IGHG3, RNF213, and GSE1 (Table 2). The top five candidate DMR-related genes with greatest hypo-methylation differences were SORCS2, COL5A1, GRID1, RGS3, and ROBO2 (Table 2). We hypothesize that the different methylation of DMR-related genes may influence the gene expression by regulating the transcription and be associated with the phenotype of hemivertebra of CS.
Table 2.
Top 5 common DMR-related genes with greatest mean hyper- and hypo-methylation differences in the CS patients
| DMR-related genes | Gene ID | DNA methylation direction in HV | Mean methylation difference |
|---|---|---|---|
| IGHG1 | ENSG00000211896 | hyper | 0.39 |
| IGHM | ENSG00000211899 | hyper | 0.33 |
| IGHG3 | ENSG00000211897 | hyper | 0.33 |
| RNF213 | ENSG00000173821 | hyper | 0.28 |
| GSE1 | ENSG00000131149 | hyper | 0.24 |
| SORCS2 | ENSG00000184985 | hypo | −0.25 |
| COL5A1 | ENSG00000130635 | hypo | −0.23 |
| GRID1 | ENSG00000182771 | hypo | −0.21 |
| RGS3 | ENSG00000138835 | hypo | −0.21 |
| ROBO2 | ENSG00000185008 | hypo | −0.20 |
SORCS2 encodes Sortilin Related VPS10 Domain Containing Receptor 2 (SORCS2), which is one family member of the vacuolar protein sorting 10 (VPS10) domain-containing receptor proteins. It is indicated that SorCS2 was prominently expressed in the vertebrae of mice embryo.15 In a genome-wide association study (GWAS), a locus of SORCS2 is significantly associated with insulin-like growth factor-binding protein-3 (IGFBP-3), which is involved in bone metabolism.16 We hypothesized that the abnormal methylation could influence the expression of SORCS2, which could induce the malformation of vertebrae.
COL5A1 encodes an alpha chain for one of the low-abundance fibrillar collagens. Mutations of this gene could lead to Ehlers-Danlos syndrome (EDS).17 EDS is a kind of connective tissue disorder, characterized by severe muscle hypotonia at birth, joint hypermobility, hyperelastic skin, osteopenia, increased bone fragility, and progressive scoliosis.18,19 Although the patients in our study manifested with only hemivertebra, the abnormal methylation of COL5A1 could lead to abnormal expression of collagen, which may increase the risk of scoliosis development.
RGS3 encodes Regulator of G-protein signaling 3, which is an antagonism of G protein and interacts with ephrin.20 It is postulated that the dislocation of RGS3 could impair the function of Ephrin-B-RGS cell fate signaling complex.21 The ephrin signaling within the osteoclast and osteoblast lineages could promote or inhibit cell differentiation in bone remodeling.22 Different methylation level of RGS3 could influence bone remodeling and have an effect on the formation of hemivertebra.
ROBO2 encodes Roundabout Guidance Receptor 2, which belongs to the ROBO family and functions in axon guidance and cell migration. The protein could function as receptor for SLIT2 and regulate bone morphogenetic proteins (BMPs) activity.23 Previous studies indicate that ROBO2 could promote chondrocyte maturation.24 In a rat animal model, mRNA of robo2 was expressed during the differentiation of osteoblast.25 The aberrant methylation of ROBO2 could regulate the differentiation of osteoblast and chondrocyte maturation, which may contribute to the manifestation of hemivertebra. Further functional studies are needed to explore the mechanism.
Discussion
In general, genetic variants and epigenetic modifications are pivotal causes to embryonic somatogenesis failure.2,26 Environmental factors, including hypoxia, are validated principal regulators of fibroblast growth factor (FGF) signaling, which is an essential pathway in the pathogenesis of CS.2 Up until now, the epigenetic modification of CS is still unclear. In this study, we present the epigenetic landscape of CS, or more specifically hemivertebra. According to previous studies, DNA methylation can predispose to a wide range of congenital developmental diseases.10,11 Some studies also indicated that DNA methylation is related to scoliosis.12, 13, 14 However, there is no study investigating the relation between the whole-genome DNA methylation pattern and CS.
Therefore, we performed whole-genome bisulfite sequencing (WGBS) to detect the whole-genome DNA methylation between the hemivertebra body and normal spinal process of CS patients. Potential pathological signal pathways and DMR-related genes were identified. DMR-related genes with differently methylated level were enriched in several signaling pathways, including the MAPK signaling pathway, calcium signaling pathway, and axon guidance in KEGG pathways, and positive regulation of cell morphogenesis involved in differentiation, regulation of cell morphogenesis involved in differentiation, and regulation of neuron projection development in GO-BP terms.
The MAPK signaling pathway is a critical pathway regulating the proliferation and differentiation of multiple cell types, including osteoprogenitor cells.27 Mutations of the genes involved in the MAPK signaling pathway are associated with multiple diseases manifesting scoliosis, such as Noonan syndrome,28 Costello syndrome,29 and cardiofaciocutaneous (CFC) syndrome.30 Transcriptional regulation, including miRNA and DNA methylation in genes involved in the MAPK signaling pathway, was also associated with scoliosis.12,31 Thus, we hypothesized that the abnormal DNA methylation of the MAPK signaling pathway-related genes could lead to the hemivertebra development.
Calcium metabolism is very important in bone formation.32,33 The abnormal calcium signaling pathway could lead to osteoblast dysfunction and scoliosis.34,35 Previous studies also proved that the abnormal transport of calcium ion is involved in scoliosis.31,36 Previous studies indicated that the proteins of axon guidance could guide growing axons during development.37 The defective axon guidance could lead to several diseases manifesting scoliosis.38,39 Thus, the abnormalities of these pathways could have potential to increase the risk for hemivertebra.
We also found several common DMR-related genes manifested with the same methylation tendency. Hyper-DMR-related genes, including IGHG1, IGHM, IGHG3, RNF213, and GSE1, and hypo-DMR-related genes, including SORCS2, COL5A1, GRID1, RGS3, and ROBO2, were the common DMR-related genes with greatest mean methylation differences.
There are several limitations in this study. First, the abnormal methylation can cause transcriptome disorder in target tissues. In our study, the transcriptome data were not detected; further studies are needed to explore the relationship between abnormal methylation modifications and unfaithful activation of key genes. Second, although this is the pilot, and to our knowledge the first study elucidating the relation between epigenetic modification and congenital scoliosis, the sample size of the study was relatively small. Further studies with larger sample size are necessary to validate our findings.
Materials and methods
Patients and materials
Four patients with CS were recruited from Peking Union Medical College Hospital (PUMCH) from July 2015 to January 2016, as a part of the Deciphering Disorders Involving Scoliosis and Comorbidities (DISCO) study (http://www.discostudy.org/). All the patients had no relationship to each other. All the CS patients manifested with hemivertebra and underwent hemivertebra resection. Parts of the hemivertebra body and normal spinal process were collected from the specimen during the surgery. Genomic DNA was extracted from those specimens using DNeasy Blood & Tissue Kits (QIAGEN, Eastwin Scientific, Beijing, China) according to the manufacturer’s instructions. Written informed consent was obtained from all the participants or their parents. The Ethical Review Board of Peking Union Medical College Hospital approved this study.
WGBS and data analysis
After the extraction of genomic DNA, WGBS was performed and data analysis was conducted as described in our previous study.12 DMRs between the hemivertebra body and the spinal process were identified using MethPipe (v.3.4.3) with significant differentially CpGs ≥5 and a minimal number of 10 CpGs that the DMR spans.
According to the methylation level difference, all the DMRs were classified into two groups, the hypermethylation group and hypomethylation group. ChIPseeker (v.1.14.0) Bioconductor package was used to evaluate the genomic position distribution of DMRs. Every DMR was assigned to a related RefSeq gene from 3 kb downstream to 3 kb upstream of the TSS. The DMR-related genes were defined as genes with at least one DMR mapped to it. In the case of multiple DMRs being mapped to the same gene, we retained the DMR with the largest absolute methylation difference to represent the gene. Hypo/hyper-DMR-related genes observed among four patients were identified as recurrent DMR-related genes. Hierarchical clustering analysis was performed to characterize the global methylation alterations between the hemivertebra group and spinal process group using the R package. Then, the recurrent DMR-related genes were ordered by the arithmetic mean value of the methylation difference of corresponding DMRs in four groups. All of the common DMR-related genes were enriched at Gene Ontology (GO) and KEGG using clusterProfiler (v.3.6.0) Bioconductor package.
Acknowledgments
We give our great thanks to the patients for their active participation. We thank ekitech Ltd. (Beijing) for providing technical support of bioinformatics analysis. This research was funded in part by the National Natural Science Foundation of China (81930068 to Z.W., 81822030 and 82072391 to N.W., 81772299 to Z.W., 81902271 to G.L., 81972037 to T.J.Z., and 81902178 to S.W.); National Key Research & Development Program of China (2017YFC1104902 to T.J.Z.); Beijing Natural Science Foundation (JQ20032 to N.W., 7191007 to Z.W., L192015 to T.J.Z., and 7184232 to S.L.); Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (2019PT320025); and Tsinghua University-Peking Union Medical College Hospital Initiative Scientific Research Program.
Author contributions
N.W. conceived the project. G.L., H.Z., and N.W. performed the research and analyzed and interpreted the data. G.L. drafted the manuscript. Z.Y., S.Z., S.L., Y.N., X.L., S.W., and Y.Y. helped sample collection. S.L., T.J.Z., and Z.W. performed phenotyping of patients. H.Z., Z.Y., and S.Z. helped with analysis and interpretation of the data. Y.N. provided technique support. Z.W. and T.J.Z. offered professional discussions and instructions. T.J.Z., Z.W., and N.W. conceived and designed the study, revised the manuscript, and provided the final approval of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.02.002.
Supplemental information
References
- 1.McMaster M.J., Ohtsuka K. The natural history of congenital scoliosis. A study of two hundred and fifty-one patients. J. Bone Joint Surg. Am. 1982;64:1128–1147. [PubMed] [Google Scholar]
- 2.Sparrow D.B., Chapman G., Smith A.J., Mattar M.Z., Major J.A., O’Reilly V.C., Saga Y., Zackai E.H., Dormans J.P., Alman B.A. A mechanism for gene-environment interaction in the etiology of congenital scoliosis. Cell. 2012;149:295–306. doi: 10.1016/j.cell.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 3.Hedequist D., Emans J. Congenital scoliosis: a review and update. J. Pediatr. Orthop. 2007;27:106–116. doi: 10.1097/BPO.0b013e31802b4993. [DOI] [PubMed] [Google Scholar]
- 4.Lin M., Zhao S., Liu G., Huang Y., Yu C., Zhao Y., Wang L., Zhang Y., Yan Z., Wang S., Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study Identification of novel FBN1 variations implicated in congenital scoliosis. J. Hum. Genet. 2020;65:221–230. doi: 10.1038/s10038-019-0698-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J., Wu N., Yang N., Takeda K., Chen W., Li W., Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study. Du R., Liu S., Zhou Y., Zhang L. TBX6-associated congenital scoliosis (TACS) as a clinically distinguishable subtype of congenital scoliosis: further evidence supporting the compound inheritance and TBX6 gene dosage model. Genet. Med. 2019;21:1548–1558. doi: 10.1038/s41436-018-0377-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao S., Zhang Y., Chen W., Li W., Wang S., Wang L., Zhao Y., Lin M., Ye Y., Lin J., Deciphering Disorders Involving Scoliosis and COmorbidities (DISCO) study Diagnostic yield and clinical impact of exome sequencing in early-onset scoliosis (EOS) J. Med. Genet. 2021;58:41–47. doi: 10.1136/jmedgenet-2019-106823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou D., Kang N., Yin P., Hai Y. Abnormalities associated with congenital scoliosis in high-altitude geographic regions. Int. Orthop. 2018;42:575–581. doi: 10.1007/s00264-018-3805-2. [DOI] [PubMed] [Google Scholar]
- 9.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 10.Jin M., Zhu S., Hu P., Liu D., Li Q., Li Z., Zhang X., Xie Y., Chen X. Genomic and epigenomic analyses of monozygotic twins discordant for congenital renal agenesis. Am. J. Kidney Dis. 2014;64:119–122. doi: 10.1053/j.ajkd.2014.01.423. [DOI] [PubMed] [Google Scholar]
- 11.Lyu G., Zhang C., Ling T., Liu R., Zong L., Guan Y., Huang X., Sun L., Zhang L., Li C. Genome and epigenome analysis of monozygotic twins discordant for congenital heart disease. BMC Genomics. 2018;19:428. doi: 10.1186/s12864-018-4814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu G., Wang L., Wang X., Yan Z., Yang X., Lin M., Liu S., Zuo Y., Niu Y., Zhao S. Whole-Genome Methylation Analysis of Phenotype Discordant Monozygotic Twins Reveals Novel Epigenetic Perturbation Contributing to the Pathogenesis of Adolescent Idiopathic Scoliosis. Front. Bioeng. Biotechnol. 2019;7:364. doi: 10.3389/fbioe.2019.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meng Y., Lin T., Liang S., Gao R., Jiang H., Shao W., Yang F., Zhou X. Value of DNA methylation in predicting curve progression in patients with adolescent idiopathic scoliosis. EBioMedicine. 2018;36:489–496. doi: 10.1016/j.ebiom.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao S.H., Qian B.P., Shi B., Zhu Z.Z., Qiu Y. Quantitative evaluation of the relationship between COMP promoter methylation and the susceptibility and curve progression of adolescent idiopathic scoliosis. Eur. Spine J. 2018;27:272–277. doi: 10.1007/s00586-017-5309-y. [DOI] [PubMed] [Google Scholar]
- 15.Boggild S., Molgaard S., Glerup S., Nyengaard J.R. Spatiotemporal patterns of sortilin and SorCS2 localization during organ development. BMC Cell Biol. 2016;17:8. doi: 10.1186/s12860-016-0085-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan R.C., Petersen A.K., Chen M.H., Teumer A., Glazer N.L., Döring A., Lam C.S., Friedrich N., Newman A., Müller M. A genome-wide association study identifies novel loci associated with circulating IGF-I and IGFBP-3. Hum. Mol. Genet. 2011;20:1241–1251. doi: 10.1093/hmg/ddq560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholls A.C., Oliver J.E., McCarron S., Harrison J.B., Greenspan D.S., Pope F.M. An exon skipping mutation of a type V collagen gene (COL5A1) in Ehlers-Danlos syndrome. J. Med. Genet. 1996;33:940–946. doi: 10.1136/jmg.33.11.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meester J.A.N., Verstraeten A., Schepers D., Alaerts M., Van Laer L., Loeys B.L. Differences in manifestations of Marfan syndrome, Ehlers-Danlos syndrome, and Loeys-Dietz syndrome. Ann. Cardiothorac. Surg. 2017;6:582–594. doi: 10.21037/acs.2017.11.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malfait F., Francomano C., Byers P., Belmont J., Berglund B., Black J., Bloom L., Bowen J.M., Brady A.F., Burrows N.P. The 2017 international classification of the Ehlers-Danlos syndromes. Am. J. Med. Genet. C. Semin. Med. Genet. 2017;175:8–26. doi: 10.1002/ajmg.c.31552. [DOI] [PubMed] [Google Scholar]
- 20.Qiu R., Wang X., Davy A., Wu C., Murai K., Zhang H., Flanagan J.G., Soriano P., Lu Q. Regulation of neural progenitor cell state by ephrin-B. J. Cell Biol. 2008;181:973–983. doi: 10.1083/jcb.200708091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geng A., Qiu R., Murai K., Liu J., Wu X., Zhang H., Farhoodi H., Duong N., Jiang M., Yee J.K. KIF20A/MKLP2 regulates the division modes of neural progenitor cells during cortical development. Nat. Commun. 2018;9:2707. doi: 10.1038/s41467-018-05152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilkinson D.G. Regulation of cell differentiation by Eph receptor and ephrin signaling. Cell Adhes. Migr. 2014;8:339–348. doi: 10.4161/19336918.2014.970007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tumelty K.E., Higginson-Scott N., Fan X., Bajaj P., Knowlton K.M., Shamashkin M., Coyle A.J., Lu W., Berasi S.P. Identification of direct negative cross-talk between the SLIT2 and bone morphogenetic protein-Gremlin signaling pathways. J. Biol. Chem. 2018;293:3039–3055. doi: 10.1074/jbc.M117.804021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H., Choi Y.J., Lee Y.S., Park S.Y., Baek J.E., Kim H.K., Kim B.J., Lee S.H., Koh J.M. SLIT3 regulates endochondral ossification by β-catenin suppression in chondrocytes. Biochem. Biophys. Res. Commun. 2018;506:847–853. doi: 10.1016/j.bbrc.2018.10.167. [DOI] [PubMed] [Google Scholar]
- 25.Sun H., Dai K., Tang T., Zhang X. Regulation of osteoblast differentiation by slit2 in osteoblastic cells. Cells Tissues Organs. 2009;190:69–80. doi: 10.1159/000178020. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y., Zhang H., Tang M., Guo C., Deng A., Li J., Wang Y., Xiao L., Yang G. High methylation of lysine acetyltransferase 6B is associated with the Cobb angle in patients with congenital scoliosis. J. Transl. Med. 2020;18:210. doi: 10.1186/s12967-020-02367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J.M., Yang Y.S., Park K.H., Oh H., Greenblatt M.B., Shim J.H. The ERK MAPK Pathway Is Essential for Skeletal Development and Homeostasis. Int. J. Mol. Sci. 2019;20:1803. doi: 10.3390/ijms20081803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kamiya N., Kim H.K., King P.D. Regulation of bone and skeletal development by the SHP-2 protein tyrosine phosphatase. Bone. 2014;69:55–60. doi: 10.1016/j.bone.2014.08.015. [DOI] [PubMed] [Google Scholar]
- 29.Detweiler S., Thacker M.M., Hopkins E., Conway L., Gripp K.W. Orthopedic manifestations and implications for individuals with Costello syndrome. Am. J. Med. Genet. A. 2013;161A:1940–1949. doi: 10.1002/ajmg.a.36047. [DOI] [PubMed] [Google Scholar]
- 30.Stevenson D.A., Schwarz E.L., Carey J.C., Viskochil D.H., Hanson H., Bauer S., Weng H.Y., Greene T., Reinker K., Swensen J. Bone resorption in syndromes of the Ras/MAPK pathway. Clin. Genet. 2011;80:566–573. doi: 10.1111/j.1399-0004.2010.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hui S., Yang Y., Li J., Li N., Xu P., Li H., Zhang Y., Wang S., Lin G., Li S. Differential miRNAs profile and bioinformatics analyses in bone marrow mesenchymal stem cells from adolescent idiopathic scoliosis patients. Spine J. 2019;19:1584–1596. doi: 10.1016/j.spinee.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Hou T., Liu Y., Kolba N., Guo D., He H. Desalted Duck Egg White Peptides Promote Calcium Uptake and Modulate Bone Formation in the Retinoic Acid-Induced Bone Loss Rat and Caco-2 Cell Model. Nutrients. 2017;9:490. doi: 10.3390/nu9050490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao R., Xie P., Zhang K., Tang Z., Chen X., Zhu X., Fan Y., Yang X., Zhang X. Selective effect of hydroxyapatite nanoparticles on osteoporotic and healthy bone formation correlates with intracellular calcium homeostasis regulation. Acta Biomater. 2017;59:338–350. doi: 10.1016/j.actbio.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 34.Qiu S., Tao Z.B., Tao L., Zhu Y. Melatonin induces mitochondrial apoptosis in osteoblasts by regulating the STIM1/cytosolic calcium elevation/ERK pathway. Life Sci. 2020;248:117455. doi: 10.1016/j.lfs.2020.117455. [DOI] [PubMed] [Google Scholar]
- 35.Doyard M., Bacrot S., Huber C., Di Rocco M., Goldenberg A., Aglan M.S., Brunelle P., Temtamy S., Michot C., Otaify G.A. FAM46A mutations are responsible for autosomal recessive osteogenesis imperfecta. J. Med. Genet. 2018;55:278–284. doi: 10.1136/jmedgenet-2017-104999. [DOI] [PubMed] [Google Scholar]
- 36.Huang C.Y., Lien C.C., Cheng C.F., Yen T.Y., Chen C.J., Tsaur M.L. K+ Channel Kv3.4 Is Essential for Axon Growth by Limiting the Influx of Ca2+ into Growth Cones. J. Neurosci. 2017;37:4433–4449. doi: 10.1523/JNEUROSCI.1076-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Battum E.Y., Brignani S., Pasterkamp R.J. Axon guidance proteins in neurological disorders. Lancet Neurol. 2015;14:532–546. doi: 10.1016/S1474-4422(14)70257-1. [DOI] [PubMed] [Google Scholar]
- 38.Placzkiewicz E., Baldys-Waligorska A. Kallmann’s syndrome: skeletal and psychological aspects of late diagnosis. Ann. Endocrinol. (Paris) 2003;64:277–280. [PubMed] [Google Scholar]
- 39.Amouri R., Nehdi H., Bouhlal Y., Kefi M., Larnaout A., Hentati F. Allelic ROBO3 heterogeneity in Tunisian patients with horizontal gaze palsy with progressive scoliosis. J. Mol. Neurosci. 2009;39:337–341. doi: 10.1007/s12031-009-9217-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.