Abstract
Learning changes the activity of neurons across multiple brain regions, but the significance of this distributed organization remains poorly understood, owing in part to the difficulty of observing brain-wide activity patterns in commonly used mammalian model systems. This review discusses the promise of using the small and optically accessible nervous system of larval zebrafish to study the brain-wide networks that encode experience. I discuss the opportunities and challenges of studying learning and memory in the larval zebrafish, the lessons learned from recent studies of brain-wide imaging during experience-dependent behavior, and the potential for using zebrafish neurotechnology to understand the physiological principles and behavioral significance of distributed memory networks.
Introduction
The capacity to store and recall information about the external world allows organisms to adjust their behavior based on their specific experiences. Studying learning and memory in diverse animal species has substantially contributed to our understanding of how nervous systems can store and recall memories. Despite the prominence of the laboratory mouse in modern neuroscience, multiple non-mammalian species have played critical roles in advancing the study of learning and memory (Yartsev 2017; Laurent 2020; Keifer and Summers 2016). For instance, the sea slug Aplysia californica has been an important model species for understanding the cellular basis of neuronal plasticity and synaptic potentiation, owing to its large identifiable neurons and robust behavioral conditioning (Hawkins, Kandel, and Bailey 2006). The vinegar fly Drosophila melanogaster has also played an essential role, by advancing our understanding of the genes and molecules that influence learning and memory, owing to its advanced genetic toolkit and short generation time (Dubnau and Tully 1998). In recent years, the study of memory has increasingly emphasized the role of distributed neural networks in the storage and recall of learned experiences (Josselyn and Tonegawa 2020; Roy et al. 2019; Vetere et al. 2017); in this review I will discuss why the unique features of another non-mammalian model system—the larval zebrafish—can provide valuable insights into the brain-wide organization of neural networks for learning and remembering.
Many research programs are presently focused on studying learning and memory at the level of neural circuits, with a particular emphasis on finding “engrams” - sets of neurons whose activity patterns and synaptic connections encode specific experiences. These neurons are identified as being active during memory formation, and whose experimental activation can induce behaviors similar to natural memory recall (Josselyn and Tonegawa 2020; Tonegawa et al. 2015; Josselyn, Köhler, and Frankland 2015). These “engram” neurons have been found in multiple locations across the rodent brain (Josselyn and Tonegawa 2020; Josselyn, Köhler, and Frankland 2015; Vetere et al. 2017; Roy et al. 2019; Tonegawa, Morrissey, and Kitamura 2018; Hebb 2005; Choi et al. 2018), providing support for the concept of “distributed engrams”, where the neurons and circuits encoding experiences are distributed across multiple interconnected brain regions.
Despite substantial progress in this field, a number of fundamental questions about distributed engrams remain unanswered: Do neurons in each brain region encode unique information, or are they redundant? What are the dynamics of engram neuron interactions within and between brain regions? Are some regions or neurons more important than others? How do distributed networks respond to localized injuries or neural circuit failures?
One challenge to answering these questions lies in the choice of model system. The majority of this research is conducted in mice, where even the most advanced modern methods in neuroscience impose limits on experimenters. For instance, one can study the entire mouse brain by visualizing the distribution of neurons with activity-dependent gene expression after a given experience (Ramirez et al. 2015; DeNardo et al. 2019; Ye et al. 2016; Josselyn, Köhler, and Frankland 2015), but the precise activity patterns of these neurons are unknown (see discussion in Box 1). Alternatively, real-time neural activity can be recorded from behaving animals with cellular-level calcium imaging or electrophysiology, but only from small numbers of neurons at a time (Figure 1a). Therefore, the critical information that can be obtained by real-time recording of all memory-encoding neurons across learning and recall cannot be obtained.
Box 1: Limitations of defining engrams by activity-dependent gene expression.
Many studies in rodents use the expression of activity-dependent genes (“immediate-early genes”) to label neurons active during a certain behavioral window with transgenic and/or viral strategies (Ramirez et al. 2015; DeNardo et al. 2019; Ye et al. 2016; Josselyn, Köhler, and Frankland 2015). Unfortunately, these transgenic and viral methods generally label neurons that are very strongly active over an hours-long temporal window (in contrast, marking neurons by endogenous mRNA expression and subcellular localization may be more precise (Brigidi et al. 2019)). Given that neurons can encode behavioral features with modest firing rates (Insanally et al. 2019), and neural activity is widespread during even very simple tasks (Allen et al. 2019; Steinmetz et al. 2019), these methods will fail to label weakly-active engram neurons but erroneously label neurons strongly encoding non-specific variables such as motor feedback and arousal state. Alternatively, using calcium imaging or electrophysiology to record activity on seconds- or milliseconds-long timescales can allow for the application of stricter thresholds for identifying experience-dependent cells (i.e.: neurons that acquire a conditioned stimulus response after classical conditioning, neurons with persistent activity during short-term motor learning, or neurons that encode outcome expectations in operant conditioning). However, these methods can only be applied to small numbers of neurons in rodents (even if distributed across multiple regions (Allen et al. 2019; Steinmetz et al. 2019)); the larval zebrafish is presently the only vertebrate where one can record the activity of (nearly) all neurons in the brain at once.
Figure 1. Whole-brain activity imaging for studies of distributed engrams.
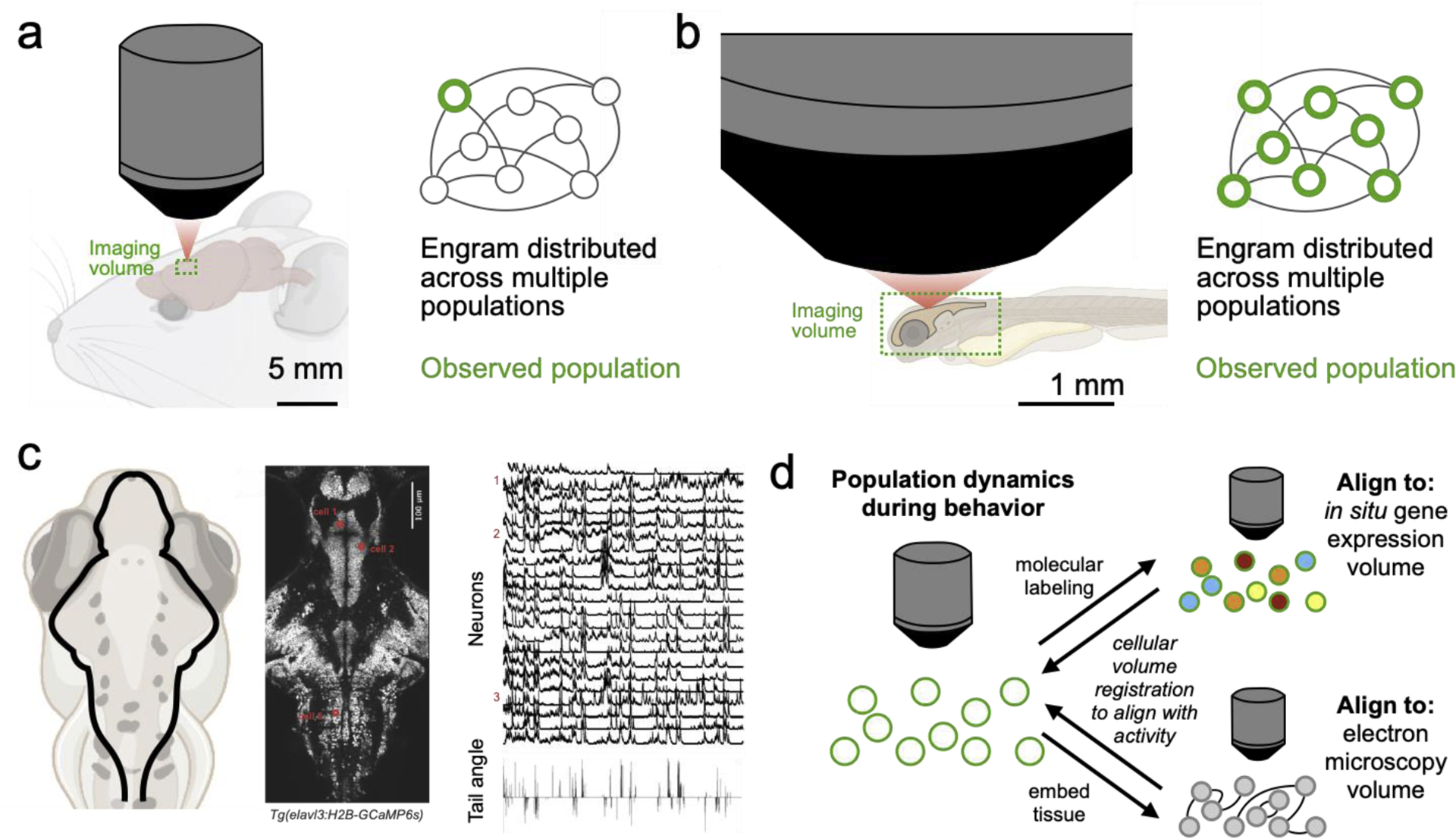
a) Schematic of limited imaging field in rodents. Only a small part of the network can be recorded during behavior b) Schematic of brain-wide imaging field in larval zebrafish. The entire network can be recorded during behavior. c) Brain-wide activity imaging in fish expressing genetically-encoded calcium indicators in all neurons. Example traces at right (Lovett-Barron, unpublished observations). d) Schematic showing how live brain neural activity imaging can be aligned to gene expression labeling (Lovett-Barron et al. 2017, 2020) or electron microscopy (Hildebrand et al. 2017; Vishwanathan et al. 2017) of the same cells, to link in vivo activity with molecular identity and connectivity.
A unique role for larval zebrafish in the study of learning and memory
As an alternative to rodents, larval zebrafish are particularly well suited to study the physiological basis and cellular mechanisms of memory storage across distributed brain-wide networks—owing to their small size, optical transparency, and shared anatomy with other vertebrates, including mammals (Chiu and Prober 2013; Lovett-Barron et al. 2017). One-to-two week old zebrafish are amenable to simultaneous recording of all neurons in the brain during behavior (Figure 1b), using light microscopy to image from behaving fish that express genetically encoded activity indicators under broad neuronal promoters (Ahrens and Engert 2015; Vanwalleghem, Ahrens, and Scott 2018; Ahrens et al. 2013) (Figure 1c). These experiments are commonly performed in head-tethered fish, though newly developed tracking microscopes have now made this possible in freely-swimming fish (Kim et al. 2017; Cong et al. 2017; Marques et al. 2020). Furthermore, recent advances have demonstrated that neural population recordings can be aligned to post hoc image-based measures of multiplexed gene expression and neural connectivity (Lovett-Barron et al. 2017, 2020; Hildebrand et al. 2017; Vishwanathan et al. 2017) (Figure 1d), allowing for simultaneous recording of many neurons with diverse gene expression and connectivity profiles. Finally, genetic targeting strategies permit imaging of specific neuron subtypes and subcellular compartments, as well as optical, chemical, and genetic manipulation of neural activity and gene expression (Vanwalleghem, Ahrens, and Scott 2018; Antinucci et al. 2020; Dal Maschio et al. 2017).
Here I will outline recent studies that leverage these experimental advantages to discover experience-dependent neural dynamics across the zebrafish brain. I will also discuss new behavioral paradigms for studying learning and memory in fish, and evaluate the challenges and opportunities for the advancement of these studies in the future.
Brain-wide imaging of experience-dependent neural dynamics
A number of studies have examined experience-dependent changes in neural activity across the zebrafish brain in response to associative or non-associative learning tasks. There is a large body of work in multiple fish species examining habituation of the acoustic startle response and associated plasticity of the giant escape-promoting Mauthner neuron, but this is reviewed elsewhere (López-Schier 2019). While this work will not be discussed here, it is worth noting that some forms of experience-dependent plasticity, such as habituation, may not involve brain-wide networks to the same extent as other forms of learning and memory. Below, we outline progress in understanding the neural basis of several other associative and non-associative learning tasks in zebrafish larvae.
Short-term motor learning
Zebrafish larvae will adjust their swimming vigor to visual feedback, in order to maintain their position in the environment (Ahrens et al. 2012; Portugues and Engert 2011). For instance, fish will swim more if each swimming action moderately moves them forward, but will swim less if each action propels them a greater distance. Kawashima and colleagues (Kawashima et al. 2016) used this paradigm to train larval zebrafish to swim with a persistently high or low motor vigor, using fictive swimming (measured as the electrical activity of motor neuron inputs to tail muscles in paralyzed fish) as a proxy for swimming intent. The authors found that such a learned motor-sensory gain is retained after a delay with no stimulus, and scales with the duration of training.
Using light sheet imaging in fish expressing a pan-neuronal calcium indicator to measure activity across the brain, the authors found that the activity of serotonin-releasing neurons in the dorsal raphe nucleus changes during the training period—increasing during high gain training (low motor vigor), or decreasing during low gain training (high motor vigor)—and is maintained over the delay period. Manipulations of these neurons demonstrated that the persistent activity of serotonin neurons serves to track the effectiveness of motor actions on sensory feedback, to guide the vigor of subsequent movements (Kawashima et al. 2016). In future studies it will be interesting to determine how the memory-related activity of serotonin-releasing neurons is able to persistently modulate motor behavior, given the complex and widely distributed expression of serotonin receptors (Norton, Folchert, and Bally-Cuif 2008).
Learned passivity
Zebrafish larvae can learn to adapt their motor actions to achieve a variety of goals, such as reducing energy expenditure when their actions are futile. Two groups have recently investigated the neural correlates of this experience-dependent behavior - a form of learned passivity or “giving up”.
In one study, Andalman and colleagues (Andalman et al. 2019) found that 1.5–2 week old zebrafish exposed to inescapable shocks would first attempt to escape from these aversive stimuli, but then transitioned into a prolonged state of behavioral passivity - no longer attempting to move because their actions were futile. Using whole brain light-field and two-photon calcium imaging, the authors found that the number of active neurons in the ventral habenula (homologue of the mammalian lateral habenula) gradually increased over this experience. Optogenetic manipulations and network modeling demonstrated that these habenula neurons control behavioral passivity through their mutual interactions with serotonergic neurons in the dorsal raphe nucleus.
In a second study, Mu and colleagues (Mu et al. 2019) examined fish in a closed-loop optomotor assay, where the fish’s fictive swimming behavior updated their visual environment (similar to (Ahrens et al. 2012; Kawashima et al. 2016)). When the authors suddenly made these swimming behaviors ineffective - no longer influencing the visual environment - fish first attempted vigorous movement before entering into an extended passive state. The authors used whole-brain calcium imaging of both neurons and glia with two-color light sheet microscopy, and discovered a complex interaction between these different circuit elements. The authors found that motor actions without visual feedback activates norepinephrine neurons in the medulla, which drive calcium accumulation in hindbrain radial glia, which in turn excite local GABAergic neurons to inhibit movement (Mu et al. 2019).
A unifying theme of these three studies is the observation that neurons and glia in different brain regions can maintain a short-term memory through persistent accumulation of evidence related to the effectiveness of motor actions. However, we note that the mechanisms of persistent activity can be ambiguous when measured by cytosolic or nuclear calcium accumulation in neurons or glia, since slow intracellular calcium changes can potentially result from mechanisms other than persistent spiking (Grienberger and Konnerth 2012). In their studies of short-term motor memory, Kawashima et al. (Kawashima et al. 2016) used single-cell electrical recordings to show that the persistent memory-related calcium signals they observed were also present at the level of phasic spiking. In future studies of short-term memory-related persistent activity, it will be important to use population voltage imaging (Villette et al. 2019; Abdelfattah et al. 2019) and intracellular electrophysiology to analyze the cell autonomous (intracellular signaling pathways and ionic conductances) and network level (recurrent connectivity) mechanisms of such persistence in neurons and glia. These approaches can determine whether persistent memory-related activity in different brain regions and learning tasks use common cellular and circuit mechanisms.
Classical conditioning
Several studies have demonstrated that zebrafish larvae are capable of associative learning in classical and operant conditioning paradigms. While adult zebrafish can reliably produce these behaviors under a number of conditions (Kenney et al. 2017; Darland and Dowling 2001; Valente et al. 2012; Frank et al. 2019; Lal et al. 2018), the optical accessibility of zebrafish generally decreases with age. Therefore, we will focus on those studies that have characterized learning tasks in larval zebrafish - particularly head-fixed fish - which hold the most promise for applying whole-brain cellular imaging and manipulation.
Two notable studies have trained head-fixed larval fish in visual classical conditioning tasks, using light flashes as a conditioned stimulus, and a tactile activation of the tail as an aversive unconditioned stimulus. Aizenberg and Schuman (Aizenberg and Schuman 2011) found that pairing a moving light with an aversive tactile stimulus over 5–10 trials produced conditioned tail movements of 5–6-fold greater velocity than pre-conditioning responses. Using calcium imaging of dye-labeled neurons, the authors found cells in the cerebellum, but not optic tectum, that acquired light responses after pairing. In a more recent study, Harmon et al. (Harmon et al. 2017) found that pairing a 5 s light stimulus with delayed electrical stimulation of the tail in paralyzed larvae resulted in the acquisition of conditioned fictive swimming to the light stimulus over 20–30 trials. Since the fish were paralyzed, the authors were able to apply whole-cell patch clamp recordings to Purkinje cells in the cerebellum, and found that the rate of complex and simple spikes change over time during learning - with substantial variability across different physiological cell types.
In both of these studies, the authors found that broad lesions of the cerebellum prevented the acquisition of conditioning. Therefore, zebrafish may be a useful model for examining the brain-wide networks involved in cerebellum-dependent learning (Raymond and Medina 2018).
Operant conditioning
In addition to classical conditioning, two groups have demonstrated that head-embedded larval zebrafish are capable of operant conditioning. Li (Li 2013), demonstrated that head-fixed fish can be trained in a closed-loop task where their directional tail movements turn off a noxious heat stimulus. Fish learned over 5–10 trials to initiate turns in the conditioned direction, and could also be trained to switch the direction of action. Lin et al. (Lin et al. 2020) expanded upon this work by performing light field imaging during this task to record from neurons primarily along the dorsal extent of the rhombencephalon, diencephalon, and telencephalon. The authors observed that neurons around the cerebellum and anterior hindbrain — ipsilateral to the reinforced tail direction — increased their activity before the movement, and that the magnitude of this pre-motor activity grew over the course of training. Future studies will benefit from studying more brain regions and cell types during this learning task, and expanding the behavioral paradigms to a variety of conditioned motor responses and stimulus modalities.
Strategies for improved behavioral assays
Despite the progress made in the studies discussed above, one critical caveat is that the expression of learning, and behavioral performance in general, is inconsistent in these behavioral tasks for larval zebrafish. Typically, only a fraction of larvae meet the behavioral criterion for inclusion in the study. For instance, the number of week-old larvae that show learning are reported to be 50% or lower by Lin et al., Harmon et al., and Mu et al. (Mu et al. 2019; Harmon et al. 2017; Lin et al. 2020). This variability presents a challenge for studies of learning and memory in head-fixed zebrafish larvae (although see (Voelkl et al. 2020) for advantages), but there are potential approaches to mitigate such concerns (see Box 2).
Box 2: Challenges and potential solutions to inconsistent learning in larval zebrafish.
An outstanding problem with studies of learning and memory in larval zebrafish is the low rate of learning in the population of larvae tested. One issue is that learning is assessed by broad, potentially ambiguous behavioral responses - typically the presence or absence of a non-specific tail movement. Future studies may be able to isolate more specific conditioned responses by monitoring and/or reinforcing specific patterns of muscle tone (Leung et al. 2019), heart rate (Matsuda et al. 2017), eye movements (Miri et al. 2011), pectoral/jaw movements (McClenahan, Troup, and Scott 2012), or specific patterns of tail kinematics (Marques et al. 2018). Additionally, learning may be impoverished when conducted in a head-fixed animal. Therefore, recent advances in cellular and near-cellular resolution microscopy in freely behaving fish (Cong et al. 2017; Kim et al. 2017; Marques et al. 2020) may permit the use of more robust learning assays. Behavioral variability in larvae may also result from differences in gene expression (Pantoja et al. 2016), developmental progression (Singleman and Holtzman 2014), and social/environmental history (Wee et al. 2020; Groneberg et al. 2020). Fluctuating levels of baseline alertness (Lovett-Barron et al. 2017) could also influence learning (Aston-Jones and Cohen 2005). Better measurement and control of these internal and external factors may increase the reliability of behavioral performance in larvae.
Applying large-scale neural activity imaging to older fish may overcome some shortcomings in behavioral performance by larvae. For instance, fish in the juvenile age range (~14–28 days old) are old enough to perform tasks such as spatial learning (Yashina et al. 2019), while being young enough to be partially embedded in agarose for imaging (Vendrell-Llopis and Yaksi 2015; Palumbo et al. 2019; Matsuda et al. 2017; Andalman et al. 2019). New methods for head-fixation (Huang et al. 2020; Torigoe et al. 2019) and deep brain imaging (Chow et al. 2020) in mature fish may further enable imaging in older animals, albeit with more limited spatial coverage of the brain.
One very promising solution to the compromise between behavioral maturity and brain size/opacity is the development of a new model system: Danionella translucida (Schulze et al. 2018; Penalva et al. 2018). These fish are genetically similar to zebrafish, but maintain a small and transparent brain and body into adulthood, where they can perform complex behaviors such as social communication (Schulze et al. 2018) and reinforcement learning (Penalva et al. 2018). The establishment of head-fixed learning and memory tasks in Danionella translucida may therefore allow for whole-brain activity imaging in older animals capable of more nuanced and reliable learning.
Opportunities for the analysis of brain-wide engrams
The key advantage of the head-fixed transparent fish preparation is optical access for imaging and manipulating of individual neurons across the entire brain during the acquisition and expression of learned behavior. Neurons with particular functional properties (such as “engram” cells defined by the acquisition of conditioned responses) can be identified online to guide precise cellular-level activation or inactivation/ablation (Vladimirov et al. 2018; Freeman et al. 2014), enabling the previously inaccessible study of the behavioral relevance, network interactions, and physiological basis of brain-wide distributed engrams (Figure 2a).
Figure 2. Opportunities for probing distributed engrams with optical studies in zebrafish.
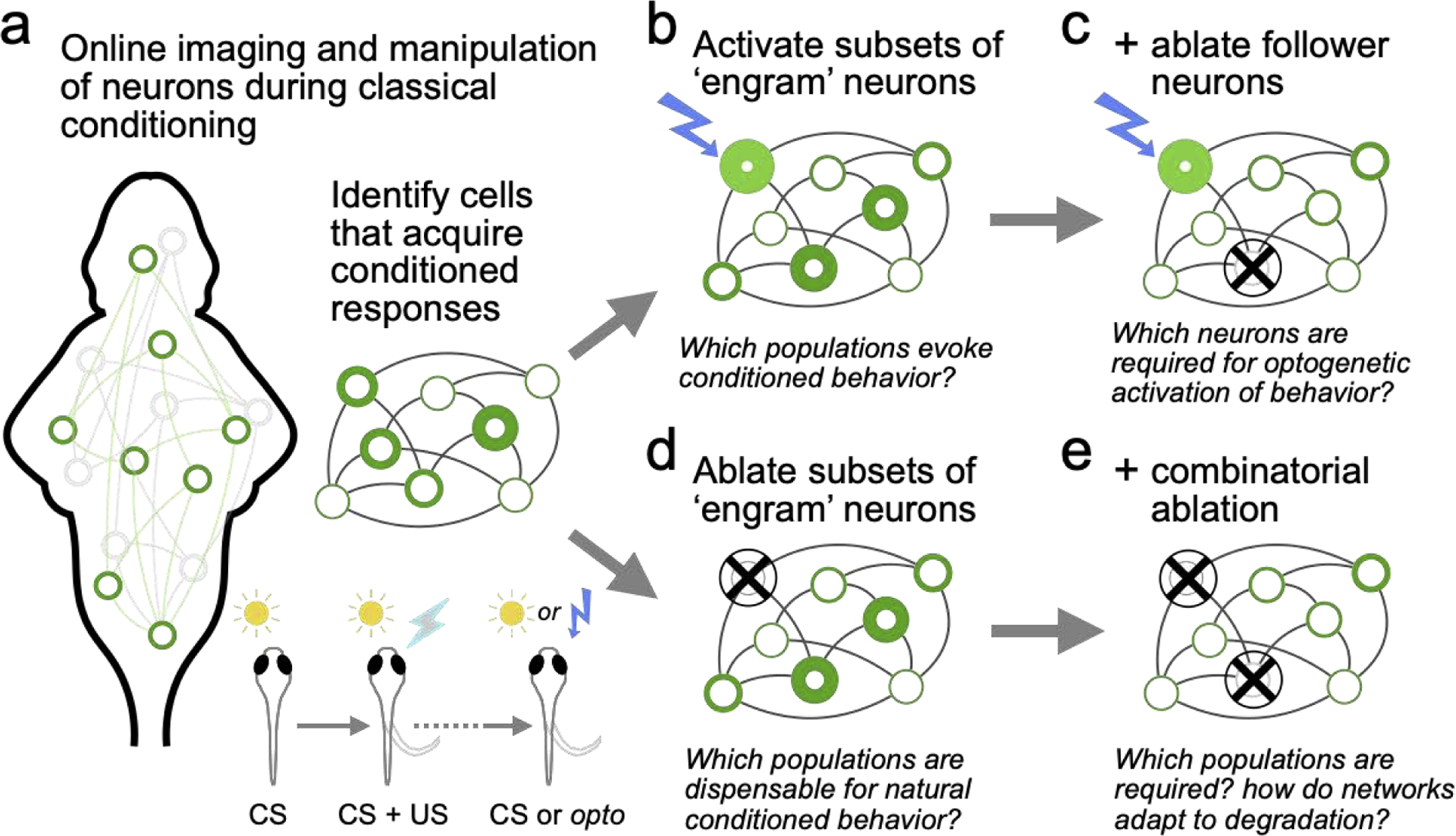
a) Schematic showing online identification of “engram” neurons across the brain during learning, to target for cellular-level manipulations.b) Optogenetic activation of “engram” neurons, while monitoring behavior and the activity of all “engram” and “non-engram” neurons.c) Repeating the experiments in panel “b”, but with selective ablation of follower neurons activated by optogenetic stimulation of engram cells.d) Selective ablation of “engram” neuron populations, while monitoring behavior and the activity of all “engram” and “non-engram” neurons.e) Testing the effects of combinatorial ablation on neural activity and behavior.
For instance, after online identification of engram neurons (Figure 2a), these cells can be activated in each brain region using two-photon optogenetics (Vladimirov et al. 2018; Jiao et al. 2018; Dal Maschio et al. 2017). These experiments can determine which sets of neurons artificially induce memory recall, and which patterns of brain-wide neural activity are evoked by such stimulation (Figure 2b). Are brain-wide neural dynamics similar between natural and artificial recall? Does partial engram stimulation preferentially recruit other engram neurons? Does activation of multiple engram neurons improve behavioral robustness? Selective ablation of those neurons (Vladimirov et al. 2018; Lovett-Barron et al. 2020) activated by engram cell stimulation can help determine the downstream circuits that drive optogenetically-induced recall (Figure 2c). Furthermore, the presence of multiple distributed engrams may imply redundancy (Edelman and Gally 2001). Therefore, it is important to selectively ablate engram neurons to determine the effects on neural activity patterns and conditioned behavior during recall (Figure 2d). Can fish show memory recall when subsets of engram neurons are ablated? Which combinations of neurons are required for memory expression (Figure 2e)? What are the differences between local (i.e.: 50 neurons in one region) and distributed (i.e.: 10 neurons in each of five regions) ablation of engram neurons? These experiments can provide important insight into the functional connectivity of memory networks, the dynamics of their interactions, and their robustness to perturbation.
Another potential approach could include efforts to measure a complete connectivity matrix before and after learning, by combining single cell optogenetic stimulation with voltage imaging (Abdelfattah et al. 2019), and/or longitudinal imaging of pre- and post-synaptic compartments (Du et al. 2018). These in vivo studies can be complemented with molecular and ultrastructural detail, by alignment to post hoc gene expression labeling or connectomics (Lovett-Barron et al. 2017, 2020; Hildebrand et al. 2017; Vishwanathan et al. 2017) (Figure 1d). These and other technical advantages of the head-fixed larval zebrafish make this a uniquely advantageous preparation for the study of memory at the cellular and subcellular resolutions and brain-wide scale.
Conclusions
Memories are stored and recalled through the concerted activity of multiple interconnected regions - a distributed organization that introduces technical challenges to its study. Here I have described how the unique features of the head-fixed behaving larval zebrafish (and related preparations) can allow for the study of these memory networks - from single cells and synapses to the whole brain. Much as the large, identifiable neurons of Aplysia californica were a boon for studying the synaptic basis of memory, the small transparent brains of zebrafish will likely provide key insights into the network basis of memory.
Highlights.
Neurons encoding experiences are distributed across the vertebrate brain
Learning and memory can be studied in the small transparent larval zebrafish
Non-associative learning can recruit persistent activity in multiple brain regions
Associative learning can drive diverse dynamics in the cerebellum
New behavioral and optical approaches can allow for analysis of distributed engrams
Acknowledgements
I thank Ritchie Chen, Susanna Bradbury, Aaron Andalman, and Tyler Benster for their comments on an earlier version of this manuscript. Some figure panels were created with BioRender. MLB is supported by funds from the NIMH (R00MH112840) and the Division of Biological Sciences at UCSD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Highlighted papers
The authors found that zebrafish larvae would enter into a passive motor state after prolonged exposure to inescapable stress. The authors then found that this learned passivity was encoded by the accumulative activity of neurons in the lateral/ventral habenula, and exert their effects on behavior through interactions with dorsal raphe serotonin neurons.
The authors describe a classical conditioning task for paralyzed larvae, allowing for in vivo whole-cell recordings of multiple cerebellar neuron classes during this task. The authors found that many classes of cerebellar neurons change their stimulus-evoked spiking properties over the course of learning.
The authors trained larvae to store a short-term memory of motor-sensory gain. Using whole-brain light sheet imaging and cellular manipulations, they found that this memory is maintained by the persistent activity of dorsal raphe serotonin neurons.
The authors trained larvae in an operant conditioning task, where fish would execute a directional tail movement to turn off a noxious thermal stimulus. Light field imaging revealed movement-predictive activity in the cerebellum that scaled with learning.
The authors found that, in the absence of sensory feedback, zebrafish will “give up” and no longer attempt to swim when such actions are in vain. Whole-brain imaging of neurons and glia revealed a circuit that links visual feedback detection in brainstem norepinephrine neurons to movement cessation via calcium accumulation and signaling in radial glial cells.
The authors establish methods to rapidly identify the functional properties of neurons in whole-brain recordings, to target these neurons for single-cell ablations and optogenetic manipulations. This platform for online cellular manipulations of any functionally-defined population of neurons across the brain will allow for the study of interacting brain-wide engrams.
COI
Matthew Lovett-Barron: Conception, funding, and writing No conflict of interest to declare
References
- Abdelfattah Ahmed S., Kawashima Takashi, Singh Amrita, Novak Ondrej, Liu Hui, Shuai Yichun, Huang Yi-Chieh, et al. 2019. “Bright and Photostable Chemigenetic Indicators for Extended in Vivo Voltage Imaging.” Science 365 (6454): 699–704. [DOI] [PubMed] [Google Scholar]
- Ahrens Misha B., and Engert Florian. 2015. “Large-Scale Imaging in Small Brains.” Current Opinion in Neurobiology 32 (June): 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens Misha B., Li Jennifer M., Orger Michael B., Robson Drew N., Schier Alexander F., Engert Florian, and Portugues Ruben. 2012. “Brain-Wide Neuronal Dynamics during Motor Adaptation in Zebrafish.” Nature 485 (7399): 471–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens Misha B., Orger Michael B., Robson Drew N., Li Jennifer M., and Keller Philipp J.. 2013. “Whole-Brain Functional Imaging at Cellular Resolution Using Light-Sheet Microscopy.” Nature Methods 10 (5): 413–20. [DOI] [PubMed] [Google Scholar]
- Aizenberg Mark, and Schuman Erin M.. 2011. “Cerebellar-Dependent Learning in Larval Zebrafish.” The Journal of Neuroscience: The Official Journal of the Society for Neuroscience 31 (24): 8708–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen William E., Chen Michael Z., Pichamoorthy Nandini, Tien Rebecca H., Pachitariu Marius, Luo Liqun, and Deisseroth Karl. 2019. “Thirst Regulates Motivated Behavior through Modulation of Brainwide Neural Population Dynamics.” Science 364 (6437): 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andalman Aaron S., Burns Vanessa M., Matthew Lovett-Barron Michael Broxton, Poole Ben, Yang Samuel J., Grosenick Logan, et al. 2019. “Neuronal Dynamics Regulating Brain and Behavioral State Transitions.” Cell 177 (4): 970–85.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinucci Paride, Dumitrescu Adna, Deleuze Charlotte, Morley Holly J., Leung Kristie, Hagley Tom, Kubo Fumi, Baier Herwig, Bianco Isaac H., and Wyart Claire. 2020. “A Calibrated Optogenetic Toolbox of Stable Zebrafish Opsin Lines.” bioRxiv. 10.1101/2020.01.13.904185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aston-Jones Gary, and Cohen Jonathan D.. 2005. “An Integrative Theory of Locus Coeruleus-Norepinephrine Function: Adaptive Gain and Optimal Performance.” Annual Review of Neuroscience 28: 403–50. [DOI] [PubMed] [Google Scholar]
- Brigidi G Stefano Michael G. B. Hayes Nathaniel P. Delos Santos Andrea L. Hartzell Lorane Texari Pei-Ann Lin Anna Bartlett, et al. 2019. “Genomic Decoding of Neuronal Depolarization by Stimulus-Specific NPAS4 Heterodimers.” Cell 179 (2): 373–91.e27. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chiu Cindy N., and Prober David A.. 2013. “Regulation of Zebrafish Sleep and Arousal States: Current and Prospective Approaches.” Frontiers in Neural Circuits 7 (April): 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Jun-Hyeok, Sim Su-Eon, Kim Ji-Il, Dong Il Choi Jihae Oh, Ye Sanghyun, Lee Jaehyun, et al. 2018. “Interregional Synaptic Maps among Engram Cells Underlie Memory Formation.” Science 360 (6387): 430–35. [DOI] [PubMed] [Google Scholar]
- Chow Dawnis M., Sinefeld David, Kolkman Kristine E., Ouzounov Dimitre G., Akbari Najva, Tatarsky Rose, Bass Andrew, Xu Chris, and Fetcho Joseph R.. 2020. “Deep Three-Photon Imaging of the Brain in Intact Adult Zebrafish.” Nature Methods, April. 10.1038/s41592-020-0819-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Lin, Wang Zeguan, Chai Yuming, Hang Wei, Shang Chunfeng, Yang Wenbin, Bai Lu, Du Jiulin, Wang Kai, and Wen Quan. 2017. “Rapid Whole Brain Imaging of Neural Activity in Freely Behaving Larval Zebrafish (Danio Rerio).” eLife 6 (September). 10.7554/eLife.28158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maschio Dal, Marco Joseph C. Donovan, Helmbrecht Thomas O., and Baier Herwig. 2017. “Linking Neurons to Network Function and Behavior by Two-Photon Holographic Optogenetics and Volumetric Imaging.” Neuron 94 (4): 774–89.e5. [DOI] [PubMed] [Google Scholar]
- Darland T, and Dowling JE. 2001. “Behavioral Screening for Cocaine Sensitivity in Mutagenized Zebrafish.” Proceedings of the National Academy of Sciences of the United States of America 98 (20): 11691–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo Laura A., Liu Cindy D., Allen William E., Adams Eliza L., Friedmann Drew, Fu Lisa, Guenthner Casey J., Marc Tessier-Lavigne, and Liqun Luo. 2019. “Temporal Evolution of Cortical Ensembles Promoting Remote Memory Retrieval.” Nature Neuroscience 22 (3): 460–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, and Tully T. 1998. “Gene Discovery in Drosophila: New Insights for Learning and Memory.” Annual Review of Neuroscience 21: 407–44. [DOI] [PubMed] [Google Scholar]
- Du Xu-Fei, Xu Bing, Zhang Yu, Chen Min-Jia, and Du Jiu-Lin. 2018. “A Transgenic Zebrafish Model for in Vivo Long-Term Imaging of Retinotectal Synaptogenesis.” Scientific Reports 8 (1): 14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelman GM, and Gally JA. 2001. “Degeneracy and Complexity in Biological Systems.” Proceedings of the National Academy of Sciences of the United States of America 98 (24): 13763–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank Thomas, Mönig Nila R., Satou Chie, Higashijima Shin-Ichi, and Friedrich Rainer W.. 2019. “Associative Conditioning Remaps Odor Representations and Modifies Inhibition in a Higher Olfactory Brain Area.” Nature Neuroscience 22 (11): 1844–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Jeremy, Vladimirov Nikita, Kawashima Takashi, Mu Yu, Sofroniew Nicholas J., Bennett Davis V., Rosen Joshua, Yang Chao-Tsung, Looger Loren L., and Ahrens Misha B.. 2014. “Mapping Brain Activity at Scale with Cluster Computing.” Nature Methods 11 (9): 941–50. [DOI] [PubMed] [Google Scholar]
- Grienberger Christine, and Konnerth Arthur. 2012. “Imaging Calcium in Neurons.” Neuron 73 (5): 862–85. [DOI] [PubMed] [Google Scholar]
- Groneberg Antonia H., Marques João C., Martins A. Lucas, de Polavieja Gonzalo G., and Orger Michael B.. 2020. “Early-Life Social Experience Shapes Social Avoidance Reactions in Larval Zebrafish.” bioRxiv. 10.1101/2020.03.02.972612. [DOI] [PubMed] [Google Scholar]
- Harmon Thomas C., Magaram Uri, McLean David L., and Raman Indira M.. 2017. “Distinct Responses of Purkinje Neurons and Roles of Simple Spikes during Associative Motor Learning in Larval Zebrafish.” eLife 6 (May). 10.7554/eLife.22537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins Robert D., Kandel Eric R., and Bailey Craig H.. 2006. “Molecular Mechanisms of Memory Storage in Aplysia.” The Biological Bulletin 210 (3): 174–91. [DOI] [PubMed] [Google Scholar]
- Hebb DO 2005. The Organization of Behavior: A Neuropsychological Theory. Psychology Press. [Google Scholar]
- Hildebrand David Grant Colburn, Cicconet Marcelo, Russel Miguel Torres Woohyuk Choi, Tran Minh Quan Jungmin Moon, Arthur Willis Wetzel, et al. 2017. “Whole-Brain Serial-Section Electron Microscopy in Larval Zebrafish.” Nature 545 (7654): 345–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Kuo-Hua, Rupprecht Peter, Frank Thomas, Kawakami Koichi, Bouwmeester Tewis, and Friedrich Rainer W.. 2020. “A Virtual Reality System to Analyze Neural Activity and Behavior in Adult Zebrafish.” Nature Methods 17 (3): 343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insanally Michele N., Carcea Ioana, Field Rachel E., Rodgers Chris C., Brian DePasquale Kanaka Rajan, DeWeese Michael R., Albanna Badr F., and Froemke Robert C.. 2019. “Spike-Timing-Dependent Ensemble Encoding by Non-Classically Responsive Cortical Neurons.” eLife 8 (January). 10.7554/eLife.42409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Zhen-Fei, Shang Chun-Feng, Wang Yu-Fan, Yang Zhe, Yang Chen, Li Fu-Ning, Xie Jin-Ze, Pan Jing-Wei, Fu Ling, and Du Jiu-Lin. 2018. “All-Optical Imaging and Manipulation of Whole-Brain Neuronal Activities in Behaving Larval Zebrafish.” Biomedical Optics Express 9 (12): 6154–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn Sheena A., Stefan Köhler, and Paul W. Frankland. 2015. “Finding the Engram.” Nature Reviews. Neuroscience 16 (9): 521–34. [DOI] [PubMed] [Google Scholar]
- Josselyn Sheena A., and Tonegawa Susumu. 2020. “Memory Engrams: Recalling the Past and Imagining the Future.” Science 367 (6473). 10.1126/science.aaw4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Takashi, Zwart Maarten F., Yang Chao-Tsung, Mensh Brett D., and Ahrens Misha B.. 2016. “The Serotonergic System Tracks the Outcomes of Actions to Mediate Short-Term Motor Learning.” Cell 167 (4): 933–46.e20. [DOI] [PubMed] [Google Scholar]
- Joyce Keifer, and Summers Cliff H.. 2016. “Putting the ‘Biology’ Back into ‘Neurobiology’: The Strength of Diversity in Animal Model Systems for Neuroscience Research.” Frontiers in Systems Neuroscience 10 (August): 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney Justin W., Scott Ian C., Josselyn Sheena A., and Frankland Paul W.. 2017. “Contextual Fear Conditioning in Zebrafish.” Learning & Memory 24 (10): 516–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Dal Hyung, Kim Jungsoo, Marques João C., Grama Abhinav, Hildebrand David G. C., Gu Wenchao, Li Jennifer M., and Robson Drew N.. 2017. “Pan-Neuronal Calcium Imaging with Cellular Resolution in Freely Swimming Zebrafish.” Nature Methods 14 (11): 1107–14. [DOI] [PubMed] [Google Scholar]
- Lal Pradeep, Tanabe Hideyuki, Suster Maximiliano L., Ailani Deepak, Kotani Yuri, Muto Akira, Itoh Mari, et al. 2018. “Identification of a Neuronal Population in the Telencephalon Essential for Fear Conditioning in Zebrafish.” BMC Biology 16 (1): 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent Gilles. 2020. “On the Value of Model Diversity in Neuroscience.” Nature Reviews. Neuroscience, June. 10.1038/s41583-020-0323-1. [DOI] [PubMed] [Google Scholar]
- Leung Louis C., Wang Gordon X., Madelaine Romain, Skariah Gemini, Kawakami Koichi, Deisseroth Karl, Urban Alexander E., and Mourrain Philippe. 2019. “Neural Signatures of Sleep in Zebrafish.” Nature 571 (7764): 198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Jennifer Mengbo. 2013. “Identification of an Operant Learning Circuit by Whole Brain Functional Imaging in Larval Zebrafish.” http://dissertations.umi.com/gsas.harvard:11032.
- Lin Qian, Manley Jason, Helmreich Magdalena, Schlumm Friederike, Li Jennifer M., Robson Drew N., Engert Florian, Schier Alexander, Nöbauer Tobias, and Vaziri Alipasha. 2020. “Cerebellar Neurodynamics Predict Decision Timing and Outcome on the Single-Trial Level.” Cell 180 (3): 536–51.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Schier Hernán. 2019. “Neuroplasticity in the Acoustic Startle Reflex in Larval Zebrafish.” Current Opinion in Neurobiology 54 (February): 134–39. [DOI] [PubMed] [Google Scholar]
- Lovett-Barron Matthew, Andalman Aaron S., Allen William E., Vesuna Sam, Kauvar Isaac, Burns Vanessa M., and Deisseroth Karl. 2017. “Ancestral Circuits for the Coordinated Modulation of Brain State.” Cell 171 (6): 1411–23.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett-Barron Matthew, Chen Ritchie, Bradbury Susanna, Andalman Aaron S., Wagle Mahendra, Guo Su, and Deisseroth Karl. 2020. “Multiple Convergent Hypothalamus-Brainstem Circuits Drive Defensive Behavior.” Nature Neuroscience, June. 10.1038/s41593-020-0655-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques João C., Lackner Simone, Félix Rita, and Orger Michael B.. 2018. “Structure of the Zebrafish Locomotor Repertoire Revealed with Unsupervised Behavioral Clustering.” Current Biology: CB 28 (2): 181–95.e5. [DOI] [PubMed] [Google Scholar]
- Marques João C., Li Meng, Schaak Diane, Robson Drew N., and Li Jennifer M.. 2020. “Internal State Dynamics Shape Brainwide Activity and Foraging Behaviour.” Nature 577 (7789): 239–43. [DOI] [PubMed] [Google Scholar]
- Matsuda Koji, Yoshida Masayuki, Kawakami Koichi, Hibi Masahiko, and Shimizu Takashi. 2017. “Granule Cells Control Recovery from Classical Conditioned Fear Responses in the Zebrafish Cerebellum.” Scientific Reports 7 (1): 11865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClenahan Phil, Troup Michael, and Scott Ethan K.. 2012. “Fin-Tail Coordination during Escape and Predatory Behavior in Larval Zebrafish.” PloS One 7 (2): e32295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miri Andrew, Daie Kayvon, Arrenberg Aristides B., Baier Herwig, Aksay Emre, and Tank David W.. 2011. “Spatial Gradients and Multidimensional Dynamics in a Neural Integrator Circuit.” Nature Neuroscience 14 (9): 1150–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Yu, Bennett Davis V., Rubinov Mikail, Narayan Sujatha, Yang Chao-Tsung, Tanimoto Masashi, Mensh Brett D., Looger Loren L., and Ahrens Misha B.. 2019. “Glia Accumulate Evidence That Actions Are Futile and Suppress Unsuccessful Behavior.” Cell 178 (1): 27–43.e19. [DOI] [PubMed] [Google Scholar]
- Norton William H. J., Folchert Anja, and Bally-Cuif Laure. 2008. “Comparative Analysis of Serotonin Receptor (HTR1A/HTR1B Families) and Transporter (slc6a4a/b) Gene Expression in the Zebrafish Brain.” The Journal of Comparative Neurology 511 (4): 521–42. [DOI] [PubMed] [Google Scholar]
- Palumbo Fabrizio, Serneels Bram, Pelgrims Robbrecht, and Yaksi Emre. 2019. “Zebrafish Dorsal Habenula Is Required for Updating Learned Behaviors.” bioRxiv. 10.1101/802256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantoja Carlos, Hoagland Adam, Carroll Elizabeth C., Karalis Vasiliki, Conner Alden, and Isacoff Ehud Y.. 2016. “Neuromodulatory Regulation of Behavioral Individuality in Zebrafish.” Neuron 91 (3): 587–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penalva Ariadne, Bedke Jacob, Cook Elizabeth S. B., Barrios Joshua P., Bertram Erin P. L., and Douglass Adam D.. 2018. “Establishment of the Miniature Fish Species Danionella Translucida as a Genetically and Optically Tractable Neuroscience Model.” bioRxiv. 10.1101/444026. [DOI] [Google Scholar]
- Portugues Ruben, and Engert Florian. 2011. “Adaptive Locomotor Behavior in Larval Zebrafish.” Frontiers in Systems Neuroscience 5 (August): 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez Steve, Liu Xu, MacDonald Christopher J., Moffa Anthony, Zhou Joanne, Redondo Roger L., and Tonegawa Susumu. 2015. “Activating Positive Memory Engrams Suppresses Depression-like Behaviour.” Nature 522 (7556): 335–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond Jennifer L., and Medina Javier F.. 2018. “Computational Principles of Supervised Learning in the Cerebellum.” Annual Review of Neuroscience 41 (July): 233–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Dheeraj S., Park Young-Gyun, Ogawa Sachie K., Cho Jae H., Choi Heejin, Kamensky Lee, Martin Jared, Chung Kwanghun, and Tonegawa Susumu. 2019. “Brain-Wide Mapping of Contextual Fear Memory Engram Ensembles Supports the Dispersed Engram Complex Hypothesis.” bioRxiv. 10.1101/668483. [DOI] [Google Scholar]
- Schulze Lisanne, Henninger Jörg, Kadobianskyi Mykola, Chaigne Thomas, Ana Isabel Faustino Nahid Hakiy, Albadri Shahad, et al. 2018. “Transparent Danionella Translucida as a Genetically Tractable Vertebrate Brain Model.” Nature Methods 15 (11): 977–83. [DOI] [PubMed] [Google Scholar]
- Singleman Corinna, and Holtzman Nathalia G.. 2014. “Growth and Maturation in the Zebrafish, Danio Rerio: A Staging Tool for Teaching and Research.” Zebrafish 11 (4): 396–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz Nicholas A., Peter Zatka-Haas Matteo Carandini, and Harris Kenneth D.. 2019. “Distributed Coding of Choice, Action and Engagement across the Mouse Brain.” Nature 576 (7786): 266–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonegawa Susumu, Liu Xu, Ramirez Steve, and Redondo Roger. 2015. “Memory Engram Cells Have Come of Age.” Neuron 87 (5): 918–31. [DOI] [PubMed] [Google Scholar]
- Tonegawa Susumu, Morrissey Mark D., and Kitamura Takashi. 2018. “The Role of Engram Cells in the Systems Consolidation of Memory.” Nature Reviews. Neuroscience 19 (8): 485–98. [DOI] [PubMed] [Google Scholar]
- Torigoe Makio, Islam Tanvir, Kakinuma Hisaya, Chi Chung Alan Fung, Isomura Takuya, Shimazaki Hideaki, Aoki Tazu, Fukai Tomoki, and Okamoto Hitoshi. 2019. “Future State Prediction Errors Guide Active Avoidance Behavior by Adult Zebrafish.” bioRxiv. 10.1101/546440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente André, Huang Kuo-Hua, Portugues Ruben, and Engert Florian. 2012. “Ontogeny of Classical and Operant Learning Behaviors in Zebrafish.” Learning & Memory 19 (4): 170–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanwalleghem Gilles C., Ahrens Misha B., and Scott Ethan K.. 2018. “Integrative Whole-Brain Neuroscience in Larval Zebrafish.” Current Opinion in Neurobiology 50 (June): 136–45. [DOI] [PubMed] [Google Scholar]
- Vendrell-Llopis Nuria, and Emre Yaksi. 2015. “Evolutionary Conserved Brainstem Circuits Encode Category, Concentration and Mixtures of Taste.” Scientific Reports 5 (December): 17825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetere Gisella, Kenney Justin W., Tran Lina M., Xia Frances, Steadman Patrick E., Parkinson John, Josselyn Sheena A., and Frankland Paul W.. 2017. “Chemogenetic Interrogation of a Brain-Wide Fear Memory Network in Mice.” Neuron 94 (2): 363–74.e4. [DOI] [PubMed] [Google Scholar]
- Villette Vincent, Chavarha Mariya, Dimov Ivan K., Bradley Jonathan, Pradhan Lagnajeet, Mathieu Benjamin, Evans Stephen W., et al. 2019. “Ultrafast Two-Photon Imaging of a High-Gain Voltage Indicator in Awake Behaving Mice.” Cell 179 (7): 1590–1608.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwanathan Ashwin, Daie Kayvon, Ramirez Alexandro D., Lichtman Jeff W., Aksay Emre R. F., and Seung H. Sebastian. 2017. “Electron Microscopic Reconstruction of Functionally Identified Cells in a Neural Integrator.” Current Biology: CB 27 (14): 2137–47.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimirov Nikita, Wang Chen, Burkhard Höckendorf Avinash Pujala, Tanimoto Masashi, Mu Yu, Yang Chao-Tsung, et al. 2018. “Brain-Wide Circuit Interrogation at the Cellular Level Guided by Online Analysis of Neuronal Function.” Nature Methods 15 (12): 1117–25. [DOI] [PubMed] [Google Scholar]
- Voelkl Bernhard, Altman Naomi S., Forsman Anders, Forstmeier Wolfgang, Gurevitch Jessica, Jaric Ivana, Karp Natasha A., et al. 2020. “Reproducibility of Animal Research in Light of Biological Variation.” Nature Reviews. Neuroscience, June. 10.1038/s41583-020-0313-3. [DOI] [PubMed] [Google Scholar]
- Wee Caroline L., Song Erin, Nikitchenko Maxim, Wong Sandy, Engert Florian, and Kunes Samuel. 2020. “Social Isolation Modulates Appetite and Defensive Behavior via a Common Oxytocinergic Circuit in Larval Zebrafish.” bioRxiv. 10.1101/2020.02.19.956854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yartsev Michael M. 2017. “The Emperor’s New Wardrobe: Rebalancing Diversity of Animal Models in Neuroscience Research.” Science 358 (6362): 466–69. [DOI] [PubMed] [Google Scholar]
- Yashina Ksenia, Álvaro Tejero-Cantero Andreas Herz, and Baier Herwig. 2019. “Zebrafish Use Visual Cues and Geometric Relationships to Form a Spatial Memory.” bioRxiv. 10.1101/620575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Li, Allen William E., Thompson Kimberly R., Tian Qiyuan, Hsueh Brian, Ramakrishnan Charu, Wang Ai-Chi, et al. 2016. “Wiring and Molecular Features of Prefrontal Ensembles Representing Distinct Experiences.” Cell 165 (7): 1776–88. [DOI] [PMC free article] [PubMed] [Google Scholar]


