Abstract
Drought is one of the major environmental stress that adversely affect the growth and development of oil seed plant, safflower. There is a limited knowledge on proteomic responses to support physiological, biochemical changes in how safflowers can regulate growth and metabolism under drought conditions and followed by re-watering. The changes in morphological, physiological, biochemical and proteomics of safflower genotypes (Carthamus tinctorius L.; Remzibey-05 and Linas, tolerant and sensitive cultivars, respectively, and C. oxyacantha M. Bieb., wild type) after exposure to drought and followed by re-watering have been examined. Drought negatively affected the shoot weight, water content, chlorophyll fluorescence, and biochemical parameters, including photosynthetic pigment, proline, MDA, and H2O2 contents and antioxidant enzyme activities in all genotypes, while the re-watering period allowed Remzibey-05 to recover, and it even provided the wild type completely recovered (approximately 100%). A total of 72 protein spots were observed as differently accumulated under treatments. The identified proteins were mainly involved in photosynthesis and carbohydrate, protein, defense, and energy metabolisms. Protein accumulation related to these metabolisms in Remzibey-05 were decreased under drought, while increased following re-watering. However, sensitive cultivar, Linas, could not exhibit an effective performance under drought and recovery when compared with other safflower genotypes.
Supplementary Information
The online version contains supplementary material available at (10.1007/s12298-021-00934-2).
Keywords: Carthamus oxyacantha M. Bieb, Drought, Morpho-physiological and biochemical traits, Proteomics, RT-qPCR, Safflower
Introduction
Drought is one of the most important factors induced by global climate change which reduces the amount of usable water, affects especially agricultural areas and causes significant yield losses in agricultural products. Therefore, the importance of irrigation increases every day; however, the resources used for agricultural irrigation are gradually decreasing in many regions of the world. Intergovernmental Panel on Climate Change (IPCC) reported that the water level in dry subtropic regions will be reduced further and the existing water cannot be used due to contamination in the twenty-first century (IPCC 2019).
Drought is a multidimensional stress factor and affects the plant at the cell, organ and organism levels. Under water-deficient conditions, the plant-water relationship is disrupted and eventually, the cell water potential and turgor are reduced (Taiz et al. 2015). The first response to the reduction of water in the plant is a reduction in photosynthesis which involves the closure of stomata and decrease in the activity of photosynthetic enzymes and ATP synthesis (Farooq et al. 2009). Another drought-induced effect exacerbates the synthesis of reactive oxygen species (ROS) to levels that are beyond the plant’s detoxification capacity, and thus trigger oxidative stress. These stress-induced changes may also interfere with plant growth and development by affecting other metabolic activities such as respiration, transport and carbohydrate metabolism. Plants regulate their adaptation strategies against drought by adjusting morphology, physiology and biochemical activities including control of water content, developing deep root systems, and accumulation of osmolytes and activating enzyme and non-enzyme systems to cope with oxidative stresses (Xin et al. 2018).
It has been reported that knowledge about molecular mechanisms generated to understand drought responses shows all stages from starting signal detection to the end product of post-translational protein modifications (PTMs) cannot be revealed by genomic or transcriptomic analysis (Nemati et al. 2019). The use of related technologies for the identification of proteins and PTMs and the detection of expression profile and protein–protein interactions facilitate research on the effects of plant development. Proteomics studies provide information about the actual fluctuations of the protein levels associated with physiological and stress responses (Budak et al. 2015; Urban et al. 2017; Nemati et al. 2019; Azra et al. 2020).
Safflower (Carthamus tinctorius L.) is one of the oldest oilseed plants in the world that has high omega-9 (16–20% oleic acid) and omega-6 (71–75% linoleic acid) content. Safflower is used mainly for the production of vegetable oil, and also used in medical, cosmetic and paint industries (Gecgel et al. 2007). Besides, safflower oil has been reported to be suitable for biodiesel production (Mihaela et al. 2013). Although, safflower has a narrow cultivation area in the world, it could be one of the important alternative oil plants especially in arid and semi-arid regions if its tolerance to stress factors such as drought can be proved (Majidi et al. 2011). The vegetative stage is reported as the most critical stage (especially before flowering) for safflower (Istanbulluoglu et al. 2009). Therefore, research on the effect of drought on safflower at this critical stage which reflects yield and production is very important. The current study aimed to determine the drought tolerance capacity of safflower cultivars and to compare the responses of these cultivars with the wild safflower Carthamus oxyacantha. To achieve this, safflower genotypes at the vegetative stage were characterized by morphological, physiological and biochemical changes, and differentially expressed protein (DEP) profiles were identified using proteomics analysis.
Materials and methods
Plant growth and treatments
Safflower (Carthamus tinctorius L.) cultivars Remzibey-05 and Linas were selected for this study according to the previous results (Çulha-Erdal et al. 2016), where Remzibey-05 and Linas were identified as tolerant and sensitive, respectively. Besides, wild type safflower genotype (Carthamus oxyacantha M.Bieb.) was used in the study. Seeds, obtained from Trakya Agricultural Research Institute, Turkey (TARI), were surface-sterilized (5% NaCIO for 3 min) and imbibed in distilled water for 2 h. After incubation, seeds were sown in PVC pots holding 800 g air-dried soil with 19.5% water holding capacity. Some characteristics of soil were clay loam texture, pH 7.52 and electrical conductivity 264 µS cm−1. Each pot contained four plants. Plants were grown in a controlled growth chamber with a temperature of 25 ± 1 °C, 16 h photoperiod, 40 ± 5% air humidity and 250 μmol m–2 s–1 light intensity. On the 40th day after sowing, pots of each genotype were randomly divided into two groups, one of which served as the control which was irrigated regularly, while the other was subjected to drought without irrigation for 7 days followed by 5 days of re-watering. Plants were harvested at the beginning of drought (C-40th day) and, at the end of drought (S-47th day) and after re-watering (R – 52nd day) periods to provide suitable analysis.
Morphological, physiological and biochemical analysis
Three plants representing each treatment were harvested to determine fresh weights. Subsequently, dry weights of plants were measured by drying the fresh plants at 80 °C for 48 h. The water status of the leaves was evaluated by calculating relative water content (RWC) (Farrant 2000).
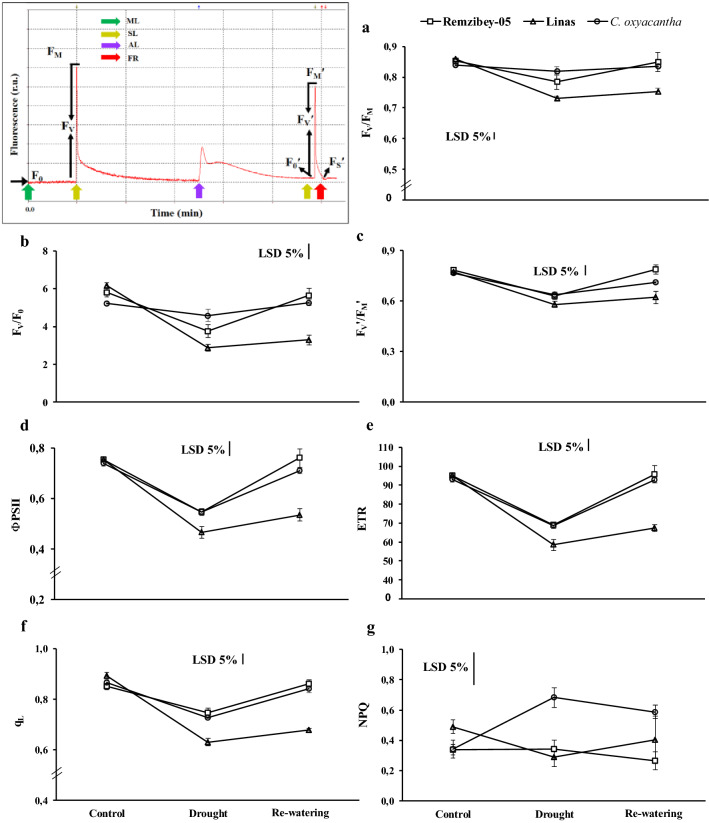
Following at least 30 min dark adaptation, the minimum (Fo) and maximum (FM) chlorophyll a fluorescence (ChlF) were measured. The maximal quantum efficiency of PSII of dark-adapted plants (FV/FM) was calculated using (FM-FO)/FM. The efficiency of the water-splitting complex on the donor side of PSII (FV/FO) was also calculated to assess PSII activity. Light induced changes in ChlF, following actinic illumination (300 μmol m−2 s−1) were recorded prior to the measurement of minimum (FO′) and maximum (FM′) fluorescence in light-adapted state. The quantum efficiency of PSII open centers in the light-adapted state, referred to as ΦPSII, (FM ′- FS/FM′) was determined from F′M and Fs (steady-state fluorescence in the light-saturated state) values. The quantum efficiency of excitation energy trapping of PSII (FV′/FM′) was calculated according to Genty et al. (1989). Later, actinic light was closed and FO′ was determined by illuminating the leaves with far-red light (7 μmol m−2 s−1). Electron transport rate (ETR) was also calculated by the following formula: (FM′-FS)/(FM′) × PAR × 0.84 × 0.5 (Genty et al. 1989). Non-photochemical quenching NPQ = FM-FM ′/FM′ were also calculated according to Bilger and Björkman (1990). The fraction of open PSII reaction centers as qL = (FM′-FS)/(FM′-FO′) × (FO′-FS) were calculated according to Kramer et al. (2004).
For each treatment, photosynthetic pigment [chlorophyll (a + b) and carotenoids (x + c)] contents were determined from leaf disks (R = 0.6 cm) and calculated as mg ml−1 cm−2 according to Lichtenthaler (1987). The anthocyanin content (mg ml–1 g FW–1) was measured as described by Mancinelli et al. (1975).
The free proline content (μmol g FW–1) was determined using the method of Bates et al. (1973) and a proline standard curve was used to determine the content of free proline. The levels of malondialdehyde (MDA) and hydrogen peroxide (H2O2) content were measured according to the method of Esterbauer and Cheeseman (1990). MDA content (nmol g FW–1) was calculated using the extinction coefficient 155 mM–1 cm–1. H2O2 content (µmol g FW–1) was calculated according to the standard curve.
Fresh leaf samples (0.5 g) were grounded with liquid nitrogen and soluble proteins were extracted by the related homogenizing buffer. In the leaf extracts, the protein concentrations were determined according to Bradford (1976). Superoxide dismutase (SOD; EC 1.15.1.1) activity was determined according to Beyer and Fridovich (1987). One unit of SOD is defined as the amount of enzyme which causes 50% decrease of the SOD-inhibited NBT reduction. Ascorbate peroxidase (APX; EC 1.11.1.11) activity was assayed according to the method of Wang et al. (1991) and the enzyme activity was calculated using ɛ = 2.8 mM cm−1 at 290 nm. Glutathione reductase (GR; EC 1.6.4.2) activity was determined according to Rao et al. (1995). The enzyme activity was calculated after subtracting the non-enzymic initial oxidation rate using the extinction coefficient of NADPH (ɛ = 6.2 mM cm−1) at 340 nm. Guaiacol peroxidase (POD; EC1.11.1.7) activity was based on the determination of guaiacol oxidation (ɛ = 26.6 mM cm−1) at 470 nm by H2O2 (Bergmeyer 1974). Catalase (CAT; EC 1.11.1.6) activity was assayed according to the method of Change and Maehly (1955) and the enzyme activity was calculated from the initial rate of decomposition of H2O2 using ɛ = 40 mM cm−1 at 240 nm. Aldose reductase (ALR; EC 1.1.1.21) activities were determined as described by Mundree et al. (2000). The enzyme activity was calculated from the initial rate of the reaction after subtracting the non-enzyme oxidation rate using the ɛ of NADPH (ɛ = 6.2 mM cm−1) at 340 nm.
Proteomic analysis
Proteins were extracted using a phenol extraction method (Hurkman and Tanaka 1986) with modifications (Ahsan et al. 2008). Leaf tissues (2 g) were ground into a fine powder with liquid nitrogen and homogenized in 8 ml pre-cooled Mg/NP-40 extraction buffer (Kim et al. 2001). The homogenates were centrifuged at 3500 g for 15 min, and fractionated with an equal volume of Tris–HCl saturated phenol (pH 8.0). After that, the top phenol phase was collected, and proteins were precipitated overnight by adding four volumes of cold 0.1 M ammonium acetate in methanol at -20 °C. The precipitated proteins were collected by centrifugation at 10,800 g for 10 min and washed with precipitation solution three times and ice-cold acetone twice. The protein pellet was air-dried and dissolved in a rehydration buffer. Protein concentration was determined using the Ramagli and Rodriguez assay (1985).
The protein extracts were subjected to two-dimensional polyacrylamide gel electrophoresis (2-D PAGE). 500 μg protein samples which were diluted with a 300 μl of rehydration buffer were loaded onto IPG (Immobilized pH gradient) strips (17 cm with 5–8 linear pH gradient; Bio-Rad, USA) and were passively rehydrated for 16 h. Isoelectric focusing (IEF) was carried out using PROTEAN®i12™ IEF System (Bio-Rad, USA) at 20 °C with the following settings: 250 V for 30 min, 10,000 V for 2 h and finally 10,000 V per hour until reaching 70,000 V. After IEF, the strips were first incubated in equilibration buffer I (6 M urea, 2% SDS, 1,5 M Tris–HCl pH 8.8, 20% glycerol and 2% DTT) for 15 min and then in equilibration buffer II (6 M urea, 2% SDS, 1,5 M Tris–HCl pH 8.8, 20% glycerol and 2.5% iodoacetamide) in the following 15 min. The second dimension SDS–PAGE was performed on running gels (PROTEAN II XL Cell, Bio-Rad; 12.5% polyacrylamide) according to Laemmli (1970). The protein spots on gels were visualized by using Oriole™ Fluorescent Gel Stain (Bio-Rad, USA). To assure reproducibility, the gels were run in 2-D PAGE three times in each experiment. Then, three of these gels obtained from each treatment were used for MALDI-TOF/TOF–MS analysis.
The stained gels were displayed using VERSADoc 4000 MP (BioRad, USA) system and images of the gels suitable for analysis were obtained using the Quantity One program (BioRad, USA). Analysis of protein spots and comparison of the gels was performed with PDQuest Advanced program (version 8.0.1, BioRad, USA). The statistical calculation of the difference in spots was conducted by Student's t-test (P< 0.05) and, changes more than two times were cut by using an automated spot cutting tool, ExQuest spot cutter (BioRad, USA).
Tryptic-digested peptides were extracted using an in-gel tryptic digestion kit (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer's protocol. Zip-Tip (Millipore, USA) cleaning was performed for each digested sample before deposition onto a MALDI plate. Protein identification was performed by using AB SCIEX MALDI-TOF/TOF 5800 system (Applied Biosystems, Framingham, 69MA, USA). Mass spectra (m/z 800–3000) were acquired in positive ion reflector mode. The 10 most intense ions were selected for subsequent MS/MS sequencing analysis in 1 kV mode. MS/MS spectra of proteins were submitted to database searching using the online MASCOT program (Matrix Science, London, UK) and GPS Explorer (Applied Biosystem).
To better understand interactions of identified proteins, the homolog of all identified proteins in Arabidopsis thaliana was determined using UniProt database, and STRING 11.0 database (http://string-db.org) was used to construct a network for protein–protein interactions (PPI). Besides, Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways have been used to schematize the drought-affected metabolic pathways.
Real-time PCR (RT-PCR) analysis
Total RNA was extracted from 0.3 g leaf samples according to Chomczynski and Sacchi (1987). RNA was purified using DNase I, RNase-free Set (Thermo Scientific, CA, USA) and Quantitation of RNA was obtained with a BioSpec-nano UV–VIS spectrophotometer (Shimadzu). The first strand cDNA was synthesized using Scientific RevertAid First-Strand cDNA Synthesis Kit (Thermo Scientific) according to the manufacturer’s instructions. Primers were designed using NCBI, Primer3 and IDT Oligo Analyzer programs (Supplementary Table S1). Expressions of genes were monitored by the real-time PCR cycler Rotor-Gene Q (Qiagen, Hilden, Germany) using Luminaris Color HiGreen qPCR Master Mix (Thermo Scientific). Elongation factor-1α (EF-1α) was used as internal control and data were normalized using the control samples.
Statistical analysis
Experiments were performed in a completely randomized design by three replicates (four plants in each replicate/pot). Differences between cultivars and treatments were analyzed by ANOVA and according to the least significant difference (LSD) test at the 5% level using the SPSS 20.0 software (IBM SPSS Statistics). qRT-PCR data were analyzed by student t-test (p ≤ 0.05).
Results and Discussion
When exposed to environmental stress such as drought, plants develop complex mechanisms and re-program their metabolism and growth as induced by morphological, physiological, biochemical, and molecular changes (Claeys and Inzé 2013). In the present study, drought and re-watering periods caused significant changes in plant morphology, physiology and biochemistry (Figs. 1, 2, 3, 4). Besides, our proteomic analysis showed that the water-stress-responsive proteins involved in photosynthesis and carbohydrate metabolism consist of the highest percentage of differentially expressed protein (DEPs) (Supplementary Table S2).
Fig. 1.
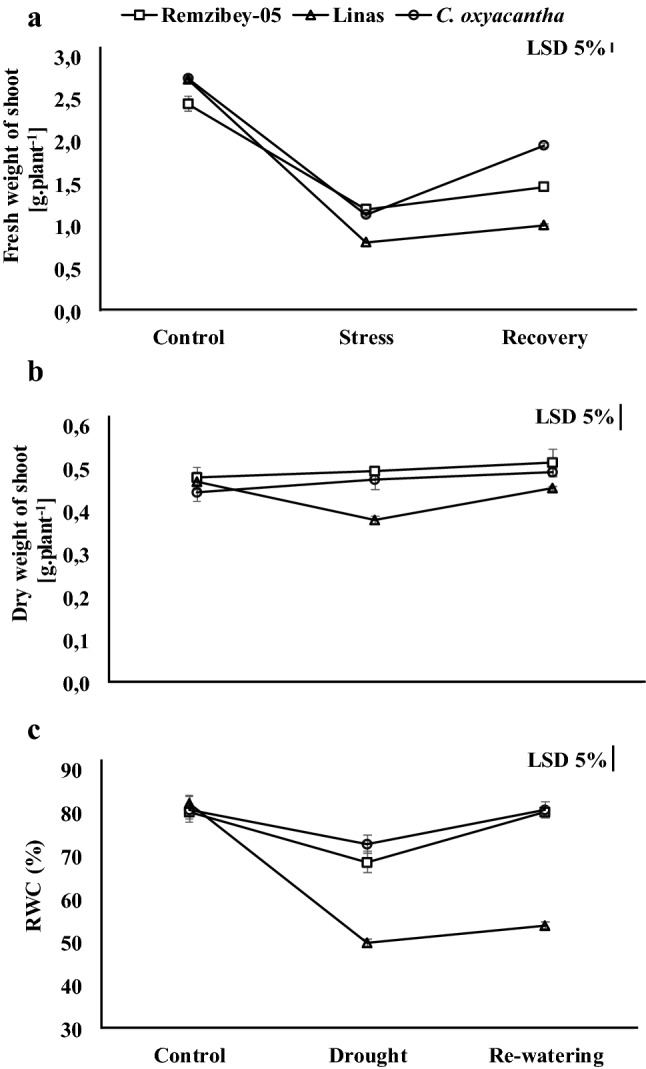
Effect of drought and re-watering periods on fresh (a) and dry (b) weight of a plant and RWC = relative water content (c) in the leaves of safflower genotypes. The values are presented as the mean ± standard error
Fig. 2.
Chlorophyll fluorescence responses of safflower leaves exposed to drought and re-watering periods: FV/FM = The maximal quantum efficiency of PSII of dark-adapted plants (a), FV/FO = The efficiency of the water-splitting complex on the donor side of PSII (b), FV′/FM′ = The quantum efficiency of excitation energy trapping of PSII (c), ФPSII = The quantum efficiency of PSII open centers in the light-adapted state, referred to as ΦPSII (d), ETR = Electron transport rate (e), qL = The fraction of open PSII reaction centers (f) and NPQ = Non-photochemical quenching (g). The values are presented as the mean ± standard error
Fig. 3.
Chlorophyll (a + b) = Chl a + b (a), chlorophyll a / b = Chl a / b ratio (b), carotenoid (c) and anthocyanin (d) content in the leaves of safflower genotypes exposed to drought and re-watering periods. The values are presented as the mean ± standard error
Fig. 4.
Drought and re-watering periods induced changes contents of proline (a), MDA = malondialdehyde (b) and H2O2 = hydrogen peroxide (c); and the activities of SOD = Superoxide dismutase (d), APX = Ascorbate peroxidase (e), GR = Glutathione reductase (f), POD = Guaiacol peroxidase (g), CAT = Catalase (h) and ALR = Aldose reductase (I) in the leaves of safflower genotypes. The values are presented as the mean ± standard error
Characterization of drought tolerance by morphological, physiological and biochemical traits
In this study, the fresh weight of shoot decreased under drought stress and increased following re-watering in all genotypes (Fig. 1a). This reduction under drought might be associated with the suppression of cell expansion and growth due to a decrease in turgor pressure (Robert et al. 2016). The dry weight of shoot was not affected in Remzibey-05 and wild safflower in treatments, whereas it was decreased under drought and increased following re-watering in Linas (about 20% decrease and increase, respectively) (Fig. 1b). Dry weight accumulation in genotypes can be connected with the accumulation of specific proteins and drought adaptability (Urban et al. 2017). The results of dry weight which directly reflect the effects of stress on biomass showed that Remzibey-05 and C. oxyacantha have higher drought tolerance potential than Linas. In addition, the RWC of genotypes was declined under drought (more than 10%) but noticeably increased after re-watering and reached control levels, except for Linas (Fig. 1c). These results showed that Remzibey-05 and C. oxyacantha can rapidly revert to the previous leaf water content and development of plants during the re-irrigation period.
Many metabolic processes including photosynthesis were negatively affected under drought conditions. Our research has indicated that the photochemical activity of genotypes was affected differently from drought (Fig. 2). FV/FM and FV/FO, were decreased in Remzibey-05 and Linas under drought (Fig. 2a, b). These reductions demonstrated that water-splitting complex and reaction centers of PSII are damaged (photochemically inactive), indicating impairment of photosynthetic electron transport (Kalaji et al. 2011). These reductions induced a decrease in FV’/FM’ for both cultivars, however to a greater extent in Linas (Fig. 2c). Besides, after re-watering, increases in FV/FM, FV/FO and FV’/FM’ of Remzibey-05 exhibited that damage to PSII and electron transport in tolerant cultivar is reversible. The decrease in FV’/FM’ also induced the decline in ΦPSII of cultivars, which indicates disruptions in electron transfer in response to stress (Fig. 2d). Huang et al. (2013) reported that the ΦPSII was related to FV’/FM’, and the parameters were decreased in ramie under drought. Moreover, drought reduced ETR and qL (Fig. 2e, f) and these reductions showed that the disruption between the excitation rate and electron transfer rate caused an increase in the reduced state of PSII reaction centers and suppression of electron transport efficiency between photosystems under drought (Efeoğlu et al. 2009). Following re-watering, ΦPSII, ETR and qL were increased in both cultivars and these values reached the level of control in Remzibey-05. In the drought tolerant wild genotype, FV’/FM’, ΦPSII, ETR and qL were decreased under drought and increased to the level of control after re-watering (Fig. 2c–f). The thermal dissipation process (NPQ) of cultivars exhibited no significant change for treatments while in C. oxyacantha, the NPQ increased 2 to 1.72 times in drought and re-watering compared to control, respectively (Fig. 2g). The results of the study indicated that drought has a direct effect on the photosynthetic mechanisms of C. oxyacantha and Remzibey-05, and the tolerant genotypes were able to improve the impairments in PSII activity, electron transport and photochemical use when water was re-supplied. In parallel, photochemical parameters showed that drought disrupted PSII activity in Linas even though plant death was not observed at stressed conditions.
Drought affected not only the photochemical apparatus but also photosynthetic pigment contents. Chlorophyll (Chl a + b) content was decreased under treatments compared to control only in Linas (Fig. 3a). The reason for the decrease in Chl content of Linas could be the destruction of chlorophyll molecules while producing ROS and causing structural damage in chloroplasts (Ahmadikhah and Marufinia 2016). At the same time, the Chl a/b ratio (antenna size) was increased during the re-watering period compared to control and drought in Linas and this result indicated a decrease in antenna size after re-watering (Fig. 3b). Also, a decrease in photosynthetic activity may be partly due to decreased Chl content in Linas. However, Chl content and Chl a/b ratio of Remzibey-05 and C. oxyacantha exhibited no significant change after treatments. The carotenoid content was significantly decreased in cultivars while its content in C. oxyacantha remained stable throughout the treatments (Fig. 3c). On the other side, the accumulation of anthocyanin is one way of protecting against photoinhibition; by masking Chls as a ROS scavenger and filter to prevent high light absorption by leaves (Farrant 2000). Anthocyanin content was increased under treatments in all genotypes (Fig. 3d). It is possible that anthocyanin may have played an effective role in protecting Chls according to the changes in the protective pigment contents.
Osmotic stress is induced due to the reduced ability of water uptake of plants under drought. In the present study, proline which has a function to maintain osmotic potential, increased 38.00, 56.66 and 2.85-times by drought compared to control, and decreased 84%, 64% and 53% following re-watering period compared to drought for Remzibey-05, Linas and C. oxyacantha, respectively (Fig. 4a). Accumulation of proline in genotypes may have been due to protein synthesis or degradation. Besides, following re-watering, the proline content of all genotypes was significantly reduced, and this decrease could be due to the use of proline as a source of nitrogen reserves during re-watering (Urban et al. 2017). Mohammadi et al. (2016) reported that a reduction in Chl content caused an increase in proline accumulation in less tolerant cultivar under drought conditions, because proline and Chl have the same precursors (synthesized from glutamate). These explanations have supported the findings of Chl and proline in Linas (Figs. 3a, 4a). Besides that, osmotic stress triggers the increase in ROS production, which causes the deterioration and peroxidation of lipid membranes in plant cells. Our results showed that MDA and H2O2 contents of safflower genotypes were markedly increased with drought while the values of these parameters were decreased after re-watering in Remzibey-05 and C. oxyacantha (Fig. 4b, c). Filippou et al. (2011) demonstrated that the content of MDA and H2O2 in leaves and roots of Medicago truncatula were increased in drought while decreased following re-watering. This recovery due to a decrease following re-watering could be explained by presence of a rapid phospholipid renewal mechanism for the maintenance of membrane integrity and that H2O2 acts as a signal molecule in these genotypes.
Plants activate antioxidant defense mechanisms to scavenge ROS caused by drought. In our study, the enzyme activities of all safflower genotypes increased within a certain level to maintain the stability of ROS due to the intensity of drought (Fig. 4d–g). Following re-watering, SOD, APX, GR and POD activities in C. oxyacantha and APX and GR in Linas decreased while enzyme activities of Remzibey-05 did not change significantly compared to drought. Also, there were significant increases in CAT activities of Remzibey-05 and C. oxyacantha under drought (Fig. 4h). These results suggested that Remzibey-05 maintained a balance of the generation and elimination of ROS during treatments. Wei et al. (2020) reported that the antioxidant enzyme activity was higher in drought-tolerant safflower varieties during drought stress. However, reduction in enzyme activities of Linas did not play an active role in the removal of ROS following re-watering. Furthermore, the higher H2O2, proline, anthocyanin and antioxidant enzyme activities in the control of wild genotype, as well as less increment in drought compared to control cultivars and return to control level following re-watering, indicated that the C. oxyacantha was always prepared for unsuitable conditions and can maintain its metabolism to the optimum level under these conditions.
Drought affects the expression levels of enzymes which has a role in other metabolic functions besides defense mechanisms in plants. ALR is one of the key enzymes of the polyol metabolic pathway that catalyze the NADPH dependent reduction of the aldehyde form of glucose to sorbitol, and have an active role in the synthesis of osmotic regulators, repression of lipid peroxidation and detoxification of ROS under stress conditions as well as holding low levels of stress-induced damage and maintenance of tolerance (Oberschall et al. 2000; Tari et al. 2010). Our results showed that ALR activities of genotypes were significantly increased with drought while the level of enzyme activities was decreased following re-watering (Fig. 4i). Also, the changes in ALR enzyme activities were supported with the results of proline, lipid peroxidation and H2O2 in genotypes (Fig. 4a–c).
Proteomic analysis of differentially abundant proteins under drought and re-watering periods
To determine the mechanism by which safflower genotypes respond to drought and re-watering periods, 2-D PAGE was utilized to compare the leaf proteome of genotypes. Protein spots obtained from 2-D PAGE gels showed high reproducibility among the three independent extractions based on the analysis of PDQuest software (version 8.0.1, BioRad, USA). Based on at least 2-fold quantitative change, 650 ± 30 spots were detected in all genotypes. When 2-DE maps of control and treated samples were compared, 23, 21 and 28 protein spots were identified as differentially expressed in Remzibey-05, Linas and C. oxyacantha, respectively (Fig. 5). These proteins were successfully identified by MALDI-TOF/TOF–MS and database searches (Uni-Prot and Swiss-Prot). The isoelectric point, molecular mass and other parameters of identified protein are shown in Supplementary Table S2 in Supplemental Materials.
Fig. 5.

2-DE gel analysis of proteins extracted from leaves of safflower genotypes during drought and re-watering periods. Representation of 2-DE protein profile for control (a, d, g); drought (b, e, h) and re-watering (c, f, i) treatments of Remzibey-05, Linas and C. oxycantha, respectively. Arrows and marked with numbers shown in red on the images of 2-D PAGE gels indicate the protein spots selected for MALDI- TOF/TOF–MS analysis
To understand how DEPS are related and how the proteins involved in different pathways cross-link to each other, we constructed a protein–protein interaction (PPI) network for DEPs both for each genotype separately and all genotypes together, using the STRING database (Supplementary Fig. S1). The Arabidopsis thaliana was used as the reference to get homolog proteins due to unavailability of PPI data of safflower proteins in the String-database (Supplementary Table S3). In genotypes, most of the interacting proteins consisted of two major groups: photosynthesis and carbohydrate metabolism. When the genotypes were evaluated together, more protein interactions were revealed compared to individual evaluations. Ribulose bisphosphate carboxylase large chain (RBCL) is the central core protein of the PPI network due to its interactions with other proteins involved in photosynthesis and carbohydrate metabolism when genotypes evaluated together and also individually (Supplementary Fig. S1a-d). Besides, proteins involved in photosynthesis and carbohydrate metabolism interacted at high levels with each other and with proteins involved in protein (CDSP32, RPS18, RPL14, PBF1) and defense (CSD2) metabolism in genotypes. Further, the functions of DEPs in metabolic pathways are discussed.
Photosynthesis and carbohydrate metabolism
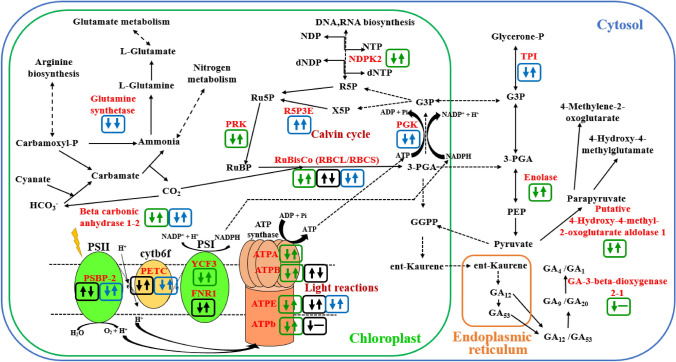
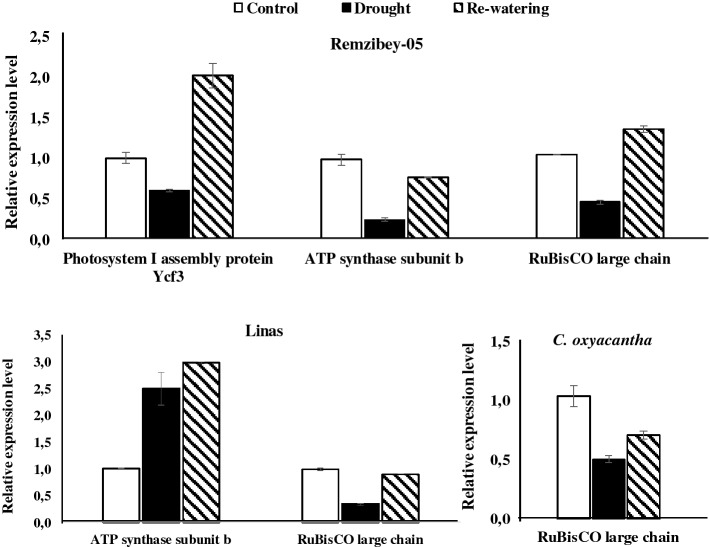
Drought has a direct impact on photosynthetic efficiency by disrupting all major components of photosynthesis and affect other primary metabolic processes such as carbohydrate metabolism (Farooq et al. 2009). In this study, 34 DEPs were associated with photosynthesis and carbohydrate metabolism after treatments (Supplementary Table S2, Fig. 7). Our proteomic analysis revealed that drought-induced changes in the expression of proteins related to photosystem reaction centers and energy transfer. Oxygen-evolving enhancer protein 2 (OEE2) which is a subunit of PSII was increased in Linas (spots 27 and 37) whereas decreased in C. oxyacantha (spot 55) under drought. The increase in abundance of OEE2 may be required to repair protein damage caused by dissociation and to maintain oxygen-evolution (Gazanchian et al. 2007), thus maintaining PSII activity and protecting the mechanism of photosynthesis (Wei et al. 2020). However, this change in protein abundance may also be an indicator of its functional suppression in the PSII complex (Azri et al. 2020). After re-watering, the abundance of OEE2 was changed in different levels in Linas (downregulation in spot 27; upregulation in spot 37) while increased in C. oxyacantha. Besides, Cytb6/f iron-sulfur subunit abundance was diminished in Linas (spot 41) and C. oxyacantha (spot 65) under drought, while increased in both genotypes following re-watering. Under drought, cyt b6/f were able to prevent excessive reductions to be limited in electron transport capacity by regulating the activity of PSII (Genty and Harbinson 1996; Li and Ma 2012), and genotypes also tried to overcome the reduction in photosynthetic electron transport activity by increasing the amount of Cyt b6/f after re-watering. One of the other components of photosynthesis, Photosystem I assembly protein ycf3 was decreased in drought while increased due to subsequent re-watering in Remzibey-05 (spot 18). Photosystem I assembly protein ycf3 interacts with at least two PSI core subunits and acts as a chaperone-like factor (Nellaepalli et al. 2018). Changes in protein abundance after treatments were supported by performance index (PItotal) values, which express the use of energy from photons absorbed by PSII to reduce PSI end receptors (Çulha-Erdal et al. 2016). These results suggest that photosystem I assembly protein in Remzibey-05 may have a role in improving the negative effects of drought under re-watering periods. In addition to these, drought stress has been shown to decrease the abundance of Ferredoxin-NADP reductase (Zadražnik et al. 2013). Similar results were obtained in this study and Ferredoxin-NADP reductase, leaf isozyme 1 accumulation was decreased under drought while increased after re-watering and reached control levels in Linas (spot 34).
Fig. 7.
The effect of drought and re-watering periods on the photosynthetic machinery, Calvin cycle, glycolysis, gluconeogenesis and nitrogen, purine, pyrimidine and hormone metabolism. The scheme was drawn by modifying KEGG metabolic pathways. Also, green, black and blue squares represent changes in Remzibey-05; Linas and C. oxyacantha, respectively. Besides, the first and second arrows indicate change according to control in drought and to drought in re-watering, respectively. The down and up arrow indicate a decrease and an increase of protein abundances, respectively. When a protein or subunit is defined in several spots in genotypes, the direction of the arrows is determined to average change levels
In this study, four subunits of ATP synthase–ATPase (alpha, beta, b and epsilon chain) showed a decrease in drought while an increase in abundance following re-watering in Remzibey-05 (spots 5, 6, 14 and 21) (Fig. 7). Also, the two isoenzymes of the ATP synthase epsilon chain described in C. oxyacantha (spot 48 and 57), and responded similarly to the subunits in Remzibey-05 (Fig. 7). A decrease of the ATPase activity in genotypes was demonstrated to mediate non-photochemical quenching that protects photosynthetic apparatus from photo-damage under drought (Tamburino et al. 2017). However, these genotypes may provide the necessary ATP synthesis by increasing cyclic electron flow despite decrease in ATPase accumulation under stress conditions (Lv et al. 2020). An increase of the ATPase activity in genotypes following re-watering, suggests their putative role in the cellular process to meet energy demand and alleviate the effect of stress by increasing ATP supply. Besides, the accumulations of ATP synthase subunit beta (spot 28), subunit b (spot 42) and epsilon chain (spot 29) differently changed in Linas under drought and re-watering. Changes in the abundance of ATPase subunits at different levels suggested that ATP cannot be synthesized from this protein.
Changes in photosynthetic proteins were consistent with ChlF parameters of genotypes (Fig. 2 and 7). Photochemical activities decreased due to the reduction of excitation energy trapping and electron transport of photosystems (PSI and PSII) under drought, whereas the photosynthetic efficiency of tolerant genotypes recovered with re-watering. This response could be due to the change in expression of photosynthetic proteins. But, Linas could not exhibit enough recovery with the changes in protein expressions.
Beta carbonic anhydrase-βCA (spot 22 and spot 71) which is one of the proteins that significantly affects the Calvin cycle in Remzibey-05 and C. oxyacantha, respectively, was diminished under drought, whereas its amount increased following re-watering. This situation in the protein amount of tolerant genotypes may indicate that βCA acts as a signal messenger under stress conditions (DiMario et al. 2017; Li et al. 2020). 10 differentially accumulated protein spots related with the large/small subunit of RuBisCO (spots 2,13, 16, 35, 36, 39, 40, 61, 63, and 68) were identified. It has been stated that RuBisCO subunits are susceptible to drought and may be caused to dissociate into subunits (Budak et al. 2013). RuBisCO large (spots 16, 2 and 63) and small (spot 13, 61 and 68) subunits were generally decreased under drought and increased following re-watering in tolerant genotypes. Under drought, the photosynthetic downregulation could be explained by a reduction in the RuBisCO subunits accumulation, and the subunit’s contents could be restored following recovery (Demirevska et al. 2009). Conversely, RuBisCO large (spots 35 and 36) and small (spot 39 and 40) subunit components were increased under drought and decreased after re-watering in Linas. The increased abundance of protein fragments could indicate that RuBisCO subunits can be cleaved by ROS generated at the metal-binding site and consequently increased protein degradation rate (Hajheidari et al. 2005). Another enzyme, phosphoribulokinase (PRK) was diminished under drought, while increased following re-watering in Remzibey-05 (spot 4). It was reported that a decrease in PRK accumulation affected the Ribulose-1,5-bisphosphate (RUBP) regeneration under drought, whereas these proteins recovered following re-watering in Kentucky bluegrass cultivars (Xu et al. 2013). However, phosphoglycerate kinase (PGK) was defined in C. oxyacantha, and accumulation of PGK 2 (spot 60) was diminished by more than 20% under drought, while this protein and Ribulose phosphate 3 epimerase (R5P3E) (spot 66) content increased by more than 65% after re-watering in C. oxyacantha. These results were compatible with the changes in photochemical activity (Supplementary Table S2, Fig. 2) and indicated that the photosynthesis mechanism was maintained by repairing the damage in light reactions and Calvin cycle that is caused by drought in wild genotype. The enzymes of the glycolytic pathway, triosephosphate isomerase – TPI (spot 67), enolase (spot 12) and Putative 4-hydroxy-4-methyl-2-oxoglutarate aldolase 1 (spot 8), exhibited similar responses with enzymes of Calvin cycle in tolerant genotypes. The reduction in protein tolerant genotypes might indicate that the transport of photoassimilates from source to sink tissues were restricted, when photosynthesis was diminished due to drought. Urban et al. (2017) reported that the changes in the proteins involved in carbohydrate metabolism were compatible with a decrease of photosynthesis-related proteins in rapeseeds. Also, an increase in the abundance of these enzymes following re-watering could be related to a need for extra energy to repair damage caused by stress.
Protein metabolism
The second largest group of protein metabolism with different accumulation in treatments was composed of proteins involved in protein synthesis (transcription and translation), storage and proteolysis. The accumulation of Cysteine synthase-CS (spot 11) which catalyzes the final step of the cysteine biosynthesis was decreased by 74% in drought while increased about 3 times following re-watering when compared to stress in Remzibey-05. The decrease in cysteine biosynthesis could support a reduction in sulphur absorption and formation of glutathione, methionine, and sulphurated secondary metabolites under drought (Tamburino et al. 2017). Another enzyme, Glutamine synthetase-GS (spot 56) was decreased under treatments in C. oxyacantha. This decrease may indicate a disruption in nitrogen assimilation that leads to the formation of organic nitrogen compounds like amino acids (Valero-Galván et al. 2013). Chloroplast stem-loop binding protein 41 kDa a-CSP41a (spot 43), associated with transcription and translation was increased under drought and decreased following re-watering in Linas. The change in the accumulation of CSP41a protein in Linas supported the changes in the accumulation of proteins encoded in chloroplast and involved in photosynthesis under treatments. As seen in the change of the proteins involved in photosynthesis and carbon metabolism in treatments, chloroplastic ribosomal proteins (spot 4, 9 and 58) were diminished in Remzibey-05 and C. oxyacantha, respectively, whereas chloroplastic ribosomal protein (spot 24) was increased in Linas under drought, and these proteins were generally increased following re-watering in genotypes. It has been reported that the accumulation of ribosomal proteins was changed in response to drought stress (Zadražnik et al. 2017; Nemati et al. 2019). In our study, changes in the amount of ribosomal proteins in genotypes support the changes in the accumulation of proteins which is encoded by the chloroplast DNA as subunits of PSI, PSII, ATPsynhase and Cyt-b6f complex and RuBisCO large subunit (Supplementary Table S2, Fig. 7). Besides, mitochondrial ribosomal protein S31 (spot 33) was decreased in treated plants in Linas. In addition to these proteins, Phaseolin alpha-type (spot 7) was increased about 3.8 times under drought whereas decreased by 53% following re-watering in Remzibey-05. It was demonstrated that some proteins may be stored in the vacuole for use as needed, rather than degradation under stress (Badowiec and Weidner 2014). Furthermore, these proteins may have been used to produce the required energy and /or to repair any damage after re-watering in Remzibey-05.
One of the proteolysis-related proteins, proteasome subunit beta type-1 (spot 20) was increased more than 2-fold under drought and following re-watering compared to control in Remzibey-05. Proteolysis-related proteins, which are necessary for the maintenance of cellular protein homeostasis, break down the proteins damaged by stress (Zadražnik et al. 2013). The accumulation of proteasome may point out to have key role in degradation of incorrectly folded, damaged or dysfunctional proteins in the tolerant cultivar. However, the accumulation of the proteasome subunit beta type-3-B (spot 31) was reduced under drought and following re-watering in Linas, and this decrease may indicate that the damaged or dysfunctional protein cannot be sufficiently proteolyzed under both conditions in this genotype.
Defense metabolism
Several DEPs that have a role in ROS detoxification and chaperone activity, have also been observed to have a role in the defense responses. The abundance of chloroplastic superoxide dismutase [Cu–Zn] (spot 15) and peroxidase 43 (spot 1) was decreased under drought and following re-watering in Remzibey-05 whereas the accumulation of SOD (spot 62) was only increased following re-watering in C. oxyacantha. Although total SOD and POD activity in the genotypes were increased after treatments, the response of these proteins may indicate that other SOD isoenzymes (MnSOD–FeSOD) and/or cellular forms (cytosolic and mitochondrial Cu/ZnSOD) have more effective function than chloroplastic superoxide dismutase and peroxidase 43 (Fig. 4d–g). Apart from these proteins, chloroplastic thioredoxin-like protein CDSP32 (spot 32) and Glycine-rich RNA-binding protein (spot 25) was decreased under drought, while increased following re-watering in Linas. Thioredoxins like (TRX-like) proteins have a role in controlling the function and structure of proteins by reducing disulfide bridges in the redox-active site (Nikkanen et al. 2014) while Glycine-rich RNA binding protein has RNA chaperone activity to protect from stress (Kim et al. 2007). The decrease in accumulation of both proteins showed that these protection systems were not functioning adequately under drought. However, the genotypes attempted to improve the damages occurring in the chloroplast, and perform biological functions of RNA by completing the processes of folding and/or assembling correctly following re-watering.
Energy metabolism
Putative cytochrome c oxidase subunit II PS17 was the only identified protein related to energy metabolism, and it was detected as multiple spots on the 2-D gel of genotypes (spots 10, 23, 26, 30, 45–47, 49, 50, 52–54, 59, 64 and 69). These variations may be caused by various factors such as protein isoforms, comigrations, degradation and partial synthesis, and protein translation from alternatively spliced mRNAs, or errors in the database sequence (partial protein sequence) due to post-translational modifications (Valero-Galván et al. 2013). The abundance of putative cytochrome c oxidase subunit II PS17 was changed in Linas after treatments (spots 26 and 30) while it was decreased under drought and increased following re-watering in Remzibey-05 (spots 10 and 23) and C. oxyacantha (spots 45–69). It has been reported that a putative cytochrome c oxidase subunit II PS17 was detected in multiple spots and decreased in wild type wheat under drought (Budak et al. 2013). Changes in the amount of cytochrome c oxidase were accompanied by changes in the amount of protein in photosynthesis and carbohydrate metabolism in genotypes after treatments and these results revealed the level of interaction between chloroplast and mitochondria, with the energy and metabolic connection between the two organelles and cytosol.
Other proteins involved in drought
Xyloglucan endotransglucosylase/hydrolase protein (spot 70), was increased under drought and did not change significantly following re-watering compared to drought in C. oxyacantha. This increase may contribute to cell wall remodeling to strengthen the wall layers and protect mesophyll cells from stress (Ye et al. 2016). Among the proteins involved in the MAP signaling pathway, Rac-like GTP-binding protein 2-RAC (spot 38) decreased and increased under drought and re-watering, respectively in Linas, whereas Serine/threonine protein phosphatase 2A – PP2A (spot 20) was accumulated more than 1.5-times in both treatments in C. oxyacantha. RAC-like proteins and protein phosphatases are involved in plant defense response to biotic/abiotic stresses as well as plant growth and development (Haider et al. 2017; Yang et al. 2018). The changes in the accumulation of a RAC-like protein indicated that cellular functions were adversely affected under drought in Linas and the genotype tried to overcome the damage with re-irrigation, whereas the accumulation of PP2A may be an indication of resistance level in the wild type genotype. Another protein, Chloroplastic NDPK II (spot 19) was diminished about by 44% under drought while increased by 53% following re-watering in Remzibey-05. Alteration in the amount of NDPK was consistent with changes in the accumulation of proteins involved in photosynthesis and protein metabolism in the chloroplast (Supplementary Table S2, Fig. 7). Besides these proteins, Gibberellin 3-beta-dioxygenase 2–1 (spot 17) was decreased under drought and was not changed following re-watering in Remzibey-05. Decreasing GA levels may have caused a limitation in plant growth under drought (Colebrook et al. 2014). Apart from these proteins, Germin-like protein 5–1 (GLPs 5–1) (spot 44) and Putative LRR disease resistance protein/transmembrane receptor kinase PS16 (spot 72) were increased under drought and decreased following re-watering in Linas and C. oxyacantha, respectively. Although, their exact role in metabolism is not fully understood, it is thought that these proteins are associated with biotic and abiotic stress resistance (Smith et al. 2006; Urban et al. 2017). Detailed explanation of the relationship between these proteins and drought may contribute in deciphering their exact functions.
Validation of selected DEP expression by RT-qPCR
Quantification of mRNA levels of three selected proteins, ATP synthase subunit beta chain (defined in Remzibey-05 and Linas), RuBisCO large chain (co-defined in genotypes) and photosystem I binding protein ycf (defined in Remzibey-05) were performed by RT-qPCR (Fig. 6). These results confirmed the correlation between the expression changes of RNA and the level of corresponding proteins (Supplementary Table S2, Fig. 6). The expression level of ATP synthase subunit beta chain and photosystem I binding protein ycf in Remzibey-05, and RuBisCO large chain in Remzibey-05 and C. oxyacantha were decreased under drought while increased following re-watering. However, the transcript level of ATP synthase subunit beta protein was observed as increased in treated plants as in the change of protein accumulation in Linas (spot 28). A discrepancy was seen between mRNA and protein abundances (spots 35 and 36) of RuBisCO large chain in Linas. The incongruence between transcript level and corresponding protein abundance of RuBisCO large chain may be associated with a time delay between mRNA production and translation of the related proteins, and possible turnover. Also the rates of translation and protein degradation differ among different genes and proteins, and post-transcriptional and translational regulatory mechanisms (Xin et al. 2018).
Fig. 6.
RT-qPCR of selected proteins in safflower genotypes exposed to drought and re-watering periods. The values are presented as the mean ± standard error
Conclusion
Drought is one of the abiotic stress factors that causes significant yield losses in crops, particularly oil crops. The present study showed that fresh and dry weights of genotypes were affected by drought, but this effect was observed to be more drastic in Linas. It has been observed that wild C. oxyacantha was more successful in drought tolerance than cultivars via activating the photochemical efficiency and defense systems, as well as protecting membrane stability and water status. Protein profiles of leaf in safflower genotypes showed that photosynthesis and carbohydrate metabolism were the most affected metabolic pathways although multiple metabolic processes altered during treatments. Furthermore, Remzibey-05 tolerant cultivar repaired the stress-induced damage by increasing the expression of the proteins involved in photosynthesis and carbohydrate metabolism during recovery, as in wild C. oxyacantha. Chlorophyll a fluorescence characteristics supported the changes seen in protein profiles related to the photosynthetic process in genotypes. Our study provides insight into the underlying response mechanism to drought tolerance in safflower genotypes which may be of value for future studies investigating drought responses of oilseed plant species. Thus, we suggest that Remzibey-05 could be grown in areas where drought is a threat and used as a parent in the safflower breeding program, together with the wild type genotype.
supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank Dr. Veli Pekcan and Dr. Metin Babaoğlu (Department of Field Crops, Trakya Agricultural Research Institute, Edirne, Turkey) for providing the seeds of safflower genotypes.
Author contributions
Conceived and designed the experiments: ŞÇE, FE and YE. Performed the experiments: ŞÇE. Analyzed the data: ŞÇE and YE. Wrote the paper: ŞÇE and YE All authors gave final approval of the submitted and published versions.
Funding
This research was financially supported by the Scientific and Technological Research Council of Turkey (TUBITAK, Project Number 1001- 214Z058). This work is part of the PhD thesis of ŞÇE (supervised by YE and FE).
Compliance with ethical standards
Conflict interest
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Ahmadikhah A, Marufinia A. Effect of reduced plant height on drought tolerance in rice. 3 Biotech. 2016;6:221. doi: 10.1007/s13205-016-0542-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahsan N, Lee D-G, Alam I, et al. Comparative proteomic study of arsenic-induced differentially expressed proteins in rice roots reveals glutathione plays a central role during as stress. Proteomics. 2008;8:3561–3576. doi: 10.1002/pmic.200701189. [DOI] [PubMed] [Google Scholar]
- Azri W, Cosette P, Guillou C, et al. Physiological and proteomic responses to drought stress in leaves of two wild grapevines (Vitis sylvestris): a comparative study. Plant Growth Regul. 2020;91:37–52. doi: 10.1007/s10725-020-00586-4. [DOI] [Google Scholar]
- Badowiec A, Weidner S. Proteomic changes in the roots of Germinating Phaseolus vulgaris seeds in response to chilling stress and post-stress recovery. J Plant Physiol. 2014;171:389–398. doi: 10.1016/j.jplph.2013.10.020. [DOI] [PubMed] [Google Scholar]
- Bates L, Waldren R, Teare I. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- Bergmeyer HU (1974) Methods for determination of enzyme activity. In: Bergmeyer HU (Ed) Methods of Enzymatic Analysis, Vol II, UK:Academic Press, London, pp 685–690.
- Beyer WF, Fridovich I. Assaying for superoxide dismutase activity: some large consequences of minor changes in conditions. Anal Biochem. 1987;161:559–566. doi: 10.1016/0003-2697(87)90489-1. [DOI] [PubMed] [Google Scholar]
- Bilger W, Björkman O. Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in Hedera canariensis. Photosynth Res. 1990;25:173–185. doi: 10.1007/BF00033159. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Budak H, Akpinar BA, Unver T, et al. Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI-MS/MS. Plant Mol Biol. 2013;83:89–103. doi: 10.1007/s11103-013-0024-5. [DOI] [PubMed] [Google Scholar]
- Budak H, Hussain B, Khan Z, et al. From genetics to functional genomics: ımprovement in drought signaling and tolerance in wheat. Front Plant Sci. 2015;6:1012. doi: 10.3389/fpls.2015.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Maehly, A Assay of catalases and peroxidases. Methods Enzymol. 1955;2:764–775. doi: 10.1002/9780470110171.ch14. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Claeys H, Inzé D. The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiol. 2013;162:1768–1779. doi: 10.1104/pp.113.220921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colebrook EH, Thomas SG, Phillips AL, et al. The role of gibberellin signalling in plant responses to abiotic stress. J Exp Biol. 2014;217:67–75. doi: 10.1242/jeb.089938. [DOI] [PubMed] [Google Scholar]
- Çulha-Erdal Ş, Nalcaiyi ASB, Eyidoğan F et al (2016) Probing the responses of safflower genotypes (Carthamus tinctorius L.) under drought stresses and re-watering using performance indexes. Plant Biology Europe EPSO/FESPB 2016 Congress.
- Demirevska K, Zasheva D, Dimitrov R, et al. Drought stress effects on Rubisco in wheat: changes in the Rubisco large subunit. Acta Physiol Plan. 2009;31:1129–1138. doi: 10.1007/s11738-009-0331-2. [DOI] [Google Scholar]
- DiMario RJ, Clayton H, Mukherjee A, et al. Plant carbonic anhydrases: structures, locations, evolution, and physiological roles. Mol Plant. 2017;10:30–46. doi: 10.1016/j.molp.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeoğlu B, Ekmekçi Y, Çiçek N. Physiological responses of three maize cultivars to drought stress and recovery. S Afr J Bot. 2009;75:34–42. doi: 10.1016/j.sajb.2008.06.005. [DOI] [Google Scholar]
- Esterbauer H, Cheeseman KH. Determination of aldehydic lipid peroxidation products: malonaldehyde and 4-hydroxynonenal. Methods Enzymol. 1990;186:407–421. doi: 10.1016/0076-6879(90)86134-H. [DOI] [PubMed] [Google Scholar]
- Farooq M, Wahid A, Kobayashi N, et al. Plant drought stress: effects, mechanisms and management. Agron Sustain Dev. 2009;29:185–212. doi: 10.1051/agro:2008021. [DOI] [Google Scholar]
- Farrant JMA. Comparison of mechanisms of desiccation tolerance among three angiosperm resurrection plant species. Plant Ecol. 2000;151:29–39. doi: 10.1023/A:1026534305831. [DOI] [Google Scholar]
- Filippou P, Antoniou C, Fotopoulos V. Effect of drought and rewatering on the cellular status and antioxidant response of Medicago truncatula plants. Plant Signal Behav. 2011;6:270–277. doi: 10.4161/psb.6.2.14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazanchian A, Hajheidari M, Sima NK, et al. Proteome response of Elymus elongatum to severe water stress and recovery. J Exp Bot. 2007;58:291–300. doi: 10.1093/jxb/erl226. [DOI] [PubMed] [Google Scholar]
- Gecgel U, Demirci M, Esendal E, et al. Fatty Acid composition of the oil from developing seeds of different varieties of safflower (Carthamus tinctorius L.) J Am Oıl Chem Soc. 2007;84:47–54. doi: 10.1007/s11746-006-1007-3. [DOI] [Google Scholar]
- Genty B, Briantais J-M, Baker NR. The relationship between the quantum yield of photosynthetic electron transport and quenching of chlorophyll fluorescence. Biochim Biophys Acta (BBA)-Gen Sub. 1989;990:87–92. doi: 10.1016/S0304-4165(89)80016-9. [DOI] [Google Scholar]
- Genty B, Harbinson J. Regulation of light utilization for photosynthetic electron transport. In: Baker ND, editor. Photosynthesis and the Environment. Netherlands: Springer; 1996. pp. 67–99. [Google Scholar]
- Hajheidari M, Abdollahian-Noghabi M, Askari H, et al. Proteome analysis of sugar beet leaves under drought stress. Proteomics. 2005;5:950–960. doi: 10.1002/pmic.200401101. [DOI] [PubMed] [Google Scholar]
- Haider MS, Kurjogi MM, Khalil-Ur-Rehman M, et al. Grapevine immune signaling network in response to drought stress as revealed by transcriptomic analysis. Plant Physiol Biochem. 2017;121:187–195. doi: 10.1016/j.plaphy.2017.10.026. [DOI] [PubMed] [Google Scholar]
- Huang C, Zhao S, Wang L, et al. Alteration in chlorophyll fluorescence, lipid peroxidation and antioxidant enzymes activities in hybrid ramie (Boehmeria Nivea L.) under drought stress. Aust J Crop Sci. 2013;7:594–599. [Google Scholar]
- Hurkman WJ, Tanaka CK. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 1986;81:802–806. doi: 10.1104/pp.81.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IPCC (2019) Global warming of 1.5°C, An IPCC Special Report on the impacts of global warming of 1.5°C above pre-industrial levels and related global greenhouse gas emission pathways, in the context of strengthening the global response to the threat of climate change, sustainable development, and efforts to eradicate poverty https://www.ipcc.ch/site/assets/uploads/sites/2/2019/06/SR15_Full_Report_High_Res.pdf
- Istanbulluoglu A, Gocmen E, Gezer E, et al. Effects of water stress at different development stages on yield and water productivity of winter and summer safflower (Carthamus tinctorius L.) Agric Water Manag. 2009;96:1429–1434. doi: 10.1016/j.agwat.2009.04.004. [DOI] [Google Scholar]
- Kalaji HM, Bosa K, Kościelniak J, et al. Effects of salt stress on photosystem II efficiency and CO2 assimilation of two syrian barley landraces. Environ Exp Bot. 2011;73:64–72. doi: 10.1016/j.envexpbot.2010.10.009. [DOI] [Google Scholar]
- Kim ST, Cho KS, Jang YS, et al. Two-dimensional electrophoretic analysis of riceproteins by poly-ethylene glycol fractionation for protein arrays. Electrophoresis. 2001;22:2103–2109. doi: 10.1002/1522-2683(200106)22:10<2103::AID-ELPS2103>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, et al. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007;35:506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer DM, Johnson G, Kiirats O, et al. New fluorescence parameters for the determination of QA redox state and excitation energy. Photosynth Res. 2004;79:209–218. doi: 10.1023/B:PRES.0000015391.99477.0d. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head ofbacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Li P, Ma F. Different effects of light irradiation on the photosynthetic electron transport chain during apple tree leaf dehydration. Plant Physiol Biochem. 2012;55:16–22. doi: 10.1016/j.plaphy.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Li P, Liu H, Yang H, et al. Translocation of drought-responsive proteins from the chloroplasts. Cells. 2020;9:259. doi: 10.3390/cells9010259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK. Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. doi: 10.1016/0076-6879(87)48036-1. [DOI] [Google Scholar]
- Lv Y, Li Y, Liu X, et al. Photochemistry and proteomics of ginger (Zingiber officinale Roscoe) under drought and shading. Plant Physiol Biochem. 2020;151:188–196. doi: 10.1016/j.plaphy.2020.03.021. [DOI] [PubMed] [Google Scholar]
- Majidi MM, Tavakoli V, Mirlohi A, et al. Wild safflower species ('Carthamus oxyacanthus' Bieb.): a possible source of drought tolerance for arid environments. Aust J Crop Sci. 2011;5:1055–1063. [Google Scholar]
- Mancinelli AL, Yang C-PH, Lindquist P, et al. Photocontrol of anthocyanin synthesis: III. The action of streptomycin on the synthesis of chlorophyll and anthocyanin. Plant Physiol. 1975;55:251–257. doi: 10.1104/pp.55.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaela P, Josef R, Monica N, et al. Perspectives of Safflower oil as biodiesel source for south eastern europe (comparative study: safflower, soybean and rapeseed) Fuel. 2013;111:114–119. doi: 10.1016/j.fuel.2013.04.012. [DOI] [Google Scholar]
- Mohammadi M, Ghassemi-Golezani K, Zehtab-Salmasi S, et al. Assessment of some physiological traits in spring safflower (Carthamus tinctorius L.) cultivars under water stress. Int J Life Sci. 2016;10:58–64. doi: 10.3126/ijls.v10i1.14512. [DOI] [Google Scholar]
- Mundree SG, Thomson WA, JA, , et al. An aldose reductase homolog from the resurrection plant Xerophyta viscosa Baker. Planta. 2000;211:693–700. doi: 10.1007/s004250000331. [DOI] [PubMed] [Google Scholar]
- Nellaepalli S, Ozawa S-I, Kuroda H, et al. The photosystem I assembly apparatus consisting of Ycf3–Y3IP1 and Ycf4 modules. Nat Commun. 2018;9:2439. doi: 10.1038/s41467-018-04823-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemati M, Piro A, Norouzi M, et al. Comparative physiological and leaf proteomic analyses revealed the tolerant and sensitive traits to drought stress in two wheat parental lines and their F6 progenie. Environ Exp Bot. 2019;158:223–237. doi: 10.1016/j.envexpbot.2018.10.024. [DOI] [Google Scholar]
- Nikkanen L, Rintamäki E. Thioredoxin-dependent regulatory networks in chloroplasts under fluctuating light conditions. Philos T R Soc B. 2014;369:1–7. doi: 10.1098/rstb.2013.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberschall A, Deák M, Török K, et al. A novel aldose/aldehyde reductase protects transgenic plants against lipid peroxidation under chemical and drought stresses. Plant J. 2000;24:437–446. doi: 10.1046/j.1365-313x.2000.00885.x. [DOI] [PubMed] [Google Scholar]
- Ramagli LS, Rodriguez LV. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis. 1985;6:559–563. doi: 10.1002/elps.1150061109. [DOI] [Google Scholar]
- Rao MV, Hale BA, Ormrod DP. Amelioration of ozone-ınduced oxidative damage in wheat plants grown under high carbon dioxide (role of antioxidant enzymes) Plant Physiol. 1995;109:421–432. doi: 10.1104/pp.109.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert GA, Rajasekar M, Manivannan P. Triazole-ınduced drought stress amelioration on growth, yield, and pigments composition of Helianthus annuus L. (Sunflower) Int Multidiscip Res J. 2016;5:6–15. [Google Scholar]
- Smith JA, Blanchette RA, Burnes TA, et al. Proteomic comparison of needles from blister rust-resistant and susceptible Pinus strobus seedlings reveals upregulation of putative disease resistance proteins. Mol Plant Microbe Interact. 2006;19:150–160. doi: 10.1094/MPMI-19-0150. [DOI] [PubMed] [Google Scholar]
- Tamburino R, Vitale M, Ruggiero A, et al. Chloroplast proteome response to drought stress and recovery in tomato (Solanum lycopersicum L.) BMC Plant Biol. 2017;17:1–14. doi: 10.1186/s12870-017-0971-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz I, Zaiger E, Møller IM, et al. Plant Physiology and Development. Sunderland: Sinauer Associates Publishers; 2015. [Google Scholar]
- Tari I, Kiss G, Deer AK, et al. Salicylic acid ıncreased aldose reductase activity and sorbitol accumulation in tomato plants under salt stress. Biol Plant. 2010;54:677–683. doi: 10.1007/s10535-010-0120-1. [DOI] [Google Scholar]
- Urban MO, Vašek J, Klíma M, et al. Proteomic and physiological approach reveals drought-ınduced changes in rapeseeds: water-saver and water-spender strategy. J Proteomics. 2017;152:188–205. doi: 10.1016/j.jprot.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Valero-Galván J, González-Fernández R, Navarro-Cerrillo RM, et al. Physiological and proteomic analyses of drought stress response in holm oak provenances. J Proteome Res. 2013;12:5110–5123. doi: 10.1021/pr400591n. [DOI] [PubMed] [Google Scholar]
- Wang SY, Jiao HJ, Faust M. Changes in ascorbate, glutathione, and related enzyme activities during thidiazuron-ınduced bud break of apple. Physiol Plant. 1991;82:231–236. doi: 10.1111/j.1399-3054.1991.tb00086.x. [DOI] [Google Scholar]
- Wang Y, Xu C, Zhang B, et al. Physiological and proteomic analysis of rice (Oryza sativa L.) in flag leaf during flowering stage and milk stage under drought stress. Plant Growth Regul. 2017;82:201–218. doi: 10.1007/s10725-017-0252-9. [DOI] [Google Scholar]
- Wei B, Hou K, Zhang H, et al. Integrating transcriptomics and metabolomics to studies key metabolism, pathways and candidate genes associated with drought-tolerance in Carthamus tinctorius L. under drought stress. Ind Crop Prod. 2020;151:112465. doi: 10.1016/j.indcrop.2020.112465. [DOI] [Google Scholar]
- Xin L, Zheng H, Yang Z, et al. Physiological and proteomic analysis of maize seedling response to water deficiency stress. J Plant Physiol. 2018;228:9–38. doi: 10.1016/j.jplph.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Xu L, Yu J, Han L, et al. Photosynthetic enzyme activities and gene expression associated with drought tolerance and post-drought recovery in kentucky bluegrass. Environ Exp Bot. 2013;89:28–35. doi: 10.1016/j.envexpbot.2012.12.001. [DOI] [Google Scholar]
- Yang Z, Liu J, Luo L. The Cotton GhRac6 Gene Encoding a Plant ROP/RAC Protein Improves the plant defense response to aphid feeding. Plant Mol Biol Rep. 2018;36:888–896. doi: 10.1007/s11105-018-1127-6. [DOI] [Google Scholar]
- Ye Z, Sangireddy S, Okekeogbu I, et al. Drought-induced leaf proteome changes in switchgrass seedlings. Int J Mol Sci. 2016;17:1–18. doi: 10.3390/ijms17081251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadražnik T, Hollung K, Egge-Jacobsen W, et al. Differential proteomic analysis of drought stress response in leaves of common bean (Phaseolus vulgaris L.) J Proteom. 2013;78:254–272. doi: 10.1016/j.jprot.2012.09.021. [DOI] [PubMed] [Google Scholar]
- Zadražnik T, Egge-Jacobsen W, Vladimir Meglič V, et al. Proteomic analysis of common bean stem under drought stress using in-gel stable isotope labeling. J Plant Physiol. 2017;209:42–50. doi: 10.1016/j.jplph.2016.10.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.