Abstract
The oomycete genus Phytophthora includes devastating plant pathogens that are found in almost all ecosystems. We sequenced the genomes of two quarantined Phytophthora species–P. fragariae and P. rubi. Comparing these Phytophthora species and related genera allowed reconstruction of the phylogenetic relationships within the genus Phytophthora and revealed Phytophthora genomic features associated with infection and pathogenicity. We found that several hundred Phytophthora genes are putatively inherited from red algae, but Phytophthora does not have vestigial plastids originating from phototrophs. The horizontally-transferred Phytophthora genes are abundant transposons that “transmit” exogenous gene to Phytophthora species thus bring about the gene recombination possibility. Several expansion events of Phytophthora gene families associated with cell wall biogenesis can be used as mutational targets to elucidate gene function in pathogenic interactions with host plants. This work enhanced the understanding of Phytophthora evolution and will also be helpful for the design of phytopathological control strategies.
Keywords: Phytophthora, Genome, Phylogenetic, Pathogenicity, Horizontally gene transfer
Phytophthora; Genome; Phylogenetic; Pathogenicity; Horizontally gene transfer.
1. Introduction
Oomycetes of the genus Phytophthora include devastating plant pathogens that are found in almost all ecosystems. These organisms often cause severe host infections and are responsible for considerable economic losses (Erwin and Ribeiro, 1996). Understanding the evolution of these pathogens and the associated virulence mechanisms are critical to the development of sustainable control strategies of the diseases caused by these organisms, some of which have become major epidemics (Tyler et al., 2006; Blair et al., 2008; Haas et al., 2009; Charley and David, 2017). There are 123 Phytophthora species (http://www.phytophthoradb.org/) which are usually soil-borne plant pathogens that cause disease in herbaceous and woody plants, particularly in dicotyledons. These diseases include root rot, basal stem rot, leaf spot or blight and fruit rot (Dodds and Rathjen, 2010). Phytophthora blights and rots are caused by oomycete pathogens having a wide host range; they infect many important crops, including red pepper, potato, tomato, alfalfa, tobacco, oaks, strawberry, and raspberry, thereby causing worldwide economic losses (Erwin and Ribeiro, 1996). Phytophthora species occurrence frequencies are the highest among all plant pathogens (Gao and Zhang, 2013). Plant pathologists have therefore conducted extensive research on these pathogens and governmental agencies have developed strict control measures (Bae et al., 2016; EPPO, 2020).
Phytophthora fragariae Hickman (Hickman, 1941) and Phytophthora rubi (W.F. Wilcox & J.M. Duncan) (Wilcox et al., 1993) Man in't Veld (Man in 't Veld, 2007) have highly similar morphology and physiology, but infect different hosts. These two species were once defined as Phytophthora fragariae C.J. Hickman and Phytophthora fragariae var. rubi W.F. Wilcox & J.M. Duncan. The principal P. fragariae hosts are the cultivated Fragaria × ananassa and Rubus ursinus var. longanobaccus (Ho and Jong, 1988). For P. rubi, the principal hosts are the cultivated Rubus idaeus var. idaeus. Several genera within Potentilleae in Rosaceae have been artificially infected by both of these species (Stewart et al., 2014). P. fragariae and P. rubi were first discovered in Scotland in 1921 and 1937, respectively (Wardlaw, 1926; Hickman, 1941). Occurrences of plant disease in the United States, Canada, Austria, France, Germany, and some other European and Mediterranean Plant Protection Organization (EPPO) regions were previously reported. P. fragariae and P. rubi outbreaks in Australia, Cyprus, Japan, Lebanon and some Asian countries, were subsequently reported (Pasiecznik et al., 2010), causing devastation to strawberry and raspberry productions. Both species were designated quarantine pests by many countries and regions, including EPPO regions (A2 list), the United States, and China. A recent study compared the genome structure and gene repertoires of the specific race types of P. fragariae and P. rubi and the function of several candidate avirulence effector genes was characterized (Adams et al., 2020). The genome structure, evolutionary relationships between species and pathogenesis-related factors within the Phytophthora genus are still limited. Here, we sequenced the P. fragariae and P. rubi genomes and compared them to other available Phytophthora genomes. We investigated the evolutionary relationships of Peronosporales and characterized gene families involved in Phytophthora pathogenesis, thus providing a basis for the prevention and control of these pathogens.
2. Results and discussion
2.1. Genome sequencing and characteristics
According to our K-mer analysis, the estimated actual genome sizes of P. fragariae 309.62 and P. rubi 109892 are 91.2 Mb and 103.4 Mb, respectively (Figure S1). The size of the final assembly of the P. fragariae genome is 75.98 Mb, with an 84.00 kb scaffold N50 and corresponding 40.50 kb contig N50 (Gao et al., 2015). The P. rubi genome assembly is 79.10 Mb, with an 83.99 kb scaffold N50 and corresponding 13.67 kb contig N50 (Tables S1 and S2). We annotated 25,250 and 24,528 protein-coding genes in P. fragariae and P. rubi, respectively. This identification was supported by transcriptomic data (Table 1; Notes S1, S2; Figures S2–5 and Tables S3, S4). The BUSCO (benchmarking universal single-copy orthologs) assessment indicated that the P. fragariae and P. rubi genome completeness was 96.1% and 95.4%, suggesting the genome assemblies are of good quality (Table S5) (Simão et al., 2015).
Table 1.
Characteristics of seven Phytophthora genomes.
|
Genome |
P. fragariae | P. rubi | P. sojae | P. ramorum | P. infestans | P. parasitica | P. nicotianae |
|---|---|---|---|---|---|---|---|
| Estimated genome size (bp) | 75,981,480 | 79,095,819 | 82,597,641 | 66,652,401 | 228,543,505 | 55,229,644 | 81,610,141 |
| Coverage (fold) | 214 | 222 | -- | -- | -- | -- | -- |
| G + C (%) | 52.0 | 44.5 | 52.4 | 44.0 | 42.4 | 49.6 | 50.0 |
| Repeat content length (bp) | 25,741,986 | 18,436,422 | 29,710,845 | 14,995,776 | 123,280,000 | 11,863,539 | 27,724,685 |
| Repeat content length (%) | 33.88 | 23.31 | 35.97 | 22.50 | 53.94 | 21.48 | 33.97 |
| Gene family |
13906 |
13838 |
12242 |
9120 |
10276 |
12053 |
10267 |
| Gene | |||||||
| Protein-coding gene number | 25250 | 24528 | 26489 | 15605 | 17785 | 23159 | 17348 |
| Average gene length (bp) | 1506.96 | 1416.35 | 1417.06 | 1642.1 | 1530.77 | 1729.53 | 1692.9 |
| Average gene gap length (bp) | 1339 | 1628 | 1706 | 1966 | 8440 | 840 | 1061 |
| Genome coverage (%) | 50.08 | 43.92 | 45.45 | 38.45 | 11.95 | 88.07 | 36.10 |
| Genome density (bp/gene) | 3009 | 3224 | 3118 | 4271 | 12850 | 2384 | 4704 |
| mRNA | 25250 | 24528 | 26489 | 15605 | 17787 | 28117 | 17348 |
| tRNA | 130 | 78 | 1631 | 140 | 1200 | 70 | 111 |
| CDS | 59680 | 54824 | 62854 | 40381 | 49146 | 69538 | 37714 |
| Pseudogenes | 13810 | 11697 | 25 | 6183 | 68 | ||
| Average exon number per gene | 2.36 | 2.24 | 2.37 | 2.59 | 2.78 | 2.47 | 2.17 |
| Average exon length (bp) | 547.55 | 523.06 | 500.62 | 554.16 | 470.72 | 648.28 | 704.30 |
| Average intron number per gene | 1.36 | 1.23 | 1.37 | 1.59 | 1.78 | 1.47 | 1.17 |
| Average intron length (bp) | 158.62 | 198.4 | 186.75 | 135.22 | 193.76 | 89.12 | 139.86 |
| Total intron number | 34255 | 30287 | 36365 | 24776 | 31748 | 41421 | 20366 |
| Average CDS length (bp) | 1290.32 | 1168.77 | 1137.48 | 1434 | 1260.66 | 1274.51 | 1531.13 |
2.2. Genome content in Phytophthora species
We analysed P. fragariae, P. rubi and five additional available Phytophthora genomes, including P. infestans T30-4 (Haas et al., 2009), P. parasitica INRA-310 (Blackman et al., 2014), P. sojae V3.0 (Tyler et al., 2006), P. nicotianae (Liu et al., 2016a, b) and P. ramorum (Tyler et al., 2006). The observed size variation among these Phytophthora genomes can be attributed to their repeat content, length of the intergenic regions, and number of pseudogenes (Table 1 and Note S1). The repeat content proportion in Phytophthora species genomes varies widely, with the highest value in P. infestans (58.42%) and the lowest in P. parasitica (22.66%). It likely explains the differences in genome sizes which are following a linear correlation (Figure S6 and Table S6). Long terminal repeat retrotransposons (LTRs) are abundant transposable elements (Note S3; Figures S7, S8 and Table S6), constituting 20.45% of the P. fragariae genome, 13.27% of the P. rubi genome (Figure S8) and 45.09% of the P. infestans genome. The distribution in the 10-kb 5′– or 3′– regions adjacent to RxLR and CRN genes in seven Phytophthora species indicated that the LTR number increases proportionally with distance from RxLR and CRN (Figure S9). This finding is inconsistent with previous studies suggesting that RxLR and CRN are likely located near LTR-rich regions in P. infestans (Judelson, 2007). The impact of these repeated elements on the genome size and structure is dramatic and they may play a role in the evolution of Phytophthora species, including in facilitating them in planta accomodation and dampening of the host plant immune mechanisms (Raffaele et al., 2010; Gilbert and Cordaux, 2013).
We scanned the seven Phytophthora genomes to identify gene-dense regions (GDRs) and gene-sparse regions (GSRs) and compared their evolutionary rates (Table S7). The GSR evolutionary rate was slightly higher than GDRs in most Phytophthora species. First, the high GSRs evolutionary rate was consistently higher than GDRs, except in P. fragariae, which facilitates coevolution and provides variation during host infection (Cooke et al., 2000). Second, P. rubi had the highest GSRs evolutionary rate, while P. fragariae had the lowest evolutionary rate, with 364 and nine genes in GSRs, respectively, and only a few single-copy genes in GDRs. Third, the two main types of effectors, RxLR and CRN, were located in GSRs for P. infestans and P. ramorum (Figures S10, S11 and Table S8). This distinctive genome organization offers a unique opportunity to identify novel candidate virulence genes (Haas et al., 2009). While these effectors are always located in GDRs for P. parasitica. In addition, we performed a functional enrichment analysis of genes located in GSRs and found 15 enriched candidate functions for P. ramorum, P. infestans, and P. fragariae (Table S9). The major enriched functional categories for P. ramorum are those related to the cell cytoskeleton and genome structure. For P. infestans, enriched functions are all associated with plant defence mechanisms and the expansion of these gene categories may partly explain the destructiveness of this species. The enriched functions identified for P. fragariae, e.g. pectinases, may explain its wide host range and ability to infect roots.
We used 2,921 conserved, single-copy protein sequences to construct the phylogenetic tree of Phytophthora and close lineages (Table S10). The results show that Plasmopara halstedii, an economically important pathogen (Runge et al., 2011), is clustered within Phytophthora (Figure 1). In addition, Hyaloperonospora arabidopsidis, an obligate parasite that causes downy mildew and grows on only Arabidopsis thaliana (Coates and Beynon, 2010), is sister to Phytophthora spp. Estimation of the divergence time shows that P. fragariae and P. rubi diverged approximately 9.1 million years ago (Mya). Although the host of P. fragariae is suggested to be cultivated strawberries (Fragaria × ananassa), this pathogen also infects loganberry in natural environments. The natural host of P. rubi is loganberry (Rubus × loganobaccus). The genus Rubus diverged from the Fragaria ancestor approximately 57 million years ago (MYA), long before P. fragariae and P. rubi diverged (Zhang et al., 2017). Thus, the host species change of P. fragariae to Fragaria might be a rare event, likely driven by divergence within Rubus.
Figure 1.
Phylogenetic tree correspondence with gene family expansion and contraction in oomycetes, and phylogenetic trees of seven Phytophthora species hosts. We constructed a phylogenetic tree (left) that included seven Phytophthora species (P. fragariae, P. rubi, P. parasitica, P. sojae, P. infestans, P. ramorum, and P. nicotianae) and other eight oomycete species, including Hyaloperonospora arabidopsidis, Plasmopara halstedii, Pythium arrhenomanes, Pythium ultimum, Pythium iwayamai, Pythium irregulare, Albugo laibachii, and Albugo candida. The phylogenetic tree of the main Phytophthora hosts species is shown on the right. The same colour represents correspondence between pathogen and host. Multiple colours indicate multiple correspondences. The phylogenetic tree shows the topology and divergence times for Phytophthora species and the corresponding hosts. Divergence times are indicated by light blue bars at the internodes. The range of these bars indicates the 95% confidence interval of the divergence time. Numbers at branches indicate the expansion and contraction of gene families.
2.3. Phytophthora genome evolution
2.3.1. Whole-genome duplication
WGD events have been detected in P. infestans, P. sojae, P. ramorum, P. nicotianae and P. parasitica genomes (Tyler et al., 2006; Haas et al., 2009; Blackman et al., 2014; Liu et al., 2016a, b). In addition, large intraspecies synteny was detected and many short homologous segments were localised on the same scaffold in P. infestans, P. sojae and P. ramorum (van Hooff et al., 2014). We analysed the Kolmogorov-Smirnov distributions of seven Phytophthora genomes. We did not observe distinct WGD signals, such as the distribution of synonymous substitutions per synonymous site (Ks), in these genomes, suggesting that recent genome duplication events did not occur in these seven species (Figure S12).
2.3.2. Phytophthora-specific gene expansion and contraction
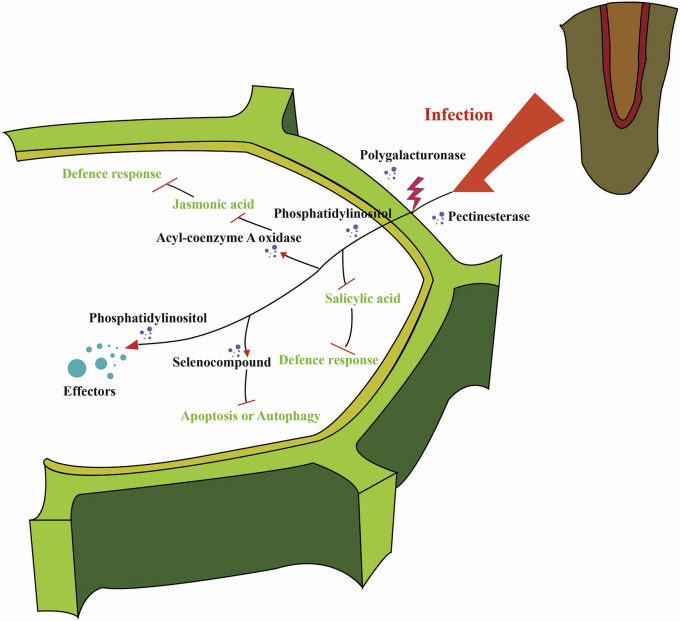
To study the unique characteristics of Phytophthora and to explore the genetic features that are possibly underlying the known pathogenicity of these species, we analysed the expansion and contraction of gene families in the Phytophthora clade. We found 131 expanding gene families and 14 contracting families (Figure 1 and Table S11). The expanding gene families comprise two main categories: 1) genes associated with pathogenicity or pathogenic processes, and 2) genes associated with maintenance of pathogenicity or common biological processes, such as energy supply or cell structure maintenance. The enriched GO terms associated with disease mechanisms are described below. For example, we found that the gene family encoding acyl-coenzyme A oxidase (ACX) is expanded in Phytophthora species compared to other genera such as Pythium, Plasmopara, and Albugo (Schneider et al., 2005). Together with the CoA synthetase, ACX is acting in the β-oxidation pathway involved in the suppression of plant resistance via signalling molecules, such as jasmonic acid. Our phylogenetic analysis showed that these ACX genes could be grouped into seven clades, including the Phytophthora-specific clades B and C (Figure S13). No conserved specific motif was found in the protein sequences suggesting that expansion of the ACX families may facilitate Phytophthora infection via the increased gene copy number rather than by a specific motif. The expansion of gene family encoding flavodoxin-like proteins in Phytophthora may also be related to the enhanced ability of the pathogen to respond to host immunity (Li et al., 2015). Expansion of tRNA uridine 5-carboxymethylaminomethyl modification enzyme (gidA) in Phytophthora may play a role in cell replication, division and weaken the host immune pattern (Figure 2) (Claussen, 2005; Shippy et al., 2011). During the Phytophthora infection process, energy supply and channeling likely facilitates efficient invasion (Giannini et al., 1988). For example, the expansion of deoxyuridine 5′-triphosphate nucleotidohydrolase (dut) is known to ensure an accurate DNA replication (Barabas et al., 2004).
Figure 2.
Schematic diagram depicting expanded gene families in Phytophthoragenomes and their putative functions during host attack. Most functions are associated with attacking plant cells. The expanded gene families where it was possible to identify a putative function are represented.
2.3.3. Evolutionary origin of MADs-Box genes
A previous study showed that the type II MADS-box genes PsMADS1 from P. sojae and PiMADS from P. infestans regulate zoosporogenesis. PsMADS1 is also involved in pathogenesis (Lin et al., 2018). The present analysis of the seven Phytophthora genomes showed that only a single MADs-box-containing gene without a K domain resembles the type II MEF2-like genes in each Phytophthora species (Figure S14). Molecular evidence indicates that in Phytophthora genomes, several hundred genes were inherited from a red alga (Tyler et al., 2006), although Phytophthora does not contain vestigial plastids originating from phototrophs. Interestingly, four HGT (horizontal gene transfer) events occurred between Phytophthora and fungi, contributing to Phytophthora genome complexity (Tyler et al., 2006). Therefore, the origin of only one type II MADs-box gene in Phytophthora genomes has been identified. Phylogenetic analysis showed that oomycete, red algae, and chlorophyte type II MEF-like MADs-box proteins form a sister clade to fungi (Figure S14). The type II MEF-like protein clade is a sister clade to the streptophyte-specific type II MIKC clade. The MRCA of streptophytes contained a protein with MADs domain similar to Type II approximately 1,000 MYA representing the ancestral MADs domain protein (Kaufmann et al., 2005). Expression analysis suggests that these genes may have a putative role in haploid reproductive cell differentiation and during the course of evolution they were recruited into a diploid generation (Thangavel, and Nayar. 2018). This result suggests that the MADs-box gene in Phytophthora might have originated from a red algae endosymbiont, but not from HGT between Phytophthora and fungi.
2.3.4. Cell wall formation
The cell wall compositions of oomycetes and fungi are chemically distinct; cellulose and chitin are the major wall components of oomycetes and fungi, respectively (Melida et al., 2013). β-1,3-glucan is another abundant polysaccharide in the oomycete wall and may play an essential role in plant pathogenesis (Raaymakers and Van den Ackerveken, 2016) as with a minor component, mannan (Hermanns and Ziegler, 1984), whose anomeric structure (α-/β-) of 1,4-linkage remains unclear (Melida et al., 2013) (Figure 3).
Figure 3.
Cell wall synthesis-related genes from Phytophthora and their association with the pathogenic interactions with host plants. Conserved gene domains and motifs are described with the NCBI conserved domain database identification number (https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). ER: endoplasmic reticulum, PM: plasma membrane, TM: transmembrane domain, CesA: cellulose synthase, UDP-Glc: UDP-glucose, GDP-Man: GDP-mannose, BcsA: conserved domain of cellulose synthase (CesA, NCBI conserved domain: COG1215), Gln: conserved β-1,3-glucan synthase domain of family 48 glycosyl transferase, GT48 (cl15642), CxxC1: cysteine repeat motif similar to the zinc finger domain in oomycete CesA3 (Blum et al., 2012), CxxC2: cysteine repeat motif similar to the zinc finger domain commonly found in methionine peptidase. CxxC2 is registered as the NCBI conserved domain motif cl33650 from Viridiplantae. Major components from Phytophthora cell walls are cellulose and β-1,3-(1,6)-glucan (Melida et al., 2013). As a minor component, mannan was detected on the cell surface via histochemistry with lectin (Hermanns and Ziegler, 1984). The major linkage in Phytophthora is a 1,4 linkage (Melida et al., 2013). Cellulose is synthesised by CesA proteins by using UDP-glucose as a substrate (UDP-Glc). Phytophthora infestans has four PiCesA genes. All PiCesAs contain the conserved CelA domain, but there are various TM domains and pleckstrin homology domains (PH) or zinc finger-like motifs (CxxC1 and 2) among the CesA genes. This difference could lead to different enzyme activities associated with CesA genes, such as the cellulose synthase-like (Csl) D proteins in Arabidopsis, which have mannan synthesis activity, although there is no report on the effects of these domains on PiCesA activities. Mandipropamid, a fungicide, targets cellulose synthase PiCesA3 to inhibit cell wall biosynthesis in P. infestans. The main 1,3-1,6-glucan backbone is synthesised by GT48 by using UDP-Glc as a substrate. The GT48 membrane protein from Phytophthora contains six TM domains in the N-terminal region, conserved glucan synthase-like domains (Gns), and eleven to twenty-two TM domains in the C-terminal regions. Phytophthora genomes also contain several mannosyl transferases that are homologous to GT32 (Och1), GT62 (Amp1), and GT71 (MNN1/2/5) enzymes identified in budding yeast. These enzymes are predicted to contain an N-terminal TM domain and may localise in Golgi membranes. The catalytic domains of these enzymes (oval shapes in the figure) are located in the Golgi lumen, and the reactions require GDP-mannose supplied by nucleotide sugar transporters such as “GONST1 homologue” from the cytoplasm. CesA genes may be required for pathogenesis in Phytophthora and are also the target of carboxylic acid amides, which are oomycete fungicides. β-1,3-1,6-glucans may be a source of glucan fragments released by host β-1,3-glucanase to elicit the synthesis of the antimicrobial substance phytoalexin in host plants as a defence response. α-mannan glycoprotein from Phytophthora megasperma has been demonstrated to suppress the defence response in host plants stimulated by the glucan elicitor.
The evolutionary trajectories of different functional classes of genes from ten Stramenopile species revealed significant expansion events in the gene classes associated with biogenesis of the cell wall, membrane and envelope in Peronosporales (Pythium and Phytophthora) but not in Saprolegniales (Seidl et al., 2012). The latter order includes mainly animal pathogens except for a few species (Gaulin et al., 2008), while plant pathogenesis of Peronosporales is thought to have evolved independently of other groups of Oomycetes (Thines and Kamoun, 2010). We conducted a phylogenetic analysis to identify cell wall synthase homologues, that is, CesA (cellulose) (Turner and Kumar, 2018), CslA, CslD (plant type-mannan) (Liepman and Cavalier, 2012), Glycosyl transferase family 48 (GT48, β-1,3-glucan) (Stone et al., 2018), GT32, GT62 and GT71 (fungal type-mannan) (Wang et al., 2017), that diverged in the ancestral lineage of Peronosporales, based on a working hypothesis that Peronosporales genes associated with plant pathogenesis should have diverged at approximately this time period (Note S4; Tables S12–15 and Figures S15–20). On the estimated phylogenetic trees, we identified a set of genes that were duplicated and already existed in the ancestral lineage of Peronosporales in CesA, GT48 and GT71 (Figures S16–20). Several duplications in these gene families occurred in the Peronosporales lineage after splitting from Saprolegniales. Some genes were duplicated more recently in different species in Peronosporales. These genes are good candidates for further functional analysis (e.g., gene inactivation) to elucidate their potential role in plant infection. Several of these genes, such as GT71 (mannan), could be used to screen fungicides, like mandipropamid (cellulose) (Blum et al., 2012) and poacic acid (β-1,3-glucan) (Piotrowski et al., 2015), targeting cell wall components.
2.3.5. Horizontal gene transfer
HGT provides a way for micro-organisms to adapt to new lifestyles, environments, and hosts (Gilbert and Cordaux, 2013). Here, we identified several HGT events in the P. fragariae and P. rubi genomes via sequence similarity comparison (Figure 4 and Table S16). Transferred genes appeared to originate from plants, fungi, bacteria, mollusc and insect donors. Most pervasive HGT genes are transposons, which is able to mobile and amplify in the host genome, which make them more prone to horizontal transfer. And horizontal transfer is an important way which wold allow the element to evade a seemingly inevitable vertical extinction in its original host lineage resulting from genetic drift, natural selection or mutational inactivation. It was result that transposition “transmitting” foreign genes to Phytophthora species from HGT. In addition, horizontal transposon transfer in genomes has been a major force propelling genomic variation and biological innovation. Here, candidatus HGT are inferred to encode secreted proteins, such as hydrolase, aspartic-type endopeptidase, integrase, chalcone-flavanone isomerase, and ubiquitin-protein transferase, which may be involved in the resistance to plant defence mechanisms or maybe effector proteins that are released into plant cells. Additional HGT candidates included repeats, such as copia-type and Tc-1-like repeats. Thus, varying degrees of expansion and contraction enlarged the Phytophthora genome complex (Tyler et al., 2006).
Figure 4.
HGT between donor and P. fragariae or P. rubi, demonstrating that most plant-, fungi-, bacteria-, mollusc-, and insect-derived genes are retained by P. fragariae and P. rubi. Using the phylogenetic analysis results in combination with alternative topology tests, we estimated the early points of transfer 23 to P. fragariae (red numbers) and 15 (black numbers) to P. rubi. The putative functions of these transferred genes are noted in the figure and listed in Table S16.
2.3.6. Pathogenesis
Phytophthora species produce apoplastic effectors that are secreted into the host extracellular space and cytoplasmic effectors that are translocated into the host cytoplasm or intracellular compartments. RxLR (where x is any amino acid) is named after an N-terminal motif involved in host cell uptake (Jiang et al., 2008). CRNs are named after a “crinkling” or necrotic phenotype that occurs when several of these proteins are overexpressed in plants (Amaro et al., 2017). We identified various candidate effectors in Phytophthora and found that RxLRs and CRNs are two main extracellular types. Phylogenetic analysis of the seven Phytophthora species provided information regarding species-specific features that may be related to pathogenesis (Figure 5 and Table S17).
Figure 5.
Phylogenetic tree with secreted RxLRs and CRN effector genes in seven Phytophthora genomes and Plasmopara halstedii. Phylogenetic trees were generated using maximum parsimony of amino acid sequences of P. fragariae, P. rubi, P. parasitica, P. sojae, P. infestans, P. ramorum, P. nicotianae, and Plasmopara halstedii.
There is a significantly higher content in RxLRs in P. sojae, P. infestans, P. parasitica, and P. nicotianae. Genome-level comparisons of RxLR genes provided evidence for diversifying selection, polymorphism, presence/absence, copy number variation, intragenic recombination, and gene conversion. CRNs are quite variable. There are far more CRNs in P. sojae and P. infestans than in other species, suggesting a potential role in the withering to death symptom observed in these two species. We screened for candidate effector gene families that were clustered into a single clade with species-level specificity during evolution (Table S18). For P. infestans, CRE4 (PITG_05910T0), CRE12 (PITG_14960T0) contributes to virulence during the early infection stage, by inhibiting plant defence responses induced by both PAMP-triggered immunity (PTI) and effector-triggered immunity (ETI) (Yin et al., 2017). PexRD1 (PITG_15287T0) and Avr-vnt11 (PITG_16294T0) enhances colonization of plant (Wang et al., 2019). Avr3a, Avr2, PexRD8 could activate innate immunity triggered by resistance protein and is a strong suppressor of the cell-death response induced by INF1 elicitin in different level (Whisson et al., 2007; Oh et al., 2009; Gilroy et al., 2011). SFI2 (PITG_04145T0) triggers converging signalling pathways recruiting MAP kinase cascades and yielding a generic antimicrobial response (Zheng et al., 2014). For P. sojae, series of Avh1b, -96 (EGZ09809), -131 (EGZ07281), -42 (EGZ29034), -13 (EGZ20804), -31 (EGZ12148), -51 (EGZ17961), -115 (EGZ29650), -7b1 (EGZ09677), -7a (EGZ09683), -7c (EGZ09679), -16 (EGZ28985), -102 (EGZ15291), -63 (EGZ26838), -81 (EGZ30372), -66 (EGZ29639), -75 (EGZ14528), -9 (EGZ19624) are specific with the similarity of Avr1 gene in P. infestans (Jiang et al., 2008). Avh 238 (EGZ19905) escapes host recognition retaining plant immunity-suppressing activity to enhance infection (Yang et al., 2018). Avh 241 (EGZ08158) locates to the membrane and triggers cell death in multiple plant species (Yu et al., 2012). However, a large number of effectors have no known function and may be involved in the infection mechanisms.
Simultaneously, functional enrichment analysis of RxLRs and CRNs in Phytophthora genomes identified several species-specific- and species-enriched candidate effectors that may be involved in the pathogenesis idiosyncrasies of each species. Enrichment was mainly reflected in several categories (Table S18 and S19): signalling pathway, receptor, ion channel, cell cytoskeleton, cellular substance, cell adhesion, motility, immunity, metabolism, cell apoptosis, and transfer. For example, with the function of signalling pathway of effectors, P. sojae, P. infestans and P. parasitica with specific G-protein-coupled receptor signalling but the different gene structures. In addition to different receptors, all these factors may be related to host specificity. The voltage-gated potassium channels of most Phytophthora species are species-specific. In P. nicotianae, the specific voltage-gated calcium channel may regulate the migration of zoosporogenesis, sporulation and mycelial growth (Liu et al., 2016a, b). G-protein-coupled receptor signalling in P. sojae and P. infestans are the same but are specific to gene structure, indicating that there may be different pathogenic mechanisms. Identification of the effector species specificity will aid the discovery and functional verification of new effectors.
3. Conclusions
We sequenced the P. fragariae and P. rubi genomes and combined the data with existing genomic data for other five Phytophthora species. Next, we conducted a comparative genomic analysis of genome structure, evolutionary relationships, and pathogenic characteristics of Phytophthora species. Our results indicate that there were no WGD events in these seven Phytophthora species. The phylogenetic relationship of Phytophthora and other oomycete species showed that Hyaloperonospora arabidopsidis was a sister to Phytophthora spp. Further, phylogenetic analysis showed that Plasmopara halstedii was a member of Phytophthora. Comparison of gene repertoires suggested that signal recognition, membrane barrier disruption, host defence weakening, and auxiliary effector molecule activity are associated with the Phytophthora infection and pathogenicity. Molecular evidence indicates that in Phytophthora genomes, several hundred genes are putatively inherited from red algae, although Phytophthora species do not have vestigial plastids originating from phototrophs. We characterized several expanding Phytophthora gene families associated with cell wall biogenesis. Further functional analysis is required to elucidate the function of these genes in pathogenic interactions. Horizontally-transferred genes in P. fragariae and P. rubi from plants, fungi, bacteria, molluscs, and insects are often transposons that impact genes involved in plant defence resistance mechanisms. Some of these genes encode effector proteins that are released in plant cells to manipulate the defence reactions. Analysis of RxLRs and CRN sequences in seven Phytophthora genomes identified genes that may be related to the pathogenesis specificity of each species. Species-specific distribution of genes involved in signalling pathways, receptors, ion channels, cell cytoskeleton, cellular substances, cell adhesion, motility, immunity, metabolism, cell apoptosis, and transfer were also identified. Our results provide a genetic basis for understanding the evolution of these pathogens and the associated virulence mechanisms, which are key to the development of sustainable disease control strategies.
4. Methods
4.1. Materials availability
The P. fragariae and P. rubi strains were imported from Westerdijk Fungal Biodiversity Institute (strain numbers 309.62 and 109892, respectively). P. fragariae 309.62 was isolated from Fragaria fruit by C.J. Hickman in Scotland, and P. rubi 109892 was isolated by C. Brasier from raspberry roots in Scotland.
4.2. Strain culture
We induced the production of motile zoospores in non-sterile pond water for up to 5 h at 13–14 °C and pH values of 6.0–6.8. Single zoospores were picked by inoculating needles under a microscope, followed by germination in V8 plates at 18 °C. After 48 h, the mycelia began to form and were collected in 5–6 days. The mycelia (~100 mg) were ground to a fine powder under liquid nitrogen. Genomic DNA was extracted using the Qiagen DNeasy Plant Mini Kit (Qiagen, Germany) according to the manufacturer's protocol.
4.3. Sequencing and assembly
We sequenced and assembled the genome of P. fragariae 309.62 and P. rubi 109892 using a strategy that combined paired-end and mate-paired libraries. Two paired-end libraries, each with a targeted insert size of 180 bp and 500 bp for P. fragariae 309.62 and of 180 bp and 300 bp for P. rubi 109892, were constructed using the TruSeq Nano DNA LT Library Prep Kit (Illumina, USA), and two mate-paired libraries (3 kb and 5 kb) were constructed for each genome using the Nextera Mate Pair Sample Prep Kit (Illumina, FC-132-1001, USA) according to the manufacturer's protocol. Both genomes were sequenced on the Illumina HiSeq 2500 platform. De novo assembly was performed using Allpaths-LG (Butler et al., 2008). The assembled genome scaffolds were aligned to the most closely related publicly available genomes using MUMmer (Edgar, 2004a, b), which included the three previously published Phytophthora complete genomes, namely, P. infestans T30-4 (Haas et al., 2009, Wang et al., 2017), P. parasitica INRA-310 (Blackman et al., 2014) and P. sojae V3.0 (Tyler et al., 2006).
Scaffolds were broken at points where non-contiguous regions of the reference genome were juxtaposed and then re-ordered so that the scaffolds were syntenic with the reference genome. All scaffolds from a given strain were concatenated into a single pseudogenome, separated by the sequence NNNNCACACACTTAATTAATTAAGTGTGTGNNNNN, which contained stop codons in all six reading frames. Scaffolds that did not match the reference genomes were concatenated in random order at the end of the genome. The pseudogenomes were annotated using the RAST automated annotation server. The genome sequences of the newly sequenced strains were deposited into GenBank.
4.4. Genome annotation
Protein-coding genes were identified by the MAKER pipeline, which integrateed ab initio gene prediction and protein and expressed sequence tag (EST) homology (Cantarel et al., 2008), and subjected to manual correction using an in-house script (HiCESAP). Gene structures were initially automatically annotated using a combination of gene models predicted by using the gene search programs Augustus (Stanke et al., 2004; Stanke and Morgenstern, 2005), GeneMark-ES (Lomsadze et al., 2005) and SNAP (Korf, 2004). Protein family and orthologue clustering were performed using TribeMCL (Dongen, 2000) and OrthoMCL (Fischer et al., 2011), respectively, using matching protein pairs (BLASTP, E<=1e-10). Protein domains and product names were assigned based on sequence homology to known proteins or Pfam domains with scores greater than the trusted cutoff scores (Bateman et al., 2004). Predicted gene models were submitted to GO (Harris et al., 2004), KEGG, Swissprot, TrEmbl, Interpro for functional annotation. Gene functional categories were computed by searching annotated proteins against the NCBI COG and KOG databases using BLASTP (E<=1e-10) and transitively assigning functional categories based on the single best match. RNA genes were predicted using both tRNA-scanSE and Rfam. Gene families corresponding to CRN proteins, NPP1-like proteins, elicitin-like proteins, small secreted cysteine-rich proteins (SCRs), and transporters were targeted for manual review. Evidence for gene identification and editing of exon boundaries was derived from protein and EST alignments based on related proteins of known function. In the case of NPP1-like proteins, elicitins, and transporters, matches to corresponding Pfam profiles were used for identification of genomic loci, and GeneWise (run with the pseudo option) was used to aid pseudogene identification. Several bioinformatics strategies have been previously employed for identification of candidate RxLR effectors within genome sequences, relying on matches to a hidden Markov model (HMM) profile or to a sequence pattern (regular expression or ‘regex’).
4.5. Repetitive elements
Repeat sequences were identified using two homologous searches (RepeatMasker version 3.3.0 and the Repbase database version 20120418) and ab initio annotations (RepeatModeler, Tandem Repeats Finder, and LTR-finder). Additionally, the high numbers of pseudogenes in both P. fragariae 309.62 and P. rubi 109892 may be a consequence of the expansion of repeat families. RIP (repeat-induced point mutation) was a self-protective mechanism developed by lower eukaryotes. Although it was generally considered to be fungus-specific, we cannot rule out that this mechanism may also occur in oomycetes. The Pearson test was used for the correlation of repeat length and repeat ration components (Venkat et al., 2017).
4.6. Comparative genomic analysis
We used OrthoMCL (version 2.0.8) to identify single-copy genes in related species, and then, MUSCLE (version 3.8.425) was used to align the sequences of the associated proteins (Edgar, 2004a, b). PAL2NAL (version 14.0) was used to convert the protein alignment to a codon alignment (Suyama et al., 2006). Gblock (version 0.91b) was used to remove the alignment results that were deemed unreliable (Castresana, 2000). Finally, the codeml program in PAML (version 4.7) was used to calculate dN (non-synonymous) and dS (synonymous) substitutions (Yang, 2007). The one-ratio model (M0) was used to estimate the overall selection pressure in the species examined, and the free-ratio model (M1) was used to estimate the dN/dS (ω) ratio in a certain branch. We then used KEGG and GoSlim for functional enrichment analysis of the obtained orthologue groups that satisfy certain criteria.
For gene family analysis, we first downloaded all the protein sequences for the studied reference genomes and used this data set as the query for all-vs-all BLASTP, with the cutoff value set at 1e−10. OrthoMCL was used to filter the query result, with an alignment length of at least 70% of the query sequences, and MCL was used to cluster the gene families, with I (inflation) set at 1.5. CAFÉ (computational analysis of gene family evolution, version 3.1) was used to calculate the expansion and contraction of these gene families (Hahn et al., 2005). A phylogenetic tree was also constructed based on the single-copy genes in the whole genome, using P. halstedii as the outgroup. After the Gblock step, ProtTest (version 3.2) (Darriba et al., 2011) was used to select the best amino acid substitution model (JTT, BIC = 17073927.92), and then, PhyML was used to build the tree. MCMCTree software in PAML (version 4.7) was used to estimate the divergence time between close species (Guindon et al., 2010). The following constraints were used for time calibrations: (i) the P. infestans and P. parasitica divergence time (2–22 (MYA)); (ii) the P. infestans and Pythium ultimum divergence time (70–80 MYA); (iii) the P. infestans and A. laibachii divergence time with a lower boundary of 135 MYA. All the data above are collected by Timetree Database (http://www.timetree.org/). Mauve (version 2.3.1) was used to align two closely related genomes for synteny analysis (Darling et al., 2010). The functional domains in each genome were identified using Pfam (http://pfam.xfam.org/) and InterProScan (Jones et al., 2014).
4.7. Whole-genome duplication
According to the method described by Martens and Van de Peer (2010), the number of synonymous substitutions per synonymous site (Ks) in whole Phytophthora genomes was analysed. Protein sequences were subjected to intra-species BLASTP alignment, and pairs of genes that were optimally aligned (RBH) were selected as homologous gene pairs for the species. MUSCLE software was used to perform multi-sequence alignment of homologous proteins and to convert the results to a nucleic acid alignment. Then, codeml in the Paml package was used to calculate the Ks value, and Ks values greater than 5 were filtered out to generate a distribution curve. The Ks distributions generated from the Phytophthora genomes all lacked a distinct peak, and it was concluded that there was no evidence of WGD or large-scale duplication in these seven genomes.
4.8. GSRs and GDRs
According to the method of Haas et al. (2009), in the study of P. infestans, with a cutoff L value of 1.5 kb, the core orthologous genes of seven Phytophthora genomes were classified as GDRs and GSRs. The gene families of GSRs and GDRs in P. fragariae and P. rubi were statistically significant (Table S7). GO functional enrichment of GSRs in P. ramorum, P. infestans and P. fragariae was performed (Table S9).
4.9. Analysis of MADS-box genes
MADS-box genes were identified by searching the InterProScan (Zdobnov and Apweiler, 2001) results of all the predicted Phytophthora proteins. The predicted genes were manually inspected, and the predicted genes were short or the MADs domains were only partially included. MADs-box domains comprising 60 amino acids, identified by SMART (Letunic et al., 2015) for all the MADs-box genes, were then aligned using ClustalW. An unrooted maximum likelihood phylogenetic tree was constructed in MEGA7 with default parameters (Kumar et al., 2016). Bootstrap analysis was performed using 1,000 iterations.
4.10. Cell wall formation
We acquired the predicted protein sequences by performing BLAST or keyword search against the following eight databases in addition to the sequences of the seven Phytophthora species (P. fragariae, P. infestans, P. parasitica, P. ramorum, P. rubi, P. nicotianae, P. sojae):
-
1)
https://bioinformatics.psb.ugent.be/plaza/versions/pico-plaza/
- 2)
- 3)
- 4)
-
5)
http://bioinformatics.psb.ugent.be/blast/moderated/?project=orcae_Chbra
- 6)
- 7)
- 8)
Data sets for NCBI protein BLAST search were selected according to taxon name—Chlorophyta (taxid: 3041), Heterokonta (taxid: 33634), Oomycota (taxid: 4762), Peronosporales (taxid: 4776), Rhodophyta (taxid: 2763), and Saprolegniales (taxid: 4763)—in addition to each species name. Accession numbers of the collected sequences were provided (Table S13).
The acquired protein sequences were analysed by NCBI conserved domain search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) to extract the conserved sequences of the CesA, GT48, GT62 and GT71 families, which were used for phylogenetic analysis. The GT32 sequences were analysed with the “phobius” program (http://phobius.sbc.su.se/) (Kall et al., 2007) for prediction of the transmembrane (TM) domains. We used the predicted catalytic regions of GT32 without the N-terminal TM domain for the phylogenetic analysis. The edited sequences were provided in Table S14, which also included the “CxxC1” and “CxxC2” motif sequences described in Figure S15. The edited sequences were analysed again with the NCBI conserved domain search, and the results are shown in Table S15.
The conserved domains of the GT protein sequences were edited as described above and used for phylogenetic tree estimation. Each set of homologous sequences was aligned using mafft (v7, G-INS-I algorithm) (Katoh and Standley, 2013). The resulting alignment, excluding gapped sites, was used to infer a maximum likelihood phylogenetic tree using PhyML v3.1 with the gamma model of heterogeneity and the estimated alpha parameter assuming the WAG amino acid substitution model. The robustness of the tree topology was assessed using 1,000 bootstrap replicates. The resulting tree was visualized with MEGA version 6 (Tamura et al., 2013).
4.11. HGT
According to the clustering results for fungi, the unique gene families of P. fragariae and P. rubi, as well as the non-clustered genes, were found to together constitute the unique genes of this species. These unique genes were not clustered with other oomycetes but may be acquired via horizontal transfer and are defined as HGT candidate genes. Subsequently, the sequences of these candidate genes were aligned to the non-redundant (nr) plant, fungal, bacterial, mollusc and insect libraries to identify a candidate gene capable of acting as donor genes that were horizontally transferred. The candidate genes generated in the second part were aligned with the NCBI nr library for optimal alignment of the donor genes, which were considered to be relatively reliable horizontal transferred genes. According to the method of Tyler et al., (2006) in the study of P. sojae and P. ramorum, we statistically analysed the horizontally transferred genes of P. fragariae and P. rubi from plants, fungi, bacteria molluscs and insects (Table S16).
4.12. Effectors
The RxLR HMM file "pf16810.hmm" was downloaded from Pfam. A protein with the same conserved region in the species as the protein set directly via hmm search was the RxLR protein of the species. For CRN prediction, multiple protein sequences of Phytophthora CRNs were downloaded from the NCBI protein database. Then, multiple sequence alignment of the Phytophthora CRN protein sequences was performed using MUSCLE 3.8 software, and an HMM of the CRN proteins was constructed using the hmmbuild program in the hmmer 3.1b2 software package.
Declarations
Author contribution statement
Rui-Fang Gao, Zhong-Jian Liu, Gui-Ming Zhang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Jie-Yu Wang: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ke-Wei Liu, Kouki Yoshida, Yu-Yun Hsiao, Yi-Xiang Shi, Kun-Chan Tsai, You-Yi Chen, Nobutaka Mitsuda, Chieh-Kai Liang, Zhi-Wen Wang, Di-Yang Zhang, Laiqiang Huang, Xiang Zhao, Wen-Ying Zhong, Ying-Hui Cheng, Ming-He Li, Wei-Hong Sun, Xia Yu, Wenqi Hu, Zhuang Zhou, Xiao-Fan Zhou: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ying Wang, Francis Martin: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Zi-De Jiang, Chuan-Ming Yeh, Kazutaka Katoh, Wen-Chieh Tsai: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This work was supported by National Key R&D Program of China (Grant No. 2019YFD1000400) and the Key Laboratory of National Forestry and Grassland Administration for Orchid Conservation and Utilization Construction Funds of Fujian, China (Grant Nos. 115/118990050 and 115/KJG18016A) for Z.-J.L., and the National Key Technology Research and Development Programme of China (Grant No. 2012BAK11B06) and National Key R&D Programme of China (Grant No. 2016YFF0203204) for G.-M. Z. The cell wall analyses were partially supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED (Grant No. JP19am0101108) for K.K.
Data availability statement
Data included in article/supp. material/referenced in article.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Contributor Information
Chuan-Ming Yeh, Email: cmyeh@mail.saitama-u.ac.jp.
Kazutaka Katoh, Email: katoh@ifrec.osaka-u.ac.jp.
Wen-Chieh Tsai, Email: tsaiwc@mail.ncku.edu.tw.
Zhong-Jian Liu, Email: zjliu@fafu.edu.cn.
Francis Martin, Email: francis.martin@inrae.fr.
Gui-Ming Zhang, Email: zgm2001cn@163.com.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- Adams T.M., Armitage An.D., Sobczyk M.K., Bates H.J., Tabima J.F., Kronmiller B.A., Tyler B.M., Grünwald N.J., Dunwell J.M., Nellist C.F. Genomic investigation of the strawberry pathogen Phytophthora fragariae indicates pathogenicity is determined by transcriptional variation in three key races. Front. Microbiol. 2020;11:1–17. doi: 10.3389/fmicb.2020.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro T.M.M.M., Thiliez G.J.A., Motion G.B., Huitema E. A perspective on CRN proteins in the genomics age: evolution, classification, delivery and function revisited. Front. Plant Sci. 2017;8:99. doi: 10.3389/fpls.2017.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S.J., Mohanta T.K., Chung J.Y., Ryu M. Trichoderma metabolites as biological control agents against Phytophthora pathogens. Biol. Contr. 2016;92:128–138. [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabas O., Pongracz V., Kovari J., Wilmanns M., Vertessy B.G. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUTPase. J. Biol. Chem. 2004;279:42907–42915. doi: 10.1074/jbc.M406135200. [DOI] [PubMed] [Google Scholar]
- Blackman L.M., Cullerne D.P., Hardham A.R. Bioinformatic characterisation of genes encoding cell wall degrading enzymes in the Phytophthora parasitica genome. BMC Genom. 2014;15:785. doi: 10.1186/1471-2164-15-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J.E., Coffey M.D., Park S.Y., Geiser D.M., Kang S. A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet. Biol. 2008;45:266–277. doi: 10.1016/j.fgb.2007.10.010. [DOI] [PubMed] [Google Scholar]
- Blum M., Gamper H.A., Waldner M., Sierotzki H., Gisi U. The cellulose synthase 3 (CesA3) gene of oomycetes: structure, phylogeny and influence on sensitivity to carboxylic acid amide (CAA) fungicides. Fungal Biol. 2012;116:529–542. doi: 10.1016/j.funbio.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Butler J., MacCallum I., Kleber M., Shlyakhter I.A., Belmonte M.K., Lander E.S., Nusbaum C., Jaffe D.B. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res. 2008;18:810–820. doi: 10.1101/gr.7337908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sánchez A.A., Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Charley G.P., David A. Phylogenomic reconstruction of the oomycete phylogeny derived from 37 genomes. Msphere. 2017;2 doi: 10.1128/mSphere.00095-17. 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen W. Proline as a measure of stress in tomato plants. Plant Sci. 2005;168:241–248. [Google Scholar]
- Cooke D.E.L., Drenth A., Duncan J.M., Wagels G., Brasier C.M. A molecular phylogeny of Phytophthora and related oomycetes. Fungal Genet. Biol. 2000;30:17–32. doi: 10.1006/fgbi.2000.1202. [DOI] [PubMed] [Google Scholar]
- Coates M.E., Beynon J.L. Hyaloperonospora arabidopsidis as a pathogen model. Annu. Rev. Phytopathol. 2010;48:329–345. doi: 10.1146/annurev-phyto-080508-094422. [DOI] [PubMed] [Google Scholar]
- Darling A.E., Mau B., Perna N.T. Progressivemauve: multiple genome alignment with gene gain, loss and rearrangement. PloS One. 2010;5 doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds P.N., Rathjen J.P. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- Dongen S.M.V. Ph.D. thesis. University of Utrecht; Utrecht: 2000. Graph Clustering by Flow Simulation. [Google Scholar]
- Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinf. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EPPO . Vol. 9. 2020. https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list (EPPO A2 list). [Google Scholar]
- Erwin D.C., Ribeiro O.K. Department of Plant Pathology, University of California; Riverside, USA: 1996. Phytophthora Disease Worldwide. [Google Scholar]
- Fischer S., Brunk B.P., Chen F., Gao X., Stoeckert C.J. Using OrthoMCL to assign proteins to OrthoMCL-DB groups or to cluster proteomes into new ortholog groups. Curr. Protoc. Bioinform. 2011 doi: 10.1002/0471250953.bi0612s35. Chapter 6, Unit 6.12.11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R.F., Zhang G.M. Potential of DNA barcoding for detecting quarantine fungi. Phytopathology. 2013;103:1103–1107. doi: 10.1094/PHYTO-12-12-0321-R. [DOI] [PubMed] [Google Scholar]
- Gao R.F., Cheng Y.H., Wang Y., Guo L.Y., Zhang G.M. Genome Sequence of Phytophthora fragariae var. fragariae, a quarantine plant-pathogenic fungus. Genome Announc. 2015;3:25–30. doi: 10.1128/genomeA.00034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaulin E., Madoui M.A., Bottin A., Jacquet C., Mathé C., Couloux A., Wincker P., Dumas B. Transcriptome of Aphanomyces euteiches: new oomycete putative pathogenicity factors and metabolic pathways. PloS One. 2008;4 doi: 10.1371/journal.pone.0001723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini J.L., Holt J.S., Briskin D.P. Isolation of sealed plasma membrane vesicles from Phytophthora megasperma f. sp. glycinea. I. Characterization of proton pumping and ATPase activity. Arch. Biochem. Biophys. 1988;265:337–345. doi: 10.1016/0003-9861(88)90136-1. [DOI] [PubMed] [Google Scholar]
- Gilbert C., Cordaux R. Horizontal transfer and evolution of prokaryote transposable elements in eukaryotes. Genome Biol. Evol. 2013;5:822–832. doi: 10.1093/gbe/evt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilroy E.M., Breen S., Whisson S.C., Squires J., Hein I., Kaczmarek M., Turnbull D., Boevink P.C., Lokossou A., Cano L.M. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 2011;191:763–776. doi: 10.1111/j.1469-8137.2011.03736.x. [DOI] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haas B.J., Sophien K., Zody M.C., Jiang R.H.Y., Handsaker R.E., Cano L.M., Grabherr M., Kodira C.D., Raffaele S., Torto-Alalibo T. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature. 2009;461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- Hahn M.W., Bie T.J., Stajich J.E., Nguyen C., Cristianini N. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 2005;15:1153–1160. doi: 10.1101/gr.3567505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris M.A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C. The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermanns R., Ziegler E. Localization of α-mannan in the hyphal wall of Phytophthora megasperma f. sp. glycinea and it's possible relevance to the host-pathogen interaction of the fungus with soybeans (Glycine max) J. Phytopathol. 1984;109:363–366. [Google Scholar]
- Hickman C.J. The red core root disease of the Strawberry caused by Phytophthora fragariae n.sp. J. Pomol. Hortic. Sci. 1941;18:89–118. [Google Scholar]
- Ho H., Jong S. Phytophthora fragariae. Mycotaxon. 1988;31:305–322. [Google Scholar]
- Jiang R.H.Y., Tripathy S., Govers F., Tyler B.M. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc. Natl. Acad. Sci. U.S.A. 2008;105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W.Z., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judelson H.S. Genomics of the plant pathogenic oomycete Phytophthora: insights into biology and evolution. Adv. Genet. 2007;57:97–141. doi: 10.1016/S0065-2660(06)57003-8. [DOI] [PubMed] [Google Scholar]
- Kall L., Krogh A., Sonnhammer E.L. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann K., Melzer R., Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005;347:183–198. doi: 10.1016/j.gene.2004.12.014. [DOI] [PubMed] [Google Scholar]
- Korf I. Gene finding in novel genomes. BMC Bioinf. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Doerks T., Bork P. SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 2015;43:D257–D260. doi: 10.1093/nar/gku949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Naseem S., Sharma S., Konopka J.B. Flavodoxin-like proteins protect candida albicans from oxidative stress and promote virulence. PLoS Pathog. 2015;11 doi: 10.1371/journal.ppat.1005147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liepman A.H., Cavalier D. The cellulose synthase-like A and cellulose synthase-like C families: recent advances and future perspectives. Front. Plant Sci. 2012;3:109. doi: 10.3389/fpls.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Ye W.W., Wu J.W., Xuan M.R., Li Y.F., Gao J., Wang L., Wang Y., Dong S.M., Wang Y.C. The MADS-box transcription factor PsMAD1 is involved in zoosporogenesis and pathogenesis of Phytophthora sojae. Front. Microbiol. 2018;9:2259. doi: 10.3389/fmicb.2018.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Ma X., Yu H.Q., Fang D.H., Li Y.P., Wang X., Wang W., Dong Y., Xiao B.G. Genomes and virulence difference between two physiological races of Phytophthora nicotianae. GigaScience. 2016;5:3. doi: 10.1186/s13742-016-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.Q., Gong J., Ding X.L., Jiang Y., Chen G.L., Li B.J., Weng Q.Y., Chen Q.H. The L-type Ca2+ channel blocker nifedipine inhibits mycelial growth, sporulation, and virulence of Phytophthora capsici. Front. Microbiol. 2016;7:1236. doi: 10.3389/fmicb.2016.01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze A., Ter-Hovhannisyan V., Chernoff Y.O., Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33:6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man in 't Veld W.A. Gene flow analysis demonstrates that Phytophthora fragariae var. rubi constitutes a distinct species, Phytophthora rubi comb. nov. Mycologia. 2007;99:222–226. doi: 10.3852/mycologia.99.2.222. [DOI] [PubMed] [Google Scholar]
- Martens C., Van de Peer Y. The hidden duplication past of the plant pathogen Phytophthora and its consequences for infection. BMC Genom. 2010;11:353. doi: 10.1186/1471-2164-11-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melida H., Sandoval-Sierra J.V., Dieguez-Uribeondo J., Bulone V. Analyses of extracellular carbohydrates in oomycetes unveil the existence of three different cell wall types. Eukaryot. Cell. 2013;12:194–203. doi: 10.1128/EC.00288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S.K., Young C., Lee M.Y., Oliva R., Bozkurt T.O., Cano L.M., Win J., Bos J.I.B., Liu H.Y., van Damme M. In planta expression screens of Phytophthora infestans RXLR effectors reveal diverse phenotypes, including activation of the Solanum bulbocastanum disease resistance protein Rpi-blb2. Plant Cell. 2009;21:2928–2947. doi: 10.1105/tpc.109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasiecznik N.M., Smith I.M., Watson G.W., Brunt A.A., Charles L.M.F. CABI/EPPO distribution maps of plant pests and plant diseases and their important role in plant quarantine. EPPO Bull. 2010;35:1–7. [Google Scholar]
- Piotrowski J.S., Okada H., Lu F., Li S.C., Hinchman L., Ranjan A., Smith D.L., Higbee A.J., Ulbrich A., Coon J.J. Plant-derived antifungal agent poacic acid targets β-1,3-glucan. Proc. Natl. Acad. Sci. U.S.A. 2015;112:e1490–1497. doi: 10.1073/pnas.1410400112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaymakers T.M., Van den Ackerveken G. Extracellular recognition of oomycetes during biotrophic infection of plants. Front. Plant Sci. 2016;7:906. doi: 10.3389/fpls.2016.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S., Farrer R.A., Cano L.M., Studholme D.J., MacLean D., Thines M., Jiang R.H.Y., Zody M.C., Kunjeti S.G., Donofrio N.M. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science. 2010;330:1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- Runge F., Telle S., Ploch S., Savory E., Day B., Sharma R., Thines M. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in Phytophthora. IMA Fungus. 2011;2:163–171. doi: 10.5598/imafungus.2011.02.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Kienow L., Schmelzer E., Colby T., Bartsch M., Miersch O., Wasternack C., Kombrink E., Stuible H.P. A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J. Biol. Chem. 2005;280:13962–13972. doi: 10.1074/jbc.M413578200. [DOI] [PubMed] [Google Scholar]
- Seidl M.F., Ackerveken G.V.D., Govers F., Snel B. Reconstruction of oomycete genome evolution identifies differences in evolutionary trajectories leading to present-day large gene families. Genome Biol. Evol. 2012;4:199–211. doi: 10.1093/gbe/evs003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippy D.C., Eakley N.M., Bochsler P.N., Chopra A.K., Fadl A.A. Biological and virulence characteristics of Salmonella enterica serovar Typhimurium following deletion of glucose-inhibited division (gidA) gene. Microb. Pathog. 2011;50:303–313. doi: 10.1016/j.micpath.2011.02.004. [DOI] [PubMed] [Google Scholar]
- Simão F.A., Waterhous R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;19:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Stanke M., Morgenstern B. AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Steinkamp R., Waack S., Morgenstern B. AUGUSTUS: a web server for gene finding in eukaryotes. Nucleic Acids Res. 2004;32:W309–W312. doi: 10.1093/nar/gkh379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart J.E., Kroese D., Tabima J.F., Larsen M.M., Grünwald J.N. Pathogenicity, fungicide resistance, and genetic variability of Phytophthora rubi isolates from raspberry (Rubus idaeus) in the western United States. Plant Dis. 2014;98:1702–1708. doi: 10.1094/PDIS-11-13-1130-RE. [DOI] [PubMed] [Google Scholar]
- Stone B.A., Jacobs A.K., Hrmova M., Burton R.A., Fincher G.B. Biosynthesis of plant cell wall and related polysaccharides by enzymes of the GT2 and GT48 families. Annu. Plant Rev. Online. 2018;14:109–165. [Google Scholar]
- Suyama M., Torrents D., Bork P. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 2006;34:W609–W612. doi: 10.1093/nar/gkl315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel G., Nayar S. A survey of mikc type mads-box genes in non-seed plants: algae, bryophytes, lycophytes and ferns. Front. Plant Sci. 2018;18:510. doi: 10.3389/fpls.2018.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines M., Kamoun S. Oomycete–plant coevolution: recent advances and future prospects. Curr. Opin. Plant Biol. 2010;13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Turner S., Kumar M. Cellulose synthase complex organization and cellulose microfibril structure. Philos. Trans. R. Soc. A. 2018;376:20170048. doi: 10.1098/rsta.2017.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler B.M., Tripathy S., Zhang X.M., Dehal P., Jiang R.H.Y., Aerts A., Arredondo F.D., Baxter L., Bensasson D., Beynon J.L. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- van Hooff J.J., Snel B., Seidl M.F. Small homologous blocks in Phytophthora genomes do not point to an ancient whole-genome duplication. Genome Biol. Evol. 2014;6:1079–1085. doi: 10.1093/gbe/evu081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkat T., Alexander S., Faheema K., Vlad D., Roger V., Magne F., Christer W., Niclas B. Rapid increase in genome size as a consequence of transposable element hyperactivity in wood-white (Leptidea) butterflies. Gen. Biol. Evol. 2017;9:2491–2505. doi: 10.1093/gbe/evx163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Wang H., Gai J.T, Tian X.L., Zhang X.X, Lv Y.Z., Jian Y. Evolution of protein N-glycosylation process in Golgi apparatus which shapes diversity of protein N-glycan structures in plants, animals and fungi. Sci. Rep. 2017;7:40301. doi: 10.1038/srep40301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.M., McLellan H., Bukharova T., He Q., Murphy F., Shi J.Y., Sun S.H., van Weymers P., Ren Y.J., Thilliez G. Phytophthora infestans RXLR effectors act in concert at diverse subcellular locations to enhance host colonization. J. Exp. Bot. 2019;70:343–356. doi: 10.1093/jxb/ery360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardlaw C. Botany Department, University of Glasgow; Glasgow: 1926. Lanarkshire Strawberry Disease. A Report for the Use of Growers. [Google Scholar]
- Whisson S.C., Boevink P.C., Moleleki L., Avrova A.O., Morales J.G., Gilroy E.M., Armstrong M.R., Grouffaud S., van West P., Chapman S. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- Wilcox W.F., Scott P.H., Hamm P.B., Kennedy D.M., Duncan J.M., Brasier C.M., Hansen E.M. Identity of a Phytophthora species attacking raspberry in Europe and North America. Mycol. Res. 1993;97:817–831. [Google Scholar]
- Yang B., Wang Y.Y., Guo B.D., Jing M.F., Zhou H., Li Y.F., Wang H.N., Huang J., Wang Y., Ye W.W. The Phytophthora sojae RXLR effector Avh238 destabilizes soybean Type2 GmACSs to suppress ethylene biosynthesis and promote infection. New Phytol. 2018;222:425–437. doi: 10.1111/nph.15581. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yin J.L., Gu B., Huang G.Y., Tian Y., Quan J.L., Lindqvist-Kreuze H., Shan W.X. Conserved RXLR effector genes of Phytophthora infestans expressed at the early stage of potato infection are suppressive to host defense. Front. Plant Sci. 2017;8:2155. doi: 10.3389/fpls.2017.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X.L., Tang J.L., Wang Q.Q., Ye W.W., Tao K., Duan S.Y., Lu C.C., Yang X.Y., Dong S.M., Zheng X.B. The RxLR effector Avh241 from Phytophthora sojae requires plasma membrane localization to induce plant cell death. New Phytol. 2012;196:247–260. doi: 10.1111/j.1469-8137.2012.04241.x. [DOI] [PubMed] [Google Scholar]
- Zdobnov E.M., Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- Zhang S.D., Jin J.J., Chen S.Y., Chase M.W., Soltis D.E., Li H.T., Yang J.B., Li D.Z., Yi T.S. Diversification of Rosaceae since the late cretaceous based on plastid phylogenomics. New Phytol. 2017;214:1355–1367. doi: 10.1111/nph.14461. [DOI] [PubMed] [Google Scholar]
- Zheng X.Z., Fraiture M., Liu X.Y., Chen Y., Brunner F. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.