Abstract
Objectives
COVID-19 might either be entirely asymptomatic or manifest itself with a large variability of disease severity. It is beneficial to identify early patients with a high risk of severe course. The aim of the analysis was to develop a prognostic model for the prediction of the severe course of acute respiratory infection.
Design
A population-based study.
Setting
Czech Republic.
Participants
The first 7455 consecutive patients with COVID-19 who were identified by reverse transcription-PCR testing from 1 March 2020 to 17 May 2020.
Primary outcome
Severe course of COVID-19.
Result
Of a total 6.2% of patients developed a severe course of COVID-19. Age, male sex, chronic kidney disease, chronic obstructive pulmonary disease, recent history of cancer, chronic heart failure, acid-related disorders treated with proton-pump inhibitors and diabetes mellitus were found to be independent negative prognostic factors (Area under the ROC Curve (AUC) was 0.893). The results were visualised by risk heat maps, and we called this diagram a ‘covidogram’. Acid-related disorders treated with proton-pump inhibitors might represent a negative prognostic factor.
Conclusion
We developed a very simple prediction model called ‘covidogram’, which is based on elementary independent variables (age, male sex and the presence of several chronic diseases) and represents a tool that makes it possible to identify—with a high reliability—patients who are at risk of a severe course of COVID-19. Obtained results open clinically relevant question about the role of acid-related disorders treated by proton-pump inhibitors as predictor for severe course of COVID-19.
Keywords: COVID-19, gastroduodenal disease, organisation of health services
Strengths and limitations of this study.
The majority of consecutive patients diagnosed with COVID-19 in the Czech Republic were included in the analysis, regardless of whether they were hospitalised or not.
The cohort covers also asymptomatic and oligosymptomatic patients identified thanks to epidemiological monitoring.
The cohort does not include strictly all COVID-19 cases in the Czech Republic because some patients are asymptomatic and have not been tested.
The proposed prediction model is a simple tool that makes it possible to identify—with a high reliability (AUC 0.893)—patients who are at risk of a severe course of COVID-19.
Flexible calibration curves based on local regression confirm the predictive model is well calibrated. The out-of-sample calibration is currently not available as data of large sample of patients from the second wave COVID-19 in the Czech Republic are still under preparation.
Due to the retrospective nature of this study, which is based on data of administrative registries, results of laboratory, clinical and X-ray examinations were not available. Conclusions regarding the influence of comorbidities and the consumption of medicinal products should be interpreted with caution and will require further validation.
Introduction
COVID-19 is caused by betacoronavirus SARS-CoV-2, which enters human cells via the membrane-bound ACE 2 (ACE2).1 The presence of infection might be entirely asymptomatic2 or manifest itself with a large variability of disease severity, a number of unspecific clinical symptoms (fever, fatigue and myalgia) and various degrees of organ dysfunction. Most frequently, the disease affects the respiratory system (manifested as dry cough, dyspnoea, haemoptysis, pneumonia or Acute Respiratory Distress Syndrome (ARDS)) and the cardiovascular system (presented as myocardial injury or myocarditis, ventricular arrhythmias, haemodynamic instability or deep vein thrombosis), while other organ systems (such as the central nervous,3 gastrointestinal system or kidneys4) are affected less frequently. In a number of patients, there is a risk of multiple organ failure, ultimately leading to death.5–9 According to the report of WHO, as of 12 November 2020, the rate of mortality among patients with COVID-19 is 2.28%.10 The management of patients with COVID-19 depends on the disease severity: patients with a mild course of the disease can be treated in their home environment.11 However, the clinical picture of patients with COVID-19 can quickly turn into an unfavourable clinical course,7 and it is therefore clinically relevant to identify patients with a high risk of severe course of the disease as early as possible.12 It was repeatedly demonstrated that older age >65 years, cancer, chronic obstructive pulmonary disease, moderate-to-severe asthma, diabetes mellitus, chronic renal disease, immunocompromised state, obesity (body mass index >30), pregnancy, sickle cell disease, smoking and cardiovascular disease are related to a high-risk course of the disease.7 13–15
In the Czech Republic, a prospective population-based and centralised collection of data on patients with COVID-19 was developed at the beginning of the pandemic, with a possibility to interconnect these data with those recorded in other population-based registries of the National Health Information System (NHIS) and thus to obtain information on each patient’s history and management.
The aim of this study was to develop a prognostic model for the prediction of the severe course of acute respiratory infection, defined by the necessity of intensive care being provided in the intensive care units (ICUs) that include mechanical ventilation, extracorporal membrane oxygenation (ECMO) support and/or death.
Methods
Population of patients
The analysis is based on data from a population-based registry containing records of all consecutive patients with COVID-19 in the Czech Republic who were identified by reverse transcription (RT)-PCR testing and validated by the National Institute of Public Health.
The monitored cohort consisted of patients who were recorded in the National Information System of Infectious Diseases (ISID) between 1 March 2020 and 17 May 2020.
As of 17 May 2020, a total of 356 515 tests were performed in the Czech Republic, and the diagnosis of COVID-19 was confirmed in 8475 cases. Of the confirmed COVID-19 cases, 464 patients with unknown history in the NHIS or the patients being foreign nationals with unknown medical history were excluded from the analysis, and further 556 patients with a follow-up period shorter than 14 days (ie, patients diagnosed after 3 May 2020) were excluded as well. Ninety per cent of events occur within 14 days (online supplemental figure S1). Analysis without censoring and with a fixed follow-up length was chosen with the objective to simplify visualisation and interpretation of results of the analysis for its practical application. The basic characteristics of patients (age and gender) were provided for all patients, and data on comorbidities were available for all patients with match between ISID and NHIS datasets; patients without match between datasets were earlier excluded from the analysis, and no other missing data handling was necessary. On top of that, characteristics of the cohort diagnosed with COVID-19 were compared with those of the population of the Czech Republic (10.6 million).
bmjopen-2020-045442supp001.pdf (111.3KB, pdf)
Diagnosis
The diagnosis of COVID-19 was based on the detection of SARS-CoV-2 by real-time reverse transcription PCR (RT-PCR). At first, the analyses were performed in the National Reference Laboratory of the National Institute of Public Health; other certified laboratories were later appointed to carry out RT-PCR testing as well.
Systematic collection of data
The analysis was done on data from the NHIS, which was supplemented with data from the Information System of Infectious Diseases (ISID). Data in the ISID are collected in compliance with Act No. 258/2000 Coll. on Protection of Public Health, whereas data in the NHIS are collected—and interconnected with data from ISID—in accordance with Act No. 372/2011 Coll., on Health Services and Conditions of Their Provision. Due to this legal mandate, the retrospective analyses did not require either approval by an ethics committee or informed consents from participants; moreover, it is a population-based analysis of all diagnosed COVID-19 cases in the Czech Republic. The cohort covers also asymptomatic and oligosymptomatic patients identified thanks to epidemiological monitoring.
The latest data on patients with COVID-19, the severity of their condition as well as the necessity of hospitalisation in an ICU, including the use of mechanical ventilation or ECMO, together with information on death, have been entered into the ISID in real time. Apart from that, data on patients with COVID-19 have been enriched with information on their comorbidities: this information is available in the National Register of Reimbursed Health Services, which contains data on all healthcare reported within the public health insurance system (accounting for almost 100% of healthcare provided in the Czech Republic). Comorbidities are determined from combinations of reported diagnoses International Classification of Diseases 10th Revision (ICD-10), ATC codes of drugs and procedures defined in code lists used by Czech health insurance companies. Only diseases and conditions with a higher prevalence in the population or those identified in literature7 9 15–17 as potential predictors of a severe course of COVID-19 were evaluated, with the aim to assess their potential influence on the resulting model.
Statistical analysis
Standard descriptive statistics were used to describe the data: categorical variables were described by absolute and relative frequencies, whereas continuous variables were described by means and SD. The Fisher’s exact test (for categorical variables) and Mann-Whitney U test (for continuous variables) were used to compare characteristics between groups depending on the monitored endpoint, unless stated otherwise. The predictive power of patient characteristics with regard to the analysed endpoint was evaluated by univariable and multivariable logistic regression and described by ORs, their 95% CIs and statistical significance; a backward stepwise algorithm was used to choose the optimal model, and a ROC analysis was employed to evaluate the overall predictive power of the model. A flexible calibration curve18 was adopted for the evaluation of goodness of fit of the model. The results of the model were expressed by a risk heat map taking account of the patients’ age, sex and comorbidities. A 10-fold cross-validation was performed to obtain estimates of model performance that are adjusted for in-sample optimism. A model was created in accordance with TRIPOD checklist for prediction model development and validation.19 The analysis was computed using the Vertica database and an MS SQL Server for data preprocessing and SPSS V.25.0.0.1 and R V.3.6.1 for the statistical analysis of data. The level of statistical significance was set at α=0.05 for all analyses.
Results
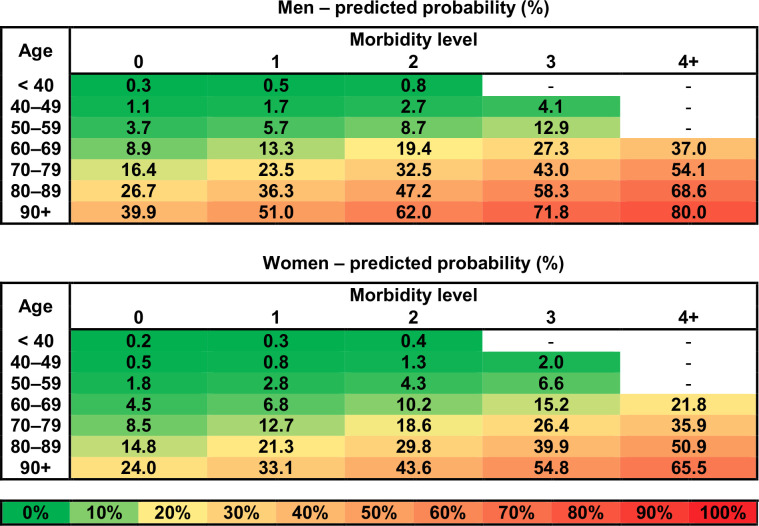
Of the total of 7455 evaluated patients, 1182 (15.9%) were hospitalised, 465 of them (6.2%) developed a severe course of the disease (ie, reached the primary endpoint—death or the necessity of intensive care being provided in the ICUs that include mechanical ventilation and/or ECMO support): 174 patients (2.3%) required mechanical ventilation, 11 patients (0.1%) were provided ECMO and 287 patients (3.8%) died. Patients with the monitored endpoint were older (74.8±13.4 vs 45.4±20.2 years), more frequently of male sex and suffered at least one of all monitored comorbidities (table 1, p<0.001 for all parameters; for univariable logistic regression results, see online supplemental table S1). Older age was determined by the multivariable logistic regression analysis to be the most significant predictor: the risk of a severe course of the disease increases progressively from the age of 40 years onwards (table 2). Male sex, chronic kidney disease, chronic obstructive pulmonary disease, recent history of cancer (in the last 5 years), chronic heart failure, acid-related disorders treated with proton-pump inhibitors and diabetes mellitus were other significant predictors; the latter six conditions are hereinafter referred to as prognostically significant comorbidities (table 2). The overall predictive power of the model, evaluated by the Receiver operating characteristic (ROC) analysis and expressed by the AUC, was 0.893 (95% CI 0.880 to 0.907; sensitivity: 85.8% and specificity: 80.3%). After performing the 10-fold cross-validation to validate the results, the average AUC of 0.891 (in the range 0.856–0.943) was obtained. For the purpose of an easier interpretation in clinical practice, a simplified version of the model was developed, taking into consideration the number of prognostically significant comorbidities obtained from the previous model (table 3). Both original and simplified models are well calibrated, as is supported by calibration curves in the supplementary figures (online supplemental figure S2A for the original model and online supplemental figure S2B for the simplified model). The results of the simplified model were visualised by risk heat maps for men and women separately (figure 1), and we called this diagram a ‘covidogram’. The diagram shows how the risk increases progressively with age and with the number of prognostically significant comorbidities.
Table 1.
Characteristics of patients with COVID-19 according to endpoint
| With severe course | Without severe course | |
| N=465 | N=6990 | |
| Basic characteristics | ||
| Men, n (%) | 260 (55.9) | 3221 (46.1) |
| Age, mean±SD | 74.8±13.4 | 45.4±20.2 |
| History, n (%) | ||
| Hypertension | 258 (55.5) | 1266 (18.1%) |
| Atrial fibrillation | 75 (16.1) | 179 (2.6) |
| History of stroke | 55 (11.8) | 131 (1.9) |
| History of MI or PCI | 32 (6.9) | 106 (1.5) |
| Chronic heart failure | 90 (19.4) | 157 (2.2) |
| Chronic kidney disease | 99 (21.3) | 215 (3.1) |
| Diabetes mellitus | 127 (27.3) | 519 (7.4) |
| Chronic obstructive pulmonary disease | 102 (21.9) | 643 (9.2) |
| Acid-related disorders | 206 (44.3) | 999 (14.3) |
| Recent history of cancer (≤5 years) | 48 (10.3) | 169 (2.4) |
| Rheumatoid arthritis | 14 (3.0) | 75 (1.1) |
| Treatment, n (%) | ||
| ACE inhibitors | 182 (39.1) | 947 (13.5) |
| ARBs | 85 (18.3) | 543 (7.8) |
| Calcium channel blockers | 121 (26.0) | 584 (8.4) |
| Beta-blockers | 212 (45.6) | 884 (12.6) |
| Diuretics | 214 (46.0) | 814 (11.6) |
| Anticoagulants/antithrombotic agents | 143 (30.8) | 502 (7.2) |
| Statins | 169 (36.3) | 882 (12.6) |
ARBs, angiotensin II receptor blockers; MI, myocardial infarction; PCI, percutaneous coronary intervention.
Table 2.
Multivariable logistic regression model using backward stepwise algorithm for selection of independent predictors of severe course of COVID-19
| Predictors | Regression coefficients | SE | OR (95% CI) | P value |
| Sex | ||||
| Women | Reference category | |||
| Men | 0.742 | 0.112 | 2.10 (1.68 to 2.62) | <0.001 |
| Age (years) | ||||
| <40 | Reference category | |||
| 40–49 | 1.227 | 0.464 | 3.41 (1.37 to 8.48) | 0.008 |
| 50–59 | 2.478 | 0.414 | 11.92 (5.30 to 26.81) | <0.001 |
| 60–69 | 3.424 | 0.399 | 30.68 (14.04 to 67.04) | <0.001 |
| 70–79 | 4.109 | 0.398 | 60.89 (27.93 to 132.73) | <0.001 |
| 80–89 | 4.725 | 0.400 | 112.68 (51.48 to 246.63) | <0.001 |
| 90+ | 5.299 | 0.428 | 200.12 (86.50 to 462.97) | <0.001 |
| Comorbidities | ||||
| Chronic kidney disease | 0.679 | 0.157 | 1.97 (1.45 to 2.68) | <0.001 |
| Chronic obstructive pulmonary disease | 0.436 | 0.144 | 1.55 (1.17 to 2.05) | 0.002 |
| Recent history of cancer (≤5 years) | 0.432 | 0.194 | 1.54 (1.05 to 2.25) | 0.026 |
| Chronic heart failure | 0.408 | 0.166 | 1.50 (1.09 to 2.08) | 0.014 |
| Acid-related disorders | 0.382 | 0.118 | 1.47 (1.16 to 1.85) | 0.001 |
| Diabetes mellitus | 0.323 | 0.129 | 1.38 (1.07 to 1.78) | 0.012 |
| Intercept | −6.448 | 0.386 | – | <0.001 |
Overall predictive power: AUC (95% CI): 0.893 (0.880 to 0.907); sensitivity: 85.8% and specificity: 80.3%.
Age, sex and comorbidities from table 1 were entered into the model.
Table 3.
Simplified multivariable logistic regression model based on predictors selected by a backward stepwise algorithm
| Predictors | Regression coefficients | SE | OR (95% CI) | P value |
| Sex | ||||
| Women | Reference category | |||
| Men | 0.744 | 0.112 | 2.10 (1.69 to 2.62) | <0.001 |
| Age (years) | ||||
| <40 | Reference category | |||
| 40–49 | 1.215 | 0.464 | 3.37 (1.36 to 8.37) | 0.009 |
| 50–59 | 2.459 | 0.414 | 11.69 (5.20 to 26.30) | <0.001 |
| 60–69 | 3.388 | 0.398 | 29.59 (13.56 to 64.59) | <0.001 |
| 70–79 | 4.082 | 0.397 | 59.27 (27.22 to 129.05) | <0.001 |
| 80–89 | 4.700 | 0.399 | 109.93 (50.31 to 240.20) | <0.001 |
| 90+ | 5.302 | 0.427 | 200.66 (86.84 to 463.63) | <0.001 |
| Morbidity level* | ||||
| 1-point increase | 0.448 | 0.049 | 1.56 (1.42 to 1.72) | <0.001 |
| Intercept | −6.453 | 0.386 | <0.001 |
Overall predictive power: AUC (95% CI): 0.893 (0.879 to 0.907); sensitivity: 85.8% and specificity: 80.3%.
*One point for each comorbidity: chronic kidney disease, chronic obstructive pulmonary disease, recent history of cancer (≤5 years), chronic heart failure, acid-related disorders and diabetes mellitus (maximum of 4 points per person; additional points were not considered due to a low prevalence in our cohort).
Figure 1.
Covidogram—predicted probability of severe course of COVID-19 for men and women—visualisation of simplified multivariable logistic regression model (for more details, see table 3).
It is obvious from the comparison of basic patient characteristics (table 1) and the results of the multivariable analysis (table 2) that although a number of conditions occur more frequently in the group of patients with a severe course of the disease, not all of them are independent predictors (coronary artery disease, history of stroke, atrial fibrillation, hypertension or treatment by ACEIs and angiotensin II receptor blockers).
The comparison of characteristics of patients with confirmed COVID-19 to those of the entire population of the Czech Republic showed that COVID-19 patients are slightly older and have the monitored comorbidities slightly more frequently (online supplemental table S2).
Discussion
New findings about COVID-19
We have developed a prognostic model for the prediction of the severe course of COVID-19 in consecutive patients with positive COVID-19 RT-PCR test. This, a simple tool called ‘covidogram’, has a very good predictive power (AUC 0.893). Age is the most significant factor, and the risk increases progressively from the age of 40 years onwards. To our knowledge, this is the first study to suggest that acid related disorders treated by proton pump inhibitors might be independent risk predictors as well. By contrast, not all cardiovascular diseases (such as uncomplicated hypertension or coronary artery disease) increase the risk of a severe course of COVID-19.
The ‘covidogram’ was designed as a model to assess the risk of unfavourable development of the patient’s condition based on his or her history of chronic disease and can serve as a tool to estimate the number of severe cases of COVID-19 in a population. When assessing the risk for an individual patient in clinical practice, it is certainly necessary to take into consideration also other pieces of information on the current condition of that patient (respiratory rate, peripheral oxygen saturation, level of consciousness, urea level, C reactive protein,20 procalcitonin, aspartate aminotransferase,17 high temperature,16 elevation of cardiac markers and lung infiltrates >50%9) as well as obesity, which can also increase the risk of a severe course of COVID-19.21
Acid-related disorders
Surprisingly, our analysis revealed that the presence of acid-related disorders might be theoretically linked to a severe course of COVID-19. Patients were predominantly treated with proton-pump inhibitors (1175 patients in total, out of which 706 were treated with omeprazole and 402 with pantoprazole as the two most frequently used drugs); only a small proportion of them were treated with H2-receptor antagonists (30 patients). The main indications for treatment with these drugs generally involve gastro-oesophageal reflux disease, functional dyspepsia, gastric and duodenal ulcers, gastric acid hypersecretory states as well as gastroprotection in patients using non-steroidal anti-inflammatory drugs, dual antiplatelet therapy, biphophonates or same selective serotonin reuptake inhibitors. The effect of inhibition of hydrochloric acid secretion is followed by an increase in the intragastric pH (to a value above 2–4), which might hypothetically decrease the physiological bactericidal/virucidal effect of gastric acid and decrease the activity of lysosomal enzymes. Published data showed that long-term use of proton-pump inhibitors could slightly increase the risk of pneumonia22 23 and enteric infections.24
Our comparison of patients with and without acid-related disorders (online supplemental table S3) showed that patients with these disorders are markedly older and have prognostically significant comorbidities more frequently. Our analysis cannot determine whether there is any causal relationship between the presence of acid-related disorders and a severe course of COVID-19 or whether it is just a coincidence. At the same time, it must be stressed out that the vast majority of patients were treated with proton-pump inhibitors, not with H2-receptor agonists. The observation is complicated also by the fact that some patients may not be adherent to their proton-pump inhibitors (PPI) regimen, and there was great variability in the amount of time that they have been on PPIs (from 1st to 12th month within 2019). Based on our analysis, we are not able to decide whether severity of disease might be theoretically explained by pharmacology or by underlying pathology of acid-related disorders. Recently, Almario et al25 demonstrated association between using proton-pump inhibitors and odds of a positive COVID-19 test. Similar trend was reported by Tarlow et al.26
Strengths and limitations
This study is based on a fully integrated national health information system covering the entire population of a country, which proposed a prediction model estimating individually based risk of a severe course of COVID-19. Because this model uses data readily available in health and administrative registries, it can be easily used for the prediction of intensive care use in the context of decision making at the national level.
However, our analysis has a number of limitations. Results of laboratory, clinical and X-ray examinations performed at the time of patient admission to hospitals were not available, and these very important pieces of information could therefore not be analysed; instead, our analysis is based on administrative data, with the exception of endpoints. Furthermore, analytical processing of a cohort of patients cannot capture the risk of less frequent conditions that might increase the risk of a severe course of COVID-19 (eg, patients with immunodeficiencies, those after organ transplantation or those undergoing immunosuppressive therapy or biological therapy). The cohort does not include strictly all COVID-19 cases in the Czech Republic because some patients are asymptomatic and have not been tested. Older people with more comorbidities are probably more likely to have a symptomatic course of COVID-19. It could be also a reason why the population of patients diagnosed with COVID-19 is older and with more comorbidities in comparison with the Czech Republic population (online supplemental table S2). Due to the retrospective nature of this study, which is based on data of administrative registries and is focused on the development of a prediction model, any conclusions regarding the influence of comorbidities and the consumption of medicinal products should be interpreted with caution and will require further validation. The in-sample callibration of the model was assessed by a flexible calibration curves that confirmed that the predictive model is well calibrated. Out-of-sample calibration is currently not available as data of large sample of patients from the second wave COVID-19 in the Czech Republic are still under preparation.
Conclusion
The proposed prediction model ‘covidogram’ is based on elementary independent variables (age, male sex and the presence of chronic disease) and represents a simple tool that makes it possible to identify—with a high reliability (AUC 0.893)—patients who are at risk of a severe course of COVID-19.
Finally, the analysis has shown, for the first time, that acid-related disorders treated with proton-pump inhibitors might also be theoretically associated with a severe course of the disease.
Supplementary Material
Footnotes
Contributors: JJ, LD, VC and JR designed the study and wrote the research plan. OM, SS, MB and HM extracted the data used for the study from the databases. JJ and KB undertook statistical analysis with feedback from LD. JP, JJ, PK and JD interpreted the results and wrote the first draft of the manuscript with critical comments and revision from VC, LD and LS.
Funding: This research was supported by grant the Czech Republic Operational Programme eHealth and Rare Disease CZ.03.4.74/0.0/0.0/15_025/.
Competing interests: None declared.
Patient consent for publication: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available on reasonable request. The anonymised data available on reasonable request. The data are deidentified participant data and available from the first author JJ (jarkovsky@uzis.cz). The reuse of the data subset is permitted only for revalidation of the results.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181:271–80. 10.1016/j.cell.2020.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sutton D, Fuchs K, D'Alton M, et al. Universal screening for SARS-CoV-2 in women admitted for delivery. N Engl J Med 2020;382:2163–4. 10.1056/NEJMc2009316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aghagoli G, Gallo Marin B, Katchur NJ. Neurological involvement in COVID-19 and potential mechanisms: a review. Neurocrit Care 2020;395:1–10. 10.1007/s12028-020-01049-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aghagoli G, Gallo Marin B, Soliman LB, et al. Cardiac involvement in COVID-19 patients: risk factors, predictors, and complications: a review. J Card Surg 2020;35:1302–5. 10.1111/jocs.14538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giacomelli A, Pezzati L, Conti F, et al. Self-Reported olfactory and taste disorders in patients with severe acute respiratory coronavirus 2 infection: a cross-sectional study. Clin Infect Dis 2020;71:889–90. 10.1093/cid/ciaa330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han C, Duan C, Zhang S, et al. Digestive symptoms in COVID-19 patients with mild disease severity: clinical presentation, stool viral RNA testing, and outcomes. Am J Gastroenterol 2020;115:916–23. 10.14309/ajg.0000000000000664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 2020;395:497–506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim I-C, Kim JY, Kim HA, et al. COVID-19-related myocarditis in a 21-year-old female patient. Eur Heart J 2020;41:1859. 10.1093/eurheartj/ehaa288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020;323:1061–9. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.WHO . Coranvirus disease (COVID-19) pandemic. Available: https://www.who.int/emergencies/diseases/novel-coronavirus-2019 [Accessed 12 Dec 2020].
- 11.Gandhi RT, Lynch JB, Del Rio C. Mild or moderate Covid-19. N Engl J Med 2020;383:1757–66. 10.1056/NEJMcp2009249 [DOI] [PubMed] [Google Scholar]
- 12.Berlin DA, Gulick RM, Martinez FJ. Severe Covid-19. N Engl J Med 2020;383:2451–60. 10.1056/NEJMcp2009575 [DOI] [PubMed] [Google Scholar]
- 13.Mehra MR, Desai SS, Kuy S. Retraction: cardiovascular disease, drug therapy, and mortality in Covid-19. N Engl J Med 2020;382:2582. 10.1056/NEJMc2021225 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Wang T, Tang C, Chen R, et al. Clinical features of coronavirus disease 2019 patients with mechanical ventilation: a nationwide study in China. Crit Care Med 2020;48:e809–12. 10.1097/CCM.0000000000004473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.CDC . Coronavirus-19. Available: https://www.cdc.gov/coronavirus/2019-ncov/need-extra-precautions/people-with-medical-conditions.html
- 16.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020;180:934–43. 10.1001/jamainternmed.2020.0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen R, Liang W, Jiang M, et al. Risk Factors of Fatal Outcome in Hospitalized Subjects With Coronavirus Disease 2019 From a Nationwide Analysis in China. Chest 2020;158:97–105. 10.1016/j.chest.2020.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Calster B, Nieboer D, Vergouwe Y, et al. A calibration hierarchy for risk models was defined: from utopia to empirical data. J Clin Epidemiol 2016;74:167–76. 10.1016/j.jclinepi.2015.12.005 [DOI] [PubMed] [Google Scholar]
- 19.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD). Circulation 2015;131:211–9. 10.1161/CIRCULATIONAHA.114.014508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knight SR, Ho A, Pius R, et al. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC who clinical characterisation protocol: development and validation of the 4C mortality score. BMJ 2020;370:m3339. 10.1136/bmj.m3339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik P, Patel U, Patel K, et al. Obesity a predictor of outcomes of COVID-19 hospitalized patients-A systematic review and meta-analysis. J Med Virol 2021;93:1188–93. 10.1002/jmv.26555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eom C-S, Jeon CY, Lim J-W, et al. Use of acid-suppressive drugs and risk of pneumonia: a systematic review and meta-analysis. CMAJ 2011;183:310–9. 10.1503/cmaj.092129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zirk-Sadowski J, Masoli JA, Delgado J, et al. Proton-Pump inhibitors and long-term risk of community-acquired pneumonia in older adults. J Am Geriatr Soc 2018;66:1332–8. 10.1111/jgs.15385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moayyedi P, Eikelboom JW, Bosch J, et al. Safety of proton pump inhibitors based on a large, Multi-Year, randomized trial of patients receiving rivaroxaban or aspirin. Gastroenterology 2019;157:682–91. 10.1053/j.gastro.2019.05.056 [DOI] [PubMed] [Google Scholar]
- 25.Almario CV, Chey WD, Spiegel BMR. Increased risk of COVID-19 among users of proton pump inhibitors. Am J Gastroenterol 2020;115:1707–15. 10.14309/ajg.0000000000000798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarlow B, Gubatan J, Khan MA, et al. Are proton pump inhibitors contributing to SARS-COV-2 infection? Am J Gastroenterol 2020;115:1920–1. 10.14309/ajg.0000000000000933 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
bmjopen-2020-045442supp001.pdf (111.3KB, pdf)