Summary
Neurons depend on proper localization of neurotrophic receptors in their distal processes for their function. The Trk family of neurotrophin receptors controls neuronal survival, differentiation, and remodeling and are well known to function as retrograde signal carriers transported from the distal axon toward the cell body. However, the mechanism driving anterograde trafficking of Trk receptors into the axon is not well established. We used microfluidic compartmental devices and inducible secretion assay to systematically investigate the retrograde and anterograde trafficking routes of TrkB receptor along the axon in rat hippocampal neurons. We show that newly synthesized TrkB receptors traffic through the secretory pathway and are directly delivered into axon. We found that these TrkB carriers associate and are regulated by Rab6. Furthermore, the combined activity of kinesin-1 and kinesin-3 is needed for the formation of axon-bound TrkB secretory carriers and their effective entry and processive anterograde transport beyond the proximal axon.
Keywords: TrkB, axon transport, kinesin-1, kinesin-3, Rab6, secretory trafficking, RUSH, anterograde transport
Graphical abstract
Highlights
-
•
Internalized TrkB receptors are sorted for retrograde axonal transport
-
•
Direct trafficking of secretory carriers delivers TrkB to the axon
-
•
These carriers associate with and depend on Rab6-GTPase for trafficking
-
•
Formation and transport of axon-bound carriers depend on kinesin-1 and kinesin-3 motors
Neurons distribute signaling receptors to distal axons to receive extracellular information. Focusing on the neurotrophic receptor TrkB, Zahavi et al. elucidate an intracellular trafficking pathway that enables neurons to drive TrkB from its site of synthesis at the cell body, via secretory transport carriers, into the distal axon.
Introduction
Neurons are uniquely structured cells with an extreme degree of morphological polarity between their dendritic and axonal domains. This segregation enables the efficient neuronal integration of multiple post-synaptic inputs in the somatodendritic compartment and the transmission of processed information via the axon to the pre-synapse. The function of the dendrites and axon depends on correct trafficking and transport of membrane proteins that comprise the pre- and post-synaptic receptor repertoire (Bentley and Banker, 2016). A particularly difficult logistic challenge for the mammalian neuron is the delivery of membrane proteins to the distal axon processes due to its sheer distance from the subcellular sites in which their biosynthesis takes place—namely that in neurons of the central nervous system the rough endoplasmic reticulum (RER) and the Golgi apparatus reside exclusively in the soma and dendritic compartment (Britt et al., 2016).
The family of tropomyosin-related kinases (Trk) receptors, including TrkA, TrkB, and TrkC, provides a prime example of membrane proteins that undergo long-distance axonal trafficking to regulate growth and survival of developing neurons. The cognate ligand of TrkB, brain-derived neurotrophic factor (BDNF), is a predominant neurotrophin in the brain that plays a major role in regulating neuronal development, survival, and remodeling by signaling in the pre- and post-synapse in the peripheral and central nervous system (Bothwell, 2016; Park and Poo, 2013). As such, the localization and activity of the TrkB receptor in the distal axon is crucial for both its local and retrograde (axon-to-soma) neurotrophic functions (Harrington and Ginty, 2013; Scott-Solomon and Kuruvilla, 2018). The cellular mechanisms underlying the retrograde delivery of the neurotrophic signal have been described by the established signaling endosome model, by which the internalized receptor-ligand complex is transported along the axon by dynein motors with its downstream signaling intact (Cosker and Segal, 2014). Nevertheless, little is understood about the molecular mechanisms responsible for the delivery Trk receptors into the axon.
Polarized trafficking of membrane proteins to the axon follows one of two main delivery routes: (1) direct path via secretory carriers that fuse with axonal membrane and (2) an indirect route, also termed the transcytosis pathway, in which these carriers are first delivered to somatodendritic membranes, followed by endocytosis re-distribution of the protein to the axon (Lasiecka and Winckler, 2011; Sampo et al., 2003; Wisco et al., 2003). Axonal delivery of TrkA, the receptor for nerve growth factor (NGF), has been shown to follow a transcytosis route from somatodendritic to axon membranes via Rab11 recycling endosomes in sympathetic cervical ganglia (SCG) neurons (Ascaño et al., 2009). The anterograde trafficking of the transcytosed TrkA cargo is regulated by a feedback mechanism induced by retrograde transport of NGF-stimulated TrkA, which activates somatic PTP1B to de-phosphorylate the receptor and re-distributes it from somatodendritic membranes to the axon (Yamashita et al., 2017). Soma-to-axon transcytosis of TrkB has not been reported; however, it is known that its sorting into Rab11 compartments regulates local recycling to the plasma membrane in dendrites and at post-synaptic domains specifically (Huang et al., 2013; Lazo et al., 2013; Song et al., 2015).
Efficient anterograde delivery of axon-bound membrane proteins depends primarily on kinesin-1 and kinesin-3 families of motors (Bentley and Banker, 2016). Kinesin-1 motors enable axon-specific transport owing to their preferential binding to acetylated and detyrosinated microtubules (MTs) that are enriched in the axon, and to the homogeneous orientation of MT plus ends pointing toward the axon tips, while in dendrites acetylated MTs are oriented toward the soma, hence, sequestering kinesin-1 entry (Fourriere et al., 2020). Kinesin-3 KIF1 motors, which drive both dendritic and axon transport of synaptic and dense core vesicles, have special axon delivery roles. In DRG neurons, KIF1A enables cargo to traverse the pre-axonal trafficking barrier for further transport via kinesin-1 (Gumy et al., 2017) and in localized targeting of synaptic vesicles to the pre-synapse in hippocampal neurons (Guedes-Dias et al., 2019). Anterograde axon transport of TrkB was shown to be mediated by kinesin-1 in Rab27 vesicles, and that of TrkA is by the kinesin-3 KIF1A motor (Arimura et al., 2009; Tanaka et al., 2016). Both studies postulated these to be secretory vesicles derived from the Golgi apparatus, which requires further examination.
Protein trafficking from the secretory pathway in neurons has focused mainly on dynamics in dendrites, highlighting the interplay between secretory organelle localization, motor proteins, and Rab proteins in regulating local trafficking for specific membrane proteins (Kennedy and Hanus, 2019). Recent studies have shown that upon exiting the secretory pathway, membrane proteins undergo selective transport or retention in order to reach the axon or dendrites in a specific manner; yet, the underlying motor and membrane sorting and trafficking proteins that regulate this selectivity remain largely unknown.
In this study, we set to elucidate the trafficking and transport mechanism that delivers TrkB to the axon in polarized hippocampal neurons. Using a combination of compartmental surface labeling and inducible secretion assays, we dissected its transport dynamics in the endosomal and secretory pathways. Our results show that delivery of TrkB into the axon is driven by direct trafficking and transport from the secretory system in Rab6 carriers and the combined activity of kinesin-1 and kinesin-3 motors.
Results
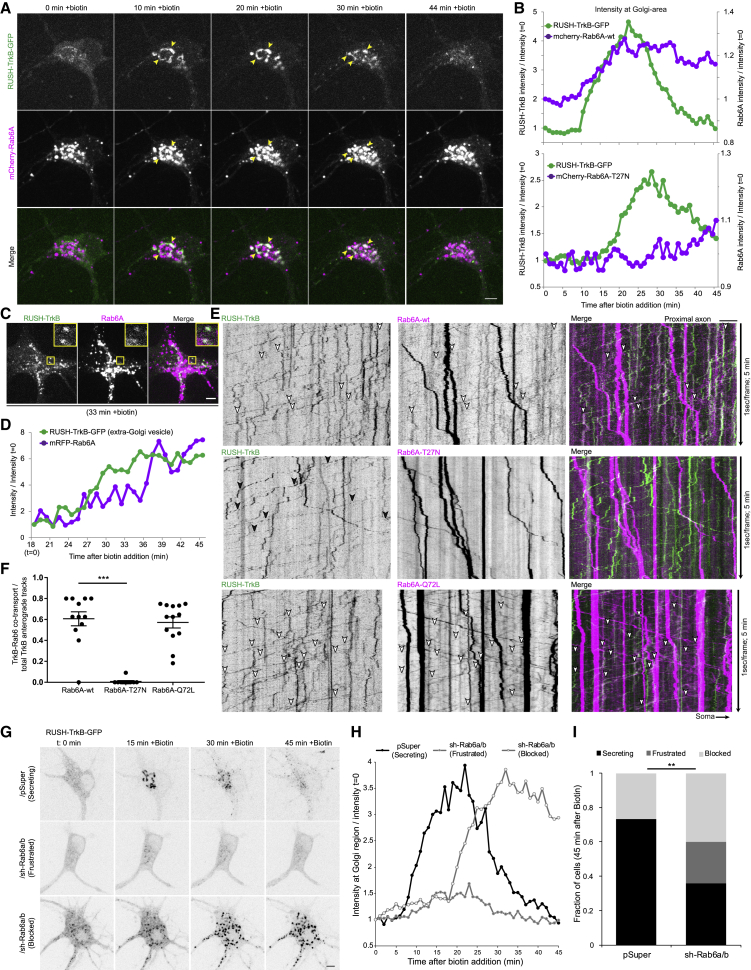
Endocytosed TrkB is coupled to retrograde transport in the axon
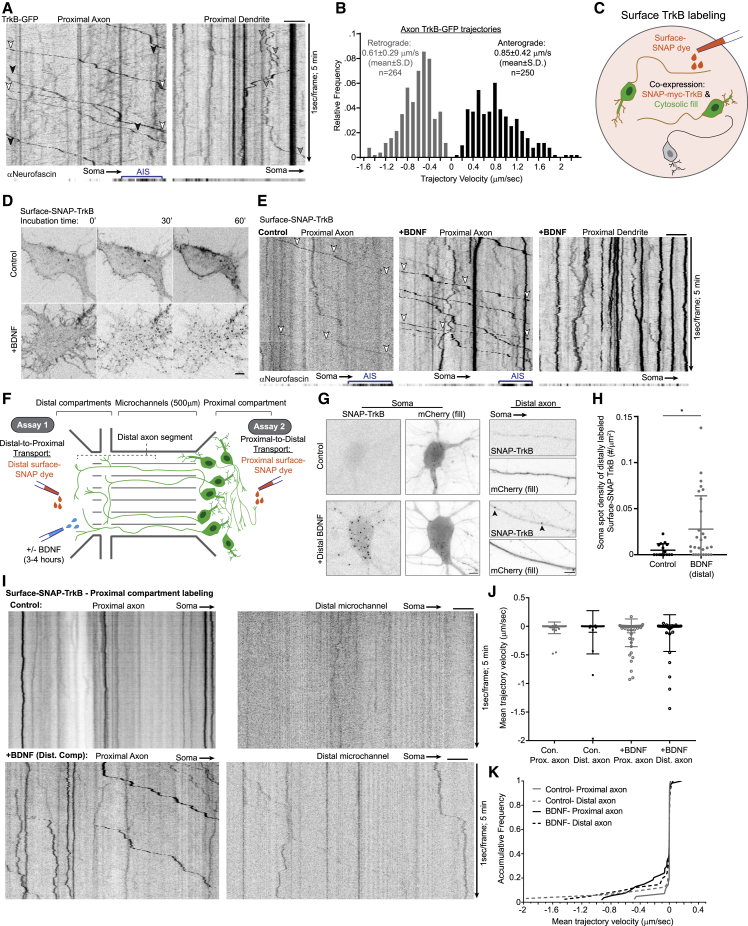
To probe the general transport dynamics of TrkB we performed live imaging of transfected TrkB-GFP in hippocampal neurons axon and dendrites. At steady state, TrkB carriers in the axon are transported toward the distal axon (anterogradely) as well into the soma (retrogradely), while in dendrites the receptor mobility is slower and with more frequent directional switches (Figure 1A; Video S1). Both the retrograde and anterograde carriers display occasional pauses between fast and processive mobility. During their processive mobility intervals, axonal TrkB-GFP moves at a mean velocity of 0.85 and 0.61 μm/s in the anterograde and retrograde directions, respectively (Figure 1B). The observed bidirectional transport in the axon and the slow dynamics of dendritic transport fit with established models of Trk receptor axonal transport driven by kinesin and dynein motors (Scott-Solomon and Kuruvilla, 2018).
Figure 1.
Axonal transport of internalized TrkB receptors is predominantly retrograde
(A) Kymographs depict bidirectional motility of TrkB-GFP in the proximal axon and dendrite. Neurofascin signal (extracellular antibody) was acquired at the first time point and is projected to localize the AIS. TrkB-GFP signal was acquired at 1 s/frame for 5 min. Examples of processively moving retrograde and anterograde TrkB cargos in the axon are marked by white and black arrowheads, respectively. Example of bidirectional trajectory of TrkB-GFP in the dendrite is marked by gray arrowheads.
(B) Histogram of TrkB-GFP trajectory velocities undergoing processive movement based on the analysis of 514 trajectories from 11 axons collected from 2 independent cultures.
(C) Scheme of surface-labeling setup used in experiments shown in (D) and (E). Neurons expressing SNAP-TrkB were labeled with surface-SNAP dye to follow the localization and transport of TrkB from the plasma membrane.
(D) BDNF induces the internalization and accumulation of cell-surface TrkB in intracellular compartments. Time-lapse images of surface-SNAP-TrkB-labeled cells treated with bath application of control media or media supplemented with BDNF. Images were taken at t = 0, 30, and 60 min after treatment.
(E) Kymographs of SNAP-TrkB motility in the proximal axon and dendrites. Neurons were pulsed labeled with surface-SNAP then treated with BDNF or control media, then imaged at a time window of 45–90 min later. Neurofascin is imaged and presented as in (A). White arrowheads mark retrograde transport of internalized TrkB.
(F) Scheme of compartmental microfluidic chamber (MFC) setup used in experiments shown in (G) and (H) depicted under (1) and (I–K) under (2). Neurons were plated in the proximal compartment, with their axon projecting into the distal compartment. Surface-SNAP labeling was carried specifically in either the proximal or the distal axon compartment to follow axon-to-soma or soma-to-axon transport. Control or BDNF supplemented media was added in the distal compartment.
(G) BDNF induces somatic accumulation of retrogradely transported TrkB. Axons of SNAP-TrkB-expressing neurons were surface labeled in the distal axon and treated with either control or BDNF supplemented media for 2 h in the distal axon then fixed. mCherry was used to label neuron morphology. Arrowheads mark SNAP-TrkB puncta appearing in the distal axon in response to BDNF.
(H) Scatter plot of mean ± SEM of density of SNAP-TrkB spots counted in somas with axons crossing into the distal compartment after surface-SNAP labeling in the distal compartment. ∗Student’s t test, p < 0.05, n = 16 and 28 somas, from 6 and 13 independent MFC for control and BDNF groups, respectively)
(I) Kymographs of surface-labeled SNAP-TrkB mobility along proximal and distal axon segments. Neurons cultured in MFC and transfected with SNAP-TrkB were surface labeled in the proximal compartment, followed by treatment with either control or BDNF media in the distal compartment for 2–4 h before live imaging.
(J) Scatter plot of mean ± SEM trajectory velocity measured for individual SNAP-TrkB trajectories.
(K) Cumulative frequency plots of mean trajectory velocity. Data in (I–K) are based on a pool of 42, 32, 54, and 41 trajectories from 6, 7, 8, and 10 axon segments for proximal control, distal control, proximal +BDNF proximal and distal +BDNF conditions, respectively, collected from 5 and 4 independent MFC cultures for control and +BDNF conditions, respectively. All scale bars, 5 μm.
Part A: live confocal imaging of a hippocampal neuron (DIV8) proximal axon expressing TrkB-GFP. The AIS is immunolabeled with antibody against cell-surface Neurofascin (shown in red overlay).
Part B: live confocal imaging of a hippocampal neuron (DIV8) dendrites expressing TrkB-GFP. The AIS of neighboring neurons are shown by neurofascin immunolabeling. Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Internalization of plasma membrane Trk receptors and subsequent endosomal sorting is coupled to receptor transport (Cosker and Segal, 2014). To understand how internalized TrkB receptors are trafficked and transported, we transfected neurons with myc-SNAP-tagged TrkB and validated its proper intracellular and surface expression using myc immunolabeling (Figure S1A).We then used the surface-SNAP tag system to follow receptor trafficking and transport in live neurons (Figure 1C). Addition of the TrkB ligand BDNF induced the internalization of surface-labeled SNAP-TrkB and its accumulation in intracellular puncta in the course of 30–60 min (Figures 1D, S1B, and S1C). Internalized TrkB is transported processively in the axon compared with erratic bidirectional mobility in dendrites, which is similar to the difference observed for TrkB-GFP (Figures 1A and 1E). An important difference is that in contrast to TrkB-GFP, the axonal transport of the internalized SNAP-TrkB is exclusively retrograde. BDNF addition enhanced SNAP-TrkB transport in the axon, doing so without altering its overall retrograde bias (Figure 1E; Video S2).
Live confocal imaging of a hippocampal neuron (DIV9) axons expressing surface labeled SNAP-TrkB. The AIS of neighboring neurons are labeled with cell-surface Neurofascin immunolabeling (shown in red overlay). Neurons were treated with BDNF or normal media as control for 60 min after SNAP-Surface labeling then promptly imaged. Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
The selectivity of axonal transport direction prompted us to further investigate the re-distribution of internalized TrkB receptors between the axonal and somatodendritic compartments of the neuron. To do this we utilized microfluidic compartmental chambers (MFCs) to specifically label and manipulate the distal axon and soma compartment. Dissociated hippocampal neurons were plated in the proximal compartment, and due to the length of the microchannels, only axons were able to traverse the distance and reach the distal compartment. We then used the MFC cultures to carry two separate experiments to follow either distal axon-to-soma or soma-to-distal axon re-distribution of internalized TrkB (Figure 1F, assay 1 and assay 2, respectively). As axonal stimulation with neurotrophin is known to activate both axon-to-soma and soma-to-axon transport of Trk receptors (Ascaño et al., 2009; Yamashita et al., 2017), we sought to examine how TrkB re-distribution is affected by application of BDNF specifically to the distal axon. To allow sufficient time for internalization and retrograde transport of axonal TrkB, BDNF incubation in the distal compartment was carried for 2 h immediately after surface-SNAP labeling in the distal axon. In BDNF treatment condition, SNAP-TrkB in the distal axon formed concentrated puncta along the axon and accumulated in the soma (Figures 1G and 1H), while in the control condition SNAP-TrkB remained diffused along the axon and did not accumulate in the soma. Thus, we confirm that in hippocampal neurons axonal TrkB undergoes internalization in distal axons, followed by retrograde transport and accumulation in the soma in response to ligand stimulation, in accordance with the retrograde signaling endosome model.
To follow anterograde re-distribution of TrkB into the axon, we carried surface labeling of SNAP-TrkB in the proximal compartment. BDNF treatment or control media was then promptly added to the distal compartment and the neurons were imaged in a time window of 3–4 h post-labeling. To follow SNAP-TrkB dynamics in the distal axon we imaged a segment of the axon that resided either at the distal compartment or in the microchannel within 100 μm from its distal opening. For the proximal axon we selected segments that were both at a distance of 100 μm from the soma and within the proximal compartment (hence these axonal segments were directly exposed to surface-SNAP labeling). We observed that proximally labeled TrkB receptors are rarely seen in the distal compartment, while in the proximal compartment more puncta can be observed (Figure 1I). Proximally labeled TrkB puncta are mostly immobile throughout the axon, while BDNF stimulation of the distal axon increased their mobility in both the distal and the proximal segments of the axon (Figures 1I and 1J). The mobility profile of proximally labeled TrkB is mainly retrograde in both proximal and distal segments in either control and in 3–4 h BDNF treatment in the distal axon compartment (Figure 1K). When BDNF treatment was carried for 1–2 h, few processive anterograde transport events of proximally labeled SNAP-TrkB were observed (Figure S1D), possibly reflecting an axon-to-soma transcytotic mechanism induced by long-distance neurotrophic signaling as previously described for TrkA (Ascaño et al., 2009). Nevertheless, our results suggest that following TrkB internalization, the endosomal TrkB is primarily sorted for retrograde transport in the axon and retention in the somatodendritic compartment.
Direct axonal trafficking of TrkB from the secretory pathway
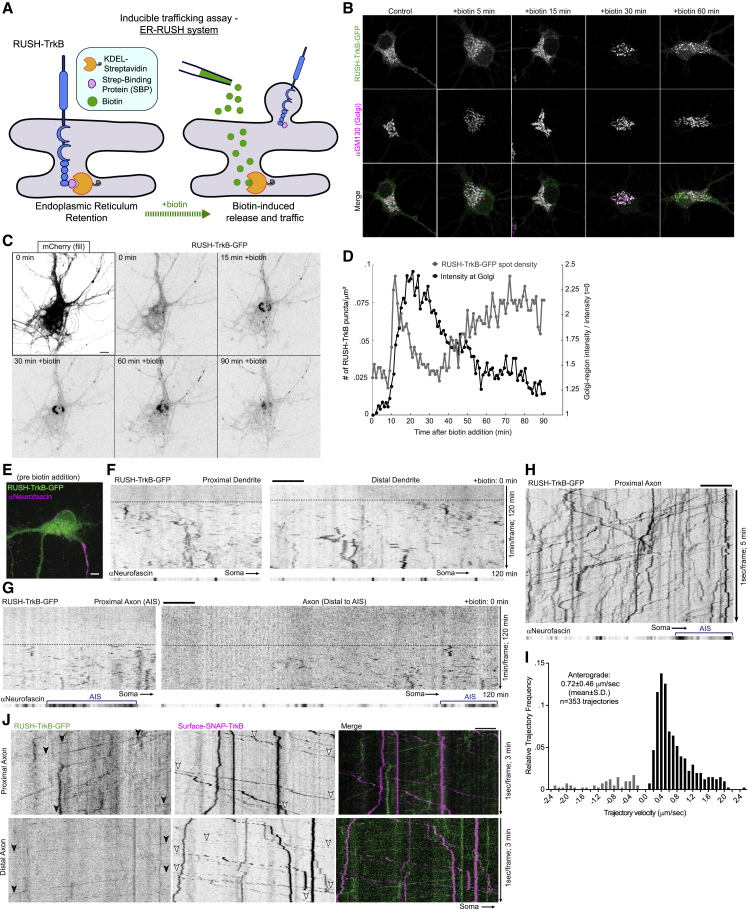
The predominance of retrograde transport of TrkB endosomes in the axon suggests that active delivery of receptors to the distal axon is via a different membrane trafficking pathway. We therefore turned our focus to the secretory pathway as a likely source for the anterogradely transported TrkB cargos. To specifically probe the trafficking and transport of TrkB originating at its site of biosynthesis, i.e., the ER, we used the inducibly released retention system (RUSH) (Boncompain et al., 2012). Briefly, we incorporated GFP- or SNAP-tagged TrkB into the KDEL-streptavidin-IRES-streptavidin-binding protein (SBP) vector. These constructs encode ER-sequestered KDEL-streptavidin and SBP conjugated TrkB tagged with GFP or SNAP (henceforth termed RUSH-TrkB-GFP and RUSH-SNAP-TrkB). Neurons were maintained with neutravidin to sequester biotin and prevent premature release of RUSH-TrkB from the ER, which is controllably induced by the addition of excess biotin (Figure 2A). Incubation with neutravidin for 48 h and treatment with biotin in naive neurons did not induce detectable changes in ER and Golgi morphologies suggesting their secretory system is intact (Figure S1I). Indeed, addition of biotin to media of RUSH-TrkB-expressing neurons induced a shift of the receptor into the peri-nuclear region and its concentration with the cis-Golgi protein GM130 in the soma within 15–30 min after treatment, followed by appearance of smaller TrkB carriers dispersed from the Golgi after 60 min (Figure 2B). Live imaging demonstrates a dynamic pattern in response to biotin: initial formation of ER-to-Golgi carriers, followed by concentration of RUSH-TrkB-GFP in a peri-nuclear region, normally within 10–30 min, then a dispersal from the Golgi concomitant with formation of post-Golgi carriers occurring within 30–45 min (Figures 2C and 2D; Video S3). RUSH-TrkB successfully traffics and fuses with the plasma membrane, as we observe an increase in the cell-surface signal of RUSH-TrkB-SNAP after treatment with biotin for 2.5 h (Figures S1E and S1F).
Figure 2.
Anterograde transport of TrkB into the axon from the secretory pathway
(A) Scheme of the RUSH-based assay used in the experiments shown in this figure. Neurons were transfected with RUSH-TrkB construct encoding a bi-cistronic mRNA for KDEL-streptavidin and an SBP-TrkB with a tag. Temporal control of tagged TrkB-SBP trafficking from the ER is enabled by the addition of biotin which dislodges TrkB-SBP from KDEL-streptavidin, resulting in the release of TrkB for further trafficking.
(B) Images of fixed neurons expressing RUSH-TrkB-GFP and immunostained with GM130 antibody after biotin addition.
(C) Live imaging series of neurons co-expressing RUSH-TrkB-GFP and cytosolic mCherry following biotin addition.
(D) Mean RUSH-TrkB-GFP signal in the peri-nuclear Golgi area (as ratio signal at t = 0) along with the density of discrete RUSH-TrkB-GFP spots in the soma of the neuron shown in (C).
(E) Soma of neuron expressing RUSH-TrkB and stained for neurofascin (AIS) imaged prior to biotin addition.
(F and G) Kymographs of RUSH-TrkB in segments along the axon (F). (G) Kymographs of RUSH-TrkB along a dendritic process. Kymographs in (F) and (G) were based on a single neuron imaged for 2 h after addition of biotin at 1 min/frame. Dashed horizontal bars highlight the onset time in which discrete RUSH-TrkB aggregates appear.
(H) Kymograph of RUSH-TrkB-GFP transport in the proximal axon after biotin-induced release costained with neurofascin. Individual axon segments were imaged at a time window of 45–90 min after biotin for 5 min at 1 s/frame.
(I) Histogram of individual RUSH-TrkB-GFP trajectories velocities in the proximal axon. Data are of 353 trajectories from 20 individual axons from 3 independent cultures.
(J) Kymograph of surface-labeled SNAP-TrkB and RUSH-TrkB-GFP motilities along the same axon segments. Neurons co-expressing SNAP-TrkB and RUSH-TrkB-GFP were labeled with surface-SNAP then stimulated with BDNF to promote TrkB internalization and transport. Biotin was then added, and neurons were imaged for 3 min between 45–90 min later. Black and white arrowheads mark trajectories of RUSH-TrkB-GFP and SNAP-TrkB, respectively. All scale bars, 5 μm.
Part A: live confocal imaging of a hippocampal neuron (DIV8) expressing RUSH-TrkB-GFP. Biotin was added in t = 0. Images were captured at 1 frame per minute.
Part B: live confocal imaging of a hippocampal neuron (DIV12) expressing RUSH-TrkB-GFP. Biotin was added 48 min prior to imaging. The AIS is labeled with cell-surface Neurofascin antibody (shown in red overlay). Images were captured at 1 frame per second. Timeseries are played at 10 frames per second. Scale bar, 5 μm.
To test the functionality of RUSH-TrkB, we transfected human embryonic kidney (HEK) cells that lack endogenous TrkB with RUSH-TrkB-GFP and stimulated them with BDNF to follow downstream extracellular signal-regulated kinases 1&2 (ERK1/2) signaling. Expression of RUSH-TrkB-GFP induced a BDNF independent ERK1/2 phosphorylation that is possibly due to ligand-independent kinase activity by overexpressed TrkB, while treatment with BDNF for 30 min activated a significant increase in ERK1/2 signaling in RUSH-TrkB-GFP cells pre-treated with biotin for 2 h (Figures S1G and S1H). Thus, release of ER sequestration by biotin-induced proper RUSH-TrkB trafficking to the plasma membrane with intact BDNF receptor function.
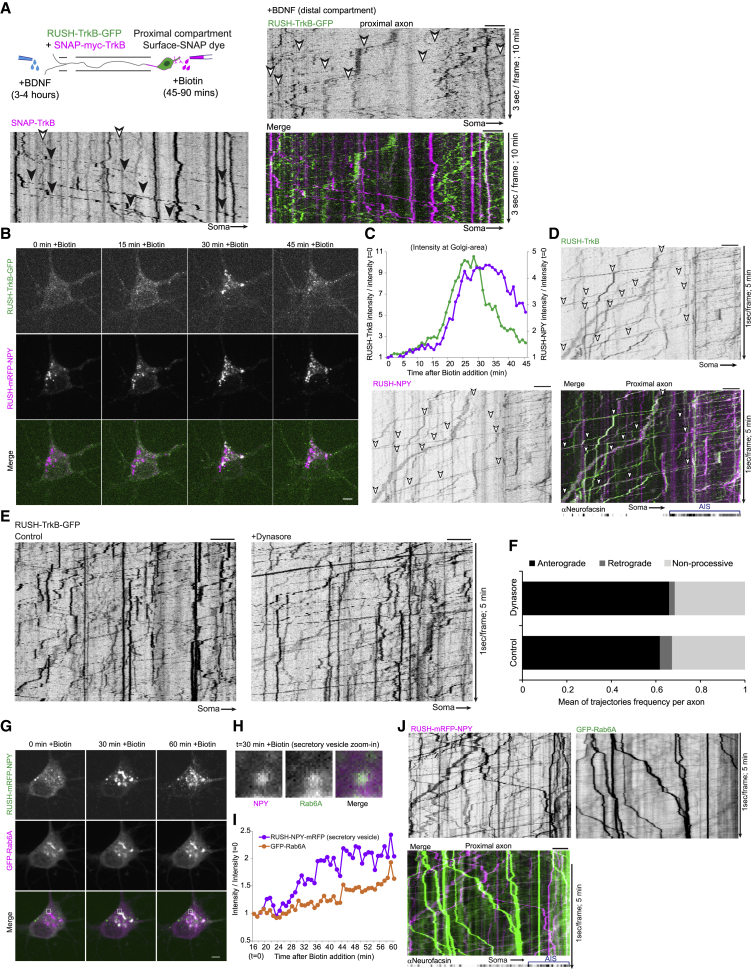
Notably, RUSH-TrkB dynamics differed in axons versus dendrites. Both proximal and distal dendritic segments contained retained RUSH-TrkB before biotin addition and responded with fast re-distribution into discreet spots 20 min after biotin addition (Figures 2E and 2F, onset time is highlighted by dashed bars). Comparatively, axonal signal is minimal before biotin addition, and the appearance of distinct punctate signal in the proximal and distal axon occurs typically after 45–60 min (Figure 2G, dashed bars). These results suggest that the trafficking of TrkB from the ER is restricted to the somatodendritic compartment and that TrkB is transported from the soma in post-Golgi carriers into the axon. Released RUSH-TrkB is transported almost exclusively in the anterograde direction toward the distal axon at a mean velocity of 0.72 μm/s, fitting with regulated motor-based transport mechanism (Figures 2H and 2I; Video S3). We further tested whether endocytosed TrkB can be co-transported with secretory TrkB in the same neuron by co-transfecting SNAP-TrkB and RUSH-TrkB-GFP. After Surface-SNAP labeling, BDNF was added for 30 min to induce TrkB endocytosis, followed by biotin addition. Similar to results shown in Figure 1, the processive mobility of SNAP-TrkB was exclusively retrograde, while the RUSH-TrkB was transported separately in the anterograde direction both at the proximal and distal axon (Figure 2J; Video S4). Separate transport was also seen using the soma-to-axon transcytosis experimental paradigm (Figure 3A), further indicating that TrkB endosomal and secretory trafficking are diametrically regulated.
Figure 3.
TrkB axonal transport from the secretory pathway is endocytosis independent
(A) RUSH-TrkB and SNAP-TrkB transport dynamics in proximal to distal transport of internalized SNAP-TrkB using microfluidic devices. Neurons were transfected with SNAP-TrkB + RUSH-TrkB-GFP as in Figure 2J, and surface-SNAP labeling was applied to the proximal compartment, followed by BDNF addition (distal compartment) and then biotin (proximal compartment). Live imaging of axons in the proximal compartment was carried 3–4 h after BDNF addition at 3 s/frame for 10 min. Kymographs are representative of 2 independent experiments. White and black arrowheads mark anterograde and retrograde transport of SNAP-TrkB or RUSH-TrkB-GFP.
(B and C) Neurons co-expressing RUSH-TrkB-GFP and RUSH-mRFP-NPY were treated with biotin and imaged for 45 min at 1 frame/min.
(C) Representative plot of RUSH-NPY and RUSH-TrkB intensities in the peri-nuclear Golgi region (normalized to t = 0) following biotin addition.
(D) Secreted TrkB and NPY co-transport along the axon.
Neurons co-transfected as in (B and C) and were immunostained for neurofascin to label the AIS. RUSH-TrkB-GFP and RUSH-mRFP-NPY were live imaged during a time window of 45–90 min after addition of biotin for 5 min at 1 frame/s. Kymographs of the proximal part of the axon show the co-transport of secreted NPY and TrkB toward the distal axon.
(E and F) Neurons expressing RUSH-TrkB-GFP were pre-treated with DMSO control or dynasore for 15 min before biotin was added to induce post-ER trafficking.
(E) Proximal axons were imaged at a time window of 45–90 min after biotin for 5 min at 1 s/frame.
(F) Kymograph trajectories were categorized and counted to measure transport dynamics in individual axons. Data are mean ratios of anterograde, retrograde, and non-processive trajectories per axon and are based on 11 (control) and 12 (dynasore) individual axons from 2 independent experiments.
(G) Secreted NPY is trafficked and transported in Rab6 carriers. Neurons were co-transfected with RUSH-mRFP-NPY and GFP-Rab6Aand were imaged during 60 min of biotin treatment.
(H) Zoom-in of a somatic RUSH-NPY puncta co-localized with Rab6A following 30 min of release.
(I) Time plot of NPY and Rab6A intensities in the puncta in (J) during the course of biotin treatment. All scale bars, 5 μm.
Live confocal imaging of a hippocampal neuron (DIV14) co-expressing RUSH-TrkB-GFP (green) and SNAP-TrkB (surface labeled in magenta). Biotin was added 66 min prior to imaging to induce trafficking of RUSH-TrkB-GFP along the secretory pathway and BDNF was added 30 minutes later (36 min before imaging) to induce internalization of surface labeled SNAP-TrkB. Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Further probing of the TrkB sorting was carried by co-expressing ER-retained TrkB and neuropeptide Y (NPY) as a soluble cargo marker, which is also delivered into the axon. RUSH-TrkB and RUSH-NPY concomitantly accumulate in the Golgi upon trafficking induction and are then transported together toward the distal axon in the same carriers (Figures 3B–3D). This affirms that the RUSH-TrkB carriers are sorted directly into the axon without an intermediate exo- and endo-cytotic step, which otherwise would result in the loss of the soluble NPY signal. Indeed, pre-incubation with dynasore, an inhibitor of dynamin-dependent endocytosis, did not affect RUSH-TrkB secretory trafficking and transport in the axon (Figures 3E and 3F). We therefore conclude that TrkB undergoes direct transport from the secretory system to the axon.
TrkB trafficking to the axon is regulated by the Rab6 GTPase
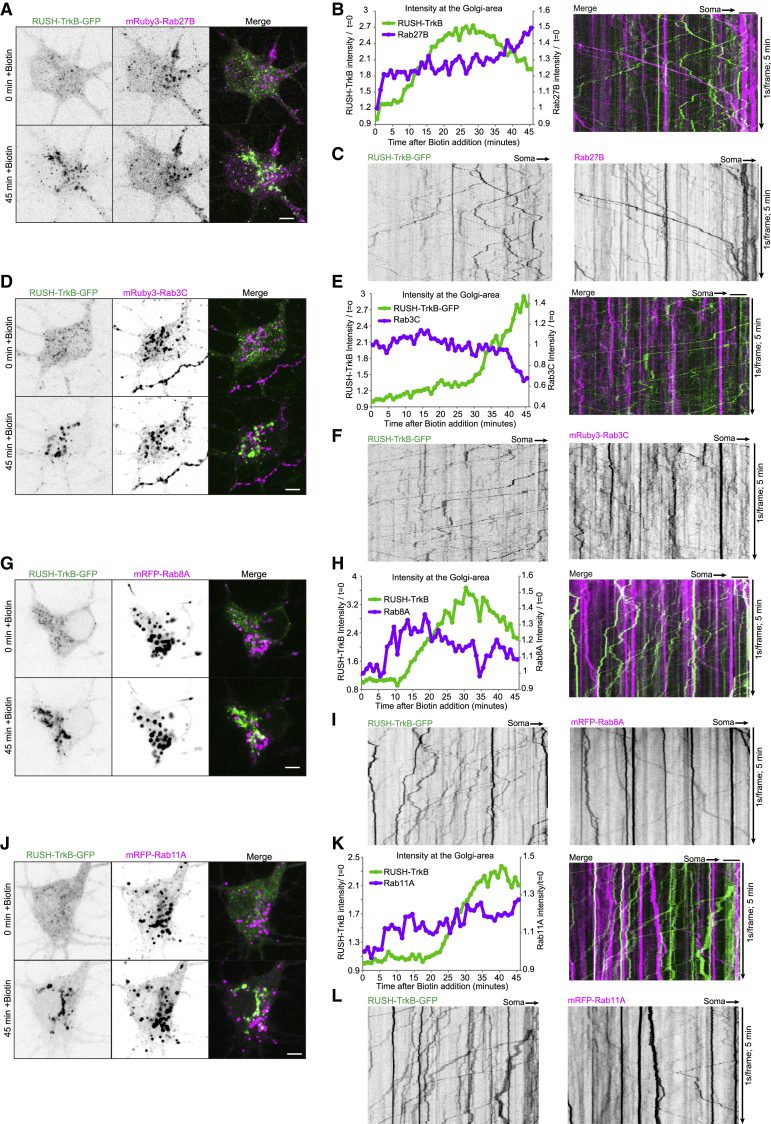
Direct trafficking of TrkB from the secretory pathway to the axon requires sorting of the receptor into anterograde-bound membrane compartment. Rab GTPase are master regulators of membranal compartment sorting and trafficking in the endosomal and secretory systems (Pfeffer, 2017; Stenmark, 2009). We decided to test the association of secretory TrkB carriers with Rab27B, Rab3C, Rab8A, Rab6A, and Rab11A as markers for membranal compartments that were associated with anterograde axonal transport or with plasma membrane delivery of secretory pathway carriers (Arimura et al., 2009; Ascaño et al., 2009; Koseki et al., 2017; Wakana et al., 2012). For this, we co-transfected RUSH-TrkB-GFP with labeled Rab proteins.
While neither Rab27B, Rab3C, Rab8A nor Rab11A co-localized with RUSH-TrkB (Figure 4), we found that RUSH-TrkB and Rab6A co-localized to a high extent in the peri-nuclear concomitant with RUSH-TrkB accumulation in the Golgi area after biotin addition (Figure 5A; Video S7). Similar co-localization of RUSH-TrkB was observed with endogenous Rab6 (Figure S2A). The spatiotemporal accumulation of RUSH-TrkB in the Golgi region is correlated with the recruitment of wild-type Rab6A, but not with GDP-locked Rab6A-T27N mutant (Figure 5B). We found that Rab6A and RUSH-TrkB are accumulating together in discrete puncta also outside the Golgi area (Figures 5C and 5D, highlighted by yellow box). Accordingly, we found that a major fraction of anterograde RUSH-TrkB carriers were co-transported with Rab6A toward the distal axon (Figure 5E, depicted by arrowheads; Video S5), while no co-transport was observed with Rab27B, Rab3C, Rab8A, or Rab11A (Figure 4). The co-transport of RUSH-TrkB and Rab6A is dependent on the GTP-binding state of Rab6A, as both wild-type and GTP-locked Rab6A-Q72L mutant co-localized with RUSH-TrkB, while the GDP-locked Rab6A-T27N mutant did not (Figures 5E and 5F). Rab6A was also recruited to and transported with RUSH-NPY carriers in the proximal axon (Figures 5G–5I), suggesting that Rab6A regulates post-Golgi trafficking and axonal sorting in neurons. Importantly, we observe a distinct co-transport of wt-Rab6A with anterogradely transported TrkB-GFP, but not with retrograde TrkB-GFP (Figures S2C and S2D). Thus, Rab6 likely has a specific role in regulating anterograde axonal trafficking of TrkB, as well as confirming that the mechanisms of TrkB axonal trafficking are not altered by the RUSH system.
Figure 4.
Anterogradely transported RUSH-TrkB carriers are negative for Rab27B, Rab3C, Rab8A, and Rab11A
(A–L) Neurons co-expressing RUSH-TrkB-GFP and labeled Rab proteins: (A–C) mRuby3-Rab27B, (D–F) mRuby3-Rab3C, (G–I) mRFP-Rab8A, and (J–L) mRFP-Rab11A. Neurons were live imaged in the soma area immediately after addition of biotin for 45 min at 1 frame/min (A, D, G, and J), and then we imaged for 5 min at 1 s/frame at the proximal axon during a time window of 45–90 min after addition of biotin (C, F, I, and L). Plots show RUSH-TrkB and Rab proteins intensities in the peri-nuclear Golgi region over time in representative neurons (B, E, H, and K). Kymographs of RUSH-TrkB-GFP and respective Rab proteins motility in individual axons (C, F, I, and L).
Figure 5.
Trafficking and anterograde transport of secreted TrkB in Rab6-positive compartments
(A–D) Neurons co-expressing RUSH-TrkB-GFP and mCherry-Rab6A-WT or Rab6A-T27N were imaged in the course of 45 min after addition of biotin at 1 min/frame.
(A) Time series of a soma in a neuron following addition of biotin. Arrowheads mark RUSH-TrkB and Rab6A co-localization in the Golgi region.
(B) Comparative intensity plots of RUSH-TrkB and Rab6A-WT or Rab6A-T27N in the peri-nuclear Golgi region (normalized to t = 0) in two separate neurons.
(C) A zoom-in of RUSH-TrkB and Rab6A in discrete tubular (upper images) and punctate (lower images) compartments, taken from the time series shown in (A). Scale bar, 2 μm.
(D) Time plot of the intensity of RUSH-TrkB and Rab6A at a somatic punctum shown in (C) following its formation and up until 45 min after addition of biotin.
(E and F) Neurons co-expressing RUSH-TrkB and indicated Rab6A constructs were immunostained against neurofascin to label the AIS, and time series were captured at a time window of 45–90 after addition of biotin, for 5 min each at 1 frame/s.
(E) Kymographs of RUSH-TrkB and Rab6A motility in proximal axons. White and black arrowheads mark Rab6A-positive (WT and Q72L) and -negative (T27N) RUSH-TrkB carriers.
(F) Ratios of anterograde TrkB traces co-transported with the indicated Rab6A mutants. Lines indicate mean and SEM. ∗∗∗Two-sided Student’s t test p < 0.001, n = 12, 11, and 13 axons of Rab6A-WT, Rab6A-T27N and Rab6A-Q72L conditions, respectively, from 3 independent cultures each.
(G–I) Neurons were transfected with RUSH-TrkB-GFP and either vector (pSuper) or shRNA constructs targeting Rab6A and Rab6B.
(G) Live image series of RUSH-TrkB-GFP in the soma following biotin addition. Neurons were manually categorized as secreting, frustrated, or blocked based on the profile of TrkB secretion in response to biotin in the course of 45 min.
(H) Time plot of RUSH-TrkB intensity in the Golgi region (normalized to t = 0) of neurons expressing either pSuper or shRab6 from different categories in (G).
(I) Frequencies of neurons categorized as secreting, frustrated or blocked. ∗∗Chi-square test, p < 0.01, n = 26 and 25 neurons for pSuper and shRab6 conditions, respectively, from 4 independent cultures each. Unless otherwise stated, all scale bars, 5 μm.
Part A: live confocal imaging of a hippocampal neuron (DIV10) co-expressing RUSH-TrkB-GFP (green) and mCherry-Rab6a. Biotin was added in t = 0. Images were captured at 1 frame per minute. Part B: live confocal imaging of a hippocampal neuron (DIV10) co-expressing RUSH-TrkB-GFP (green) and mCherry-Rab6a (magenta). Biotin was added 61 minutes prior to imaging. The AIS is labeled with cell-surface Neurofascin antibody (shown in red overlay). Images were captured at 1 frame per second and are played at 10 frames per second. Scale bar, 5μm.
To effectively disrupt Rab6 functions we depleted both Rab6A and Rab6B proteins in neurons using shRNA (Figure S2A). Depletion of Rab6 disrupted RUSH-TrkB trafficking, causing either a block in the Golgi following the early phase of ER-to-Golgi re-distribution (termed “blocked”) or a retrograde flow from condensates at the Golgi back to diffuse ER signal (termed “frustrated”) (Figures 5G and 5H; Video S6). The ratio of neurons displaying successful, frustrated, or blocked secretion process after 45 min of biotin treatment shows that Rab6 function is required for efficient formation of TrkB secretory carriers (Figure 5I). As a previous study suggested the role of Rab27B in anterograde axon transport of TrkB (Arimura et al., 2009), we sought to re-examine whether it plays a regulatory role in TrkB trafficking and transport using shRNA against Rab27A and Rab27B. In agreement with our observation that secretory pathway derived TrkB is not associated with Rab27B and in contrast with Rab6 depletion, we found no effect of Rab27 knockdown on secretory trafficking and axon transport of released RUSH-TrkB (Figures S2E–S2G).
Live confocal imaging of a hippocampal neurons (DIV9-10) co-expressing RUSH-TrkB-GFP and either pSuper control or shRab6a/b vectors demonstrating proper trafficking (pSuper) and frustrated and blocked trafficking (shRab6a/b). Biotin was added in t = 0. Images were captured at 1 frame per minutes and are played at 7 frames per second. Scale bar, 5 μm
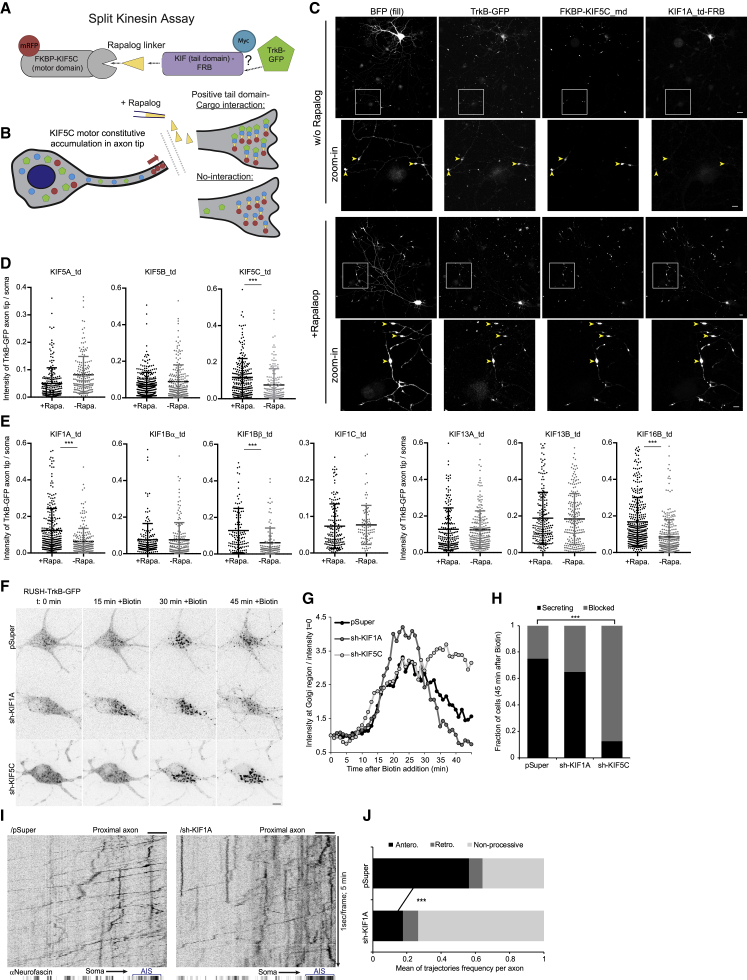
KIF5C and KIF1A regulate formation and axonal transport of TrkB carriers
Kinesin-1 and kinesin-3 families are the major drivers of anterograde axonal transport of various cargos (Maday et al., 2014). For a non-biased identification of specific kinesins that interact with TrkB, we applied an imaging-based split-kinesin assay previously described (Jenkins et al., 2012). We co-transfected hippocampal neurons with FRB-conjugated kinesin cargo-binding tail domain tagged with Myc as bait (KIF-td-FRB), TrkB-GFP as the probed cargo and an mRFP-tagged KIF5C motor domain fused to FKBP (KIF5C-md-FKBP). In this assay, association between kinesin tail domain and TrkB-GFP results in accumulation of the receptor in axon tips following rapalog-induced FKBP-FRB coupling (Figures 6A and 6B). We initially tested TrkB-GFP distribution in neurons expressing KIF1A-td-FRB and KIF5C, observing TrkB-GFP enrichment in the axon tips following rapalog addition (Figure 6C). We therefore systematically tested kinesin tail domains interaction with TrkB by quantifying the ratio between mean TrkB-GFP signal at the axon tip and its respective soma for different kinesin tail domains and thus identified significant TrkB interaction for the KIF5C kinesin-1 motor (Figure 6D) and for the kinesin-3 motors: KIF1A, KIF1Bβ, and KIF16B (Figure 6E). We also tested several kinesin-2 and kinesin-4 family members, observing a positive interaction of TrkB and the tail domain of kinesin-4 KIF4B (Figure S3A). To validate our measurements of TrkB-GFP accumulation, we confirmed that upon addition of rapalog all the KIF tail domains examined were successfully recruited to the axon tips (Figure S3B). Notably, while some KIF-td constructs were expressed at lower amounts relative to TrkB-GFP in comparison with others (Figure S3C), these differences could not explain positive or negative interaction results with TrkB-GFP—e.g., KIF5A-td and KIF5B-td both were expressed efficiently but did interact with TrkB, while KIF13B-td and KIF1A-td were similarly expressed, however, only KIF1A-td showed clear interaction with TrkB (Figures 6D and 6E).
Figure 6.
The kinesins KIF5C and KIF1A regulate the axonal trafficking of TrkB
(A and B) Schemes of the split-kinesin assay (SKA) used in the experiments presented in (C–E).
(A) The components of the SKA with TrkB-GFP as cargo reporter.
(B) Rapalog addition induces the mobilization of the kinesin tail domain and accumulation at the axon tips due to tight interaction with the constitutively motile motor KIF5C. Interaction between the kinesin tail domain and the TrkB cargo will result in accumulation of TrkB in the axon tips together with the KIF-td and KIF5C-md where rapalog is added.
(C–E) Neurons were transfected with SKA components described in (A) and treated either with control (−Rapa) or rapalog treatment (+Rapa) for 16–24 h before fixation, followed by immunostaining for myc-tagged KIF-td.
(C) Images of the soma and axon tips (highlighted in zoom-in insets) of neurons assayed for TrkB-KIF1A interaction with and without rapalog addition. Scale bars, 10 and 5 μm (insets).
(D) SKA results for TrkB-GFP and Kinesin-1 family. (E) SKA results for TrkB-GFP and Kinesin-3 family. Data in (D–E) are TrkB-GFP signal in individual axon tips, normalized to the signal in the soma of each axon tip. Lines depict mean and SD. ∗∗∗Two-sided Student’s t test, p < 0.001. Data points were collected from the following number of axon tip: KIF5A: 221 and 181; KIF5B: 303 and 214; KIF5C: 239 and 176; KIF1A: 270 and 219; KIF1Bα: 167 and 162; KIF1Bβ: 118 and 110; KIF1C: 158 and 86; KIF13A: 187 and 230; KIF13B: 204 and 208; and KIF16B: 357 and 288 for +Rapa and −Rapa conditions, respectively.
(F–H) Neurons co-transfected with RUSH-TrkB-GFP and shRNA constructs against KIF1A or KIF5C were treated with biotin and RUSH-TrkB-GFP at the soma was live imaged for 45 min at 1 frame/min.
(F) Time series images of representative neurons expressing RUSH-TrkB-GFP with specified shRNA constructs.
(G) Time plots of RUSH-TrkB intensity at the Golgi (normalized to t = 0) of neurons shown in (F).
(H) Frequency of neuron categorized as secreting or blocked in response to 45 min of biotin. ∗∗∗Chi-square test p < 0.001, n = 28, 20, and 16 neurons from 5, 3, and 3 experiments for pSuper, sh-KIF1A, and sh-KIF5C conditions, respectively.
(I and J) Neurons expressing RUSH-TrkB-GFP and pSuper or shKIF1A were stained for neurofascin (AIS). Time-lapse imaging was carried in a time window of 45–90 min after addition of biotin. Images series were captured at 1 frame/s for 5 min each.
(I) Kymographs of proximal axon segments of representative neurons.
(J) Frequency of trajectory directionality in the proximal segment of the axon. Trajectories with net displacement of less than 15 μm in the course of 5 min were counted as non-processive. The processive traces were classified based on their directionality as either retrograde or anterograde. Bars depict mean ratio of trajectories per each time series (axon). ∗∗∗Two-sided Student’s t test (anterograde fraction) p < 0.001, n = 12 and 10, based on 234 and 180 trajectories of pSuper and sh-KIF1A conditions, respectively, from 3 independent experiments. Scale bars, 5 μm.
To validate the identified TrkB-kinesin interactions we carried a streptavidin pull-down in HEK cells by co-expressing Bio-tagged kinesin tail domain (Bio-Kif-td), BirA biotin ligase, and TrkB-GFP. In accordance with the split-kinesin-positive interaction, Bio-KIF5C-td and Bio-KIF1A-td efficiently interacted with TrkB-GFP (Figure S4A). KIF1Bβ and KIF16B showed weak and barely detectable TrkB binding, respectively, while KIF5A, KIF5B, and KIF21B co-precipitated with TrkB but did not associate with the receptor in the neuronal split-kinesin assay (Figures 6D, 6E, and S4A). To validate and further examine the interactions between KIF5C, KIF1A, and TrkB we carried a mass-spectrometry-based proteomic analysis using Bio-Kif-td as bait. We probed the interactome of Bio-KIF5C-td, Bio-KIF1A-td in HEK cells either expressing TrkB-GFP or GFP alone and used Bio-KIF17-td in TrkB-GFP expressing cells as a negative control, as KIF17-td did not interact with TrkB according to both split-kinesin and pull-down assays. The full list of detected proteins in the different pull-down conditions and their peptide spectrum matches (PSM) value is presented in Table S1. In accordance with the split-kinesin and western blot assays, we find higher spectral counts and fold-enrichment of TrkB peptides (PSM) in KIF1A and KIF5C compared with KIF17 and normalized to GFP-bait control (Figures S4B and S4C). Expected interactors of Bio-KIF5C-td and Bio-KIF1A-td, respectively, e.g., MAP7-family proteins and calmodulin were unaffected by TrkB co-expression. An additional proteomic analysis of KIF5C, KIF1A, and KIF16B tail domain baits confirmed that KIF5C- and KIF1A-specific interactors were not altered by TrkB co-expression (Figure S4D; Table S1).
To further test the function of KIF5C and KIF1A in the anterograde axonal trafficking of TrkB, we used shRNA to deplete either motor in neurons co-transfected with RUSH-TrkB-GFP (Figure 6F). Depletion of either KIF5C or KIF1A did not disrupt the initial ER-to-Golgi trafficking of released RUSH-TrkB, which accumulated in the peri-nuclear region similar to the control. Nevertheless, KIF5C depletion blocked the dispersal of RUSH-TrkB carriers from the Golgi area into discrete carriers, while depletion of KIF1A did not affect ER-to-Golgi and post-Golgi trafficking, which was similar to control (Figures 6G and 6H; Video S7). While vesicular carriers of RUSH-TrkB formed normally in KIF1A-depleted neurons, their anterograde transport in the proximal axon was inefficient, evident by a reduction in the ratio of processive anterograde-bound vesicles, and more pausing and bidirectional trajectories (Figures 6I and 6J; Video S8). Altogether these data show that the kinesin-1 motor KIF5C and kinesin-3 motor KIF1A co-operate in the formation and transport of TrkB carriers from the secretory compartment into and along the axon.
Live confocal imaging of a hippocampal neurons (DIV10) co-expressing RUSH-TrkB-GFP and shRNA against Kif5c (Part A) or shRNA against Kif1a (Part B). Biotin was added in t = 0. Images were captured at 1 frame per minutes and are played at 7 frames per second. Scale bar, 5 μm.
Live confocal imaging of a hippocampal neurons (DIV9-10) co-expressing RUSH-TrkB-GFP and shRNA against Kif1a (Part A) or pSuper control vector (Part B). Biotin was added 48 min prior to imaging. The AIS is labeled with cell-surface Neurofascin immunolabeling (shown in red overlay). Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Discussion
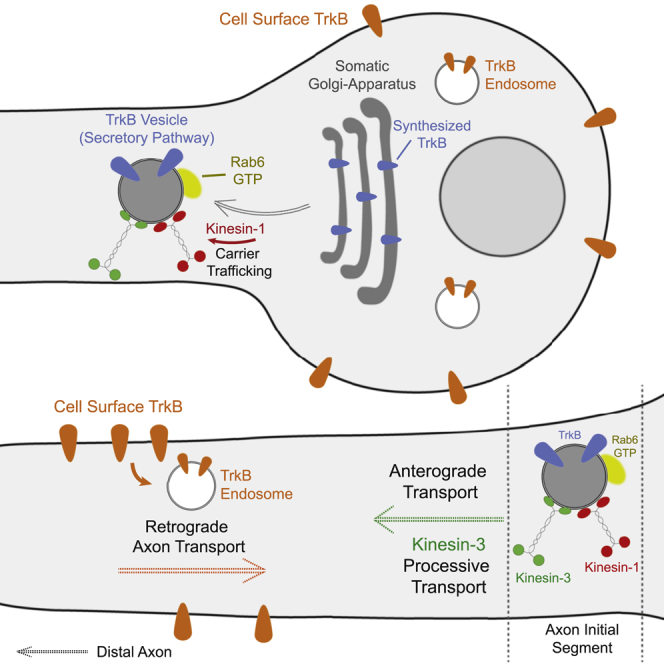
In this study, we visualized TrkB receptor trafficking and transport in hippocampal neurons and describe the molecular mechanisms that control its delivery in the axon. We showed that endocytosed TrkB undergoes retrograde transport in the axon, in accordance with the established model of the neurotrophin signaling endosome (Cosker and Segal, 2014). We also found that anterogradely transported TrkB is not derived from endosomal compartments. Instead, TrkB is sorted directly from the Golgi and secretory pathway to the axon in Rab6 carriers. The formation and anterograde transport of these TrkB-positive carriers depend on kinesin-1 KIF5C and kinesin-3 KIF1A motor proteins.
Direct versus indirect trafficking of Trk receptors into the axon
TrkA receptors were previously shown to take an indirect, transcytotic route in SCG neurons from somatodendritic membrane to the distal axon via recycling endosomes (Ascaño et al., 2009; Yamashita et al., 2017). In comparison, we found that the transport of endocytic TrkB in the axons of hippocampal neurons is almost entirely retrograde and that BDNF stimulation in the axon enhances the overall transport flux without affecting this retrograde bias. Interestingly, soma-to-axon transport of internalized TrkB was more apparent after short BDNF stimulation of the distal axon, suggesting that long-distance signaling induces TrkB transcytosis, at least to some extent. Polarized sorting of endosomal cargos in the axon may differ between types of neurons and during their maturation, which may explain the sparsity of anterograde transcyctotic transport of TrkB in hippocampal axons compared with that of TrkA in SCG neurons. In DRG and SCG neurons Rab11-positive recycling endosomes are transported into the axon and regulate insertion of membrane proteins in the growth cone (Ascaño et al., 2009; Eva et al., 2010). In contrast, CNS neurons display a shift in Rab11 trafficking dynamics during neuronal development, from localization throughout the neuron in early stages to a somatodendritic-specific localization as neurons mature (Eva et al., 2010). These differences in endosomal trafficking mechanisms merit further investigation, as they may underlie differences in axon growth and regeneration capacity in the PNS versus CNS in developing and mature neurons. (Fawcett, 2020).
Axonal trafficking of proteins in the secretory pathway
TrkB is a glycoprotein membrane receptor and as such undergoes N-linked glycosylation in the ER and subsequent maturation in the Golgi before it is transported to the plasma membrane (Haniu et al., 1995; Rajagopal et al., 2004; Watson et al., 1999). Accordingly, here, we show that TrkB’s axonal trafficking follows a canonical course within the secretory pathway. The receptor leaves the ER and re-distributes in discrete compartments in dendrites and soma, which is followed by its transition into the somatic Golgi. The immediate re-distribution into discrete puncta in dendrites prior to as well as concomitant with accumulation in the Golgi suggests that post-ER trafficking of TrkB takes place in neurites—however, we did not assess whether these events represented Golgi-independent trafficking to the plasma membrane as has been demonstrated for AMPAR and GABABR in dendrites (Bowen et al., 2017; Valenzuela et al., 2014). Interestingly, in a screen for atypically glycosylated membrane proteins, TrkB was found to also be inserted into the plasma membrane without undergoing Golgi-dependent trimming of its glycan chains, suggesting that it can take a Golgi-independent route to the plasma membrane via ER-to-plasma membrane carriers such as Rab11 (Bowen et al., 2017; Hanus et al., 2016). While this trafficking route is mostly linked to local synthesis and trafficking in dendrites, it was also speculated that a similar Golgi-bypass route could be utilized for local biosynthesis of certain membrane proteins in axons (González et al., 2018). Nevertheless, considering that in CNS neurons the secretory organelles (i.e., ribosomal ER, ERGIC, and Golgi) are confined to the somatodendritic domain and the sparse evidence for local membrane protein synthesis, it seems likely that the vast majority of the membrane protein cargos designated to the distal axon originate from the somatodendritic-localized secretory compartments and are transported there by microtubule-based motors, as our data show in the case of TrkB (Farías et al., 2015; González et al., 2018; Krijnse-Locker et al., 1995; Maday et al., 2014).
Motor-cargo interactions for anterograde axonal transport of TrkB
Here, we found that the Rab6 small GTPase regulates the formation of post-Golgi TrkB carriers and that its GTP-binding state controls their formation and subsequent transport into the distal axon. While early studies ascribed Rab6 to regulate retrograde Golgi-to-ER trafficking (Martinez et al., 1994; White et al., 1999), more recent studies established its role in the exocytic pathway (Fourriere et al., 2019; Grigoriev et al., 2007, 2011; Schlager et al., 2010). Our data suggest that Rab6 is a master regulator of trafficking in the secretory pathway, particularly for post-Golgi axon-bound cargos. Rab6 has been shown to interact with both anterograde and retrograde motor proteins (Lee et al., 2015; Matanis et al., 2002). In HeLa cells, KIF5B kinesin-1 motor was described to drive Rab6 vesicle transport from the Golgi to the plasma membrane (Grigoriev et al., 2007). Here, we found that the formation post-Golgi Rab6-positive vesicles containing TrkB is dependent on kinesin-1 motor KIF5C. Kinesin-1 was previously shown to be critical for TrkB anterograde transport in hippocampal neurons via a Rab27B-dependent mechanism (Arimura et al., 2009). However, we did not find Rab27B to associate nor to regulate TrkB direct transport of TrkB into the axon. This discrepancy could result from the use of more mature DIV7-11 neurons in our experiments, compared with DIV3 neurons, whose Ftial segment (AIS) is still maturing and therefore could differ in their axonal sorting mechanisms in which Rab27B participates. Also, we used inducible release RUSH system to focus on TrkB transport directly from the secretory pathway, which we identified as the prominent mechanism for axonal delivery of TrkB, while they had imaged the total TrkB signal, which may represent different endomembranal compartments whose anterograde axon transport is regulated by Rab27B. Here, we show that KIF5C is required for the formation of discrete secretory TrkB compartments but is yet not sufficient to drive TrkB transport beyond the initial part of the proximal axon. Our data indicate that TrkB trafficking to the distal axon also depends on KIF1A motor activity. We suggest that distinct kinesin family members differentially regulate axonal TrkB trafficking. Interestingly, a recent study dissected the collaborative roles of the kinesin-1 and kinesin-3 pair KIF5B and KIF13B in peripheral transport of secretory Rab6 carrier in HeLa cells (Serra-Marques et al., 2020). A previous work from our lab described a mechanism for selective cargo axon entry in DRG neurons that is based on the coordinated action of the coordinated action of kinesin-1 and kinesin-3 motors at the pre-axonal filtering zone (Gumy et al., 2017). That study showed that the microtubule-associated protein MAP2, which localizes at the pre-axonal zone, inhibits microtubule binding of kinesin-1 but not of KIF1 motors, hence, requiring association of KIF1 motors on the cargo for its proper transport into the axon. While the structures of the pre-axonal zone and the initial segment of DRG and hippocampal neurons are altogether different, our two studies suggest that the cooperative action of kinesin-1 and kinesin-3 motors is a general mechanism for axonal delivery. KIF1A was recently shown to have weaker binding to GTP-tubulin that is needed for KIF1A synaptic vesicle precursor cargo release at pre-synaptic sites, which are rich in GTP-tubulin, dynamic microtubules (Guedes-Dias et al., 2019). It is intriguing to speculate that an opposite mechanism takes effect in the AIS, by which stable microtubules attract KIF1A binding and processive transport.
In summary, our study proposes a model for the anterograde transport of TrkB by direct trafficking from the biosynthetic system to the axon in Rab6 carriers that is driven by the mutual activity of two kinesin motors: kinesin-1 KIF5C, which is required for the trafficking and transport of secretory carriers, and kinesin-3 KIF1A that facilitates passage through the AIS and processive transport along the axon.
STAR★methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-Neurofascin (extracellular domain) | NeuroMab/Antibodies Incorporated | Cat# 75–172; RRID: AB_2282826 |
| Rabbit polyclonal anti-TRIM46 | Synaptic Systems | Cat. # 377 003; RRID: AB_2631232 |
| Mouse monoclonal anti-GM130 | BD Bioscience | Cat. # 610823; RRID: AB_398142 |
| Rabbit polyclonal anti-RTN4a (Nogo) | Novus Biologicals | Cat. # NB100-56681; RRID: AB_838641 |
| Mouse monoclonal anti-ERK1/2 (p44/42 MAPK) | Cell Signaling | Cat. # 4696; RRID: AB_390780 |
| Rabbit polyclona anti-phospho-ERK1/2 (phospho-Thr202/Tyr204) | Cell Signaling | Cat. # 9101; RRID: AB_331646 |
| Mouse monoclonal anti-Rab6; Clone: 5B10 | Prof. Angelika Barnekow (Münster University) | N/A |
| Rabbit polyclonal anti-myc | Santa Cruz | Cat. # SC-40; RRID: AB_2857941 |
| Rabbit polyclonal anti-GFP | Abcam | Cat. # ab290; RRID: AB_303395 |
| Goat Anti-Rabbit IgG Antibody, IRDye 800CW Conjugated | LI-COR Biosciences | Cat. # 827-08364; RRID: AB_10793856 |
| Goat Anti-Mouse IgG Antibody, IRDye 680LT Conjugated | LI-COR Biosciences | Cat. # 827-11081; RRID: AB_10795015 |
| anti-mouse Alexa647 | Thermo Fisher | Cat. # A21236; RRID: AB_2535805 |
| anti-rabbit Alexa647 | Thermo Fisher | Cat. # A21245; RRID: AB_2535813 |
| anti-mouse Alexa405 | Thermo Fisher | Cat. # A-31553; RRID: AB_221604 |
| anti-mouse Alexa488 | Thermo Fisher | Cat. # A11029; RRID: AB_138404 |
| anti-rabbit Alexa488 | Thermo Fisher | Cat. # A11034; RRID: AB_2576217 |
| anti-mouse Alexa568 | Thermo Fisher | Cat. # A11031; RRID: AB_144696 |
| anti-rabbit Alexa568 | Thermo Fisher | Cat. # A11036; RRID: AB_10563566 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Recombinant mouse Brain Derived Neurotrophic Factor (BDNF) | Sino Biological | 50240-MNAS |
| Biotin | Sigma | B4639 |
| Dynasore | Sigma | D7693 |
| Rapalog (A/C Heterodimerizer) | Takara Bio | 635056 |
| Lipofectamine 2000 | Thermo Fisher | 11668019 |
| Surface-SNAP-Alexa-488 | NEB | S9129S |
| Surface-SNAP-Alexa-546 | NEB | S9132S |
| Surface-SNAP-Alexa-647 | NEB | S9136S |
| SYLGARD 184 Silicone Elastomer Kit | Dow | N/A |
| Experimental Models: Cell Lines | ||
| Human embryonic kidney 239T (HEK293T) | ATCC | CRL-3216 |
| Experimental Models: Organisms/Strains | ||
| Rat (Wistar) | Janvier | N/A |
| Oligonucleotides | ||
| Rab27A (rat) shRNA targeting sequence: CCAGTGTACTGTACCAGTA | This study, based on (Arimura et al., 2009) | N/A |
| Rab27B (rat) shRNA targeting sequence: CAACATTTCTTTACCGATA | This study, based on (Arimura et al., 2009) | N/A |
| KIF1A (rat) shRNA targeting sequence: CGAATATGCTGACATCTTC | (Kevenaar et al., 2016) | N/A |
| KIF5C (rat) shRNA targeting sequence: TGAGATCACTTGGACAAA | (Pan et al., 2019) | N/A |
| Rab6A (rat) shRNA targeting sequence: CACCTATCAGGCAACAATT |
(Schlager et al., 2010) | N/A |
| Rab6B (rat) shRNA targeting sequence: CACCTACCAGGCAACCATC | (Schlager et al., 2010) | N/A |
| Rab6A mutagenesis primer Q72L-Fw: ACAGCAGGTCTAGAGCGGTTC | This study | N/A |
| Rab6A mutagenesis primer Q72L-Rev: GTCCCATAATTGCAATCGTAC | This study | N/A |
| Rab6A mutagenesis primer T27N-Fw: GTTGGAAAGAACTCTTTGATC ACCAGATTCATGTATGACAG |
This study | N/A |
| Rab6A mutagenesis primer T27N-Rev: GCTTTGCTCCCCCAGGAA | This study | N/A |
| Recombinant DNA | ||
| pGW1-CMV | (Hoogenraad et al., 2005) | N/A |
| pSuper vector | (Schlager et al., 2010) | N/A |
| pBio-GFP | Hoogenraad lab | N/A |
| pBirA | Hoogenraad lab | N/A |
| TrkB-GFP | Prof. Rosalind Segal (Harvard University) | N/A |
| SNAP-myc-TrkB | This study | N/A |
| RUSH-TrkB-GFP | This study | N/A |
| RUSH-SNAP-TrkB | This study | N/A |
| RUSH-mRFP-NPY | This study | N/A |
| GFP-Rab6A | Hoogenraad lab | N/A |
| mCherry-Rab6A | Hoogenraad lab | N/A |
| mCherry-Rab6A-Q72L | This study | N/A |
| mCherry-Rab6A-T27N | This study | N/A |
| mRuby3-Rab3C | This study | N/A |
| mRuby3-Rab27B | This study | N/A |
| mRFP-Rab11A | Hoogenraad lab | N/A |
| mRFP-Rab8A | Hoogenraad lab | N/A |
| mRFP-KIF5Cmd_(aa 1-559)-FKBP | Hoogenraad lab | N/A |
| FRB-myc-KIF5Atd_(aa 375-1032) | Hoogenraad lab | N/A |
| FRB-myc-KIF5Btd_(aa 374-963) | Hoogenraad lab | N/A |
| FRB-myc-KIF5Ctd_(aa 376-955) | Hoogenraad lab | N/A |
| FRB-myc-KIF17td_(aa 376-1028) | Hoogenraad lab | N/A |
| FRB-myc-KIF3Atd_(aa 386-702) | Hoogenraad lab | N/A |
| FRB-myc-KIF1Atd_(aa 395-1698) | Hoogenraad lab | N/A |
| FRB-myc-KIF1Bα_td_(aa 390-1153) | Hoogenraad lab | N/A |
| FRB-myc-KIF1Bβ_td_(aa 390-1770) | Hoogenraad lab | N/A |
| FRB-myc-KIF1Ctd_(aa 395-1103) | Hoogenraad lab | N/A |
| FRB-myc-KIF13Atd_(aa 397-1770) | Hoogenraad lab | N/A |
| FRB-myc-KIF21Atd_(aa 377-1661) | Hoogenraad lab | N/A |
| FRB-myc-KIF21Btd_(aa 410-1624) | Hoogenraad lab | N/A |
| Bio-KIF5Atd_(aa 375-1032) | This study | N/A |
| Bio-KIF5Btd_(aa 374-963) | This study | N/A |
| Bio-KIF5Ctd_(aa 376-955) | This study | N/A |
| Bio-KIF17td_(aa 376-1028) | This study | N/A |
| Bio-KIF3Atd_(aa 386-702) | This study | N/A |
| Bio-KIF1Atd_(aa 395-1698) | This study | N/A |
| Bio-KIF1Bβ_td_(aa 390-1770) | This study | N/A |
| Bio-KIF1Ctd_(aa 395-1103) | This study | N/A |
| Bio-KIF13Atd_(aa 397-1770) | This study | N/A |
| Bio-KIF21Atd_(aa 377-1661) | This study | N/A |
| Bio-KIF21Btd_(aa 410-1624) | This study | N/A |
| Software and Algorithms | ||
| Fiji/ImageJ | NIH | Imagej.net |
| Kymoreslicewide | Dr. Eugene Kathrukha (Utrecht University) | https://github.com/ekatrukha/KymoResliceWide |
| Prism 7.0 | Graphpad | https://www.graphpad.com/scientific-software/prism/ |
| R-Software | R Project | https://www.r-project.org/ |
| Other | ||
| 3-Compartment Microfluidic Device (MFC) mold | Prof. Frederic Saudou (Virlogeux et al., 2018) | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Casper Hoogenraad (c.hoogenraad@uu.nl).
Materials availability
All plasmids generated in this study are available upon request.
Data and code availability
Additional datasets, data analysis and visualization algorithms generated in this study are available upon request.
Experimental model and subject details
Animal handling
Pregnant Wistar Rat dames at day 18 of pregnancy were supplied from Janvier Labs (France) at the same day they were used for preparing neuronal cultures. Handling and euthanization of the animals were performed in compliance with the guidelines for the welfare of experimental animals issued by the Federal Government of the Netherlands, and were approved by the Animal Ethical Review Committee (DEC) of Utrecht University. The hippocampal cultures where prepared from groups of 7-14 embryos of undetermined sex in each independent experiment.
Cell line and primary hippocampal neurons cultures
HEK293T (HEK) cells were maintained in DMEM media supplemented with FBS 10% and Penicillin-Streptomycin 1%. Cultures were kept in humidified incubator at 37°C and CO2 5%. Cells were passaged by vigorous pipetting and re-plated at 1:10 density every 3-4 days and kept at sub-confluent density.
Primary hippocampal neuron culture was prepared from E18 rat brains using a modified protocol based on the procedure of Kaech & Banker (Kaech and Banker, 2006). Dissociated hippocampal neurons were plated on 18mm coverslips coated with poly-L-Lysine (37.5μg/ml) and (laminin 5μg/ml) without a glial feeder layer. For normal cultures on cover slips, 100,000 viable cells were seeded on 18mm coverslip placed in 12-well multiwell plate, resulting in a cell density of 28,000 /cm2. Neurons were maintained in Neurobasal media supplemented with B27 2%, Glutamine 0.5mM, Glutamate 15.6μM and Penicillin-Streptomycin 1% (Complete NBM). Cultures were maintained in humidified incubator at 37°C and CO2 5%.
Method details
Antibodies, probes and reagents
The following antibodies and probes were used in Immunofluorescence (IF) and Western-Blot (WB) assays at the specified dilutions: Neurofascin, IF: 1:250; TRIM46, IF:1:500; GM130, IF: 1:500; RTN4a (NOGO), IF: 1:500; ERK1/2, WB: 1:5000; phospho-ERK1/2, WB: 1:2500; Rab6, IF: 1:200; GFP, WB: 1:10000; myc-tag, IF: 1:200; WB:1:1000. Alexa-dye conjugated secondary antibodies were purchased from Life Technologies/Thermo Fisher and used at 1:250-1:1000 dilution. For WB, secondary antibodies conjugated with IRdye680LT- or IRdye800CW were used at 1:10000 dilution, for detection of biotinylted proteins, Streptavidin-IR680LT (Li-Cor) was used at 1:10000 dilution. Surface SNAP-dyes (New England Biolabs) were dissolved at 2mM concentration in DMSO and was freshly diluted to 1μM for labeling. BDNF was reconstituted at 10μg/ml and was applied to cells at a 50ng/ml concentration. Dynasore was dissolved in DMSO at stock concentration of 60mM and applied at a concentration of 60uM.
Compartmental neuronal culture in Microfluidic Chamber (MFC)
Polydimethylsiloxane (PDMS) microfluidic devices were produced in-house using a mold designed by the Saudou lab for in-vitro cortico-striatal connectivity studies (Virlogeux et al., 2018). Briefly, the MFC was composed of 3 compartments: proximal, intermediate and distal compartments (respectively corresponding to cortical, synaptic and striatal in the original paper). Two arrays of parallel microchannels separated the compartments, a 500μm long microchannel between the proximal and intermediate compartment and a 75μm long between the intermediate and the distal compartment. Microchannels were 5μm high and 3μm wide, thereby permitting neurite extension while blocking the passage of cell somas. MFC devices were attached to 25mm glass coverslips and then coated with Poly-L-lysine and Laminin as specified above. Following hippocampal neuron preparation, 50,000 cells were seeded in the proximal compartment and were maintained in complete NBM. Due to the length of the 500μm channel, only axons could cross it and reach the intermediate and distal compartments. As we used the device to compartmentalize the distal axon from the somatodendritic domains, we refer here to both the intermediate and distal compartments as the distal compartment, and refer to the section of the axon 250μm and further from the proximal opening of the 500μm-long microchannel as the distal axon.
DNA constructs for protein and shRNA expression
pGW-CMV was used as an expression backbone vector unless otherwise specified. shRNA constructs were cloned in pSuper backbone (Brummelkamp et al., 2002). TrkB-GFP was previously constructed by insertion of Rat full-length TrkB CDS (NTRK2) into pEGFP-N1 backbone and gifted by R. Segal (Harvard University). SNAP-myc-TrkB was cloned using Gibson assembly by fusing into pGW1 the following fragments: TrkB signal-peptide, myc-SNAP-tag fusion from myc-SNAP-mGluR2 (gifted by Harold McGillavry, Utrecht University) and the rest of TrkB CDS from TrkB-GFP. RUSH-TrkB-GFP, RUSH-SNAP-TrkB and RUSH-mRFP-NPY were cloned by inserting PCR amplified TrkB-EGFP CDS (from TrkB-GFP), SNAP-TrkB CDS (from SNAP-myc-TrkB) and mRFP-NPY CDS (from previously in-house constructed N1-mRFP-NPY plasmid) into SbfI/XbaI restricted IRES3-STR-KDEL-SBP-EGFP-Ecadherin (Boncompain et al., 2012).
GFP-Rab6A and mCherry-Rab6A were previously cloned in-house by inserting human Rab6A CDS into pGW2-GFP and pGW2-mCherry (modified pGW1) backbones. mCherry-Rab6A-Q72L and mCherry-Rab6A-T27N were constructed by site-directed mutagenesis of mCherry-Rab6A using circular PCR with the following primers: Q72L-Fw: ACAGCAGGTCTAGAGCGGTTC, Q72L-Rev: GTCCCATAATTGCAATCGTAC, T27N-Fw: GTTGGAAAGAACTCTTTGATCACCAGATTCATGTATGACAG and T27N-Rev: GCTTTGCTCCCCCAGGAA.
mRuby3-Rab3C and mRuby3-Rab27B were constructed by cloning Rab3C CDS (from in-house cloned GW1-GFP-Rab3C) and Rab27B CDS (from GFP-Rab27B, Addgene # 89447) into BglII/BamHI restricted mRuby3-Tubulin (Addgene # 74256); mRFP-Rab11A, mRFP-Rab8A was previously constructed in-house by inserting respective human Rab CDS into pGW1-mRFP backbone.
Constructs for Split-Kinesin Assay were prepared as follows: mRFP-KIF5Cmd-FKBP was cloned by fusing mRFP CDS, human KIF5C (aa 1-559) CDS and FKBP sequences in pGW1 backbone. Kinesin-tail domain constructs were prepared by fusing into pGW1 a FRB-3xMyc sequence with respective human kinesin CDS fragments: KIF5A (aa 375-1032), KIF5B (aa 374-963), KIF5C (aa 376-955), KIF17 (aa 376-1028), KIF3A (aa 386-702), KIF1A (aa 395-1698), KIF1Bα (aa 390-1153), KIF1Bβ (aa 390-1770), KIF1C (aa 395-1103), KIF13A (aa 397-1770), KIF13B (aa 443-1826), KIF16B (aa 395-1266), KIF4A (aa 374-1129), KIF4B (aa 374-1234), KIF21A (aa 377-1661) and KIF21B (aa 410-1624). Biotin-tagged kinesin-tail domains were prepared by inserting their sequences, without the FRB-3xMyc sequence into a modified pBio-GFP backbone (previously made in house by addition of E. Coli BirA biotin-ligase recognition sequence into pEGFP-C1 (Schatz, 1993)) resulting in Bio-KIF-td constructs without GFP, FRB and Myc tags. Plasmid for BirA expression was previously formed by subcloning BirA CDS from the original pEV-Puro backbone (De Boer et al., 2003) into a pCI-NEO vector;
shRNA constructs against rat Rab27A and Rab27B were based on published siRNA sequences (Arimura et al., 2009): CCAGTGTACTGTACCAGTA (Rab27A) and CAACATTTCTTTACCGATA (Rab27B), which were inserted into pSuper vector. The shRNA constructs targeting rat transcripts were previously described: shRab6A and shRab6B (Schlager et al., 2010); shKIF5C (Pan et al., 2019) and shKIF1A (Kevenaar et al., 2016).
Transfection of protein expression and shRNA constructs
Hippocampal neurons cultured on coverslips and in MFC were transfected on days in-vitro (DIV) 7-10 post culturing using Lipofectamine. A total of 1.8μg DNA was used for the transfection of each sample (coverslip/MFC). The ratios of the different constructs in the mix were varied depending on the specific assays, which are further detailed in their respective sections. The transfection mix (1.8μg plasmid DNA and 3.3μl Lipofectamine) were mixed in Neurobasal media at a total volume of 200μl or 25μl per coverslip or MFC respectively. Neurons in coverslips were refreshed with Neurobasal media supplemented only with Glutamine. The mix was incubated for 30 minutes then applied on the neurons and incubated for 90 minutes. Neurons were washed with Neurobasal media once, then refreshed with a mix of 1:1 (v/v) of fresh Complete NBM and conditioned media (removed from the samples before transfection). For neurons transfected with RUSH constructs, the post-transfection incubation media was supplemented with Neutravidin. Neurons were then incubated for 1-3 days before being assayed, depending on specific experiments. Transfection of HEK cells was carried on cells plated 1-2 days before transfection, at a confluence of 60-90%, using Polyethylenimine (PEI) at a ratio of 3:1 (μl/μg) PEI/DNA. HEK cells were replaced with DMEM only media before transfection, and the transfection mix was incubated in DMEM for 15 minutes before applying on cells. For assaying ERK1/2 phosphorylation, HEK cells in 12-well plates transfected with 1μg DNA of either pCDNA3-GFP, TrkB-GFP and RUSH-TrkB-GFP. For kinesin tail-domain pulldown cells were transfected with a mix of BirA (4μg), Bio-Kif-td (6μg) and TrkB-GFP (6μg).
Retention Using Selective Hooks (RUSH) assay
In order to deplete extracellular biotin before experimental induction of ER-release, complete Neurobasal media supplemented with Neutravidin 100μg/ml and HEK cells were kept in serum-free DMEM after transfection. To induce trafficking from the ER, media was replaced with normal growth media supplemented with biotin at 40μM.
Immunofluorescent labeling in fixed cells
Neurons were fixed by incubating in Paraformaldehyde 4% + Sucrose 4% in PBS for 25 minutes at room-temperature and then washed 3x in PBS. Samples were placed in humidified dark chamber. Cells were permeabilized in Triton X-100 0.2% in Blocking solution (BSA 1% in PBS) for 5 minutes in room termperature, then washed out and incubated with Blocking solution for 15 minutes at 37°C. Primary antibody was added and incubated for 60 minutes at 37°C for samples on coverslips or overnight at 4°C for samples in MFC. Primary antibody mix was washed out 3x with PBS and replaced with secondary antibody which was incubated for 60 minutes at 37°C (for coverslips) or 120 minutes at room-temperature (for MFC). For fluorescence preservation coverslips were mounted on slides using either Vectashield or Fluoromount-G. For MFC samples, wells were filled with Vectashield.
Surface immunolabeling and internalization assay of SNAP-TrkB
Cell surface expression of SNAP-TrkB was probed by applying myc antibody (1:200) on live neurons for 15 minutes in room-temperature, followed by 2 washouts in NBM. Cells were fixed and secondary antibody (Alexa-647-anti-mouse IgG, 1:200) was applied for 30 minutes followed by 3 washouts with PBS. Labeling of intracellular SNAP-TrkB was done by permeabilizing for 5 minutes, blocking for 15 minutes at 37°C, and incubating with myc antibody (1:200) for 30 minutes followed by 3 washouts and labeling with Alexa-488-anti mouse IgG. For internalization assay, a pulse labeling with myc antibody (1:200) was carried for 15 minutes at 37°C. Neurons were then treated with control or BDNF supplemented media for 30 minutes followed by fixation. Alexa-647-anti mouse IgG was applied for 30 minutes prior to permeabilization to label SNAP-TrkB that remained at the cell surface. Samples were then permeabilized and blocking was carried, followed by a second incubation with Alexa-488-secondary antibody. Confocal imaging of surface (Alexa-647) and internalized (Alexa-488) was carried along the volume of the soma. To quantify the total signal, a Sum slices projection was carried and the mean 647 and 488 signals at the soma region per neuron were measured using Fiji software.
Confocal imaging of fixed cells
Zeiss LSM700 microscopes were used to perform confocal imaging of fixed cells. Samples were imaged with 60x oil-immersion NA 1.4 objective equipped with automated stage. Gain and laser power was adjusted according to intensity to obtain non-saturated signals. Zoom level were adjusted to reach a resolution of 90-110 nm /pixel and z-stack images were taken at 0.5μm intervals. For imaging of neurons in compartmental SNAP-TrkB labeling experiments, only labelled neurons with axons crossing through the microchannels from proximal to distal compartment were selected. Tile-scans were manually registered and automatically taken to capture individual neurons and their axons extensions from the proximal to the distal compartments and along the microchannels.
Live-cell confocal imaging
Live neurons were mounted on coverslip holders and placed under a Nikon Ti microscope inside a Tokai HIT on-stage incubator and objective heater maintaining humidified ambience of 37°C and CO2 5%. A Nikon 60x oil-immersion objective (NA 1.4) and Nikon Perfect Focus System were used in all experiments. Individual neurons were located using widefield fluorescence imaging via the eyepieces. Confocal imaging was obtained using Yokogawa CSU-X1 unit and images were captured with either EMCCD or sCOMS cameras. Control of microscopy system, imaging protocol and image capture was done with Metamorph software. EMCCD gain was set to 950 in all channels. Laser was adjusted to obtain sufficient emission signal for tracking of individual vesicles in all imaging channels while minimizing phototoxicity. Imaging of neuronal transport was carried at 1 frame per second. Imaging of RUSH trafficking after biotin-induced release was carried at 1 min per frame. Dual-color imaging was done by sequential switching between 488 & 561nm laser excitation and fast GFP and mCherry filter. Multi-position imaging of RUSH trafficking and SNAP-TrkB internalization was controlled via automated stage.
Imaging based Split-Kinesin Assay
Neurons at DIV 7-10 were transfected with a combination of TrkB-GFP, mRFP-KIF5C-md-FKBP and variable tagged Kinesin tail domain (FRB-3xMyc-KIF-td). Neurons were incubated with either normal culture media as control or media supplemented with rapalog 100nM. Cells were fixed 16-24 hours after transfection and processed for immunofluorescent labeling for myc-tag detection. Samples where imaged using Nikon Eclipse Ni80 upright fluorescent microscope equipped with CoolSNAP HQ2 CCD (Photometrics). Axon tips were imaged based on focal accumulation of KIF5C-md signal, and respective somas were imaged by tracing TrkB-GFP signal. Image analysis was carried using Fiji software. To quantify the enrichment of fluorescence signal in axon tips, circular region of interest (ROI) were manually marked on axon tips and corresponding somas. The average signal of KIF-td and TrkB-GFP signal was then measured and used to calculate the tip/soma enrichment ratio.
Preparation of HEK cell lysates for Pull-down and Western Blot (WB)
HEK cells were harvested 16-24 hours after transfection by vigorous pipetting. Cell suspension was centrifuged at 700g’ for 1 min, then washed once with cold PBS and spun again. For direct WB analysis of cell lysate, pellet was lysed in RIPA buffer: NaCl 150mM, TX-100 1%, sodium deoxycholate (SDC) 0.5% and sodium dodecyl sulfate (SDS) 0.1%, Tris 50mM, pH 8.0 supplemented with protease and phosphatase Inhibitors. For pull-down experiments, a milder detergent buffer was used instead: NaCl 150mM, TX-100 1% and Tris 25mM pH 8.0. Cells were mixed 10x by pipetting and incubated in the lysis buffer for 5 minutes on ice then centrifuged 1000xg’ for 5’ at 4°C. Cleared supernatant lysate was collected into a fresh tube and used for pull-down or direct analysis. For SDS-PAGE, samples were mixed 1:3 with Sample Buffer x4 (Tris 500mM pH 6.8, SDS 8%, glycerol 2.5%, DTT 10mM and bromophenol Blue), then heated to 95°C for 5’.
Pull-down assay for Kinesin-tail domain and TrkB interaction
Cell lysate of HEK cells expressing TrkB-GFP, BirA and Bio-Kif-td was incubated with Dynabeads Streptavidin magnetic beads (Thermo Fisher). Dynabeads (25μl per 10cm plate of lysed HEK cells) were pre-incubated with blocking buffer (Tris 20 mM pH 8.0, KCl 150 mM, BSA 0.2%, Glycerol 0.2%). Lysates were then incubated with beads for 1 hour at 4°C and the beads were washed 3 times with wash buffer (Tris 25mM PH 8.0, NaCl 150mM, TX-100 0.1%). For western blot analysis, bound proteins were eluted by addition of 50μl SDS-PAGE sample buffer and heated to 95°C for 5 minutes.
SDS-PAGE and Western Blot assay
Samples were run on polyacrylamide gels (8-10%) and transferred to nitrocellulose membranes using Mini-Protean apparatus (Bio-Rad). Membranes were blocked in BSA 1% and incubated with primary antibodies for 2 hour in room-temperature or overnight at 4°C. Secondary antibodies conjugated with IRdyes (Li-Cor) were incubated for 2 hours in room temperature. Blots were scanned with Odyssey and analyzed with Fiji Gel Analyzer application.
Proteomic analysis of Kinesin-tail domain interactome
Beads were re-suspended in 20 μL of Laemmli Sample buffer (Biorad) and supernatants were loaded on a 4-12% gradient Criterion XT Bis-Tris precast gel (Biorad). The gel was fixed with 40% methanol/10% acetic acid and then stained for 1 h using colloidal coomassie dye G-250 (Gel Code Blue Stain Reagent, Thermo Scientific). After in-gel digestion (Stucchi et al., 2018), all samples were analyzed on a Orbitrap Q-Exactive Plus mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) coupled to an Agilent 1290 Infinity LC (Agilent Technologies). Peptides were loaded onto a trap column (Reprosil C18, 3 μm, 2 cm × 100 μm; Dr. Maisch) with solvent A (0.1% formic acid in water) at a maximum pressure of 800 bar and chromatographically separated over the analytical column (Zorbax SB-C18, 1.8 μm, 40 cm × 50 μm; Agilent) using 90 min linear gradient from 7-30% solvent B (0.1% formic acid in acetonitrile) at a flow rate of 150 nL/min. The mass spectrometer was used in a data-dependent mode, which automatically switched between MS and MS/MS. After a survey scan from 350-1500 m/z the 10 most abundant peptides were subjected to HCD fragmentation. For data analysis, raw files were processed using Proteome Discoverer 1.4 (version 1.4.1.14, Thermo Scientific, Bremen, Germany). Database searches were performed using Mascot as search engine (version 2.5.1, Matrix Science, UK) on the Human Uniprot database. Carbamidomethylation of cysteines was set as a fixed modification and oxidation of methionine was set as a variable modification. Trypsin was set as cleavage specificity, allowing a maximum of 2 missed cleavages. Data filtering was performed using percolator, resulting in 1% false discovery rate (FDR). Additional filters were search engine rank 1 and mascot ion score >20. To infer protein abundancy of each individual protein co-purified with the bait protein, we relied on total numbers of Peptide Spectrum Matches (PSM). Crapome (Mellacheruvu et al., 2013) was used to analyze interacting binding proteins. The Fold Change calculation (FC-B, Crapome; by averaging the spectral counts compared to GFP-bait control) and PSM values were used for data visualization. Bubble plot of PSM and fold-enrichment presentation was created for selected proteins using custom made R script.
Quantification and statistical analysis
Statistical analysis and data visualization
All statistical details including the definitions of n, numbers of n and statistical tests performed can be found in each figure and figure legends. No statistical methods were used to pre-determine sample sizes. Data distribution was assumed to be normal but this was not formally tested. Statistical testing was perfromed using unpaired Student’s T-test unless noted otherwise. p-value<0.05 was considered significant. Quantitative data was organized, statistically analyzed and visualized using Excel (Microsoft) and Prism (Graphpad) software, except for where otherwise noted.
Kymograph preparation and analysis of transport dynamics
Live imaging time-lapse series of transport dynamics were processed using Fiji. Image series were background subtractied using rolling ball filter radius of 50 pixels (5.5μm). Neurites were traces using 11 pixel wide (1.21μm) segmented line ROI tool and Kymographs were created using KymoResliceWide plugin (https://github.com/ekatrukha/KymoResliceWide). To measure velocity of traces, straight line ROIs were manually traced along linear segments of individual trajectories. The angle of the lines was used to calculate the average velocity of the mobility segments. To analyze the processivity of secreted RUSH-TrkB cargos along the axon, segmented line ROIs were traced. Trajectories that could be traced for 30 seconds or longer, or that crossed the entire distance of the traced axon were selected for further analysis. Trajectories covering more than 15μm in the duration of imaging were counted as either retrograde or anterograde, depending on the vector of movement, while trajectories that were immobile or erratic were counted as non-processive.
Acknowledgments
We thank Prof. Frederic Saudou and Amelie Grenoux from Grenoble Institute for Neuroscience for providing the molds and the training for the production of the microfluidic devices used in this study. We thank Dr. Maud Martin for kindly providing guidance, plasmids, and reagents for the RUSH experiments. This work was supported by EMBO Long-Term Fellowship (EMBO-LTF, E.E.Z.), the Netherlands Organization for Scientific Research (NWO-ALW-VICI, 865.10.010, C.C.H.), the Netherlands Organization for Health Research and Development (ZonMw-TOP, 912.16.058, C.C.H.), the European Research Council (ERC) (ERC-consolidator, 617050, C.C.H.).
Author contributions
E.E.Z. initiated the study, designed and performed experiments, analyzed and formatted data for presentation in the manuscript, and wrote the manuscript. J.J.A.H. established experimental procedure, performed experiments, and guided experimental work. Y.H. and C.B. performed experimental work. R.S. performed the mass spectrometry experiment and analysis. M.A. gave valuable advice on mass spectrometry; C.C.H. designed the overall experimental plan, supervised the research, and wrote the manuscript. All authors contributed to the formatting and editing of the manuscript.
Declaration of interests
C.C.H. is an employee of Genentech, a member of the Roche group. The authors declare no additional competing interests.
Published: February 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.devcel.2021.01.010.
Supplemental information
Detected proteins in different pull-down conditions described in the results section and presented in summary in Figures S4B and S4C.
Detected proteins in different pull-down conditions described in the results section and presented in summary in Figure S4D.
References
- Arimura N., Kimura T., Nakamuta S., Taya S., Funahashi Y., Hattori A., Shimada A., Ménager C., Kawabata S., Fujii K. Anterograde transport of TrkB in axons is mediated by direct interaction with Slp1 and Rab27. Dev. Cell. 2009;16:675–686. doi: 10.1016/j.devcel.2009.03.005. [DOI] [PubMed] [Google Scholar]
- Ascaño M., Richmond A., Borden P., Kuruvilla R. Axonal targeting of Trk receptors via transcytosis regulates sensitivity to neurotrophin responses. J. Neurosci. 2009;29:11674–11685. doi: 10.1523/JNEUROSCI.1542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M., Banker G. The cellular mechanisms that maintain neuronal polarity. Nat. Rev. Neurosci. 2016;17:611–622. doi: 10.1038/nrn.2016.100. [DOI] [PubMed] [Google Scholar]
- Boncompain G., Divoux S., Gareil N., De Forges H., Lescure A., Latreche L., Mercanti V., Jollivet F., Raposo G., Perez F. Synchronization of secretory protein traffic in populations of cells. Nat. Methods. 2012;9:493–498. doi: 10.1038/nmeth.1928. [DOI] [PubMed] [Google Scholar]
- Bothwell M. Recent advances in understanding neurotrophin signaling. F1000Res. 2016;5:1885. doi: 10.12688/f1000research.8434.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen A.B., Bourke A.M., Hiester B.G., Hanus C., Kennedy M.J. Golgi-Independent secretory trafficking through recycling endosomes in neuronal dendrites and spines. eLife. 2017;6:e27362. doi: 10.7554/eLife.27362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt D.J., Farías G.G., Guardia C.M., Bonifacino J.S. Mechanisms of polarized organelle distribution in neurons. Front. Cell. Neurosci. 2016;10:88. doi: 10.3389/fncel.2016.00088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Cosker K.E., Segal R.A. Neuronal signaling through endocytosis. Cold Spring Harb. Perspect. Biol. 2014;6:a020669. doi: 10.1101/cshperspect.a020669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer E., Rodriguez P., Bonte E., Krijgsveld J., Katsantoni E., Heck A., Grosveld F., Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eva R., Dassie E., Caswell P.T., Dick G., Ffrench-Constant C., Norman J.C., Fawcett J.W. Rab11 and its effector Rab coupling protein contribute to the trafficking of β1 integrins during axon growth in adult dorsal root ganglion neurons and PC12 cells. J. Neurosci. 2010;30:11654–11669. doi: 10.1523/JNEUROSCI.2425-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farías G.G., Guardia C.M., Britt D.J., Guo X., Bonifacino J.S. Sorting of dendritic and axonal vesicles at the pre-axonal exclusion zone. Cell Rep. 2015;13:1221–1232. doi: 10.1016/j.celrep.2015.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett J.W. The struggle to make CNS axons regenerate: why has it been so difficult? Neurochem. Res. 2020;45:144–158. doi: 10.1007/s11064-019-02844-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourriere L., Jimenez A.J., Perez F., Boncompain G. The role of microtubules in secretory protein transport. J. Cell Sci. 2020;133:jcs237016. doi: 10.1242/jcs.237016. [DOI] [PubMed] [Google Scholar]
- Fourriere L., Kasri A., Gareil N., Bardin S., Bousquet H., Pereira D., Perez F., Goud B., Boncompain G., Miserey-Lenkei S. RAB6 and microtubules restrict protein secretion to focal adhesions. J. Cell Biol. 2019;218:2215–2231. doi: 10.1083/jcb.201805002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González C., Cornejo V.H., Couve A. Golgi bypass for local delivery of axonal proteins, fact or fiction? Curr. Opin. Cell Biol. 2018;53:9–14. doi: 10.1016/j.ceb.2018.03.010. [DOI] [PubMed] [Google Scholar]
- Grigoriev I., Splinter D., Keijzer N., Wulf P.S., Demmers J., Ohtsuka T., Modesti M., Maly I.V., Grosveld F., Hoogenraad C.C., Akhmanova A. Rab6 regulates transport and targeting of exocytotic carriers. Dev. Cell. 2007;13:305–314. doi: 10.1016/j.devcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Grigoriev I., Yu K.L., Martinez-Sanchez E., Serra-Marques A., Smal I., Meijering E., Demmers J., Peränen J., Pasterkamp R.J., van der Sluijs P. Rab6, Rab8, and MICAL3 cooperate in controlling docking and fusion of exocytotic carriers. Curr. Biol. 2011;21:967–974. doi: 10.1016/j.cub.2011.04.030. [DOI] [PubMed] [Google Scholar]
- Guedes-Dias P., Nirschl J.J., Abreu N., Tokito M.K., Janke C., Magiera M.M., Holzbaur E.L.F. Kinesin-3 responds to local microtubule dynamics to target synaptic cargo delivery to the presynapse. Curr. Biol. 2019;29:268–282.e8. doi: 10.1016/j.cub.2018.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gumy L.F., Katrukha E.A., Grigoriev I., Jaarsma D., Kapitein L.C., Akhmanova A., Hoogenraad C.C. MAP2 defines a pre-axonal filtering zone to regulate KIF1- versus KIF5-dependent cargo transport in sensory neurons. Neuron. 2017;94:347–362.e7. doi: 10.1016/j.neuron.2017.03.046. [DOI] [PubMed] [Google Scholar]
- Haniu M., Talvenheimo J., Le J., Katta V., Welcher A., Rohde M.F. Extracellular domain of neurotrophin receptor trkB: disulfide structure, N-glycosylation sites, and ligand binding. Arch. Biochem. Biophys. 1995;322:256–264. doi: 10.1006/abbi.1995.1460. [DOI] [PubMed] [Google Scholar]
- Hanus C., Geptin H., Tushev G., Garg S., Alvarez-Castelao B., Sambandan S., Kochen L., Hafner A.S., Langer J.D., Schuman E.M. Unconventional secretory processing diversifies neuronal ion channel properties. eLife. 2016;5:e20609. doi: 10.7554/eLife.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington A.W., Ginty D.D. Long-distance retrograde neurotrophic factor signalling in neurons. Nat. Rev. Neurosci. 2013;14:177–187. doi: 10.1038/nrn3253. [DOI] [PubMed] [Google Scholar]
- Hoogenraad C.C., Milstein A.D., Ethell I.M. GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci. 2005;8:906–915. doi: 10.1038/nn1487. [DOI] [PubMed] [Google Scholar]
- Huang S.H., Wang J., Sui W.H., Chen B., Zhang X.Y., Yan J., Geng Z., Chen Z.Y. BDNF-dependent recycling facilitates TrkB translocation to postsynaptic density during LTP via a Rab11-dependent pathway. J. Neurosci. 2013;33:9214–9230. doi: 10.1523/JNEUROSCI.3256-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins B., Decker H., Bentley M., Luisi J., Banker G. A novel split kinesin assay identifies motor proteins that interact with distinct vesicle populations. J. Cell Biol. 2012;198:749–761. doi: 10.1083/jcb.201205070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech S., Banker G. Culturing hippocampal neurons. Nat. Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- Kennedy M.J., Hanus C. Architecture and dynamics of the neuronal secretory network. Annu. Rev. Cell Dev. Biol. 2019;35:543–566. doi: 10.1146/annurev-cellbio-100818-125418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevenaar J.T., Bianchi S., Van Spronsen M., Olieric N., Lipka J., Frias C.P., Mikhaylova M., Harterink M., Keijzer N., Wulf P.S. Kinesin-binding protein controls microtubule dynamics and cargo trafficking by regulating kinesin motor activity. Curr. Biol. 2016;26:849–861. doi: 10.1016/j.cub.2016.01.048. [DOI] [PubMed] [Google Scholar]
- Koseki H., Donegá M., Lam B.Y.H., Petrova V., van Erp S., Yeo G.S.H., Kwok J.C.F., Ffrench-Constant C., Eva R., Fawcett J.W. Selective rab11 transport and the intrinsic regenerative ability of CNS axons. eLife. 2017;6:e26956. doi: 10.7554/eLife.26956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J., Parton R.G., Fuller S.D., Griffiths G., Dotti C.G. The organization of the endoplasmic reticulum and the intermediate compartment in cultured rat hippocampal neurons. Mol. Biol. Cell. 1995;6:1315–1332. doi: 10.1091/mbc.6.10.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasiecka Z.M., Winckler B. Mechanisms of polarized membrane trafficking in neurons - focusing in on endosomes. Mol. Cell. Neurosci. 2011;48:278–287. doi: 10.1016/j.mcn.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo O.M., Gonzalez A., Ascaño M., Kuruvilla R., Couve A., Bronfman F.C. BDNF regulates Rab11-mediated recycling endosome dynamics to induce dendritic branching. J. Neurosci. 2013;33:6112–6122. doi: 10.1523/JNEUROSCI.4630-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P.L., Ohlson M.B., Pfeffer S.R. The Rab6-regulated KIF1C kinesin motor domain contributes to Golgi organization. eLife. 2015;4:e06029. doi: 10.7554/eLife.06029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A.E., Moughamian A.J., Holzbaur E.L.F. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O., Schmidt A., Salaméro J., Hoflack B., Roa M., Goud B. The small GTP-binding protein rab6 functions in intra-Golgi transport. J. Cell Biol. 1994;127:1575–1588. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matanis T., Akhmanova A., Wulf P., Del Nery E., Weide T., Stepanova T., Galjart N., Grosveld F., Goud B., De Zeeuw C.I. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat. Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Cao Y., Stucchi R., Hooikaas P.J., Portegies S., Will L., Martin M., Akhmanova A., Harterink M., Hoogenraad C.C. MAP7D2 localizes to the proximal axon and locally promotes kinesin-1-mediated cargo transport into the axon. Cell Rep. 2019;26:1988–1999.e6. doi: 10.1016/j.celrep.2019.01.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H., Poo M.M. Neurotrophin regulation of neural circuit development and function. Nat. Rev. Neurosci. 2013;14:7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- Pfeffer S.R. Rab GTPases: master regulators that establish the secretory and endocytic pathways. Mol. Biol. Cell. 2017;28:712–715. doi: 10.1091/mbc.E16-10-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R., Chen Z.Y., Lee F.S., Chao M.V. Transactivation of Trk neurotrophin receptors by G-protein-coupled receptor ligands occurs on intracellular membranes. J. Neurosci. 2004;24:6650–6658. doi: 10.1523/JNEUROSCI.0010-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampo B., Kaech S., Kunz S., Banker G. Two distinct mechanisms target membrane proteins to the axonal surface. Neuron. 2003;37:611–624. doi: 10.1016/s0896-6273(03)00058-8. [DOI] [PubMed] [Google Scholar]
- Schatz P.J. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Schlager M.A., Kapitein L.C., Grigoriev I., Burzynski G.M., Wulf P.S., Keijzer N., de Graaff E., Fukuda M., Shepherd I.T., Akhmanova A., Hoogenraad C.C. Pericentrosomal targeting of Rab6 secretory vesicles by bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 2010;29:1637–1651. doi: 10.1038/emboj.2010.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Solomon E., Kuruvilla R. Mechanisms of neurotrophin trafficking via Trk receptors. Mol. Cell. Neurosci. 2018;91:25–33. doi: 10.1016/j.mcn.2018.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Marques A., Martin M., Katrukha E.A., Grigoriev I., Peeters C.A., Liu Q., Hooikaas P.J., Yao Y., Solianova V., Smal I. Concerted action of kinesin-1 KIF5B and kinesin-3 KIF13B promotes efficient transport of exocytotic vesicles to microtubule plus ends. eLife. 2020;9:e61302. doi: 10.7554/eLife.61302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M., Giza J., Proenca C.C., Jing D., Elliott M., Dincheva I., Shmelkov S.V., Kim J., Schreiner R., Huang S.-H. Slitrk5 mediates BDNF-dependent TrkB receptor trafficking and signaling. Dev. Cell. 2015;33:690–702. doi: 10.1016/j.devcel.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stucchi R., Plucińska G., Hummel J.J.A., Zahavi E.E., Guerra San Juan I., Klykov O., Scheltema R.A., Altelaar A.F.M., Hoogenraad C.C. Regulation of KIF1A-driven dense core vesicle transport: Ca2+/CaM controls DCV binding and liprin-α/TANC2 recruits DCVs to postsynaptic sites. Cell Rep. 2018;24:685–700. doi: 10.1016/j.celrep.2018.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Niwa S., Dong M., Farkhondeh A., Wang L., Zhou R., Hirokawa N. The molecular motor KIF1A transports the TrkA neurotrophin receptor and is essential for sensory neuron survival and function. Neuron. 2016;90:1215–1229. doi: 10.1016/j.neuron.2016.05.002. [DOI] [PubMed] [Google Scholar]
- Valenzuela J.I., Jaureguiberry-Bravo M., Salas D.A., Ramírez O.A., Cornejo V.H., Lu H.E., Blanpied T.A., Couve A. Transport along the dendritic endoplasmic reticulum mediates the trafficking of GABAB receptors. J. Cell Sci. 2014;127:3382–3395. doi: 10.1242/jcs.151092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virlogeux A., Moutaux E., Christaller W., Genoux A., Bruyère J., Fino E., Charlot B., Cazorla M., Saudou F. Reconstituting corticostriatal network on-a-chip reveals the contribution of the presynaptic compartment to Huntington’s disease. Cell Rep. 2018;22:110–122. doi: 10.1016/j.celrep.2017.12.013. [DOI] [PubMed] [Google Scholar]
- Wakana Y., van Galen J., Meissner F., Scarpa M., Polishchuk R.S., Mann M., Malhotra V. A new class of carriers that transport selective cargo from the trans Golgi network to the cell surface. EMBO J. 2012;31:3976–3990. doi: 10.1038/emboj.2012.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson F.L., Porcionatto M.A., Bhattacharyya A., Stiles C.D., Segal R.A. TrkA glycosylation regulates receptor localization and activity. J. Neurobiol. 1999;39:323–336. doi: 10.1002/(sici)1097-4695(199905)39:2<323::aid-neu15>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- White J., Johannes L., Mallard F., Girod A., Grill S., Reinsch S., Keller P., Tzschaschel B., Echard A., Goud B., Stelzer E.H. Rab6 coordinates a novel Golgi to ER retrograde transport pathway in live cells. J. Cell Biol. 1999;147:743–760. doi: 10.1083/jcb.147.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisco D., Anderson E.D., Chang M.C., Norden C., Boiko T., Fölsch H., Winckler B. Uncovering multiple axonal targeting pathways in hippocampal neurons. J. Cell Biol. 2003;162:1317–1328. doi: 10.1083/jcb.200307069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N., Joshi R., Zhang S., Zhang Z.Y., Kuruvilla R. Phospho-regulation of soma-to-axon transcytosis of neurotrophin receptors. Dev. Cell. 2017;42:626–639.e5. doi: 10.1016/j.devcel.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Part A: live confocal imaging of a hippocampal neuron (DIV8) proximal axon expressing TrkB-GFP. The AIS is immunolabeled with antibody against cell-surface Neurofascin (shown in red overlay).
Part B: live confocal imaging of a hippocampal neuron (DIV8) dendrites expressing TrkB-GFP. The AIS of neighboring neurons are shown by neurofascin immunolabeling. Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Live confocal imaging of a hippocampal neuron (DIV9) axons expressing surface labeled SNAP-TrkB. The AIS of neighboring neurons are labeled with cell-surface Neurofascin immunolabeling (shown in red overlay). Neurons were treated with BDNF or normal media as control for 60 min after SNAP-Surface labeling then promptly imaged. Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Part A: live confocal imaging of a hippocampal neuron (DIV8) expressing RUSH-TrkB-GFP. Biotin was added in t = 0. Images were captured at 1 frame per minute.
Part B: live confocal imaging of a hippocampal neuron (DIV12) expressing RUSH-TrkB-GFP. Biotin was added 48 min prior to imaging. The AIS is labeled with cell-surface Neurofascin antibody (shown in red overlay). Images were captured at 1 frame per second. Timeseries are played at 10 frames per second. Scale bar, 5 μm.
Live confocal imaging of a hippocampal neuron (DIV14) co-expressing RUSH-TrkB-GFP (green) and SNAP-TrkB (surface labeled in magenta). Biotin was added 66 min prior to imaging to induce trafficking of RUSH-TrkB-GFP along the secretory pathway and BDNF was added 30 minutes later (36 min before imaging) to induce internalization of surface labeled SNAP-TrkB. Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Part A: live confocal imaging of a hippocampal neuron (DIV10) co-expressing RUSH-TrkB-GFP (green) and mCherry-Rab6a. Biotin was added in t = 0. Images were captured at 1 frame per minute. Part B: live confocal imaging of a hippocampal neuron (DIV10) co-expressing RUSH-TrkB-GFP (green) and mCherry-Rab6a (magenta). Biotin was added 61 minutes prior to imaging. The AIS is labeled with cell-surface Neurofascin antibody (shown in red overlay). Images were captured at 1 frame per second and are played at 10 frames per second. Scale bar, 5μm.
Live confocal imaging of a hippocampal neurons (DIV9-10) co-expressing RUSH-TrkB-GFP and either pSuper control or shRab6a/b vectors demonstrating proper trafficking (pSuper) and frustrated and blocked trafficking (shRab6a/b). Biotin was added in t = 0. Images were captured at 1 frame per minutes and are played at 7 frames per second. Scale bar, 5 μm
Live confocal imaging of a hippocampal neurons (DIV10) co-expressing RUSH-TrkB-GFP and shRNA against Kif5c (Part A) or shRNA against Kif1a (Part B). Biotin was added in t = 0. Images were captured at 1 frame per minutes and are played at 7 frames per second. Scale bar, 5 μm.
Live confocal imaging of a hippocampal neurons (DIV9-10) co-expressing RUSH-TrkB-GFP and shRNA against Kif1a (Part A) or pSuper control vector (Part B). Biotin was added 48 min prior to imaging. The AIS is labeled with cell-surface Neurofascin immunolabeling (shown in red overlay). Images were captured at 1 frame per second and are played at 20 frames per second. Scale bar, 5 μm.
Detected proteins in different pull-down conditions described in the results section and presented in summary in Figures S4B and S4C.
Detected proteins in different pull-down conditions described in the results section and presented in summary in Figure S4D.
Data Availability Statement
Additional datasets, data analysis and visualization algorithms generated in this study are available upon request.