Abstract
Recently, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of coronavirus disease 2019 (COVID-19), has led to a worldwide pandemic with millions of infected patients. Alteration in humans' microbiota was also reported in COVID-19 patients. The alteration in human microbiota may contribute to bacterial or viral infections and affect the immune system. Moreover, human's microbiota can be altered due to SARS-CoV-2 infection, and these microbiota changes can indicate the progression of COVID-19. While current studies focus on the gut microbiota, it seems necessary to pay attention to the lung microbiota in COVID-19. This study is aimed at reviewing respiratory microbiota dysbiosis among COVID-19 patients to encourage further studies on the field for assessment of SARS-CoV-2 and respiratory microbiota interaction.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as a novel coronavirus, is spreading from China and is known to be the etiologic agent for coronavirus disease 2019 (COVID-19) [1–3]. The SARS-CoV-2 belongs to betacoronavirus genera and phylogenetically is relevant to SARS-CoV [4]. The SARS-CoV-2 can exploit the angiotensin-converting enzyme 2 (ACE2) for priming Spike (S) protein [5, 6]. The ACE2 is expressed in the esophagus, lungs, liver, and intestinal epithelium [7, 8]. SARS-CoV-2 infection can be asymptomatic or can cause a wide spectrum of signs and symptoms: fever, dry cough, shortness of breath, pneumonia, pulmonary edema, acute respiratory distress syndrome (ARDS), multiple organ failure, and death [9]. In some patients, common symptoms include headache, nausea, and vomiting, and diarrhea is also reported [10].
At the infancy age, various bacteria, fungi, and viruses colonize in the skin, oral cavity, and gut. These microorganisms are known as the human microbiota [11–13]. The predominant human oral microbiota is summarized in Table 1. The microbiome plays an essential role in human physiology, and it is considered an important factor for the maintenance of human health [14]. Typically, these microbes are commensal or mutualists, and they help to digest food and even provide immunity [15]. As mentioned before, microbial communities are found throughout the human body; there are specialized bacterial communities in certain regions of the respiratory system that are believed to play a significant role in preserving human health [16].
Table 1.
The predominant human oral microbiota.
| Sites | Microbiota | Ref |
|---|---|---|
| Lips | Streptococcus spp., C. Albicans | [30] |
| Hard palate | Streptococcus spp., Uncl. Pasteurellaceae, Mogibacterium Veillonella, Catonella Prevotella, Uncl. Lactobacillales, Gemella | [21] |
| Tongue | Front two-thirds of the tongue: Streptococcus mutans | [25, 31] |
| Tongue dorsum: Streptococcus salivarius, S. oralis, S. mitis, Actinomyces naeslundii, Haemophilus spp., Rothia mucilaginosa | ||
| Gingival sulcus | Proteobacteria (genus Acinetobacter, Haemophilus, Moraxella), Firmicutes (Streptococcus, Granulicatella, Gemella) | [32] |
| Buccal mucosa | Firmicutes (Streptococcus sanguinis, S. oralis, S. mitis) | [31, 155] |
| Palatine tonsils | Streptococcus, Prevotella, Neisseria, Fusobacterium, Veillonella | [156] |
| Saliva | Firmicutes (genus Streptococcus and Veillonella), Bacteroidetes (genus Prevotella), and Betaproteobacteria (genus Neisseriaceae) | [25, 31, 157] |
| Teeth (dental plaque) | Tooth crown: Firmicutes (genus Streptococcus and Veillonella) | [25, 31, 32] |
| Supragingival plaque: Firmicutes and Actinobacteria (genus Corynebacterium and Actinomyces) | ||
| Subgingival plaque: Obsidian Pool OP11, TM7, Deferribacteres, Spirochaetes, Fusobacteria, Actinobacteria, Firmicutes, Proteobacteria, Bacteroidetes, C. albicans |
The essential factor for upper respiratory tract (URT), lower respiratory tract (LRT), or disseminated respiratory infections is colonization in the URT [17]. Variations in lung microbiota could potentially improve immune response against viral and secondary bacterial infection [18]. Recent studies have shown the lung's microbiota contributed to the immunologic homeostasis and potentially altered viral infection susceptibility [19]. The ARDS is a severe complication of COVID-19 [19]. Studies showed that the lung microbiota of the patients with ARDS is different from those without ARDS [20]. This fact could be an essential issue in COVID-19 progress.
2. Respiratory Bacterial and Fungal Microbiota
The oral cavity can be considered as the main route of entry for different pathogens. Various microorganisms, including bacteria, fungi, viruses, archaea, are colonized in the oral cavity and termed oral microbiota [21, 22]. Temperature (37°C), saliva pH (6.5-7), and humidity of the oral cavity make an appropriate environment for microorganism survival and maintenance [23, 24]. Furthermore, oxygen availability and consuming different food with acidic or alkaline pH can influence oral organism's growth pattern. Bacterial and fungal are primary microbiota communities of the oral cavity. Six strains of bacteria include Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes, and Fusobacteria, make up 94% of the oral bacteria community, while the major fungal population includes Candida species followed by Cladosporium spp., Aureobasidium spp., and Saccharomycetales in healthy cases [25, 26].
Commensal, symbiotic, and potentially pathogenic bacteria and fungi are in equilibrium. Poor oral hygiene such as periodontitis and dental caries, also pathogens like the Epstein–Barr virus (EBV), cytomegalovirus (CMV), smoking, drinking, and antibiotic consumption can compromise this ecological balance [27–30]. Microbial either plankton or biofilm habitats are found in the oral cavity; for instance, lingual microbiota contains stable multilayers of biofilms. Microbiota in the saliva is considered plankton and cannot be due to saliva being fluid and swallowed continuously [31]. On the other hand, saliva contains proteins such as mucins, agglutinin, and proline-rich proteins that help microbial adhesions to hard tissue like teeth [32]. Using high-precision sequencing methods introduces the human oral microbiome as a part of the Human Microbiome Project. This particular field is divided into two parts: (i) core: shared in all individuals. Among all the microbiota in the body, four-strains were found more frequently than others: Actinobacteria, Firmicutes, Proteobacteria, and Bacteroidetes [33]. (ii) Variable is dependent on lifestyle and environmental determinants and is variable between individuals [23].
Moreover, the diversity of microbiota changes is highly influenced by age. Alteration in the microbiota begins at birth, for instance, the delivery route of the baby. This change in the types of microbiota in infants is less than in adults (due to the absence of hard dental tissues, only feeding by breast milk/formula and so on) and is observed until later ages [32, 34]. Microbiota maturation by biological or passive changes due to vaccines, antibiotics, viral infection, teeth decay/filling, and different disease alerts gradually [28, 35–37]. Common oral diseases like dental caries, gingivitis, and oral mucosal disease are caused by endogenous bacteria [38]. Pathogenic viruses act as exogenous factors to make dysbiosis. Ling et al. indicated that hepatitis B virus (HBV) infection elevated Fusobacterium, Filifactor, Eubacterium, Parvimonas, and Treponema in the oral cavity leading to the unpleasant smell of mouth [39]. Also, dysbiosis of bacterial colonization in the respiratory tract and oral cavity was induced by the H1N1 influenza virus, leading to secondary bacterial infection [18].
2.1. The Microbiota of the Oral Cavity
The oral cavity consists of soft tissues (including lips, soft palate, tonsil, and tongue), saliva, and hard tissue, e.g., teeth. It harbors a high diversity of microbial organisms, and each tissue contains its specialized microbial community. Mucosal surfaces have monolayers of microorganisms compared with the tongue that has thick biofilms [40].
2.2. The Microbiota of the Oropharynx
The oropharynx is located in soft palate and upper of the epiglottis. Microbiota of the oropharynx in healthy adults is similar to other mucosal surfaces in the oral cavity and colonized by members of Firmicutes, Proteobacteria, and Bacteroidetes (including Streptococcus, Neisseria, Haemophilus, and Lachnospira spp.) [18, 41–43].
2.3. The Microbiota of the Laryngopharynx
The salivary microbiota after the oropharynx drain into the laryngopharynx. Indeed, it connects the upper aerodigestive tract to the digestive tract. The Firmicutes, Fusobacteria, Proteobacteria, Actinobacteria, and Bacteroidetes were reported as the primary bacterial population in this site [44].
3. Physiologic Features of Respiratory Microbiota
Over the past two decades, many studies have examined the impact of oral microbiota on disease or human health. The oral tissues use some mechanisms and molecules to balance the oral flora and potential pathogens. Microbial communities are tissue-specific, which can tolerate the dominant physicochemical environment. The microbiota adhere to the epithelial surfaces' mucosal membrane and can resist the saliva flow [38]. However, the saliva flow plays a role in host defense and contains antimicrobial peptides, lysozyme, lactoferrin, defensins, and lactoperoxidase to prevent microbial overgrowth [45–48]. Immunomodulation of commensals is another mechanism to maintain the oral host-microbe balance. The epithelial cells are natural physical barriers against pathogens, and they secrete antimicrobial mediators like IL-6, IL-8, TNF-α, IL-1β/α, defensins, and cathelicidin LL-37 [49]. The formation of pores on the bacterial cytoplasmic membrane is considered as a significant role of defensins and LL-37. α/β–defensins are found in all oral tissues, saliva, and gingival crevicular fluid.
Defensins as antimicrobial functions can induce chemotactic ability to recruit monocytes, macrophages, and even T cells [50, 51]. Among immune cells that are involved in healthy oral immunity responses, neutrophils serve the main role. In healthy junctional epithelial tissue, LL-37 and defensins attract neutrophils. This attraction leads to migrated neutrophils that lie in the gingival margin to make a barrier against dental plaque germs [52]. Commensals also control neutrophil migration in gingival tissues through modulating intracellular adhesion molecule 1 (ICAM-1) and E-selectin expression [53]. Neutrophils can generate nitric oxide and nitrogen intermediates with protective effects against bacteria [54]. IL-17-mediated immunity contributes to mucosal fungal surveillance, especially Candida spp. In parallel, IL-17 enhances the epithelium integrity via regulation of claudin, promotes the antimicrobial peptides expressed by epithelial cells, and elicits the secretion of neutrophil chemotaxis [49, 55]. The point to consider is that the commensal bacteria inhibit IL-17 family members' overexpression in a negative feedback manner to keep the oral homeostasis [56].
The other mechanism is bacteriophages that regulate the oral ecosystem as biocontrollers. Endodontic infection caused by Enterococcus faecalis could be healed through bacteriophages [57, 58]. Lytic bacteriophages can lyse bacteria and alleviate the bacterial pathogen numbers. The released substances from lysed bacteria also activate the immunity responses. These findings led to defining a concept called “immunophage synergy” [59]. Besides, bacteriophages have a direct impact on host immunity, either adaptive or innate immunity. Macrophages and dendritic cells can take up the bacteriophages as a virus or with their hosts and, consequently, induce cytokine responses. They also act as opsonin molecules to cover bacterial cytoplasmic membrane to stimulate phagocytosis. Commensal bacteriophages induce specific anti-phage antibodies. Specific anti-T4 phage IgG against viral gp24 and gp23 proteins was found in sera of healthy subjects [60, 61].
The presence of multiple species can give balance to populations of microorganisms in the body, e.g., Pichia in the oral cavity has an antagonistic relation with Aspergillus, Fusarium, and particularly Candida. Sometimes competition for nutrient uptake can limit germination and adhesion. A decrease in Pichia amount accompanies by increase in the growth of opportunistic fungi [62]. Bacteria use quorum sensing to communicate with other bacteria. Antagonistic interactions occur between Porphyromonas gingivalis (Pg), a periodontal pathogen and normal flora Streptococcus Gordonii (S. Gordonii), Streptococcus intermedius, and Streptococcus mitis. Arginine deiminase, encoded by the ArcA gene in these commensals, decreases expression of FimA that is a virulence factor in Pg. Hydrogen peroxidase produced by these streptococci can limit P. gingivalis growth in oral cavity [63]. Due to the lack of catalases in S. Gordonii, Actinomyces naeslundi breaks down the H2O2 generated by S. Gordonii. A symbiotic relationship is present between these two bacteria while competes with other possible pathogens [64, 65]. A competition between commensals and Streptococcus mutans (S. mutans) was suggested. Commensals overcome S. mutans by alkali components like urea to nearly provide a neutral environment [66]. Further, serine protease challisin derived from S. Gordonii interferes and degrades S. mutans bacteriocin production [67].
4. Pathogenesis of Respiratory Microbiota
Periodontitis, defined as destructive gum infection with tooth attachment loss and severe inflammation, is mainly caused by Porphyromonas gingivalis (P. gingivalis). Pg's adherence is mediated by a virulence gene known as FimA [68]. P. gingivalis also harbors dpp genes, which code dipeptidyl peptidases (DPP) [69, 70]. Interestingly, dpp genes present in subgingival crevice colonized bacteria, but not in mucosal surfaces and tongue isolated bacteria [71]. The high DPP4 activity was observed in the saliva of patients with chronic periodontitis [72]. DPP4 can degrade incretin hormones released in response to fat and glucose ingestion by increasing insulin secretion. However, the effect of insertion is not seen in people who have type 2 diabetes [71, 73, 74]. P. gingivalis through α5β1-integrin expressed on the epithelial cells, crosses the epithelial barrier, and enters the bloodstream [75]. LPS from P. gingivalis activates the TLR-4 signaling and triggers the secretion of IL-1β and IL-6 [76, 77]. TLR-4 signaling activated by Pg is also reported to be associated with human pancreatic tumors [78]. Moreover, anti-P. gingivalis antibodies in mouse model of periodontitis were able to prevent mice developing metabolic diseases [69]. Viral infections such as Herpes simplex virus-1, cytomegalovirus, and EBV virus can impair or suppress the immune system and induce aggressive periodontitis. A cooperative complex of Pg, S. aureus, and Herpes simplex-1 accelerates aggressive periodontitis [79]. Kaposi's sarcoma-associated herpesvirus (KSHV) is known as the most common AIDS-associated tumor [80]. The lipoteichoic acid (LTA) of S. aureus and lipopolysaccharide (LPS) of Pg can facilitate entry of KSHV through upregulation of heparan sulfate and heparan sulfate proteoglycans (viral receptors) and induce reactive oxygen species production (ROS). The LTA and LPS established viral latency by increasing viral latency-associated nuclear antigen (LANA) expression [81]. These findings suggested the role of Pg as a periodontal microbiota on the immune system and systemic diseases.
On the other hand, other periodontal pathogens, Fusobacterium, Prevotella, and Alloprevotella were enriched in HPV-negative in nonsmokers patients with oral cavity squamous cell cancer (OC-SCC) while commensal Streptococcus spp. was decreased. These oral pathogens were the primary source for transcriptional stimulation of genes encoding HSP90A, TLR-1/2/4 ligands [82]. Kim et al. indicated that HSP90 could increase telomerase expression through promoter activation of human oral cancer cells. This expression can interact with the human telomerase reverse transcriptase (hTERT) promoter [83].
Dental caries is much dependent on dietary carbohydrates. The S. mutans can alter these carbohydrates to organic acids and reduce the pH [84].
Mucosal candidiasis, known as thrush [85], is a common disease in patients receiving high doses of chemotherapy or immunosuppressive agents and caused by Candida albicans (C. albicans) [49, 86]. A key point of C. albicans diseases is the yeast-to-hyphal transformation by phospholipases (PLs). This phospholipase is capable of destroying the junctions between epithelial cells and cell membranes [45]. The C. albicans penetrates the epithelial cells of mucosal membranes directly or by binding Als3 and Ssa1 of hypha to E-cadherin, epidermal growth factor receptors, and HER2 of cells [87]. Furthermore, aspartic proteinase 2 (Sap2), another C. albicans's lytic enzyme, can protect the organism from immune system proteins such as salivary lactoferrin and immunoglobulins. Saps can activate inflammatory factor IL-1β in mucosal lesions [88]. Also, some external factors like antifungals can help to elevate dysbiosis. In immunocompromised patients, fluconazole can enhance C. dubliniensis. The C. dubliniensis is known as another germ in oral candidiasis and candidemia, which can increase, Saps expression [88–90].
Hepatocytes are the hepatitis B virus's primary host cells. The HBV infection is transmitted by blood or sexual activity [91]. Interestingly, the diversity of oral microbiota was decreased in HBV chronic liver disease (HBV-CLD) patients. In HBV-CLD, patients' Fusobacterium, Treponema, Eubacterium, Parvimonas, Pseudomonas, and Filifactor could be detected, which can induce an increased risk of periodontal disease. Indeed, the long-term course of HBV infection and gut-liver axis microbiome changes were the probable causes of oral microbiota alteration. This reduction led to dysbiosis in gut microbiota. In HBV-CLD patients, a high level of inflammation factors like IL-6 and IL-1β impaired the oral immunity system by increasing the abundance of Fusobacterium and Treponema, which attacked gut microbiota as opportunistic pathogens [39]. Immunodeficiency disorders or infections dysregulate the immune system and influence the balance of oral microbiota. In HIV patients, dominant oral organisms are correlated with CD4 T cell count [92].
5. Respiratory Microbiota and COVID-19
The primary transmission route of COVID-19 is respiratory droplets. It can also be transmitted through close contact [93, 94]. Human microbiota comprises viruses, phages, bacteria, and fungi [95]. It is believed that bacteria and fungi' coinfection play a notable role during COVID-19 [10]. For instance, comorbidity associated with severe COVID-19 is a chronic pulmonary disease (CPD) [96]. The airway microbiota composition is altered in CPD patients [97].
Zhou et al. reported the secondary infections and coinfections in COVID-19 patients [98]. Regularly, the human microbiota influences susceptibility to respiratory infections [99]. Microbiota compounds in the lung are altered in COVID-19 patients, and the changes may have an essential role in the COVID-19 immunity and severity [100]. Commensal bacteria can affect antiviral immunity activation, and probiotics can reduce the time duration and degree of respiratory viral infections [101]. Some Gram-positive bacterial microbiota like Staphylococcus aureus has been shown to prevent influenza virus infections [102]. In patients with influenza A and B admitted to the ICU, the percentage of invasive pulmonary aspergillosis (IPA) is higher than patients with severe pneumonia caused by other pathogens except for flu (19% versus 5%) [103]. Schauwvlieghe et al. reported that the 3-month mortality rate of influenza patients with and without the IPA is 51% and 28%, respectively [103]. Regarding the epidemiological data to decrease morbidity and mortality in COVID-19 patients, antifungal chemoprophylaxis and environmental measures could be proposed [104].
Oral health deterioration in COVID-19 patients due to external ventilation and subsequent complexities can be caused by hyposalivation, even affecting the lower respiratory tract, similar to aspiration pneumonia [105]. Impaired balance of oral microbiota arises from systemic treatments and changes in the intraoral environment and may lead to other problems [105]. The large populations in the oral and upper respiratory tract microbiotas are from the Streptococcus spp. [106]. Streptococci can metabolize carbohydrates in the fermentation process and yield acids, which has a role in dental caries progress by species like S. mutans [106]. Patients with COVID-19 have notable lung microbiota, especially with potential dysbiosis and divergence from healthy individuals [107]. Streptococcus salivarius (S. salivarius) is a predominant oral cavity microbiota [108]. Colonization of S. salivarius K12 strain reduces the occurrence of some viral upper respiratory tract infections; in SARS-CoV-2 patients, this field needs further investigation [107]. In a study published in 2003, the severe acute respiratory syndrome (SARS) patients had a secondary infection, including a high percentage of the Pseudomonas aeruginosa, Staphylococcus spp., Stenotrophomonas maltophilia, Klebsiella terrigena, and fungal [109]. Further research is needed to confirm how microbiota communication is changing post-COVID-19 infection, inter- and intrapersonally. The results of current studies related to microbiota in the COVID-19 patients are shown in Table 2.
Table 2.
The predominant microbiota in the COVID-19 patients reported from current studies.
| Type | Outcome | Ref |
|---|---|---|
| Acinetobacter, Chryseobacterium, Burkholderia, Brevundimonas, Sphingobium | The critical impact of mucosal microbiota on the susceptibility to SARS-CoV2 infection and severity of COVID-19 patients | [158] |
| Cutaneotrichosporon, Issatchenkia, Wallemia, Cladosporium, Alternaria, Dipodascus, Mortierella, Aspergillus, Naganishia, Diutina, and Candida | ||
| Firmicutes (42%), Bacteroidetes (25), Proteobacteria (18%), Actinobacteria (8%), and Fusobacteria (5%) | No statistically significant differences in nasopharyngeal microbiota of SARS-CoV-2 infection. | [144] |
| Acinetobacter (80.70%), Chryseobacterium (2.68%), Burkholderia (2.00%), Brevundimonas (1.18%), Sphingobium (0.93%), Mycobacterium (3.59%), and Prevotella (0.56%) | COVID-19 mortality is associated with complex mixed bacterial and fungal infections in the lungs, and microbiota monitoring is necessary in the lower respiratory tract for on-time personalized therapy. | [159] |
| Cutaneotrichosporon (Cryptococcus, 28.14%), followed by Issatchenkia (8.22%), Wallemia (4.77%), Cladosporium (4.67%), Alternaria (4.46%), Dipodascus (4.01%), Mortierella (3.22%), Aspergillus (2.72%), Naganishia (2.53%), Diutina (2.15%), and Candida (1.42%) |
6. Respiratory Microbiota Dysbiosis and COVID-19
A neglected function of lung microbiota is the maintenance of immune tolerance, which leads to the prevention of inflammatory responses, helps lung homeostasis, and can also be supposed as lung health status [110]. The oral cavities are known as a notable reservoir of SARS-CoV-2 [111]. Since the oral microbiota interacts with SARS-CoV-2, efficient oral health care efforts are needed to reduce severe SARS-CoV-2 infections [112]. The microbiota in the human body, such as nasal channels, oral cavities, skin, gastrointestinal tract, and urogenital tract, are important in physiological process, immunity, and nourishment [113]. By recognizing crucial microbiota functions in human health and disease, it could be found that many complicated human disorders are correlated with microbiota [113, 114]. The schematic view of lung microbiota changes in disease and health conditions is conducted in Figure 1 [100]. With new insight into microbiota's role in human diseases and health, these findings can be implemented as a novel therapeutic target [115]. The healthy oral cavity's microbiota is distinct from bacterial inhabitants of other organs in human body. The human oral cavity comprises a distinct set of niches containing the tongue, tonsils, saliva, and teeth [116]. The same bacteria population organizes the oral microbiome in each healthy oral cavity niche [113, 116].
Figure 1.
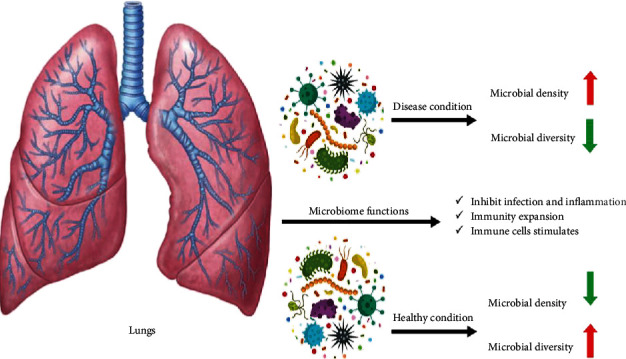
The lung microbiome in disease and health condition.
However, the microbiota is not uniform in different oral cavity circumstances. Bacterial diversity varies significantly between other sampling sites, including saliva, buccal mucosa, and back of the tongue supragingival plaque, and subgingival plaque [117].
Lung microbiota contributes to immunological homeostasis [110]. Viral infection may have considerable interplays with the commensal microbiota. Commensal microbiota can be altered by viral infections or even be reduced during infection [118].
Concerning COVID-19, a highly significant difference in the lung microbiota composition has been observed between patients with SARS-CoV-2 pneumonia and healthy population, implying a dysbiosis in patient's lung microbiota [119]. The Corynebacterium spp., Staphylococcus spp., Propionibacterium spp., and several Malassezia spp. have been recognized as the core nasal members microbiome already [120]. Chonmaitree et al. collected nasopharyngeal microbiota samples longitudinally during health and disease in infants [121]. The results suggested that bacterial otopathogen genera (Haemophilus spp., Streptococcus spp., and Moraxella spp.) were highly abundant in nasopharyngeal microbiota. These bacteria appear to correlate with upper respiratory tract infection (URI) symptoms during viral infection. Chonmaitree et al. mentioned the probiotic bacterium Staphylococcus spp. and Bifidobacterium spp. played a crucial role in inhibiting the otopathogens' harmful effects [121].
Respiratory microorganisms were widely characterized [42, 113, 122]. Balance in three factors, microbial immigration, microbial elimination, and relative reproduction rates, can determine lung microbiome characteristics [123]. The human respiratory tract harbors a homogenous microbiota that reduces biomass from the upper to the lower tract [42]. The nasopharynx core microbiome remains indistinct because it varies extensively from person to person in seasons [122]. One study reported that the upper respiratory tract's microbial balance is typically unique to each person, changing little over time [124]. However, the antimicrobial prophylaxis and treatment may induce dysbiosis in airway microbiota and increase the Haemophilus parainfluenzae and yeast colonization [125].
By increasing mucosal function and the ability to differentiate structure, stimulating in both the innate and adaptive immune systems, and giving “colonization resistance” against pathogen invasion, the human microbiota is regarded to benefit the host [126]. The commensal microbiota's importance was described in viral infection, with the commensal microbiota composition critically regulating host immune response following respiratory infections such as influenza A virus [127]. A wide range of respiratory tract infections is caused by viruses, including coronavirus, rhinovirus, respiratory syncytial virus, and influenza virus [128]. Infection by respiratory viruses has a pathological effect on the respiratory tract caused by the viral invasion or immunopathogenesis process and induced microbiome alterations and secondary infection [18, 129, 130]. Lei et al. reported that monitoring fungal infection in patients with SARS-CoV-2 should be considered due to the high positive rate of fungal antigenemia [131]. Also, Chen et al. reported fungal coinfections, including C. albicans and C. glabrata, between patients with COVID-19 [10]. Preliminary reports showed further investigations need to evaluate fungal coinfection among COVID-19 patients [10, 131]. In one study beginning in the outbreak and with the fast spread of the SARS-CoV, the first few cases were treated with a mixture of ribavirin and corticosteroids, with good results. Long-term treatment with high-dose steroids and the lack of an active antimicrobial agent can cause difficulties such as disseminated fungal infection in patients [132]. Corticosteroid therapy, which is usually sufficient to modulate immune reaction in severe inflammatory conditions, seems harmful in some of the COVID-19 cases [133, 134]. Fungal and bacterial infections are common complications of viral pneumonia in seriously ill patients [135]; a comprehensive investigation is needed in COVID-19 patients.
7. Respiratory Microbiota and COVID-19 Transmission
Yildiz et al., in an experience of influenza A virus infection on a mouse model, indicate qualitative dysbiosis and bacterial superinfection sensitivity in the lower respiratory tract microbiota compounds [136]. Observing overall shifts in the bacterial and fungal community of sinus diversity was shown to be attributed to a compound of personal, seasonal, and annual changes [120]. Oral opportunistic pathogens like Capnocytophaga and Veillonella were found in the bronchoalveolar lavage (BAL) sample of the COVID-19 patients [112]. The poor oral hygiene, cough, raised inhalation conditions, and ventilation cause a transmission route for oral microbiota to penetrate the lower respiratory tract and cause respiratory disorders [112].
During COVID-19, some pathological oral conditions could be aggregated, especially in the compromised immune system and prolonged therapeutic approach [105]. Appearing evidence submits that the nasopharyngeal microbiota's composition is correlated with susceptibility to acute respiratory infections and, importantly, the host immune response in children [137]. It has been shown that respiratory tract bacteria are not inactive during severe respiratory infections but rather have a complex interaction with the host immune response and infecting viruses [138, 139]. Ecosystem imbalance may cause overgrowth and invasion by bacterial pathogens and beginning respiratory or invasive diseases [140]. Respiratory bacteria and respiratory viruse colonization is frequently competitive interspecies interactions and can induce microbiota dysbiosis at the nasopharyngeal niche [140].
SARS-CoV-2 infection likely occurs in patients already colonized with bacteria. Besides, the very reasonable possibility exists that severe COVID-19 patients could be subsequently or coincidentally infected by bacteria and fungi [10]. In COVID-19, detecting bacterial or fungal infection based on the clinical and radiological form could be challenging. The microbiological techniques can help diagnose, mainly sputum culture [135]. The bacterial composition of the nasal microbiota varies between stages of life [141]. A cross-sectional study focused on this transition indicates that puberty has a significant impact on nasal microbiota composition. There are statistically significant differences in nostril microbiota compounds, in which Actinobacteria spp. and particularly Corynebacterium spp., Propionibacterium spp., and Turicella spp. are overrepresented in some conditions [142]. By affecting COVID-19 on most of the ciliated cells in the alveoli and disturbance on clearing the airways, progressive debris and fluid accumulation could be expected [143].
8. Respiratory Microbiota and COVID-19 Severity
The human upper respiratory tract is the leading entrance for aerosol-transmitted microorganisms, including SARS-CoV-2 [144]. The complex interactive oral microbiota has an expansive biofilm configuration. Besides the bacteria, Candida is a typical microbiota. Also, 100 recognized species of pathogenic fungi, including Cryptococcus spp., Aspergillus spp., and Fusarium spp., appear to reside in some individuals [145]. The microbiota of healthy lungs overlaps with that found in the mouth [146]. In bronchoalveolar lavage fluid samples from healthy adults, the well-known genera consist of Streptococcus spp., Prevotella spp., and Veillonella spp. are detected [146, 147]. Strain K12 of Streptococcus salivarius has been clinically demonstrated to play a role in creating a stable upper respiratory tract microbiota due to the ability to stimulate IFN-γ release and to activate natural killer cells (NK) without triggering aggressive inflammatory responses. Also, strain K12 is capable of protecting the host from pathogenic viral infections. The proposed antiviral capability of strain K12 has been attributed to the observed development of an adaptive immune response, as revealed by the detection of enhanced IFN-γ levels in human saliva [107]. More investigation needs to evaluate the impact of strain K12 on SARS-CoV-2 and COVID-19 severity.
The innate and adaptive immune systems are active against the SARS-CoV-2 infection. Lymphopenia, with an enormously decreased of B cells, CD4+ and CD8+ T cells, NK cells, and monocytes, is associated with the increased severity of COVID-19 [148, 149]. The regulatory T cells can affect microbiota and microbiota regulating the immune system and play an essential role in maintaining homeostasis [150, 151]. Gathering obtained evidence with different mediations such as antibiotic exposure, and microbiota transfer showed that the microbiota could enhance antiviral immunity, a new perspective for efficient treatments in COVID-19 patients [19]. The SARS-CoV-2 mutations could cause alterations in virus pathogenicity [152]. Hence, it is crucial to investigate the pattern and rate of mutations that happened [153].
Lung microbiota is associated with disease susceptibility and severity [154]. Shen et al. analyzed changes in the lung microbiota composition in SARS-CoV-2-infected patients and showed the microbial balance in these patients' BAL. Commensal and pathogenic bacteria dominate this communication, and this composition is also different from the healthy control group [119]. Few studies have been performed on the interaction between lower respiratory tract (LRT) microbiota and viral infections. Alterations in the microbiota in the LRT during viral infection were variable and might result from the reduced capability to remove pathogens in the upper respiratory tract [19]. Probiotics can develop immunity against influenza infection. The microbiota can probably work as a target for antiviral therapy [19]. It needs to be understood how microbiota could help assess clinical status and serve as a target for anti-SARS-CoV-2 therapies [19].
9. Conclusion
Microbiota communities play critical roles in immune system homeostasis. Therefore, any alteration in the healthy humans' microbiota can have detrimental impacts on health and may lead to an infection or the progression of the disease. It seems that the microbiota balance differs between the healthy group and COVID-19 patients. Dysbiosis in certain microbiota species' populations may alter the pathogenesis of COVID-19 in patients. Therefore, tracking these changes is useful as a prognostic approach during COVID-19 treatment. Further studies are needed to determine significant cellular changes resulting from SARS-CoV-2 and microbiota interactions.
Acknowledgments
The present study is funded by Hormozgan University of Medical Sciences, Bandar Abbas, Iran (grant no. 990699). We gratefully acknowledge the Vice-Chancellor for Research and Technology, Hormozgan University of Medical Sciences, Bandar Abbas, Iran. Also, we are sincerely thankful to our counsellors in Clinical Research Development Center of Shahid Mohammadi Hospital, Hormozgan University of Medical Sciences, Bandar Abbas, Iran.
Data Availability
All data associated with this manuscript is inclusive in this paper.
Additional Points
All tables and figure in this study are original.
Disclosure
The supporting organization has no role in the design of the study, collection, analysis, and interpretation of data and in writing the manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Saber Soltani and Abbas Farahani conceptualized and designed the review and interpretation of data for the work, wrote the manuscript, and finally approved the version to be published. Armin Zakeri, Milad Zandi, Mahsa Dastranj, Mina Mobini Kesheh, Samireh Faramarzi, Mojtaba Didehdar, Hossein Hafezi, Parastoo Hosseini, Samireh Faramarzi, and Alireza Tabibzadeh collected the data and wrote the manuscript. Saber Soltani and Abbas Farahani supervised the collection of the data and wrote the manuscript. All authors reviewed and approved the manuscript.
References
- 1.Guo Y.-R., Cao Q.-D., Hong Z.-S., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Military Medical Research. 2020;7(1):p. 11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soltani S. The hemagglutinin-esterase gene in human coronaviruses SARS-CoV-2, HKU1 and OC43. European Review for Medical and Pharmacological Sciences. 2020;24(12):6484–6485. doi: 10.26355/eurrev_202006_21630. [DOI] [PubMed] [Google Scholar]
- 3.Soltani S., Zandi M., Aghbash P. S., et al. A review of COVID‐19 vaccines and major considerations for diabetic patients. Biotechnology and Applied Biochemistry. 2020;67(6):1–11. doi: 10.1002/bab.2076. [DOI] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., et al. A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belouzard S., Chu V. C., Whittaker G. R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hasan A., Paray B. A., Hussain A., et al. A review on the cleavage priming of the spike protein on coronavirus by angiotensin-converting enzyme-2 and furin. Journal of Biomolecular Structure and Dynamics. 2020;38:1–9. doi: 10.1080/07391102.2020.1754293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Amico F., Baumgart D. C., Danese S., Peyrin-Biroulet L. Diarrhea During COVID-19 Infection: Pathogenesis, Epidemiology, Prevention, and Management. Clinical Gastroenterology and Hepatology. 2020;18(8):1663–1672. doi: 10.1016/j.cgh.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang D., Hu B., Hu C., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N., Zhou M., Dong X., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. The Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reyes A., Haynes M., Hanson N., et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.NIH HMP Working Group, Peterson J., Garges S., et al. The NIH human microbiome project. Genome Research. 2009;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rooks M. G., Veiga P., Wardwell-Scott L. H., et al. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. The ISME Journal. 2014;8(7):1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ley R. E. Obesity and the human microbiome. Current Opinion in Gastroenterology. 2010;26(1):5–11. doi: 10.1097/MOG.0b013e328333d751. [DOI] [PubMed] [Google Scholar]
- 16.Lloyd-Price J., Abu-Ali G., Huttenhower C. The healthy human microbiome. Genome Medicine. 2016;8(1):p. 51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogaert D., de Groot R., Hermans P. W. M. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. The Lancet Infectious Diseases. 2004;4(3):144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 18.Hanada S., Pirzadeh M., Carver K. Y., Deng J. C. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Frontiers in Immunology. 2018;9:p. 2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y., Wang J., Li F., Shi Y. Main clinical features of COVID-19 and potential prognostic and therapeutic value of the microbiota in SARS-CoV-2 infections. Frontiers in Microbiology. 2020;11:p. 1302. doi: 10.3389/fmicb.2020.01302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kyo M., Nishioka K., Nakaya T., et al. Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respiratory Research. 2019;20(1):p. 246. doi: 10.1186/s12931-019-1203-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zaura E., Nicu E. A., Krom B. P., Keijser B. J. F. Acquiring and maintaining a normal oral microbiome: current perspective. Frontiers in Cellular and Infection Microbiology. 2014;4:p. 85. doi: 10.3389/fcimb.2014.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wade W. G. The oral microbiome in health and disease. Pharmacological Research. 2013;69(1):137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Deo P. N., Deshmukh R. Oral microbiome: unveiling the fundamentals. Journal of Oral and Maxillofacial Pathology: JOMFP. 2019;23(1):122–128. doi: 10.4103/jomfp.JOMFP_304_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamarre A., Talbot P. J. Effect of pH and temperature on the infectivity of human coronavirus 229E. Canadian Journal of Microbiology. 1989;35(10):972–974. doi: 10.1139/m89-160. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y., Wang X., Li H., Ni C., Du Z., Yan F. Human oral microbiota and its modulation for oral health. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2018;99:883–893. doi: 10.1016/j.biopha.2018.01.146. [DOI] [PubMed] [Google Scholar]
- 26.Ghannoum M. A., Jurevic R. J., Mukherjee P. K., et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathogens. 2010;6(1, article e1000713) doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamashita Y., Takeshita T. The oral microbiome and human health. Journal of Oral Science. 2017;59(2):201–206. doi: 10.2334/josnusd.16-0856. [DOI] [PubMed] [Google Scholar]
- 28.Tonoyan L., Vincent-Bugnas S., Olivieri C.-V., Doglio A. New viral facets in oral diseases: the EBV paradox. International Journal of Molecular Sciences. 2019;20(23):p. 5861. doi: 10.3390/ijms20235861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin Y. L., Li M. Human cytomegalovirus and Epstein-Barr virus inhibit oral bacteria-induced macrophage activation and phagocytosis. Oral Microbiology and Immunology. 2009;24(3):243–248. doi: 10.1111/j.1399-302X.2009.00504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jia G., Zhi A., Lai P. F. H., et al. The oral microbiota – a mechanistic role for systemic diseases. British Dental Journal. 2018;224(6):447–455. doi: 10.1038/sj.bdj.2018.217. [DOI] [PubMed] [Google Scholar]
- 31.Arweiler N. B., Netuschil L. The oral microbiota. Advances in Experimental Medicine and Biology. 2016;902:45–60. doi: 10.1007/978-3-319-31248-4_4. [DOI] [PubMed] [Google Scholar]
- 32.Costalonga M., Herzberg M. C. The oral microbiome and the immunobiology of periodontal disease and caries. Immunology Letters. 2014;162(2):22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costello E. K., Lauber C. L., Hamady M., Fierer N., Gordon J. I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lif Holgerson P., Harnevik L., Hernell O., Tanner A. C. R., Johansson I. Mode of birth delivery affects oral microbiota in infants. Journal of Dental Research. 2011;90(10):1183–1188. doi: 10.1177/0022034511418973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagan T., Cortese M., Rouphael N., et al. Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell. 2019;178(6):1313–28.e13. doi: 10.1016/j.cell.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ozkan B., Topuk S. N., Taser N., Filik L. Oral microbiota change, tooth decay and hemorrhoidal disease. Anaerobe. 2015;33:p. 137. doi: 10.1016/j.anaerobe.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 37.Siqueira J. F., Jr., Rôças I. N. The oral microbiota in health and disease: an overview of molecular findings. Methods in Molecular Biology. 2017;1537:127–138. doi: 10.1007/978-1-4939-6685-1_7. [DOI] [PubMed] [Google Scholar]
- 38.Lamont R. J., Koo H., Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nature Reviews Microbiology. 2018;16(12):745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ling Z., Liu X., Cheng Y., et al. Decreased diversity of the oral microbiota of patients with hepatitis B virus-induced chronic liver disease: a pilot project. Scientific Reports. 2015;5(1):p. 17098. doi: 10.1038/srep17098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilbert S. A., Mark Welch J. L., Borisy G. G. Spatial ecology of the human tongue dorsum microbiome. Cell Reports. 2020;30(12):p. 4003. doi: 10.1016/j.celrep.2020.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemon K. P., Klepac-Ceraj V., Schiffer H. K., Brodie E. L., Lynch S. V., Kolter R. Comparative analyses of the bacterial microbiota of the human nostril and oropharynx. MBio. 2010;1(3) doi: 10.1128/mbio.00129-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Charlson E. S., Bittinger K., Haas A. R., et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. American Journal of Respiratory and Critical Care Medicine. 2011;184(8):957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yi H., Yong D., Lee K., Cho Y.-J., Chun J. Profiling bacterial community in upper respiratory tracts. BMC Infectious Diseases. 2014;14(1):p. 583. doi: 10.1186/s12879-014-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gong H., Wang B., Shi Y., et al. Composition and abundance of microbiota in the pharynx in patients with laryngeal carcinoma and vocal cord polyps. Journal of Microbiology. 2017;55(8):648–654. doi: 10.1007/s12275-017-6636-8. [DOI] [PubMed] [Google Scholar]
- 45.Vila T., Sultan A. S., Montelongo-Jauregui D., Jabra-Rizk M. A. Oral candidiasis: a disease of opportunity. Journal of Fungi. 2020;6(1):p. 15. doi: 10.3390/jof6010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suomalainen K., Saxen L., Vilja P., Tenovuo J. Peroxidases, lactoferrin and lysozyme in peripheral blood neutrophils, gingival crevicular fluid and whole saliva of patients with localized juvenile periodontitis. Oral Diseases. 1996;2(2):129–134. doi: 10.1111/j.1601-0825.1996.tb00213.x. [DOI] [PubMed] [Google Scholar]
- 47.Salazar M. G., Jehmlich N., Murr A., et al. Identification of periodontitis associated changes in the proteome of whole human saliva by mass spectrometric analysis. Journal of Clinical Periodontology. 2013;40(9):825–832. doi: 10.1111/jcpe.12130. [DOI] [PubMed] [Google Scholar]
- 48.Lis M., Bhatt S., Schoenly N. E., Lee A. Y., Nislow C., Bobek L. A. Chemical genomic screening of a Saccharomyces cerevisiae genomewide mutant collection reveals genes required for defense against four anti-microbial peptides derived from proteins found in human saliva. Anti-Microbial Agents and Chemotherapy. 2013;57(2):840–847. doi: 10.1128/AAC.01439-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verma A., Gaffen S. L., Swidergall M. Innate immunity to mucosal Candida infections. Journal of Fungi. 2017;3(4):p. 60. doi: 10.3390/jof3040060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Machado L. R., Ottolini B. An evolutionary history of defensins: a role for copy number variation in maximizing host innate and adaptive immune responses. Frontiers in Immunology. 2015;6(115) doi: 10.3389/fimmu.2015.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doss M., White M. R., Tecle T., Hartshorn K. L. Human defensins and LL-37 in mucosal immunity. Journal of Leukocyte Biology. 2010;87(1):79–92. doi: 10.1189/jlb.0609382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greer A., Zenobia C., Darveau R. P. Defensins and LL-37: a review of function in the gingival epithelium. Periodontology 2000. 2013;63(1):67–79. doi: 10.1111/prd.12028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devine D. A., Marsh P. D., Meade J. Modulation of host responses by oral commensal bacteria. Journal of Oral Microbiology. 2015;7(1):p. 26941. doi: 10.3402/jom.v7.26941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hezel M. P., Weitzberg E. The oral microbiome and nitric oxide homoeostasis. Oral Diseases. 2015;21(1):7–16. doi: 10.1111/odi.12157. [DOI] [PubMed] [Google Scholar]
- 55.Abusleme L., Moutsopoulos N. M. IL-17: overview and role in oral immunity and microbiome. Oral Diseases. 2017;23(7):854–865. doi: 10.1111/odi.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zaph C., Du Y., Saenz S. A., et al. Commensal-dependent expression of IL-25 regulates the IL-23–IL-17 axis in the intestine. Journal of Experimental Medicine. 2008;205(10):2191–2198. doi: 10.1084/jem.20080720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Górski A., Weber-Dabrowska B. The potential role of endogenous bacteriophages in controlling invading pathogens. Cellular and Molecular Life Sciences. 2005;62(5):511–519. doi: 10.1007/s00018-004-4403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bachrach G., Leizerovici-Zigmond M., Zlotkin A., Naor R., Steinberg D. Bacteriophage isolation from human saliva. Letters in Applied Microbiology. 2003;36(1):50–53. doi: 10.1046/j.1472-765X.2003.01262.x. [DOI] [PubMed] [Google Scholar]
- 59.Roach D. R., Leung C. Y., Henry M., et al. Synergy between the host immune system and bacteriophage is essential for successful phage therapy against an acute respiratory pathogen. Cell Host & Microbe. 2017;22(1):38–47.e4. doi: 10.1016/j.chom.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 60.van Belleghem J., Dąbrowska K., Vaneechoutte M., Barr J., Bollyky P. Interactions between bacteriophage, bacteria, and the mammalian immune system. Viruses. 2018;11(1):p. 10. doi: 10.3390/v11010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Majewska J., Beta W., Lecion D., et al. Oral application of T4 phage induces weak antibody production in the gut and in the blood. Viruses. 2015;7(8):4783–4799. doi: 10.3390/v7082845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukherjee P. K., Chandra J., Retuerto M., et al. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathogens. 2014;10(3, article e1003996) doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanel A. N., Herzog H. M., James M. G., Cuadra G. A. Effects of oral commensal streptococci on Porphyromonas gingivalis invasion into oral epithelial cells. Dentistry Journal. 2020;8(2):p. 39. doi: 10.3390/dj8020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Avila M., Ojcius D. M., Yilmaz O. The oral microbiota: living with a permanent guest. DNA and Cell Biology. 2009;28(8):405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jakubovics N. S., Gill S. R., Vickerman M. M., Kolenbrander P. E. Role of hydrogen peroxide in competition and cooperation between Streptococcus gordonii and Actinomyces naeslundii. FEMS Microbiology Ecology. 2008;66(3):637–644. doi: 10.1111/j.1574-6941.2008.00585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowen W. H., Burne R. A., Wu H., Koo H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends in Microbiology. 2018;26(3):229–242. doi: 10.1016/j.tim.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mohammed W. K., Krasnogor N., Jakubovics N. S. Streptococcus gordonii Challisin protease is required for sensing cell--cell contact with Actinomyces oris. FEMS Microbiology Ecology. 2018;94(5) doi: 10.1093/femsec/fiy043. [DOI] [PubMed] [Google Scholar]
- 68.Enersen M., Nakano K., Amano A. Porphyromonas gingivalis fimbriae. Journal of Oral Microbiology. 2013;5(1):p. 20265. doi: 10.3402/jom.v5i0.20265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blasco-Baque V., Garidou L., Pomié C., et al. Periodontitis induced byPorphyromonas gingivalisdrives periodontal microbiota dysbiosis and insulin resistance via an impaired adaptive immune response. Gut. 2017;66(5):872–885. doi: 10.1136/gutjnl-2015-309897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clais S., Boulet G., Kerstens M., et al. Importance of biofilm formation and dipeptidyl peptidase IV for the pathogenicity of clinical Porphyromonas gingivalis isolates. Pathogens and Disease. 2014;70(3):408–413. doi: 10.1111/2049-632X.12156. [DOI] [PubMed] [Google Scholar]
- 71.Ohara-Nemoto Y., Shimoyama Y., Nakasato M., et al. Distribution of dipeptidyl peptidase (DPP) 4, DPP5, DPP7 and DPP11 in human oral microbiota—potent biomarkers indicating presence of periodontopathic bacteria. FEMS Microbiology Letters. 2018;365(22) doi: 10.1093/femsle/fny221. [DOI] [PubMed] [Google Scholar]
- 72.Aemaimanan P., Sattayasai N., Wara-aswapati N., et al. Alanine aminopeptidase and dipeptidyl peptidase IV in saliva of chronic periodontitis patients. Journal of Periodontology. 2009;80(11):1809–1814. doi: 10.1902/jop.2009.090233. [DOI] [PubMed] [Google Scholar]
- 73.Holst J. J., Orskov C. The incretin approach for diabetes treatment: modulation of islet hormone release by GLP-1 agonism. Diabetes. 2004;1(53, Supplement 3):s197–s204. doi: 10.2337/diabetes.53.suppl_3.s197. [DOI] [PubMed] [Google Scholar]
- 74.Ohara-Nemoto Y., Nakasato M., Shimoyama Y., et al. Degradation of incretins and modulation of blood glucose levels by periodontopathic bacterial dipeptidyl peptidase 4. Infection and Immunity. 2017;85(9) doi: 10.1128/IAI.00277-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amano A. Disruption of epithelial barrier and impairment of cellular function by Porphyromonas gingivalis. Front Biosci. 2007;1(12):p. 74. doi: 10.2741/2363. [DOI] [PubMed] [Google Scholar]
- 76.de Andrade K. Q., Almeida-da-Silva C. L. C., Coutinho-Silva R. Immunological pathways triggered by Porphyromonas gingivalis and Fusobacterium nucleatum: therapeutic possibilities? Mediators of Inflammation. 2019;2019:20. doi: 10.1155/2019/7241312.7241312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nativel B., Couret D., Giraud P., et al. Porphyromonas gingivalis lipopolysaccharides act exclusively through TLR4 with a resilience between mouse and human. Scientific Reports. 2017;7(1):p. 15789. doi: 10.1038/s41598-017-16190-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Michaud D. S., Izard J. Microbiota, oral microbiome, and pancreatic cancer. Cancer Journal. 2014;20(3):203–206. doi: 10.1097/PPO.0000000000000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miccoli G., Gaimari G., Seracchiani M., Morese A., Khrenova T., di Nardo D. Evaluation of microbiota associated with Herpesviruses in active sites of generalized aggressive periodontitis. Annali di Stomatologia. 2017;8(2):p. 59. doi: 10.11138/ads/2017.8.2.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Goncalves P. H., Ziegelbauer J., Uldrick T. S., Yarchoan R. Kaposi sarcoma herpesvirus-associated cancers and related diseases. Current Opinion in HIV and AIDS. 2017;12(1):47–56. doi: 10.1097/COH.0000000000000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dai L., DeFee M. R., Cao Y., et al. Lipoteichoic acid (LTA) and lipopolysaccharides (LPS) from periodontal pathogenic bacteria facilitate oncogenic Herpesvirus infection within primary oral cells. PLoS One. 2014;9(6, article e101326) doi: 10.1371/journal.pone.0101326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ganly I., Yang L., Giese R. A., et al. Periodontal pathogens are a risk factor of oral cavity squamous cell carcinoma, independent of tobacco and alcohol and human papillomavirus. International Journal of Cancer. 2019;145(3):775–784. doi: 10.1002/ijc.32152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim R. H., Kim R., Chen W., et al. Association of hsp90 to the hTERT promoter is necessary for hTERT expression in human oral cancer cells. Carcinogenesis. 2008;29(12):2425–2431. doi: 10.1093/carcin/bgn225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang X., Browngardt C. M., Jiang M., Ahn S. J., Burne R. A., Nascimento M. M. Diversity in antagonistic interactions between commensal oral streptococci and Streptococcus mutans. Caries Research. 2018;52(1-2):88–101. doi: 10.1159/000479091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Millsop J. W., Fazel N. Oral candidiasis. Clinics in Dermatology. 2016;34(4):487–494. doi: 10.1016/j.clindermatol.2016.02.022. [DOI] [PubMed] [Google Scholar]
- 86.Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., White T. C. Hidden killers: human fungal infections. Science Translational Medicine. 2012;4(165):p. 165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 87.Zhu W., Phan Q. T., Boontheung P., Solis N. V., Loo J. A., Filler S. G. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(35):14194–14199. doi: 10.1073/pnas.1117676109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schaller M., Borelli C., Korting H. C., Hube B. Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses. 2005;48(6):365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- 89.Khan Z., Ahmad S., Joseph L., Chandy R. Candida dubliniensis: an appraisal of its clinical significance as a bloodstream pathogen. PLoS One. 2012;7(3, article e32952) doi: 10.1371/journal.pone.0032952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Borg-von Zepelin M., Niederhaus T., Gross U., Seibold M., Monod M., Tintelnot K. Adherence of different Candida dubliniensis isolates in the presence of fluconazole. AIDS. 2002;16(9):1237–1244. doi: 10.1097/00002030-200206140-00005. [DOI] [PubMed] [Google Scholar]
- 91.Kwon S. Y., Lee C. H. Epidemiology and prevention of hepatitis B virus infection. The Korean Journal of Hepatology. 2011;17(2):87–95. doi: 10.3350/kjhep.2011.17.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hegde M. C., Kumar A., Bhat G., Sreedharan S. Oral microflora: a comparative study in HIV and normal patients. Indian Journal of Otolaryngology and Head & Neck Surgery. 2014;66(S1):126–132. doi: 10.1007/s12070-011-0370-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sun C., Zhai Z. The efficacy of social distance and ventilation effectiveness in preventing COVID-19 transmission. Sustainable Cities and Society. 2020;62, article 102390 doi: 10.1016/j.scs.2020.102390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fani M., Zandi M., Soltani S., Abbasi S. Future developments in biosensors for field-ready SARS-CoV-2 virus diagnostics. Biotechnology and Applied Biochemistry. 2020;2020 doi: 10.1002/bab.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gilbert J. A., Blaser M. J., Caporaso J. G., Jansson J. K., Lynch S. V., Knight R. Current understanding of the human microbiome. Nature Medicine. 2018;24(4):392–400. doi: 10.1038/nm.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.He Y., Xie M., Zhao J., Liu X. Clinical characteristics and outcomes of patients with severe COVID-19 and chronic obstructive pulmonary disease (COPD) Medical Science Monitor: International Medical Journal of Experimental and Clinical Research. 2020;26, article e927212 doi: 10.12659/MSM.927212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Huang Y. J., Lynch S. V. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Review of Respiratory Medicine. 2014;5(6):809–821. doi: 10.1586/ers.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou F., Yu T., du R., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Libertucci J., Young V. B. The role of the microbiota in infectious diseases. Nature Microbiology. 2019;4(1):35–45. doi: 10.1038/s41564-018-0278-4. [DOI] [PubMed] [Google Scholar]
- 100.Khatiwada S., Subedi A. Lung microbiome and coronavirus disease 2019 (COVID-19): possible link and implications. Human Microbiome Journal. 2020;17, article 100073 doi: 10.1016/j.humic.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abt M. C., Osborne L. C., Monticelli L. A., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37(1):158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J., Li F., Sun R., et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nature Communications. 2013;4(1, article 2106) doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schauwvlieghe A. F., Rijnders B. J., Philips N., et al. Invasive aspergillosis in patients admitted to the intensive care unit with severe influenza: a retrospective cohort study. The Lancet Respiratory Medicine. 2018;6(10):782–792. doi: 10.1016/S2213-2600(18)30274-1. [DOI] [PubMed] [Google Scholar]
- 104.Gangneux J.-P., Bougnoux M. E., Dannaoui E., Cornet M., Zahar J. R. Invasive fungal diseases during COVID-19: we should be prepared. Journal De Mycologie Medicale. 2020;30(2):p. 100971. doi: 10.1016/j.mycmed.2020.100971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dziedzic A., Wojtyczka R. The impact of coronavirus infectious disease 19 (COVID-19) on oral health. Oral Diseases. 2020;27(2):1–4. doi: 10.1111/odi.13359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abranches J., Zeng L., Kajfasz J. K., et al. Biology of oral streptococci. Gram-Positive Pathogens. 2019;1:426–434. doi: 10.1128/9781683670131.ch26. [DOI] [Google Scholar]
- 107.Di Pierro F. A possible probiotic (S. salivarius K12) approach to improve oral and lung microbiotas and raise defenses against SAR S-CoV-2. Minerva Medica. 2020;111(3):281–283. doi: 10.23736/s0026-4806.20.06570-2. [DOI] [PubMed] [Google Scholar]
- 108.Wescombe P. A., Hale J. D., Heng N. C., Tagg J. R. Developing oral probiotics from Streptococcus salivarius. Future Microbiology. 2012;7(12):1355–1371. doi: 10.2217/fmb.12.113. [DOI] [PubMed] [Google Scholar]
- 109.Zheng Z., Chen R., Li Y. The clinical characteristics of secondary infections of lower respiratory tract in severe acute respiratory syndrome. The Chinese Journal of Respiratory and Critical Care Medicine. 2003;2:270–274. [Google Scholar]
- 110.Sommariva M., le Noci V., Bianchi F., et al. The lung microbiota: role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cellular and Molecular Life Sciences. 2020;77(14):2739–2749. doi: 10.1007/s00018-020-03452-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xiang Z., Koo H., Chen Q., Zhou X., Liu Y., Simon-Soro A. Potential implications of SARS-CoV-2 oral infection in the host microbiota. Journal of Oral Microbiology. 2020;13(1, article 1853451) doi: 10.1080/20002297.2020.1853451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bao L., Zhang C., Dong J., Zhao L., Li Y., Sun J. Oral microbiome and SARS-CoV-2: beware of lung co-infection. Frontiers in Microbiology. 2020;11:p. 1840. doi: 10.3389/fmicb.2020.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ling Z., Liu X., Luo Y., et al. Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates. BMC Genomics. 2013;14(1):p. 390. doi: 10.1186/1471-2164-14-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Koren O., Spor A., Felin J., et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Supplement_1):4592–4598. doi: 10.1073/pnas.1011383107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Belizário J. E., Napolitano M. Human microbiomes and their roles in dysbiosis, common diseases, and novel therapeutic approaches. Frontiers in Microbiology. 2015;6:p. 1050. doi: 10.3389/fmicb.2015.01050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pflughoeft K. J., Versalovic J. Human microbiome in health and disease. Annual Review of Pathology: Mechanisms of Disease. 2012;7(1):99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 117.Aas J. A., Paster B. J., Stokes L. N., Olsen I., Dewhirst F. E. Defining the normal bacterial flora of the oral cavity. Journal of Clinical Microbiology. 2005;43(11):5721–5732. doi: 10.1128/JCM.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li N., Ma W. T., Pang M., Fan Q. L., Hua J. L. The commensal microbiota and viral infection: a comprehensive review. Frontiers in Immunology. 2019;10:p. 1551. doi: 10.3389/fimmu.2019.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shen Z., Xiao Y., Kang L., et al. Genomic Diversity of Severe Acute Respiratory Syndrome–Coronavirus 2 in Patients With Coronavirus Disease 2019. Clinical Infectious Diseases. 2020;71(15):713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wagner Mackenzie B., Chang K., Zoing M., et al. Longitudinal study of the bacterial and fungal microbiota in the human sinuses reveals seasonal and annual changes in diversity. Scientific Reports. 2019;9(1):p. 17416. doi: 10.1038/s41598-019-53975-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chonmaitree T., Jennings K., Golovko G., et al. Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One. 2017;12(7, article e0180630) doi: 10.1371/journal.pone.0180630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bogaert D., Keijser B., Huse S., et al. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6(2, article e17035) doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dickson R. P., Erb-Downward J. R., Martinez F. J., Huffnagle G. B. The microbiome and the respiratory tract. Annual Review of Physiology. 2016;78:481–504. doi: 10.1146/annurev-physiol-021115-105238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Charlson E. S., Chen J., Custers-Allen R., et al. Disordered microbial communities in the upper respiratory tract of cigarette smokers. PLoS One. 2010;5(12, article e15216) doi: 10.1371/journal.pone.0015216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kosikowska U., Biernasiuk A., Rybojad P., Łoś R., Malm A. Haemophilus parainfluenzae as a marker of the upper respiratory tract microbiota changes under the influence of preoperative prophylaxis with or without postoperative treatment in patients with lung cancer. BMC Microbiology. 2016;16(1):p. 62. doi: 10.1186/s12866-016-0679-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Blaser M. J., Falkow S. What are the consequences of the disappearing human microbiota? Nature Reviews Microbiology. 2009;7(12):887–894. doi: 10.1038/nrmicro2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ichinohe T., Pang I. K., Kumamoto Y., et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eccles R. Understanding the symptoms of the common cold and influenza. The Lancet Infectious Diseases. 2005;5(11):718–725. doi: 10.1016/S1473-3099(05)70270-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aherne W., Bird T., Court S. D. M., Gardner P. S., McQuillin J. Pathological changes in virus infections of the lower respiratory tract in children. Journal of Clinical Pathology. 1970;23(1):7–18. doi: 10.1136/jcp.23.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chin M., de Zoysa M., Slinger R., et al. Acute effects of viral respiratory tract infections on sputum bacterial density during CF pulmonary exacerbations. Journal of Cystic Fibrosis. 2015;14(4):482–489. doi: 10.1016/j.jcf.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lei Y., Song Y., Shu Y., et al. Fungal antigenemia in patients with severe coronavirus disease 2019 (COVID-19): the facts and challenges. Journal of Microbiology, Immunology, and Infection. 2020;53(4):657–659. doi: 10.1016/j.jmii.2020.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tai D. Y. Pharmacologic treatment of SARS: current knowledge and recommendations. Annals-Academy of Medicine Singapore. 2007;36(6):p. 438. [PubMed] [Google Scholar]
- 133.Mehta P., McAuley D. F., Brown M., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. The Lancet (London, England) 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Russell C. D., Millar J. E., Baillie J. K. Clinical evidence does not support corticosteroid treatment for 2019-nCoV lung injury. The Lancet. 2020;395(10223):473–475. doi: 10.1016/S0140-6736(20)30317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhou P., Liu Z., Chen Y., Xiao Y., Huang X., Fan X.-G. Bacterial and fungal infections in COVID-19 patients: a matter of concern. Infection Control & Hospital Epidemiology. 2020;41(9):1124–1125. doi: 10.1017/ice.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yildiz S., Mazel-Sanchez B., Kandasamy M., Manicassamy B., Schmolke M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. 2018;6(1):p. 9. doi: 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Man W. H., van Houten M. A., Mérelle M. E., et al. Bacterial and viral respiratory tract microbiota and host characteristics in children with lower respiratory tract infections: a matched case-control study. The Lancet Respiratory Medicine. 2019;7(5):417–426. doi: 10.1016/S2213-2600(18)30449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Biesbroek G., Tsivtsivadze E., Sanders E. A., et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. American Journal of Respiratory and Critical Care Medicine. 2014;190(11):1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 139.Følsgaard N. V., Schjørring S., Chawes B. L., et al. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. American Journal of Respiratory and Critical Care Medicine. 2013;187(6):589–595. doi: 10.1164/rccm.201207-1297OC. [DOI] [PubMed] [Google Scholar]
- 140.Bosch A. A. T. M., Biesbroek G., Trzcinski K., Sanders E. A. M., Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathogens. 2013;9(1, article e1003057) doi: 10.1371/journal.ppat.1003057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Bomar L., Brugger S. D., Lemon K. P. Bacterial microbiota of the nasal passages across the span of human life. Current Opinion in Microbiology. 2018;41:8–14. doi: 10.1016/j.mib.2017.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Oh J., Conlan S., Polley E. C., Segre J. A., Kong H. H. Shifts in human skin and nares microbiota of healthy children and adults. Genome Medicine. 2012;4(10):p. 77. doi: 10.1186/gm378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Perico L., Benigni A., Remuzzi G. Should COVID-19 concern nephrologists? Why and to what extent? The emerging impasse of angiotensin blockade. Nephron. 2020;144(5):213–221. doi: 10.1159/000507305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Maio F., Posteraro B., Ponziani F. R., Cattani P., Gasbarrini A., Sanguinetti M. Nasopharyngeal microbiota profiling of SARS-CoV-2 infected patients. Biological Procedures Online. 2020;22(1) doi: 10.1186/s12575-020-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Baumgardner D. J. Oral fungal microbiota: to thrush and beyond. Journal of Patient-Centered Research and Reviews. 2019;6(4):252–261. doi: 10.17294/2330-0698.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Beck J. M., Young V. B., Huffnagle G. B. The microbiome of the lung. Translational Research. 2012;160(4):258–266. doi: 10.1016/j.trsl.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang J., Li F., Tian Z. Role of microbiota on lung homeostasis and diseases. Science China Life Sciences. 2017;60(12):1407–1415. doi: 10.1007/s11427-017-9151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cao X. COVID-19: immunopathology and its implications for therapy. Nature Reviews Immunology. 2020;20(5):269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Diao B., Wang C., Tan Y., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Frontiers in Immunology. 2020;11:p. 827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cebula A., Seweryn M., Rempala G. A., et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature. 2013;497(7448):258–262. doi: 10.1038/nature12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hepworth M. R., Fung T. C., Masur S. H., et al. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria–specific CD4+ T cells. Science. 2015;348(6238):1031–1035. doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Abdullahi I. N., Emeribe A. U., Ajayi O. A., Oderinde B. S., Amadu D. O., Osuji A. I. Implications of SARS-CoV-2 genetic diversity and mutations on pathogenicity of the COVID-19 and biomedical interventions. Journal of Taibah University Medical Sciences. 2020;15(4):258–264. doi: 10.1016/j.jtumed.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Berngruber T. W., Froissart R., Choisy M., Gandon S. Evolution of virulence in emerging epidemics. PLoS Pathogens. 2013;9(3, article e1003209) doi: 10.1371/journal.ppat.1003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.O’Dwyer D. N., Dickson R. P., Moore B. B. The lung microbiome, immunity, and the pathogenesis of chronic lung disease. The Journal of Immunology. 2016;196(12):4839–4847. doi: 10.4049/jimmunol.1600279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Willis J. R., Gabaldón T. The human oral microbiome in health and disease: from sequences to ecosystems. Microorganisms. 2020;8(2):p. 308. doi: 10.3390/microorganisms8020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Fukui Y., Aoki K., Ishii Y., Tateda K. The palatine tonsil bacteriome, but not the mycobiome, is altered in HIV infection. BMC Microbiology. 2018;18(1):p. 127. doi: 10.1186/s12866-018-1274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Jo R., Nishimoto Y., Umezawa K., et al. Comparison of oral microbiome profiles in stimulated and unstimulated saliva, tongue, and mouth-rinsed water. Scientific Reports. 2019;9(1):p. 16124. doi: 10.1038/s41598-019-52445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.AlKhater S. A. Dynamic interplay between microbiota and mucosal immunity in early shaping of asthma and its implication for the COVID-19 pandemic. Journal of Asthma and Allergy. 2020;13:369–383. doi: 10.2147/JAA.S272705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fan J., Li X., Gao Y., et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. Journal of Infection. 2020;81(3):e64–e67. doi: 10.1016/j.jinf.2020.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data associated with this manuscript is inclusive in this paper.


