Rapidly spreading coronavirus disease 2019 (COVID-19) is currently affecting the world. Specifically, cytokine storms are a key feature in a substantial number of COVID-19 patients,1 and studies from our group and others suggest that the IL-6/IL-6R cascade plays a dominant role in symptom-correlated cytokine storms.2,3 Cell-free circulating RNAs (cfRNAs) in plasma carry information from pathologic sites, and they have been reported to play important roles in disease development;4 however, their involvement in COVID-19 has not yet been clarified. Here, we report the characteristics of plasma cfRNA profiles of COVID-19 patients, and we found that no SARS-CoV-2 RNA is present in the plasma of COVID-19 patients. Compared with healthy donors, significantly higher mRNA expression of IL-6R was observed; miR-451a, a known negative regulator of IL-6R translation, was downregulated, which may promote IL-6R expression at the protein level. In addition, three upregulated long noncoding RNAs (lncRNAs) carrying miR-451a binding sites might function as miRNA sponges to compete with IL-6R for miR-451a in COVID-19 patients. Taken together, we provide the cfRNA landscape of COVID-19 patient plasma and describe the possible mechanisms underlying elevated cytokine storms in COVID-19 patients. These findings will contribute to the identification of drug targets for this new disease.
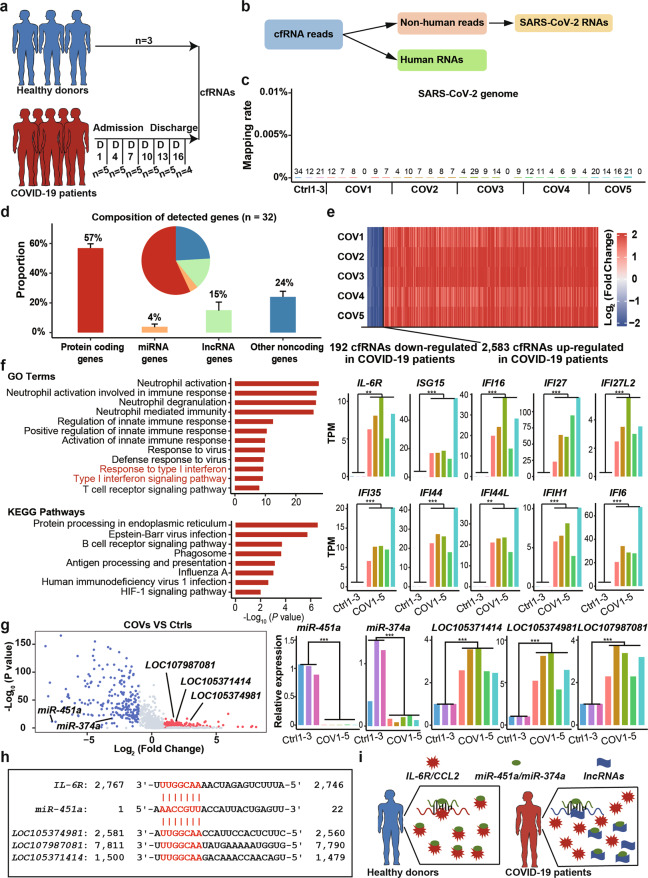
We analyzed the cfRNAs from three healthy donors and five COVID-19 patients (Fig. 1a), and no SARS-CoV-2 RNA was detected (Fig. 1b, c). We detected 33,562 human genes, including 57% protein-coding genes, 4% miRNA genes, 15% lncRNA genes, and 24% other noncoding genes (Fig. 1d and Fig. S1a). Compared with healthy donors, we identified 2583 upregulated and 192 downregulated cfRNA genes in all COVID-19 patients (Fig. 1e). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses revealed that the functions of the upregulated genes are significantly enriched in antivirus-related pathways such as “type I interferon signaling pathway” and “innate immune response and response to virus” (Fig. 1f). In particular, many interferon-stimulated genes (ISGs) were upregulated in COVID-19 patients, including ISG15, IFI6, IFI16, and IFI27L (Fig. 1f), which is consistent with a previous study reporting that IFNα expression is increased in COVID-19 patients.5 Among these ISGs, ISG15 may be related to prolonged viral latency of SARS-CoV-2.6 A previous study found that the IL-6 concentration in COVID-19 patient plasma was higher than the normal range (0–7 pg/ml).5 We also detected high expression of IL-6R in COVID-19 patients compared with healthy donors (Fig. 1f). this high level of IL-6R transcription might be a result of increased type I interferon signaling.7 Moreover, regarding stages of disease progression, expression levels of interferon-stimulated genes, and IL-6R showed a downward trend from stage 1 to stage 4 (Fig. S1b and Supplementary Table S2).
Fig. 1.
The landscape characteristics of cell-free circulating RNAs (cfRNAs) in the plasma of COVID-19 patients; no SARS-CoV-2 RNA was detected in COVID-19 patient plasma. a Schematic diagram showing the sample preparation procedure in this study. cfRNAs were collected from 3 healthy donors and 5 COVID-19 patients. b The computational pipeline to detect human RNAs and SARS-CoV-2 RNAs. c Bar plot showing the mapping rate and number of mapped reads against the SARS-CoV-2 genome (n = 32). d Bar plot and pie plot showing the composition of detected genes in all samples. Data are shown as the mean ± SD (n = 32). e Heatmap showing fold changes (relative to healthy donors) of 2,583 upregulated cfRNA genes and 192 downregulated cfRNA genes in COVID-19 patients. Blue and red represent log2-transformed fold changes < 0 and > 0, respectively. f GO and KEGG enrichment analyses of 2,583 upregulated cfRNA genes in COVID-19 patients (left). Expression levels (TPMs) of IL-6R and representative interferon-stimulated genes (ISGs) are shown on the right. g Volcano plot showing up- and downregulated microRNA (miRNA) and long noncoding RNA (lncRNA) genes in COVID-19 patients relative to healthy donors (left). Relative expression of miR-451a, LOC105371414, LOC105374981 and LOC107987081 is shown on the right. h Base-pairing interaction between miR-451a and IL-6R (top) and the three upregulated lncRNAs (bottom). miR-451a target sites (seed sequences) are highlighted in red. i A proposed model for the regulatory network of miR-451a, IL-6R and lncRNAs in healthy donors and COVID-19 patients. Asterisks indicate statistically significant differences: **P < 0.01; ***P < 0.001
In addition to increased RNA expression of IL-6R, we also found miR-451a, a reported translational repressor of IL-6R,8 to be one of the top five downregulated microRNA (miRNA) genes in COVID-19 patients (Fig. 1g, Fig. S1c, d, Supplementary Table S1 and Supplementary Table S2). Moreover, expression levels of miR-451a showed an upward trend from stage 1 to stage 4 (Fig. S1b and Supplementary Table S3), suggesting that decreased miR-451a may promote expression of IL-6R in COVID-19 patients at the protein level. LncRNAs can act as miRNA sponges to inhibit miRNA function.9 We identified in COVID-19 patients three upregulated lncRNAs, LOC105371414, LOC105374981, and LOC107987081, carrying miR-451a binding sites (Fig. 1g, h and Fig. S1d), which may compete with IL-6R for miR-451a binding. We also found that miR-374a, the target of which is CCL2,10 was downregulated in COVID-19 patients (Fig. 1g). Derepression of CCL2 may confer acute respiratory distress syndrome (ARDS) and cause cytokine storms in COVID-19 patients.11 We also detected 16 upregulated lncRNAs carrying miR-374a binding sites (Fig. 1g and Fig. S2b). In healthy donors, miR-451a/miR-374a can maintain the normal level of IL-6R/CCL2 by targeting IL-6R/CCL2 mRNAs. However, in COVID-19 patients, decreased expression of miR-451a/miR-374a and its binding to lncRNAs may promote expression of IL-6R/CCL2 at the protein level (Fig. 1i). These results suggest that decreased miR-451a/miR-374a and enhanced lncRNA levels may exacerbate IL-6-induced cytokine storms by promoting IL-6R/CCL2 translation in COVID-19 patients.
The cytokine storm in COVID-19 patients is characterized by increased IL-62. However, the mechanisms of cytokine storm-correlated symptoms from the perspective of cfRNAs remain unclear. Our study identified obvious differences in cfRNA molecules between COVID-19 patients and healthy donors. We found activation of type I interferon-responsive genes and low expression of miR-451a, which may lead to uncontrolled expression of IL-6R at both mRNA and protein levels, enhancing cytokine storms in COVID-19 patients. Furthermore, the three lncRNAs identified as upregulated in COVID-19 patients may compete with IL-6R for miR-451a to reverse overexpression of IL-6R at the protein level. Collectively, our work provides the cfRNA landscape of plasma in COVID-19 patients, offers insight into the potential mechanism to understand the elevated cytokine storms caused by IL-6 in COVID-19 patients and may shed light on drug development for this new disease.
Supplementary information
Acknowledgements
The study was supported by CAMS Research Units of Adaptive Evolution and Control of Emerging Viruses (2018RU009) and Beijing New-star Plan of Science and Technology (Z181100006218080). WJL is supported by the Excellent Young Scientist Program of the National Natural Science Foundation of China (81822040) and the National Youth Talent Support Program. L.L. is funded by the National Natural Science Foundation of China (31900466). C.L. is funded by the China Postdoctoral Science Foundation (2020T130080ZX).
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Penghui Yang, Yingze Zhao, Jie Li, Chuanyu Liu, Linnan Zhu.
Contributor Information
Shaogeng Zhang, Email: zhangsg302@hotmail.com.
George F. Gao, Email: gaofu@chinacdc.cn
Longqi Liu, Email: liulongqi@genomics.cn.
William J. Liu, Email: liujun@ivdc.chinacdc.cn
Hai-Xi Sun, Email: sunhaixi@genomics.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00652-5.
References
- 1.Liu Y, et al. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl Sci. Rev. 2020;7:1003–1011. doi: 10.1093/nsr/nwaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang L, et al. Sepsis-associated severe interleukin-6 storm in critical coronavirus disease 2019. Cell. Mol. Immunol. 2020;17:1092–1094. doi: 10.1038/s41423-020-00522-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang, X. et al. PALM-Seq: integrated sequencing of cell-free long RNA and small RNA. bioRxiv10.1101/686055 (2019).
- 5.Zhu L, et al. Single-cell sequencing of peripheral mononuclear cells reveals distinct immune response landscapes of COVID-19 and influenza patients. Immunity. 2020;53:685–696.e683. doi: 10.1016/j.immuni.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perng Y-C, Lenschow D. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018;16:423–439. doi: 10.1038/s41579-018-0020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lasfar A, Wietzerbin J, Billard C. Differential regulation of interleukin-6 receptors by interleukin-6 and interferons in multiple myeloma cell lines. Eur. J. Immunol. 1994;24:124–130. doi: 10.1002/eji.1830240119. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Zhang A, Xiang J, Lv Y, Zhang X. miR-451 acts as a suppressor of angiogenesis in hepatocellular carcinoma by targeting the IL-6R-STAT3 pathway. Oncol. Rep. 2016;36:1385–1392. doi: 10.3892/or.2016.4971. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Developmental cell. 2013;25:69–80. doi: 10.1016/j.devcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Hu Y, Lu R, Ge M, Zhang L. MicroRNA-374a-5p inhibits neuroinflammation in neonatal hypoxic-ischemic encephalopathy via regulating NLRP3 inflammasome targeted Smad6. Life Sci. 2020;252:117664. doi: 10.1016/j.lfs.2020.117664. [DOI] [PubMed] [Google Scholar]
- 11.Ye Q, Wang B, Mao J. The pathogenesis and treatment of the Cytokine Storm’in COVID-19. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.