Abstract
L-Tryptophan (Trp) is a high-value aromatic amino acid with diverse applications in food and pharmaceutical industries. Although production of Trp by engineered Escherichia coli has been extensively studied, the need of multiple precursors for its synthesis and the complex regulations of the biosynthetic pathways make the achievement of a high product yield still very challenging. Metabolic flux analysis suggests that the use of a phosphoenolpyruvate:sugar phosphotransferase system (PTS) independent glucose uptake system, i.e. the galactose permease/glucokinase (GalP/Glk) system, can theoretically double the Trp yield from glucose. To explore this possibility, a PTS− and GalP/Glk-dependent E. coli strain was constructed from a previously rationally developed Trp producer strain S028. However, the growth rate of the S028 mutant was severely impaired. To overcome this problem, promoter screening for modulated gene expression of GalP/Glk was carried out, following by a batch mode of adaptive laboratory evolution (ALE) which resulted in a strain K3 with a similar Trp yield and concentration as S028. In order to obtain a more efficient Trp producer, a novel continuous ALE system was developed by combining CRISPR/Cas9-facilitated in vivo mutagenesis with real-time measurement of cell growth and online monitoring of Trp-mediated fluorescence intensity. With the aid of this automatic system (auto-CGSS), a promising strain T5 was obtained and fed-batch fermentations showed an increase of Trp yield by 19.71% with this strain compared with that obtained by the strain K3 (0.164 vs. 0.137 g/g). At the same time, the specific production rate was increased by 52.93% (25.28 vs. 16.53 mg/g DCW/h). Two previously engineered enzyme variants AroGD6G−D7A and AnTrpCR378F were integrated into the strain T5, resulting in a highly productive strain T5AA with a Trp yield of 0.195 g/g and a specific production rate of 28.83 mg/g DCW/h.
Keywords: L-Tryptophan, GalP/Glk-dependent, Adaptive laboratory evolution, CRISPR/Cas9-facilitated in vivo mutagenesis, Auto-CGSS
Highlights
-
•
A PTS negative and GalP/Glk-dependent E. coli strain constructed from a rationally developed Trpptophan producer.
-
•
An automatic and continuous adaptive laboratory evolution system (ALE) developed and used.
-
•
ALE combined with CRISPR/Cas9-facilitated in vivo mutagenesis and on-line monitoring.
-
•
A highly productive strain with a Trp yield of 0.20 g/g and a specific production rate of 28.83 mg/g DCW/h.
1. Introduction
L-Tryptophan (Trp), one of the high-value aromatic amino acids, is widely used in feed additive and as intermediate for pharmaceutics such as neurotransmitters (Mateos et al., 2009; Williams et al., 2014) and antitumor drugs (Fang et al., 2016; Rodrigues et al., 2013). Escherichia coli has been extensively exploited for effective bioproduction of Trp from renewable feedstock, e.g., glucose, and various rational strategies have been applied in order to achieve a high Trp productivity (Liu et al., 2019; Li et al., 2020), including (i) alleviation of restrictive regulations (Chen et al., 2018; Ikeda, 2003; Oldiges et al., 2004); (ii) deletion of competing pathways; (iii) enhancement and balancing of precursor supplements (Ikeda, 2006); and (iv) removal of degradation pathways (Aiba et al., 1980). For instance, Zhao et al. (2012) rationally engineered an E. coli and made it able to produce 14.70 g/L of Trp with a yield of 0.120 g/g. Another rationally engineered E coli strain S028 constructed by Chen and Zeng (2017) was capable of producing 40.30 g/L of Trp with a yield of 0.15 g/g. Besides pure rational engineering, Du et al. (2019) constructed a strain TRP07 by modifying the central metabolic pathway of a Trp-producing E. coli strain which had been undergone random mutagenesis, and the strain TRP07 could accumulate 49 g/L of Trp with a yield of 0.186 g/g.
Despite these achievements, biosynthesis of Trp in strains harboring the phosphoenolpyruvate:sugar phosphotransferase system (PTS) has a limited maximum theoretical yield of 0.227 g Trp/g glucose (Fig. S1A). The availability of phosphoenolpyruvate (PEP) has been proposed as a restriction for the yield of Trp (Li et al., 2020). It was suggested that inactivation of PTS in E. coli could foster a higher glucose conversion rate (Floras et al., 1996). This is because PTS requires the consumption of 1 mol PEP for each mol of internalized glucose; in a PTS-defective strain over 80% of PEP can theoretically be utilized for Trp biosynthesis (Fig. S1B). Therefore, inactivation of the cytoplasmic components of PTS (e.g., the ptsHIcrr operon) has been a representative strategy to repeal the PTS-facilitated glucose transport system (Carmona et al., 2015; Chen et al., 2018b; Lu et al., 2012). However, the PTS-defective strain is normally seriously impaired in its growth capability. To restore the glucose transport in a PTS-defective strain, activation of other potential sugar transporters or carriers, e.g., the glucose facilitator Glf from Zymomonas mobilis and the housekeeping enzyme glucokinase (Glk) (Knop et al., 2001; Chandran et al., 2003), or the galactose permease (GalP) and Glk pathway (Carmona et al., 2015, 2020), can serve as a promising strategy. As shown in Fig. S1B, the maximum theoretical yield in a GalP/Glk-dependent strain is calculated to be 0.454 g/g, which is approximately twice that of a PTS-dependent strain.
Besides the rational engineering strategies mentioned above, non-rational approaches, e.g., adaptive laboratory evolution (ALE), are also widely used in recent years for development of super-producers. ALE can be conducted in either batch or continuous mode. The batch mode is easy to be established, but it deals with each manipulation, such as construction of gene variant library and screening of candidates, separately and thus quite time- and labor-consuming. To the contrary, continuous ALE seamlessly integrates all manipulations into an intact evolutionary cycle (d’Oelsnitz and Ellington, 2018), which enables not only precise control of cultivation parameters (e.g., pH and temperature) but also online detection and monitoring of parameters related to cell growth or productivity (e.g., optical density and fluorescence intensity). As for generation of mutations, this can be achieved either by introducing random mutagenesis in vitro at user-defined genes (e.g., error-prone PCR and site-saturation mutagenesis) or by genome-wide mutagenesis in vivo, e.g., PACE (Bryson et al., 2017; Song and Zeng, 2017) and OrthoRep (Ravikumar et al., 2018). Especially, mutations can occur not only within but also outside of user-defined genes when plasmid mutators are applied (Badran and Liu, 2015). This is helpful to address a wide range of targets that require simultaneous modulation of multiple genes.
With the advent of gene-targeting technologies, e.g., CRISPR/Cas9 technique, more precise technologies for engineering of genes at a specific locus have been developed, such as CasPER (Jakočiūnas et al., 2018) and EvolvR (Halperin et al., 2018). We had previously developed a method that integrates CRISPR/Cas9-facilitated engineering of the target gene(s) with Growth-coupled and Sensor-guided in vivo Screening (CGSS), which was successfully used to engineer one of the key enzymes 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase AroG (Chen et al., 2019). Also, a novel screening approach was applied to engineer another anthranilate-activated trifunctional enzyme AnTrpC from Aspergillus niger (Chen et al., 2018). In this work, based on the GalP/Glk-dependent PTS-defective E. coli, we attempted to screen a Trp producer with better performance, e.g. with higher yield, by applying batch/continuous ALE together with CGSS. To this end, the advantages of CGSS are further exploited by integrating it into an automatic evolutionary system (auto-CGSS). Specifically, auto-CGSS is coupled with a real-time measurement of cell growth and online monitoring of the fluorescence intensity as an indicator of intracellular Trp concentration. With the aid of such a novel evolutionary system, a Trp producer with high yield and specific production rate was obtained. This evolutionarily developed strain T5 was able to produce 38.77 g/L of Trp with a yield of 0.164 g/g. And after further integration of the two favorable enzyme variants AroGD6G−D7A and AnTrpCR378F (Chen et al., 2018, 2019), we constructed a strain which achieved a yield of 0.195 g/g and a specific production rate of 28.83 mg/g DCW/h and thus represents a highly productive strain for Trp biosynthesis. It should be mentioned that during the final revision of this paper, we noticed the latest development reported by Xiong et al. (2021). By redirecting the fluxes of the central carbon metabolism of E. coli with a PTS-independent glucose uptake these authors could increase the Trp yield from 0.164 g/g to 0.227 g/g in a fed-batch fermentation with quite high Trp concentration (41.7 g/L Trp) and volumetric productivity (>1 g/L.h). The performance of our recombinant strain could be further improved in a similar way.
2. Results
2.1. Modulated expression of galP/glk recovers glucose utilization in a PTS-defective E. coli strain
To examine the potential Trp yield of a PTS-defective Trp-producing E. coli, a PTS-defective strain was first constructed. To this end, PTS was deactivated in a rationally constructed Trp-producing strain S028TS/pCas9 (Table S1) by knocking out the gene ptsI (encoding PTS Enzyme I) (details in Supplementary Materials). The resulting strain is named as G028. To evaluate the glucose uptake rate and cell growth rate of the PTS-defective strain G028, an overnight culture grown on LB medium was washed three times with M9 medium, and it was then cultivated in M9 medium with glucose as the sole carbon source. After almost 60 h of cultivation, the growth of G028 only reached an OD600 of 0.21 ± 0.02 (the initial OD600 was 0.10). This result demonstrated that in the PTS-defective strain, the glucose uptake was seriously blocked and thus the cell growth was severely hindered. Therefore, other potential system(s) for the uptake and phosphorylation of glucose, e.g., the GalP/Glk-assisted uptake, need to be activated in the PTS-defective strain.
To alleviate the decreased glucose utilization rate and the growth retardation caused by disruption of PtsI, our previously developed CGSS approach (Chen et al., 2019) was applied to engineer the promoters of galP and glk, resulting in combinatorial promoter mutants with reliable modulated expression (for details of the method see Fig. S2). To this end, a host strain G028Δglk::CmR (G028Cm, Table S1) was constructed by inserting the chloramphenicol resistance gene CmR into the native glk gene with the aim to offer the sgRNA target sequence for CRISPR/Cas9 application in further promoter engineering. Afterwards, a mutagenesis library of the promoter variants was generated in vitro by using the plasmid pCmN20-GGMT (Fig. S3) and integrated into the chromosome of host strain using the CRISPR/Cas9 technique (details in Supplementary Materials). Finally, the mutants (G028a) were screened and selected by using the complementary growth-coupled screening and Trp biosensor-based in vivo characterization (Fig. S2).
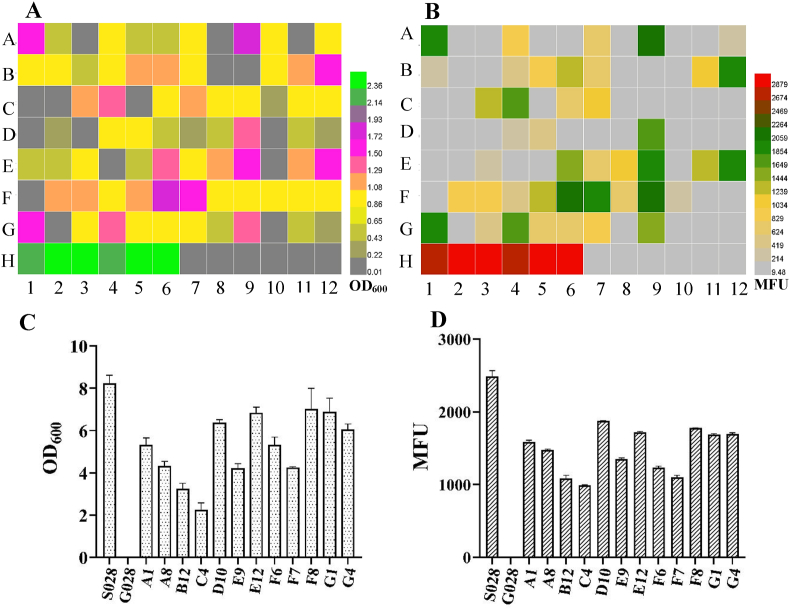
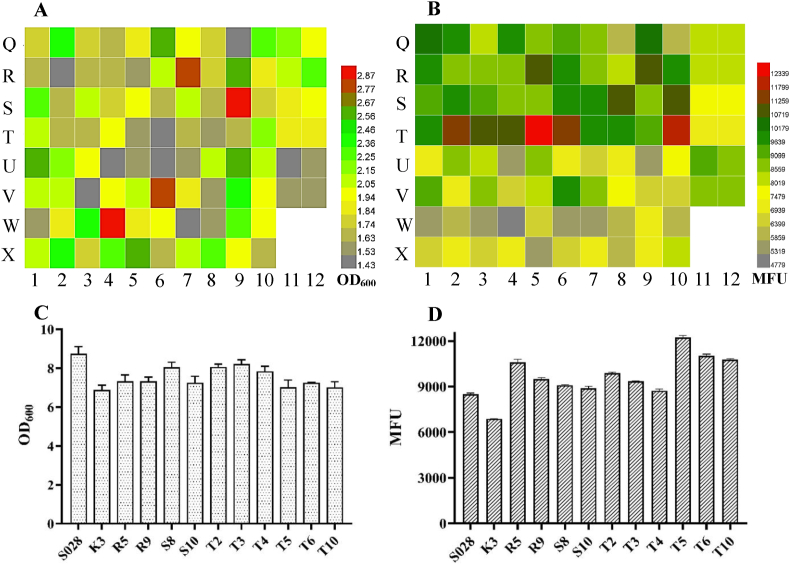
After confirming the phenotype of mutants on M9 agar plate, a total of 84 independent mutants with relatively bigger colony size and higher fluorescence signal were selected and re-checked in M9 medium with a 96 deep well plate. A total of six samples in the H1–H6 wells and H7–H12 wells were used as the positive control of strain S028 with a functional PTS and the strain G028GG with non-mutated synthetic promoters (ptac-galP-pJ23119-glk), respectively (Fig. 1A and B). The cell growth (OD600) and the fluorescence intensity (MFU) of all mutants were presented in heat maps. As shown in Fig. 1A and B, 12 of the mutants with a higher cell growth (e.g., over 1.25) were coupled with a stronger fluorescence intensity (colored in green in Fig. 1B), corresponding to those in the A1, A8, B12, C4, D10, E9, E12, F6, F7, F8, G1 and G4 wells respectively. Although other mutants, such as the mutants in the E11 and F12 wells in Fig. 1A also exhibited a high cell growth (around 1.0), their fluorescence intensity was not detectable owing to the fact that the accumulation of Trp was inconspicuous (Fig. 1B). These results indicated the advantage of the growth and biosensor-coupled in vivo screening and characterization approach compared to the dialogue-oriented approaches, e.g., cell growth complementation-assisted library screening (Zhu et al., 2017).
Fig. 1.
Heat maps of cell growth (OD600) (A) and fluorescence intensity (MFU) (B) of selected E. coli strains with mutated promoters for the enzymes GalP/Glk. A total of six samples in H1–H6 wells are positive controls (strain S028 with a functional PTS). The other six samples in the H7–H12 wells are reference controls (strain G028GG with non-mutated synthetic promoters). Measurements of OD600 (C) and fluorescence intensity (D) of the S028 and selected mutants from the second round of screening. The cells were cultured with FM-III medium in shake flasks. All the data are from two independent biological samples.
Batch-fermentations of the 12 mutants and the positive control (S028) were then carried out with FM-III medium in 50 mL conical tubes. Among these mutants, four of them, D10, E12, F8, and G1, showed a significant advantage of growth compared with the others (Fig. 1C). However, the growth and MFU of these mutants were still lower than that of the positive control (Fig. 1C and D). These results suggested that these mutants should be further optimized in order to achieve a better Trp production.
2.2. Batch mode adaptive laboratory evolution of the GalP/Glk-dependent E. coli strain
To improve the performance of the GalP/Glk-dependent strain further, 12 selected mutants with a relatively high MFU were subjected to a batch mode of adaptive evolution (for details see Supplementary Materials). A strain K3 with a relative high cell growth and a seemly good Trp production was chosen for further evaluation in fed-batch fermentation. Its performance is compared with that of the strain S028.
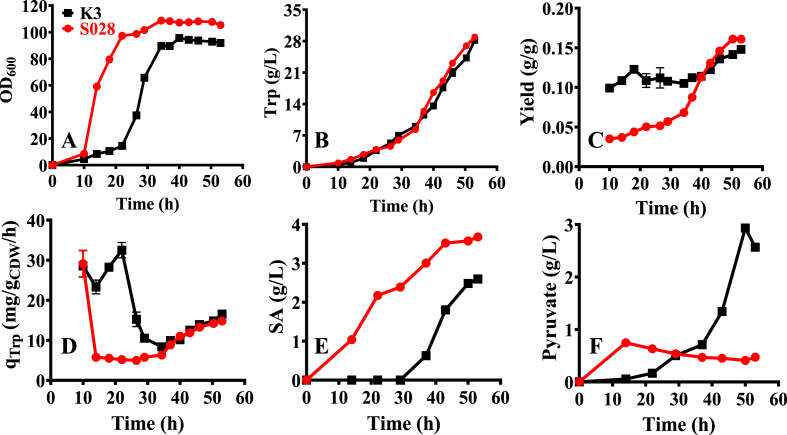
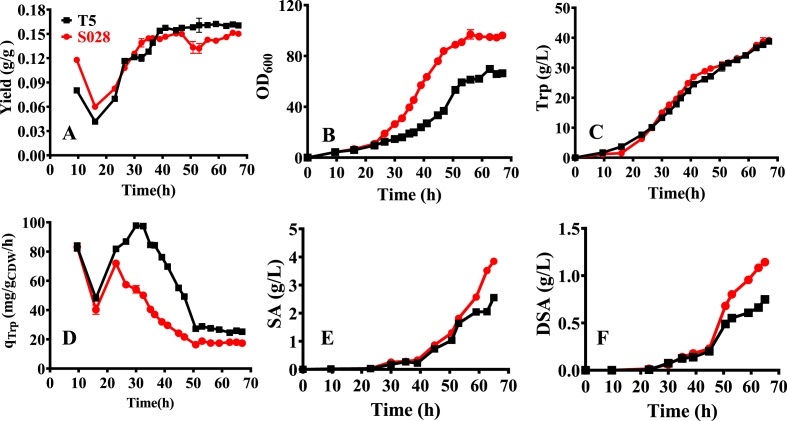
As depicted in Fig. 2, fed-batch fermentation of the strain K3 suffered from growth retardation severely before the stationary phase (within 30 h, Fig. 2A). However, after the mid-exponential phase, the strain gradually regained its ability to utilize glucose and restored its growth to the level of S028 (Fig. 2A). Considering that K3 still had a higher glucose uptake rate during the stationary phase (4.10 g/L/h from 40 to 50 h) but produced a similar amount of Trp as the strain S028 (Fig. 2B), this resulted in a reduction of Trp yield of the strain K3 (e.g., 0.137 g/g vs. 0.150 g/g at 50 h, Fig. 2C). According to the previous report, the increase in glucose consumption rate is because of the sensory mutation introduced into the GalP and Glk enzymes or the deletion of GalR repressor during the adaptive evolution (Aguilar et al., 2018; Alva et al., 2020). Therefore, the genes galP, glk, and galR were sequenced. The results revealed that no mutation was generated in the galP, glk, and galR genes. These results demonstrated that other potential glucose uptake pathways might be activated instead of the GalP/Glk bypass pathway. For instance, Carmona et al. (2020) found that adaptive laboratory evolution resulted in the selection of MglB as the primary glucose transporters in the PTS− mutant of E. coli PB11.
Fig. 2.
Fed-batch fermentations with the strains S028 (red and square) and K3 (black and circle). (A) Cell growth; (B) Trp production; (C) Overall yield; (D) Specific production rate of Trp production (qTrp). Accumulation of the intermediates shikimate (E) and pyruvate (F). The initial OD600 after inoculation was 0.45. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Overall, increasing glucose uptake rate of the strain K3 did not significantly improve the yield of Trp, but it showed benefits in other aspects such as the accumulation of intermediates. As seen in Fig. 2E, lower accumulation of extracellular shikimate was observed in the strain K3 during the entire fermentation process. It was believed that a lower accumulation of shikimate indicates an increase in Trp production, especially during the stationary phase. However, the pyruvate production was dramatically increased to 2.95 g/L after the exponential phase (Fig. 2F), which is approximately five-fold higher than that obtained with strain S028 and indicates the potential importance of this intermediate in supporting its growth. We hypothesize that the strain K3 was evolved to activate the pathways of pyruvate synthesis for supporting its growth instead of Trp production. Therefore, a more efficient evolutionary method should be established for further development of the GalP/Glk-dependent E. coli strain.
2.3. Improvement of GalP/Glk-dependent E. coli in an automatic continuous evolutionary system
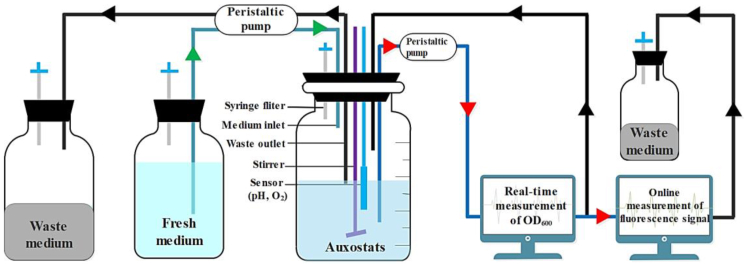
To obtain a better GalP/Glk-dependent Trp producer with a higher product yield, the strain G028 was optimized by integrating the CRISPR/Cas9-facilitated in vivo mutagenesis with an automatic continuous evolutionary system (Fig. 3).
Fig. 3.
Improvement of GalP/Glk-dependent E. coli in an automatic continuous evolutionary system. . During the continuous evolution, the flow rates of the fresh medium inlet and the waste culture outlet are dynamically regulated by peristaltic pumps in response to the real-time measured optical density of the culture. The real-time fluorescence intensity monitored by the flow cytometer is used as an indicator for intracellular concentration of Trp.
As illustrated in Fig. S3, the promoter library generated by pCmN20-GGMT was integrated into the chromosome of strain G028Cm/pJC184/pCas9MT (G028JC, Table S1) in the same way as outlined in the aforementioned CGSS approach (Chen et al., 2019). The resulting cells were then inoculated into a cultivation vessel containing a synthetic medium (SynM) with corresponding antibiotics. The growth of the mutants was monitored in real-time by a cell density meter. Noteworthy, the OD600 of the strain S028JC reached around 0.8 in the mid-exponential phase when it was cultivated in the synthetic medium. As soon as the OD600 reached a set point of 0.8, the cultivation system was operated in an auxostat mode, in which fresh medium supplemented with antibiotics and 10% w/v L-arabinose was supplied via a dedicated inlet pump, and depleted medium and cells were pumped out to a waste container (Fig. 3). The feeding of L-arabinose was aimed to induce the expression of mutators in the mutagenesis plasmid (pJC184) to conduct a genome-context random mutagenesis and thus to accelerate the mutation rate during the continuous evolution. Moreover, the flow rates of the fresh medium (inlet) and the waste (outlet) were dynamically and automatically controlled in response to the real-time monitored cell density so that a constant cell density at around 0.8 was maintained. The real-time fluorescence intensity measured by a flow cytometer was used as an indicator of the intracellular concentration of Trp.
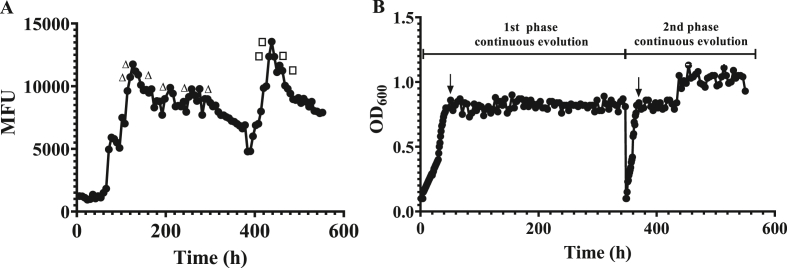
As observed in Fig. 4, the fluorescence intensity reached to the maximum level (~11,446 MFU) when the evolution continued up to 126 h and then decreased to a stable level (~8000–9000 MFU, Fig. 4A). Six samples from the first phase (labeled with triangles in Fig. 4A) were selected and their fluorescence intensity and Trp production were characterized via batch fermentation in shake flasks. The best-characterized mutant G028c was then forced to the second round of continuous evolution for further improvement of its performances. In this round, the fluorescence intensity was increased to the maximum level (~13,541 MFU) in a shorter period (~390–438 h), but encountered the same issue as the first phase, in which the fluorescence intensity decreased significantly in the final stage (Fig. 4A). Therefore, four samples from the second phase (labeled with squares in Fig. 4A) were further characterized. To this end, the samples from different time points were plated on the SynM agar medium and grown for 30 h. Afterwards, a total of 20 single colonies from each sample were selected and together with the positive controls (S028, K3, and G028c) cultivated in a 96 deep-well plate with SynM medium. After 10 h of cultivation, cell growth and fluorescence intensity were measured.
Fig. 4.
Real-time measurements of fluorescence intensity (A) and OD600 (B) of the mutants during the whole process of continuous evolution. The process was performed in two stages, in which the best-characterized mutant from the first phase (0–380 h) was forced to the second phase of continuous evolution (381–550 h) for further evolution. The data points in (A) marked with open triangles or squares indicate that samples at those time points were selected for off-line characterization. The data points in (B) labeled with arrows indicate the time points for introduction of 0.1% w/v L-arabinose for induction.
It was found that, after 10 h of cultivation, the average growth of mutants sampled at the time point of 462 h (G/H1-G/H10) was higher than those sampled at an early time point (C/D1-C/D10), but the average fluorescence intensity (MFU) was much lower (6480.30 ± 872.70 vs. 10230.70 ± 1136.60) (Fig. 5A and B). The results indicated that the PTS-defective cells are preferentially evolved to synthesize fundamental molecules to support its growth rather than Trp synthesis. Moreover, in comparison to the strain G028c (E/F11-E/F12) and the reference strain S028 (A/B11-A/B12), a total of 10 mutants shared a higher fluorescence intensity (over 10000, Fig. 5B). They were again subjected to further characterization by cultivation in FM-III medium in shake flasks for 20 h. As shown in Fig. 5C and D, batch fermentation of all those mutants resulted in a comparable growth rate but with a higher fluorescence intensity than that of the positive strains (S028 and K3). Obviously, the strain T5 showed the best performance in Trp production and therefore it was selected as the candidate for further evaluation by fed-batch fermentation.
Fig. 5.
Heat maps of cell growth (OD600) (A) and fluorescence intensity (MFU) (B) of the selected mutants from the second phase of continuous evolution. A total of twenty samples in Q1-Q10 and R1-R10 wells, S1–S10 and T1-T10 wells, U1–U10 and V1–V10 wells, and W1–W10 and X1-X10 wells are presented as single colonies from four different time points. A total of four samples in Q11-Q12 and R11-R12 wells, S11–S12 and T11-T12 wells, and U11–U12 and V11–V12 wells are presented as the positive controls: S028, K3 and G028c, respectively. Measurements of the growth (C) and the fluorescence intensity (D) of S028, K3 and the selected mutants. The cells were cultured with FM-III fermentation in shake flasks. All the data are from two independent biological samples.
2.4. Fed-batch fermentation with strain T5 resulted in a high specific production rate of Trp
To examine the performance of the promising mutant T5 for the biosynthesis of Trp, fed-batch fermentation was carried out in bioreactors. In the meanwhile, the fermentation with strain S028 was used as the reference. The plasmid pJC184 was removed from the strain to avoid undesired mutations in subsequent experiment.
As shown in Fig. 6A, the Trp yield of T5 is calculated to be 0.164 g/g (Table 1), which is 9.33% higher than that achieved by the strain S028 (0.150 g/g, Table 1) and 19.71% higher than that obtained by the strain K3 (0.137 g/g, Fig. 2C). It was noteworthy that the growth of T5 was remarkably impaired compared to that of the reference strains S028 (Fig. 6B) and K3 (Fig. 2B). The biomass formation of T5 (22.89 ± 0.20 g/L) was 32.02% less than that of S028 (33.67 ± 0.10 g/L, Table 1). However, T5 resulted in almost the same amount of Trp as that obtained by S028 at the end of fermentation (Fig. 6C and Table 1). Reasonably, the specific production rate of Trp (qTrp) was significantly higher than that of S028, especially in the exponential phase (Fig. 6D). As shown in Table 1, the overall specific production rate was increased by 45% (25.28 vs. 17.38 mg/g DCW/h) calculated at the end of the fermentation. In comparison to T5, K3 shared the same qTrp with the strain S028 (Fig. 2C). Therefore, the improvement in qTrp and production yield indicate that the evolved mutant T5 has much favorable traits for Trp production.
Fig. 6.
Fed-batch fermentations with strains S028 (black and circle) and T5 (red and square). (A) Production yield of Trp; (B) Cell growth; (C) Trp production; (D) Specific production rate of Trp production (qTrp). Accumulation of the intermediates shikimate (SA) (E) and dehydroshikimate (DSA) (F). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 1.
Results of fed-batch fermentations of different Trp-producing strains.
| Strains | OD600 | DCW (g/L) |
L-Trp (g/L) |
qTrp (mg/gDCW/h) |
Yield (g/g) |
Vp (g/L/h) |
|---|---|---|---|---|---|---|
| S028 | 96.20 ± 0.28 | 33.67 ± 0.10 | 39.20 ± 0.78 | 17.38 | 0.150 | 0.59 |
| T5 | 65.40 ± 0.57 | 22.89 ± 0.20 | 38.77 ± 0.02 | 25.28 | 0.164 | 0.58 |
| S028AA | 92.20 ± 0.28 | 32.27 ± 0.10 | 48.27 ± 0.29 | 23.74 | 0.185 | 0.77 |
| T5AA | 66.20 ± 0.84 | 23.17 ± 0.29 | 41.49 ± 0.03 | 28.83 | 0.195 | 0.67 |
Fed-batch fermentations were performed in a highly instrumented and automated 4-paralleled 1.5 L bioreactors system DASGIP for 65 h; the initial glucose concentration was 25 g/L; the initial inoculation OD600 was 0.45. The means ± standard deviations are calculated from two independent measurements.
It is noted that the Trp yield achieved by T5 is still significantly below the theoretical maximum yield (0.227 and 0.454 g/g for PTS-based glucose uptake and GalP/Glk-based glucose transport, respectively) (Fig. S1). In this context, key intermediates of the Trp synthesis pathway such as dehydroshikimate (DSA) and shikimate (SA) were found to be accumulated in the culture. Fig. 6E and F revealed that the trends of SA and DSA formation in T5 were similar to those in S028, indicating limitations in the downstream metabolic pathways as previously found for the strain S028 (Chen et al., 2018a, 2019).
2.5. Further improvement of Trp production by integration of enzyme variants AroGD6G−D7A and AnTrpCR378F
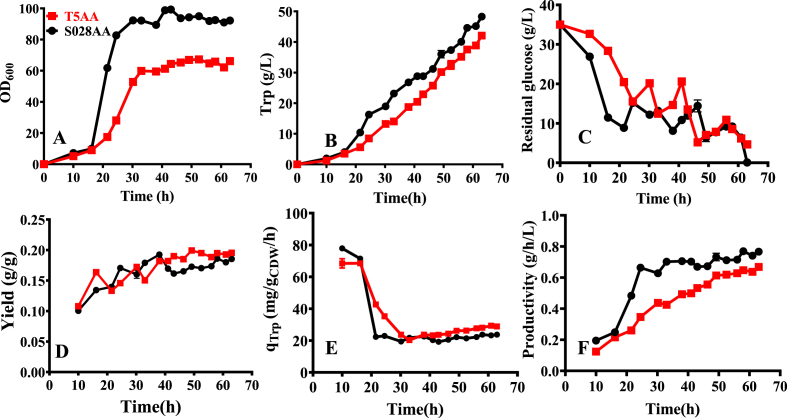
In previous studies with the strain S028 we showed that the incorporation of a L-phenylalanine-resistant enzyme variant AroGD6G−D7A and an anthranilate-activated enzyme variant AnTrpCR378F can significantly increase Trp production and glucose conversion yield (Chen et al., 2018a, 2019). To further enhance Trp synthesis of T5, aroGD6G-D7A and AntrpCR378F (Figs. S6 and S7) were subsequently integrated into its chromosome, resulting in a strain designated as T5AA (details in Supplementary Materials). In the meanwhile, both of the gene variants were integrated into the chromosome of S028, resulting in a strain named as S028AA. Thereafter, fed-batch fermentations of the strains T5AA and S028AA were carried out and compared (Fig. 7).
Fig. 7.
Fed-batch fermentations with strains T5AA (red and square) and S028AA (black and circle). (A) Cell growth; (B) Trp production; (C) Glucose concentration; (D) Production yield of Trp; (E) Specific formation rate of Trp (qTrp); and (F) Overall productivity. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
As seen from Figs. 6B and 7A, similar to the parent strain T5, the growth of T5AA reached up to the stationary phase (60.12 ± 0.74) in 30 h of cultivation with obvious retardation. Biomass achieved in the fed-batch cultivation of T5AA was 23.17 ± 0.29 gDCW/L (Table 1), which was 28.20% lower than that of the strain S028AA (32.27 ± 0.10 gDCW/L, Table 1). However, the Trp concentration (41.49 ± 0.03 g/L, Table 1) achieved by T5AA was significantly improved by 7.03% compared to that of T5. These results again demonstrate that additional expression of AnTrpCR378F and AroGD6G-D7A facilities the biosynthesis of Trp. Also, expression of the enzyme variants in T5 contributed to the glucose conversion yield. The results showed that Trp yield of T5AA (0.195 g/g, Table 1) is 18.90% and 5.41% higher than that of T5 (0.164 g/g, Table 1) and S028AA (0.185 g/g, Table 1), respectively. Since the strains T5AA and S028AA contained the enzyme variants AnTrpCR378F and AroGD6G−D7A, the improved Trp yield could be mainly resulted from the increased availability of PEP.
It is also interesting to note that the specific production rate of Trp (qTrp) of T5AA (28.83 ± 0.35 mg/gDCW/h) is also significantly higher than that of S028AA (23.74 ± 0.07 mg/gDCW/h), especially during the production phase (Table 1 and Fig. 7E). Besides, the final Trp production of T5AA is lower than that of S028AA (41.49 ± 0.03 g/L vs. 48.27 ± 0.29 g/L, Table 1). Thus, the overall productivity of T5AA was lower than that of S028AA (Fig. 7F). One direct reason for this is the lower cell growth and glucose consumption. As also shown in previous studies (Chen et al., 2018a, 2019), the biomass formation level is one of the key parameters determining the final concentration and volumetric productivity of Trp. Thus, further development of T5AA should try to identify and remove the limiting factor(s) involved in biomass formation.
2.6. Genomic analysis of the strain T5
In order to elucidate potential mutations that resulted in the behaviors of T5, the whole genome of T5 was sequenced by using the Next-Generation Sequencing (NGS) approach. With the genomic information of E. coli W3110 as a reference, pairwise genome alignment of E. coli W3110 and T5 was conducted and the results revealed that 15 genes in the T5 strain were modified and 5 genes were deleted (Table 2). Among those assigned with KEGG Orthology (KO) numbers, seven are involved in energy, amino acid, nucleotide and carbohydrate metabolisms (folD, aroG, cmk, trpE, zwf, serA, rpsL) and four take part in genetic information processing (ycbW, umuC, dgsA, rrlD). The transporter encoded by galP belongs to the group of signaling and cellular processes and fliC contributes to cell motility by encoding a flagellar filament structural protein. Particularly, three genes are of great interest because of their functions in glucose utilization and cell growth. To further confirm the mutations identified by the high-throughput NGS method, the key mutations such as those in the genes galP, zwf, and mlc were validated by direct PCR product sequencing in both the strains T5 and S028 (data not shown).
Table 2.
Summary of modified ORFs in the strain T5 in alignment with the reference strain E. coli W3110.
| Number | Gene | Coded Protein | Mutation | EcoCycID1//KEGG number2//UniPro ID3 | Function | KEGG Orthology (KO) | |
|---|---|---|---|---|---|---|---|
| 1 | galP | Galactose:H+ symporter | Lys137Thr | EG12148//b2943//P0AEP1 | Major facilitator superfamily (MFS) | Signaling and cellular processes | |
| 2 | galP | Galactose:H+ symporter | Pro195Ala | EG12148//b2943//P0AEP1 | Major facilitator superfamily (MFS) | Signaling and cellular processes | |
| 3 | galP | Galactose:H+ symporter | Phe204Leu | EG12148//b2943//P0AEP1 | Major facilitator superfamily (MFS) | Signaling and cellular processes | |
| 4 | folD | Bifunctional 5,10-methylene-tetrahydrofolate dehydrogenase/5,10-methylene-tetrahydrofolate cyclohydrolase | Leu36Gln | EG10328//b0529//P24186 | (6R)-5,10-methylene-5,6,7,8-tetrahydrofolate + NADP+ = 5,10-methenyltetrahydrofolate + NADPH | Energy metabolism | |
| 5 | rzoD | DLP12 prophage | Arg34Pro | G0-10436//b4510//P58041 | Polypeptide; Putative prophage lysis lipoprotein RzoD | – | |
| 6 | aroG∗ | 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase; Phenylalanine repressible | Ser180Phe | EG10079//b0754//P0AB91 | D-erythrose 4-phosphate + H2O + phosphoenolpyruvate = 7-phospho-2-dehydro-3-deoxy-D-arabino-heptonate + phosphate | Amino acid metabolism | |
| 7 | cmk | Cytidylate kinase | Glu153Gly | EG11265//b0910//P0A6I0 | ATP + CMP = ADP + CDP | Nucleotide metabolism | |
| 8 | ycbW | Cell division protein ZapC | Gln168Leu | G6486//b0946//P75862 | Contributes to the efficiency of the cell division process | Genetic information processing | |
| 9 | ycdT | Probable diguanylate cyclase DgcT | Val130Ala | G6532//b1025//P75908 | 2 GTP = cyclic di-3′,5′-guanylate + 2 diphosphate | – | |
| 10 | hflD | Lysogenization regulator | Trp181Arg | EG11345//b1132//P25746 | Negative regulator of phage lambda lysogenization | – | |
| 11 | umuC | DNA polymerase V catalytic protein | Phe287Leu | EG11056//b1184//P04152 | Poorly processive, error-prone DNA polymerase involved in translesion repair | Genetic information processing | |
| 12 | trpE∗ | Anthranilate synthase subunit TrpE | Ser40Phe | EG11028//b1264//P00895 | chorismate + L-glutamine = anthranilate + H+ + L-glutamate + pyruvate | Amino acid metabolism | |
| 13 | zwf | NADP + -dependent glucose-6-phosphate dehydrogenase | Glu165 Stop codon | EG11221//b1852//P0AC53 | D-glucose 6-phosphate + NADP+ = 6-phospho-D-glucono-1,5-lactone + H+ + NADPH | Carbohydrate metabolism | |
| 14 | fliC | Flagellar filament structural protein | Glu115Lys | EG10321//b1923//P04949 | Polymerizes to form the filaments of bacterial flagella | Cell motility | |
| 15 | yohC | Putative inner membrane protein | Val33Ala | EG12016//b2135//P0AD17 | Composes of five predicted transmembrane domains | – | |
| 16 | yeiB | DUF418 domain-containing protein YeiB | Ser28Pro | EF11290//b2152//P25747 | Involves in transport | – | |
| 17 | yfcO | DUF2544 domain-containing protein YfcO | Met218Ile | G7203//b2332//P76498 | Contributes to adhesion to various surfaces in specific environmental niches | – | |
| 18 | serA∗ | Phosphoglycerate dehydrogenase | Asp364Ala | EG10944//b2913//P0A9T0 | (2R)-3-phosphoglycerate + NAD+ = 3-phosphooxypyruvate + H+ + NADH | Energy metabolism/Amino acid metabolism | |
| 19 | serA∗ | Phosphoglycerate dehydrogenase | His344Ala | EG10944//b2913//P0A9T0 | (2R)-3-phosphoglycerate + NAD+ = 3-phosphooxypyruvate + H+ + NADH | Energy metabolism/Amino acid metabolism | |
| 20 | rpsL∗ | 30S ribosomal subunit protein S12 | Lys43Arg | EG10911//b3342//P0A7S3 | With S4 and S5 plays an important role in translational accuracy | Carbohydrate metabolism/Nucleotide metabolism | |
| 21 | yhaC | uncharacterized protein YhaC | Glu134 Stop codon | EG11174//b3121//P11864 | – | – | |
| 22 | yhaC | uncharacterized protein YhaC | Thr216Ala | EG11174//b3121//P11864 | – | – | |
| 23 | yibJ | Putative uncharacterized protein YibJ | Lys35Gln | EG11766//b3595//P32109 | – | – | |
| 24 | ykfF | CP4-6 prophage; protein YkfF | Deletion4 | G6124//b0249//P75677 | – | – | |
| 25 | ypjJ | CP4-57 prophage; DUF987 domain-containing protein YpjJ | Insertion5 | G0-10464//b4548//P58033 | – | – | |
| 26 | ykfH | DUF987 domain-containing protein YkfH | Deletion | G0-10432//b1504//Q9XB42 | – | – | |
| 27 | ybfD | H repeat-associated putative transposase YbfD | Deletion | EG11524//b0706//P28916 | Expression of ybfD is upregulated in long-term stationary phase cultures | – | |
| −28 |
dgsA (mlc) |
DNA-binding transcriptional repressor Mlc | Deletion | G6852//b1594//P50456 | Regulates the expression of proteins involved in phosphotransferase system for sugar uptake | Genetic information processing | |
| 29 | rrlD | 23S ribosomal RNA | Deletion | EG30080//b3275//-- | Belongs to the large subunit (50S subunit) of ribosome | Genetic information processing | |
Note: EcoCyc ID1, https://biocyc.org/organism-summary?object=ECOLI; KEGG number2, https://www.genome.jp/dbget-bin/www_bget?eco; Unipro ID3, https://ebi14.uniprot.org/uniprot; ∗ the gene variants aroGS180F, serAH344A/N364A, trpES40F and rpsLK43R were generated by rational protein design (Chen, 2017); Deletion4 or insertion5 indicate that part of sequences of gene is deleted or inserted.
3. Discussion
3.1. Enhancing expression of various PTS genes by inactivation of the Mlc repressor
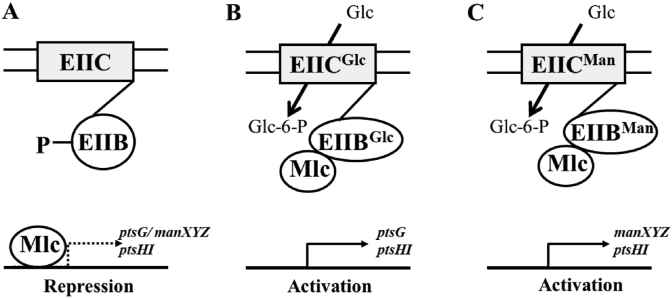
As shown in Table 2, one of the genes that are directly related to PTS is found to be deleted in the mutant T5: mlc (also known as dgsA, EcoCyc ID: G6852).
Mlc is a dual transcriptional regulator that controls the expression of a number of genes encoding enzymes of PTS (Kim et al., 1999; Plumbridge, 2001). In particular, Mlc represses the expression of two genes ptsG and manXYZ which encode respectively the two transporters EIICBGlc and EIIABCDMan involved in the uptake of glucose (Plumbridge, 2001; Plumbridge and Kolb, 1993). In E. coli, PtsG-facilitated PTS is the major glucose transporter. As shown in Fig. 8A and B, in the absence of glucose, PtsG (EIICBGlc) is predominantly phosphorylated and Mlc is bound to its operators; in the presence of glucose, the dephosphorylated form of PtsG (EIICBGlc) interacts directly with Mlc and induces the transcription of Mlc-regulated genes (ptsGH). As the major uptake system for mannose, the manXYZ operon, which encodes three proteins EIIABCDMan to form the enzyme II of the mannose PTS, the latter also transports glucose efficiently (Plumbridge, 1998) (Fig. 8C). Plumbridge found that a mutation in the mlc gene resulted in a threefold activation of manX expression (Plumbridge, 1998). It is assumed that the deletion of mlc gene in T5 could also result in derepression of manX and ptsG (Fig. 8B and C). As shown in Fig. 8B, an activation of manX and ptsG expression also leads to the production of glucose-6-phosphate (Glc-6-P), and the accumulated Glc-6-P can be subsequently converted to PEP by the glycolysis pathway. This indicates that deletion or inactivation of the repressor Mlc could be one of the important strategies to increase the availability of PEP, and it also explains why the fed-batch fermentation of T5 resulted in a higher Trp yield (Table 1). However, further experiments are required to confirm this hypothesis.
Fig. 8.
Models for PtsG and ManXYZ regulations. PtsG (EIICBGlc) and ManXYZ (EIIABCDMan) are shown as a membrane-anchored EIIC domain and a soluble EIIB domain (Plumbridge, 2001; Plumbridge and Kolb, 1993). (A) In the absence of glucose, PtsG or ManXYZ is predominately phosphorylated and Mlc is bound to its operators. In the presence of glucose, EIIBGlc (B) or EIIBMan (C) becomes dephosphorylated; Mlc binds to the EIIB domain and Mlc controlled genes are activated.
3.2. Structure-based analysis of the enzyme variant of GalP for glucose permeation
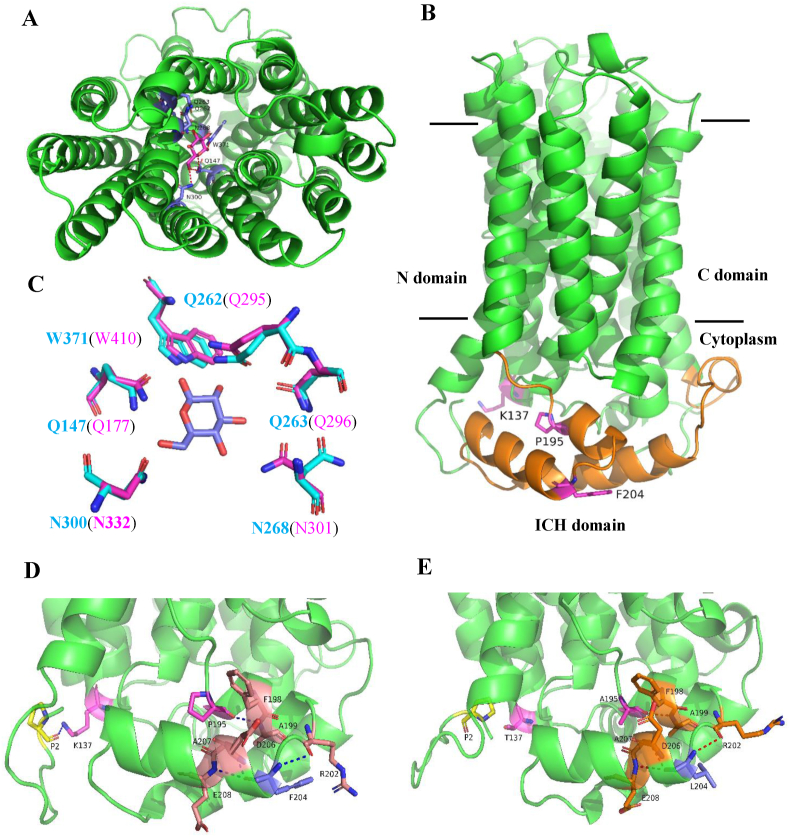
The galP gene in T5 was found to have three mutations (K137T, P195A, and F204L) compared with that of the strain W3110 (Table 2). In this regard, it is interesting to explore how these mutations affect the structure of the GalP protein and its transport capability. As depicted in Fig. 9A and B, the modeled GalP symporter consists of the N- and C-terminal domains. Each domain contains six transmembrane helices and both domains are connected by a long loop, namely the intracellular helical bundle (ICH) domain (Fig. 9B, in orange). In the homology model of GalP, the glucose binding sites are located at the interior hydrophilic cavity surrounded by the N- and C-domains (Fig. 9C), and the mutation site K137T is located at the Helix V in the N-domain (Fig. 9B). As shown in Fig. 9D, the backbone ε-ammonium group (NH3+) of the wild-type K137 is bound to the carboxyl group of residue P2, while the location of the mutated residue K137T is out of the H-bond distance of the P2 residue (Fig. 9E). It is assumed that although the mutated residue K137T is far away from the predicated glucose binding sites, the hydrophilic amino acid Thr may have an indirect effect on the hydrophilic cavity, e.g., an increase in the affinity of glucose-binding.
Fig. 9.
A 3D model of E. coli GalP generated based on the crystal structure of Arabidopsis thaliana symporter STP10 (PDB: 6H7D). (A) The structure (viewed from the periplasmic face) represents an outward-facing conformation in complex with D-glucose. (B) Side view of the modeled GalP structure in a ribbon representation. The intracellular helical bundle (ICH) domain is shown in orange, where the mutated residues in GalP are presented in pink sticks. (C) Comparison of the glucose binding sites of E. coli GalP (blue) and STP10 (pink). (D) For the wild-type residue K137, its backbone ε-ammonium group (NH3+) is attached to the carboxyl group of P2, while (E) the mutated residue K137T is out of H-bond distance of the P2 residue. In comparison to the wild type, the other two mutated residues P195A and F204L are still bound to the adjacent residues to form the flexible ICH domain. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Moreover, Fig. 9B and E shows that the other two mutated residues P195A and F204L are located in the ICH domain. According to early studies, although it has two short helical segments, the ICH domain is an extended and flexible structure that determines the change between inward and outward conformations (Iancu et al., 2013; Paulsen et al., 2019). Compared to the wild-type model of GalP, there is no significant conformation change of the helical bundle in the mutated GalP (Fig. 9E). Looking into the mutated residues, proline at the position of 195 was mutated to Ala and the aromatic acid Phe at the position of 204 was changed to a branched-chain amino acid Leu, which is supposed to result in increase of the flexibility of the helical bundle and lower the energetic barrier of the transition between the inward and outward conformations. Collectively, the mutated residues K137T, P195A, and F204L are supposed to be conducive to the flexibility of the ICH domain and the affinity of glucose-binding, which should be confirmed by further experiments.
3.3. Effect of cell growth and Trp production by interruption of the pentose phosphate pathway
As a key enzyme in the pentose phosphate pathway (PP pathway), the NADP+-dependent glucose-6-phosphate dehydrogenase (Zwf) was found to be mutated in T5, in which the residue E165 was replaced by a stop codon. PP pathway is a metabolic pathway parallel to glycolysis, and it is mainly used for generation of ribose 5-phosphate (R5P). R5P is mainly used in the synthesis of nucleotides and nucleic acids, and the deficiency of R5P therefore will lead to the insufficiency of nucleotides and nucleic acids synthesis. The process of DNA duplication and cell division will consequently be influenced. This might partially explain why T5 exhibited a severe growth deficiency. Moreover, blocking the PP pathway could affect the intracellular supplies of D-erythrose 4-phosphate and phosphoribosyl 5-pyrophosphate, the direct precursors of Trp biosynthesis. Besides, glucose-6-phosphate dehydrogenase (Zwf) functions in the regeneration of NADPH which is required for many enzymes, such as shikimate dehydrogenase (AroE) in shikimate biosynthesis. Thus, inactivation of the enzyme Zwf could result in an insufficient supply of NADPH in the Trp biosynthetic pathway. These might partially explain why T5 produced a similar amount of Trp as the strain S028.
3.4. Further developments of the Trp-producing strain
As presented in Fig. S1, disruption of the PTS system in E. coli aims to block the conversion of PEP to pyruvate and switch PEP to the Trp pathway. Since pyruvate is an essential precursor for many intermediates and for energy metabolism via acetyl-CoA in the TCA cycle, activation of the enzymes involved in pyruvate formation is one of the promising strategies to increase the growth rate of PTS-defective strain. Possible targets include pyruvate kinases encoded by pykA and pykF, anthranilate synthase encoded by TrpE, carboxylase encoded by ppc and phosphoenolpyruvate carboxykinase encoded by pck. However, a balance should be established for PEP between cell growth and Trp biosynthesis (Fig. S1). Therefore, a dynamic regulation at the key node of PEP in a PTS-defective strain is desired. With this dynamic regulation, enzymes like PykAF could be activated during the growth phase. Once the cell enters into the stationary phase, PykAF could be inactivated and another enzyme involved in the Trp pathway such as AroG could be switched on. In this case, PEP is mainly utilized for Trp biosynthesis during the stationary phase. So far, various well-characterized models for stationary phase-responsive regulation of gene expression have been developed and applied for microbial production, e.g., stationary phase sigma factor RpoS (Hengge-Aronis, 1993), quorum sensing (QS) (Miller and Bassler, 2001), and growth/stationary phase-dependent promoter and degradation degron (Hou et al., 2020). Thus, applying these stationary phase-responsive regulators at the node of PEP for dynamically controlled expression of enzymes PykAF and AroG in PTS-defective strain is a promising strategy for further development of Trp-producing strain.
4. Methods
4.1. Strains, plasmids, and primers
All bacterial strains and plasmids used in this study are listed in Supplementary Table S1. Primers used for the genetic modifications are listed in Supplementary Table S2.
4.2. Implementation of CRISPR/Cas9 facilitated promoter engineering
4.2.1. Construction of the host strain for in vivo screening
To realize CRISPR/Cas9-facilitated promoter engineering followed by in vivo screening for modulated expression of GalP/Glk enzymes, we first constructed a PTS-defective strain in which a Trp biosensor was integrated into the chromosome and the ptsI gene encoding the PtsI enzyme (also known as PTS Enzyme I) was removed. Specifically, the Trp-biosensor (Ptac-tnaC-eGFP) was integrated into the chromosome of a previously developed Trp-producing strain S028/pCas9 (Chen and Zeng, 2017) using the CRISPR/Cas9 technique, resulting in a strain designated as S028TS/pCas9. Then, the ptsI gene was removed from S028TS/pCas9, generating a strain designated as G028. Afterwards, the native Glk enzyme of the strain G028 was inactivated by replacing the first part (from −50 bp to 0 bp) of its promoter with chloramphenicol resistance gene (CmR, as a selection marker), generating the strain G028Δglk::CmR (Table S1). The insertion of gene CmR offers single guide RNA (sgRNA) targets for further genome-editing using CRISPR/Cas9 technique.
4.2.2. Construction of promoter variant library
To achieve a high efficiency, it is necessary to provide donor DNA via a plasmid. Therefore, the plasmid pCmN20-GGWT was constructed, which contains the parts expressing the sgRNA targeting the CmR gene and the DNA fragment ptac-galp-pJ23119-glk flanked by the corresponding homologous arms for recombination. It was then used as the template for preparing the plasmid library containing promoter variants for both galP and glk genes. Please refer to the Supplementary Material for more details.
To engineer the promoter sequences of galP/glk genes, −10 and −35 elements of their promoter sequences were chosen for random mutagenesis in this study (Fig. S2). The mutagenesis was introduced with a pair of primers XhoI-galP-MT and galP-XbaI-MT (Table S2) by amplifying the plasmid pCmN20-GGWT using the Phusion High-Fidelity PCR Master Mix (Thermo Scientific). Besides, the plasmid backbone was amplified with a pair primers glk-out-FR and glk-out-RF (Table S2) by amplifying the same plasmid. After digestion of the template DNA with DpnI, two PCR products were fused and then the fused product was transformed into chemically competent E. coli Top10 cells. The plasmids were extracted from these cultivations and eluted in water as promoter variant library: pCmN20-GGMT (Fig. S3).
4.2.3. Genome editing with CRISPR/Cas9 technique
The procedures for preparation of electroporation-competent cells and cell transformation were modified from the protocol reported by Chen and Zeng (2017)(Chen and Zeng, 2017). Specifically, an overnight culture (grown at 30 °C) of the desired strain was inoculated (2%, vol/vol) into 10 mL fresh SOB medium containing 10 mM L-arabinose. When OD600 reached to 0.4–0.5, the cells were then put on ice immediately for 10 min. After that, the cells were harvested by centrifugation at 4 °C and washed three times with precooled 10% glycerol or distilled water. Competent cells were re-suspended in 400 μL precooled 10% glycerol and divided into 200 μL for each reaction. sgRNA plasmid eluted in water was transformed into the corresponding electrocompetent cells by electroporation. Which was done in the 0.2 cm cuvette at 2.5 kV, and the cells were suspended in 1 mL SOB medium and recovered for 2 h at 30 °C before plating. Plates were then incubated for more than 24 h at 30 °C.
4.3. Continuous in vivo mutagenesis of PTS-defective strains for efficient glucose utilization
The continuous in vivo mutagenesis experiment was performed as described in (Packer et al., 2017) with minor modifications (Packer et al., 2017). Briefly, the donor plasmid pCmN20-GGMT was first transformed into G028Cm/pJC184/pCas9MT (G028JC) by electroporation, and then the resultant cells were inoculated into the SOB medium with 10% w/v L-arabinose and cultured for 3 h at 30 °C. Auxostat vessel containing SynM (Chen, 2017) with 15 μg/mL chloramphenicol and 25 μg/mL kanamycin was inoculated with the pre-cultures (resuspended three times with SynM medium) and grown at 30 °C while mixing via a magnetic stirrer. Once the OD600 = 0.8, the inlet flow of fresh media (SynM with 10% w/v arabinose, 15 μg/mL Cm, and 25 μg/mL Kan) was set at a rate of 60–80 mL/h, and the outlet flow of the auxostat culture was set accordingly to maintain the volume of the vessel at a constant of 30 mL. To realize the real-time measurement of OD600 and monitoring of the fluorescence signal, a cell density meter (Ultrospec 10, Biochrom) and a flow cytometer (CytoFLEX, Beckman Coulter) were connected to the vessel. The final evolutionary samples were taken from the auxostat vessel and further screened on the SynM agar plates. After incubation at 30 °C for more than 24 h, transformants with a bigger size and a stronger fluorescent signal were picked and re-checked by streaking them on the SynM agar plates. Finally, the candidate strains were tested by cultivation in 5 mL FM-III medium3 at 30 °C for 24 h. The mutants with a higher medium fluorescent (MFU) were selected for further evaluation by fed-batch fermentations.
4.4. Measurements of fluorescent intensities
Single colonies (or 3 μL of cryo-stock) of interest were inoculated into LB medium and grown at 37 °C, 220 rpm overnight. Cells from each overnight culture were harvested by centrifugation (4 °C, 5000 rpm for 10 min) and followed by washing three times with M9 medium. They were finally re-suspended in fresh M9 medium and then inoculated with the same initial OD600 into 10 mL of fresh M9 medium in a 50 mL conical tube. After 10 h of cultivation at 37 °C and 220 rpm, fluorescence analysis was carried out for each sample using CytoFLEX Flow Cytometer (Beckman Coulter, USA). To do so, cells from each culture were washed three times with PBS buffer and diluted 100-fold before measurements. The eGFP fluorescence (MFI of 10,000 events) was detected with an excitation wavelength of 488 nm and an emission peak wavelength of 509 nm. All data were processed with the Beckman Flow software, and electronic gating was used to separate positive signals from the instrument and the PBS buffer background. For fluorescence intensities, a medium fluorescence unit (MFU) was calculated for each culture.
4.5. Fermentation conditions
The condition for batch fermentations in shake flasks was reported previously (Chen and Zeng, 2017). All the batch fermentations were carried out at 37 °C and 250 rpm. For fed-batch fermentation in the bioreactor, the pre-culture and seed culture were performed under the same conditions as reported previously (Gu et al., 2012). Since the mutagenesis plasmid pJC184 could cause undesired mutations during the fermentation, it was removed from the strain T5/pJC184 and a plasmid-free engineering strain, T5, was obtained. To this end, the specific sgRNA-pMB1 sequence in the pCas9 plasmid was altered to the sgRNA-CmR sequence, by which the Cas9MT protein (Table S1) was able to recognize the target gene CmR located in the plasmid pJC184 and facilitate the cleavage of pJC184 plasmid when induced by IPTG. Fed-batch fermentations were carried out in a highly instrumented and automated 4-parallel 1.5L bioreactors system (DASGIP Parallel Bioreactor System, Jülich, Germany) with an initial working volume of 500 mL. If not indicated otherwise, the fermentation medium in bioreactor, feeding solution, and fermentation conditions were the same as reported previously (Chen and Zeng, 2017).
4.6. Genome DNA sequence analysis of the strain T5
Whole genome sequencing of T5 was done by using a BGISEQ system at BGI Hong Kong Company (Hong Kong, China). A total of 8,764,210 reads were generated for T5, and it was assembled into contig using SOAPdenovo v1.05. Subsequent multiple genome alignment and identification of single nucleotide polymorphisms (SNPs) were conducted using alignment software MUMmer (http://mummer.sourceforge.net/) and BLAT. BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was also conducted to confirm the modified gene sequences in the engineered strain. Moreover, with LASTZ (http://www.bx.psu.edu/miller_lab/dist/README.lastz-1.02.00/), the reference sequence and query sequence were aligned to get the alignment results, and the insertion or deletion (InDel) results were preliminarily obtained. The alignment results were verified with BWA (http://bio-bwa.sourceforge.net/) and samtools (http://samtools.sourceforge.net/).
4.7. Structure analysis and docking study
The prototypical H+/Galactose symporter GalP, a specialized membrane channel for the transport of sugars into and out of cells, are members of the Major Facilitator Superfamily (MFS) (Zheng et al., 2010). To date, many of the crystal structures are available for the MFS, e.g., symporter STP10 form Arabidopsis thaliana (PDB: 6H7D), xylose transporter XylE from E. coli (PDB: 4JA3), and glucose/H+ symporter GlcPSe from Staphylococcus epidermidis (PDB: 4LDS), but the crystal structure for GalP symporter is not yet available. Therefore, a homology model of GalP was carried out using Modeller v9.24 with the crystal structure of A. thaliana STP10 as the structural template (Paulsen et al., 2019; Webb and Sali, 2016). Structural alignment was carried out with PyMoL. Docking of flexible ligand glucose into the GalP-symporter was performed with AutoDock Vina (Trott and Olson, 2010), which is integrated into Chimera (Pettersen et al., 2004).
4.8. Analytic methods
The quantification of glucose, 3-dehydroshikimate (DSA), and shikimate (SA) were done using HPLC as reported previously (Bommareddy et al., 2014; da Luz et al., 2014). The determination of Trp was carried out by using a sensitive spectrophotometric method (Nagaraja et al., 2003).
Compliance with ethical standards ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Support of the Chinese Scholarship Council for M. Chen is acknowledged. We acknowledge support for the Open Access fees by Hamburg University of Technology (TUHH) in the funding programme Open Access Publishing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2021.e00167.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Aguilar C., Martínez-Batallar G., Flores N., Moreno-Avitia F., Encarnación S., Escalante A., Bolívar F. Analysis of differentially upregulated proteins in ptsHIcrr− and rppH− mutants in Escherichia coli during an adaptive laboratory evolution experiment. Appl. Microbiol. Biotechnol. 2018;102:10193–10208. doi: 10.1007/s00253-018-9397-3. [DOI] [PubMed] [Google Scholar]
- Aiba S., Imanaka T., Tsunekawa H. Enhancement of tryptophan production by Escherichia coli as an application of genetic engineering. Biotechnol. Lett. 1980;2:525–530. [Google Scholar]
- Alva A., Sabido-Ramos A., Escalante A., Bolívar F. New insights into transport capability of sugars and its impact on growth from novel mutants of Escherichia coli. Appl. Microbiol. Biotechnol. 2020;104:1463–1479. doi: 10.1007/s00253-019-10335-x. [DOI] [PubMed] [Google Scholar]
- Badran A.H., Liu D.R. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat. Commun. 2015;6:1–10. doi: 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommareddy R.R., Chen Z., Rappert S., Zeng A.-P. A de novo NADPH generation pathway for improving lysine production of Corynebacterium glutamicum by rational design of the coenzyme specificity of glyceraldehyde 3-phosphate dehydrogenase. Metab. Eng. 2014;25:30–37. doi: 10.1016/j.ymben.2014.06.005. [DOI] [PubMed] [Google Scholar]
- Bryson D.I., Fan C., Guo L.-T., Miller C., Söll D., Liu D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017;13:1253–1260. doi: 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmona S.B., Flores N., Martínez-Romero E., Gosset G., Bolívar F., Escalante A. Evolution of an Escherichia coli PTS− strain: a study of reproducibility and dynamics of an adaptive evolutive process. Appl. Microbiol. Biotechnol. 2020;104:9309–9325. doi: 10.1007/s00253-020-10885-5. [DOI] [PubMed] [Google Scholar]
- Carmona S.B., Moreno F., Bolívar F., Gosset G., Escalante A. Inactivation of the PTS as a strategy to engineer the production of aromatic metabolites in Escherichia coli. J. Mol. Microbiol. Biotechnol. 2015;25:195–208. doi: 10.1159/000380854. [DOI] [PubMed] [Google Scholar]
- Chandran S.S., Yi J., Draths K., Daeniken R.v., Weber W., Frost J. Phosphoenolpyruvate availability and the biosynthesis of shikimic acid. Biotechnol. Prog. 2003;19:808–814. doi: 10.1021/bp025769p. [DOI] [PubMed] [Google Scholar]
- Chen L., Chen M., Ma C., Zeng A.-P. Discovery of feed-forward regulation in L-tryptophan biosynthesis and its use in metabolic engineering of E. coli for efficient tryptophan bioproduction. Metab. Eng. 2018;47:434–444. doi: 10.1016/j.ymben.2018.05.001. [DOI] [PubMed] [Google Scholar]
- Chen L., Zeng A.-P. Rational design and metabolic analysis of Escherichia coli for effective production of L-tryptophan at high concentration. Appl. Microbiol. Biotechnol. 2017;101:559–568. doi: 10.1007/s00253-016-7772-5. [DOI] [PubMed] [Google Scholar]
- Chen M., Chen L., Zeng A.-P. CRISPR/Cas9-facilitated engineering with growth-coupled and sensor-guided in vivo screening of enzyme variants for a more efficient chorismate pathway in E. coli. Metab. Eng. Commun. 2019;9 doi: 10.1016/j.mec.2019.e00094. e00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu Y., Ding D., Cong L., Zhang D. Rational design and analysis of an Escherichia coli strain for high-efficiency tryptophan production. J. Ind. Microbiol. Biotechnol. 2018;45:357–367. doi: 10.1007/s10295-018-2020-x. [DOI] [PubMed] [Google Scholar]
- d’Oelsnitz S., Ellington A. Continuous directed evolution for strain and protein engineering. Curr. Opin. Biotechnol. 2018;53:158–163. doi: 10.1016/j.copbio.2017.12.020. [DOI] [PubMed] [Google Scholar]
- da Luz J.A., Hans E., Zeng A.P. Automated fast filtration and on-filter quenching improve the intracellular metabolite analysis of microorganisms. Eng. Life Sci. 2014;14:135–142. [Google Scholar]
- Du L., Zhang Z., Xu Q., Chen N. Central metabolic pathway modification to improve L-tryptophan production in E. coli. Bioengineered. 2019;10:59–70. doi: 10.1080/21655979.2019.1592417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Wang T., Zhang C., Bai J., Zheng X., Zhao X., Lou C., Xing X.-H. Intermediate-sensor assisted push–pull strategy and its application in heterologous deoxyviolacein production in Escherichia coli. Metab. Eng. 2016;33:41–51. doi: 10.1016/j.ymben.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Floras N., Xiao J., Berry A., Bolivar F., Valle F. Pathway engineering for the production of aromatic compounds in Escherichia coli. Nat. Biotechnol. 1996;14:620–623. doi: 10.1038/nbt0596-620. [DOI] [PubMed] [Google Scholar]
- Gu P., Yang F., Kang J., Wang Q., Qi Q. One-step of tryptophan attenuator inactivation and promoter swapping to improve the production of L-tryptophan in Escherichia coli. Microb. Cell Factories. 2012;11:30. doi: 10.1186/1475-2859-11-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin S.O., Tou C.J., Wong E.B., Modavi C., Schaffer D.V., Dueber J.E. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature. 2018;560:248–252. doi: 10.1038/s41586-018-0384-8. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993;72:165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- Hou J., Gao C., Guo L., Nielsen J., Ding Q., Tang W., Hu G., Chen X., Liu L. Rewiring carbon flux in Escherichia coli using a bifunctional molecular switch. Metab. Eng. 2020;61:47–57. doi: 10.1016/j.ymben.2020.05.004. [DOI] [PubMed] [Google Scholar]
- Iancu C.V., Zamoon J., Woo S.B., Aleshin A., Choe J.-y. Crystal structure of a glucose/H+ symporter and its mechanism of action. Proc. Natl. Acad. Sci. Unit. States Am. 2013;110:17862–17867. doi: 10.1073/pnas.1311485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M. Springer; 2003. Amino Acid Production Processes. In Microbial Production of L-Amino Acids; pp. 1–35. [DOI] [PubMed] [Google Scholar]
- Ikeda M. Towards bacterial strains overproducing L-tryptophan and other aromatics by metabolic engineering. Appl. Microbiol. Biotechnol. 2006;69:615. doi: 10.1007/s00253-005-0252-y. [DOI] [PubMed] [Google Scholar]
- Jakočiūnas T., Pedersen L.E., Lis A.V., Jensen M.K., Keasling J.D. CasPER, a method for directed evolution in genomic contexts using mutagenesis and CRISPR/Cas9. Metab. Eng. 2018;48:288–296. doi: 10.1016/j.ymben.2018.07.001. [DOI] [PubMed] [Google Scholar]
- Kim S.-Y., Nam T.-W., Shin D., Koo B.-M., Seok Y.-J., Ryu S. Purification of Mlc and analysis of its effects on the pts expression in Escherichia coli. J. Biol. Chem. 1999;274:25398–25402. doi: 10.1074/jbc.274.36.25398. [DOI] [PubMed] [Google Scholar]
- Knop D.R., Draths K.M., Chandran S.S., Barker J.L., von Daeniken R., Weber W., Frost J.W. Hydroaromatic equilibration during biosynthesis of shikimic acid. J. Am. Chem. Soc. 2001;123:10173–10182. doi: 10.1021/ja0109444. [DOI] [PubMed] [Google Scholar]
- Li M., Liu C., Yang J., Nian R., Xian M., Li F., Zhang H. Common problems associated with the microbial productions of aromatic compounds and corresponding metabolic engineering strategies. Biotechnol. Adv. 2020:107548. doi: 10.1016/j.biotechadv.2020.107548. [DOI] [PubMed] [Google Scholar]
- Liu L., Bilal M., Luo H., Zhao Y., Iqbal H. Metabolic engineering and fermentation process strategies for L-tryptophan production by Escherichia coli. Processes. 2019;7:213. [Google Scholar]
- Lu J., Tang J., Liu Y., Zhu X., Zhang T., Zhang X. Combinatorial modulation of galP and glk gene expression for improved alternative glucose utilization. Appl. Microbiol. Biotechnol. 2012;93:2455–2462. doi: 10.1007/s00253-011-3752-y. [DOI] [PubMed] [Google Scholar]
- Mateos S.S., Sánchez C.L., Paredes S.D., Barriga C., Rodríguez A.B. Circadian levels of serotonin in plasma and brain after oral administration of tryptophan in rats. Basic Clin. Pharmacol. Toxicol. 2009;104:52–59. doi: 10.1111/j.1742-7843.2008.00333.x. [DOI] [PubMed] [Google Scholar]
- Miller M.B., Bassler B.L. Quorum sensing in bacteria. Annu. Rev. Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Nagaraja P., Yathirajan H.S., Vasantha R.A. Highly sensitive reaction of tryptophan with p-phenylenediamine. Anal. Biochem. 2003;312:157–161. doi: 10.1016/s0003-2697(02)00504-3. [DOI] [PubMed] [Google Scholar]
- Oldiges M., Kunze M., Degenring D., Sprenger G., Takors R. Stimulation, monitoring, and analysis of pathway dynamics by metabolic profiling in the aromatic amino acid pathway. Biotechnol. Prog. 2004;20:1623–1633. doi: 10.1021/bp0498746. [DOI] [PubMed] [Google Scholar]
- Packer M.S., Rees H.A., Liu D.R. Phage-assisted continuous evolution of proteases with altered substrate specificity. Nat. Commun. 2017;8:1–11. doi: 10.1038/s41467-017-01055-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen P.A., Custódio T.F., Pedersen B.P. Crystal structure of the plant symporter STP10 illuminates sugar uptake mechanism in monosaccharide transporter superfamily. Nat. Commun. 2019;10:1–8. doi: 10.1038/s41467-018-08176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. Control of the expression of the manXYZ operon in Escherichia coli: mlc is a negative regulator of the mannose PTS. Mol. Microbiol. 1998;27:369–380. doi: 10.1046/j.1365-2958.1998.00685.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. DNA binding sites for the Mlc and NagC proteins: regulation of nagE, encoding the N-acetylglucosamine-specific transporter in Escherichia coli. Nucleic Acids Res. 2001;29:506–514. doi: 10.1093/nar/29.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J., Kolb A. DNA loop formation between Nag repressor molecules bound to its two operator sites is necessary for repression of the nag regulon of Escherichia coli in vivo. Mol. Microbiol. 1993;10:973–981. doi: 10.1111/j.1365-2958.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- Ravikumar A., Arzumanyan G.A., Obadi M.K., Javanpour A.A., Liu C.C. Scalable, continuous evolution of genes at mutation rates above genomic error thresholds. Cell. 2018;175:1946–1957. doi: 10.1016/j.cell.2018.10.021. e1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A.L., Trachtmann N., Becker J., Lohanatha A.F., Blotenberg J., Bolten C.J., Korneli C., de Souza Lima A.O., Porto L.M., Sprenger G.A. Systems metabolic engineering of Escherichia coli for production of the antitumor drugs violacein and deoxyviolacein. Metab. Eng. 2013;20:29–41. doi: 10.1016/j.ymben.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Song L., Zeng A.-P. Engineering ‘cell robots’ for parallel and highly sensitive screening of biomolecules under in vivo conditions. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-15621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016;54 doi: 10.1002/cpbi.3. 5.6.1-5.6.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B.B., Van Benschoten A.H., Cimermancic P., Donia M.S., Zimmermann M., Taketani M., Ishihara A., Kashyap P.C., Fraser J.S., Fischbach M.A. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe. 2014;16:495–503. doi: 10.1016/j.chom.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong B., Zhu Y., Tian D., Jiang S., Fan X., Ma Q., Wu H., Xie X. Flux redistribution of central carbon metabolism for efficient production of L-tryptophan in Escherichia coli. Biotechnol. Bioeng. 2021 doi: 10.1002/bit.27665. [DOI] [PubMed] [Google Scholar]
- Zhao Z., Chen S., Wu D., Wu J., Chen J. Effect of gene knockouts of L-tryptophan uptake system on the production of L-tryptophan in Escherichia coli. Process Biochem. 2012;47:340–344. [Google Scholar]
- Zheng H., Taraska J., Merz A.J., Gonen T. The prototypical H+/galactose symporter GalP assembles into functional trimers. J. Mol. Biol. 2010;396:593–601. doi: 10.1016/j.jmb.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Zhao D., Qiu H., Fan F., Man S., Bi C., Zhang X. The CRISPR/Cas9-facilitated multiplex pathway optimization (CFPO) technique and its application to improve the Escherichia coli xylose utilization pathway. Metab. Eng. 2017;43:37–45. doi: 10.1016/j.ymben.2017.08.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.