Abstract
Objective
The ventromedial nucleus of the hypothalamus (VMH) is a critical component of the forebrain pathways that regulate energy homeostasis. It also plays an important role in the metabolic response to fasting. However, the mechanisms contributing to these physiological processes remain elusive. Autophagy is an evolutionarily conserved mechanism that maintains cellular homeostasis by turning over cellular components and providing nutrients to the cells during starvation. Here, we investigated the importance of the autophagy-related gene Atg7 in Sf1-expressing neurons of the VMH in control and fasted conditions.
Methods
We generated Sf1-Cre; Atg7loxP/loxP mice and examined their metabolic and cellular response to fasting.
Results
Fasting induces autophagy in the VMH, and mice lacking Atg7 in Sf1-expressing neurons display altered leptin sensitivity and impaired energy expenditure regulation in response to fasting. Moreover, loss of Atg7 in Sf1 neurons causes alterations in the central response to fasting. Furthermore, alterations in mitochondria morphology and activity are observed in mutant mice.
Conclusion
Together, these data show that autophagy is nutritionally regulated in VMH neurons and that VMH autophagy participates in the control of energy homeostasis during fasting.
Keywords: Atg7, Ventromedial nucleus, Fasting, Energy balance, Hypothalamus, Mitochondria
Graphical abstract

Highlights
-
•
Fasting induces autophagy in the ventromedial nucleus of the hypothalamus.
-
•
Genetic loss of Atg7 in the VMH impairs metabolic response to fasting.
-
•
Mice lacking Atg7 in the VMH display impaired mitochondria morphology and activity.
1. Introduction
Autophagy has received considerable attention over the past decade because dysfunction of this cellular process, which degrades cytoplasmic materials including organelles and misfolded proteins, causes cellular alterations and contributes to a variety of diseases such as cancer, muscular disorders, and neurodegeneration [[1], [2], [3]]. Autophagy has also been implicated in physiological processes, such as metabolic regulations. For example, the deletion of essential autophagy genes, such as the autophagy-related gene (Atg) 7, in the liver, pancreas, or adipose tissue produces alterations in body weight, adiposity, and glucose homeostasis [[4], [5], [6], [7]]. More recent studies have shown the importance of autophagy in the central regulation of feeding behavior and energy homeostasis. Downregulation of Atg7 in the mediobasal hypothalamus increases energy intake and reduces energy expenditure, resulting in weight gain and insulin resistance when animals are fed a high-fat diet (HFD) [8]. More specific conditional deletion of autophagy-related genes in pro-opiomelanocortin (POMC) and agouti-related peptide (AgRP) neurons revealed the importance of autophagy in the arcuate nucleus of the hypothalamus (ARH). Loss of Atg7 in POMC neurons causes weight gain, hyperphagia, as well as leptin and insulin resistance [[9], [10], [11]]. Mice lacking Atg7 in POMC neurons also exhibit increased sensitivity to weight gain when exposed to an HFD [10,11]. Similarly, the loss of Atg12 in POMC neurons promotes weight gain and perturbations in glucose homeostasis induced by HFD [12]. In contrast, deficiency in Atg4 in POMC neurons or Atg7 in AgRP neurons causes leanness, suggesting that autophagy induces distinct physiological effects based on the pathways and cell types engaged [13,14].
Most of the research on hypothalamic autophagy has focused on neurons in the ARH thus far. However, although constitutive autophagy is also found in other hypothalamic nuclei, including in the ventromedial nucleus of the hypothalamus (VMH) [9], the importance of autophagy in VMH neurons has yet to be delineated. The VMH has long been associated with body weight regulation and glucose homeostasis. Physical lesions of the VMH induce hyperphagia, obesity, and diabetes [15]. Moreover, genetic studies have shown the importance of VMH neurons, particularly a subpopulation of VMH neurons expressing the steroidogenic factor-1 (SF-1), in relaying leptin and insulin action on energy balance and glucose metabolism [[16], [17], [18], [19]]. In the present study, we examined the metabolic importance of autophagy in VMH neurons in fed and fasted conditions by generating mice that lack Atg7 in Sf1 neurons.
2. Material and methods
2.1. Animals
Mice were housed in individual cages under specific pathogen-free conditions, maintained in a temperature-controlled room with a 12-h light/dark cycle, and provided ad libitum access to water and standard laboratory chow (Special Diet Services). Animal studies were approved by the Institutional Ethics Committees of Care and Use of Experimental Animals of the University of Lille 2 (France). All experiments were performed in accordance with the guidelines for animal use specified by the European Union Council Directive of September 22, 2010 (2010/63/EU) and the approved protocol (APAFIS#13387–2017122712209790 v9) by the Ethical Committee of the French Ministry of Education and Research. Mice in which microtubule-associated protein one chain light chain (LC3) had been fused to the green fluorescent protein (GFP) were kindly provided by Dr. Mizushima [20]. To generate VMH-specific Atg7 knockout (Sf1-Cre; Atg7loxP/loxP) mice, Sf1-Cre mice (Jax mice stock# 006364) [16] were mated to mice carrying a loxP-flanked Atg7 allele (Atg7loxP/loxP) (C57BL/6 background, kindly provided by Dr. Komatsu) [21]. Breeding colonies were maintained by mating Sf1-Cre; Atg7loxP/+ mice to Atg7loxP/loxP mice. All mice were generated in a C57BL/6 background. Animals were genotyped by polymerase chain reaction (PCR) as described previously [16,21]. Cre-negative Atg7loxP/loxP were used as controls.
2.2. Fasting experiments
Mice were divided into the following groups: a first group of mice was fed standard chow ad libitum (also referred to as “fed” mice. A second group of mice was deprived of food for 12 h (during the dark phase) before the experiment (also referred to as “fasted” mice). In some experiments, a third group of mice was deprived of food for 12 h and then given free access to food for an additional 24 h (also referred to as “refed” mice).
2.3. Physiological measures
One day after birth, the litter size was adjusted to 7 pups to ensure adequate and standardized nutrition until weaning. Male and female mice were weighed weekly from P7 through P119 using an analytical balance. Food intake, energy expenditure, and heat production were monitored in 2- to 3-month-old mice using a combined indirect calorimetry system (TSE Systems). Briefly, O2 consumption and CO2 production were measured for 24 h to determine the energy expenditure. In addition, food intake was determined continuously by the integration of weighing sensors fixed at the top of the cage from which the food containers were suspended into the sealed cage environment. Heat production was also measured through the experiment. At the end of the experiment, body composition analysis (fat/lean mass) was performed using an NMR (EchoMRI). These physiological measures were performed at the University of Cincinnati Mouse Metabolic Phenotyping Center. In addition, glucose and insulin tolerance were performed in 2- to 3-month-old mice through the intraperitoneal (i.p.) administration of glucose (1.5 mg/g body weight) or insulin (0.75 U/kg body weight) after overnight fasting, and the blood glucose levels were measured 0, 15, 30, 45, 60, 90, 120, and 150 min following glucose challenge, as previously described [9]. Leptin sensitivity tests were performed in 2- to 3-month-old male mice. Briefly, the mice underwent i.p. injection with vehicle (5 mM of sodium citrate buffer) or leptin (3 mg/kg body weight, Peprotech) according to the following scheme: vehicle injections for 5 days, followed by leptin injections for 3 days. Body weight was measured during the injection period. Serum leptin and insulin levels were also assayed in 2- to 3-month-old mice using leptin and insulin enzyme-linked immunosorbent assay (ELISA) kits, respectively (Millipore).
2.4. Immunohistochemistry
Anesthetized male mice were perfused transcardially with 4% paraformaldehyde at 2–3 months of age. The brains were then frozen and sectioned at 30-μm-thick and processed for immunofluorescence using standard procedures. The primary antibodies used for immunohistochemistry (IHC) were as follows: rabbit anti-GFP (1:10,000, Invitrogen), rabbit anti-cFos (1:2,000, Oncogene), rabbit anti-p62/SQSTM1 (1:1,000, Abcam), and rabbit anti-ubiquitin (1,000, Dako). The primary antibodies were visualized with Alexa Fluor 488 or Alexa Fluor 568 goat anti-rabbit IgGs (1:200, Invitrogen) and Alexa Fluor 568 donkey anti-sheep IgGs (1:200, Invitrogen). Sections were counterstained using bis-benzamide (1:10,000, Invitrogen) to visualize cell nuclei and coverslipped with Fuoromount-G (SouthernBiotech).
2.5. Hypothalamic primary cultures
Hypothalami were collected from P2 wild-type mouse pups and dissociated by incubation with activated papain, followed by trituration accordingly to the manufacturer's protocol (Worthington). Dissociated cells were pelleted, then re-suspended in supplemented Neurobasal-A medium and cultured onto cell culture slides coated with poly-l-lysine (Sigma–Aldrich). After one day in vitro, cells were transfected (TransIT-TKO® Transfection Reagent, Mirus) with siRNA against Atg7 (50 nM, Cell Signaling). Scramble siRNA (Cell Signaling) was used as control. After 72 h, cells were incubated with 250 nM of MitoTracker® (Invitrogen), fixed in paraformaldehyde, and stained with MAP2 (1:500, Sigma). Experiments were independently repeated 3–4 times.
2.6. Image analysis
For the histological experiments, two sections through the VMH (for cFos, p62, and ubiquitin stainings and LC3-GFP fluorescence), the ARH and DMH (for cFos staining), and the ARH and PVH (for LC3-GFP fluorescence), from animals of each experimental group were acquired using a Zeiss LSM 710 confocal system equipped with a 20X objective. Slides were numerically coded to obscure the treatment group. Image analysis was performed using ImageJ analysis software (NIH) as described previously [9].
For the quantitative analysis of cell number, the number of cFos-immunopositive cells in the VMH, ARH, and DMH were manually counted. The average number of cells counted in two sections from each mouse was used for statistical comparisons.
For the quantification of ubiquitin- and p62-immunoreactivity and Mitotracker staining, each image plane was binarized to isolate labeled fibers from the background and compensate for differences in fluorescence intensity. The integrated intensity was then calculated for each image, which reflects the total number of pixels in the binarized image. This procedure was conducted on each image plane in the stack, and the values for all of the image planes in a stack were summed. The resulting value is an accurate index of the density of the processes in the volume sampled.
The NIH ImageJ macro called GFP-LC3 (http://imagejdocu.tudor.lu/author/rkd8/) [22] was used to quantify the number of LC3-GFP puncta.
2.7. Quantitative reverse transcription PCR (RT-PCR)
VMH of 2- to 3-month-old male mice fed ad libitum and fasted for 12 h were microdissected. Total RNA was isolated using the Arcturus PicoPure RNA Isolation kit (Applied Biosystem). cDNAs were generated with the high-capacity cDNA Reverse Transcription Kit (Applied Biosystem). Quantitative real-time PCR analysis was performed using the TaqMan Fast universal PCR Mastermix. mRNA expression was calculated using the 2-DDCt method after normalization with Gapdh as a housekeeping gene. Inventoried TaqMan® Gene expression assays Atg5 (Mm00504340_m1), Atg7 (Mm00512209_m1), Atg12 (Mm00503201_m1), InsR (Mm01211875_m1), Pomc (Mm00435874_m1), Npy (Mm03048253_m1), Cpt1 (Mm01231183_m1), Pgc-1α (Mm01208835_m1), Acaca (Mm01304257_m1), Ucp2 (Mm00627598_m1), and Gapdh (Mm99999915_g1) were used. All assays were performed using an Applied Biosystems Prism 7900HT fast sequence detection system.
2.8. Electron microscopy
Two- to 3-month-old mice were perfused with 4% paraformaldehyde, and their brains were processed for electron microscopy examination. Briefly, ultrathin sections were then cut on a Leica ultra-microtome, collected on Formvar-coated single-slot grids, and analyzed with a Tecnai 12 Biotwin electron microscope (FEI).
2.9. Mitochondria activity
Mediobasal hypothalami from 2- to 3-month-old male mice were dissected, and mitochondrial oxygen consumption was determined using high-resolution respirometry (Oxygraph-2K, Oroboros Instruments), as described previously [23]. Briefly, basal O2 flux of the respiratory chain was measured at 37 °C without substrate or with a substrate for complex I (glutamate, 20 mM) and (D) complex II (succinate, 20 mM). States 3 and 4 respiration rates were achieved by adding to the incubation medium ADP (2.5 mM) and carboxy-atractyloside (CAtr, 0.75 μM), respectively. Measurement of maximal mitochondrial respiration was performed using carbonyl cyanide m-chlorophenylhydrazone (CCCP, 0.5 μM), a chemical uncoupling molecule. Rotenone (0.5 μM), a CI inhibitor, was used to measure the maximal respiration capacity due to complex II only. The respiratory control ratio (RCR) was calculated by dividing the state respiration 3 rate by the state 4 respiration rate. The experimental data were analyzed using DataGraph software (Oroboros Instruments).
2.10. Statistical analysis
All values were expressed as mean ± standard error of the mean (SEM). Statistical analyses were conducted using GraphPad PRISM (version 5.0d). Statistical significance was determined using unpaired two-tailed Student's t-tests, one-way analysis of variance (ANOVA) followed by the Tukey post-hoc test, and two-way ANOVA followed by the Bonferroni post-hoc test when appropriate. P-values less than 0.05 were considered statistically significant.
3. Results
3.1. Fasting induces autophagy in the ventromedial nucleus of the hypothalamus
We previously reported that autophagy is constitutively activated in the hypothalamus of normal fed mice, including in the VMH [9]. However, how autophagy is regulated in specific hypothalamic nuclei remains largely unknown. To explore whether nutrition influences autophagy in the VMH, we first examined the effect of food deprivation in transgenic mice in which microtubule-associated protein 1 light chain 3 (LC3) has been fused to the green fluorescent protein (GFP). Because LC3 is a reliable marker of autophagosomes, LC3-GFP mice can be used to monitor autophagy in vivo [24]. LC3-GFP puncta were readily detectable throughout the VMH of fed mice, and fasting markedly increased the density of LC3-GFP puncta found in both the dorsomedial and ventrolateral parts of the VMH (Figure 1A,B). Fasting-induced autophagy was not restricted to the VMH; the density of LC3-GFP puncta was also significantly higher in the ARH of fasted mice than in fed mice and elevated in the PVH as well, although it did not reach statistical significance (Figure S1A–B). We next examined whether fasting caused changes in autophagy-related genes. Quantitative PCR analyses revealed that Atg7 and Atg12 mRNA levels were increased and decreased, respectively, in the VMH of fasted animals compared to fed mice (Figure 1C). However, mRNA expression of Atg5 was unchanged (Figure 1C). Together, these data indicate that hypothalamic autophagy is nutritionally regulated and that fasting triggers autophagy in the VMH.
Figure 1.
Fasting promotes autophagy in the ventromedial nucleus of the hypothalamus. (A) Representative images and (B) quantification of LC3-GFP immunofluorescence in the dorsomedial (dm) and ventrolateral (vl) parts of the ventromedial nucleus of the hypothalamus (VMH) of adult fed or 12 h-fasted mice (n = 3 per group). (C) Autophagy-related gene mRNA levels in the VMH of adult fed or 12 h-fasted mice (n = 3–5 per group). V3, third ventricle. Scale bar, 10 μm. Values are shown as mean ± SEM. ∗P ≤ 0.05 and ∗∗P ≤ 0.01 versus fed.
3.2. Deletion of Atg7 in SF1 neurons impairs metabolic response to fasting
To investigate the functional role of autophagy in VMH neurons, we generated mice lacking Atg7 in cells that express Sf1, a factor predominantly expressed in VMH neurons (Sf1-Cre; Atg7loxP/loxP mice) [16]. We first confirmed that the VMH is indeed autophagy-defective by analyzing the formation of ubiquitin- and polyubiquitin-binding protein p62/SQSTM1 because the accumulation of these aggregates is a hallmark of autophagy deficiency [25]. As expected, Sf1-Cre; Atg7loxP/loxP mice displayed a 9- and 4-fold increase in the density of ubiquitin- and p62-immunoreactive aggregates, respectively, in the VMH (Figure S2A, B). Importantly, induction of ubiquitin and p62 immunoreactivity was not found in any other brain regions, confirming that the loss of autophagy in mutant mice occurs exclusively in the VMH and not in other brain sites expressing Sf1.
We next investigated the role of VMH autophagy in energy homeostasis and response to fasting. We first assessed food intake, locomotor activity, and energy expenditure (including O2 consumption, and CO2 and heat production) in fed and fasted mice. When fed ad libitum, Sf1-Cre; Atg7loxP/loxP mice were comparable to their control littermates with regard to food intake (Figure 2B), locomotor activity (data not shown), oxygen consumption (VO2), carbon dioxide production (VCO2), and heat production (Figures 2G,I, J, L and S3A, C, D, F). However, when fasted overnight, Sf1-Cre; Atg7loxP/loxP mice displayed reduced VO2, VCO2, and heat production (Figures 2H,I, K, L and S3E, F) as well as reduced cumulative food intake during a refeeding following an overnight fast (Figure 2C) compared to Atg7loxP/loxP mice. However, this altered metabolic response was not accompanied by changes in carbohydrates versus lipid oxidation rates, as the RQ was normal in fed and fasted mutants (Figure S3A–C). Moreover, loss of Atg7 in Sf1 neurons did not result in changes in body weight (Figure 2A). Serum leptin levels were reduced in Sf1-Cre; Atg7loxP/loxP mice (Figure 2F) despite an elevated fat mass (Figure 2D). Moreover, mutant mice showed an attenuated weight loss after exogenous leptin treatment (Figure 2E).
Figure 2.
Altered energy homeostasis in mice lacking autophagy in SF1 neurons. (A) Body weight and (B) average food intake of Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP (n 4–6 per group) male mice. (C) Food intake of refed of adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice after fasting (n = 5 for each group). (D) Body composition and (E) leptin sensitivity of adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice (n = 5–6 for each group). (F) Serum leptin levels in fed and 12 h-fasted adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP (n = 3–5 per group) male mice. (G–I) Oxygen (O2) consumption and (J–L) carbon dioxide (CO2) production in (G, J) fed and (H, K) 12 h-fasted Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice (n = 5 for each group). Values are shown as mean ± SEM. ∗P ≤ 0.05, and P ≤ 0.01 versus each group.
Previous studies reported a role for SF1 neurons in glucose metabolism [16,19,26,27]. We therefore measured several indices of glucose metabolism in Sf1-Cre; Atg7loxP/loxP mice and found significantly lower fed plasma insulin concentrations compared to Atg7loxP/loxP mice (Figure 3B) without changes in plasma glucose levels (Figure 3A). In addition, insulin receptor (Insr) mRNA levels in the VMH of fed Sf1-Cre; Atg7loxP/loxP mice were significantly higher than those in control mice (Figure 3C). Fasting equally reduced glucose and insulin levels in Sf1-Cre; Atg7loxP/loxP and Atg7loxP/loxP mice (Figure 3A–B). We also performed glucose and insulin tolerance tests and found that Sf1-Cre; Atg7loxP/loxP mice responded to a glucose or an insulin challenge normally (Figure 3D–E).
Figure 3.
Glucose metabolism in Sf1-Cre; Atg7loxP/loxPmice. (A) Serum glucose and (B) insulin levels and (C) VMH insulin receptor (InsR) mRNA expression in fed and 12h-fasted adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP (n = 4–7 per group) male mice. (D) Glucose and (E) insulin tolerance tests of adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP (n = 4–6 per group) male mice. Values are shown as mean ± SEM. ∗P ≤ 0.05, ∗∗P ≤ 0.01, and ∗∗P ≤ 0.001 versus each group.
3.3. Altered hypothalamic response to fasting in Sf1-Cre; Atg7loxP/loxP mice
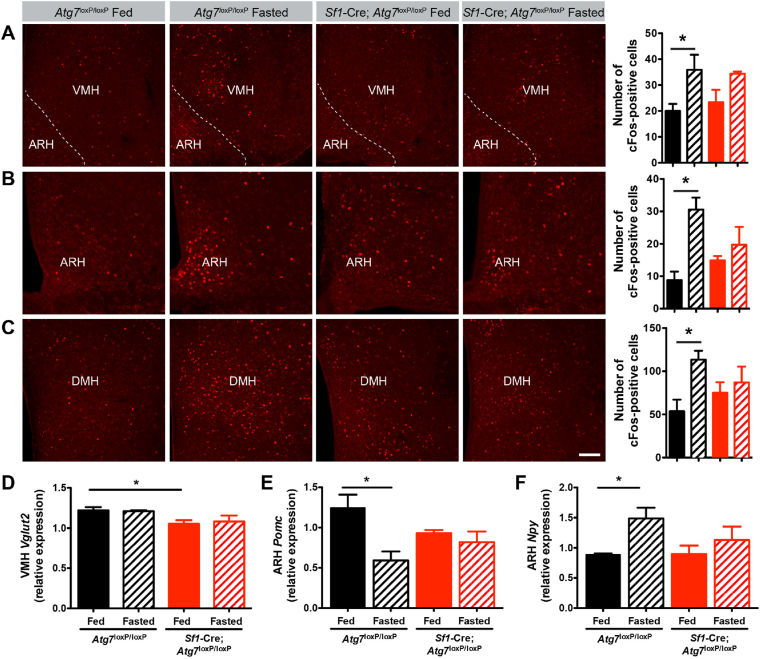
The data described above suggest impairments in the physiological response to fasting in Sf1-Cre; Atg7loxP/loxP mice. To determine whether these impairments are centrally mediated, we evaluated the ability of fasting to induce cFos expression (a surrogate marker of neuronal activation) in Sf1-Cre; Atg7loxP/loxP and Atg7loxP/loxP mice. Fed control and mutant mice had only a low density of cFos-immunoreactive cells in the ventromedial, arcuate (ARH), and dorsomedial (DMH) nuclei of the hypothalamus (Figure 4A–C). Fasting caused a 2- to 3-fold increase in the density of cFos-immunoreactive cells in the VMH, ARH, and DMH of Atg7loxP/loxP mice; however, this induction was abrogated in Sf1-Cre; Atg7loxP/loxP mice (Figure 4A–C). The exact phenotype of these cFos+ cells remains to be investigated. VMH neurons are predominantly glutamatergic and express synaptic vesicular transporter 2 (VGLUT2) [26]. Sf1-Cre; Atg7loxP/loxP mice displayed lower Vglut2 mRNA levels in the VMH than control mice, but Vglut2 mRNA levels are unaffected by fasting in both control and mutant mice (Figure 4 D). However, fasting induced down- and upregulation of Pomc and Npy gene expression, respectively, in control mice, but this response was blunted in mutant mice (Figure 4 E-F).
Figure 4.
Fasting-induced hypothalamic neuronal activation is impaired in Sf1-Cre; Atg7loxP/loxPmice. (A–C) Representative images and quantification of cFos-immunoreactive cells in the (A) ventromedial (VMH), (B) arcuate (ARH), and (C) dorsomedial (DMH) nuclei of the hypothalamus of fed and fasted adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice (n = 3–7 per group). (D) Vglut2, (E) Pomc, and (F) Npy mRNA expression in the ARH of adult fed and fasted Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice (n = 3–4 per group). Values are shown as mean ± SEM. ∗P ≤ 0.05 versus each group.
Because autophagy has been shown to maintain cell homeostasis and survival [21,28], we also assessed whether Atg7 deletion in Sf1 neurons could affect the survival of these neurons using activated-caspase 3-immunoreactivity. The number of nuclei (stained by bisbenzimide) in the VMH of Sf1-Cre; Atg7loxP/loxP mice was not different from that of Atg7loxP/loxP mice (data not shown). We also counted the number of activated-caspase 3-positive cells (a surrogate marker of cell death), and we did not find any evidence of neuronal cell death (data not shown).
3.4. Loss of Atg7 in VMH neurons alters mitochondrial morphology and function
Because during fasting mitochondrial efficiency must increase dramatically [[29], [30], [31], [32]] and autophagy is an important process for mitochondrial elimination [[33], [34], [35]], we next investigated whether the deletion of Atg7 in Sf1 neurons causes mitochondria structural alterations using electron microscopy. Ultrastructural analysis showed a normal density of mitochondria related to cell size in VMH neurons of mutant mice (data not shown). However, there was a reduction in the number of mitochondria-autophagosome contacts in Sf1-Cre; Atg7loxP/loxP mice compared to control mice (Figure 5A). Moreover, the relative aspect ratio, which is an indicator of mitochondrial length, was significantly increased in mutant mice compared to control mice (Figure 5A). To further examine the consequences of Atg7 deficiency on mitochondria, we transfected primary cultures of hypothalamic cells with adenovirus-containing siRNA directed against Atg7 and stained mitochondria using MitoTracker. We found a 1.3-fold increase in the intensity of MitoTracker labeling in Atg7 KD cells compared with control cells (Figure 5B). Collectively, these results indicate that both mitochondrial morphology and their association with autophagosomes are altered in the VMH of mutant mice.
Figure 5.
Loss of autophagy in SF1 neurons causes altered mitochondrial morphology and function. (A) Representative electron microscopy images of VMH neurons, percentage of autophagy-mitochondria contacts, and mitochondria aspect ratio in adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice (n = 4 per group). (B) Representative images and quantification of MitoTracker labeling in primary cultures of hypothalamic neurons 1 day after transfection with Atg7 siRNA or scrambled siRNA (n = 3–4 per group). (C) Mitochondria-related gene mRNA levels in the VMH of adult (10-week-old) fed or 12 h-fasted mice (n = 3–5 per group). (D) Measurement of mitochondrial respiration in the mediobasal hypothalamus of adult Atg7loxP/loxP and Sf1-Cre; Atg7loxP/loxP male mice (n = 4–8 per group). Values are shown as mean ± SEM. ∗P ≤ 0.05 versus each group.
We next measured mitochondria-related genes and found that Cpt1a and Pgc1a expression was downregulated in fed and fasted Sf1-Cre; Atg7loxP/loxP mice, respectively, compared to controls (Figure 5C). Moreover, fasting induced Ucp2 mRNA levels in the VMH of Atg7loxP/loxP mice, but this response was blunted in mutant mice (Figure 5C). Cpt1c and Acaca mRNA levels were normal in both fed and fasted Sf1-Cre; Atg7loxP/loxP and Atg7loxP/loxP mice (Figure 5C). To explore the functional impact of Atg7 deletion in Sf1 neurons, we assessed mitochondrial respiration in the mediobasal hypothalamus of control and mutant mice. Basal oxygen consumption was approximately 20% lower in Sf1-Cre; Atg7loxP/loxP mice compared to control animals (Figure 5D). A significant reduction in mitochondrial oxygen consumption was also observed after the addition of complex I (glutamate) and II (succinate) substrates (Figure 5D). Moreover, Sf1-Cre; Atg7loxP/loxP mice displayed a reduction in coupled (state 3) and uncoupled (state 4) mitochondrial respiration (Figure 5D). CCCP-induced maximal respiration was also attenuated in mutant mice, and this effect appears unaffected by the complex I inhibitor rotenone (Figure 5D).
Together, these data indicate that basal mitochondria morphology and activity are altered in mice lacking Atg7 in Sf1 neurons.
4. Discussion
Autophagy is involved in a variety of physiological and pathological pathways, and more recent studies have demonstrated the importance of hypothalamic autophagy in energy balance regulation and glucose homeostasis. Here, we show that in fed conditions, loss of the autophagy gene Atg7 in Sf1 VMH neurons has no effects on body weight, food intake, energy expenditure, or glucose metabolism. However, when fasted, Sf1-Cre; Atg7loxP/loxP mice display abnormalities in O2 consumption, CO2 and heat production, and they consume less food after refeeding. These findings differ from other studies showing that loss of Atg7 in arcuate POMC neurons increases body weight and fat mass, decreases O2 consumption, and impairs glucose metabolism [[9], [10], [11]]. These data indicate that the autophagy Atg7 pathway exerts distinct physiological functions depending on the neuronal cell type that is engaged. Consistent with this idea, mice lacking Atg7 in AgRP neurons are leaner and display an opposite phenotype than mice lacking Atg7 in the neighboring POMC neurons [14].
The mediobasal hypothalamus, including the VMH, plays a critical role in sensing and integrating metabolic and nutrient signals from the periphery and engaging neural circuits to control feeding and glucose regulation. Autophagy is an essential component of the cellular response to starvation for energy provision [36]. In the present study, we show that fasting triggers autophagy in the VMH as well as in other hypothalamic nuclei, such as the ARH. These findings are in good agreement with a previous study from Kaushik et al. that showed that starvation increases LC3-II/I in the mediobasal hypothalamus, a region comprising both the ARH and the VMH [14]. The finding that fasting causes cFos activation in the VMH could be surprising given that opto- or chemo-genetic stimulation of VMH Sf1 neurons reduces feeding [37,38], and photostimulation of VMH neurons stimulates anorexigenic POMC neurons [39]. However, like many other hypothalamic nuclei, the VMH contains heterogeneous populations of neurons that play distinct physiological functions. For example, in addition to containing anorexigenic Sf1 neurons, the VMH also contains glucose-inhibited (GI) neurons that are stimulated during hypoglycemia. We found that fasting (which causes hypoglycemia) induces cFos activation primarily in the ventrolateral part of the VMH where GI neurons are located [40], and we observed less cFos-positive neurons in the central or dorsomedial parts of the VMH where Sf1 neurons reside [16]. Leptin and insulin are likely candidates to mediate the effect of fasting on VMH autophagy. Fasting reduces leptin and insulin levels, and we recently reported that leptin-deficiency induces autophagy in the hypothalamus [41]. In addition, the VMH expresses high levels of leptin and insulin receptors [42,43], and their loss in VMH Sf1 neurons influences the response to HFD-induced metabolic disturbances [16,19]. Moreover, electrophysiological studies indicated that the effects of leptin and insulin on VMH SF1 neurons are mediated through PI3K [44], which is a critical signaling pathway involved in autophagy activation [45]. Future studies are needed to determine the role of VMH leptin and insulin signaling in the physiological phenotype of Sf1-Cre; Atg7loxP/loxP mice, particularly in response to fasting. We also cannot rule out the possibility that the reduced feeding response in mutant mice is independent of leptin or insulin but may be the result of a hypersensitivity to other anorexigenic signals, such as estrogens. In addition, it is possible that the lack of autophagy in Sf1-expressing neurons indirectly impacts neurons outside of the VMH. For example, a previous study showed that POMC neurons in the ARH receive strong excitatory inputs from the VMH and that the strength of the excitatory input from the VMH to POMC neurons is diminished by fasting [39]. Interestingly, in the present study, we show that mutant mice exhibit a reduction in Vgltu2 expression associated with an impaired response of POMC neurons to fasting. However, different nutritional signals might trigger a distinct autophagy response. For example, HFD reduces autophagy in the hypothalamus, as evidenced by a reduction in Atg5, Atg7, and LC3-II/I expression [8]. Moreover, loss of Atg7 in the mediobasal hypothalamus or POMC neurons exacerbates the effects of HFD on adiposity and glucose homeostasis [8]. Although beyond the scope of this study, it would be interesting to examine whether HFD exerts similar or opposite effects in mice lacking Atg7 in VMH Sf1 neurons.
Neuronal function is energetically costly, and mitochondria almost exclusively provide this energy need. In addition, to produce energy in the form of adenosine triphosphate via oxidative phosphorylation, mitochondria also play many other important roles, such as buffering cytosolic Ca2+, synthesizing lipids, and producing signaling molecules, such as reactive oxygen species (ROS) [46]. To perform these functions effectively, mitochondria undergo morphological changes and localize to specific cellular compartments as required, brought about by fission and fusion. A series of studies have highlighted the importance of the hypothalamic mitochondria dynamic. Disruption of mitochondria fusion in arcuate AgRP neurons protects against HFD-induced obesity [32]. In contrast, it causes obesity when it is blocked in POMC neurons [31]. Moreover, dynamin-related protein1-dependent mitochondrial fission is a key mechanism in nutrient sensing in the ventromedial part of the hypothalamus [23]. It is also essential to have cellular processes in place to ensure the degradation of mitochondria that are damaged beyond repair, and this process is called mitophagy. In the present study, we report that loss of Atg7 in Sf1 neurons causes a reduction in mitochondria-autophagosome contacts, suggesting impairments in mitophagy in mutant mice. Notably, this abnormal mitophagy appears to be associated with alterations in mitochondria morphology and respiration. UCP2 has been linked with mitophagy [47], and VMH UCP2 expression regulates mitochondria fission, which in turn controls glucose regulation [48]. Consistent with these findings, we report here that Ucp2 expression is dysregulated in Sf1-Cre; Atg7loxP/loxP mice. Together, these observations suggest that the UCP2-autophagy pathway is a critical component of VMH mitochondria dynamic and function. Because during fasting mitochondrial efficiency must increase dramatically [31], these mitochondria defects observed in basal conditions will likely cause reduced mitochondria efficiency when animals are subjected to fasting contributing to an inadequate metabolic response. In addition to playing a role in the mitochondrial dynamic, autophagy is also involved in the lipolytic process to breakdown cellular lipid stores and provide energy to cells, including hypothalamic neurons, in fasting conditions [14,49,50]. However, whether VMH autophagy controls mitochondria oxidative metabolism and avoids fatty acid toxicity in starved cells remains to be investigated.
Author contributions
Conceptualization, B.C. and SGB; Methodology, BC, CL, JM, and TLH; Formal Analysis, BC, JM, CL, and TLH; Writing - Original Draft, BC; Writing - Review & Editing, SGB; Supervision, LP and SGB. All authors contributed to editing the manuscript and approved the manuscript for publication.
Acknowledgments
We thank Drs. Masaaki Komatsu and Noboru Mizushima for the generous gift of the Atg7loxP/loxP and LC3-GFP mice, respectively. We also would like to thank the University of Cincinnati Mouse Metabolic Phenotyping Center (supported by grant U24DK059630).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101186.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Shintani T., Klionsky D.J. Autophagy in health and disease: a double-edged sword. Science. 2004;306(5698):990–995. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132(1):27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim K.H., Lee M.-S. Autophagy–a key player in cellular and body metabolism. Nature Reviews Endocrinology. 2014;10(6):322–337. doi: 10.1038/nrendo.2014.35. [DOI] [PubMed] [Google Scholar]
- 4.Ebato C., Uchida T., Arakawa M., Komatsu M., Ueno T., Komiya K. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metabolism. 2008;8(4):325–332. doi: 10.1016/j.cmet.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 5.Jung H.S., Chung K.W., Won Kim J., Kim J., Komatsu M., Tanaka K. Loss of autophagy diminishes pancreatic [beta] cell mass and function with resultant hyperglycemia. Cell Metabolism. 2008;8(4):318–324. doi: 10.1016/j.cmet.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 6.Singh R., Xiang Y., Wang Y., Baikati K., Cuervo A.M., Luu Y.K. Autophagy regulates adipose mass and differentiation in mice. The Journal of Clinical Investigation. 2009;119(11):3329–3339. doi: 10.1172/JCI39228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Goldman S., Baerga R., Zhao Y., Komatsu M., Jin S. Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proceedings of the National Academy of Sciences. 2009;106(47):19860–19865. doi: 10.1073/pnas.0906048106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng Q., Cai D. Defective hypothalamic autophagy directs the central pathogenesis of obesity via the IκB kinase β (IKKβ)/NF-κB pathway. Journal of Biological Chemistry. 2011;286(37):32324–32332. doi: 10.1074/jbc.M111.254417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coupe B., Ishii Y., Dietrich M.O., Komatsu M., Horvath T.L., Bouret S.G. Loss of autophagy in pro-opiomelanocortin neurons perturbs axon growth and causes metabolic dysregulation. Cell Metabolism. 2012;15(2):247–255. doi: 10.1016/j.cmet.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaushik S., Arias E., Kwon H., Lopez N.M., Athonvarangkul D., Sahu S. Loss of autophagy in hypothalamic POMC neurons impairs lipolysis. EMBO Reports. 2012;13(3):258–265. doi: 10.1038/embor.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan W., Kim H.-K., Moon E.-Y., Kim S.S., Choi C.S., Komatsu M. Role of hypothalamic proopiomelanocortin neuron autophagy in the control of appetite and leptin response. Endocrinology. 2012;153(4):1817–1826. doi: 10.1210/en.2011-1882. [DOI] [PubMed] [Google Scholar]
- 12.Malhotra R., Warne J.P., Salas E., Xu A.W., Debnath J. Loss of Atg12, but not Atg5, in pro-opiomelanocortin neurons exacerbates diet-induced obesity. Autophagy. 2015;11(1):145–154. doi: 10.1080/15548627.2014.998917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao Y., Deng Y., Yuan F., Xia T., Liu H., Li Z. ATF4/ATG5 signaling in hypothalamic proopiomelanocortin neurons regulates fat mass via affecting energy expenditure. Diabetes. 2017;66(5):1146–1158. doi: 10.2337/db16-1546. [DOI] [PubMed] [Google Scholar]
- 14.Kaushik S., Rodriguez-Navarro J.A., Arias E., Kiffin R., Sahu S., Schwartz G.J. Autophagy in hypothalamic AgRP neurons regulates food intake and energy balance. Cell Metabolism. 2011;14(2):173–183. doi: 10.1016/j.cmet.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hetherington A.W., Ranson S.W. Hypothalamic lesions and adiposity in the rat. The Anatomical Record. 1940;78:149–172. [Google Scholar]
- 16.Dhillon H., Zigman J.M., Ye C., Lee C.E., McGovern R.A., Tang V. Leptin directly activates SF1 neurons in the VMH, and this action by leptin is required for normal body-weight homeostasis. Neuron. 2006;49(2):191–203. doi: 10.1016/j.neuron.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 17.Kim K.W., Donato J., Jr., Berglund E.D., Choi Y.-H., Kohno D., Elias C.F. FOXO1 in the ventromedial hypothalamus regulates energy balance. The Journal of Clinical Investigation. 2012;122(7):2578–2589. doi: 10.1172/JCI62848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiappini F., Catalano K.J., Lee J., Peroni O.D., Lynch J., Dhaneshwar A.S. Ventromedial hypothalamus–specific Ptpn1 deletion exacerbates diet-induced obesity in female mice. The Journal of Clinical Investigation. 2014;124(9):3781–3792. doi: 10.1172/JCI68585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klockener T., Hess S., Belgardt B.F., Paeger L., Verhagen L.A.W., Husch A. High-fat feeding promotes obesity via insulin receptor/PI3K-dependent inhibition of SF-1 VMH neurons. Nature Neuroscience. 2011;14(7):911–918. doi: 10.1038/nn.2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. Vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Molecular Biology of the Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 22.Dagda R.K., Zhu J., Kulich S.M., Chu C.T. Mitochondrially localized ERK2 regulates mitophagy and autophagic cell stress. Autophagy. 2008;4(6):770–782. doi: 10.4161/auto.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carneiro L., Allard C., Guissard C., Fioramonti X., Tourrel-Cuzin C., Bailbé D. Importance of mitochondrial dynamin-related protein 1 in hypothalamic glucose sensitivity in rats. Antioxidants and Redox Signaling. 2012;17(3):433–444. doi: 10.1089/ars.2011.4254. [DOI] [PubMed] [Google Scholar]
- 24.Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. (3rd ed.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komatsu M., Waguri S., Koike M., Sou Y.-s., Ueno T., Hara T. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131(6):1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- 26.Tong Q., Ye C., McCrimmon R.J., Dhillon H., Choi B., Kramer M.D. Synaptic glutamate release by ventromedial hypothalamic neurons is part of the neurocircuitry that prevents hypoglycemia. Cell Metabolism. 2007;5(5):383–393. doi: 10.1016/j.cmet.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramadori G., Fujikawa T., Fukuda M., Anderson J., Morgan D.A., Mostoslavsky R. SIRT1 deacetylase in POMC neurons is required for homeostatic defenses against diet-induced obesity. Cell Metabolism. 2010;12(1):78–87. doi: 10.1016/j.cmet.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T., Nakamura K., Matsu i.M., Yamamoto A., Nakahara Y., Suzuki-Migishima R. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 29.Leloup C., Magnan C., Benani A., Bonnet E., Alquier T., Offer G. Mitochondrial reactive oxygen species are required for hypothalamic glucose sensing. Diabetes. 2006;55(7):2084–2090. doi: 10.2337/db06-0086. [DOI] [PubMed] [Google Scholar]
- 30.Benani A., Troy S., Carmona M.C., Fioramonti X., Lorsignol A., Leloup C. Role for mitochondrial reactive oxygen species in brain lipid sensing: redox regulation of food intake. Diabetes. 2007;56(1):152–160. doi: 10.2337/db06-0440. [DOI] [PubMed] [Google Scholar]
- 31.Raefsky S.M., Mattson M.P. Adaptive responses of neuronal mitochondria to bioenergetic challenges: roles in neuroplasticity and disease resistance. Free Radical Biology and Medicine. 2017;102:203–216. doi: 10.1016/j.freeradbiomed.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez S., Gomez-Valades A.G., Schneeberger M., Varela L., Haddad-Tovolli R., Altirriba J. Mitochondrial dynamics mediated by Mitofusin 1 is required for POMC neuron glucose-sensing and insulin release control. Cell Metabolism. 2017;25(6):1390–1399. doi: 10.1016/j.cmet.2017.05.010. e1396. [DOI] [PubMed] [Google Scholar]
- 33.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of Cell Biology. 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Novak I. Mitophagy: a complex mechanism of mitochondrial removal. Antioxidants and Redox Signaling. 2012;17(5):794–802. doi: 10.1089/ars.2011.4407. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y., Qi H., Taylor R., Xu W., Liu L.F., Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3(4):337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 36.He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annual Review of Genetics. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coutinho E.A., Okamoto S., Ishikawa A.W., Yokota S., Wada N., Hirabayashi T. Activation of SF1 neurons in the ventromedial hypothalamus by DREADD technology increases insulin sensitivity in peripheral tissues. Diabetes. 2017;66(9):2372–2386. doi: 10.2337/db16-1344. [DOI] [PubMed] [Google Scholar]
- 38.Viskaitis P., Irvine E.E., Smith M.A., Choudhury A.I., Alvarez-Curto E., Glegola J.A. Modulation of SF1 neuron activity coordinately regulates both feeding behavior and associated emotional states. Cell Reports. 2017;21(12):3559–3572. doi: 10.1016/j.celrep.2017.11.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sternson S.M., Shepherd G.M., Friedman J.M. Topographic mapping of VMH→ arcuate nucleus microcircuits and their reorganization by fasting. Nature Neuroscience. 2005;8(10):1356–1363. doi: 10.1038/nn1550. [DOI] [PubMed] [Google Scholar]
- 40.He Y., Xu P., Wang C., Xia Y., Yu M., Yang Y. Estrogen receptor-α expressing neurons in the ventrolateral VMH regulate glucose balance. Nature Communications. 2020;11(1) doi: 10.1038/s41467-020-15982-7. 2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park S., Aintablian A., Coupe B., Bouret S.G. The endoplasmic reticulum stress-autophagy pathway controls hypothalamic development and energy balance regulation in leptin-deficient neonates. Nature Communications. 2020;11(1):1914. doi: 10.1038/s41467-020-15624-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caron E., Sachot C., Prevot V., Bouret S.G. Distribution of leptin-sensitive cells in the postnatal and adult mouse brain. Journal of Comparative Neurology. 2010;518(4):459–476. doi: 10.1002/cne.22219. [DOI] [PubMed] [Google Scholar]
- 43.Scott M.M., Lachey J.L., Sternson S.M., Lee C.E., Elias C.F., Friedman J.M. Leptin targets in the mouse brain. The Journal of Comparative Neurology. 2009;514(5):518–532. doi: 10.1002/cne.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sohn J.W., Oh Y., Kim K.W., Lee S., Williams K.W., Elmquist J.K. Leptin and insulin engage specific PI3K subunits in hypothalamic SF1 neurons. Molecular Metabolism. 2016;5(8):669–679. doi: 10.1016/j.molmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heras-Sandoval D., Pérez-Rojas J.M., Hernández-Damián J., Pedraza-Chaverri J. The role of PI3K/AKT/mTOR pathway in the modulation of autophagy and the clearance of protein aggregates in neurodegeneration. Cellular Signalling. 2014;26(12):2694–2701. doi: 10.1016/j.cellsig.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 46.Leloup C., Casteilla L., Carrière A., Galinier A., Benani A., Carneiro L. Balancing mitochondrial redox signaling: a key point in metabolic regulation. Antioxidants and Redox Signaling. 2011;14(3):519–530. doi: 10.1089/ars.2010.3424. [DOI] [PubMed] [Google Scholar]
- 47.Hass D.T., Barnstable C.J. Mitochondrial uncoupling protein 2 knock-out promotes mitophagy to decrease retinal ganglion cell death in a mouse model of glaucoma. The Journal of neuroscience : The Official Journal of the Society for Neuroscience. 2019;39(18):3582–3596. doi: 10.1523/JNEUROSCI.2702-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Toda C., Kim J.D., Impellizzeri D., Cuzzocrea S., Liu Z.-W., Diano S. UCP2 regulates mitochondrial fission and ventromedial nucleus control of glucose responsiveness. Cell. 2016;164(5):872–883. doi: 10.1016/j.cell.2016.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh R., Kaushik S., Wang Y., Xiang Y., Novak I., Komatsu M. Autophagy regulates lipid metabolism. Nature. 2009;458(7242):1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambold A.S., Cohen S., Lippincott-Schwartz J. Fatty acid trafficking in starved cells: regulation by lipid droplet lipolysis, autophagy, and mitochondrial fusion dynamics. Developmental Cell. 2015;32(6):678–692. doi: 10.1016/j.devcel.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.