Summary
Zinc (Zn2+) is a trace element, playing pivotal roles during host-pathogen interactions. Macrophages can sequester Zn2+ and restrict bioavailability or increase phagolysosomal Zn2+ to kill pathogens. This method quantifies Zn2+-mediated clearance of the human fungal pathogen C. glabrata after phagocytosis by innate immune cells. Double staining with propidium iodide and a zinc-specific fluorescence dye allows for discrimination of live versus dead pathogens inside phagolysosomes. Moreover, elevated phagolysosomal Zn2+ decreases fungal viability as a function of intracellular Zn2+ concentrations in macrophages.
For complete details on the use and execution of this protocol, please refer to Riedelberger et al. (2020).
Subject areas: Flow cytometry/mass cytometry, Cell-based assays, Immunology
Graphical Abstract
Highlights
-
•
A host-pathogen interaction system of primary macrophages and fungal pathogens
-
•
Flow cytometric assay to quantify Zn2+ intoxication of fungal pathogens by macrophages
-
•
Fungal viability depends on intra-phagosomal Zn2+ changes during immune responses
-
•
Zn2+ levels in reisolated pathogens correlate with fungal killing by macrophages
Zinc (Zn2+) is a trace element, playing pivotal roles during host-pathogen interactions. Macrophages can sequester Zn2+ and restrict bioavailability or increase phagolysosomal Zn2+ to kill pathogens. This method quantifies Zn2+-mediated clearance of the human fungal pathogen C. glabrata after phagocytosis by innate immune cells. Double staining with propidium iodide and a zinc-specific fluorescence dye allows for discrimination of live versus dead pathogens inside phagolysosomes. Moreover, elevated phagolysosomal Zn2+ decreases fungal viability as a function of intracellular Zn2+ concentrations in macrophages.
Before you begin
Preparation of L929-conditioned cell supernatant
Timing: 16 days
-
1.
Thaw two vials of L929 cells (0.5 mL of freezing medium containing 1 × 106 cells) in a water bath (37°C) for several seconds; fill up to 1.5 mL with pre-warmed (37°C) DMEM medium.
-
2.
Transfer thawed cells into a 100 × 20 mm tissue culture (TC)-treated dish (Starlab) and adjust to a final volume of 12 mL with DMEM medium; incubate at 37°C, 5% CO2.
-
3.
On the next day, split cells in a 1:4 ratio by aspirating the old medium and gently scraping them with a natural-rubber spatula (Deutsch & Neumann) into 12 mL fresh DMEM medium. Split 3 mL of the cell suspension into four new dishes, already containing 9 mL of fresh pre-warmed DMEM medium. Incubate at 37°C, 5% CO2.
CRITICAL: The natural-rubber spatula must be sterilized in freshly prepared 70% Ethanol for at least 5 min before use to avoid contaminations. Only use a spatula that has a visibly intact edge. Even small spatula unevenness or damages can destroy cells during the harvest.
-
4.
After 2 days, transfer cells into a tissue-culture-treated T-150 flask with a vented filter cap (Starlab). Gently scrape the cells off the dish into fresh DMEM medium and transfer the cell suspension from each dish into one new flask. Fill up with fresh DMEM medium to a final volume of 80 mL. Incubate at 37°C, 5% CO2.
-
5.
After 3 days, check for confluency of cells. If the cells are more than 80% confluent, aspirate the medium.
-
6.
Add 100 mL supplement-free DMEM (Thermo Fisher Scientific), as antibiotics could interfere with the macrophage-colony stimulating factor (M-CSF) production.
-
7.
Incubate cells for 10 days at 37°C, 5% CO2.
-
8.
On day 10, the M-CSF containing supernatants can be harvested. Pool supernatants of all flasks and filter-sterilize through a 0.22 μm filter system (Millipore Express PLUS).
-
9.
Aliquot the M-CSF-containing supernatant into 50 mL conical tubes (Starlab) to avoid freeze-thaw cycles and store at −20°C until needed.
-
10.
A quality control step is suggested after each harvest for reproducible results. Test and compare fresh L929-conditioned supernatants with previous stocks for efficacy and yield.
-
11.Prepare macrophage medium (as listed in the Materials and equipment section) containing several dilutions of fresh L929-conditioned supernatant (10%–20% final concentration). Prepare the same dilutions using a reference L929-conditioned supernatant stock. These media are used to differentiate bone marrow-derived macrophages (BMDMs) for final analysis on day 10 (see Culture of bone marrow-derived macrophages).
-
a.Test the cell yield in a CASY cell counter (Innovatis) or similar cell counter.
-
b.Test the differentiation capacity by surface marker expression of CD11b (eBioscience antibody) and F4/80 (BioLegend antibody). Use both antibodies with a final concentration of 0.25 μg per million cells in 100 μL volume.
-
a.
Heat-inactivation of fetal calf serum (hiFCS)
Timing: 2 days
-
12.
Thaw a 500 mL bottle of FCS (Sigma-Aldrich) for 17 h at 4°C.
-
13.
On the next day, complete thawing the serum by placing the bottle in a 37°C water bath. Make sure that the water level is higher than the serum level in the bottle; mix every 5–10 min by inversion.
-
14.
Once serum is completely thawed, incubate for an additional 15 min to allow serum to equilibrate to 37°C.
-
15.
Raise the temperature of the water bath to 56°C. This will usually take 15–20 min.
-
16.
During this incubation, invert the bottle every 10 min.
-
17.
Once the bath reaches 56°C, incubate serum bottle for another 30 min. Invert the bottle every 10 min.
-
18.
Remove serum bottle from the water bath and allow to cool at room temperature (RT) for 30 min.
Note: Room temperature (RT) refers to a temperature range from 20°C–25°C.
-
19.
Prepare aliquots of the heat-inactivated (hi) serum and store at −20°C.
Murine bone marrow isolation
Timing: 3–4 h
-
20.
Euthanize mice in accordance with institutional guidelines and national laws.
Note: Male and/or female mice from different mouse strains or knock-out mice may be used, following testing and perhaps optimization of macrophage isolation / differentiation. Particular consideration should be given to possible gender-specific differences and impact of different euthanasia methods. We experienced the highest bone marrow yield from 8- to 12-week-old mice.
-
21.
Spray the fur of the mouse with 70% ethanol.
-
22.
Use needles to fix the mouse with the belly facing up on a styrofoam working bed, keeping the body under tension.
-
23.
Make an incision on the lower abdomen and cut along the leg until you reach the knee joint. Cut away the skin and separate the leg from the body by displacing the femoral joint from the hip. Avoid breaking the bone during this step.
-
24.
Most of the muscle tissue should be removed to free femur and tibia before bones are quickly rinsed in 70% ethanol.
-
25.
Store one leg in sterile PBS (Sigma) at 4°C, while preparing the second one.
-
26.
Take both hind legs and rinse them again with 70% ethanol before transferring them into an ice-cold sterile porcelain mortar filled with 10 mL sterile DMEM (Thermo Fisher Scientific).
-
27.
Grind the bones thoroughly with an ice-cold porcelain pestle until the bone marrow is completely released.
Note: Before usage, mortar and pestle are completely covered with aluminum wrap and baked at 120°C for 3 h in a sterilizing oven.
-
28.
Filter the suspension containing the bone marrow cells though a 40 μm cell strainer (Nylon Corning) and centrifuge at 300 × g for 6.5 min at 4°C (Rotina 38R Hettich) in a 50 mL conical tube (Starlab).
-
29.
Aspirate the medium and lyse red blood cells with 0.5 mL sterile lysis buffer (BioLegend) per leg for 30 s at RT.
-
30.
Stop red blood cell lysis by adding at least 5× the volume of sterile ice-cold DMEM (Thermo Fisher Scientific).
-
31.
Centrifuge the cells at 300 × g for 6.5 min at 4°C (Rotina 38R Hettich) in a 50 mL conical tube (Starlab).
-
32.
Take off supernatants and resuspend bone marrow cells in macrophage medium for differentiation (see Culture of bone marrow-derived macrophages) or sterile hiFCS and 10% DMSO for freezing cells in cryogenic vials (Starlab) at −150°C.
Culture of primary bone marrow-derived macrophages
Timing: 10 days
-
33.
To obtain BMDMs, bone marrow cells from one femur and tibia after harvest are differentiated in a 10 cm2 TC-treated square Petri dish (Thermo Fisher Scientific) in 9 mL macrophage medium at 37°C, 5% CO2.
-
34.
On day 3, add 5 mL of fresh pre-warmed (to 37°C) sterile macrophage medium.
-
35.
Split macrophage culture after 7 days in a 1:2 ratio by aspiration of the old medium; use a natural-rubber spatula (Deutsch & Neumann) to gently scrape cells into 12 mL of fresh macrophage medium pre-warmed to 37°C.
-
36.
On day 10, macrophages are ready to be used for assays.
Note: Although frozen bone marrow stocks (in hiFCS and 10% DMSO at −80°C) can be used, freshly isolated bone marrow yields more BMDMs. Differentiated macrophages appear on day 7, but day 10 provides an optimal yield of BMDMs. Healthy macrophages are growing adherent to plastic surface and should look as shown in Figure 1.
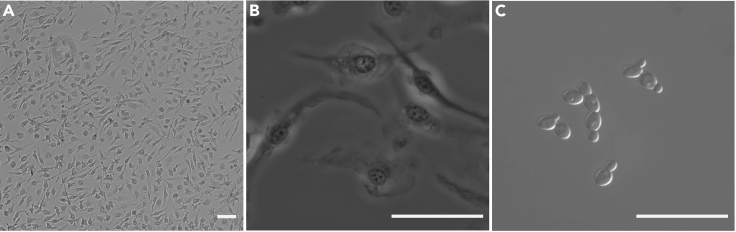
Figure 1.
Morphology of differentiated BMDMs
Representative DIC pictures of BMDMs after 10 days of differentiation in conditioned medium. (A) 10× magnification, (B) a 40× magnification and (C) a representative picture of yeast-form C. glabrata cells at 64× magnification. Scale bars, 10 μm.
Preparation of Candida glabrata cells
Timing: 3 days
-
37.
At least 3 days before the experiment, streak out C. glabrata from a frozen stock (at −80°C in PBS and 15% glycerol) on solid yeast peptone extract (YPD) agar plates (Sarstedt); incubate plates for 3 days at RT until fungal colonies appear.
CRITICAL:Candida spp. fungal pathogens are classified as biosafety level 2 (BL2 or S2) in most countries. A biological safety level S2 laboratory and a laminar flow safety cabinet (Class I or II) is needed. All surfaces must be disinfected after work with 70% Ethanol or fungicidal disinfectants. As required by Good Laboratory Practice (GLP) protocols, the use of personal protective gear (gloves, laboratory coat) is necessary. In the case of a spillage, disinfect the area by soaking the contaminated area for 10–15 min with 70% ethanol before further cleanup. Dispose paper towels used for the cleanup as biohazard waste.
Prepare buffer, media, and solutions
-
a.Macrophage medium can be stored for up to 4 weeks at 4°C after addition of L929-conditioned supernatant.
-
b.SDS solution can be prepared in advance and stored up to 6 months at RT.
-
c.Zinpyr-1 and Propidium iodide (PI) solutions should be freshly prepared on the day of use.
-
a.
Note: All buffers and solutions used are sterile-filtered through a 0.22 μm filter system (Millipore Express PLUS).
Key resources table
| REAGENTS or RESOURCES | SOURCE | IDENTIFIER |
|---|---|---|
| Fungal strains | ||
| Candida glabrata ATCC 2001 (wild-type) | American Type Culture Collection (ATCC) | ATCC 2001 |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM | Thermo Fisher Scientific | Cat#11584486 |
| Fetal calf serum (FCS) | Sigma-Aldrich | Cat#F7524 |
| Penicillin/streptomycin | Sigma-Aldrich | Cat#P4333 |
| PBS | Gibco | Cat#21600069 |
| Zinpyr-1 | Santa Cruz | Cat#sc-213182 |
| Propidium iodide (PI) | Sigma-Aldrich | Cat#P4864 |
| Yeast extract | BD Biosciences | Cat#212720 |
| Tryptone | AppliChem | Cat#A1553,1000 |
| D-Glucose | VWR Chemicals | Cat#97061-172 |
| Sodium dodecyl sulfate solution (SDS) | Sigma-Aldrich | Cat#71736 |
| Glycerol | Thermo Fisher Scientific | Cat#10296200 |
| L929 fibroblast cells | ATCC | ATCC CCL-1 |
| CD11b-FITC | eBioscience | Cat#11-0112-85 |
| F4/80-PE | BioLegend | Cat#123110 |
| DMSO | Sigma | Cat#D2438-10ML |
| Experimental models: organisms/strains | ||
| C57BL/6J | The Jackson Laboratory | Cat#000664; RRID; IMSR_JAX;0000664 |
| Software and algorithms | ||
| FlowJo software, version 7.6.5 | Tree Star | https://www.flowjo.com/ |
| GraphPad Prism version 5.04 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Equipment | ||
| 50 mL conical tube | Starlab | Cat#E1450-0200 |
| 15 mL conical tube | Starlab | Cat#E1415-0200 |
| T-150 flask, vented filter cap | Starlab | Cat#CC7682-4815 |
| 24-well lid plates, flat bottom, TC-treated | Starlab | Cat#CC7682-7506 |
| 100 × 20 mm dish with lid, TC-treated | Starlab | Cat#CC7682-3394 |
| 10 cm2 square Petri dish, TC-treated | Thermo Fisher Scientific | Cat#11349273 |
| 1.5 mL microcentrifuge tube | Eppendorf | Cat#0030120086 |
| 5 mL serological pipet, sterile | Starlab | Cat#E4860-0005 |
| 10 mL serological pipet, sterile | Starlab | Cat#E4860-0010 |
| 25 mL serological pipet, sterile | Starlab | Cat#E4860-0025 |
| Culture tube and cap, 14 mL, sterile | Starlab | Cat#I1485-0810 |
| CASY cups | OMNI Life Science | Cat#5651794 |
| Cryogenic vial | Starlab | Cat#E3110-6122 |
| Petri dish 92 × 16 mm with cams | Sarstedt | Cat#82.1473 |
| Spectrophotometer | Hitachi | U-2000 |
| CASY counter | Innovatis | n/a |
| Microscope | Zeiss | Axiovert 200M |
| Centrifuge | Hettich | Rotina 38R |
| Tabletop centrifuge | Eppendorf | 5702R |
| Thermomixer | Eppendorf | Thermomixer comfort |
| Flow cytometer | BD Bioscience | LSRFortessa |
| Lamina flow class II | Szabo Scandic | Safefast Premium |
| CO2 incubator | Binder | CB-S 170 |
| Incubator shaker | New Brunswick Scientific | Innova 44 |
| Vortex | Heidolph | Reax 2000 |
| Mortar and pestle (porcelain) | Thermo Fisher Scientific | Cat#11970897 |
| Scissor | FST | Cat#91460-11 |
| Forceps (Dumont mini forceps) | FST | Cat#11200-14 |
| Autoclave | Tuttnauer | 5075 ELV-D |
| Pipets | Gilson | Pipetman |
| Pipette tips | TipOne | Cat#S1161-1700-C |
| Pipetboy | Integra | Pipetboy2 |
| Polycarbonate cryogenic storage boxes | Starlab | Cat#I2381-2305 |
Materials and equipment
SDS solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | n/a |
| SDS solution | 0.005% (w/v) | n/a |
| Total | n/a | 300 μL/condition |
Propidium iodide (PI) solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | n/a |
| PI solution | 2 μg/mL | n/a |
| Total | n/a | 200 μL/condition |
Zinpyr-1 stock solution
| Reagent | Final concentration | Amount |
|---|---|---|
| DMSO 100% | 100% | 607 μL |
| Zinpyr-1 | 10 mM | 5 mg |
| Total | n/a | ~607 μL |
Note: Store 10 μL aliquots of the Zinpyr-1 stock solution at −20°C until use. After thawing an aliquot, do not freeze again.
Zinpyr-1 solution
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | n/a |
| Zinpyr-1 stock solution (10 mM) | 10 μM | n/a |
| Total | n/a | 300 μL/condition |
Note: Zinpyr-1 is light sensitive! Keep Zinpyr-1 solution in the dark until use.
DMEM medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | n/a | 44.5 mL |
| Heat-inactivated FCS (hiFCS) | 10% | 5 mL |
| Penicillin/streptomycin (P/S) | 100 μg/mL | 0.5 mL |
| Total | n/a | 50 mL |
Macrophage medium
| Reagent | Final concentration | Amount |
|---|---|---|
| DMEM | n/a | 37 mL |
| Heat-inactivated FCS (hiFCS) | 10% | 5 mL |
| Penicillin/streptomycin (P/S) | 100 μg/mL | 0.5 mL |
| L929-conditioned supernatant | 15% | 7.5 mL |
| Total | n/a | 50 mL |
Note: hiFCS and L929-conditioned supernatant are sterile-filtered through a 0.22 μm filter system (Millipore Express PLUS).
Alternatives: For differentiation of BMDMs, recombinant murine M-CSF (20 ng/mL final concentration) can also be used instead of L929-conditioned supernatant.
Freezing medium for bone marrow cells
| Reagent | Final concentration | Amount |
|---|---|---|
| hiFCS | n/a | 4.5 mL |
| DMSO | 10% | 0.5 mL |
| Total | n/a | 5 mL |
Note: Freezing medium can be stored at −20°C for up to 1 year.
Solid YPD medium
| Reagent | Final concentration | Amount |
|---|---|---|
| Yeast peptone (YP) | n/a | 250 mL |
| Glucose 20% | 2% (w/v) | 50 mL |
| Agar 4% | 2% | 250 mL |
| Total | n/a | 500 mL |
Note: YP, glucose, and agar are sterilized separately by autoclaving before mixing. Solid YPD plates can be stored at 4°C for up to 6 months.
Liquid YPD medium
| Reagent | Final concentration | Amount |
|---|---|---|
| YP | n/a | 450 mL |
| Glucose 20% | 2% (w/v) | 50 mL |
| Total | n/a | 500 mL |
Note: YP and glucose are sterilized separately by autoclaving before mixing. Liquid YPD medium and components can be stored at RT for up to 6 months.
Freezing medium for Candida
| Reagent | Final concentration | Amount |
|---|---|---|
| PBS | 1× | 42.5 mL |
| Glycerol | 15% | 7.5 mL |
| Total | n/a | 50 mL |
Note: Mix components in aseptic working conditions.
Step-by-step method details
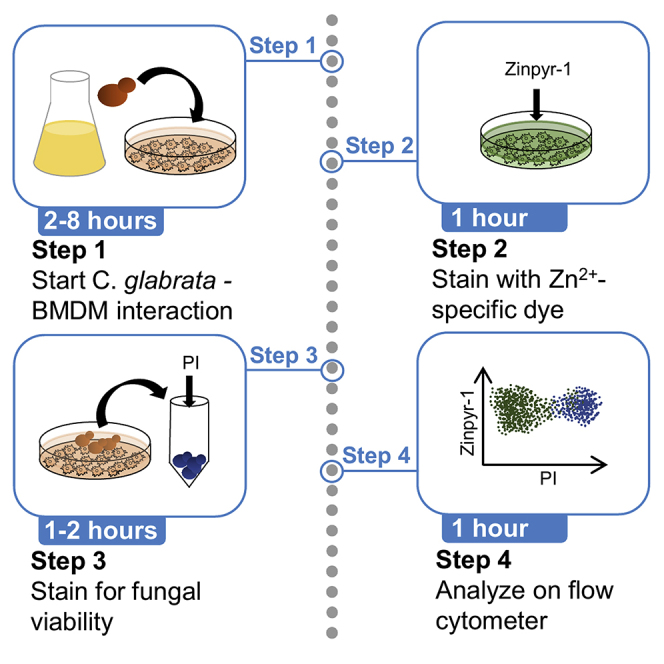
Preparation of immune cells and fungal cells: day 1
Timing: 2–3 h
BMDMs are seeded into wells and Candida cultures are started for next-day interaction.
-
1.
In the morning, pick one fungal colony with a sterile tip and inoculate into 5 mL liquid YPD medium in culture tubes (Starlab). Let the cells grow in the tube for 4–8 h at 30°C with agitation (200 rpm) at an angle of 45° (Innova 44 New Brunswick Scientific).
-
2.
Measure the optical density at 600 nm (OD600) in a spectrophotometer (U-2000 Hitachi) to follow growth. To stay in the linear range of the spectrophotometer for the measurement, dilute the fungal cell suspension 1:10 with fresh YPD.
-
3.
Adjust fungal cells to reach logarithmic growth phase (OD600 = 0.8–1) at the time of harvest on the next day with the formula:
Note: This formula works if the doubling time of C. glabrata is around 1 h. It is highly discouraged to use fungal cell suspensions with OD600 above 2. In this case, new fungal cultures should be set up.
-
4.
Let cells grow in the tube for 14–17 h at 30°C with agitation (200 rpm) at an angle of 45° (Innova 44 New Brunswick Scientific).
Note: 1 mL YPD culture at OD600 = 1 contains 1 × 107C. glabrata cells. Hence, set up enough culture volume to have sufficient numbers on the next day. It is advisable to include a tube filled with liquid YPD only, when inoculating Candida to exclude media contamination.
-
5.
Harvest differentiated BMDMs by gently scraping them off the culture dish with a natural-rubber spatula (Deutsch & Neumann) into a 50 mL conical tube (Starlab); pellet BMDMs at 300 × g for 6 min at RT (Rotina 38R Hettich).
-
6.
Resuspend macrophages in 1 mL fresh macrophage medium and take out an aliquot to count in a CASY cell counter. Seed 2 × 105 macrophages/well into a 24-well plate (Starlab) at least in triplicates for each experimental condition. Troubleshooting 1
-
7.
Add macrophage medium at a final volume of 1 mL/well; let BMDMs equilibrate 17 h at 37°C, 5% CO2 (CB-S 170 Binder).
Note: Macrophage medium should be pre-warmed to 37°C to minimize temperature stress.
Interaction and staining: day 2
Timing: 6–18 h
Immune cells are infected with fungal cells, harvested, and stained.
-
8.Harvest Candida cells when OD600 = 1 is reached by transferring the fungal suspension into a 50 mL conical tube (Starlab).
-
a.Take a small aliquot to check for morphology of the fungal cells and possible contaminations under the microscope.
-
a.
-
9.
Pellet fungal cells at 3,000 × g for 2 min (5702R Eppendorf).
-
10.
Discard the supernatant and resuspend the cells in the same volume of PBS.
-
11.
Repeat the centrifugation and remove supernatant.
-
12.
Resuspend the fungal pellet in 1 mL PBS.
-
13.
Take a small aliquot of 10 μL to determine the cell count in a CASY cell counter.
-
14.
Suspend fungal cells in macrophage medium at a final cell number of 4 × 106 counts/mL. This gives a multiplicity of infection (MOI) = 1 when adding 50 μL/well of this suspension to immune cells.
-
15.
For a heat-killed Candida control (see Expected outcomes), take a culture aliquot of 150–200 μL and transfer it into a new 1.5 mL microcentrifuge tube (Eppendorf). Heat-stress fungal cells by placing them on a thermomixer (Eppendorf Thermomixer comfort) at 65°C for 1 min.
Note: All centrifugation steps and reagents are at RT.
-
16.
Infect macrophages by adding 50 μL of Candida suspension/well (2 × 105 C. glabrata cells).
-
17.
Incubate host-pathogen mixtures in the incubator (CB-S 170 Binder) at 37°C, 5% CO2.
CRITICAL: The time, BMDMs are outside the incubator should be kept to a minimum to avoid major temperature changes.
-
18.
15 min before the end of the infection, prepare a 10 μM Zinpyr-1 solution (as described in Materials and equipment) and store at RT in the dark.
CRITICAL: Zinpyr-1 is a light sensitive probe and the prepared Zinpyr-1 solution should be stored in the dark until use.
-
19.
After 2–12 h of infection, aspirate culture medium.
Note: Optimal endpoints can vary, depending on the macrophages and Candida strains used and should be tested for every new setup.
-
20.
Gently wash each well 3× with 1 mL PBS at RT.
Note: At least three washing steps are needed to get rid of extracellular Candida cells, which would interfere with the readout of the assay. Discarding the PBS can be done by inverting plates and applying reasonable force to decant, as macrophages are very adherent to the plastic.
-
21.
Add 300 μL of the Zinpyr-1 solution/well and incubate cells for 30 min at 37°C, 5% CO2.
-
22.
Decant supernatant and wash 3× with 1 mL PBS at RT.
-
23.
To lyse macrophages and liberate phagosomal Candida cells, put the plate on ice, add 300 μL of the SDS solution/well and incubate for 15 min at 4°C.
Note: SDS lysis to remove immune cells like macrophages is only possible for pathogens that are not susceptible to the applied SDS concentrations. This should be tested before the assay.
-
24.
During step 23, pre-cool a tabletop centrifuge (5424 R Eppendorf) to 4°C.
-
25.
Fill up every well with ice-cold PBS to 1 mL and harvest C. glabrata via pipetting. Transfer volume of each well into a separate 1.5 mL microcentrifuge tube (Eppendorf).
CRITICAL: Pipet bottom of the wells vigorously and make sure to include the entire bottom of each well. Candida spp adhere easily to plastic surfaces and thus need gentle force for removal.
-
26.
Pellet fungal cell suspension at 21,000 × g for 10 min at 4°C (5424 R Eppendorf).
-
27.
During step 26, prepare a fresh PI solution (2 μg/mL) as described in Materials and equipment.
-
28.
Aspirate supernatant and resuspend pellet in 200 μL PI solution.
-
29.
Incubate for 5 min at RT in the dark.
-
30.
Add 800 μL ice-cold PBS to a final volume of 1 mL and pellet at 21,000 × g for 10 min at 4°C (5424 R Eppendorf).
-
31.
Carefully aspirate supernatant and wash fungal pellet 2× with 1 mL ice-cold PBS.
-
32.
Resuspend fungal cell pellet after the last washing step in 300 μL PBS.
Note: To avoid loss of fungal cells in steps 30 and 31, centrifuge each time with 21,000 × g for 10 min.
-
33.Analyze Candida cells right away on a flow cytometer.
-
a.Discriminate Candida cells from debris by morphology and granularity, by setting proper forward (FSC-A) and side scatter (SSC-A) gates surrounding the dense cell population (see Expected outcomes). Troubleshooting 2
-
b.Exclude doublets by plotting FSC-A against FSC-H and gate on the dense population.
-
c.Analyze PI+ (dead) population against Zinpyr-1+ population. Troubleshooting 3
-
a.
Note: Zinpyr-1 has an excitation and emission spectrum peak wavelengths of approximately 488 nm and 525 nm, respectively. PI is excited at 561 nm and detected at 610 nm.
Expected outcomes
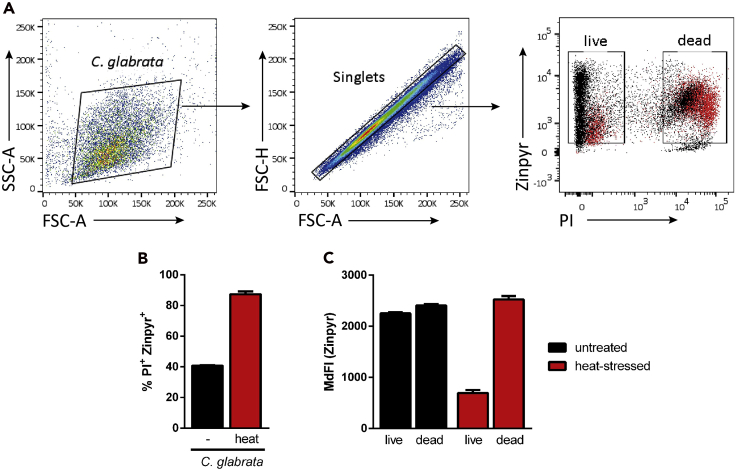
A successful interaction assay should show two distinct fungal populations after flow cytometry analysis. One population of viable Candida (PI-) with a relatively low median fluorescence intensity (MdFI) of Zinpyr-1, and a population of dead Candida (PI+) cells with a higher Zinpyr-1 MdFI. The percentage of PI+/Zinpyr-1+ double-positive cells of the total Candida population allows for estimating the killing efficiency by BMDMs (Figure 2).
Figure 2.
Visualization of viable Candida glabrata inside phagolysosomes
(A) Gating strategy for analysis of C. glabrata by flow cytometry. The PI-Zinpyr panel compares untreated (black) with heat-stressed (red) C. glabrata.
(B) Percentage of dead C. glabrata, comparing untreated and heat-stressed fungal cells before interaction with BMDMs.
(C) Live and dead C. glabrata populations are analyzed for their zinc-dependent signal intensity (MdFI) equaling the intracellular amount of labile zinc in fungal cells. MdFI, median fluorescence intensity.
Means and SD are shown.
Infection with heat-killed C. glabrata is an important positive control, showing, a larger PI+/Zinpyr-1+ double-positive population and a higher MdFI of Zinpyr-1 within the dead population when compared to the live one. When setting up the assay, this control should be included to obtain a reference population of dead Candida cells (Figure 2).
Quantification and statistical analysis
Events were acquired on a LSR Fortessa (BD Bioscience); raw data was exported as FCS 3.0 files. Files were then analyzed using FlowJo 7.6.5 (Tree Star Inc.) as follows: A morphology gate was set around the Candida population, to exclude cell debris. Further, only singlets were selected and then plotted against propidium iodide (x axis) and Zinpyr-1 (y axis). Gates were set to distinguish 2 populations: PI+ and PI-. Each population was analyzed as a percentage from the parent population (all Candida singlets) to quantify the relative abundance of dead versus live cells. Finally, each population was analyzed for their MdFI (Zinpyr-1). Data values were exported as Excel files (Microsoft Office 2009) and visualized with GraphPad Prism 5.04 (GraphPad Software Inc.). Unpaired Student t test was used to check for significance (α = 0.05) between treatment groups.
Limitations
Zinpyr-1 is a small membrane-permeable fluorescence dye, which increases its fluorescence intensity upon binding of Zn2+. It is a member of the Zinpyr family showing a very high affinity to free zinc (KD = 7 × 10−10 M), with an intermediate increase in fluorescence intensity of about 3-fold when binding free Zn2+. The probe measures primarily the labile, freely accessible and loosely-bound intracellular Zn2+ pools (Burdette et al., 2001; Figueroa et al., 2014). However, a large fraction of Zn2+ is tightly-bound to proteins, both in mammalian and fungal cells due to a tight control of metal ion homeostasis (Maret, 2006). Therefore, the fluorescent probe Zinpyr-1 can be used to trace the labile Zn2+ fraction, which is a mere part of the global zinc response during complex host-pathogen interactions. Further, Zinpyr-1 does not determine the absolute intracellular zinc concentrations, as the fluorescent signal is influenced by additional factors such as the dye concentration inside cells, the extent of metal ion buffering by the cellular milieu, the pH as well as membrane permeabilities (Carter et al., 2014). Hence, it should only be used as a relative quantification method to compare live-dead phagocytosed fungal populations within the same experiment, along with appropriate controls.
This assay detects and visualizes viable fungal cells inside phagosomes as a function of intracellular Zn2+ concentrations in BMDMs, since host cells exploit Zn2+ intoxication to elicit fungal clearance (Riedelberger et al., 2020). Indeed, certain bacterial pathogens are subject to Zn2+ intoxication by immune cells through nutritional defense responses (Botella et al., 2011; Subramanian Vignesh and Deepe, 2016). Moreover, macrophage-mediated zinc toxicity constitutes a defense response against S. typhimurium and E. coli (Kapetanovic et al., 2016; Stocks et al., 2019). Hence, this assay was developed to investigate the viability of intracellular C. glabrata, the first Candida spp subject to heavy metal intoxication by macrophages. We suggest that this assay is broadly applicable to other intracellular fungal pathogens, particularly in combination with microscopy-based total zinc quantification assays such as inductively coupled plasma mass spectrometry (ICP-MS) and parallel assessment of fungal zinc stress responses.
Troubleshooting
Problem 1
Poor quality of BMDMs (step 6).
Potential solution
Avoid contaminations by working exclusively in under sterile conditions. Use freshly isolated bone marrow for macrophage differentiation to increase yield and viability. If yield of BMDMs is low, this could be an indicator that M-CSF in macrophage medium is degraded. Prepare medium with freshly thawed L929-conditioned supernatant.
Problem 2
Only very low number of events can be detected on the flow cytometer or it is not possible to distinguish a clearly defined Candida population (step 33).
Potential solution
Make sure to pellet fungal cells at 21,000 × g for 10 min to minimize loss of cells during washing steps. Using PBS without divalent ions (Ca2+ and Mg2+) may help to reduce clumping and loss of Candida cells. If this problem persists, the number of interacting cells can be scaled up, when using a plate with appropriate surface area and medium volume, keeping the same MOI (step 6).
Problem 3
The percentage of killing and Zn2+ staining has a high variability between experiments (step 33).
Potential solution
Try to keep the protocol as standardized as possible. Use differentiated bone marrow-derived macrophages always on the same day of differentiation and minimize temperature changes or other disturbances leading to cell stress (step 5). Small differences in the handling can have unexpectedly huge effects on the efficiency of clearance mechanisms like phagocytic killing of fungal cells.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Karl Kuchler (karl.kuchler@meduniwien.ac.at).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate new datasets or analyze new code.
Acknowledgments
This work was supported by the Austrian Science Fund project InnateFun (FWF-I3319-B22) to K.K. and in part by the FWF doc-funds TissueHome PhD Program (I.T., H.A., K.K.), and the FWF Special Research Area SFB-F70-HIT (P.P, K.K).
Author contributions
M.R. and K.K. developed the assay; M.R. and P.P. performed the assay and experiments; P.P., I.T., H.A., and S.J. optimized the FACS assay; P.P. and K.K. wrote the manuscript and interpreted data.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Philipp Penninger, Email: philipp.penninger@meduniwien.ac.at.
Karl Kuchler, Email: karl.kuchler@meduniwien.ac.at.
References
- Botella H., Peyron P., Levillain F., Poincloux R., Poquet Y., Brandli I., Wang C., Tailleux L., Tilleul S., Charrière G.M. Mycobacterial P1-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host Microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette S.C., Walkup G.K., Spingler B., Tsien R.Y., Lippard S.J. Fluorescent sensors for Zn2+ based on a fluorescein platform: synthesis, properties and intracellular distribution. J. Am. Chem. Soc. 2001;123:7831–7841. doi: 10.1021/ja010059l. [DOI] [PubMed] [Google Scholar]
- Carter K.P., Young A.M., Palmer A.E. Fluorescent sensors for measuring metal ions in living systems. Chem. Rev. 2014;114:4564–4601. doi: 10.1021/cr400546e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa J.A.L., Vignesh K.S., Deepe G.S., Caruso J. Selectivity and specificity of small molecule fluorescent dyes/probes used for the detection of Zn2+ and Ca2+ in cells. Metallomics. 2014;6:301–315. doi: 10.1039/c3mt00283g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic R., Bokil N.J., Achard M.E.S., Ong C.Y., Peters K.M., Stocks C.J., Phan M., Monteleone M., Schroder K., Irvine K.M. Salmonella employs multiple mechanisms to subvert the TLR-inducible zinc-mediated antimicrobial response of human macrophages. FASEB J. 2016;30:1901–1912. doi: 10.1096/fj.201500061. [DOI] [PubMed] [Google Scholar]
- Maret W. Zinc coordination environments in proteins as redox sensors and signal transducers. Antioxid. Redox Signal. 2006;8:1419–1441. doi: 10.1089/ars.2006.8.1419. [DOI] [PubMed] [Google Scholar]
- Riedelberger M., Penninger P., Tscherner M., Hadriga B., Brunnhofer C., Jenull S., Stoiber A., Bourgeois C., Petryshyn A., Glaser W. Type I interferons ameliorate zinc intoxication of Candida glabrata by macrophages and promote fungal immune evasion. iScience. 2020;23:101121. doi: 10.1016/j.isci.2020.101121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks C.J., Phan M.D., Achard M.E.S., Nhu N.T.K., Condon N.D., Gawthorne J.A., Lo A.W., Peters K.M., McEwan A.G., Kapetanovic R. Uropathogenic Escherichia coli employs both evasion and resistance to subvert innate immune-mediated zinc toxicity for dissemination. Proc. Natl. Acad. Sci. U S A. 2019;116:6341–6350. doi: 10.1073/pnas.1820870116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian Vignesh K., Deepe G.S. Immunological orchestration of zinc homeostasis: the battle between host mechanisms and pathogen defenses. Arch. Biochem. Biophys. 2016;611:66–78. doi: 10.1016/j.abb.2016.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate new datasets or analyze new code.