Highlights
-
•
64 of 69 patients’ samples could be profiled. Median alterations: 4 (0 - 23), most frequently TP53, KRAS & CDKN2A/B.
-
•
In 13 patients (20% of 64 successful profiles), personalized therapies could be initiated.
-
•
Effectiveness was seen in 8/13 patients (61,5%) of the precision oncology cohort vs 3/22 (13,5%) treated with chemotherapy.
-
•
Kaplan-Meier curves show significant PFS benefit for personalized treated patients (p = 0,0165; median 151 days vs 83 days).
-
•
Personalized cancer therapy is effective and feasible also in the setting of a middle-sized oncologic center.
Keywords: Precision oncology, Comprehensive genomic profiling, Retrospective cohort study
Abbreviations: CUP, Cancer of Unknown Primary; ECOG, Eastern Cooperative Oncology Group; EGFR, Epidermal Growth Factor Receptor; ESMO, European Society for Medical Oncology; ESCAT, ESMO Scale for Clinical Actionability of molecular Targets; FISH, Fluorescence In-Situ Hybridization; HER-2, Human Epidermal Growth Factor Receptor-2; HRD, Homologous Recombination Deficiency; IHC, Immunohistochemistry; MSI, Microsatellite Instability; NGS, Next Generation Sequencing; TMB, Tumor Mutational Burden
Abstract
Background and Aim
To successfully apply personalized cancer therapies, thorough understanding of the patient's tumor is needed. In-depth, comprehensive genomic profiling systems allow gathering this knowledge by testing hundreds of cancer-related genes. Several large institutions have established precision oncology programs in recent years with promising results for patients. However, especially middle-sized oncologic institutions face challenges to implement such programs.
This study aims to retrospectively analyze the effects of comprehensive genomic tumor profiling with respect to feasibility and effectiveness in a middle-sized oncologic center in Austria.
Methods
From May 1st, 2016 to December 31st, 2019 patients at the University Clinic Krems, who suffered from CUP-syndromes plus patients, who were resistant to conventional therapy or have progressed after all available therapy lines, were offered to get their tumors analyzed by comprehensive genomic profiling in order to establish a customized therapy.
Results
Of 69 considered patients, 64 patients’ samples could be profiled. The median number of detected alterations was 4 (minimum 0; maximum 23). Most frequent alterations were reported for TP53, KRAS and CDKN2A/B.
In 13 patients (20% of 64 successful profiles), personalized therapies could be initiated. 22 patients were treated with another line of chemotherapy as no actionable alteration could be detected. Effectiveness, determined by a PFS of the newly initiated therapy longer than 130% of the last conventional therapy line, could be seen in 8 of 13 patients (61,5%) of the precision oncology cohort compared to only 3 of 22 patients (13,5%) in the chemotherapy group. Additionally, Kaplan-Meier curves of PFS demonstrate a significant benefit for personalized treated patients (p = 0,0165 with a median PFS of 151 days, compared to 83 days in the chemotherapy group).
Conclusion
In summary, personalized cancer therapy based on comprehensive genomic profiling is effective and feasible also in the setting of a middle-sized oncologic center.
Introduction
"Precision Oncology" treatments aim to specifically destroy cancer cells. Thus, in-depth characterization and understanding of malignant tissue is of high importance. Throughout the last decades, cancer research could identify key factors of malignant growth [1] that differentiate normal from aberrant cells.
An exemplary mechanism often employed by cancer cells is overexpression of growth factor receptors. Malignant cells can accumulate up to 25–50 copies of the human epidermal growth factor receptor-2 (HER-2) gene, meaning 40–100-fold increase of HER-2 protein, resulting in 2 million HER-2 molecules expressed at the cancer cell surface [2, 3]. These extreme ratios allow relatively exact targeting of tumor cells with only minor side effects on healthy tissues [4, 5]. Overexpression of HER-2 protein can be detected via immunohistochemistry (IHC). Therefore, specific IHC-kits were developed and are routinely applied in clinical pathology. Notably, the first anti-HER-2 drug, trastuzumab (HerceptinⓇ, Roche, Basel, Switzerland) and the first IHC-test kit (HercepTestⓇ, DAKO, Glostrup, Denmark) were co-approved as specific treatment and companion-diagnostic [6]. Results of the HercepTestⓇ are decisive for therapy with anti-HER-2 agents [7]. As it could be observed, that IHC-testing for HER-2 demonstrated to a certain amount equivocal results (i.e. 2+ in HercepTestⓇ), a second, "reflex"-testing, of these samples via fluorescent in-situ hybridization (FISH) should be performed [8], where gene amplification is determined. To a small percentage even HER-2 IHC-negative/FISH-positive patients exist, who also benefit from anti-HER-2 treatment [9], underlying the need for in-depth characterization of tumor samples.
For the epidermal growth factor receptor (EGFR), it could be revealed that point mutations such as T790M or mutations of the downstream-signal proteins KRAS or NRAS are decisive for success or failure of EGFR-targeted therapies [9, 10]. Therefore, all these factors have to be determined before the initiation of an anti-EGFR therapy.
Clinical oncologists currently face the challenge, that in order to successfully apply targeted therapies, not only thorough understanding of the target and its role inside a cancer cell is needed, but also information and knowledge about up- or downstream signaling cascades, signaling partners and the tumor microenvironment.
As more and more of these interactions have been unveiled, the need for multiple testings can exceed the biopsy material. Instead of choosing between tests with the risk of missing important information from omitted tests, parallel sequencing with next generation sequencing (NGS) systems allow to test hundreds of genes. This so-called "comprehensive genomic tumor profiling" enables detection of genomic alterations, including base substitutions, indels, copy number alterations, genomic rearrangements plus description of genomic signatures. The limitations of this technique are its' costs and the time needed as oncologic treatment should normally be initiated fast. Thus, reasonable compromises between testing a plethora of genes and a reasonable turnaround time resulted in analysis of 300–500 tumor-related genes. Such panel testings have been established in academic institutions but are meanwhile also commercially available.
A best practice example of a university precision oncology program is the UC San Diego Moores Cancer Center. A retrospective analysis yielded that patients receiving personalized treatment had a longer median PFS survival than patients receiving chemotherapy [11]. However, as more than 85% of all cancer patients in the United States are treated in the community seeting, the American Society of Clinical Oncology published an educational book on how to implement precision medicine programs in the community-based oncology practice and which barriers are still to overcome [12]. Our work aims to complement these efforts by sharing data of a middle-sized oncologic center in Europe.
One factor determining if an institution offers genetic testing in its own molecular pathology department or uses commercially available services is its size. Especially middle-sized institutions face the challenge, that these institutions offer oncologic treatment for a broad range of tumor patients and should offer the best possible therapy in all of these indications. However, these institutions are not big enough to implement a specific team of molecular pathologists, molecular biologists and bio-informaticians in order to generate these results, analyze the huge amount of data and last, maybe most difficult to achieve, monitor all new published information on gene alterations and their implications for clinical treatment.
In our institution, the University Clinic Krems with 460 beds, approximately 1200 employees, an oncological department with 20 beds reserved for hemato-oncologic patients, and approximately 11.000 outpatient oncological contacts per year, we could not yet establish in-house comprehensive molecular tumor profiling. A feasible solution would be to organize it in a bigger setting within the Association of Hospitals of the Federal State of Lower Austria (i.e. 27 hospitals serving 1,6 million people).
However, until such structures are established, there are many clinical situations in which comprehensive genomic profiling of tumor samples would be highly warranted. These are patients with cancer of unknown primary (CUP)-syndromes, patients, in which conventional therapy lines did not yield successful responses or patients, who already underwent all available therapy lines. In these instances, we performed comprehensive genomic profiling via the commercially available services of Foundation Medicine (Cambridge, Massachusetts, USA).
This study aims to retrospectively analyze the effects of comprehensive genomic tumor profiling with respect to feasibility in the setting of a middle-sized oncologic center in Europe. Furthermore, success rates of personalized therapies based on molecular alterations detected in the patient's tumor are evaluated.
Methods
Patients
This retrospective study analyzed patients’ records, whose tumors underwent comprehensive genomic profiling (or were considered) between May 1st, 2016 and December 31st, 2019. Outcomes were analyzed for a longer observational period until March 31st, 2020. Demographic data and disease specific parameters were analyzed. Progression was defined as cumulative endpoint of either radiologically assessed, biochemically assessed (tumor marker progress) or clinically assessed (i.e. novel tumor-related symptoms such as ascites, pleural effusion or an ileus occurred).
Exclusion criteria were age < 18 years or patients with primary CNS tumors.
This study followed the principles of the Declaration of Helsinki and was approved by the ethics committee of the Karl-Landsteiner University ("EK Nr: 1009/2018″).
Informed consent for comprehensive genomic profiling was obtained from the patient participants in oral and written form.
Comprehensive genomic profiling methods
All tumor samples were sent to Foundation Medicine (Cambridge, Massachusetts, USA). Initially testing could only be performed in Cambridge, Massachusetts. From 2019 on, tests were performed in Penzberg, Germany. Minimal requirements for testing a specimen are a sample size of 25 mm2 and a tumor content of >20% (i.e. number of tumor cells divided by total number of all cells with nuclei in the sample).
Whenever available, testing of solid tumor specimens was endeavored.
Here, initially Foundation OneⓇ and later the refined Foundation OneⓇ CDx test was used.
The Foundation OneⓇ test was designed for sequencing the coding region of 315 cancer-related genes plus introns from 28 genes. The refined Foundation OneⓇ CDx test is a diagnostic device for detection of substitutions, insertion and deletion alterations (indels), and copy number alterations in 324 genes and selected gene rearrangements, as well as genomic signatures including microsatellite instability (MSI), tumor mutational burden (TMB) and homologous recombination deficiency (HRD) using DNA isolated from formalin-fixed paraffin embedded tumor tissue specimens.
From 2019 on also comprehensive genomic profiling of circulating cell free tumor DNA could be performed. Here, Foundation OneⓇ Liquid was employed. This test identifies base substitutions, insertions and deletions, copy number alterations, genomic rearrangements and reports high microsatellite instability. In total 70 commonly altered oncogenes are analyzed with this test.
Statistical analysis
To analyze our data, Microsoft Excel and GraphPad Prism 9 (GraphPad Software, San Diego, California, USA) were used. Through descriptive measures like median, mean, minimum and maximum values data was evaluated.
Patients included in the PFS-ratio analysis: to reject the Null-hypothesis, PFS-ratio needs to be >1,3 in at least 15% of the patients with an alpha risk of 5% and a power of 90%.
For analysis of PFS and OS Kaplan-Meier curves were plotted and Log-rank (Mantel Cox) test was performed. Significance was accepted at p < 0,05 (*).
Results
Between May 1st, 2016 and December 31st, 2019 69 patients treated at our department were considered for comprehensive genomic tumor profiling. The demographic data is depicted in Table 1. Most patients suffered from metastatic disease, patients with locally advanced disease had either tumors of the head and neck region (n = 3), pancreatic carcinoma (n = 1) or an adenocarcinoma of the gall bladder (n = 1).
Table 1.
Demographic data of included patients.
| Age - Median (Min; Max) | 61 (34; 81) |
|---|---|
| Male Sex n (%) | 34 (49,28) |
| Female Sex n (%) | 35 (50,72) |
| Metastatic Disease n (%) | 64 (92,75) |
| Locally Advanced n (%) | 5 (7,25) |
| No of previous therapy lines - Median (Min; Max) | 2 (0; 10) |
Anatomically, 14 tumors were of gynecologic origin (20%), 10 occurred inside the gastrointestinal tract (14,5%), 8 at the head and neck region (11,5%), 7 were genito-urinary (10%), 6 breast cancers (8,5%), 4 of hepato-biliary origin (6%), 4 pancreatic (6%), 2 lung cancers (3%) and 1 soft tissue sarcoma (1,5%). Notably, 13 tested tumor samples were of patients, who suffered from a CUP-syndrome (19%). Exact histological diagnoses are listed in Supplementary Table 1. Whenever possible, testing of solid tumor samples was preferred. However, in 10 cases (14,5%) no suitable specimen could be obtained. In these instances, a liquid biopsy of blood specimen was investigated.
Out of 59 solid tumors samples, 56 could be investigated (94,92%). Reasons for failure were in 1 case too less material, in 1 specimen a second DNA-signature was found, and 1 testing was canceled, as the patient died within the transport period of the sample to the lab overseas. Concerning liquid biopsy specimens, 8 out of 10 could be processed (80%). Here, two sample failures occurred. Overall, in our study 64 of the 69 intended patients’ samples could be molecularly profiled (92,75%, Table 2).
Table 2.
Success rates and numbers of detected alterations by Comprehensive Genomic Profiling.
| Successful Genomic Profiling - total n (%) | 64 of 69 (92,75) |
| Successful Genomic Profiling - solid tumor samples n (%) | 56 of 59 (94,92) |
| Successful Genomic Profiling - liquid biopsies n (%) | 8 of 10 (80) |
| Detected Alterations - total Median (Min; Max) | 4 (0; 23) |
| Detected Alterations - solid tumor samples Median (Min; Max) | 5 (1; 23) |
| Detected Alterations - liquid biopsies Median (Min; Max) | 1 (0; 4) |
The median number of detected genetic alterations with known implications on tumor growth was 4, as listed in Table 2. Foundation Medicine Reports also describe molecular variations of yet unknown significance. Here, the median number of these variations was 7 overall (see Supplementary Table 2).
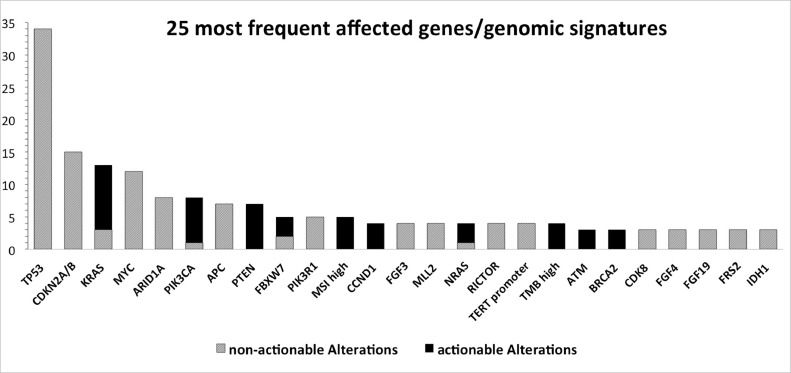
By far the most alterations in a single gene were reported for TP53, followed by KRAS and CDKN2A/B. In total, in 121 different genes alterations were found. An overview of the 25 most frequent affected genes/genomic signatures is presented in Fig. 1. Supplementary Table 3 describes all detected genetic alterations in detail. Concerning genomic signatures, 5 of 64 profiled tumors (8%) displayed MSI and 4 (6%) had a high TMB (Supplementary Table 3). Actionable alterations could be detected in 40 patients. Considering 64 successful genomic profiles, in 63% actionable alterations were detected. In 20 patients 1 actionable mutation was found, in 14 patients 2, in 5 patients 3 and 1 patient even 4.
Fig. 1.
Overview of the 25 most affected genes/genomic signatures.
Gray bars represent the number of tumor samples, in which a genetic alteration in the respective gene was detected. The number of detected actionable alterations is shown as black bars within.
In 13 patients (20% of 64 successful profiles), personalized therapies could be initiated. Notably, 1 of these patients could receive more than 1 targeted therapy line based on the NGS result. Table 3 lists all initiated therapies. Table 4 depicts initiated therapies, patients’ characteristics and outcomes for each patient treated with personalized therapies.
Table 3.
Detected actionable alterations and initiated therapy.
| Detected actionable alteration | Initiated treatment |
|---|---|
| ATM K2213fs*22, BRCA1 K654*fs47 | Olaparib |
| BRCA2 loss | Olaparib |
| BRCA2 S1848fs*15 | Olaparib |
| BRCA2 T3033fs*11 | Olaparib |
| CCND1 amplification, CDK4 amplification | Palbociclib |
| EGFR amplification | Erlotinib |
| FLT3 amplification | Sorafenib |
| KRAS amplification | Trametinib |
| MSI high | Atezolizumab |
| MSI high | Pembrolizumab |
| MSI high, TMB-high (23 Muts/Mb) | Atezolizumab |
| MSI high, TMB-high 36 Muts/Mb | Pembrolizumab |
| NF1 I679fs*21 | Trametinib |
| PIK3CA H1047L | Alpelisib |
Table 4.
Detailed Information of patients treated with personalized therapies.
| Pat. ID | Sex | Age+ | Diagnosis | Previous Lines of Treatment | Actionable Mutations§ | Initiated Treatment | Time to Treatment (days)& | PFS 1 (days) | PFS 2 (days) | ESCAT Tier |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 48 | Adenocarcinoma of the head and neck, G 2–3 (origin: ethmoid bone) | 3 | EGFR amplification | Erlotinib | 9 | 14 | 70 | III |
| 3 | F | 56 | esophageal Adenocarcinoma G2 | 3 | KRAS amplification | Trametinib | 22 | 57 | 18 | III |
| 10 | F | 40 | endometrial Carcinoma G2–3 | 1 | MSI high | Pembrolizumab | 46 | 50 | 92 | I |
| 13 | F | 54 | poor differentiated adenocarcinoma G3 of the esophagus | 2 | CCND1 amplification, CDK4 amplification | Palbociclib | 80 | 93 | 132 | III |
| 22 | M | 68 | invasiv ductal mammary Carcinoma G3, ER positive | 6 | BRCA2 S1848fs*15 | Olaparib | 85 | 141 | 432# | II |
| 28 | M | 68 | CUP - Adenocarcinoma G2 | 1 | MSI high, TMB-high 36 Muts/Mb; ATM K2213fs*22, BRCA1 K654*fs47 | Pembrolizumab; Olaparib | 173 | 179 | 438# | I III |
| 41 | M | 64 | Prostate acinar adenocarcinoma | 6 | BRCA2 T3033fs*11 | Olaparib | 91 | 109 | 151 | II |
| 47 | M | 70 | Colon Adenocarcinoma | 3 | MSI high, TMB-high (23 Muts/Mb) | Atezolizumab | 17 | 179 | 309# | III |
| 49 | M | 75 | Colon Adenocarcinoma | 5 | FLT3 amplification | Sorafenib | 49 | 246 | 132 | III |
| 52 | F | 67 | endometrioid Adenocarcinoma G3 | 9 | MSI high | Atezolizumab | 45 | 602 | 179# | III |
| 54 | M | 68 | Prostate acinar adenocarcinoma | 8 | BRCA2 loss | Olaparib | 39 | 59 | 183# | II |
| 60 | F | 76 | Breast cancer (NOS) | 6 | PIK3CA H1047L | Alpelisib | 3 | 96 | 98# | I |
| 62 | F | 63 | CUP - Adenocarcinoma G2 | 7 | NF1 I679fs*21 | Trametinib | 78 | 162 | 45 | III |
PFS1 indicates the progression free survival of the last standard-of-care therapy line, PFS2 the progression free survival of the treatment based on the comprehensive genomic profile.
… Age at Comprehensive Genomic Profiling.
… only actionable Mutations that led to treatment initiation are reported here.
… Time from Test Report to Treatment Initiation.
… ongoing treatment
ESCAT… ESMO Scale for Clinical Actionability of molecular Targets.
Additionally, in 16 patients, targeted therapy was considered, but as patients were stable in an initiated chemotherapy line while waiting for the test report, targeted therapy was held as option for later requirements. This strategy to keep patients in successful therapy lines is also reflected by the time to treatment initiation in treated patients in Table 4.
Unfortunately, the condition of 7 patients in the study cohort deteriorated too rapidly to initiate a further therapy line based on the NGS result.
Finally, 22 patients were treated with another round of chemotherapy, as no actionable alteration could be detected.
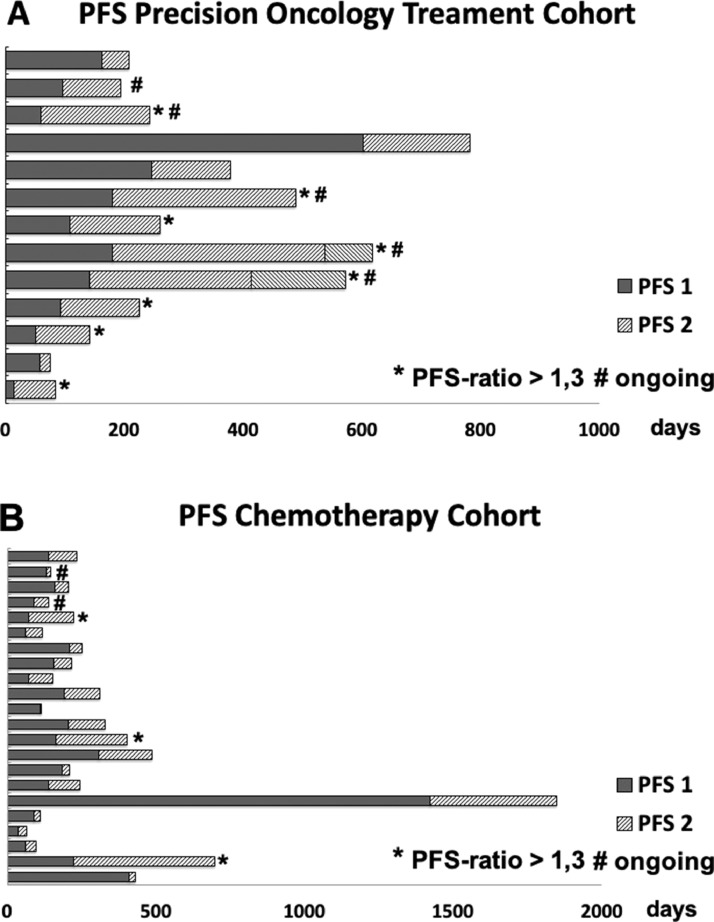
As functional outcome parameter, PFS-ratio was chosen. Here, the progression-free survival (PFS) of the last conventional therapy line is compared to the PFS of the newly initiated therapy. In our personalized therapy cohort, 8 of 13 patients (61,5%) displayed a PFS ratio >1,3 (Fig. 2A), compared to only 3 of 22 patients (13,5%) in the chemotherapy group (Fig. 2B). As mentioned above, 1 patient could receive more than 1 targeted therapy line based on the NGS result (Fig. 2A, Table 4). Furthermore, at the end of the observational period, 5 patients were still on treatment (labeled with ‘#’ in Fig. 2A). On the contrary, only 2 patients were still on treatment in the chemotherapy group (Fig. 2B).
Fig. 2.
Overview of PFS of diffent therapy lines.
A: Overview of PFS in the Precision Oncology Treatment Cohort.
B: Overview of PFS in the Chemotherapy Cohort.
Dark gray bars represent PFS of the last conventional chemotherapy line (PFS 1) in days. Light shaded gray bars represent PFS of the initiated Precision Oncology Treatment (PFS 2) in days. When 2 subsequent Precision Oncology therapies were initiated, both are depicted, divided by an upright separating line.
*… indicates patients, in whom the ratio of PFS 2/PFS 1 is > 1,3.
#… indicates patients, in whom the initiated treatment is still applied at the end of the observational period.
Comparison of PFS-curves of the personalized therapy group with the chemotherapy group shows a significant PFS-benefit for patients treated with personalized treatment (p = 0,0165; Fig. 3A). Median PFS of the personalized therapy group was 151 days, compared to 83 days in the chemotherapy group.
Fig. 3.
Kaplan-Meier survival curves.
A: Kaplan-Meier curve of Progression Free Survival.
B: Kaplan-Meier curve of Overall Survival.
Full Lines indicate survival rates in the Precision Oncology Treatment Cohort.
Dashed Lines indicate survival rates in the Chemotherapy Cohort.
*… p < 0.05
ns… not significant.
With regard to overall survival (OS), no significant difference could be observed (p = 0,6305; median OS personalized therapy: 713 days; median OS chemotherapy: 519 days; Fig. 3B).
At the end of the observational period, 8 of 13 patients in the personalized therapy cohort were still alive (61,5%) compared to 10 of 22 patients in the chemotherapy cohort (45,5%).
Discussion
Recent developments in cancer research have led to better understanding of carcinogenesis, tumor biology, host-tumor interactions and therapy-resistance formation. These findings translated into a flood of novel drugs, leading to more specific and less toxic therapies. In order to customize these therapies specifically to a genetic tumor signature, in-depth characterization of the tumor is the key requirement. According to estimates, medical knowledge currently doubles every 73 days [13]. The biggest challenge to offer "personalized" therapy for cancer patients is to keep all testing methods state-of-the-art and have an up-to-date overview of all novel findings. Here especially medium-sized oncologic centers face big challenges as mentioned above. Two of the biggest benefits commercial comprehensive genomic profiling services can provide are constant evolution of their test systems and provision of an up-to-date database curating the incoming knowledge of cancer research.
Our center initiated comprehensive genomic profiling in May 2016 for certain patients. Until the end of 2019 in total 69 patients underwent comprehensive genomic tumor profiling. As this patient cohort is very vulnerable, rapid turnaround time is of high importance, underlined by the fact, that 1 patient died even before the sample arrived at the testing facility overseas. Another crucial fact is the time point of testing. Since only in CUP-syndromes comprehensive genomic profiling is recommended at first line treatment in international guidelines [14], often patients are considered too late. In very late stages, new lesions are often difficult to reach for biopsy or the condition of the patient is to frail. In our cohort, in 7 patients comprehensive genomic profiling could be performed but no new therapy line could be established due to frailty after getting the report.
In 64 of 69 patients, a test report could be generated. Here, the introduction of the liquid biopsy test facilitated successful testing enormously. The first liquid biopsy test was performed in January 2019, and since then 13 of 32 tests (41%) used this principle. Before that, many eligible patients could not be tested, as no large enough specimen of their tumors could be harvested. Nonetheless, our institution still tries to prioritize testing from solid biopsies of newly occurred or progressing lesions, whenever possible. This is because the applied test for solid tumor specimens investigates 324 genes, whereas the applied liquid biopsy test can profile only 70 genes. However, also liquid biopsy tests get continously improved and can investigate larger gene sets from year to year.
As our patient cohort comprises challenging tumor entities, especially here, an in-depth understanding of the biology of these tumors is important. The best way to generate such knowledge is of course participation in clinical trials, where high-level evidence is collected. It is important to state that the approach of finding personalized therapies through comprehensive genomic profiling does not compete with participation in clinical trials. Whenever a patient is eligible for clinical trials, we strongly recommend participation. Recently, many trials have been designed for patients with specific genetic alterations. Here, comprehensive genomic profiling is often the first step to identify eligible patients.
In clinical trials patients‘ responses are usually compared with matched controls. In our cohort, evaluation of clinical success was not feasible via this approach, as patient characteristics and genomic signatures of the investigated tumors differed considerably. We thus went for a novel approach to define successful therapies for each individual patient: determination of the PFS-ratio. Here, the treated patient is his own control, as PFS of the newly initiated therapy is compared to PFS of the last conventional therapy line [15]. This is based on the finding, that in conventional chemotherapy, normally later therapy lines have less effect, as the tumor could acquire resistance mechanisms like e.g. overexpression of multi-drug resistance genes [16].
In general, with conventional chemotherapy only 15% of patients display a PFS of the later initiated therapy (PFS 2) that is longer than 130% of the PFS from the previous therapy line (PFS 1) [17]. This means, that in order to determine an effective therapy, the PFS-ratio (PFS 2/PFS 1) is > 1,3. Thus, to reject the Null-hypothesis that only random events are observed, the PFS-Ratio needs to be >1,3 in at least 15% of the patients.
In our patients, in whose tumors no targetable alterations could be identified and that received another line of chemotherapy, only 3 of 22 (13,5%) had a PFS-Ratio >1,3 (Fig. 2B), confirming the Null-hypothesis. By contrast, when personalized therapy was applied, 8 of 13 patients (61,5%) had a PFS-Ratio of >1,3 (Fig. 2A). Notably, at the end of the observational period, 5 patients in the personalized therapy cohort were still on treatment compared to 2 patients in the chemotherapy group.
Also, conventional comparison of PFS-curves demonstrated a PFS-benefit for the precision-oncology treated group (Fig. 3A). With regard to OS, no significant difference could be observed (Fig. 3B). Here, the observational period is too short, and the data is certainly too preliminary.
The observed differences were not due to better performance status of the personalized therapy patients, as in both groups median ECOG score of the patients was 1 (range in the precision oncology cohort: 0–4; in the chemotherapy group: 0–3). With regard to the site of metastases, both groups where also comparable. In the precision oncology cohort 30,8% of patients had bone metastasis (chemotherapy group: 27,3%) and 84,6% of patients suffered from visceral metastases (chemotherapy group: 72,3%). Only with regard to cerebral metastases, the proportion in the precision oncology group was notably higher with 15,4% compared to 4,5%.
An analysis of possible differences between initiated targeted therapies and immunotherapies with checkpoint inhibitors did not show a significant difference with regard to PFS and OS (Supplementary Fig. 1), but a trend towards better PFS for patients treated with immunotherapies. As immunotherapies for MSI-high cancers are emerging for many tumor types and this treatment can be considered as "extended" standard of care in Western countries, it was interesting to evaluate if the PFS-benefit of the precision oncogy cohort was "carried" by these interventions. However, these comparisons were done in very small patient cohorts and are for sure too preliminary. This is a topic we want to address in future studies with more patients and a longer observational period. Nonetheless, the data of clinical trials of immune checkpoint inhibitor therapy for MSI-high cancers are very promising and we want to stress the importance of broad MSI testing also in order to identify patients with Lynch Syndrome. As MSI can be also determined via immunohistochemistry, this should be feasible in many settings.
One general limitation of this study is the relatively small sample size. This is a challenge, medium-sized oncologic centers face in many aspects. A promising way to overcome this challenge is collaboration within bigger associations. Unfortunately, such solutions need long time due to the high complexity of decision making and allocation of resources.
Until then, a feasible solution is to get access to up-to-date testing facilities and an up-to-date knowledge database via commercial comprehensive genomic profiling services. These companies provide high-quality test results, compared with a curated report summarizing the most recent findings of cancer research. This report facilitates clinical decision making to a great extent.
A good supporting tool for clinical decision making is the ESMO Scale for Clinical Actionability of molecular Targets (ESCAT). Here molecular targets are ranked based on available evidence supporting their value as clinical targets. This Scale ranges (amongst other classications) from tier I: “targets ready for implementation in routine clinical decisions” to tier IV with only “preclinical evidence of actionability” [18]. The ESCAT tiers of our decisions are depicted in Table 4. Based on the ESCAT Scale, the European Society for Medical Oncology (ESMO) has issued in August 2020 Recommendations on using NGS for advanced cancers. This guideline suggests that NGS should be applied routinely in advanced lung adenocarcinoma, prostate cancer, ovarian cancer and cholangiocarcinoma. Moreover, it should be also applied on an individual basis in patients with other advanced cancers and in order to speed up drug development [19].
In our practice CGP reports are discussed interdisciplinary and decision support tools are used.
However, ideally, every patient should still be discussed in a molecular tumor board to find the optimal treatment. This is especially important if more than one actionable alteration is detected. Here, factors that determine the decision should be noted, curated and re-evaluated in regular intervals.
Unfortunately, patients from small and middle-sized oncologic centers are not regularly followed-up and analyzed in retrospective studies. However, especially such "real-world evidence" is important complementary information to knowledge gathered in clinical trials. We thus investigated our patient cohort of the last 3 1/2 years and aimed to share our experience and data.
In summary, personalized cancer therapy based on comprehensive genomic profiling is effective and feasible also in the setting of a middle-sized oncologic center. Patients benefit from in-depth profiling of their tumor samples and this service should be offered to a broader group of patients, especially to patients with high clinical need, such as patients with cancers of unknown primary, patients, whose tumors are resistant to conventional therapy, or patients that have progressed after all available therapy lines. Ideally, comprehensive genomic profiling facilitates participation in clinical studies in order to generate more high-level evidence. If participation in clinical trials is not possible, test results as well as patients' outcomes should nonetheless be analyzed as important real-world evidence of personalized therapy.
Declaration of Competing Interest
Josef Singer
Consulting or Advisory Role: Amgen, Gilead, Janssen, Merck, Merck Sharp & Dohme, Roche, Novartis, Pfizer
Speakers' Bureau: Abbvie, Amgen, Gilead, Janssen, Roche,
Travel, Accommodations, Expenses: Amgen, AOP Orphan, Merck, Roche, Pfizer, Takeda
Elias Brauneck:
A part of this work was the bachelor's thesis of Elias Brauneck.
Elisabeth Zwickl-Traxler:
None.
Martin Pecherstorfer:
Research Funding: Roche
Funding
There was no funding for this work.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101021.
Appendix. Supplementary materials
References
- 1.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Kallioniemi O.P., Kallioniemi A., Kurisu W. Erbb2 amplification in breast cancer analyzed by fluorescence in situ hybridization. Proc. Natl. Acad. Sci. U S A. 1992;89:5321–5325. doi: 10.1073/pnas.89.12.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iqbal N., Iqbal N. Human epidermal growth factor receptor 2 (her2) in cancers: overexpression and therapeutic implications. Mol. Biol. Int. 2014;2014 doi: 10.1155/2014/852748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudis C.A. Trastuzumab–mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 5.Harbeck N., Wuerstlein R. Optimal sequencing of anti-her2 therapy throughout the continuum of her2-positive breast cancer: evidence and clinical considerations. Drugs. 2013;73:1665–1680. doi: 10.1007/s40265-013-0118-z. [DOI] [PubMed] [Google Scholar]
- 6.Love D., Stratton E., Stocum M. Best practices for companion diagnostic and therapeutic development: translating between the stakeholders. N. Biotechnol. 2012;29:689–694. doi: 10.1016/j.nbt.2012.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Waks A.G., Winer E.P. Breast cancer treatment: a review. JAMA. 2019;321:288–300. doi: 10.1001/jama.2018.19323. [DOI] [PubMed] [Google Scholar]
- 8.Tchrakian N., Flanagan L., Harford J. New asco/cap guideline recommendations for her2 testing increase the proportion of reflex in situ hybridization tests and of her2 positive breast cancers. Virchows Arch. 2016;468:207–211. doi: 10.1007/s00428-015-1871-z. [DOI] [PubMed] [Google Scholar]
- 9.Gibbons-Fideler I.S., Nitta H., Murillo A. Identification of her2 immunohistochemistry-negative, fish-amplified breast cancers and their response to anti-her2 neoadjuvant chemotherapy. Am. J. Clin. Pathol. 2019;151:176–184. doi: 10.1093/ajcp/aqy136. [DOI] [PubMed] [Google Scholar]
- 10.Lo Nigro C., Ricci V., Vivenza D. Prognostic and predictive biomarkers in metastatic colorectal cancer anti-egfr therapy. World J. Gastroenterol. 2016;22:6944–6954. doi: 10.3748/wjg.v22.i30.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwaederle M., Parker B.A., Schwab R.B. Precision oncology: the uc san diego moores cancer center predict experience. Mol. Cancer Ther. 2016;15:743–752. doi: 10.1158/1535-7163.MCT-15-0795. [DOI] [PubMed] [Google Scholar]
- 12.Ersek J.L., Black L.J., Thompson M.A. Implementing precision medicine programs and clinical trials in the community-based oncology practice: barriers and best practices. Am. Soc. Clin. Oncol. Educ. Book. 2018;38:188–196. doi: 10.1200/EDBK_200633. [DOI] [PubMed] [Google Scholar]
- 13.Densen P. Challenges and opportunities facing medical education. Trans. Am. Clin. Climatol. Assoc. 2011;122:48–58. [PMC free article] [PubMed] [Google Scholar]
- 14.Fizazi K., Greco F.A., Pavlidis N. Cancers of unknown primary site: esmo clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2015;26(Suppl 5):v133–v138. doi: 10.1093/annonc/mdv305. [DOI] [PubMed] [Google Scholar]
- 15.Mick R., Crowley J.J., Carroll R.J. Phase ii clinical trial design for noncytotoxic anticancer agents for which time to disease progression is the primary endpoint. Control Clin. Trials. 2000;21:343–359. doi: 10.1016/s0197-2456(00)00058-1. [DOI] [PubMed] [Google Scholar]
- 16.Robey R.W., Pluchino K.M., Hall M.D. Revisiting the role of abc transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Von Hoff D.D., Stephenson J.J., Jr., Rosen P. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J. Clin. Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 18.Mateo J., Chakravarty D., Dienstmann R. A framework to rank genomic alterations as targets for cancer precision medicine: the esmo scale for clinical actionability of molecular targets (escat) Ann. Oncol. 2018;29:1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosele F., Remon J., Mateo J. Recommendations for the use of next-generation sequencing (ngs) for patients with metastatic cancers: a report from the esmo precision medicine working group. Ann. Oncol. 2020;31:1491–1505. doi: 10.1016/j.annonc.2020.07.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.