Abstract
Background:
Gestational hyperglycemia is associated with adverse perinatal outcomes and long-term offspring metabolic programming, likely through dysregulation of DNA methylation (DNAm).
Materials & methods:
We tested associations between maternal HbA1c and cord blood DNAm among 412 mother–child pairs in the genetics of glucose regulation in gestation and growth (Gen3G) and implemented Mendelian randomization to infer causality. We sought replication in an independent sample from Healthy Start.
Results:
Higher second trimester HbA1c levels were associated with lower DNAm at cg21645848 (p = 3.9 × 10-11) near URGCP. Mendelian randomization and replication analyses showed same direction of effect between HbA1c and DNAm at cg21645848, but did not reach statistical significance.
Conclusion:
We found that higher maternal glycemia reflected by HbA1c is associated with cord blood DNAm at URGCP, a gene related with inflammatory pathways.
Keywords: : cord blood, DNA methylation, HbA1c, hyperglycemia, Mendelian randomization, pregnancy
Lay abstract
An adverse metabolic environment in utero, such as elevated maternal blood glucose levels, may have long-lasting impacts on child development. Studies show that maternal glucose can cross the placenta and may impact regulatory markers acting upon genes through epigenetics. We investigated associations between 3 month maternal glucose levels, reflected by HbA1c, during pregnancy and cord blood DNA methylation (one type of epigenetic marker) in two prospective cohorts of mother–child pairs. We found that higher maternal blood glucose levels were associated with lower cord blood DNA methylation near a gene called URGCP, suggesting potential influence of elevated maternal glucose on inflammatory pathways in the newborn.
Hyperglycemia during pregnancy is associated with impaired glucose tolerance and adverse cardiometabolic outcomes in the exposed offspring later in life [1–3]. Current evidence indicates that maternal glycemia, as a continuous measure, is linearly associated with perinatal complications for the mother and her child [1,4–6], and with adverse metabolic programming in the offspring later in life [6–8]. Glycated hemoglobin (HbA1c) can be used to diagnose diabetes outside of pregnancy and to monitor glycemia, as it provides a standardized measure of average blood glucose over approximately 3 months period [9,10]. In a normal pregnancy, levels of HbA1c are generally lower than non pregnant women due to a faster red cell turnover, and a physiologic decrease in fasting blood glucose [9,11,12]. Still, higher levels of maternal HbA1c during pregnancy, in the range below the threshold for pre-existing diabetes diagnosis, have been associated with various adverse perinatal outcomes, including higher risk of pre-eclampsia, preterm birth, shoulder dystocia, congenital anomalies and perinatal loss [13,14]. Additionally, higher prenatal maternal HbA1c has also been associated with higher adiposity in the offspring at 11 years [7]. Thus, maternal hyperglycemia reflected by HbA1c may affect fetal programming at key stages during development.
Animal models and human studies with careful control for genetic effects [15,16] have demonstrated that maternal hyperglycemia and gestational diabetes mellitus (GDM) may influence fetal programming via epigenetic mechanisms [16–21]. The most studied of these mechanisms is DNA methylation (DNAm) [22,23]. Current evidence of associations between GDM and DNAm levels in fetal tissues (cord blood and placenta), suggests dysregulation of DNAm in or near genes involved in placental function, fetal development [19,24] and in disease-related metabolic pathways [25]. We have recently demonstrated that elevated 2 h glucose after a 75 g oral glucose tolerance test (OGTT) is associated with alteration in DNAm levels in placental tissue at key inflammatory genes [21]. Apart from this study, few others have addressed the association between maternal hyperglycemia along the continuous spectrum of glucose, and genome-wide DNAm variation in fetal tissues, particularly in cord blood. We hypothesize that maternal HbA1c would capture longer integrated hyperglycemic exposure during in utero development compared with traditional glycemic markers, and reveal novel epigenetic markers associated with offspring DNAm.
Therefore, we aimed to identify novel DNAm markers of fetal adaptation to intrauterine glycemia by conducting an epigenome-wide association study (EWAS) of HbA1c measured during pregnancy, and cord blood DNAm. In the genetics of glucose regulation in gestation and growth (Gen3G) cohort, we measured HbA1c in the first trimester (capturing glycemic exposure from pre conception to early pregnancy) and at the end of the second trimester (capturing the mid-pregnancy period). Additionally, we sought to replicate markers from our EWAS in an independent study (Healthy Start), and we implemented the Mendelian randomization (MR) approach [26] to investigate causality in the association between maternal HbA1c and DNAm at top differentially methylated sites. Robust DNAm markers detected at birth may inform a later in life risk of adverse metabolic programming and provide markers for early intervention.
Materials & methods
Study population
In this study, we included mother–child pairs from Gen3G, a Canadian prospective prebirth cohort study. Gen3G was designed to investigate the determinants of glucose regulation in pregnancy and their influence in fetal development [27]. Expecting mothers enrolled in the study were ≥18 years of age, mainly of Caucasian origin, had singleton pregnancies, and no records of pre-existing diabetes or diabetes detected in the first trimester [27]. Gen3G research staff collected biological samples and sociodemographic, lifestyle and anthropometric variables from the mothers in the first and second trimesters of pregnancy, at delivery, and from the newborns at birth. These variables have been described in detail elsewhere [27]. For this study, we selected a sample of mother–child pairs based on the availability of purified DNA from full-term (i.e., gestational age at birth ≥37 weeks) cord blood samples to conduct the DNAm analyses (Supplementary Figure 1). All participants provided written informed consent prior to enrollment in agreement with the Declaration of Helsinki. The institutional review board from the Centre Hospitalier Universitaire de Sherbrooke (CHUS) approved all study protocols.
Phenotype measures
We measured maternal anthropometrics at each visit using standardized procedures. Weight was measured with an electronic calibrated scale, and height with a wall stadiometer. We calculated BMI as weight/height2 (in kg/m2). Mothers reported their smoking status in the first trimester (coded as non smoker vs current smoker). We screened all women for GDM in the second trimester using a 75 g OGTT (mean ± SD weeks of gestation 26.4 ± 1.0). GDM was diagnosed following the International Association of Diabetes and Pregnancy Study Groups (IADPSG) criteria [27]. For our primary analyses, we excluded mothers who were diagnosed with GDM from these cord blood DNAm analyses to avoid bias by GDM treatment between time of HbA1c measurement and DNAm assessment (birth). We quantified HbA1c (%) from whole blood samples collected during the first and second trimesters using HPLC (Bio-Rad VARIANT, CA, USA) [27], a method certified by the National Glycohemoglobin Standardization Program [28], at the CHUS biomedical laboratory. We used the multiplexed particle-based flow cytometric assays (Human MILLIPLEX map kits, EMD Millipore, MA, USA) to measure TNF-α concentrations (pg/ml) from cord blood samples collected at delivery (within 30 min postpartum) by trained research staff.
Cord blood DNA extraction
Trained personnel collected cord blood samples from each delivery using a standardized protocol, and then stored the samples at -80°C until analysis. We extracted DNA from cord blood buffy coats using a protocol previously described [21].
Epigenome-wide DNAm measurement
We performed bisulfite conversion of genomic DNA from cord blood using the EZ-Methylation Kit (Zymo Research, CA, USA), followed by DNAm quantification utilizing the Infinium Methylation EPIC BeadChip (Illumina, CA, USA). To minimize batch effects, we randomly allocated samples to different plates and chips. We applied quality control (QC) of DNAm data at the sample and probe levels. At the sample level, we removed samples with low quality (n = 3), technical replicates (n = 10) and samples with genotype mismatch (n = 6 samples; based on SNPs on EPIC array from paired cord blood and placenta samples). At the probe level, we excluded probes with detection p > 0.05 in at least 5% of the samples [29] (n = 1,754), probes with null variance [30] (n = 125), probes in sex chromosomes [31,32] (n = 19,128 sites), non-CpG probes [31] (n = 2,836 sites), SNP-associated probes at the single base extension (n = 5,547 sites) [32] or at the target CpG site with MAF ≥5% [31] (n = 5668 sites), and cross-reactive probes [33] (n = 40,454 sites). In total, we retained for analyses 791,324 CpG sites in 412 cord blood samples from non-GDM mothers that passed all QC steps. We corrected for non specific background signals using the minfi R package [34], and controlled for additional unwanted technical variation using functional normalization with two principal components derived from control probes [35]. To correct for probe-type bias, we implemented the regression on correlated probes method, which uses the correlation between consecutive probes to adjust the distribution of type II probes [36]. Finally, we used ComBat [37] in the sva R package [38] to adjust for sample plate. We performed all the statistical analyses using DNAm on the M-value scale (logit transformation of methylation β-values) considering the statistical validity of this method [39]. However, to ease interpretability, we also showed effect estimates at our top findings in the β-value scale. Associations at the individual CpG sites were interpreted as the mean percent difference in DNAm (in a range between 0% or completely unmethylated, and 100% or completely methylated), per 1%-unit increase in HbA1c.
Replication in an independent study: the Healthy Start cohort
The Healthy Start cohort is a prospective prebirth cohort including mother–infant pairs of multiple ethnicities (non-Hispanic white, Hispanic and non-Hispanic African–American). Women in the study were recruited from the obstetric clinic at the University of Colorado Anschutz Medical Campus (CO, USA) from 2009 to 2014 [40]. Eligible women had singleton-pregnancies, were aged ≥16 years, and had no history of preterm births, stillbirths or chronic conditions including diabetes, cancer, steroid-dependent asthma or psychiatric conditions [40]. The standard clinical care was to offer women screening for GDM using a 1 h, 50 g glucose challenge test at about 26 weeks of gestation. If screening values of glucose during the glucose challenge test were ≥7.8 mmol/l, mothers were asked to undergo a 3 h, 100 g glucose test, with confirmation of GDM when Carpenter–Coustan criteria were met [41]. For this analysis, mothers with physician diagnosis of GDM were excluded to avoid confounding by GDM or its treatment. HbA1c was quantified from whole blood drawn in women that were in their 20–34th week of pregnancy (second–third trimester) using the DCA Vantage analyzer (Siemens, PA, USA) system, a method certified by the National Glycohemoglobin Standardization Program [28]. HbA1c quantification was performed at the University of Colorado Hospital Clinical and Translational Science Institute Core Laboratory. The study protocol was approved by the Colorado Multiple Institutional Review Board. Maternal participants provided written informed consent prior to study enrollment.
In the Healthy Start cohort, DNAm levels in cord blood were measured using the Illumina Infinium HumanMethylation 450K array. We excluded probes with detection p > 0.05, bead-count <3 in at least 5% of the samples, and samples with mismatch between predicted sex (by DNAm) and reported sex. Normalization of DNAm was conducted using the preprocessQuantile function in the minfi R package [34], with correction for batch effects using ComBat from the sva R package. We investigated in silico replication of CpG sites discovered in Gen3G with association p < 1.0 × 10-6. In total, we performed replication analyses in 506 mother–child pairs (58% non-Hispanic white) with complete measures of second trimester maternal HbA1c, using normalized DNAm (M-values) from cord blood samples.
Statistical analyses
EWAS of maternal HbA1c versus cord blood DNAm
We described baseline characteristics of 412 mother–child pairs in Gen3G included in this analysis using the mean and standard deviation (SD), or frequency and proportions. We compared baseline characteristics of participants in our study sample, with that of 456 individuals in Gen3G for whom HbA1c levels were measured at the second visit, but who were not included in our DNAm analysis. We conducted epigenome-wide assessment of differential methylation at the individual CpG sites in relation to levels of maternal HbA1c (%) using fitted robust linear regressions (RLR). For these analyses, DNAm in the M-value scale was modeled as the response variable, and maternal HbA1c at first or second trimester as the predictor. RLR was used to account for potential heteroskedasticity in the distribution of DNAm [21,42]. To control for differences in DNAm arising from cellular heterogeneity, we estimated cell counts for six white-cells (CD4+T, CD8+T, monocytes, granulocytes, natural killer cells & B cells) and nucleated red blood cells in cord blood using the Balkulski reference panel [43]. Models were adjusted for maternal age, maternal BMI, gestational week at HbA1c measurement, gravidity, gestational age at delivery, child sex, smoking status during pregnancy and estimated cord blood cellular composition. CpG-by-CpG associations were controlled for multiple testing using Bonferroni (α = 6.32 × 10-8), but we considered as ‘suggestive’ associations CpG sites identified with p < 1.0 × 10-6, and we took forward for replication and MR analyses this list of CpGs. To illustrate results of the EWAS, we used genome-wide Manhattan plots, volcano plots and Q-Q plots of p-values. We calculated the genomic inflation factor (γ) using the median values of the observed versus the expected chi-squared test statistic for the distribution of p-values in the EWAS. Finally, we used regional association plots to illustrate our findings at top differentially methylated CpGs. All analyses were conducted in R (version 3.5.1) [44].
To explore the effect of timing of glycemic exposure on our results, we took CpGs identified in the analysis with second trimester HbA1c and inspected their association with maternal HbA1c values in the first trimester among 410 mother–child pairs in Gen3G with complete measures of HbA1c, offspring DNAm and covariates. Regressions were adjusted for the same covariates as in the initial analysis with second trimester HbA1c. To determine if associations observed with offspring DNAm were driven by inflammatory processes, we tested the associations of maternal HbA1c and of DNAm at CpGs identified in the main EWAS with predicted cord blood cell types, and with the inflammatory marker TNF-α measured in a subset of cord blood samples (n = 406). For this analysis, HbA1c and DNAm were independently modeled as the exposure, while predicted count for cord blood cells and TNF-α were considered as the outcomes in multivariable linear regressions adjusted for maternal (age, gestational week at HbA1c measurement, gravidity, BMI and smoking) and newborn covariates (sex and gestational age at delivery). TNF-α values were log-transformed before the analysis. Associations between cells and HbA1c were considered statistically significant at p < 0.01 (α = 0.05/7 cell types), between cells and the CpGs at p < 0.002 (α = 0.05/4 CpGs tested*7 cell types), and between TNF-α and the CpGs at p < 0.01 (α = 0.05/4 CpGs tested).
Sensitivity analyses
We performed a secondary analysis to investigate if a non linear relationship was present between HbA1c and cord blood DNAm. For this analysis, we categorized HbA1c into quartiles and measured difference in DNAm at the individual CpG site (in M-values) when comparing the first (Q1 or lower HbA1c) versus the top quartile (Q4 or higher HbA1c) of HbA1c distribution, adjusting for same covariates as in our main analysis with continuous HbA1c. To rule out potential overfitting of the model by adjusting for maternal BMI, we tested correlations between BMI and HbA1c using Pearson correlations, and we inspected regression estimates for our top hits identified in the main EWAS using a model without adjustment for BMI.
Association between discovered CpGs & OGTT glucose levels
To determine robustness and specificity of our EWAS signals to HbA1c levels, we investigated associations between top differentially methylated CpGs and additional glycemic markers measured during the OGTT (i.e., fasting glucose, 1- and 2-h glucose levels). For this analysis, we used multiple RLRs with DNAm at the identified CpGs in the EWAS (in M-values) as the outcomes, and the glycemic markers as independent exposures. Associations with glucose levels were adjusted for the same covariates as in the discovery analysis using second trimester maternal HbA1c. To allow comparability in the interpretation of effect estimates obtained in the analysis with OGTT glucose (mmol/l) and HbA1c (%), we transformed values for these measures of glycemia to z-scores (mean = 0 and SD = 1) before the analysis. We interpreted association estimates at the individual CpGs as the mean percent difference in DNAm β-values, per SD increase in the glycemic measure. We considered associations with other glucose values at p < 0.02 (α = 0.05/3 CpGs tested).
Replication of top differentially methylated CpGs in the Healthy Start cohort
In the replication analysis, we adjusted associations for self-reported maternal ethnicity, maternal age, child sex, gestational age at visit, gestational age at delivery, gravidity (primigravid vs multiparous), smoking status during pregnancy (smoker vs non smoker), predicted count for seven cell types in cord blood [43], and maternal BMI (kg/m2) at the time of HbA1c measurement. Multivariable RLRs modeled DNAm as the outcome on the M-value scale, and continuous HbA1c (%) as the exposure. Effect estimates were reported using DNAm in the β-value scale to ease interpretability. In a sensitivity analysis, we used only samples of non-Hispanic white origin (n = 291) to investigate potential ethnic-specific effects at CpGs previously discovered. We considered associations to be replicated at p < 0.05/#CpGs tested.
Using MR to infer causality in the association between maternal HbA1c & cord blood DNAm
Considering our observational associations may be biased by unknown confounders, we implemented a MR approach to test if higher maternal glycemia reflected by HbA1c levels may be causally implicated in epigenetic changes in cord blood DNAm. Briefly, MR uses genetic variants as instrumental variables (IVs) to proxy levels of the exposure (HbA1c), and to provide a causal estimate of the exposure-outcome association unaffected by reverse causation and unmeasured confounding [26,45,46]. Strong IVs for MR analysis are selected based on a prespecified F-statistic threshold >10 [45]. In this study, genetic variants used to construct a polygenic score were extracted from a recent transethnic GWAS meta-analysis of HbA1c measured outside pregnancy (n = 60 SNPs at p < 5.0 × 10-8) [47]. We built the score for each participant by adding the number of HbA1c raising alleles for every SNP in the score, weighted by the GWAS effect for each SNP. To fulfill with specific assumptions of MR studies [26,45,46], we conducted several unadjusted linear and logistic regressions between the score as the exposure, and HbA1c, top differentially methylated CpGs and potential confounders of the main association as the outcome. To ease interpretability, we used the score in z-values (mean = 0, SD = 1), and association estimates were interpreted as the effect of one SD increase in the score, on a unit change in the trait. In total, we extracted information for 57 out of 60 HbA1c SNPs (three other SNPs were monomorphic in our sample) from 384 mothers in Gen3G with the availability of genetic, phenotype, and offspring DNAm data. All SNPs included in the score met our QC thresholds including MAF >0.01, genotype calling rate >0.8, r2 <0.5 and correct allele assignment.
We conducted the MR using a two stage least square instrumental variable (2SLS-IV) analysis implemented in the AER R package [48]. We applied correction for multiple testing in MR results using the Bonferroni method (p < 0.05/#CpGs tested). Results were interpreted as the mean percent difference in DNAm β-values, per 1%-unit increase in genetically predicted values of maternal HbA1c. Additionally, we reported MR diagnostic tests to indicate strength of the IV (Weak instrument test), model fit (Wald test) and consistency in magnitude and direction of associations between the observational and causal estimates (Endogeneity test). Power of the MR was calculated using the online MR power calculator mRnd (http://cnsgenomics.com/shiny/mRnd/) [49]. For CpGs analyzed, we visually inspected consistency in effect-size and direction of effect between the MR and EWAS estimates using linear plots.
Results
Participant characteristics
All mothers in Gen3G included in these analyses were of European descent, the mean age was 28 years, 34% were primigravid and 92% were non smokers (Table 1). We observed very similar baseline characteristics between study participants (n = 412) and individuals in Gen3G not included in the DNAm analysis (n = 456 mother–child pairs without cord blood DNAm or relevant covariates), except for differences in the proportion of non-white participants. Based on the fact that we excluded women with GDM from our current analyses, we observed higher glucose values in the OGTT (1 & 2 h post glucose load) and more frequent GDM among excluded participants (Supplementary Table 1). In our study sample, levels of HbA1c were slightly higher in the first compared with the second trimester (mean difference = 0.3%; p < 0.001), consistent with previous literature showing that in normal pregnancies, levels of HbA1c gradually decrease from the first to the second trimester, when they reach their lowest values in pregnancy [9,12]. Maternal HbA1c was normally distributed in the two visits, mean ± SD for HbA1c in visit one was 5.2 ± 0.2 and for HbA1c in visit two was 4.9 ± 0.3. For this study, we included only term deliveries (i.e., gestational age at delivery >37 weeks); 53% of the offspring were males.
Table 1. . Characteristics of participants in genetics of glucose regulation in gestation and growth included in the cord blood DNA methylation analysis of maternal HbA1c.
| Variable | n = 412 Mean (SD) or n [%] |
|---|---|
| Maternal age (years) | 28.1 (4.1) |
| Gravidity [% primigravid] | 138.0 [33.5] |
| Ethnicity [% Caucasian] | 412 [100] |
| Smoking during pregnancy [% non smoker] | 378 [91.7] |
| First trimester | |
| Gestational age (weeks) | 9.8 (2.3) |
| BMI (kg/m2) | 24.6 (5.6) |
| 1 h glucose challenge 50 g (mmol/l)† | 5.4 (1.3) |
| Hemoglobin A1c (%)† | 5.2 (0.2) |
| Second trimester | |
| Gestational age (weeks) | 26.4 (1.0) |
| Body mass index (kg/m2) | 27.8 (5.3) |
| Glucose challenge 75 g | |
| Fasting glucose (mmol/l) | 4.2 (0.31) |
| 1 h glucose (mmol/l) | 6.9 (1.4) |
| 2 h glucose (mmol/l) | 5.6 (1.1) |
| Hemoglobin A1c (%) | 4.9 (0.3) |
| Offspring variables | |
| Gestational age at delivery (weeks) | 39.6 (1.1) |
| Child sex [% males] | 218 [52.9] |
| Birth weight (g) | 3437.2 (431.1) |
Smaller sample size was observed for measures of 1 h, 50 g glucose test (n = 384) and maternal HbA1c in first visit (n = 410).
Association between maternal HbA1c & cord blood DNAm (discovery EWAS)
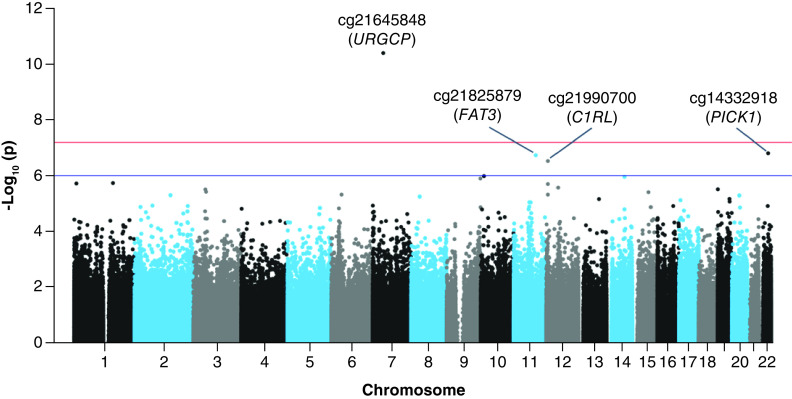
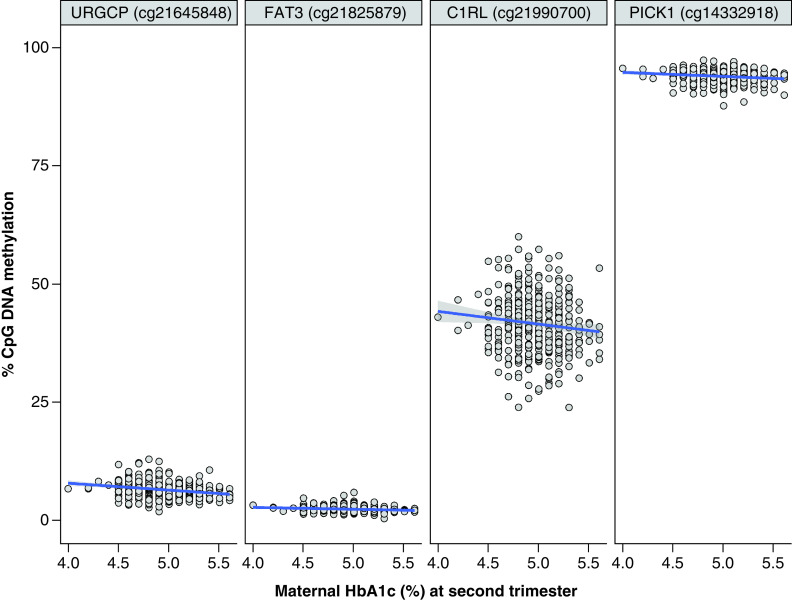
In our CpG-by-CpG analysis adjusted for covariates (λ = 1.18; Supplementary Figure 2), we found one CpG site strongly associated with second trimester HbA1c after Bonferroni correction (p < 6.3 × 10-8) (Figure 1 & Supplementary Figure 2). At cg21645848, we observed that a 1%-unit increase in maternal HbA1c was associated with an estimated 1.7% lower cord blood DNAm (95% CI: -2.2, -1.2%, p = 3.9 × 10-11) (Figure 2 & Table 2). This CpG mapped within the first exon of the upregulator of cell proliferation or URGCP gene, located within a CpG island but showing a weak correlation in DNAm β-values with surrounding CpGs (Supplementary Figure 3).
Figure 1. . Manhattan plot for the epigenome-wide association study of second trimester maternal HbA1c and cord blood DNA methylation.
Horizontal red line is the Bonferroni significant threshold at p < 6.3 × 10-8 or -log10(p) > 7.3; blue line is a borderline significance threshold set at p ≤ 10-7 or -log10(p) > 6.0.
Figure 2. . Scatter plots for the correlation between second trimester maternal HbA1c and cord blood DNAm at the top hit identified in cg21645848 (URGCP), and at three other CpGs identified with suggestive association in the epigenome-wide association study.
Table 2. . Top differentially methylated CpGs in cord blood identified in the epigenome-wide association study of second trimester maternal HbA1c and inspected for their association with first trimester maternal HbA1c.
| CpG | DNAm (%), mean (SD) | Mapped gene | Genomic context | Maternal HbA1c second trimester (n = 412) | Maternal HbA1c first trimester (n = 410) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Effect estimates† | 95% CI | p-value | Effect estimates‡ | 95% CI | p-value | ||||
| cg21645848 | 6.48 (1.64) | URGCP | Body | -1.7 | (-2.2, -1.2) | 3.97 × 10-11 | -0.7 | (-1.2, -0.1) | 1.22 × 10-2 |
| cg21825879 | 2.40 (0.60) | FAT3 | Intergenic | -0.5 | (-0.7, -0.3) | 1.83 × 10-7 | -0.1 | (-0.4, 0.1) | 2.96 × 10-1 |
| cg21990700 | 41.67 (5.98) | C1RL | TSS200, Body | -4.5 | (-6.2, -2.8) | 2.97 × 10-7 | -2.9 | (-4.9, -0.9) | 3.72 × 10-3 |
| cg14332918 | 94.0 (1.36) | PICK1 | TSS1500 | -0.9 | (-1.3, -0.6) | 1.56 × 10-7 | -0.6 | (-0.9, -0.2) | 4.59 × 10-3 |
Highlighted in bold is the association at cg21645848 (URGCP) surpassing the Bonferroni significance threshold at p < 6.3 × 10-8. Associations with first trimester HbA1c were regarded statistically significant at p < 0.01.
Effect estimates interpreted as the mean percent difference in DNAm (β-values) per 1%-unit increase in second (or first) trimester maternal HbA1c.
Regressions adjusted for similar covariates as in the discovery analysis with second trimester HbA1c, except for gestational age and BMI that corresponded to measures in the first visit.
In addition, results of our main analysis suggested associations of second trimester HbA1c with three other CpGs (with p < 1.0 × 10-6) at cg21825879 (near FAT3), cg21990700 (C1RL) and cg14332918 (PICK1). Association estimates for these CpGs are reported in Table 2. In general, we observed small effect sizes in the associations with maternal HbA1c (second trimester); among top four differentially methylated CpGs, median estimated percent difference in DNAm was 1.3 (estimated percent change in DNAm ranged from 0.5 to 4.5) in response to a 1%-unit increase in maternal HbA1c.
We found modest correlation between maternal BMI and HbA1c in the second visit (r = 0.2, p = 1.5 × 10-5). When excluding BMI from our regression models, we observed that the association at cg21645848 (URGCP) showed similar effect estimates and statistical significance as in the main model (p = 5.69 × 10-11 in BMI-unadjusted model, and p = 3.97 × 10-11 in main model). No other CpGs surpassed the threshold for Bonferroni significance in this sensitivity analysis. Overall, for all four differentially methylated CpGs previously identified in our initial analysis, excluding BMI from the models changed the absolute magnitude of the effect estimates by less than 10% (range = 3.0–8.8% decrease).
In sensitivity analyses, we used quartiles of HbA1c (second visit) as the exposure to investigate the assumption of a non linear relationship between HbA1c and cord blood DNAm. We found that an increase in HbA1c levels from Q1 (mean ± SD = 4.7 ± 0.1) to Q4 (mean ± SD = 5.3 ± 0.1) was associated with lower DNAm at CpGs in cg21645848 (URGCP), cg22322019 (intergenic) and cg04289992 (PALM) after correction for multiple testing (Supplementary Table 2). Thus, we confirmed our top hit in cg21645848 (URGCP) and found two other CpG sites that were not identified in our primary analysis, yet we recognized that this sensitivity EWAS showed greater inflation (λ = 1.4). Therefore, we remained prudent in the interpretation of these findings.
In multivariable linear regressions with predicted cord blood cells as the outcomes, we found no association between cell proportions and maternal HbA1c (p > 0.01), but specific cell types were associated with DNAm levels at cg21645848 (URGCP) (associated with CD8T, NK, Mono & Gran), cg21990700 (C1RL) (associated with CD4T, CD8T, B cells & Gran) and cg14332918 (PICK1) (associated with CD4T, Gran & nucleated red blood cell) at p < 0.002. In addition, none of the top four differentially methylated CpGs identified in the EWAS were associated with the inflammatory marker TNF-α measured in cord blood (p > 0.01).
Additionally, we investigated specificity of our top differentially methylated CpGs identified in the EWAS to timing of glycemic exposure by measuring their association with first trimester HbA1c. We observed that per 1%-unit increase in first trimester HbA1c was associated with lower DNAm in cg21645848 (URGCP), cg21990700 (C1RL) and cg14332918 (PICK1) (p < 0.01), consistent in the direction of association with the initial analysis using second trimester HbA1c, but effect sizes were smaller than for associations in the discovery analysis (Table 2).
For the EWAS conducted with first trimester maternal HbA1c (λ = 1.11), we did not identify any CpG sites where cord blood DNAm was robustly associated with HbA1c after adjusting for covariates and applying correction for multiple testing using Bonferroni (Supplementary Figure 4), but we found three CpGs with association p < 1.0 × 10-6 (Supplementary Table 3). These three CpGs did not overlap with top associations identified by p-value in our main EWAS of second trimester HbA1c. We also assessed the impact of adjusting (or not) for maternal BMI in this EWAS. We did not find correlation between BMI and HbA1c in the first visit (r = 0.1, p = 0.18), and in regressions excluding adjustment for BMI we obtained similar results at top three CpGs identified in the main model. In adjusted regressions between quartiles of HbA1c (first visit) and genome-wide cord blood DNAm (λ = 1.12), we did not find evidence of a non linear association between a change in HbA1c levels from Q1 (mean ± SD = 4.9 ± 0.1) to Q4 (mean ± SD = 5.5 ± 0.1), and difference in DNAm.
Association of OGTT glucose levels with cord blood DNAm at CpGs identified in EWAS of second trimester HbA1c
For glucose values measured during the second trimester OGTT, we found that greater maternal fasting glucose was nominally associated (at p < 0.05) with lower DNAm at previously discovered CpGs in cg21825879 (FAT3) and cg21990700 (C1RL) (p < 0.05) (Table 3). Associations between fasting or 1 h post-load maternal glucose and DNAm at cg21645848 (URGCP), cg21825879 (FAT3), cg21990700 (C1RL) and cg14332918 (PICK1), were in the same direction as in the discovery analysis with second trimester HbA1c, but only suggestive of associations (no significant p-values). Only cg21645848 (URGCP) and cg21990700 (C1RL) showed association in the same expected direction of effect with 2 h glucose values exposure, yet also non significant. The effect estimates identified in association with standardized glucose values measured during the OGTT were smaller than for associations with HbA1c in the second trimester at the four CpGs previously identified. All the glucose values standardized before the analysis were normally distributed. The correlations between second trimester HbA1c levels and OGTT glucose measures were weak (r range 0.2–0.3), but statistically significant (p < 0.001).
Table 3. . Association of maternal glucose values during a second trimester oral glucose tolerance test with top four CpG sites identified in the epigenome-wide association study of second trimester maternal HbA1c (all z-scores).
| CpG | Gene | Fasting glucose z-score (n = 412) | 1 h glucose z-score (n = 412) | 2 h glucose z-score (n = 412) | HbA1c second trimester z-score (n = 412) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Effect estimates (95% CI) | p-value | Effect estimates (95% CI) | p-value | Effect estimates (95% CI) | p-value | Effect estimates (95% CI) | p-value | ||
| cg21645848 | URGCP | -0.09 (-0.23, 0.05) | 0.24 | -0.06 (-0.20, 0.09) | 0.47 | -0.13 (-0.28, 0.03) | 0.10 | -0.43 (-0.55, -0.30) | 3.97 × 10-11 |
| cg21825879 | FAT3 | -0.05 (-0.11, -0.0001) | 0.04 | -0.001 (-0.05, 0.05) | 0.83 | 0.003 (-0.04, 0.05) | 0.99 | -0.12 (-0.17, -0.07) | 1.83 × 10-7 |
| cg21990700 | C1RL | -0.56 (-1.06, -0.07) | 0.03 | -0.40 (-0.91, 0.12) | 0.13 | -0.34 (-0.83, 0.15) | 0.18 | -1.13 (-1.56, -0.7) | 2.97 × 10-7 |
| cg14332918 | PICK1 | -0.08 (-0.18, 0.02) | 0.10 | -0.03 (-0.13, 0.08) | 0.44 | 0.002 (-0.10, 0.10) | 0.98 | -0.23 (-0.32, -0.14) | 1.56 × 10-7 |
Measures of glycemia were transformed to z-scores (mean = 0 and SD = 1) before the analysis to allow comparability in the effect estimates among traits. CpGs analyzed were identified in the epigenome-wide association study of second trimester maternal HbA1c (n = 412). Highlighted in bold are associations identified with p < 0.05 with one measure of glycemia.
Replication in the Healthy Start cohort
In the multi-ethnic external replication cohort (n = 506), we sought to replicate top four CpGs identified with p < 1.0 × 10-6 in the discovery analysis of second trimester HbA1c. Study characteristics of the replication cohort are presented in Supplementary Table 4. Of the four CpGs analyzed in the Healthy Start cohort, we identified a consistent direction of association with Gen3G analyses only at the top hit previously discovered in cg21645848 (URGCP) (effect estimate in Healthy Start = 0.84% lower DNAm β-values per 1%-unit increase in maternal HbA1c), but this association was not statistically significant (p = 0.09) (Supplementary Table 5). To address potential confounding by ethnicity at the CpGs investigated, we repeated the analysis using only samples of non-Hispanic white origin in the independent cohort (n = 291), but results were similar to those identified in the complete sample (data not shown).
MR analyses to support causality of associations between maternal HbA1c & cord blood DNAm
In an additional analysis, we evaluated the possibility that CpGs identified in our fully adjusted model were influenced by unmeasured confounders. For this analysis, we used a polygenic score (PS) with 57 SNPs as an IV for maternal HbA1c. In unadjusted regressions, we found that each SD increase in the PS was associated with a 0.06%-unit increase (95% CI: 0.03, 0.08%-unit) in maternal HbA1c in the second trimester, and this association was robust by statistical testing (p = 2.47 × 10-6, n = 384). Distribution of the PS in the Gen3G sample and its correlation with measured maternal HbA1c is illustrated in Supplementary Figure 5. We did not identify a substantial change in the association of the PS with measured maternal HbA1c after including maternal age and the first five genetic PCs as covariates in the regression model (Supplementary Table 6). In the raw model, the PS explained 5.4% (r2) of the variation in maternal HbA1c in our sample, and this result is consistent with previous findings in Europeans [47]. The F-statistic of the regression model including the PS was 22.9, indicating that the genetic score was a strong IV for maternal HbA1c. In additional analyses, we demonstrated that the PS was independent of measured confounders of the exposure-outcome association (p > 4.0 × 10-3 or α = 0.05/14 traits tested), and the PS was not directly associated with top four differentially methylated CpGs identified in the EWAS of HbA1c (p > 0.01 or α = 0.05/4 CpGs) (data not shown).
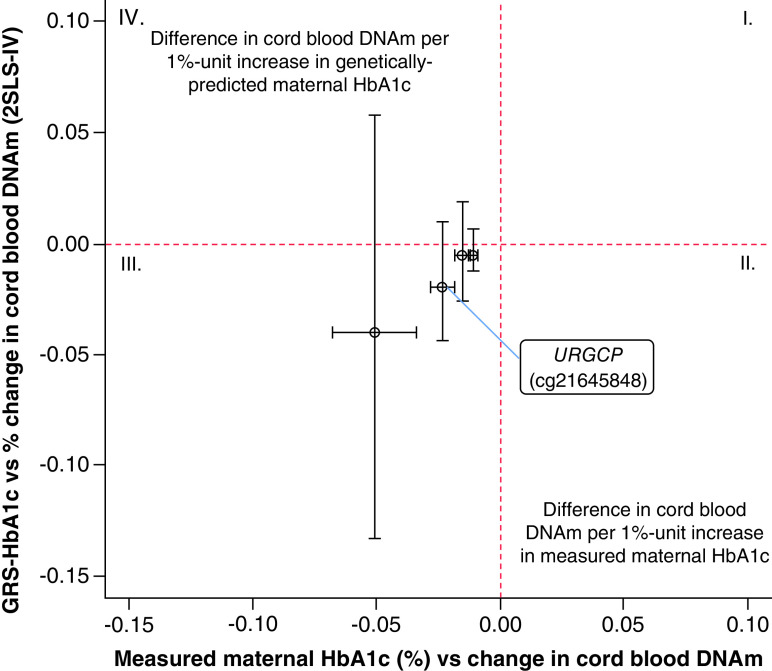
Our MR analyses demonstrated that the predicted effects were all in line with the observed effect in size and direction (Supplementary Table 7). Even though the effect size predicted was very similar to estimates obtained in the adjusted EWAS model, none of the MR estimates reached statistical significance (p > 0.01). Figure 3 shows a linear plot comparing effect-sizes and direction of association between predicted estimates of the MR and the observational analysis. We suspected that our MR analyses were underpowered to detect a causal effect of maternal HbA1c on difference in cord blood DNAm at top four CpGs identified in the EWAS. This assumption was supported by the results of a post hoc power calculation [49] that revealed that our analysis had 12% power to detect a 1.9% causal effect of maternal HbA1c on lower DNAm at cg21645848 (URGCP), our strongest signal of the EWAS, with statistical significance (α = 0.01). Furthermore, the endogeneity test (Hausman test) was underpowered to detect confounding in estimates of the observational analysis (p > 0.05). Genetically predicted values of maternal HbA1c explained between 2% and 5% of the total variation in DNAm at top four CpGs identified in the EWAS.
Figure 3. . Linear plot comparing association estimates between the observational and the Mendelian randomization analysis (2SLS-IV) for top four CpG sites identified in association with second trimester maternal HbA1c in the epigenome-wide association study.
X-axis represents estimates of the fully adjusted EWAS using measured maternal HbA1c as the exposure. Y-axis represents estimates of the MR analysis (unadjusted) using genetically predicted values of HbA1c as the exposure. Vertical error-bars represent the 95% CI for the MR estimates, while horizontal error-bars are the 95% CI for the observational estimates. CIs of the MR estimates overlapped with the red-dashed line of null associations, indicating no evidence of causality at these CpGs.
DNAm: DNA methylation; EWAS: Epigenome-wide association study; MR: Mendelian randomization.
Discussion
In this study we report, to our knowledge, the first EWAS of cord blood DNAm in relation to maternal hyperglycemic exposure reflected by HbA1c levels in the second trimester. We found one CpG in URGCP at which DNAm levels were strongly inversely associated with maternal HbA1c. The direction of association at URGCP was consistent with results we found in our replication analyses in an independent cohort. Three other epigenomic loci mapping to the genes FAT3, C1RL and PICK1 were suggestively associated with second trimester HbA1c in our discovery EWAS at a less stringent statistically significant threshold, but did not replicate. Our MR analyses predicted effects that were consistent with the observed effects in size and direction. However, we did not find statistical support for a causal effect of second trimester maternal HbA1c on cord blood DNAm at loci identified in the EWAS, likely due to limited power.
Cord blood DNAm in association with second trimester HbA1c
We demonstrated that maternal HbA1c in mid-pregnancy (as continuous or categorical) was associated with lower DNAm in cord blood at cg21645848 in URGCP. DNAm at this CpG was also associated with specific cord blood cells, indicating that HbA1c association with DNAm at cg21645848 may still reflect differences in the proportion of cells, despite our models being adjusted for cell types. URGCP or URG4 is the upregulator of cell proliferation, a recently discovered gene induced by hepatitis-B virus-encoded X-antigen (HbxAg) that has been identified as a putative oncogene involved in multistep carcinogenesis, and in various cellular processes [50]. In hepatocellular carcinogenic cells, overexpression of URGCP/URG4 correlates with upregulation of target genes for the NF-κB signaling pathway, including TNF-α, IL-6, IL-8 and MYC proto-oncogene [51].TNF-α is a proinflammatory cytokine that reduces insulin sensitivity by facilitating the phosphorylated state of the insulin receptor [52]. In our cohort, maternal TNF-α levels in pregnancy have been previously found independently associated with a higher maternal insulin resistance [53]. Similarly, our study previously identified an association between elevated 2-h glucose exposure in mid-pregnancy and increased placental methylation of a CpG mapping to the TNF receptor superfamily member 1b gene (TNFRSF1B), a high affinity receptor for TNF-α [21]. In non pregnant populations, overexpression of TNF-α in peripheral blood mononuclear cells was characteristic of patients with T2D [54], and expression of TNF was strongly correlated with HbA1c levels in the same study [54]. We demonstrated that cg21645848 was associated with cell types, but not with TNF-α in cord blood. Thus, our results provide some evidence that lower DNAm of cg21645848 (URGCP) in response to maternal hyperglycemia may be related with proinflammatory markers, yet further studies are needed to elucidate exact pathways and cytokines involved.
The CpG cg21645848 (URGCP) has not been reported before in EWAS of glucose traits [55] or T2D [56,57] in peripheral blood DNAm of middle-age adults, or in EWAS of GDM in fetal tissues (cord blood or placenta) [19,24,58]. However, we demonstrated the same direction of effect of HbA1c on cg21645848 (URGCP) in our replication study, and we showed a robust association of cg21645848 with first trimester HbA1c, and a consistent direction of association with other glycemic measures. Using a publicly available dataset, we found a methylation quantitative trait loci (meQTL) associated with DNAm at cg21645848 in cord blood according to data from the mQTL database (www.mqtldb.org/) [59]. This finding indicates that methylation at cg21645848 may be partially under fetal genetic control.
Lower DNAm at cg21990700 (C1RL), cg21825879 (FAT3) and cg14332918 (PICK1), showed some association with increased mid-pregnancy HbA1c in our discovery EWAS, but none of these signals were found with consistent effect size and direction of association in our replication study. Of these epigenetic loci, the largest effect size was observed at cg21990700 in the C1RL gene. In addition to second trimester HbA1c, cg21990700 was also associated with first trimester HbA1c, cord blood cell types, and there was a trend towards association with fasting glucose (second trimester). Previous EWAS have shown associations of cg21990700 with differential DNAm of fetal versus adult liver tissue [60], and with CRP [61], age [62] and smoking [63] in peripheral blood DNAm. The C1RL protein mediates the proteolytic cleavage of haptoglobins (cellular secretory proteins) in the endoplasmic reticulum [64]. C1RL is also a member of the complement system, which includes many proteins relevant to the innate immune response, and to the regulation of inflammation in adipose tissue [65]. Upregulation of C1RL expression was previously found in association with acquired obesity in subcutaneous adipose tissue of BMI-discordant twin pairs (lean vs heavy) [65]. In addition, C1RL expression was positively correlated with adiposity and hyperinsulinemia, and negatively correlated with the expression of insulin-signaling related genes in the same study [65].
Cord blood DNAm in association with 1st trimester HbA1c
Early pregnancy is a critical period for the effect of adverse exposures on embryonic and placental development [66]. In early pregnancy, higher maternal glucose levels impair the development of the placenta and restrict fetal growth [67]. Some studies have reported an association of higher maternal HbA1c levels in early pregnancy with a lower rate of fetal growth in mid-pregnancy in the offspring of non-GDM mothers [68], and with an increased risk of adverse birth outcomes independent of GDM diagnosis later in pregnancy [69]. Because first trimester HbA1c covers hyperglycemic exposure during the peri-conception and early pregnancy period, we initially hypothesized that first trimester HbA1c will have more profound programming effects on DNAm at birth. However, our results do not support that first trimester maternal HbA1c levels were strongly associated with changes in cord blood DNAm. Several factors can explain this result. First, HbA1c levels in later pregnancy may be a better reflection of the exposure to hyperglycemic episodes, while first trimester HbA1c levels may be influenced to a greater extent by erythrocyte cells biology (faster turnover). Second, large-scale DNAm remodeling in fetal tissues occurs primarily during mid-pregnancy (from week 9th to 22nd) [70], with a gain of DNAm in genes related to developmental functions, but lowering DNAm in genes involved in tissue-specific functions [70]. Finally, cord blood DNAm may not reflect epigenetic changes that may have occurred in more target tissues early in the embryonic development.
Findings from MR analyses
We attempted to support causality in the association between maternal HbA1c in pregnancy and cord blood DNAm by implementing MR. We observed a strong consistency in results of the MR and the observational analysis with respect to effect sizes and direction of effect for the associations analyzed. However, findings from the MR were underpowered, even though an F-statistic >10 from the IV-exposure association confirmed the strength of our instrument to proxy HbA1c levels in our sample. We estimated that more than 2361 mother–child pairs would have been required in our study to detect a similar causal effect at our top signal of the EWAS in cg21645848 (URGCP), with statistical significance (α = 0.01) and 80% power.
Putting our findings in the context of other epigenetic studies of glycemic traits in pregnancy
We did not find overlap between our cord blood CpGs associated with maternal HbA1c, and CpGs previously reported for GDM in cord blood [19,71,72] and placenta tissues [19,72]. Also, these CpGs were not overlapping with those previously identified in the placenta in association with maternal 2 h glucose [21] and insulin sensitivity (Matsuda index) [73] in our cohort.
Strengths & limitations
Our study has both strengths and limitations. Our strengths include our relatively large sample size, our prospective study design and the use of the EPIC array that allowed us to inspect >790,000 CpGs across the genome. We controlled for many potential confounders, including cord blood cells. In addition, we tested for epigenome-wide associations with maternal HbA1c measured at two time points in pregnancy. Finally, we attempted to validate results in an independent study.
We also had limitations. First, we acknowledge that cord blood is not considered as a tissue with ‘direct’ metabolic relevance (i.e., adipose tissue, muscle, liver, pancreas) to reveal DNAm involved in metabolic programming in response to maternal hyperglycemia. Second, DNAm at some of the CpGs identified showed associations with cells proportions even though we adjusted for cellular heterogeneity in our main analysis, suggesting incomplete correction for cells types or reflection of underlying inflammation. Third, despite including multiple potential confounders in our main analysis, unmeasured confounders may still have biased the associations identified. Finally lack of power due to the small sample-size used in the MR analysis, limited our detection of a causal effect between maternal HbA1c in mid-pregnancy and DNAm at CpGs identified in the EWAS.
Conclusion
We found that exposure to higher maternal glycemia in mid-pregnancy reflected by HbA1c was associated with lower cord blood DNAm at cg21645848 in URGCP. The URGCP gene is implicated in cell survival and growth, and in inflammatory response. These functions relate to genes previously identified with differential DNAm in response to GDM and HbA1c in fetal and adult tissues, respectively. Future research should aim to confirm the association between maternal glycemia in pregnancy and cord blood DNAm at the CpG in URGCP in European and non-European populations, while functional and longitudinal analyses could help to elucidate whether this locus has a role in gene regulation in specific tissues, and in long-term offspring metabolic programming.
Future perspective
The identification and validation of DNAm signatures in cord blood associated with exposure to maternal hyperglycemia in pregnancy may help to identify candidate biomarkers for early intervention and prevention of adverse cardiometabolic outcomes in the offspring. These biomarkers will provide insights into the mechanisms dysregulated by exposure to hyperglycemia early during development. Future validation of these markers can be achieved through target-tissue studies, longitudinal studies and the use of well-powered causal inference methods.
Summary points.
We identified a novel signal at cg21645848 in URGCP at which cord blood DNA methylation levels were inversely associated with maternal HbA1c levels in mid-pregnancy.
We found consistency in the direction of association at URGCP in our replication study.
We identified three other epigenetic loci mapping to C1RL, FAT3 and PICK1 with ‘suggestive’ association with mid-pregnancy maternal HbA1c levels, although none were replicated.
Our Mendelian randomization analysis showed estimates consistent in direction of effect and in effect size with observational estimates for all four loci tested. However, the Mendelian randomization was underpowered and did not support a causal effect of maternal HbA1c on cord blood DNA methylation levels.
We showed consistent direction of association with first trimester HbA1c, and with some of the glucose measures during the oral glucose tolerance test, for our top four CpGs identified by p-value in the epigenome-wide association study of second trimester maternal HbA1c.
We hypothesized that for some of the epigenetic loci identified in association with mid-pregnancy maternal HbA1c levels (i.e., URGCP & C1RL), adverse metabolic programming in the offspring may involve inflammatory pathways.
Acknowledgments
We thank participants of the Gen3G and the Healthy Start cohorts who contributed to this study, as well as clinical research nurses and research assistants in each study for recruiting women and obtaining consent. We also thank the CHUS biomedical laboratory for performing some assays in the Gen3G study. Last, we thank the Harvard Medical School for providing the computational resources required to conduct the analyses.
Footnotes
Supplementary data
To view the supplementary data that accompany this paper please visit the journal website at: www.futuremedicine.com/doi/suppl/10.2217/epi-2020-0279
Author contributions
DL Juvinao-Quintero and MF Hivert participated in the study concept and design. DL Juvinao-Quintero conducted analyses for Gen3G and wrote the manuscript. D Dabelea provided access to data in the Healthy Start cohort and AP Starling conducted the replication analyses for this cohort. A Cardenas, CE Powe, P Perron and L Bouchard contributed with interpretation of analyses and discussion. All authors read the manuscript, contributed to the discussion with important intellectual content, and agreed with its final version. DL Juvinao-Quintero has access to data presented in this study and is responsible for the integrity of the data and accuracy of the data analysis.
Financial & competing interests disclosure
Gen3G work presented in this study was supported by an American Diabetes Association Pathways Award #1-15-ACE-26 (MFH); Gen3G has also been supported by Fonds de recherche du Québec en santé #20697 (MFH); Canadian Institute of Health Research #MOP 115071 (MFH), and Diabète Québec grants (P Perron and L Bouchard). DLJQ is also supported by a Pyle fellowship in the Department of Population Medicine at Harvard Pilgrim Health Care Institute. The Healthy Start study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK076648), the National Institutes of Health Office of the Director (UH3OD023248), and the National Institute of Environmental Health Sciences (R01ES022934). AP Starling was supported by a grant from the National Institute of Environmental Health Sciences (R00ES025817) as well as AC (R01ES031259). CE Powe is supported by NIH K23DK113218 and the Robert Wood Johnson Foundation’s Harold Amos Medical Faculty Development Program. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Data sharing statement
Access to data will be available by the authors upon reasonable request.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.The HAPO Study Cooperative Research Group, Metzger BE, Lowe LP Hyperglycemia and adverse pregnancy outcomes. N. Engl. J. Med. 358(19), 1991–2002 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Lowe WL Jr, Scholtens DM, Kuang A et al. Hyperglycemia and adverse pregnancy outcome follow-up study (HAPO FUS): maternal gestational diabetes mellitus and childhood glucose metabolism. Diabetes Care 42(3), 372–380 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pedersen J. The pregnant diabetic and her newborn. Problems and Management. Munksgaard, Copenhagen, Denmark: (1967). [Google Scholar]
- 4.Metzger BE, Persson B, Lowe LP et al. Hyperglycemia and adverse pregnancy outcome study: neonatal glycemia. Pediatrics 126(6), e1545–1552 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Sesmilo G, Meler E, Perea V et al. Maternal fasting glycemia and adverse pregnancy outcomes in a Mediterranean population. Acta Diabetol. 54(3), 293–299 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Tam WH, Ma RCW, Ozaki R et al. In utero exposure to maternal hyperglycemia increases childhood cardiometabolic risk in offspring. Diabetes Care 40(5), 679–686 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowe WL Jr, Lowe LP, Kuang A et al. Maternal glucose levels during pregnancy and childhood adiposity in the hyperglycemia and adverse pregnancy outcome follow-up study. Diabetologia 62(4), 598–610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Prospective study shows the influence of higher maternal HbA1c levels in pregnancy on adiposity in the offspring at adolescence.
- 8.Scholtens DM, Kuang A, Lowe LP et al. Hyperglycemia and adverse pregnancy outcome follow-up study (HAPO FUS): maternal glycemia and childhood glucose metabolism. Diabetes Care 42(3), 381 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes RC, Rowan J, Florkowski CM. Is there a role for HbA1c in pregnancy? Curr. Diab. Rep. 16(1), 5 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Hinkle SN, Tsai MY, Rawal S, Albert PS, Zhang C. HbA(1c) measured in the first trimester of pregnancy and the association with gestational diabetes. Sci. Rep. 8(1), 12249–12249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Gonzalez CM, Castillo-Mora A, Alvarado-Maldonado IN et al. Reference intervals for hemoglobin A1c (HbA1c) in healthy Mexican pregnant women: a cross-sectional study. BMC Pregnancy Childbirth 18(1), 424 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelson PK, James KE, Leong A et al. Longitudinal changes in the relationship between hemoglobin A1c and glucose tolerance across pregnancy and postpartum. J. Clin. Endocrinol. Metab. 105(5), e1999–2007 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hughes RC, Moore MP, Gullam JE, Mohamed K, Rowan J. An early pregnancy HbA1c >/=5.9% (41 mmol/mol) is optimal for detecting diabetes and identifies women at increased risk of adverse pregnancy outcomes. Diabetes Care 37(11), 2953–2959 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Lowe LP, Metzger BE, Dyer AR et al. Hyperglycemia and adverse pregnancy outcome (HAPO) study: associations of maternal A1C and glucose with pregnancy outcomes. Diabetes Care 35(3), 574–580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; • Observational study shows the association of maternal HbA1c in pregnancy and below the threshold for gestational diabetes mellitus (GDM) diagnosis, with adverse perinatal outcomes.
- 15.Lindsay RS, Nelson SM, Walker JD et al. Programming of adiposity in offspring of mothers with Type 1 Diabetes at age 7 years. Diabetes Care 33(5), 1080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauguier D, Bihoreau MT, Ktorza A, Berthault MF, Picon L. Inheritance of diabetes mellitus as consequence of gestational hyperglycemia in rats. Diabetes 39(6), 734–739 (1990). [DOI] [PubMed] [Google Scholar]
- 17.Cleal JK, Lewis RM. Chapter 22 the placenta and developmental origins of health and disease. : The Epigenome and Developmental Origins of Health and Disease. Rosenfeld CS (). Elsevier Science, London, UK, 439–461 (2016). [Google Scholar]
- 18.Dabelea D, Hanson RL, Lindsay RS et al. Intrauterine exposure to diabetes conveys risks for type 2 diabetes and obesity: a study of discordant sibships. Diabetes 49(12), 2208–2211 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Finer S, Mathews C, Lowe R et al. Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum. Mol. Genet. 24(11), 3021–3029 (2015). [DOI] [PubMed] [Google Scholar]; • Evidence of differential DNA methylation (DNAm) in fetal tissues (placenta & cord blood) in response to GDM.
- 20.Franzago M, Fraticelli F, Stuppia L, Vitacolonna E. Nutrigenetics, epigenetics and gestational diabetes: consequences in mother and child. Epigenetics 14(3), 215–235 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cardenas A, Gagne-Ouellet V, Allard C et al. Placental DNA methylation adaptation to maternal glycemic response in pregnancy. Diabetes 67(8), 1673–1683 (2018). [DOI] [PubMed] [Google Scholar]; •• First epigenome-wide association study shows the influence of increased maternal glycemia in pregnancy (non-GDM) along the continues spectrum of glucose, on changes in placental DNAm.
- 22.Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med. 7(10), e1000356 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mill J, Heijmans BT. From promises to practical strategies in epigenetic epidemiology. Nat. Rev. Genet. 14, 585 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Howe CG, Cox B, Fore R et al. Maternal gestational diabetes mellitus and newborn DNA methylation: findings from the pregnancy and childhood epigenetics consortium. Diabetes Care 43(1), 98 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Recent meta-analysis of epigenome-wide association study of GDM in cord blood DNAm.
- 25.Hjort L, Novakovic B, Grunnet LG et al. Diabetes in pregnancy and epigenetic mechanisms-how the first 9 months from conception might affect the child's epigenome and later risk of disease. The Lancet. Lancet DiabetesEndocrinol. 2019) (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 26.Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23(R1), R89–R98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillemette L, Allard C, Lacroix M et al. Genetics of glucose regulation in gestation and growth (Gen3G): a prospective prebirth cohort of mother-child pairs in Sherbrooke, Canada. BMJ Open 6(2), e010031 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.National Glycohemoglobin Standardization Program. List of NGSP certified methods and Laboratories. 1–21 (2020). www.ngsp.org/docs/methods.pdf
- 29.Bibikova M, Barnes B, Tsan C et al. High density DNA methylation array with single CpG site resolution. Genomics 98(4), 288–295 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Meng H, Joyce AR, Adkins DE et al. A statistical method for excluding non-variable CpG sites in high-throughput DNA methylation profiling. BMC Bioinformatics 11, 227–227 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Touleimat N, Tost J. Complete pipeline for Infinium® Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 4(3), 325–341 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Wu X, Wang Y. A framework for analyzing DNA methylation data from Illumina Infinium Human Methylation 450 BeadChip. BMC Bioinformatics 19(5), 115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pidsley R, Zotenko E, Peters TJ et al. Critical evaluation of the Illumina Methylation EPIC BeadChip microarray for whole-genome DNA methylation profiling. Genome Biol. 17(1), 208 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aryee MJ, Jaffe AE, Corrada-Bravo H et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30(10), 1363–1369 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fortin J-P, Labbe A, Lemire M et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 15(12), 503–503 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niu L, Xu Z, Taylor JA. RCP: a novel probe design bias correction method for Illumina Methylation BeadChip. Bioinformatics 32(17), 2659–2663 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics (Oxford, England) 8(1), 118–127 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28(6), 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du P, Zhang X, Huang C-C et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11(1), 587 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauder Katherine A, Stamatoiu Alexandra V, Leshchinskaya E, Ringham Brandy M, Glueck Deborah H, Dabelea D. Cord blood Vitamin D levels and early childhood blood pressure: the healthy start study. J. Am. Heart Assoc. 8(9), e011485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carpenter MW, Coustan DR. Criteria for screening tests for gestational diabetes. Am. J. Obstet. Gynecol. 144(7), 768–773 (1982). [DOI] [PubMed] [Google Scholar]
- 42.Sharp GC, Salas LA, Monnereau C et al. Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum. Mol. Genet. 26(20), 4067–4085 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakulski KM, Feinberg JI, Andrews SV et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics 11(5), 354–362 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Viennna, Austria, (2018). www.R-project.org/ [Google Scholar]
- 45.Lawlor DA, Harbord RM, Sterne JA, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27(8), 1133–1163 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32(1), 1–22 (2003). [DOI] [PubMed] [Google Scholar]; • Reference paper outlines the principles of Mendelian randomization and its application in epidemiological studies.
- 47.Wheeler E, Leong A, Liu CT et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med. 14(9), e1002383 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Recent GWAS meta-analysis of HbA1c in non pregnant populations. Reference paper used to extract genetic variants associated with HbA1c levels.
- 48.Christian K, Zeileis A. Applied Econometrics with R. Robert G, Kurt H, Parmigiani G (). Springer-Verlag, New York, USA: (2008). [Google Scholar]
- 49.Brion M-JA, Shakhbazov K, Visscher PM. Calculating statistical power in Mendelian randomization studies. Int. J. Epidemiol. 42(5), 1497–1501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dodurga Y, Secme M, Lale Satiroglu-Tufan N. A novel oncogene URG4/URGCP and its role in cancer. Gene 668, 12–17 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Xing S, Zhang B, Hua R et al. URG4/URGCP enhances the angiogenic capacity of human hepatocellular carcinoma cells in vitro via activation of the NF-kappaB signaling pathway. BMC Cancer 15, 368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hameed I, Masoodi SR, Mir SA, Nabi M, Ghazanfar K, Ganai BA. Type 2 diabetes mellitus: from a metabolic disorder to an inflammatory condition. World J. Diabetes 6(4), 598–612 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guillemette L, Lacroix M, Battista MC et al. TNFalpha dynamics during the oral glucose tolerance test vary according to the level of insulin resistance in pregnant women. J. Clin. Endocrinol. Metab. 99(5), 1862–1869 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Slieker RC, Van Der Heijden A, Van Leeuwen N et al. HbA1c is associated with altered expression in blood of cell cycle- and immune response-related genes. Diabetologia 61(1), 138–146 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kriebel J, Herder C, Rathmann W et al. Association between DNA methylation in whole blood and measures of glucose metabolism: KORA F4 study. PLoS ONE 11(3), e0152314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cardona A, Day FR, Perry JRB et al. Epigenome-wide association study of incident Type 2 Diabetes in a British population: EPIC-Norfolk study. Diabetes 2019) (Epub ahead of print). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Juvinao-Quintero DL, Hivert M-F, Sharp GC, Relton CL, Elliott HR. DNA methylation and Type 2 Diabetes: the use of mendelian randomization to assess causality. Curr. Genet. Med. Rep. 7(4), 191–207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruchat SM, Houde AA, Voisin G et al. Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 8(9), 935–943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gaunt TR, Shihab HA, Hemani G et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol. 17, 61 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonder MJ, Kasela S, Kals M et al. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genomics 15, 860 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ligthart S, Marzi C, Aslibekyan S et al. DNA methylation signatures of chronic low-grade inflammation are associated with complex diseases. Genome Biol. 17(1), 255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li S, Christiansen L, Christensen K et al. Identification, replication and characterization of epigenetic remodelling in the aging genome: a cross population analysis. Sci. Rep. 7(1), 8183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeilinger S, Kuhnel B, Klopp N et al. Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 8(5), e63812 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wicher KB, Fries E. Prohaptoglobin is proteolytically cleaved in the endoplasmic reticulum by the complement C1r-like protein. Proc. Natl Acad. Sci. USA 101(40), 14390–14395 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaye S, Lokki AI, Hanttu A et al. Upregulation of early and downregulation of terminal pathway complement genes in subcutaneous adipose tissue and adipocytes in acquired obesity. Front Immunol. 8, 545–545 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Murphy VE, Smith R, Giles WB, Clifton VL. Endocrine regulation of human fetal growth: the role of the mother, placenta, and fetus. Endocr. Rev. 27(2), 141–169 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Sletner L, Jenum AK, Yajnik CS et al. Fetal growth trajectories in pregnancies of European and South Asian mothers with and without gestational diabetes, a population-based cohort study. PLoS ONE 12(3), e0172946 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geurtsen ML, Van Soest EEL, Voerman E, Steegers EaP, Jaddoe VWV, Gaillard R. High maternal early-pregnancy blood glucose levels are associated with altered fetal growth and increased risk of adverse birth outcomes. Diabetologia 62(10), 1880–1890 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mañé L, Flores-Le Roux JA, Benaiges D et al. Role of first-trimester HbA1c as a predictor of adverse obstetric outcomes in a multiethnic cohort. J. Clin. Endocrinol. Metab. 102(2), 390–397 (2016). [DOI] [PubMed] [Google Scholar]
- 70.Slieker RC, Roost MS, Van Iperen L et al. DNA methylation landscapes of human fetal development. PLoS Genet 11(10), e1005583–e1005583 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Haertle L, El Hajj N, Dittrich M et al. Epigenetic signatures of gestational diabetes mellitus on cord blood methylation. Clin. Epigenetics 9, 28–28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weng X, Liu F, Zhang H et al. Genome-wide DNA methylation profiling in infants born to gestational diabetes mellitus. Diabetes Res. Clin. Pract. 142, 10–18 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Hivert M-F, Cardenas A, Allard C et al. Interplay of placental DNA methylation and maternal insulin sensitivity in pregnancy. Diabetes 69(3), 484 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]