Abstract
Fungus-growing ants engage in a multilateral symbiosis: they cultivate a fungal garden as their primary food source and host symbiotic actinobacteria (Pseudonocardia spp.) that provide chemical defenses. The bacterial symbionts produce small specialized metabolites that protect the fungal garden from specific fungal pathogens (Escovopsis spp.), and in return, they are fed by the ant hosts. Multiple studies on the molecules underlying this symbiotic system have led to the discovery of a large number of structurally diverse antifungal molecules, but somewhat surprisingly no shared structural theme emerged from these studies. A large systematic study of Brazilian nests led to the discovery of the widespread production of a potent but overlooked antifungal agent, which we named attinimicin, by nearly two-thirds of all Pseudonocardia strains from multiple sites in Brazil. Here we report the structure of attinimicin, its putative biosynthetic gene cluster, and the evolutionary relationship between attinimicin and two related peptides, oxachelin A and cahuitamycin A. All three nonribosomal peptides are structural isomers with different primary peptide sequences. Attinimicin shows iron-dependent antifungal activity against specific environmental fungal parasites but no activity against the fungal cultivar. Attinimicin showed potent in vivo activity in a mouse Candida albicans infection model comparable to clinically used azole-containing antifungals. In situ detection of attinimicin in both ant nests and on worker ants supports an ecological role for attinimicin in protecting the fungal cultivar from pathogens. The geographic spread of the attinimicin biosynthetic gene cluster in Brazilian Pseudonocardia spp. marks attinimicin as the first specialized metabolite from ant-associated bacteria with broad geographic distribution.
Short abstract
Attinimicin, the product of an evolutionarily conserved combinatorial three-module nonribosomal peptide biosynthetic pathway, provides iron-dependent chemical defenses to fungus-growing ants over a broad geographical swath.
Introduction
Several insect families—ants,1 termites,2 beetles,3 and bees4,5—cultivate fungi as their primary food source. The fungus-growing attine ants, which have been tending fungal gardens for 50 million years, represent one of the most successful.1 Attine ants had a single origin in the Amazon6 followed by dispersion to Central America and the rest of the Neotropics.7 This model is supported in part by finding close relatives of the ants’ fungal cultivars only in wet tropical forest environments.7,8 It is believed that the ants moved to Central America through multiple migration events, with the earliest event occurring about 25 million years ago, prior to the closure of the Panamanian isthmus.9−11
Specialized fungal parasites, Escovopsis spp., pose existential threats to fungus-growing ant colonies, and the ants employ several mechanisms, including the mechanical removal of fungal spores from the nest12 and the formation of symbiotic relationships with actinobacteria belonging to the genera Pseudonocardia and Streptomyces, to survive.13 The bacterial symbionts protect the fungal cultivar by producing antifungal metabolites that selectively kill fungal pathogens but leave the cultured fungal crop intact (Figure 1). Since the discovery of this specialized symbiosis between fungus-growing ants and their actinobacterial symbionts, many studies led to the discovery of structurally diverse biologically active compounds isolated from actinomycetes associated with attine ants. Examples include candicidin,14,15 valinomycin,16 actinomycins,16,17 antimycins,16 isolated from Streptomyces, and dentigerumycins,18 pseudonocardones,19 selvamicin,20 and 9-methoxyrebeccamycin,21 isolated from Pseudonocardia. One curious feature shared by all of those examples (and others) was their limited distribution. The highly diverse suite of molecular structures seemed to reflect a highly fragmented and geographically limited evolutionary history. No unifying structural motif—a molecular framework that is geographically widespread—was observed for any of the molecules reported to date.
Figure 1.
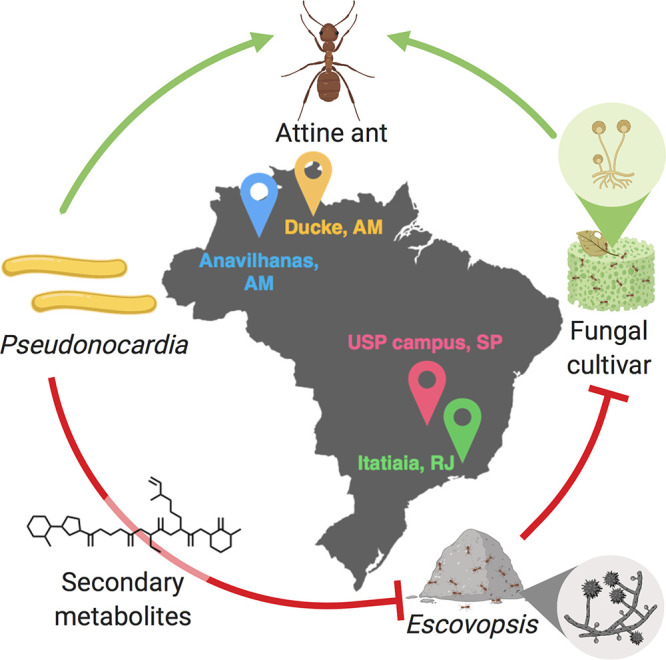
Symbiotic interactions in the attine ant system. Green arrows represent mutualistic relationships, and red T-bars indicate antagonism. Collecting sites in Brazil are shown.
Here, we describe a nonribosomal peptide, attinimicin, that is produced by more than 70% of all Pseudonocardia spp. isolated across four geographical regions in Brazil with an aggregate area of almost two-million square kilometers. Attinimicin selectively inhibits growth of the fungal pathogen Escovopsis spp. but shows no activity against the fungal cultivar. In addition, attinimicin shows potent, iron-dependent bioactivity in a mouse Candidiasis model.
Results and Discussion
Forty-two ant-associated Pseudonocardia strains were isolated from four different geographical locations in Brazil: Adolpho Ducke Reserve (Amazonas), Anavilhanas National Park (Amazonas), Itatiaia National Park (Rio de Janeiro), and the University of São Paulo campus at Ribeirão Preto (São Paulo) (Supporting Information, Table S1). The 16S rRNA gene as well as three housekeeping genes (atpD, dnaA, and gyrA) were amplified, sequenced, and used for phylogenetic analysis along with the same set of genes from 47 Pseudonocardia spp. that were previously isolated from ant nests in Panama (Supporting Information, Table S2). Our phylogenetic studies show that the Pseudonocardia species isolated in Brazil form a monophyletic clade with the exception of strains isolated from Itatiaia National Park in the state of Rio de Janeiro that form an additional monophyletic clade, and two other Brazilian isolates that group with the Panamanian strains (Figure 2). Motivated by the likelihood that these ant-associated Pseudonocardia isolates were involved in protecting the fungal cultivar from invading pathogenic fungi, we subjected all 42 Brazilian Pseudonocardia strains to a binary interaction screen against fungal pathogens (Supporting Information, Figure S1). In total, 16 strains (38% of all strains tested) showed antifungal activity, and these strains were subsequently cultured, extracted, and subjected to untargeted LC-MS/MS metabolomics analysis.
Figure 2.
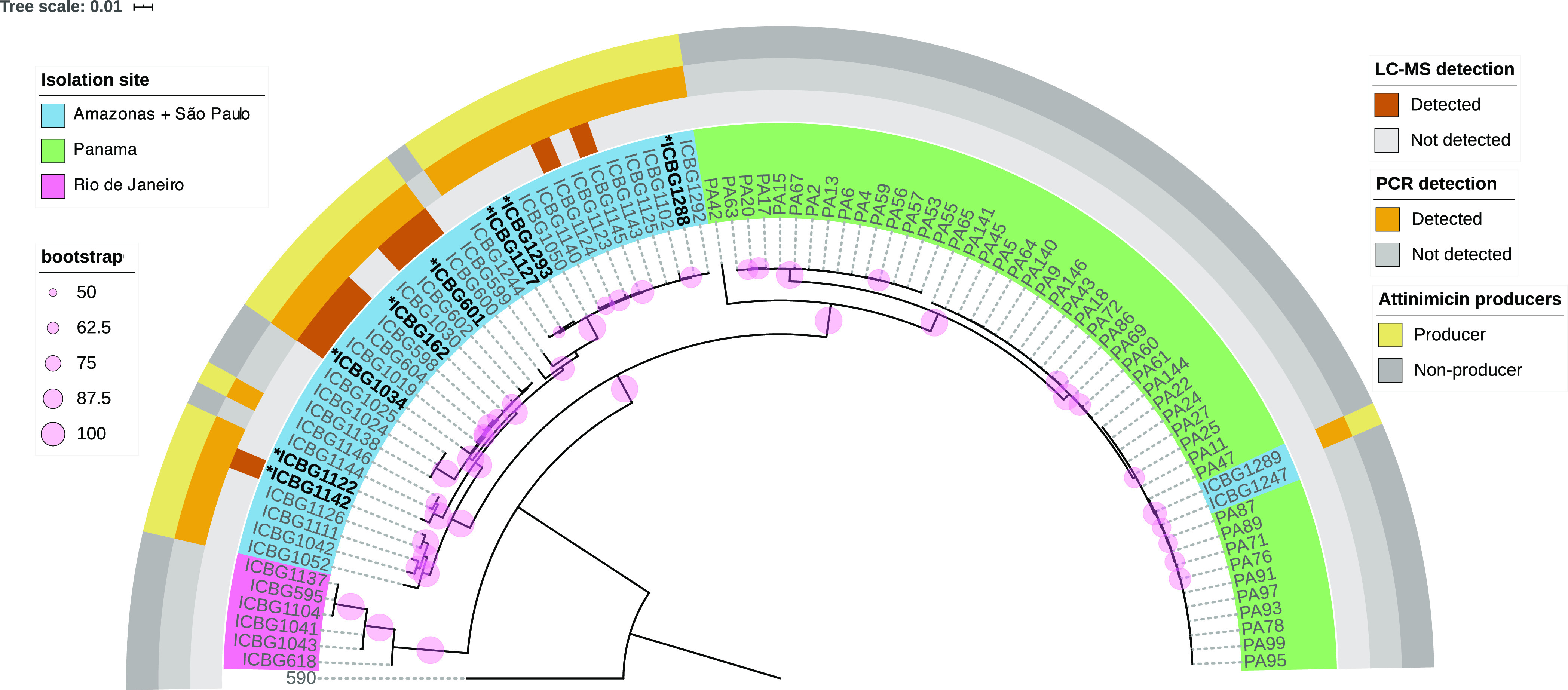
Phylogenetic representation of ant-associated Pseudonocardia spp. from Brazil and Panama. Strains are colored according to their collecting sites. Ant-associated Amycolatopsis sp. ICBG590 was chosen as the outgroup. The phylogeny is based on the concatenated sequence of the 16S rRNA gene and three housekeeping genes (atpD, dnaA, and gyrA). Bootstrap support values (pink circles) are based on 1000 bootstrap replicates. * genome sequenced strains; Brown squares: LC-MS-based detection of attinimicin in the extract of the corresponding strain; orange squares: PCR-based detection of the att biosynthetic gene cluster; yellow squares: attinimicin producers based on either LC-MS or PCR detection. Gray squares: no detection of attinimicin or the corresponding gene cluster.
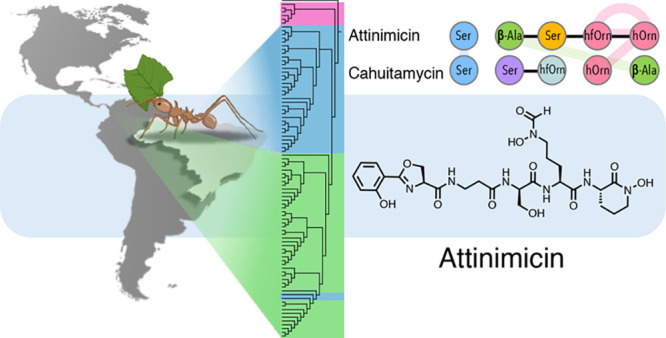
Surprisingly, 62.5% of the strains that showed initial activity in our binary interaction screen produced a metabolite with a mass of m/z 636.2318 [M + H]+ (Supporting Information, Figure S2, Table S3), which indicated a molecular formula of C27H37N7O11 and a classical peptidic fragmentation pattern (Supporting Information, Figure S3). This observation stood out as all previously reported active metabolites from insect-associated bacteria are restricted to one or a few producing strains.14−21 Widespread distribution of a single active product had not been reported. In order to investigate the production of this metabolite, 89 Pseudonocardia spp.—42 from Brazil and 47 from Panama—were grown in small scale, and individual chemical extracts were analyzed by LC-MS to determine if the metabolite is constitutively produced. Intriguingly, the molecule was not detected in any of the extracts from Panamanian strains but was detected in 12 of the 42 Brazilian strains analyzed. Database search (Antibase, NPAtlas, SciFinder) identified two known compounds with an identical exact mass: oxachelin A, produced by Streptomyces sp. GW9/125822 (1), and cahuitamycin A (2), isolated from Streptomyces gandocaensis(23) (Figure 3). Both have the same amino acid composition, albeit a different primary sequence. Intriguingly, tandem mass spectra from the compound produced by the Brazilian ant-associated Pseudonocardia isolates was not consistent with either of the two amino acid sequences reported for 1 and 2, indicating that Pseudonocardia spp. produced a distinct but related metabolite.
Figure 3.
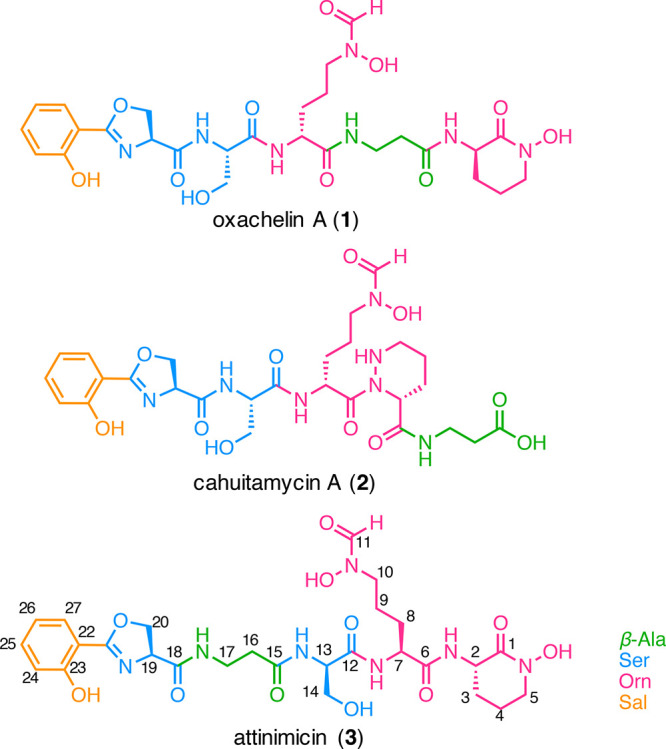
Structures of oxachelin A (1), cahuitamycin A (2), and attinimicin (3).
Two consecutive rounds of reverse phase HPLC fractionation guided by the antifungal activity and the detected mass (m/z 636.2318 [M + H]+) of the compound identified in our binary interactions screen led to the isolation of the novel peptide that we named attinimicin (3) after the tribe Attini to which the ant hosts belong (Figure 3). The 1H NMR spectrum revealed a peptide, with signals corresponding to four amide protons (δH 8.33, 8.23, 8.21, and 7.64) and five α-protons (δH 4.96, 4.92, 4.36, 4.30, and 2.37). In the case of the β-alanine (β-Ala) residue, the α-protons are more shielded as the nitrogen atom is linked to the β-carbon rather than the α-carbon. 1D and 2D NMR spectra suggested the presence of an aromatic ABCD-spin system, showing signals at δH 7.63 (dd), 7.46 (dt), 7.00 (d), and 6.94 (t), with δC 128.1, 134.0, 116.6, and 119.1, respectively. Analysis of the COSY correlations revealed six spin systems, and interpretation of HMBC signals allowed us to elucidate the structure of attinimicin as a linear peptide with an ortho-substituted aromatic ring, an oxazoline ring, a β-Ala residue, a Ser residue, and two modified ornithines—a Nδ-hydroxy-Nδ-formylornithine (N-OH-N-formyl-Orn) and a cyclic Nδ-hydroxyornithine (N-OH-Orn) (Supporting Information, Figures S4–S10, Table S3).
The absolute configuration of the individual amino acids in 3 was determined by Marfey’s method24 (Supporting Information, Figure S11, Table S5). Hydrolysis, derivatization with l-FDAA, and analysis of the derivatized hydrolysate by LC-MS revealed the presence of equal amounts of d-Ser and l-Ser as well as exclusively l-Orn residues. In comparison, oxachelin A and cahuitamycin A exclusively harbor l-Ser, and d-Orn residues, and this pattern is almost completely reversed in attinimicin.
The observed differences between attinimicin, oxachelin A, and cahuitamycin A are likely resulting from different biosynthetic origins. Identifying the putative biosynthetic gene cluster (BGC) for attinimicin biosynthesis would therefore likely not only reveal insights into the evolution of the biosynthetic pathways that give rise to 1, 2, and 3 but also open up the possibility to bioinformatically determine the position of both l/d-configured Ser residues identified by Marfey’s analysis.
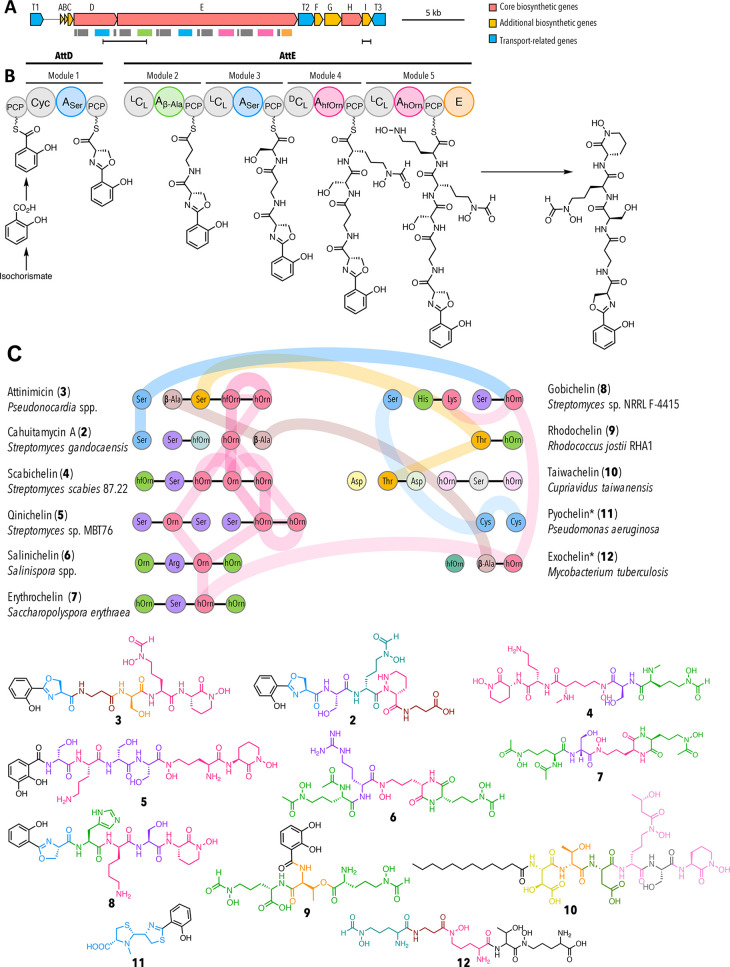
We therefore isolated genomic DNA from eight selected attinimicin producers (* in Figure 2), subjected them to PacBio sequencing, and assembled the genomes using the Canu pipeline (Supporting Information, Table S6).25 The draft genome sequences were mined for a conserved candidate attinimicin BGC present in all sequenced producers using antiSMASH5.0.26 A putative BGC for attinimicin biosynthesis, further referred to as the att BGC, was identified as part of a large biosynthetic island that also harbors a modular polyketide synthase BGC. The involvement of the conserved att BGC in attinimicin biosynthesis is furthermore supported by BiG-SCAPE27 analysis (Supporting Information, Figure S12), which groups gene clusters according to their relatedness in a similarity network. This analysis revealed the att BGC as the only conserved peptide BGC (ribosomal and nonribosomal peptide BGCs) present in the genomes of all sequenced producers. In addition, it shows strong similarity to the BGC responsible for cahuitamycin A biosynthesis. The putative att BGC encodes a nonribosomal peptide synthetase (NRPS) that is the only conserved peptide BGC identified in all sequenced attinimicin producers (Figure 4). The att BGC comprises three NRPS genes that are flanked by genes encoding transporters, and enzymes involved in precursor biosynthesis and tailoring reactions (Figure 4a, Supporting Information, Table S7). The three NRPS proteins harbor in total five elongating modules each of which incorporates one amino acid into the peptide backbone, which is in agreement with the characterized structure of 3. In-silico adenylation domain substrate specificity predictions were furthermore in good agreement with the structure of 3 (Supporting Information, Table S8). The biosynthesis of 3 involves the production of salicylate from isochorismate, likely catalyzed by the salicylate synthase homologue AttG, that serves as the first building block loaded onto the first peptidyl-carrier protein (Figure 4b). The first module putatively elongates the nascent peptide with a l-Ser residue as determined by the adenylation domain specificity and in-silico analysis of the corresponding heterocyclization domain. The heterocyclization domain is likely responsible for oxazoline ring formation. Modules 2 and 3, encoded on attE, are putatively involved in incorporating the β-Ala and d-Ser residues, respectively. The presence of a condensation domain belonging to the DCL class in the subsequent module indicates the incorporation of a d-Ser residue, which is in agreement with our Marfey’s analysis. The lack of a module-encoded epimerization domain in all sequenced strains, and the absence of a dual condensation-epimerization domain in module 3, suggests that l-Ser is either epimerized prior to its incorporation into the nascent peptide or alternatively that the C-terminal epimerization domain remotely epimerizes l-Ser to d-Ser. The final two modules are likely responsible for the incorporation of two l-Orn residues. Both residues are N-hydroxylated to form the characteristic hydroxamate residue putatively catalyzed by the ornithine N-monooxygenase AttA followed by formylation likely catalyzed by the formyl transferase AttF in the case of the first Orn residue and lactam ring formation in the case of the second Orn residue. Intramolecular lactamization is likely the mode of chain release that could be catalyzed by AttI that belongs to the α/β hydrolase superfamily. A similar reaction has been postulated for the biosynthesis of other NRPS-derived siderophores (amychelin, coelichelin, and taiwachelin).28−30 As an alternative, and since the C-terminal E-domain of AttE is not responsible for epimerization of the terminal N–OH-Orn, it is tempting to speculate that it is involved in the release from the assembly line. This scenario is supported by the model for scabichelin biosynthesis.31 The scabichelin BGC likewise encodes an NRPS that harbors a seemingly superfluous C-terminal epimerization domain but does not contain a gene encoding an α/β hydrolase or any other gene involved in peptide chain release. As a consequence, it was speculated that the epimerization domain might be involved in peptide release.31
Figure 4.
Bioinformatic analysis of the putative attinimicin biosynthetic gene cluster. (A) Attinimicin biosynthetic gene cluster. Colored rectangles indicate the domains shown in B. Horizontal black bars represent regions amplified for PCR-based detection of the att BGC (left) and att-like BGC (right); (B) proposed model for attinimicin biosynthesis. A, adenylation domain–amino acids in subscript indicate the monomers incorporated into the nascent peptide; PCP, peptidyl carrier protein; Cyc, cyclization domain; DCL, condensation domain that catalyzes the peptide bond between the terminal d-amino acid of the growing peptide chain and an l-amino acid to be incorporated; LCL, condensation domains condense two l-amino acids; E, epimerization domain; (C) Proposed evolutionary relatedness between the attinimicin and other siderophore NRPSs based on phylogenetic analysis of adenylation domains (for a detailed representation see Supporting Information, Figure 14). Only relationships between attinimicin and other clusters are shown. Horizontal black lines represent domains encoded on the same gene; colored vertical lines and colored horizontal connections represent domains that are evolutionarily related and/or duplicated.
The similarity of attinimicin with cahuitamycin A and oxachelin A suggests that all three corresponding BGCs have evolved from a common ancestor. While the cahuitamycin BGC has recently been reported,23 the BGC responsible for oxachelin is still elusive. Phylogenetic analysis of all domains encoded in the attinimicin and cahuitamycin BGCs along with a representative set of homologous domains retrieved from the antiSMASH database32 and all publicly available NRPS hydroxamate-siderophore BGCs point to the following evolutionary relationship between both pathways: (1) modules of the att and cah NRPSs have been swapped. This is the case for both the β-Ala incorporating module 2 of the att NRPS, which corresponds to module 5 in the cah NRPS as well as the Ser-incorporating module 1 in the att NRPS corresponding to module 1 in the cah NRPS (Figure 4c). (2) The modules responsible for Orn incorporation appear to have different origins: While the module responsible for the incorporation of the first Orn residue is conserved between both NRPSs, the second module has a different origin in both pathways. In the case of the cah NRPS, the second Orn-incorporating module was likely acquired from elsewhere as indicated by our phylogenetic analysis. In the case of the att NRPS, however, the second Orn-incorporating module is a result of a likely duplication event in which the adenylation domain of the second Orn-incorporating module is a duplication of the upstream Orn-specific adenylation domain (92% sequence identity) (Supporting Information, Figure S13). The condensation domain of module 5, on the other hand, seems to have evolved from the duplication of the condensation domain encoded in module 3 rather than the other Orn-incorporating module 4 (Supporting Information, Figure S14).
On the basis of the biosynthetic insights gained, we next explored whether the number of producing strains identified in our initial metabolomic analysis is in agreement with the number of strains that harbor the attinimicin or a related BGC. We therefore designed two sets of degenerate primers for the detection of genes that are (1) likely conserved between the attinimicin and related BGCs such as the α/β hydrolase or (2) unique for the biosynthesis of attinimicin, such as the NRPS fragment that spans the adenylation domain with substrate specificity for β-Ala two modules upstream of the adenylation domain specific for l-Orn. Both primer pairs were used to screen all of the Brazilian and Panamanian Pseudonocardia isolates. No att-like BGC was detected in the Panamanian strains. This result is in agreement with genome mining studies of 44 publicly available ant-associated Pseudonocardia genomes from bacteria isolated in Panama that were likewise devoid of an att or cah-like BGC (Figure 2). In total, 73% of Brazilian strains were positive for both primer pairs suggesting the presence of att-like BGCs. This result indicates that the att BGC is present in significantly more genomes than indicated by our metabolomic analysis, suggesting that some att-like BGCs might be silent under standard laboratory culture conditions or produce attinimicin at concentrations below our detection limit (Figure 2). Finally, we wanted to determine whether 3 could be detected in situ, on collected Acromyrmex ants and within fungal gardens at Itatiaia National Park (RJ, Brazil). Both sample types were extracted and subjected to LC-MS/MS analysis, and attinimicin was detected in both sample types in 12 out of the 31 samples (38.7%) (Supporting Information, Table S9), as revealed by comparison with an authentic standard and matching fragmentation patterns. The in situ detection of 3 in more than one-third of all samples indicates that attinimicin is produced in its natural habitat.
Such high conservation of a secondary metabolite and clear in situ production indicates that attinimicin likely plays an important ecological role for either the producing bacterium or ant host. We therefore explored two likely ecological functions: (1) Attinimicin is an iron chelator (siderophore) based on its structural similarity to reported siderophores with 2-hydroxybenzoyl-oxazoline and hydroxamate residues. Iron chelation could provide either the ant host or its fungal cultivar with iron. And (2) Attinimicin’s antifungal activity protects the fungal cultivar from infections by pathogenic fungi.
The affinity of attinimicin toward iron was quantified by EDTA titration33 resulting in a moderate pFeIII value of 18.57 ± 0.81 (Supporting Information, Figure S15), which is in the range of structurally related compounds such as cahuitamycin but significantly lower than siderophores produced by other bacteria or used to treat acute iron poisoning (Supporting Information, Table S10). To test the metal ion selectivity of 3, attinimicin was incubated with a suite of different metals. Attinimicin chelates Ga3+ (Supporting Information, Figure S16), but does not coordinate Ca2+, Zn2+, Cu2+, and Mg2+. When incubated with a mixture containing all metals in equimolar ratio, attinimicin showed a strong selectivity for iron (Supporting Information, Figure S17).
To test our second hypothesis, we evaluated the biological activity of attinimicin against three different pathogens of the fungal cultivar. While iron-bound attinimicin had no inhibitory activity against any of three Escovopsis spp. tested, iron-depleted attinimicin showed moderate activity (MIC 12.5 μg/mL) against all three pathogens (Figure 5, Supporting Information, Figure S18). Notably, it did not show any activity against the fungal cultivar Leucoagaricus gongylophorus ICBG107 independent of the presence or absence of iron (Supporting Information, Figure S19).
Figure 5.
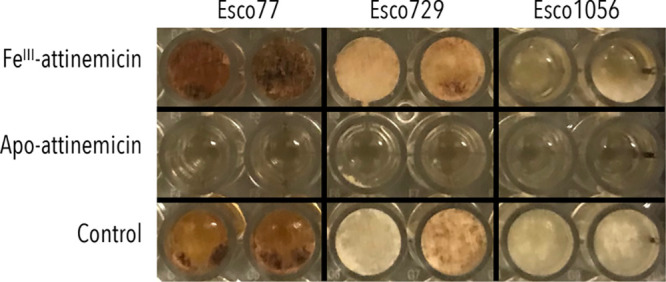
In vitro antifungal activity of FeIII-attinimicin and apo-attinimicin (100 μg).
Since attinimicin’s inhibitory activity against ant nest pathogens was dependent on the absence of iron, we reasoned that it might show in vivo activity as iron is almost exclusively bound to hemoglobin, myoglobin, transferrin, or ferritin in mammals, all of which have higher iron-binding affinities than attinimicin. We therefore tested both the iron-bound as well as iron-depleted attinimicin in a neutropenic mouse disseminated candidiasis model.34,35 In short, neutropenia was chemically induced, the compromised mice injected with Candida albicans, and the infected mice treated with attinimicin, clinically relevant antifungals or buffer. Mice were subsequently sacrificed, and C. albicans CFUs were determined from kidneys. These studies confirmed our in vitro studies and showed that the iron-bound form of attinimicin is inactive. Treatment with the iron-depleted attinimicin formulation, on the other hand, resulted in a significant reduction of fungal burden in the range of the clinically relevant antifungal triazoles (Supporting Information, Figure S20).36,37
Conclusion
The fungus-growing ant symbiotic system is a remarkable evolutionary success story beginning with a single species some 50 million years ago and expanding to over 200 species today. Fungus-growing ants have also become the dominant herbivores in the planet’s neotropical regions. Studies on the molecular exchanges that maintain the multilateral symbiotic system have both informed chemical ecology and provided new molecules and potential therapeutic agents. This study was designed to provide a systematic molecularly based survey of Brazilian attine ants, and the unexpected but major finding is a broadly dispersed and previously unreported antifungal agent: attinimicin.
Attinimicin was discovered through a phenotypic screen for antifungal compounds produced by the bacterial symbionts of fungus-growing ant nests. Correlation studies in combination with in depth bioinformatic analysis suggest that it is produced by the att NRPS pathway, and its structure and biosynthetic pathway are related to several previously reported NRPS-derived molecules, especially cahuitamycin, which also has a defined biosynthetic gene cluster. Attinimicin’s wide geographic distribution distinguishes it from other antifungal metabolites that have been identified from the fungus-growing ant system.
Attinimicin’s broad geographic distribution appears to be limited to Brazil. Its absence in Pseudonocardia isolates from Panama suggests, as does our phylogenetic analyses, that Brazilian and Panamanian ants have established and coevolved symbiotic relationships with different subfamilies of the genus Pseudonocardia. Comparison of the att NRPS with the cah NRPS suggests a common biosynthetic ancestor and that the differences between both pathways are likely a result of module swapping, the duplication of domains, and the loss of a single module or the gain of a module from elsewhere.
There were two findings in our studies on attinimicin that, while not germane to the main story, are worth noting. The first is attinimicin’s potent in vivo activity but negligible in vitro activity. This differential activity is probably related to the second unexpected finding in our study: attinimicin’s antifungal activity is iron-dependent. It is only active when not bound to iron. There are many examples of iron-dependent antibiotic activity, but in most of them the antibiotic functions as a siderophore; it binds iron tightly and prevents the target organism from accessing it. The structural conservation of residues involved in iron-chelation between cahuitamicin, oxachelin, and attinimicin suggests that iron binding is an important feature, but its moderate iron binding affinity (pFeIII ≈ 19) does not seem consistent with that mechanism. Enterobactin, a typical bacterial siderophore, has a pFeIII of 36.6, making it a 1017 tighter iron binder.30 It is possible that attinimicin retards growth rather than kills as part of a strategy to limit resistance development by decreasing selective pressure.38 In addition, attinimicin is likely one component in a suite of antifungal molecules. Determining attinimicin’s mechanism of action will require further studies, but its in vivo ability to reduce fungal burdens in mammals as well as clinically used therapeutics could justify such studies.
Acknowledgments
This work was supported by the São Paulo Research Foundation (FAPESP) #2015/26349-5 (T.T.H.F.), #2017/17305-0 (T.T.H.F.), #2015/01001-6 (W.G.P.M.) #2013/50954-0 (M.T.P.), and the Coordenação de Aperfeiçoamento de Pessoal de Nivel Superior, Brasil (CAPES), Finance Code 001. This work was funded by #U19TW009872 and R01AT009874 (J.C.). We thank Munhyung Bae for the help with the mass spectrometry data and Allison Walker for the help with the bioinformatics. We thank the Information Technology Superintendence at the University of São Paulo for HPC resources. We thank both the Analytical Chemistry Core (ACC) facility in the Biological Chemistry and Molecular Pharmacology Department within Harvard Medical School for analytical support and the East Quad NMR facility at Harvard Medical School for NMR support. We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Shared Resource, which provided sequencing service. We thank Zhu Zhuo, Shannan Ho Sui, and the Harvard Chan Bioinformatics Core, for the genome assembly. E.J.N.H. acknowledges funding from a Postdoc Mobility fellowship granted by the Swiss National Science Foundation. M.T.P. also thanks CNPq for research fellowship #303792/2018-3.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscentsci.0c00978.
Experimental Methods, MS and NMR data, derivatization, bioassays, and other biological assays (PDF)
Author Contributions
∇ T.T.H.F. and E.J.N.H. contributed equally to this work.
Author Contributions
T.T.H.F., E.J.N.H., E.E.M., W.G.P.M., D.R.A., C.R.C., M.T.P., and J.C. designed the research, T.T.H.F., E.J.N.H., E.E.M., W.G.P.M., and E.B.V.A. conducted experiments and analyzed data, and T.T.H.F., E.J.N.H., E.E.M., M.T.P., and J.C. wrote the manuscript with contributions from all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Mueller U. G.; Schultz T. R.; Currie C. R.; Adams R. M. M.; Malloch D. The Origin of the Attine Ant-Fungus Mutualism. Q. Rev. Biol. 2001, 76 (2), 169–197. 10.1086/393867. [DOI] [PubMed] [Google Scholar]
- Sands W. A. Some Factors Affecting the Survival of Odontotermes Badius. Insectes Soc. 1956, 3 (4), 531–536. 10.1007/BF02226456. [DOI] [Google Scholar]
- Klepzig K. D.; Wilkens R. T. Competitive Interactions among Symbiotic Fungi of the Southern Pine Beetle. Appl. Environ. Microbiol. 1997, 63 (2), 621–627. 10.1128/AEM.63.2.621-627.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menezes C.; Vollet-Neto A.; Marsaioli A. J.; Zampieri D.; Fontoura I. C.; Luchessi A. D.; Imperatriz-Fonseca V. L. A Brazilian Social Bee Must Cultivate Fungus to Survive. Curr. Biol. 2015, 25 (21), 2851–2855. 10.1016/j.cub.2015.09.028. [DOI] [PubMed] [Google Scholar]
- Paludo C. R.; Menezes C.; Silva-Junior E. A.; Vollet-Neto A.; Andrade-Dominguez A.; Pishchany G.; Khadempour L.; Do Nascimento F. S.; Currie C. R.; Kolter R.; Clardy J.; Pupo M. T. Stingless Bee Larvae Require Fungal Steroid to Pupate. Sci. Rep. 2018, 8 (1), 1–10. 10.1038/s41598-018-19583-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz T. R.; Meier R. A Phylogenetic Analysis of the Fungus-growing Ants (Hymenoptera: Formicidae: Attini) Based on Morphological Characters of the Larvae. Syst. Entomol. 1995, 20 (4), 337–370. 10.1111/j.1365-3113.1995.tb00100.x. [DOI] [Google Scholar]
- Branstetter M. G.; Ješovnik A.; Sosa-Calvo J.; Lloyd M. W.; Faircloth B. C.; Brady S. G.; Schultz T. R.. Dry Habitats Were Crucibles of Domestication in the Evolution of Agriculture in Ants. Proc. R. Soc. London, Ser. B 2017, 284 ( (1852), ).20170095. 10.1098/rspb.2017.0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S. E.; Rabeling C.; Sosa-Calvo J.; Lopes C. T.; Rodrigues A.; Vasconcelos H. L.; Bacci M.; Mueller U. G.; Schultz T. R. The Molecular Phylogenetics of Trachymyrmex Forel Ants and Their Fungal Cultivars Provide Insights into the Origin and Coevolutionary History of ‘Higher-Attine’ Ant Agriculture. Syst. Entomol. 2019, 44 (4), 939–956. 10.1111/syen.12370. [DOI] [Google Scholar]
- Winston M. E.; Kronauer D. J. C.; Moreau C. S. Early and Dynamic Colonization of Central America Drives Speciation in Neotropical Army Ants. Mol. Ecol. 2017, 26 (3), 859–870. 10.1111/mec.13846. [DOI] [PubMed] [Google Scholar]
- Montes C.; Cardona A.; Jaramillo C.; Pardo A.; Silva J. C.; Valencia V.; Ayala C.; Pérez-Angel L. C.; Rodriguez-Parra L. A.; Ramirez V.; Niño H. Middle Miocene Closure of the Central American Seaway. Science 2015, 348 (6231), 226–229. 10.1126/science.aaa2815. [DOI] [PubMed] [Google Scholar]
- Bacon C. D.; Silvestro D.; Jaramillo C.; Smith B. T.; Chakrabarty P.; Antonelli A. Biological Evidence Supports an Early and Complex Emergence of the Isthmus of Panama. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (19), 6110–6115. 10.1073/pnas.1423853112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C. R.; Stuart A. E. Weeding and Grooming of Pathogens in Agriculture by Ants. Proc. R. Soc. London, Ser. B 2001, 268 (1471), 1033–1039. 10.1098/rspb.2001.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie C. R.; Scott J. a.; Summerbell R. C.; Malloch D. Fungus-Growing Ants Use Antibiotic-Producing Bacteria to Control Garden Parasites. Nature 1999, 398 (April), 701–704. 10.1038/19519. [DOI] [Google Scholar]
- Haeder S.; Wirth R.; Herz H.; Spiteller D. Candicidin-Producing Streptomyces Support Leaf-Cutting Ants to Protect Their Fungus Garden against the Pathogenic Fungus Escovopsis. Proc. Natl. Acad. Sci. U. S. A. 2009, 106 (12), 4742–4746. 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barke J.; Seipke R. F; Gruschow S.; Heavens D.; Drou N.; Bibb M. J; Goss R. J.; Yu D. W; Hutchings M. I A mixed community of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol. 2010, 8 (1), 109. 10.1186/1741-7007-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenian I.; Spiteller M.; Ghaste M.; Wirth R.; Herz H.; Spiteller D. Chemical Basis of the Synergism and Antagonism in Microbial Communities in the Nests of Leaf-Cutting Ants. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (5), 1955–1960. 10.1073/pnas.1008441108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipke R. F.; Barke J.; Brearley C.; Hill L.; Yu D. W.; Goss R. J. M.; Hutchings M. I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS One 2011, 6 (8), e22028. 10.1371/journal.pone.0022028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D.-C.; Poulsen M.; Currie C. R.; Clardy J. Dentigerumycin: A Bacterial Mediator of an Ant-Fungus Symbiosis. Nat. Chem. Biol. 2009, 5 (6), 391–393. 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr G.; Derbyshire E. R.; Caldera E.; Currie C. R.; Clardy J. Antibiotic and Antimalarial Quinones from Fungus-Growing Ant-Associated Pseudonocardia sp. J. Nat. Prod. 2012, 75 (10), 1806–1809. 10.1021/np300380t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arnam E. B.; Ruzzini A. C.; Sit C. S.; Horn H.; Pinto-Tomás A. A.; Currie C. R.; Clardy J. Selvamicin, an Atypical Antifungal Polyene from Two Alternative Genomic Contexts. Proc. Natl. Acad. Sci. U. S. A. 2016, 113 (46), 12940–12945. 10.1073/pnas.1613285113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Arnam E. B.; Ruzzini A. C.; Sit C. S.; Currie C. R.; Clardy J. A Rebeccamycin Analog Provides Plasmid-Encoded Niche Defense. J. Am. Chem. Soc. 2015, 137 (45), 14272–14274. 10.1021/jacs.5b09794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag B.; Gerlitz M.; Paululat T.; Rasser H.-F.; Grun-Wollny I.; Hansske F. G. Oxachelin, a Novel Iron Chelator and Antifungal Agent from Streptomyces sp. GW9/1258. J. Antibiot. 2006, 59 (10), 659–663. 10.1038/ja.2006.88. [DOI] [PubMed] [Google Scholar]
- Park S. R.; Tripathi A.; Wu J.; Schultz P. J.; Yim I.; McQuade T. J.; Yu F.; Arevang C. J.; Mensah A. Y.; Tamayo-Castillo G.; Xi C.; Sherman D. H. Discovery of Cahuitamycins as Biofilm Inhibitors Derived from a Convergent Biosynthetic Pathway. Nat. Commun. 2016, 7, 1–11. 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfey P. Determination of D-Amino Acids. II. Use of a Bifunctional Reagent, 1,5-Difluoro-2,4-Dinitrobenzene. Carlsberg Res. Commun. 1984, 49 (6), 591–596. 10.1007/BF02908688. [DOI] [Google Scholar]
- Koren S.; Walenz B. P.; Berlin K.; Miller J. R.; Bergman N. H.; Phillippy A. M. Canu: Scalable and Accurate Long-Read Assembly via Adaptive κ-Mer Weighting and Repeat Separation. Genome Res. 2017, 27 (5), 722–736. 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Steinke K.; Villebro R.; Ziemert N.; Lee S. Y.; Medema M. H.; Weber T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47 (W1), W81–W87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Munoz J. C.; Selem-Mojica N.; Mullowney M. W.; Kautsar S. A.; Tryon J. H.; Parkinson E. I.; De Los Santos E. L. C.; Yeong M.; Cruz-Morales P.; Abubucker S.; Roeters A.; Lokhorst W.; Fernandez-Guerra A.; Cappelini L. T. D.; Goering A. W.; Thomson R. J.; Metcalf W. W.; Kelleher N. L.; Barona-Gomez F.; Medema M. H. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 2020, 16 (1), 60–68. 10.1038/s41589-019-0400-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R.; Traxler M. F.; Zheng S. L.; Kolter R.; Clardy J. Structure and Biosynthesis of Amychelin, an Unusual Mixed-Ligand Siderophore from Amycolatopsis sp. AA4. J. Am. Chem. Soc. 2011, 133 (30), 11434–11437. 10.1021/ja203577e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis G. L.; Ravel J. Coelichelin, a New Peptide Siderophore Encoded by the Streptomyces coelicolor Genome: Structure Prediction from the Sequence of Its Non-Ribosomal Peptide Synthetase. FEMS Microbiol. Lett. 2000, 187 (2), 111–114. 10.1111/j.1574-6968.2000.tb09145.x. [DOI] [PubMed] [Google Scholar]
- Kreutzer M. F.; Nett M. Genomics-Driven Discovery of Taiwachelin, a Lipopeptide Siderophore from Cupriavidus taiwanensis. Org. Biomol. Chem. 2012, 10 (47), 9338–9343. 10.1039/c2ob26296g. [DOI] [PubMed] [Google Scholar]
- Kodani S.; Bicz J.; Song L.; Deeth R. J.; Ohnishi-Kameyama M.; Yoshida M.; Ochi K.; Challis G. L. Structure and Biosynthesis of Scabichelin, a Novel Tris-Hydroxamate Siderophore Produced by the Plant Pathogen Streptomyces scabies 87.22. Org. Biomol. Chem. 2013, 11 (28), 4686–4694. 10.1039/c3ob40536b. [DOI] [PubMed] [Google Scholar]
- Blin K.; Medema M. H.; Kottmann R.; Lee S. Y.; Weber T. The AntiSMASH Database, a Comprehensive Database of Microbial Secondary Metabolite Biosynthetic Gene Clusters. Nucleic Acids Res. 2017, 45 (D1), D555–D559. 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abergel R. J.; Zawadzka A. M.; Raymond K. N. Petrobactin-Mediated Iron Transport in Pathogenic Bacteria: Coordination Chemistry of an Unusual 3,4-Catecholate/Citrate Siderophore. J. Am. Chem. Soc. 2008, 130 (7), 2124–2125. 10.1021/ja077202g. [DOI] [PubMed] [Google Scholar]
- Zhao M.; Lepak A. J.; Andes D. R. Animal Models in the Pharmacokinetic/Pharmacodynamic Evaluation of Antimicrobial Agents. Bioorganic. Bioorg. Med. Chem. 2016, 24 (24), 6390–6400. 10.1016/j.bmc.2016.11.008. [DOI] [PubMed] [Google Scholar]
- Pfaller M. A.; Diekema D. J. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 2007, 20 (1), 133–163. 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepak A. J.; Marchillo K.; VanHecker J.; Diekema D.; Andes D. R. Isavuconazole Pharmacodynamic Target Determination for Candida Species in an in Vivo Murine Disseminated Candidiasis Model. Antimicrob. Agents Chemother. 2013, 57 (11), 5642–5648. 10.1128/AAC.01354-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andes D. In Vivo Pharmacodynamics of Antifungal Drugs in Treatment of Candidiasis. Antimicrob. Agents Chemother. 2003, 47 (4), 1179–1186. 10.1128/AAC.47.4.1179-1186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baym M.; Stone L. K.; Kishony R. Multidrug Evolutionary Strategies to Reverse Antibiotic Resistance. Science 2016, 351 (6268), aad3292. 10.1126/science.aad3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.