Abstract
The standard anatomical brain template provides a common space and coordinate system for visualizing and analyzing neuroimaging data from large cohorts of subjects. Previous templates and atlases for the common marmoset brain were either based on data from a single individual or lacked essential functionalities for neuroimaging analysis. Here, we present new population-based in-vivo standard templates and tools derived from multi-modal data of 27 marmosets, including multiple types of T1w and T2w contrast images, DTI contrasts, and large field-of-view MRI and CT images. We performed multi-atlas labeling of anatomical structures on the new templates and constructed highly accurate tissue-type segmentation maps to facilitate volumetric studies. We built fully featured brain surfaces and cortical flat maps to facilitate 3D visualization and surface-based analyses, which are compatible with most surface analyzing tools, including FreeSurfer, AFNI/SUMA, and the Connectome Workbench. Analysis of the MRI and CT datasets revealed significant variations in brain shapes, sizes, and regional volumes of brain structures, highlighting substantial individual variabilities in the marmoset population. Thus, our population-based template and associated tools provide a versatile analysis platform and standard coordinate system for a wide range of MRI and connectome studies of common marmosets. These new template tools comprise version 3 of our Marmoset Brain Mapping Project and are publicly available via marmosetbrainmapping.org/v3.html.
Keywords: Magnetic resonance imaging, Diffusion tensor imaging, Computed tomography, Callithrix jacchus, Population templates, Brain surfaces
1. Introduction
The common marmoset (Callithrix jacchus), a New World primate, has rapidly emerged as a promising animal model for biomedical and neuroscience research (Okano et al., 2012). Marmosets are one of the smallest non-human primates, featuring lissencephalic brains, which provide practical advantages for comprehensively mapping brain circuits both in health and disease models (Lin et al., 2019; Majka et al., 2016). Because they are phylogenetically closer to humans, marmosets allow for studying higher cognitive brain function and human brain disorders that rodents cannot adequately model (Buckner and Margulies, 2019). With a shorter life span and higher reproduction rates than macaques, the marmoset offers strategic advantages in the development of non-human primate models for human brain diseases, such as autism (Zhao et al., 2018), multiple sclerosis (Lee et al., 2018), stroke (Le Gal et al., 2018), Parkinson’s disease (Hikishima et al., 2015), and Alzheimer’s disease (Philippens et al., 2017). Neuroimaging techniques, including magnetic resonance imaging (MRI) (Silva, 2017), are essential for translational and preclinic studies with animal models. However, as a relatively new animal model, neuroimaging tools currently available for the marmoset are far from well developed.
One of the essential tools in urgent need of development for studies of the marmoset brain is a population-based template and surface-based data analysis tool. The human brain has been well-known for its large inter-individual variabilities (Mueller et al., 2013; Reardon et al., 2018). Because each individual has a different brain size, shape, and gyrification, cross-subject analyses require the spatial normalization of individual brains into a common template space. The template is usually constructed by averaging the images of many subjects to represent the neuroanatomical features of the population. For humans and macaques, various kinds of population-based templates and atlases were developed to serve different purposes, ranging from volumetric structural templates (Evans et al., 1993; Lv et al., 2020; Seidlitz et al., 2018) to surface-based templates (Glasser et al., 2016; Van Essen, 2005). Compared to the abundantly available template tools designed for humans and macaques, population-based tools for marmosets are scarce (Hikishima et al., 2011). Because they are outbred, marmosets have a diverse genetic background and present substantial individual variability in brain shape, size, and regional structural volumes (Fig. 1). While one population-averaged template was developed for common marmosets (Hikishima et al., 2011), this template lacks many essential features that limit its applications, including few MRI contrasts, inaccurate tissue segmentation, and no surface-based analysis tools. Thus, a comprehensive population template and surface-based tool for marmosets is of pressing need to facilitate data analyses in neuroimaging studies.
Fig. 1. Individual variabilities of adult common marmosets.
(A) Sagittal view of the marmoset brain size distribution in our marmoset population (N = 27). The top row shows the intracranial volume mask (red) of the population-averaged template overlaid on CT images, and the bottom row shows the template brain mask (red) overlaid on T2w MRI. From left to right, the underlay images are population-averaged templates, the images from the animal with the smallest brain (a 4-year-old male), and the images from the animal with the largest brain (also a 4-year-old male). (B) Violin and box plots of brain volume distributions across the population. The box plot shows the first quartile to the third quartile and the median. The red point represents the mean value, and the gray points represent the volume of each animal. Plots are drawn for all animals (gray violin plots), as well as for females (red violins) and males (blue violins).
The Marmoset Brain Mapping Project (marmosetbrainmapping.org) aims to develop comprehensive brain atlases and tools to promote neuroimaging and connectome studies of marmosets. The project has already released two versions of the atlases, one focused on the cortex (Liu et al., 2018) and the other on white matter pathways (Liu et al., 2020). However, these two previous versions were based on only a few marmoset brain samples and are more suited for high-detailed neuroanatomical analyses than for in-vivo studies of large cohorts of animals. To address this problem, we present here the “populational multi-modal standard volumetric and surface-based templates.” We collected in-vivo multi-modal MRI and CT data of 27 marmosets from our colony at the NIH and constructed population-averaged templates and surface-based analyzing tools. This new template set features the most comprehensive image modalities, accurate (manually corrected) tissue segmentation maps, multi-atlas labeled template space, and fully featured brain surface and flat maps for surface-based analysis. These templates and associated tools comprise version 3 of our Marmoset Brain Mapping Project. They are publicly available via https://marmosetbrainmapping.org/atlas.html and will significantly aid in a wide range of neuroimaging and connectome studies that involve across-subject analysis.
2. Materials and methods
2.1. Animals and data collection
All procedures were approved by the Animal Care and Use Committee of the National Institute of Neurological Disorders and Stroke. Twenty-seven healthy adult marmosets (20 males and 7 females) were recruited for the study (detailed information shown in Supplementary Table S1). The animals fasted for 12 h before MRI scanning. The animals were anesthetized with an intramuscular injection of 10 mg/kg ketamine, orally intubated, and ventilated with 1.5% - 2% isoflurane. The animals were placed in an MR-compatible cradle, and their heads were held to a stereotaxic frame by two ear bars coated with 2% lidocaine jelly. Vital signs, including heart rate, end-tidal CO2, SPO2, and rectal temperature (maintained at 38.5°C with a water heating pad), were monitored and maintained at physiological values throughout the MRI scanning.
MRI scanning was performed in a 7T/300 mm magnet (Bruker, Billerica, USA) equipped with a 150 mm gradient set (450 mT/m strength and 150 μs rising time; Resonance Research Inc., Billerica, USA), a 16-rung high-pass birdcage radiofrequency coil for transmission, and a custom-built 8-channel phased-array coil for reception (Silva, 2017). Multi-modal images were collected using the following sequences:
T1-weighted images were collected with a 3D MP3RAGE sequence: TR = 6 s, TE = 2.8 ms, flip angle = 12°, FOV = 36 × 28 × 24 mm, matrix size = 144 × 112 × 96, three inversion times (TI1 = 1200 ms, TI2 = 2600 ms, and TI3 = 4000 ms), and the scanning duration was 9 min 42 s. The image with T1 = 1200 ms was used as the T1w contrast. Five to six images were scanned and averaged for each animal. Thus, the total scanning time for each animal was about 50 min to 60 min.
T2-weighted images were collected with a 2D RARE sequence: TR = 30 s, TE = 8 s with three effective TE (TE1 = 16 ms, TE2 = 48 ms and TE3 = 80 ms), FOV = 36 × 28 × 24 mm, matrix size = 144 × 112, slice thickness = 0.25 mm, and the scanning duration was about 10 min 30 s. Three different effective TEs provided different levels of T2-w contrasts. Five to six images were scanned and averaged for each animal. Thus, the total scanning time for each animal was about 50 min to 60 min.
Multishell diffusion MRI data were collected with a 2D diffusion-weighted spin-echo EPI sequence: TR = 5.1 s, TE = 38 ms, number of segments = 88, FOV = 36 × 28 mm, matrix size = 72 × 56, slice thickness = 0.5 mm, a total of 400 DWI images for two phase encodings (blip-up and blip-down) and each has three b values (8 b = 0, 64 b =2400, and 128 b = 4800), and the scanning duration was about 34 min. The multishell gradient sampling scheme was generated by the Q-shell sampling method (Caruyer et al., 2013).
Large-FOV whole head images were collected with a 3D FLASH sequence: TR = 10 s, TE = 1.6 ms, flip angle = 6°, FOV = 64 × 51.2 × 64 mm, matrix size = 160 × 128 × 160, and the scanning duration was 27 min 18 s.
Also, whole head CT images were collected with a SkyScan 1276 Micro-CT (Bruker, Billerica, USA), with an isotropic resolution of 0.288 mm and scanning time of one minute per animal.
2.2. Construction of volumetric templates
2.2.1. Creation of population-averaged templates
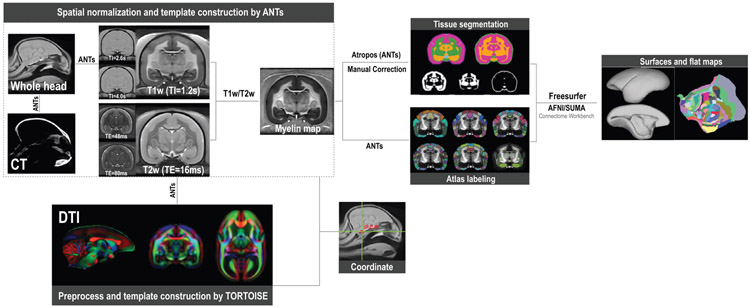
Fig. 2 shows the processing pipeline for creating the multi-modal population-averaged templates. The population-averaged templates of the MRI and CT data were constructed using the ANTs software (Avants et al., 2009). A multimodal template of the marmoset brain was first created by all T1w (TI = 1200 ms) and T2w (TE = 16 ms) images using the antsMultivariateTemplateConstruction2.sh function. The function created initial templates by averaging all raw individual images and then updated the templates iteratively using multivariate information from different image modalities. The final outputs included the multivariate templates and the transformation information from subject spaces to the template space. The transformation information was applied to all MP3RAGE images (TI = 1200 ms, 2600 ms, and 4000 ms) and all RARE (TE = 16 ms, 48 ms, and 80 ms) images, and the transformed images were averaged to create the template for each modality. The default averaging method of the ANTs script involved inversion of both the average diffeomorphism and the blurring induced by intensity averaging (soft sharpening), which produced a sharper template (Avants et al., 2010).
Fig. 2. Pipeline for creating the population multi-modal volumetric and surface-based templates.
We used ANTs to create the population-averaged templates of structural MRI images and CT images, and TORTOISE to preprocess (eddy-current distortion and EPI distortion correction) and construct the template of DTI data. Different template modalities are co-registered to the same coordinate space. Initial automatic segmentation (by ANTs/Atropos) followed by manual correction provides accurate tissue-type segmentation, and multiple atlases were fused into the template space. Finally, a complete set of brain surfaces and flat maps was generated by a customized FreeSurfer pipeline as well as by AFNI/SUMA and connectome workbench.
The MRI and CT whole head templates were constructed separately by the same function as above. The brain of the whole head MRI template (FLASH sequence) was manually extracted and nonlinearly registered to the brain of the MP3RAGE-TI2 template, which shared similar contrasts. The resulting transformation was applied to the whole head MRI template, including a linear transformation on the whole image and a non-linear warp on the brain part so that the whole head image would be in the same template space as the T1w and T2w brain templates. We then reversed the contrast of the whole head MRI templates and manually delineated the bone area to create a sham CT-like image, to which the CT whole head template was registered.
The DTI template was created using TORTOISE (Pierpaoli et al., 2010), one of the few software packages that provide a complete pipeline of DTI data preprocessing and tensor-based template construction. In brief, all DTI data were preprocessed by DIFF_PREP and DR_BUDDI, which incorporated eddy-current distortion and EPI distortion correction. The DR_BUDDI routine utilized pairs of diffusion data sets acquired with opposite phase encoding (blip-up and blip-down) and the T2w image for the EPI distortion correction and merged the preprocessed pairs into one dataset. After data preprocessing, the DTI template was created by DR_TAMAS and non-linearly transformed into the T1w template.
All templates were resampled to 0.2 mm isotropic resolution and co-registered to the same coordinate space, which was defined by the “eye-bars and ear-bars” axis. The origin was set at the intersection between the “eye-bars and ear-bars” plane and the mid-line plane. The two ear bars were visible in MRI whole head images, and the location of the ear bars was estimated based on the skull CT images. By combining information from MRI and CT data, the templates were rotated so that the eye-bars and the ear-bars were on the same plane.
2.2.2. Tissue segmentation and probability maps
A “myelin map ” was created from the ratio of T1w (TI = 1200 ms)/T2w (TE = 16 ms) templates (Glasser and Van Essen, 2011), which corrected intensity inhomogeneity and enhanced contrasts between the gray matter (GM) and the white matter (WM). The initial automatic segmentation was conducted on the “myelin map” by the Atropos algorithm of the ANTs software (Avants et al., 2011b), which segmented the image into three tissue types – the GM, the WM, and the cerebrospinal fluid (CSF). As the automatic method was inaccurate for the marmoset data, we had to correct the initial results manually and segment images into three tissue types (the GM, the WM, and the CSF) and six tissue types (the cortical GM, the subcortical and cerebellar GM, the WM, the CSF, the major vasculature, and the skull). These manually corrected tissue segments were then transformed into the image space of each animal by using the transformation files generated from the previous template creation. Automatic segmentation was performed on the “myelin map” of each animal by the Atropos algorithm with the manually corrected segments as the prior probability images (weight = 0.25). For each tissue type, the resulting probability maps of all animals were transformed back into the template space and averaged to generate the final template tissue probability maps.
2.2.3. Atlases fusion
We fused different brain atlases into the template space, including the Marmoset Brain Mapping Atlas V1 (MBM-V1) atlas (Liu et al., 2018) and the Riken Brain/MINDS cortical brain atlas (Riken) (Woodward et al., 2018). The MBM-V1 included four different types of cortical atlases (cortical parcellation into 13-, 54-, and 106-areas, as well as the Paxinos parcellation). It also included a coarse subcortical atlas (Liu et al., 2018). The MTR template of the MBM-V1 atlas and the T2w template of the Riken atlas were transformed to the T1w and T2w template of this study, respectively, by the antsRegistrationSyn.sh function of ANTs (Avants et al., 2011a). Based on the resulting transformation files, atlas labeling images were fused into the template space by the antsApplyTransforms function with the MultiLabel interpolation method.
2.3. Construction of brain surfaces and flat maps
2.3.1. Brain surfaces by the AFNI/SUMA and Connectome Workbench
The cortical GM mask, the WM mask, and the skull mask of the CT template were used to generate the pial surface, the WM surface, and the skull surface, respectively, by the Isosurface function of AFNI/SUMA (Saad et al., 2004). The pial and WM surfaces were further smoothed and inflated by the Connectome Workbench Command (Marcus et al., 2011). All surfaces were converted into GIFTI format to provide compatibility with different software.
2.3.2. Brain surfaces and flat maps by FreeSurfer
The T1w templates, tissue segmentation maps, and atlases were modified to conform to the requirements of FreeSurfer (Fischl, 2012), including i. resampling the orientation to RSP, ii. adding planes to make the image matrix to 256 × 256 × 256, and iii. modifying the resolution information in the image header to 1 mm x 1 mm x 1 mm.
We combined the WM segment and the subcortical GM, manually created hemispheric cutting planes, and filled WM with specific values for subsequent surface tessellation (equivalent to mri_fill). The original WM surfaces were created from the manually filled image. FreeSurfer, which is optimized for the human brain, could not act appropriately on our marmoset data with the default settings for the creation of the pial surface. Thus, we had to manually combine the GM and WM segments, assign specific values for each tissue type, and smooth the combined image. We then called the mris_make_surfaces command (Dale et al., 1999) with modified parameters to allow the WM surface to expand to the pial surface of the combined image. The WM surface and the pial surface were smoothed (by mris_smooth) and inflated (by mris_inflate) to create the inflated version of each surface. For the flat map creation, we mapped the atlas labels to the surface as a guide for the surface cutting. We followed a similar cutting strategy as the flat map of the marmoset brain connectivity atlas (Majka et al., 2016), where one cut was placed along the calcarine sulcus, and the other cut was along the edge of the orbitofrontal cortex until these cuts met the middle wall. With the cut patches, flat maps were created by the mris_flatten function (Fischl et al., 1999).
Generally, the average inter-vertex spacing of a surface should match or exceed the spatial resolution of its corresponding volume image (Autio et al., 2020; Glasser et al., 2013). Our FreeSurfer-based surfaces had about 38K vertices per hemisphere. The spacing of the pial and the white surface (estimated by Connectome Workbench “wb_command - surface-information”) was about 0.2 mm and 0.18 mm, respectively, which matched the spatial resolution of the volumetric templates (0.2 mm isotropic).
We converted all FreeSurfer-based surfaces back to the original template space using the ConvertSurface function of AFNI. Thus, users can use these surfaces directly from our original multi-modal volumetric template space “without” modifying their data into FreeSurfer-conformed formats. GIFTI format for these surfaces was provided for software compatibility.
2.3.3. Examples of surface-mapping applications
Our surfaces were made compatible with multiple software for surface-based analysis and visualization. In the first example, we used the “Ball & Box” approach (“@measure_bb_thick”) of AFNI to estimate the cortical thickness from our cortical template mask. The resulting volumetric thickness map was mapped to AFNI/SUMA-based pial surfaces by AFNI’s “3dVol2Surf” (using the “max” mapping function and 3 normal lengths) and visualized in SUMA software. In the second example, we used AFNI/SUMA to analyze and visualize visual task fMRI data of the marmosets. The detailed information of the data and fMRI preprocessing pipeline was described in our previous study (Liu et al., 2019). The resulting T-statistical map was mapped to AFNI/SUMA-based pial surfaces by AFNI’s “3dVol2Surf” (using the “nzave” mapping function and 3 normal lengths). In the third example, we projected the Paxinos parcellation of the MBM_V1 atlas to CT skull surface by AFNI’s “3dVol2Surf” (using the “nzmax” mapping function and 6 normal lengths). In the fourth example, we extracted the cortical part of the myelin-map template and converted it to a FreeSurfer-conforming format. The converted myelin map was mapped to the FreeSurfer-based surfaces by the “myelin-style” method of the Connectome Workbench command (wb_command - volume-to-surface-mapping) and visualized in the Connectome Workbench GUI.
3. Results
3.1. Multimodal volumetric templates
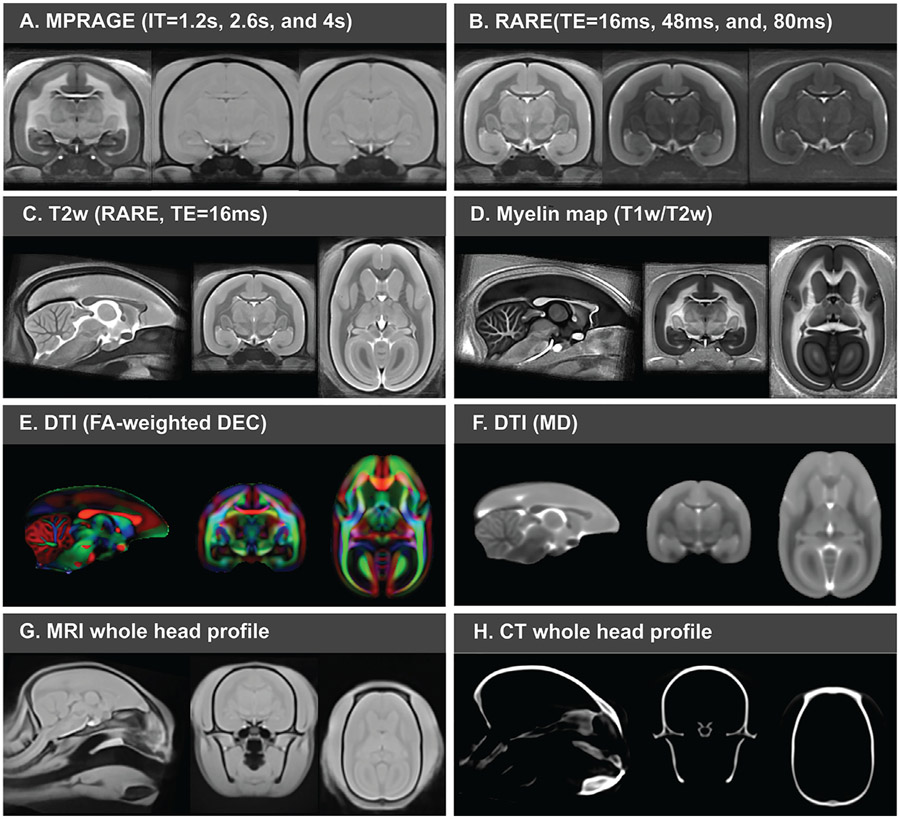
We constructed multimodal population-averaged templates by using the Multivariate Template Construction method of ANTs (for MRI and CT data) and the tensor-based method of the TORTOISE software (for DTI data). The template set includes multiple modalities (Fig. 3). The MP3RAGE (TI = 1200 ms) provides T1w contrast (Fig. 3A), and the other TIs (TI = 2600 ms and 4600 ms) can be used for the registration of images with low contrast. With different effective TEs, we provide templates with varying levels of T2w contrast (Fig. 3B). We can estimate the T1 map based on the multi-TI MP3RAGE images and the T2 map on multi-echo RARE images, respectively. Based on the ratio between T1w and T2w images, we generated the “myelin map” template, which enhances contrasts between the GM and the WM (Fig. 3D). The diffusion tensor template allows for tensor-based registration and can generate multiple DTI contrasts, such as fractional anisotropy (FA) and mean diffusivity (MD) (Fig. 3E and F). Also, we include two types of large FOV whole head template: one is MRI-based (Fig. 3G), and the other is CT-based (Fig. 3H). The rich template modalities facilitate a wide range of neuroimaging applications, as we can choose the matched template contrast for spatial normalization.
Fig. 3. Multi-modal population-based standard templates.
The standard templates include the MP3RAGE with TI = 1.2 s (T1w contrast), 2.6 s, and 4 s (A), RARE with TE = 16 ms, 48 ms, and 80 ms for different T2w contrasts (B and C), myelin map (T1w/T2w), contrasts from diffusion tensor images (E and F), the MRI whole head profiles (G) and CT whole head profiles (H).
We co-registered all templates to the same coordinate space that was defined by the “eye-bars and ear-bars” (Fig. 4). During the MRI scanning, we used two ear-bars to constrain animals, and these ear-bars were visible in the MRI whole head template (Fig. 4A). As there were no real eye-bars, we estimated the eye-bars plane based on the skull CT head template (Fig. 4A). We rotated templates so that ear-bars and estimated eye-bars were in the same plane (the “eye-bar and ear-bar” plane) (Fig. 4B). The origin of the template space is defined at the intersection of the “eye-bars and ear-bars ” plane (z = 0), the mid-line plane (x = 0), and the center plane of the ear bars (y = 0). Combining information from the MRI with that of the CT dataset, the template space allows a more accurate estimation of stereotaxic coordinates of different brain regions than the AC-PC coordinates used in our previous ex-vivo brain atlases (Liu et al., 2020; Liu et al., 2018).
Fig. 4. The coordinate space of our templates.
(A) The ear bar can be directly identified in the MRI head profile template, and the CT skull head profile can estimate the eye-bar location. (B) The “eye-bars and ear-bars” plane in 3D view of marmoset head. (C) The origin is located at the intersection between the mid-line plane and the ear-bars-eye-bars plane.
3.2. Tissue-type segmentation
Tissue-type segmentation is an essential procedure for volumetric analysis of MRI data, such as voxel-based morphometry, and a prerequirement for brain surface reconstruction. Although state-of-art automatic segmentation algorithms can produce reasonable results, the final segmentation highly depends on the MRI image quality and may not always guarantee accuracy. Additionally, most MRI segmentation tools are optimized and tested for the human brain, but not for small marmoset brains. Thus, it is hard to avoid segmentation errors by automatic segmentation.
After initial automatic segmentation, we manually segmented the marmoset brain template to correct many common segmentation errors existing in previously published ones. Here, we demonstrate examples of these errors (Fig. 5). (1) Due to its relatively higher intensity than the cortical GM, parts of subcortical GM were commonly mislabeled as the WM (Fig. 5, red arrow and yellow arrow). (2) The WM in the temporal pole, including parts of the uncinate fasciculus, was too thin to be classified as the WM (Fig. 5, blue arrow). (3) Due to the high intensity, major blood vessels were misclassified as the WM (Fig. 5, green arrow, and purple arrow). (4) The cerebellum WM was poorly segmented (Fig. 5, orange arrow).
Fig. 5. Examples of common segmentation errors.
Compared to the previously available marmoset population template (Hikishima et al., 2011), our new templates provide images with not only better contrast but also more accurate tissue-type segmentation. Examples are highlighted by arrows, including the subcortical GM misclassified as WM (red arrow and yellow arrow), the WM misclassified as the GM (blue arrow), hyper-intensive tissue misclassified as the WM (green arrow and purple arrow), and cerebellum WM segmentation (orange arrow). These commonly misclassified regions are corrected in our tissue-type segmentation.
We provided two versions of manual hard segmentation (Fig. 6A-B). The three-tissue-type segmentation includes the GM, the WM, and the other tissues (including CSF). The six-tissue segmentation consists of the cortical GM, the subcortical and cerebellum GM, the WM, the CSF, the blood vasculatures, and the skull. We transformed these manually corrected segments into each animal and used them as the prior images for automatic segmentation. The resulting tissue probability maps were converted back to the template space and averaged to form the template probability maps (Fig. 6C). Without the errors mentioned above, our new template tissue segmentation maps are more accurate and reliable than other published maps.
Fig. 6. Fine-detailed tissue-type hard segmentations and probability maps.
(A) Hard segmentations of three-tissue types include the GM (magenta), the WM (orange), and the others (green). (B) Six-tissue segmentation consists of the cortical GM (magenta), the subcortical and cerebellum GM (orange), the WM (green), the cerebrospinal fluid (blue), the blood vasculatures (azure), the skull (dark purple). (C) Probability maps of three tissue types include the GM (top), the WM (middle), and the others (bottom). We also provide probability maps of six tissue types in our release (images not shown).
3.3. Atlases, brain surfaces, and flat maps
As brain atlases are indispensable tools for defining regions of interest (ROI), the usability of a population template highly depends on the atlas-labeling of its template space. Currently, several different 3D digital brain atlases are available for the marmoset brain, such as the Marmoset Brain Mapping Atlas (MBM) and the Riken Brain/MINDS atlas (Riken). We fused them into our new template space (Fig. 7), including four cortical parcellations (A-D) and subcortical labels (F) from the MBM-V1 atlas and the cortical parcellation from the Riken atlas (E). We did not fuse our previous WM atlas (MBM-V2) to the template because the in-vivo template reported here had insufficient resolution to capture all WM structures defined in our ex-vivo atlas. By registering image data to our template space, we can directly access these GM atlases for region identification and ROI-based analysis.
Fig. 7. Atlas labeling of the templates.
The following atlases label our template space: Four different cortical parcellations from MBM-V1 atlas (Liu et al., 2018), including 13- (A), 54- (B), and 106 regions (C), and the Paxinos parcellation (D). The cortical parcellation from the Riken atlas (Woodward et al., 2018), which also follows the Paxinos nomenclature (Paxinos et al., 2012) (E); and subcortical labels from MBM-V1 atlas (F).
Brain surfaces and flat maps are fundamental tools for surface-based morphometry and fMRI analysis, as well as 3D visualization of MRI results. However, there were no fully-featured surface-based atlas tools available for the marmoset neuroimaging studies. Here,
We present two sets of brain surfaces and flat maps. The first set of surfaces was generated by FreeSurfer (Fig. 8A). As the original FreeSurfer pipeline is highly optimized for human data and incompatible with the marmoset data, we modified the marmoset templates to conform to the FreeSurfer requirement (see Materials and Methods for details). The FreeSurfer-based set includes original, smoothed, and inflated white matter surfaces, pial surfaces, surface spheres, surface cutting patches, flat maps, and all annotation files of cortical atlases. By following the FreeSurfer stream, all these elements have node-to-node correspondence and can be used simultaneously for surface-based analysis.
Fig. 8. Brain surfaces and flat maps.
(A) Surfaces generated by FreeSurfer. From left to right are the white matter surface, the mid-thickness cortical surface, the pial surface, and the flat map. Inflated versions of these surfaces are also available. (B) Surfaces generated by AFNI/SUMA. From left to right are the pial surface, the inflated pial surface, the white matter surface, and the skull surface (from CT data). (C) Cortical atlases mapped onto the flat map. The left image shows the Paxinos parcellation of the MBM_V1 atlas (Liu et al., 2018). The middle image shows the cortical parcellation from the Riken atlas (Woodward et al., 2018), which also follows the Paxinos nomenclature. The third image shows the outlines of the two atlases, where green outlines represent the Paxinos parcellation of the MBM_V1 and red outlines represent that of the Riken atlas. (D) Surface-based analysis and visualization. The left images show cortical thickness (estimated by the “Ball & Box” approach of AFNI) and the visual task fMRI results (Liu et al., 2019) mapped to the pial surface, and the Paxinos parcellation of the MBM_V1 atlas projected to CT skull surface by AFNI/SUMA. The right images show the myelin map of our template, which is mapped to the mid-thickness cortical surfaces and the flat map by the “myelin-style” approach of the Connectome Workbench command. The outlines on the flat map display the “106-region” parcellation of the MBM_V1 atlas.
The second set of surfaces, which was generated directly from segmentation images by AFNI/SUMA (Fig. 8B), includes the pial and white matter surfaces, the inflated pial and white matter surfaces, and the skull surface (based on CT data). Due to the limitation of AFNI/SUMA, we created these surfaces separately, and thus these surfaces cannot be used simultaneously as pairs for volume-to-surface mapping. However, AFNI/SUMA provides flexible functions (for example, 3dVol2Surf) to map volumetric data to brain surfaces by using the surface normal to solve this practical issue.
We mapped all cortical atlases onto the surfaces and flat maps as annotation (label) files, which improved the functionality of our template in data visualization. For example, in our previous study (Liu et al., 2018), we had estimated the inconsistency rate between two different Paxinos-style atlases. Now, with our new surface tools, we can directly visualize and compare both atlases on the flat map (Fig. 8C), highlighting their differences. With the support for generic GIFTI formats, our surfaces are compatible with multiple surface-analyzing software, including FreeSurfer, Connectome Workbench, and AFNI/SUMA (Fig. 8D). For example, we used AFNI/SUMA to analyze and map the cortical thickness, the visual task fMRI results, the cortical atlas onto different surfaces, and adopted Connectome Workbench to visualize the myelin map on brain surfaces and flat maps. With the completeness and compatibility, our surface set will be a versatile tool for surface-based analysis and visualization of the marmoset neuroimaging data.
4. Discussion
The population templates and surface-analyzing tools comprise the third version of the Marmoset Brain Mapping project. Previously, we constructed full 3D brain atlases of the marmoset by ultra-high resolution MRI: version 1 about the cortical parcellation (Liu et al., 2018), and version 2 about the white matter pathways (Liu et al., 2020). The two previous versions focused on the fine-detailed neuroanatomy of the marmoset brain, and thus we conducted long-duration scans on only a few brain samples, which allowed much higher resolution than the in-vivo data. However, ex-vivo data impose limitations on their application in neuroimaging studies, in particular, to analyze large cohorts of in-vivo MRI data. To overcome this limitation, we developed here “population multi-modal standard volumetric and surface-based templates”, which will significantly facilitate a wide range of volumetric and surface-based analysis of group-based marmoset neuroimaging data.
Brain extraction, or skull-stripping, is usually an early preprocessing step for most neuroimaging studies. Most existing automatic brain extraction tools, e.g., BET of FSL (Smith, 2002) and 3dSkullStrip of AFNI (Cox, 1996), were mainly tailored for the human brain and do not perform robustly for animal data. Template-based brain extraction is another popular (semi-) automatic approach for skull-stripping (Lohmeier et al., 2019). By spatial normalization of individual images to a template, one can warp the brain mask of the template nonlinearly to the individual data for brain extraction. This brain extraction method requires head templates (with skulls) and a brain mask, and the accuracy highly depends on the spatial normalization. Our previous atlases were based on ex-vivo data, and no head template was available for brain extraction (Liu et al., 2020; Liu et al., 2018). The previous in-vivo population template only provided low-contrast T1w and T2w modalities (Hikishima et al., 2011), which restricts their applications in other modalities. Here, we provided complete multi-modal templates, including multiple T1w and T2w templates, multiple contrasts from the DTI template, and large FOV head templates from MRI and CT data. The users can find the matched template modality to conduct template-based brain extraction, which ensures the accuracy of the spatial normalization and skull-stripping.
In addition to the rich modality, our population templates are associated with accurate tissue-type segmentation. Brain segmentation is an essential preprocessing step for MRI data analysis, such as voxel-based morphometry and brain surface reconstruction. The performance of modern segmentation algorithms depends on image contrasts, and the accuracy varies in different areas. For example, the highly myelinated GM area and many subcortical regions have a relatively high intensity in T1w images, which may be misclassified as white matter. The thin white matter may be misclassified as gray matter due to insufficient resolution. It is also hard for algorithms to distinguish high-intensity blood vasculatures from the brain white matter. Thus, prior knowledge is required for the algorithms to avoid these errors. Our manually-corrected tissue segmentation will be useful as prior maps to guide brain segmentation and improve the accuracy of structural image analysis.
The population template could serve as a standard coordinate space for adopting different brain atlases and reporting neuroimaging results. For humans, the MNI space, originally defined by the MNI305 MRI template (Evans et al., 1993), has become the standard template space for human MRI studies. By referring to the MNI templates, we could access multiple human brain atlases and neuroimaging results. For marmosets, as no such a template space was available previously. Here, by fusing existing digital atlases into the same template space, we no longer have to register our data to distinct template spaces for using or comparing different brain atlases. By combining information of MRI and CT datasets, the template space also allows a more accurate estimation of stereotaxic coordinates of different brain regions, which can be used, for example, for a more precise surgical planning than our previous versions.
In our previous V1 atlas, we released a basic version of a cortical surface (Liu et al., 2018), which has already been widely used to visualize MRI results (Hori et al., 2020; Liu et al., 2019; Schaeffer et al., 2019a; Schaeffer et al., 2019b; Schaeffer et al., 2019c; Schaeffer et al., 2019d; Selvanayagam et al., 2019). However, this surface is not compatible with most modern software, and its functions are limited to visualization. Due to the high demand for better surface tools, we provide here a comprehensive set of brain surfaces and flat maps, which are fundamental tools missing in existing marmoset templates or atlases. Because of the inherent limitations of surface-generation tools, we generated two different surface sets: one generated by the AFNI/SUMA and the other by the FreeSurfer. The AFNI-based set was generated directly from the segmentation images, while the FreeSurfer-based set provided node-to-node correspondence of all surface elements. The two surfaces sets will meet different requirements of most neuroimaging applications, from basic 3D visualization to advanced functional connectivity analysis on surfaces.
The Marmoset Brain Mapping V3 solves many existing issues in previously published marmoset brain templates by providing comprehensive population templates and surface tools. However, the present version also faces limitations that require attention. First, the spatial resolution of the in-vivo data was insufficient to capture all anatomical details of the brain. For example, our previous V2 revealed fine-detailed white matter structures of the marmoset brain by ultra-high resolution ex-vivo dMRI (Liu et al., 2020), but many of which cannot be reflected in the in-vivo data. Thus, there will be much to be gained by improving the resolution of in-vivo data. Second, existing standard surface creation pipelines (e.g., FreeSurfer) are not tailored for small-animal data, including marmosets. Here, we circumvented many of the default settings for humans by manually modifying our marmoset templates, but it was not done in a fully automatic or robust way. Future work is needed to optimize the existing surface-based software packages to accommodate marmoset data. Finally, we were not able to obtain a balanced gender distribution in our marmoset population. Due to other competing experimental priorities at the NIH, we could only recruit seven females in our study, far fewer than the twenty males used. Future efforts to scan a larger cohort of females will significantly improve our understanding of individual variability and gender differences of the marmoset population.
Supplementary Material
Acknowledgments
We thank Roger Depaz, Xianfeng (Lisa) Zhang, and Madeline Marcelle for their assistance in animal scanning. This work utilized the computational resources of the NIH HPC Biowulf cluster (hpc.nih.gov), and the CT images were collected using the scanner provided by the NIH Mouse Imaging Facility. This research was supported, in part, by the Intramural Research Program of the NIH, NINDS (ZIA NS003041), and by the PA Department of Health SAP #4100083102 to ACS.
Footnotes
Data and code availability statement
All data used to generate the population-based template, along with the software code used to analyze the data and generate the marmoset brain templates and associated tools comprise version 3 of our Marmoset Brain Mapping Project and are publicly available via marmosetbrainmapping.org/v3.html.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2020.117620.
References
- Autio JA, Glasser MF, Ose T, Donahue CJ, Bastiani M, Ohno M, Kawabata Y, Urushibata Y, Murata K, Nishigori K, Yamaguchi M, Hori Y, Yoshida A, Go Y, Coalson TS, Jbabdi S, Sotiropoulos SN, Kennedy H, Smith S, Van Essen DC, Hayashi T, 2020. Towards HCP-Style macaque connectomes: 24-Channel 3T multi-array coil, MRI sequences and preprocessing. Neuroimage 215, 116800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison N, Song G, 2009. Advanced normalization tools (ANTS). Insight J. 2, 1–35. [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC, 2011a. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC, 2011b. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9, 381–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC, 2010. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 49, 2457–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Margulies DS, 2019. Macroscale cortical organization and a default-like apex transmodal network in the marmoset monkey. Nat. Commun 10 1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruyer E, Lenglet C, Sapiro G, Deriche R, 2013. Design of multishell sampling schemes with uniform coverage in diffusion MRI. Magn. Reson. Med 69, 1534–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res 29, 162–173. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI, 1999. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 9, 179–194. [DOI] [PubMed] [Google Scholar]
- Evans AC, Collins DL, Mills S, Brown E, Kelly R, Peters TM, 1993. 3D statistical neuroanatomical models from 305 MRI volumes. In: Proceedings of the IEEE Conference Record Nuclear Science Symposium and Medical Imaging Conference. IEEE, pp. 1813–1817. [Google Scholar]
- Fischl B, 2012. FreeSurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Dale AM, 1999. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 9, 195–207. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Coalson TS, Robinson EC, Hacker CD, Harwell J, Yacoub E, Ugurbil K, Andersson J, Beckmann CF, Jenkinson M, Smith SM, Van Essen DC, 2016. A multi-modal parcellation of human cerebral cortex. Nature 536, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, Consortium, W.U.-M.H., 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Van Essen DC, 2011. Mapping human cortical areas in vivo based on myelin content as revealed by T1-and T2-weighted MRI. J. Neurosci 31, 11597–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikishima K, Ando K, Yano R, Kawai K, Komaki Y, Inoue T, Itoh T, Yamada M, Momoshima S, Okano HJ, Okano H, 2015. Parkinson disease: diffusion MR imaging to detect nigrostriatal pathway loss in a marmoset model treated with 1-Methyl-4-phenyl-1,2,3,6-tetrahydropyridine. Radiology 275, 430–437. [DOI] [PubMed] [Google Scholar]
- Hikishima K, Quallo MM, Komaki Y, Yamada M, Kawai K, Momoshima S, Okano HJ, Sasaki E, Tamaoki N, Lemon RN, Iriki A, Okano H, 2011. Population-averaged standard template brain atlas for the common marmoset (Callithrix jacchus). Neuroimage 54, 2741–2749. [DOI] [PubMed] [Google Scholar]
- Hori Y, Schaeffer DJ, Gilbert KM, Hayrynen LK, Cléry JC, Gati JS, Menon RS, Everling S, 2020. Comparison of resting-state functional connectivity in marmosets with tracer-based cellular connectivity. Neuroimage 204, 116241. [DOI] [PubMed] [Google Scholar]
- Le Gal R, Bernaudin M, Toutain J, Touzani O, 2018. Assessment of behavioural deficits following ischaemic stroke in the marmoset. Behav. Brain Res 352, 151–160. [DOI] [PubMed] [Google Scholar]
- Lee NJ, Ha SK, Sati P, Absinta M, Luciano NJ, Lefeuvre JA, Schindler MK, Leibovitch EC, Ryu JK, Petersen MA, Silva AC, Jacobson S, Akassoglou K, Reich DS, 2018. Spatiotemporal distribution of fibrinogen in marmoset and human inflammatory demyelination. Brain 141, 1637–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MK, Takahashi YS, Huo BX, Hanada M, Nagashima J, Hata J, Tolpygo AS, Ram K, Lee BC, Miller MI, Rosa MG, Sasaki E, Iriki A, Okano H, Mitra P, 2019. A high-throughput neurohistological pipeline for brain-wide mesoscale connectivity mapping of the common marmoset. Elife 8, e40042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ye FQ, Newman JD, Szczupak D, Tian X, Yen CC, Majka P, Glen D, Rosa MGP, Leopold DA, Silva AC, 2020. A resource for the detailed 3D mapping of white matter pathways in the marmoset brain. Nat. Neurosci 23, 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ye FQ, Yen CC, Newman JD, Glen D, Leopold DA, Silva AC, 2018. A digital 3D atlas of the marmoset brain based on multi-modal MRI. Neuroimage 169, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Yen CC, Szczupak D, Ye FQ, Leopold DA, Silva AC, 2019. Anatomical and functional investigation of the marmoset default mode network. Nat. Commun 10 1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmeier J, Kaneko T, Hamm B, Makowski MR, Okano H, 2019. atlasBREX: automated template-derived brain extraction in animal MRI. Sci. Rep 9, 12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Yan M, Shen X, Wu J, Yu W, Yan S, Yang F, Zeljic K, Shi Y, Zhou Z, Lv L, Hu X, Menon R, Wang Z, 2020. Normative analysis of individual brain differences based on a population MRI-Based atlas of cynomolgus macaques. Cereb Cortex, bhaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majka P, Chaplin TA, Yu HH, Tolpygo A, Mitra PP, Wójcik DK, Rosa MG, 2016. Towards a comprehensive atlas of cortical connections in a primate brain: Mapping tracer injection studies of the common marmoset into a reference digital template. J. Compar. Neurol 524, 2161–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus D, Harwell J, Olsen T, Hodge M, Glasser M, Prior F, Jenkinson M, Laumann T, Curtiss S, Van Essen D, 2011. Informatics and data mining tools and strategies for the human connectome project. Front. Neuroinformat 5 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Wang D, Fox MD, Yeo BT, Sepulcre J, Sabuncu MR, Shafee R, Lu J, Liu H, 2013. Individual variability in functional connectivity architecture of the human brain. Neuron 77, 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Hikishima K, Iriki A, Sasaki E, 2012. The common marmoset as a novel animal model system for biomedical and neuroscience research applications. In: Seminars in Fetal and Neonatal Medicine. Elsevier, pp. 336–340. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C, Petrides M, Rosa M, Tokuno H, 2012. The Marmoset Brain in Stereotaxic Coordinates. Elsevier Academic Press. [Google Scholar]
- Philippens IH, Ormel PR, Baarends G, Johansson M, Remarque EJ, Doverskog M, 2017. Acceleration of amyloidosis by inflammation in the amyloid-beta marmoset monkey model of Alzheimer’s disease. J. Alzheimer’s Dis 55, 101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C, Walker L, Irfanoglu M, Barnett A, Basser P, Chang L, Koay C, Pajevic S, Rohde G, Sarlls J, 2010. TORTOISE: an integrated software package for processing of diffusion MRI data. In: Proceedings of the 18th Scientific Meeting of the International Society for Magnetic Resonance in Medicine, p. 1597. [Google Scholar]
- Reardon PK, Seidlitz J, Vandekar S, Liu S, Patel R, Park MTM, Alexander-Bloch A, Clasen LS, Blumenthal JD, Lalonde FM, Giedd JN, Gur RC, Gur RE, Lerch JP, Chakravarty MM, Satterthwaite TD, Shinohara RT, Raznahan A, 2018. Normative brain size variation and brain shape diversity in humans. Science 360, 1222–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad ZS, Reynolds RC, Argall B, Japee S, Cox RW, 2004. SUMA: an interface for surface-based intra-and inter-subject analysis with AFNI. In: Proceedings of the 2nd IEEE International Symposium on Biomedical Imaging: Nano to Macro (IEEE Cat No. 04EX821). IEEE, pp. 1510–1513. [Google Scholar]
- Schaeffer DJ, Gilbert KM, Gati JS, Menon RS, Everling S, 2019a. Intrinsic functional boundaries of lateral frontal cortex in the common marmoset monkey. J. Neurosci 39, 1020–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer DJ, Gilbert KM, Ghahremani M, Gati JS, Menon RS, Everling S, 2019b. Intrinsic functional clustering of anterior cingulate cortex in the common marmoset. Neuroimage 186, 301–307. [DOI] [PubMed] [Google Scholar]
- Schaeffer DJ, Gilbert KM, Hori Y, Gati JS, Menon RS, Everling S, 2019c. Integrated radiofrequency array and animal holder design for minimizing head motion during awake marmoset functional magnetic resonance imaging. Neuroimage 193, 126–138. [DOI] [PubMed] [Google Scholar]
- Schaeffer DJ, Gilbert KM, Hori Y, Hayrynen LK, Johnston KD, Gati JS, Menon RS, Everling S, 2019d. Task-based fMRI of a free-viewing visuo-saccadic network in the marmoset monkey. Neuroimage 202, 116147. [DOI] [PubMed] [Google Scholar]
- Seidlitz J, Sponheim C, Glen D, Ye FQ, Saleem KS, Leopold DA, Ungerleider L, Messinger A, 2018. A population MRI brain template and analysis tools for the macaque. Neuroimage 170, 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvanayagam J, Johnston KD, Schaeffer DJ, Hayrynen LK, Everling S, 2019. Functional localization of the frontal eye fields in the common marmoset using microstimulation. J. Neurosci 39, 9197–9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AC, 2017. Anatomical and functional neuroimaging in awake, behaving marmosets. Dev. Neurobiol. 77, 373–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, 2002. Fast robust automated brain extraction. Hum. Brain Map. 17, 143–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, 2005. A population-average, landmark- and surface-based (PALS) atlas of human cerebral cortex. Neuroimage 28, 635–662. [DOI] [PubMed] [Google Scholar]
- Woodward A, Hashikawa T, Maeda M, Kaneko T, Hikishima K, Iriki A, Okano H, Yamaguchi Y, 2018. The Brain/MINDS 3D digital marmoset brain atlas. Sci Data 5, 180009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Jiang YH, Zhang YQ, 2018. Modeling autism in non-human primates: opportunities and challenges. Autism. Res 11, 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.