Abstract
Declining tissue nicotinamide adenine dinucleotide (NAD) levels are linked to ageing and its associated diseases. However, the mechanism for this decline is unclear. Here, we show that pro-inflammatory M1-like macrophages, but not naive or M2 macrophages, accumulate in metabolic tissues, including visceral white adipose tissue and liver, during ageing and acute responses to inflammation. These M1-like macrophages express high levels of the NAD-consuming enzyme CD38 and have enhanced CD38-dependent NADase activity, thereby reducing tissue NAD levels. We also find that senescent cells progressively accumulate in visceral white adipose tissue and liver during ageing and that inflammatory cytokines secreted by senescent cells (the senescence-associated secretory phenotype, SASP) induce macrophages to proliferate and express CD38. These results uncover a new causal link among resident tissue macrophages, cellular senescence and tissue NAD decline during ageing and offer novel therapeutic opportunities to maintain NAD levels during ageing.
NAD is an oxidation–reduction (redox) coenzyme that is central to energy metabolism and is an essential cofactor for non-redox NAD-dependent enzymes, including sirtuins and poly-ADP-ribose polymerases (PARPs)1. Recently, a progressive decrease in NAD levels during ageing in both rodents and humans has been documented in multiple tissues2. Remarkably, restoration of NAD levels with the NAD precursor vitamins nicotinamide riboside (NR), nicotinamide (NAM) and nicotinic acid (NA), in addition to the biosynthetic NAD precursor nicotinamide mononucleotide (NMN), appears to mitigate several age-associated diseases2-4. These observations have stimulated much research activity aiming to better understand how NAD levels affect the ageing process and how or why NAD levels decline during ageing, with the goal of developing therapeutics to combat ageing-related diseases.
NAD can be synthesized from tryptophan through the de novo pathway, and by salvage of the three NAD precursor vitamins and NMN4. Although dietary precursors can contribute to NAD pools in a manner that depends on which pathways are expressed in each tissue5, the prevailing thought is that the recycling of NAM via nicotinamide phosphoribosyltransferase (NAMPT) is the predominant pathway used by most cells to maintain intracellular NAD levels6. The rate of NAD synthesis is countered by the rate of consumption by NAD-consuming enzymes, including sirtuins, PARPs and the CD38 and bone-marrow stromal cell antigen 1 (BST1, also known as CD157) NAD hydrolases. Importantly, it is not clear whether depressed de novo NAD biosynthesis, depressed NAM salvage, enhanced NAD consumption or a combination of these processes is the main driver of the NAD decrease observed during ageing and conditions of metabolic stress.
Interestingly, a recent report has shown an increased expression of CD38 during ageing in visceral white adipose tissue7. CD38 is a transmembrane protein that consumes NAD to form cyclic ADP-ribose (cADPR), ADP-ribose (ADPR) and NAM7. Importantly, mice lacking CD38 (Cd38 KO) were protected from age-related NAD decline and had enhanced metabolic health and sirtuin 3 (SIRT3)-dependent mitochondrial function, supporting the idea that CD38 is the primary NAD-consuming enzyme responsible for age-related NAD decline in this tissue7. However, these data did not identify which cells express CD38 in aged tissues or the mechanism(s) driving aberrant CD38 expression during ageing.
CD38 is ubiquitously expressed by immune cells, and its expression increases during inflammatory conditions8-10. Chronic low-grade inflammation, a feature of ageing termed ‘inflammaging’11, is a leading mechanism behind many ageing-associated diseases and is a significant risk factor for morbidity and mortality12. Sustained activation of the immune system is energetically costly and requires sufficient metabolites to fuel effector immune functions13. Thus, the immune system and metabolism are highly integrated. Despite this knowledge, it is unclear how age-related inflammation affects NAD metabolism and the ageing process.
Here, we report that pro-inflammatory M1 macrophages show increased CD38 expression, enhanced NADase activity and production of the NAD-degradation byproducts NAM and ADPR. Using macrophages from wild-type (WT) and Cd38 KO mice, we show that the high NADase activity of M1 macrophages is completely dependent on CD38 and not other NAD-consuming enzymes. Moreover, CD38 expression levels are elevated in resident macrophages from epididymal white adipose (eWAT) and liver tissues from old mice compared with those from young mice and from mice treated with pro-inflammatory toll-like receptor (TLR) ligands, such as lipopolysaccharide (LPS). Lastly, we show that enhanced CD38 expression by tissue-resident macrophages during ageing is driven by the SASP of senescent cells14. Because senescent cells increase progressively in adipose tissue and liver during ageing, our results identify a new causal link between senescence in visceral tissue cells and tissue NAD decline during ageing.
Results
M1 macrophages have increased expression of NAD hydrolases CD38 and CD157 and enhanced degradation of NAD.
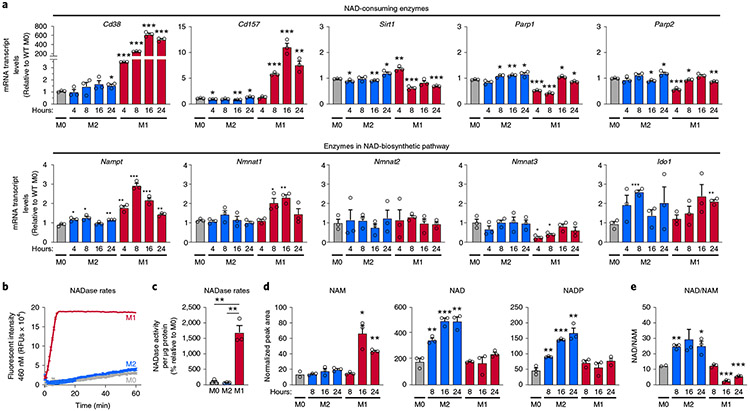
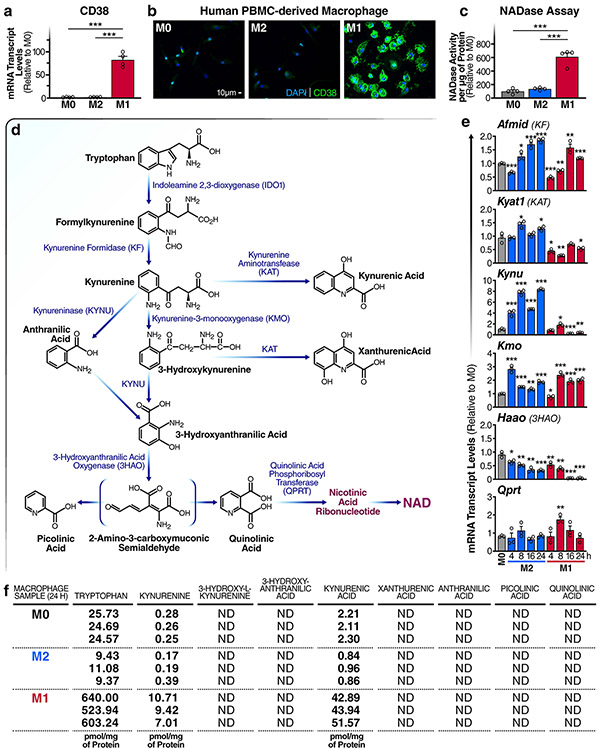
Despite renewed interest in both NAD metabolism and immunometabolism, little is known about how NAD levels are regulated by immune cells and whether NAD levels influence immune-cell function. To better understand how tissue-resident macrophages contribute to ageing-associated NAD changes, we first surveyed the levels of messenger RNAs encoding enzymes that consume or are involved in the biosynthesis of NAD during pro- and anti-inflammatory macrophage polarization. We polarized naive (M0) primary mouse bone-marrow-derived macrophages (BMDMs) to classical pro-inflammatory M1 macrophages and to anti-inflammatory alternatively activated M2 macrophages. In M1, relative to M0 and M2, macrophages, the only significantly upregulated NADase-encoding mRNAs were Cd38 (600-fold increase) and, to a lesser degree, its homologue Cd157; Sirt1, Parp1 and Parp2 mRNAs were not highly upregulated in M1 relative to M0 and M2 macrophages (Fig. 1a). A similar increase in CD38 expression was observed in human M1 macrophages (Extended Data Fig. 1a,b). The data suggest that pro-inflammatory M1 macrophages may exhibit higher NADase activity than M2 or M0 macrophages. To test this hypothesis, we utilized nicotinamide 1,N6-etheno-adenine dinucleotide (ε-NAD), a modified version of NAD in which cleavage by an NAD hydrolase yields a time-dependent increase in fluorescence at 460 nm. Cellular lysates from M1 macrophages showed a significant and robust increase in NADase activity (Fig. 1b,c), whereas those from M0 or M2 macrophages showed very little (Fig. 1b).
Fig. 1 ∣. M1 macrophages are characterized by increased NADase activity.
a, mRNA levels of NAD-consuming and NAD-biosynthetic enzymes were measured using quantitative PCR and untreated BMDMs (M0) or BMDMs treated with IL-4 (M2) or LPS (M1) for the indicated times. b, NADase rates were measured in cell lysates from naive BMDMs (M0) or BMDMs treated with IL-4 (M2) or LPS (M1) 16 h after activation. c, Quantification of NADase activity rates. d, LC-MS was used to quantify NAD and NAD-related metabolites in naive M0, M1 and M2 BMDMs for the indicated times. e, NAD/NAM ratios from the LC-MS data above for each indicated timepoint. Data are shown as the mean ± s.e.m. (n = 3 independent biological experiments). *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test.
Because mouse and human macrophage phenotypes can differ substantially15, we measured NADase activity in macrophages derived from human peripheral blood monocytes (PBMCs). Human M1 lysates showed increased NAD hydrolysis compared with that in M0 and M2 subsets (Extended Data Fig. 1c). Together, our data show that enhanced NAD hydrolysis is a conserved feature of inflammatory M1 macrophages in both humans and mice.
The difference in NAD hydrolase activity in the different macrophage subtypes shown above suggests that M1 and M2 macrophages might have different NAD levels. Little is known about how NAD levels are linked to the reprogramming of macrophage metabolism, or how NAD levels differ among macrophage polarization states. To address this knowledge gap, we used targeted liquid chromatography–mass spectrometry (LC–MS) to quantify the NAD metabolome in M1 and M2 macrophages. Importantly, other NAD-consuming enzymes, such as PARPs, sirtuins, CD38 and CD157, produce NAM as a byproduct of NAD hydrolysis. Consistent with the high NADase activity of M1 macrophages, we observed significantly elevated levels of NAM in M1, but not M0 or M2, macrophages (Fig. 1d). We also observed elevated levels of NAD and NADP in M2, but not M1, macrophages (Fig. 1d), and NAD/NAM ratios were significantly lower in M1 macrophages (Fig. 1e).
Despite the high NADase activity associated with M1 macrophages, NAD levels appeared to be stably maintained in these cells compared with M0 macrophages (Fig. 1d). This finding suggests that M1 macrophages might upregulate biosynthetic and NAM-salvage pathways to compensate for increased NAD degradation. The culture medium used in our experiments lacks NA but contains NAM, so NAD can be derived de novo from tryptophan or recycled from NAM via the NAM-salvage pathway. Therefore, we measured mRNA levels of indoleamine 2,3-dioxygenase 1 (Ido1 mRNA) and other key enzymes in the de novo NAD-synthetic pathway (Fig. 1a and Extended Data Fig. 1d,e), and mRNAs encoding NAM-salvage-pathway enzymes (Nampt mRNA) and nicotinamide mononucleotide adenylyltransferases 1–3 (Nmnat1–3 mRNAs) (Fig. 1a). In M1 macrophages, we observed increased levels of Nampt or Nmnat1 mRNAs, which maintain nuclear NAD levels (Fig. 1a), but no significant changes in the levels of Nmnat2 or Nmnat3 mRNAs, which regulate NAM salvage in the cytoplasm or mitochondria, respectively, or Ido1 mRNA (Fig. 1a). Interestingly, mRNAs encoding several key enzymes in the de novo synthesis pathway declined during M1 polarization, including those encoding kynureninase (Kynu mRNA) and 3-hydroxy-anthranilic acid oxygenase (Haao mRNA) (Extended Data Fig. 1e). Furthermore, LC–MS analyses showed increased levels of tryptophan, kynurenine and kynurenic acid in M1, but not M0 or M2, macrophages (Extended Data Fig. 1f). We were unable to detect tryptophan-pathway metabolites downstream of kynurenine in M0, M1 or M2 macrophages, which suggests that tryptophan may not contribute to de novo NAD synthesis in macrophages, regardless of polarization state.
The NAM-salvage pathway is a crucial regulator of macrophage polarization and fine-tunes gene expression.
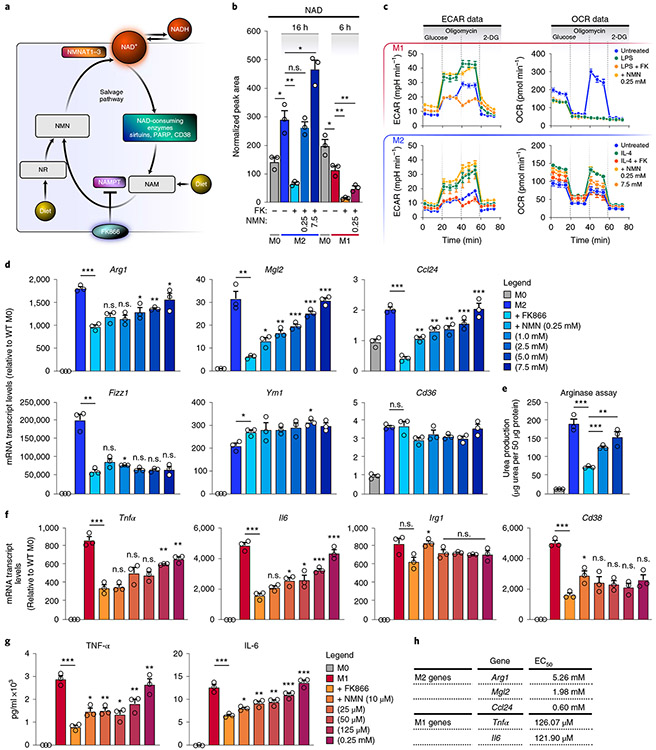
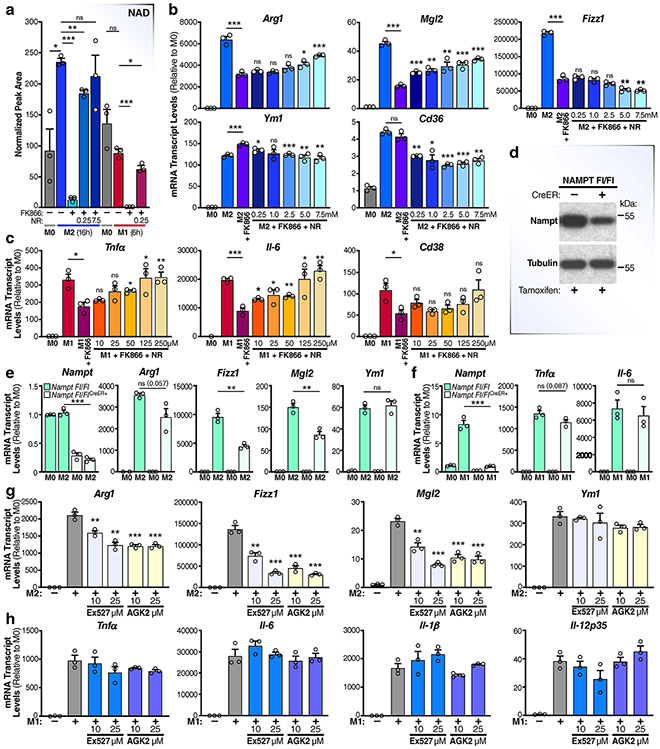
Our observation that de novo NAD synthesis from tryptophan may not contribute to maintenance of NAD levels in M1 and M2 macrophages predicts a critical role for the NAM-salvage pathway in maintaining NAD levels. Indeed, M1 and M2 macrophages pretreated with FK866, a specific NAMPT inhibitor (Fig. 2a), had significantly lower NAD levels (Fig. 2b and Extended Data Fig. 2a). The data indicate that macrophages rely almost exclusively on the NAM-salvage pathway to sustain NAD levels and that M1 macrophages compensate for the enhanced NADase activity by increasing mRNA levels of Nampt and Nmnat1, encoding NAD salvage pathway enzymes (Fig. 1a). To confirm the role of NAD depletion, we supplemented the culture medium of macrophages treated with FK866 with NMN or NR16, NAD precursors that feed into the NAM-salvage pathway downstream of NAMPT17 (Fig. 2a,b and Extended Data Fig. 2a); both significantly restored NAD levels in M2, but not M1, macrophages, perhaps due to their quick turnover of NAD (Fig. 1). Interestingly, a recent report has shown that targeting NAMPT affected glycolytic rates of M1, but not M2, macrophages18. Thus, we asked whether altering NAD levels by targeting NAMPT influences macrophage metabolism by measuring the extracellular acidification rate (ECAR) and oxygen-consumption rate (OCR), indicative of glycolysis and oxidative phosphorylation, respectively. Consistent with this prior report, FK866-treated M1 macrophages showed reduced glycolytic capacity, which was rescued by addition of NMN (Fig. 2c); however, in contrast to the report, the glycolytic capacity of M2 macrophages was sensitive to NAMPT-dependent fluctuations in NAD levels (Fig. 2c). Furthermore, FK866-treated M2 macrophages had reduced oxidative phosphorylation that was rescued by addition of NMN (Fig. 2c), whereas the OCR of M1 macrophages declined, as expected19, regardless of NAD levels. Thus, our data suggest that both M1 and M2 macrophage metabolic shifts depend on NAD derived from the NAM-salvage pathway.
Fig. 2 ∣. The NAM-salvage pathway controls NAD levels to regulate macrophage activation and polarization.
a, Schematic representation of the NAM-salvage pathway. b, NAD levels were measured by LC-MS in M0, M1 and M2 BMDMs pretreated with or without 50 nM FK866 and NMN for 6 h prior to stimulation with LPS for an additional 6 h or IL-4 for 16 h. n.s., not significant. c, Seahorse assay measuring ECAR and OCR for the conditions and timepoints described in b in M1 and M2 BMDMs (n = 4 independent biological experiments per condition). 2-DG, 2-deoxy-d-glucose. d, mRNA levels of M2 genes in BMDMs pretreated with or without FK866 and NMN for 6 h prior to stimulation with IL-4 for 16 h. e, Arginase assay for M2 macrophages pretreated with or without FK866 and NMN for 6 h prior to stimulation with IL-4 for 16 h. f,g, mRNA and supernatant protein (ELISA) levels of TNF-α and IL-6 in BMDMs pretreated with or without FK866 and NMN for 6 h prior to stimulation with LPS for 6 h. h, Quantification of the half maximal effective concentration (EC50) of NMN needed to rescue M1 and M2 macrophage gene expression. Data are shown as the mean ± s.e.m. (n = 3 independent biological experiments). *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test. Unless noted with a bar, statistical comparisons are relative to M2/M1 + FK866.
In the experiments above, we consistently observed rising NAD levels during M2, but not M1 or M0, polarization (Figs. 1d and 2b and Extended Data Fig. 2a). This result suggests that M2 polarization requires higher NAD levels. To test this idea, we pretreated BMDMs with the NAMPT inhibitor FK866 to lower NAD levels prior to treatment with interleukin-4 (IL-4). This treatment decreased mRNA levels of multiple M2 markers (arginase 1 (Arg1), galactose N-acetyl-galactosamine-specific lectin 2 (Mgl2), C–C motif chemokine ligand 24 (Ccl24) and resistin-like beta (Retnlb, also known as Fizz1)) (Fig. 2d), although the M2 markers Ym1 and Cd36 were unaffected. In agreement with reduced expression of Arg1, arginase activity, a key anti-inflammatory effector function of M2 macrophages, was also attenuated by FK866 (Fig. 2e). To confirm the role of NAD in regulating M2 gene expression, we supplemented FK866-treated macrophages with different doses of NMN or NR in the culture medium. Both rescued the mRNA levels of M2 genes inhibited by FK866 (Fig. 2d and Extended Data Fig. 2b) and arginase activity (Fig. 2e) in a dose-dependent manner, with the exception of Fizz1. In contrast to previous reports18,20, pretreatment of macrophages with FK866 also suppressed mRNA levels and secretion of M1 inflammatory cytokines, such as IL-6 and tumour necrosis factor-α (TNF-α), but not the metabolic enzyme Irg1 (Fig. 2f,g). NMN and NR also restored mRNA and protein levels of inflammatory cytokines in M1 macrophages (Fig. 2f,g and Extended Data Fig. 2c). However, much higher concentrations of NMN (Fig. 2d-f) and NR (Extended Data Fig. 2b,c) were needed to rescue FK866-sensitive M2 gene expression than were needed to rescue that in M1 cells (Fig. 2h).
These results show that the NAM-salvage pathway regulates macrophage NAD levels, and that NAD is a crucial regulator of macrophage polarization and mRNA abundance of a subset of M2 and M1 genes. Notably, they show that M1 and M2 macrophages have different requirements for NAD levels, with M2 macrophages requiring higher levels to support M2 gene expression and immune effector functions (Fig. 2h). To further investigate the role of NAMPT in M2 polarization, and to rule out potential off-target effects of FK866, we polarized BMDMs from mice expressing a floxed NAMPT gene and oestrogen receptor Cre-recombinase that can be activated with tamoxifen, leading to a significant and acute partial deletion of the NAMPT gene (Nampt Fl/FlCreER) (Extended Data Fig. 2d-f). As expected, IL-4-treated Nampt Fl/FlCreER (but not Nampt Fl/Fl) M2 macrophages had reduced expression of multiple M2 genes, including Arg1, Fizz1 and Mgl2, but not Ym1, similar to Fk866-treated BMDMs (Extended Data Fig. 2e). Nampt Fl/FlCreER BMDM M1 gene expression was only slightly affected (Extended Data Fig. 2f), probably due to incomplete deletion of NAMPT (Extended Data Fig. 2d). Interestingly, NAD levels are unlikely to control M2 polarization, owing to their impact on metabolic reprogramming; even smaller doses of NMN were able to fully restore both the glycolytic and oxidative-phosphorylation rates of M2 macrophages (Fig. 2c), whereas higher doses were necessary to restore M2 gene expression (Fig. 2d). These data suggest that NAD regulates M2 gene expression independently of its effects on glycolysis and oxidative phosphorylation.
Considering that sirtuin proteins have been shown to support macrophage polarization21,22, and sirtuin deacylase activity is directly linked to NAD levels, we hypothesized that the higher NAD levels in M2 macrophages would favour greater sirtuin activity and a role in supporting M2 polarization. In contrast, the increased degradation of NAD by CD38 and increased NAM/NAD ratio would likely suppress sirtuin function in M1 macrophages, making it unlikely that activation of sirtuins play a major role in M1 polarization. In support of this hypothesis, treatment of M2 macrophages with the sirtuin inhibitors EX527 and AGK2 suppressed expression of the FK866-sensitive M2 genes (Arg1, Fizz1 and Mgl2), but not the FK866-insensitive M2 gene Ym1 (Extended Data Fig. 2g). Further, the sirtuin inhibitors had no effect on the expression of inflammatory cytokines in M1 macrophages (Extended Data Fig. 2h). Thus, while the NAD-dependent processes that support M1 and M2 polarization are likely to be pleiotropic, we provide evidence that NAD is necessary to support macrophage metabolic reprogramming and fine-tunes the expression of a subset of M1 and M2 genes. Additionally, increased NAD levels in M2 macrophages may be necessary to support or enhance sirtuin function to control M2 activation and gene expression.
CD38 is exclusively expressed by M1 macrophages and is responsible for enhanced NAD hydrolase activity.
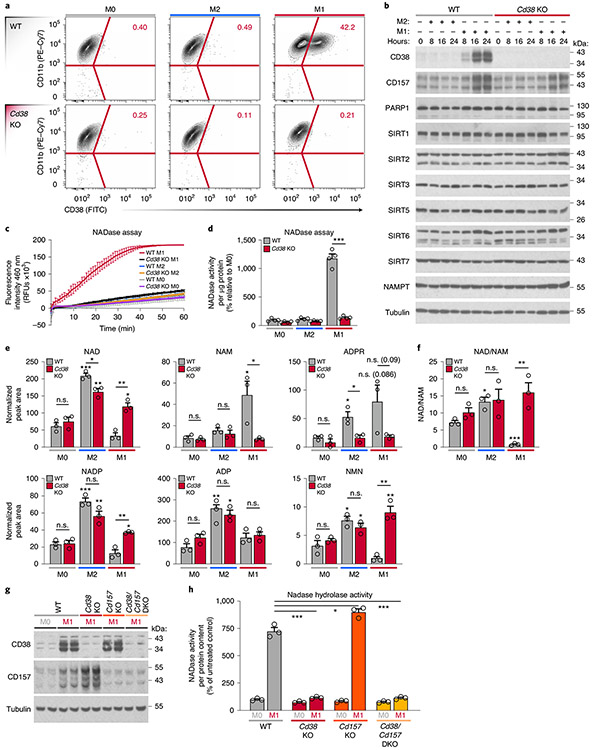
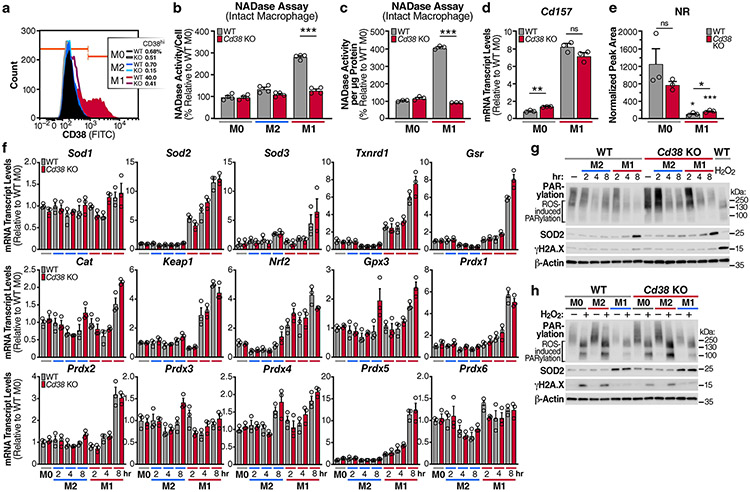
We showed that M1 macrophage polarization entails enhanced NADase activity and enhanced expression of the NAD hydrolases Cd38 and its homologue Cd157 (Fig. 1). To test the hypothesis that the high NADase activity in M1 macrophages is CD38/CD157 dependent, we first analysed CD38 surface expression using flow cytometry. We compared M0, M1 and M2 BMDMs from WT mice and mice lacking CD38 (Cd38 KO)23 (Fig. 3a). Consistent with high Cd38 mRNA levels in M1 macrophages (Fig. 1a), cell-surface CD38 was exclusively expressed by WT M1 macrophages (Fig. 3a, b and Extended Data Fig. 3a), a finding recently reported24. In contrast, PARP and sirtuin expression was unaffected by Cd38 KO or M1 or M2 polarization (Fig. 3b). In support of our hypothesis that NADase activity in M1 macrophages is CD38 dependent, lysates from Cd38 KO M1 macrophages showed near basal NADase activity compared to that in WT M1 macrophages (Fig. 3c, d). These data indicate that NADase activity in M1 macrophages requires CD38, and not any other NAD-consuming enzyme. CD38 can localize to cell membranes in a type II and type III orientation, with its enzymatic domain facing outside (type II) or inside (type III) the cell25-27. To determine the orientation of the CD38 enzymatic activity in M1 macrophages, we measured degradation of the non-cell permeable εNAD in intact cells. Extracellular εNAD was hydrolysed only by M1 macrophages (Extended Data Fig. 3b, c), although to a lesser extent than whole cell lysates (Fig. 3d). These data demonstrate that CD38 cleaves NAD outside and inside cells, and that a substantial fraction of CD38 NADase activity in macrophages is intracellular.
Fig. 3 ∣. High NADase activity in M1 macrophages is CD38 dependent.
a, Representative flow-cytometry plots comparing CD38 surface staining in naive (M0) WT and Cd38 KO BMDMs or BMDMs treated with IL-4 (M2) and LPS (M1) for 16 h. b, Western analysis of NADase enzymes in M0, M1 and M2 WT and Cd38 KO BMDMs for the indicated times. c, NADase rates measured in WT and Cd38 KO M0, M2 and M1 BMDMs activated for 16 h. d, Quantification of the NADase activity rate. e, LC-MS was used to quantify NAD and NAD-related metabolites in M0, M2 and M1 WT and Cd38 KO BMDMs activated for 16 h. f, NAD/NAM ratios from LC-MS data in e. g, Western analysis of CD38 and CD157 in M0 and M1 BMDMs from WT, Cd38 KO, Cd157 KO and Cd38/Cd157 DKO mice stimulated for 16 h. h, NADase rates measured in M0 and M1 BMDMs from WT, Cd38 KO, Cd157 KO and Cd38/Cd157 DKO mice stimulated for 16 h. Data are shown as the mean ± s.e.m. (n = 3 independent biological experiments, but n = 4 in d). *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test. Unless noted with a bar, statistical comparisons are relative to the appropriate M0 WT or Cd38 KO sample of the same genotype.
To further investigate how CD38 regulates intracellular NAD levels in macrophages, we quantified NAD and its metabolites in WT and Cd38 KO BMDMs. WT M1 macrophages had more NAM and ADPR, two products resulting from CD38 activity on NAD (Fig. 3e), whereas these metabolites were suppressed to basal levels in M1 macrophages from Cd38 KOs (Fig. 3e). Cd38 KO M1 macrophages also had significantly higher levels of NAD and NADP and a higher NAD/NAM ratio than did WT M1 macrophages (Fig. 3e,f). Additionally, there was no significant difference in the level of ADP in WT versus Cd38 KO M1 macrophages (Fig. 3e). Therefore, CD38 specifically consumes and regulates NAD levels, without affecting the levels of other adenine-containing nucleotides. Unexpectedly, despite M2 macrophages expressing only a small amount of basal Cd38 (Figs. 1a and 3b), we observed a subtle but significant decline in NAD and ADPR in M2 Cd38 K0 macrophages, compared with WT M2 macrophages (Fig. 3e), which suggests that basal expression of CD38 may impact NAD levels in quiescent and M2 macrophage subsets, perhaps due to compensation from another NAD-consuming enzyme. However, given the strong changes in NAD and NAD-related metabolites in Cd38 KO M1 macrophages compared with WT M1 macrophages, these data support the model that CD38 is a key regulator of NAD and its metabolites NAM and ADPR in M1 macrophages. These results seem to exclude a major role for CD157. Indeed, whereas Cd38 KO and Cd38/Cd157 double-KO (DKO) M1 macrophages have attenuated NADase activity, Cd157 KO M1 macrophages retain NADase activity, similarly to WT M1 macrophages (Fig. 3g,h). Interestingly, recent reports have suggested that the preferred CD157 substrate is the NAD precursor NR28,29. In indirect support of this idea, CD157 expression, which increased in response to LPS treatment, was associated with significantly reduced NR levels in both WT and Cd38 KO M1 macrophages (Extended Data Fig. 3d,e). Thus, although CD157 may indirectly influence NAD levels in M1 macrophages by consuming NR, the enhanced NADase activity of pro-inflammatory M1 macrophages is mediated by CD38.
A recent report has suggested that during M1 macrophage polarization, reactive oxygen species derived from mitochondrial electron-transport chain complex III causes DNA damage and enhanced NAD degradation as a result of a PARP1-dependent DNA-repair response18. However, consistent with this report and another recent report30, we observed an early increase in the expression of antioxidant genes in M1, but not M0 or M2, macrophages (Extended Data Fig. 3f) in both WT and Cd38 KO BMDMs. These data are consistent with the hypothesis suggesting that M1 macrophages upregulate antioxidant genes as a survival mechanism and are protected from the DNA-damaging effects of ROS31. Indeed, we found no detectable DNA damage, as measured by PARylation and phosphorylated histone H2AX (γH2AX), a marker of DNA double-strand breaks, after early or later timepoints in either WT or Cd38 KO M1 and M2 macrophages (Extended Data Fig. 3g). In fact, of all macrophage subsets, M1 macrophages in particular seem to down-regulate PARP1 activity as measured by reduced basal PARylation, which is more pronounced in Cd38 KO M1 macrophages (Extended Data Fig. 3g). Furthermore, treatment of macrophages with H2O2 induced detectable DNA damage in M0 and M2 macrophages (Extended Data Fig. 3h); however, both WT and Cd38 KO M1 macrophages showed less DNA damage, as evidenced by reduced PARylation and very little γH2AX in M1, compared with M0 and M2, macrophages (Extended Data Fig. 3h). These findings support the idea that CD38 and not PARP1 is the primary NAD-consuming enzyme in M1 macrophages.
Ageing is associated with enhanced adipose-tissue inflammation and the accumulation of CD38+ macrophages.
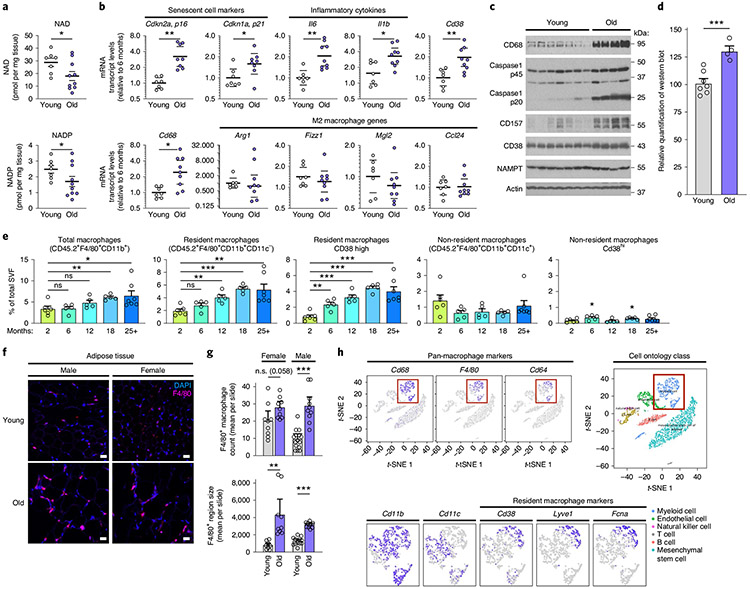
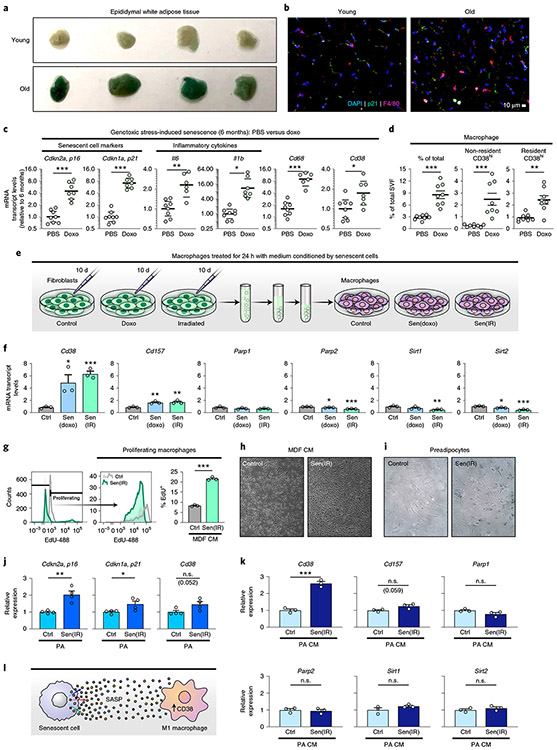
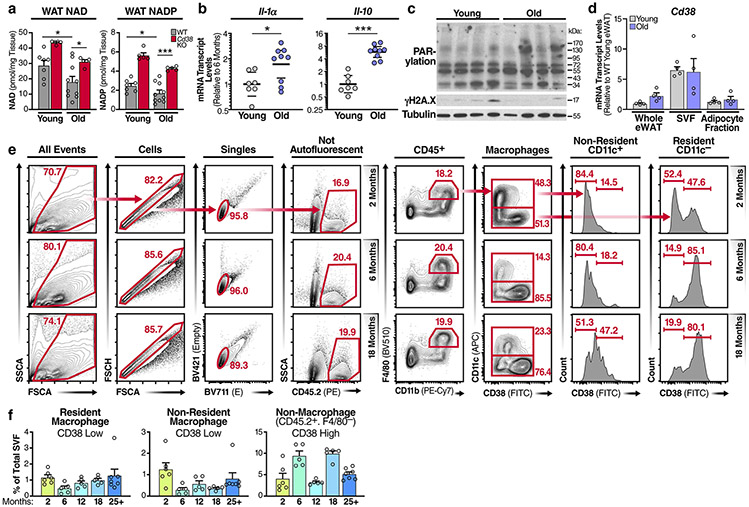
To verify that NAD levels decline in adipose tissue during ageing, we measured NAD levels in the eWAT from young and old WT mice. NAD and NADP levels were nearly 50% lower in old WT mice than in young WT mice (Fig. 4a), whereas Cd38 K0 mice were protected from significant NAD and NADP decline, as compared with their WT counterparts (Extended Data Fig. 4a), consistent with previous reports7,32. Our results showing that M1 macrophages feature enhanced CD38-dependent NAD degradation suggest that M1 macrophages may be a key driver of NAD loss during the chronic inflammation that occurs during ageing. To determine whether declining NAD levels during ageing are associated with chronic inflammation, we measured mRNA levels of several genes in old versus young eWAT. mRNAs encoding CD38, senescence markers (Cdkn2a mRNA, encoding p16Ink4a, and Cdkn1a mRNA encoding p21Cip1) and the inflammatory cytokines that comprise the SASP (Il6, Il1b, ll1a and Il10 mRNAs) increased in aged eWAT, as compared with their levels in young eWAT (Fig. 4b and Extended Data Fig. 4b). Conversely, M2 macrophage markers (such as Arg1, Fizz1 and Mgl2) trended down or were unchanged (Ccl24) (Fig. 4b). Furthermore, mRNA and protein levels of the macrophage marker CD68 increased in old mice, consistent with more activated macrophage in old tissues33 (Fig. 4b,c). Interestingly, the ageing-related decline in NAD levels was not associated with increased markers of DNA damage or PARP activity (Extended Data Fig. 4c). Rather, old eWAT tissue contained greater protein levels of CD38 and CD157 (Fig. 4c,d), and noticeably elevated levels of inflammasome activation, as indicated by higher levels of the 20-kDa processed form of caspase-1 (Fig. 4c). These data suggest that the CD38 expression in eWAT and ageing-related decline in NAD levels are associated with inflammation.
Fig. 4 ∣. NAD decline during ageing is associated with increased CD38+ tissue-resident macrophages in eWAT.
a, LC-MS was used to quantify NAD and NADP in visceral epididymal white adipose tissue (eWAT) from 6- and 25-month-old WT male mice. NAD and NADP concentrations are shown as pmol mg−1 of tissue (young n = 7 mice per group, old n = 10 mice per group). b, mRNA levels of senescence markers, inflammatory genes, macrophage marker Cd68 and M2 genes in eWAT from young (6 months old) and old (25 months old) WT male mice (young n = 7 mice per group, old n = 9 mice per group). c, Western analysis of the indicated proteins in eWAT from young (3 months old) and old (30 months old) WT male mice. Each lane represents one mouse (young n = 7 mice per group, old n = 4 mice per group). d, Quantification of CD38 protein levels in c, relative to actin levels, in eWAT from young (3 months old) and old (30 months old) WT mice (young n = 7 mice per group, old n = 4 mice per group). e, Quantification of total macrophages, CD38+ resident macrophages and CD38+ non-resident macrophages isolated from eWAT of WT male mice at the indicated ages (2 months n = 6 mice per group, 6 months n = 5 mice per group, 12 months n = 5 mice per group, 18 months n = 5 mice per group, 25+ months n = 7 mice per group). f, IF of the macrophage marker/ antigen F4/80 (magenta) and DAPI-stained nuclei (blue) in eWAT in young (4 months old) and old (26 months old) male and female WT mice. Scale bars, 10 μm. Images are representative of 4-5 mice per group. g, Results from trained neural network analysis of IF images, quantifying the mean number of macrophages (F4/80 colocalized to DAPI) per slide, and mean F4/80+ region size for old and young eWAT, graphed as mean cell count or region size (arbitrary units) per slide. Each column is based on two mice per group, except young/male which has three mice. Multiple images were taken from each mouse, with each image/slide represented as a dot; 9,11,9,13 images/slides for old/f, old/m, young/f, young/m, respectively. h, Analysis of CD38 and other macrophage markers in eWAT from single-cell transcriptome data using the Tabula Muris database (https://tabula-muris.ds.czbiohub.org). Data from individual mice are shown for in vivo experiments. t-SNE, t-distributed stochastic neighbour embedding. Data are shown as the mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test except, for one-sided t-test in a.
In both young and old eWAT, CD38 expression was most abundant in the stromal vascular fraction (SVF), instead of in adipocytes and whole tissue (Extended Data Fig. 4d). Therefore, we analysed CD38 expression in macrophages, the predominant immune cell in visceral adipose tissue34, isolated from the eWAT SVF from old and young mice. As reported, flow-cytometry analysis of the SVF can distinguish resident and non-resident macrophages on the basis of cell-surface markers, including F4/80 (EMR1) and CD11b (ITGAM), which are expressed by both resident and non-adipose-tissue-resident macrophages, and the integrin CD11c (ITGAX), which is expressed by non-resident adipose-tissue macrophages34,35. Using a similar gating strategy35 and as outlined in Extended Data Fig. 4e, the proportion of macrophages in the SVF increased significantly with age, particularly after 12 months of age (Fig. 4e). Additionally, we used immunofluorescence (IF) histological staining for the macrophage-specific marker F4/80, coupled with an unbiased neural network to quantify F4/80+ macrophages in old and young eWAT. Visually and with the trained neural network, F4/80+ macrophages were more abundant in old female and male mice than in younger mice (Fig. 4f,g). This increase was accounted for by an increase in the proportion of macrophages bearing a signature of resident macrophages (F4/80+CD11b+CD11c−), but not non-resident macrophages (F4/80+CD11b+CD11c+) (Fig. 4e). Interestingly, resident macrophages from older animals, but not non-resident macrophages, showed progressively increased CD38 expression (Fig. 4e and Extended Data Fig. 4f).
To further investigate in an unbiased fashion which macrophage populations express CD38 in adipose tissue, we interrogated the Tabula Muris single-cell RNA-sequencing (RNA-seq) database36 to dissect gene expression in single-cell populations in multiple organs and tissues. CD38 expression was specific to adipose-tissue-resident macrophages (CD68+CD64+F4/80+CD11b+CD11c−), not the non-resident population (CD68+CD64+F4/80+CD11b+CD11c+) (Fig. 4h), consistent with our flow-cytometry analysis results (Fig. 4e). Recent reports have characterized gene expression in tissue-resident immune cells and found that Lyve1 and Fcna are expressed along with Cd38 by a subset of tissue-resident macrophages37,38, a finding also confirmed in the Tabula Muris database (Fig. 4h).
Although CD38 is also expressed by B cells and activated T cells, we saw no significant increase in CD38 expression during ageing in non-macrophage immune cells in the SVF (Extended Data Fig. 4f). Thus, compared with that from young mice, adipose tissue from old animals was highly inflamed, had more senescent cells and macrophages and showed a noticeable shift in tissue-resident macrophage polarization from a M2 to a pro-inflammatory M1-like state with increased CD38 expression.
The senescence-associated secretory phenotype promotes CD38 expression.
Senescent cells are a key source of sterile inflammation in aged tissues and are characterized by an essentially permanent cell-cycle arrest, increased senescence-associated β-galactosidase (SA-Bgal) activity39 and a complex SASP that includes pro-inflammatory chemokines, damage-associated molecular patterns (DAMPs) and cytokines40.
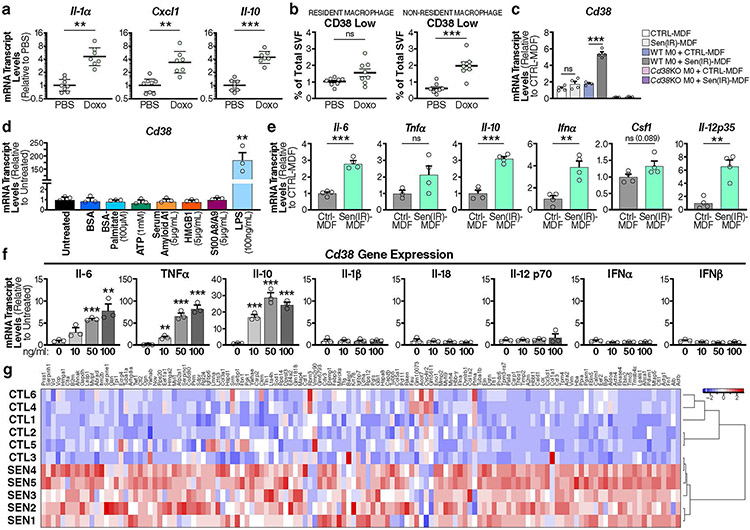
Visceral fat has emerged as a tissue that is most susceptible to accumulating senescence cells during ageing41, and this accumulation can impact the healthspan of mice42. Accordingly, we observed an abundance of SA-Bgal activity in old versus young eWAT (Fig. 5a). Further IF analysis showed an increase in both F4/80+ macrophages and p21+ senescent cells in close proximity to each other in old, compared with young, eWAT (Fig. 5b). We therefore investigated whether senescent cells influence macrophage polarization and CD38 expression by injecting young mice with the chemotherapeutic agent doxorubicin (doxo), which promotes DNA damage and cellular senescence in both cultured cells and in vivo43,44. In the eWAT of doxo-treated mice, Cd38 mRNA increased in parallel with mRNAs encoding the senescence markers (p16Ink4a, p21Cip1) and inflammatory cytokines and chemokines (Fig. 5c and Extended Data Fig. 5a), similar to that observed during ageing (Fig. 4b). Flow-cytometry analysis of eWAT from doxo-treated mice showed increased macrophage accumulation compared with that in control mice (Fig. 5d). Furthermore, doxo treatment also increased CD38+ resident and non-resident macrophages (Fig. 5d and Extended Data Fig. 5b). These results suggest that senescent cells and the SASP might be responsible for increasing CD38 expression in adipose-tissue macrophages in ageing eWAT.
Fig. 5 ∣. Cytokines secreted by senescent cells promote macrophage CD38 expression and proliferation.
a, SA-Bgal staining in young (3 months old) and old (19 months old) eWAT from WT male mice. n = 4 mice per group. b, IF images of the macrophage marker F4/80 (magenta) and DAPI-stained nuclei (blue) in eWAT in young (4 months old) and old (26 months old) WT male mice. Scale bars, 10 μm. Representative of 4-5 mice/group. c, mRNA levels in eWAT from 6-month-old WT male mice, which were intraperitoneally (i.p.) injected with PBS or doxo, of senescence markers, inflammatory cytokines, macrophage markers Cd68 and Cd38 in total tissue and isolated macrophages (PBS n = 8 mice per group, doxo n = 7 mice per group). d, Flow-cytometry analysis and quantification of CD38+ macrophages isolated from 6-month-old WT male mice i.p. injected with PBS or doxo (PBS n = 8 mice per group, doxo n = 8 mice per group). e, Conditioned medium (CM) was isolated from non-senescent control mouse dermal fibroblasts (CTRL-MDF), doxo-treated senescent MDFs (sen(doxo)-MDF) or irradiated senescent MDFs (sen(IR)-MDF) at 10 d following treatment, and then was used to stimulate BMDMs for 24 h. f, mRNA levels of Cd38 and those encoding other NAD-consuming enzymes in BMDMs treated for 24 h with CM from CTRL-MDFs, sen(doxo)-MDFs or sen(IR)-MDFs. g, Results from flow cytometry of EdU+ BMDMs treated with sen(IR)-MDF CM or CM from CTRL-MDFs for 24 h. h, Representative bright-field microscopy image of BMDMs treated with CTRL-MDF CM or sen(IR)-MDF CM for 24 h. i, SA-Bgal staining in control (CTRL-PA) or irradiated senescent primary mouse preadipocytes (sen(IR)-PA). j, mRNA levels of the indicated genes in CTRL-PA or sen(IR)-PA. k, mRNA levels of Cd38 and other NAD-consuming enzymes in BMDMs treated with CM from CTRL-PA sen(IR)-PA for 24 h. l, Model showing that inflammatory cytokines (SASP) derived from senescent cells can promote macrophage expression of CD38. Data from individual mice are shown for in vivo experiments. Data are shown as the mean ± s.e.m. (n = at least 3 independent biological experiments). *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test.
To more critically test the hypothesis that SASP drives CD38 expression in macrophages, we induced senescence in cultured mouse dermal fibroblasts (MDFs) by ionizing radiation (sen(IR)) or treatment with doxo (sen(doxo)) and collected the conditioned medium43,45 (Fig. 5e). Macrophages that were cultured for 24 h in conditioned media from senescent cells upregulated Cd38, and to a lesser extent Cd157, but not other genes encoding NAD-consuming enzymes (Fig. 5f). This upregulation of CD38 did not occur in macrophages co-cultured with non-senescent fibroblasts (Extended Data Fig. 5c). Furthermore, Cd38 mRNA levels did not increase in sen(IR)-fibroblasts alone compared with non-senescent fibroblasts (Extended Data Fig. 5c).
To investigate the SASP components responsible for the rise in CD38 expression in macrophages, we treated macrophages with DAMPs, endogenous alarmins derived from damaged or dead cells, that can activate TLRs46. Surprisingly, treatment of BMDMs with multiple DAMPs, including HMGB1, did not activate Cd38 expression (Extended Data Fig. 5d). In addition to DAMPs, cytokines such as TNF-α and type 1 interferons (IFN-α, IFN-β) were reported to drive CD38 expression in mouse and human macrophages47,48. Compared with non-senescent MDFs, sen(IR)-MDFs significantly upregulated Ifna and other inflammatory cytokine mRNAs (Il12, Il6, Il10 and Csf1; Extended Data Fig. 5e). Therefore, we treated BMDMs with recombinant cytokines and measured Cd38 mRNA levels (Extended Data Fig. 5f). Notably, although many of these cytokines including type 1 interferons did not promote Cd38 expression, TNF-α, IL-6 and IL-10 significantly increased Cd38 expression (Extended Data Fig. 5f). Thus, sterile sources of inflammation, including inflammatory cytokines that comprise the SASP, are sufficient to increase the expression of Cd38 in macrophages.
One of the most striking features of ageing eWAT in mice is the accumulation of resident macrophages (Fig. 4). Adipose-tissue-resident macrophages are seeded during development and have the capacity to self-renew; this renewal is driven by CSF1 and other growth factors, including MCP-1 and IL-4 during helminth infections21,49-52. Interestingly, sen(IR)-fibroblasts had increased expression of Csf1 transcripts (Extended Data Fig. 5e) and other growth factors40, which we confirmed were significantly increased at the protein level by proteomic analysis of the conditioned medium (Extended Data Fig. 5g). To determine whether the SASP can promote macrophage proliferation, we treated BMDMs with conditioned medium from sen(IR) and non-senescent fibroblasts and measured cell proliferation using a fluorescent thymidine analogue (EdU), which is incorporated into DNA during active synthesis (Fig. 5g). On treatment with conditioned medium from sen(IR), but not non-senescent, fibroblasts, macrophages incorporated threefold more EdU (Fig. 5g). This enhanced proliferation was evident after just 24 h of SASP exposure (Fig. 5h).
Finally, because CD38+ macrophages were elevated in aged eWAT, we asked whether the SASP from a senescent adipose progenitor cell can promote CD38 expression in macrophages. Preadipocytes are one of the most abundant adipose progenitor cells that can undergo senescence, and were recently implicated in reducing healthspan and lifespan in ageing mice42. As expected, sen(IR)-preadipocytes, compared with non-senescent preadipocytes, showed elevated SA-Bgal activity (Fig. 5i) and higher levels of mRNAs encoding p16Ink4a and p21Cip, but only modest expression of Cd38 (Fig. 5j). In comparison, BMDMs treated with conditioned medium from sen(IR)-preadipocytes showed significantly elevated Cd38 expression (Fig. 5k). These data provide cell-culture and in vivo evidence that sterile sources of inflammation, such as inflammatory cytokines that comprise the SASP of senescent cells, are a key driver of CD38 expression by macrophages (Fig. 5l).
Ageing is associated with increased CD38 expression by Kupffer cells in response to cellular senescence.
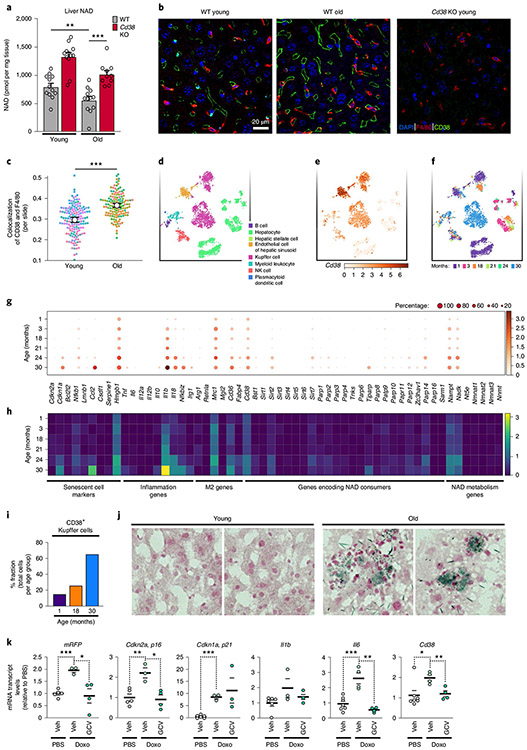
Ageing is associated with a decline in liver NAD levels, which we can confirm is CD38 dependent (Fig. 6a) as has recently been reported7. However, it is not known which cells in the aged-liver express CD38, or whether they are a key source of CD38 NADase activity. Liver-resident macrophages, known as Kupffer cells, comprise up to 15% of total liver cells and 80–90% of all tissue-resident macrophages53. Activation of Kupffer cells to a pro-inflammatory state is thought to be a driver not only of ageing-related liver disease, such as non-alcoholic liver disease, cirrhosis and fibrosis, but also in the pathogenesis of infectious diseases such as hepatitis54. Given the connection among inflammation, senescence and CD38 expression by adipose-tissue-resident macrophages, we investigated whether CD38 expression was altered in Kupffer cells in aged mice.
Fig. 6 ∣. CD38+ Kupffer cells accumulate in the livers of ageing mice.
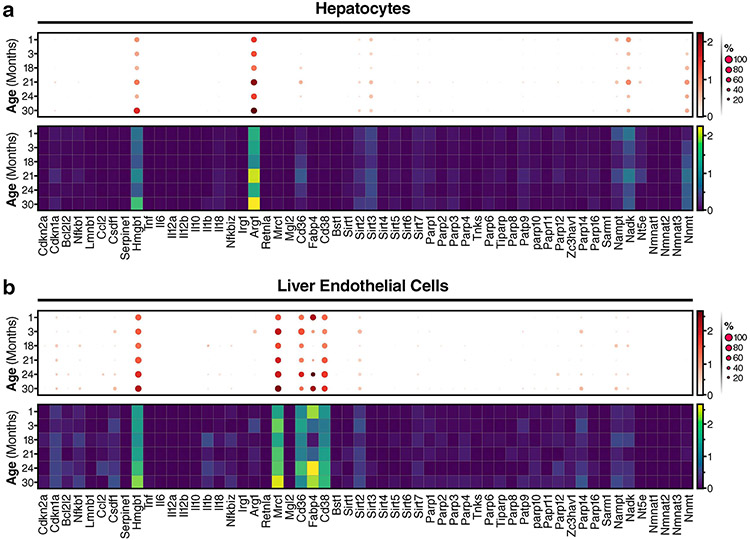
a, LC-MS quantification of NAD in the liver from young (4 months old) and old (26 months old) WT male and WT female mice (old n = 6 male and 6 female mice per group, young n = 6 male and 6 female mice per group), and Cd38 KO young (3 months old) and old (26 months old) male and female mice (old Cd38 KO n = 5 male and 5 female mice per group, young Cd38 KO n = 5 male and 5 female mice per group) NAD concentrations are shown as pmol per mg of tissue. b, IF images of the macrophage marker/antigen F4/80 (red), CD38 (green) and nuclei with DAPI (blue) in liver from WT young (4 months old) and WT old (26 months old) male mice, and young (3 months old) Cd38 KO male mice. Scale bars, 10 μm. Representative of 7-8 mice per group. c, Analysis of IF images above (a trained neural network to identify macrophage regions and colocalization of F4/80 and CD38, measured by Pearson correlation) for WT old and young liver slides (each dot represents 1 slide), n = 20 slides per mouse (young n = 8 mice per group, old n = 7 mice per group). d, t-SNE plot of annotated cell populations found in the livers of old and young male and female mice using single-cell transcriptome data from the Tabula Muris database (https://tabula-muris-senis.ds.czbiohub.org; used for d-i). e, t-SNE plot of CD38 expression in liver-cell populations in aged mice. f, t-SNE plot of liver cells annotated on the basis of mouse age. Note: Kupffer cells cluster by age. g, Dot plot of the indicated genes in Kupffer cells, with data sorted by mouse age. Logarithmic axes, base 10. h, Heatmap of the indicated genes in Kupffer cells, with data sorted by mouse age. Logarithmic axes, base 10. i, Percentage of total CD38+ Kupffer cells per total amount of cells per age group. j, SA-Bgal staining in liver sections from young (3 months old) and old (19 months old) WT male mice. Representative images of two out of four mice per group. k, mRNA levels of the p16+ senescent-cell reporter mRFP, senescent-cell markers (Cdkn2a (p16Ink4a) and Cdkn1a (p21Cip1)), inflammatory cytokines (I1b and Il6) and Cd38 in liver from 4- to 6-month-old p16-3MR male mice treated with PBS (vehicle); n = 7 mice per group, doxo (vehicle) n = 4 mice per group, and doxo (GCV) n = 4 mice per group. Data from individual mice are shown for in vivo experiments. Data are shown as the mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test, except one-tailed t-test in a.
IF staining for F4/80+ Kupffer cells and CD38 revealed increased expression of CD38 in liver sinusoids and greater numbers of F4/80+ macrophages near the sinusoids in old than in young mice (Fig. 6b). Because CD38 can also be expressed by liver endothelial cells29, and endothelial cells and Kupffer cells are in close proximity in the sinusoid capillary, we used an unbiased neural network to analyse coexpression of CD38 and F4/80. We found a significant increase in colocalization of F4/80+ and CD38 (Fig. 6c), indicative of greater amounts of CD38+ Kupffer cells in the livers of old mice. To explore this finding further, we collaborated with the Tabula Muris Consortium55. Annotation of single-cell RNA-seq data from the livers of young and old (1–30 months of age) mice revealed 8 distinct cell populations in the liver (Fig. 6d). However, Cd38 was expressed in the liver primarily by endothelial and Kupffer cells, whereas other immune cells and hepatocytes showed little Cd38 expression (Fig. 6e). Additionally, annotated Kupffer cells tended to cluster in distinct populations by age, suggesting that there are substantial differences in their pattern of gene expression during ageing (Fig. 6f).
Next, we measured mRNA levels of Cd38, as well as other NAD-consuming enzymes, M2 macrophage markers, inflammatory cytokines and senescent-cell markers in multiple liver-cell types (Fig. 6g,h). Cd38 gene expression was higher in Kupffer cells from old mice (24–30 months old), and the number of Kupffer cells that expressed Cd38 also increased with age (Fig. 6g-i), in accordance with our IF data. Notably, increased expression of Cd38 in aged Kupffer cells was paralleled by increased mRNA expression of the nicotinamide-salvage-pathway enzyme (Nampt mRNA), inflammatory cytokines (Il1b and Il18 mRNAs) and an inflammatory transcription factor (Nfkbiz mRNA), which is activated downstream of nuclear factor-κB, suggesting that Kupffer cells from old mice undergo a pro-inflammatory M1-like polarization. In contrast to Kupffer cells, expression of Cd38 in endothelial cells from old animals increased only modestly, and hepatocytes did not express Cd38 at any age (Extended Data Fig. 6a,b). Thus, similar to results in the aged eWAT (Fig. 4), pro-inflammatory M1-like CD38+ Kupffer cells also increase in the livers of aged mice.
In aged mice, senescent cells have recently been implicated in liver inflammation, characterized by fat deposition and greater numbers of macrophages56. Consistent with this finding, SA-Bgal activity increased in the livers of old versus young mice, indicative of an increase in senescent cells (Fig. 6j). Because increased inflammation and senescent-cell burden is sufficient to drive CD38 expression in adipose-tissue macrophages, we investigated whether senescent cells also promote CD38 expression in the liver. To test this hypothesis, we used the mouse strain p16-3MR, which contains a transgene in which the Cdkn2a (encoding p16Ink4a) promoter drives expression of monomeric red fluorescent protein (mRFP), Renilla luciferase and the HSV-1 thymidine kinase (HSV-TK). This transgene allows both the detection of p16+ senescent cells and the ability to selectively kill them by treating the mice with ganciclovir (GCV)57. We treated p16-3MR mice with doxo, which, as expected, increased expression of the senescent-cell markers p16Ink4a and p21Cip1 along with mRFP reporter (Fig. 6k). Notably, this increase was paralleled by increased CD38 expression, which was reversed after deleting senescent cells with GCV (Fig. 6k). Thus, the accumulation of senescent cells in the liver of doxo-treated mice is both sufficient and necessary to drive enhanced CD38 expression.
Chronic and acute inflammation promotes macrophage CD38 expression and NAD decline in metabolic tissues.
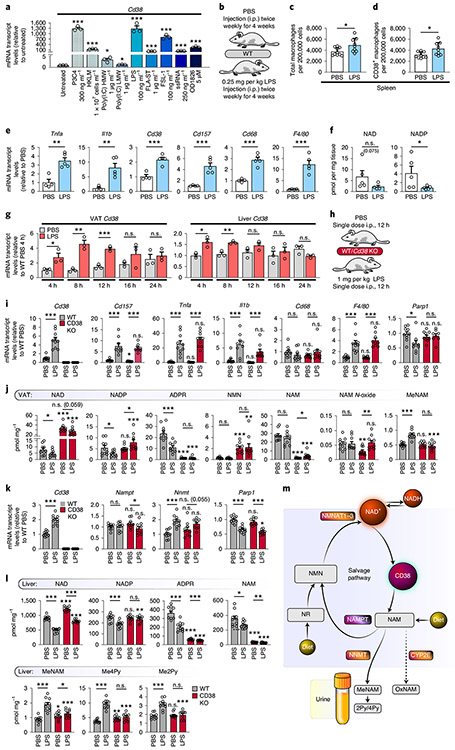
A recent study has shown that intestinal-barrier permeability increases during ageing, leading to higher levels of endotoxins, including LPS, in ageing tissues58,59. Therefore, we asked whether TLR ligands besides LPS increase Cd38 mRNAs in macrophages. We treated BMDMs with multiple pathogen-associated molecular patterns (PAMPs) that act as TLR ligands. All the PAMPs greatly increased Cd38 mRNA levels (Fig. 7a), with the exception of poly(I:C) (Fig. 7a). Together, these observations show that multiple microbial sources of inflammation are sufficient to promote the expression of Cd38 in macrophages.
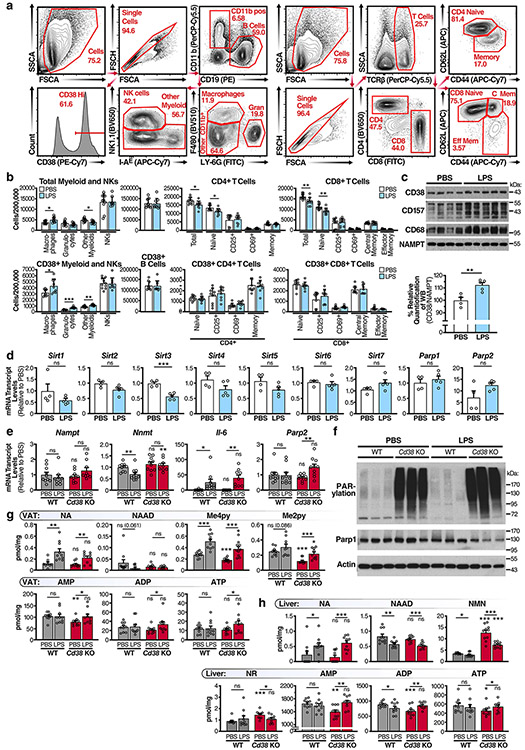
Fig. 7 ∣. Acute and chronic LPS treatment causes a CD38-dependent decrease in tissue NAD levels.
a, mRNA levels of Cd38 in WT BMDMs treated with the indicated TLR ligands for 16 h (n = 3 independent biological experiments). LMW, low molecular weight; HMW, high molecular weight. b, Treatment summary for WT male mice (2 months old) treated with 0.25 mg per kg (body weight) LPS or PBS for 4 weeks. c,d, Quantification of total and CD38+ macrophages in the spleen of 2-month-old WT male mice treated with LPS or PBS as above, and Cd38 KO mice, by flow cytometry. PBS n = 10 mice per group, LPS n = 9 mice per group. e, mRNA levels in eWAT from 3-month-old WT male mice i.p. injected with PBS or LPS for 4 weeks (PBS n = 4 mice per group, LPS n = 5 mice per group). f, LC-MS quantification of NAD and NADP in eWAT from 3-month-old WT male mice i.p. injected with PBS or LPS for 4 weeks. NAD and NADP concentrations are shown as pmol per mg of tissue (PBS n = 5 mice per group, LPS n = 5 mice per group). g, Cd38 mRNA levels in visceral adipose tissue (VAT)/eWAT and liver of WT male mice (4 months old) treated with 1 mg per kg (body weight) LPS or PBS over a 24-h period (PBS n = 3 mice per group, LPS n = 3 mice per group). h, Treatment summary for WT and Cd38 KO male mice (4 months old) treated with 1 mg per kg (body weight) LPS or PBS for 12 h. i, mRNA levels in eWAT from 4-month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 h (n = 10 mice per group). j, LC-MS quantification of NAD and other metabolites in VAT/eWAT from 4-month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 h (n = 10 mice per group). k, mRNA levels in livers from 4-month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 h (n = 10 mice per group). l, LC-MS was used to quantify NAD and other metabolites in livers from 4-month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 h (n = 10 mice per group). m, Diagram showing how excess NAM, derived from CD38, is methylated by NNMT and shunted away from the NAM-salvage pathway. Data from individual mice are shown for in vivo experiments. Data are shown as the mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001; two-sided Student’s t-test, except for one-tailed t-test in j and l.
As described above, ageing-related inflammation is associated with enhanced expression of CD38 by macrophages. To test whether CD38+ macrophages influence tissue NAD levels, we treated mice for 4 weeks with a non-lethal dose of LPS (Fig. 7b). Similar to what happened in aged mice (Fig. 4), LPS significantly elevated the numbers of total and CD38+ macrophages and myeloid cells in the spleen (Fig. 7c,d), whereas CD38 expression was not increased, and was even decreased, in other immune-cell populations (see Extended Data Fig. 7a for gating strategies), including B and T cells (Extended Data Fig. 7b). Further analysis of eWAT from mice injected with LPS showed higher levels of inflammatory cytokine (Tnfa and Il1b) (Fig. 7e), macrophage marker (Cd68 and F4/80) and NAD hydrolase (Cd38 and Cd157) mRNAs (Fig. 7e), which also increased at the protein level (Extended Data Fig. 7c), than in eWAT of mice injected with PBS. In contrast, the expression of other NAD-consuming enzymes, including sirtuins and PARPs, were unchanged or down-regulated in LPS-treated mice (Extended Data Fig. 7d). Metabolomic analysis of the eWAT also revealed decreased NAD and NADP levels in the inflamed LPS-treated eWAT compared with those in PBS-treated mice (Fig. 7f). These data provide evidence that exposure to low levels of endotoxins and PAMPs is sufficient to promote a low-grade chronic inflammatory state associated with increased expression of CD38 and lower NAD levels (Fig. 8).
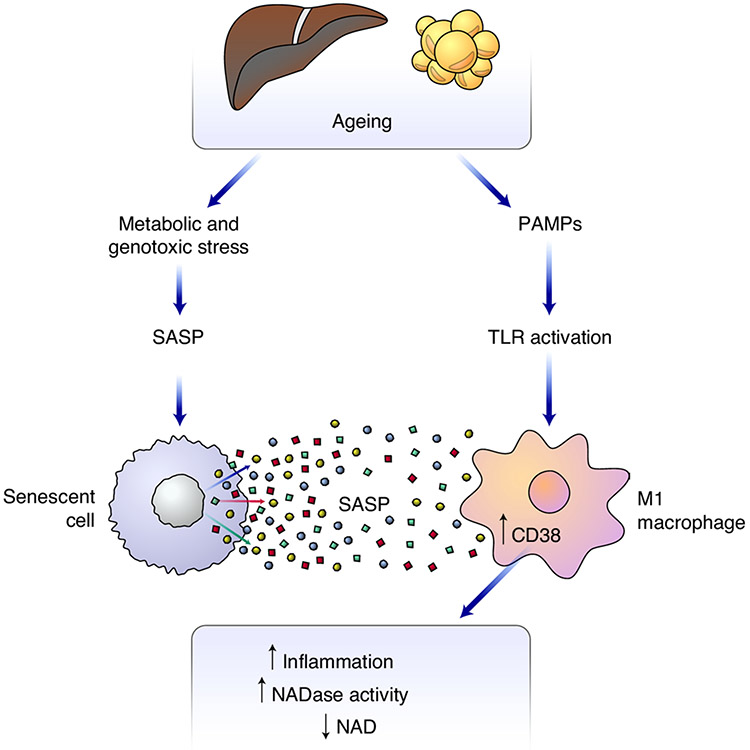
Fig. 8 ∣. Proposed model for how ageing-related inflammation enhances NAD degradation.
Cellular stressors such as DNA damage lead to an accumulation of senescent cells over time. Using in vivo and cell-culture models, we show that the accumulation of senescent cells and accompanying inflammatory cytokines of the SASP is necessary and sufficient to promote CD38 expression and proliferation in macrophages. In addition, increased intestinal permeability occurs during ageing, increasing serum levels of endotoxins and other PAMPS, which activate innate immune cells. Chronic and acute exposure to LPS promotes CD38 expression in macrophages in the eWAT and liver, and decreases tissue NAD levels. Collectively, the SASP and PAMPs promote an inflammatory state associated with increased expression of CD38 by tissue-resident M1-like macrophages, and hence enhanced NADase activity.
To determine whether the lower tissue NAD levels in mice treated with LPS were CD38 dependent, we treated WT mice with an acute sublethal dose of LPS and measured CD38 expression over 24 h. CD38 expression peaked in WT eWAT and liver around 8–12 h after exposure to LPS (Fig. 7g); thus 12 h was used to measure NAD in Wt and Cd38 KO mice (Fig. 7h). Similar to that of mice treated with chronic LPS, the eWAT of mice treated with an acute LPS dose also had significantly elevated expression of inflammatory cytokines (Fig. 7i and Extended Data Fig. 7e) and macrophage markers (F4/80 but not Cd68), and significant expression of Cd38 and Cd157 (Fig. 7i). Consistent with Extended Data Fig. 3, we did not detect enhanced expression of Parp1 or Parp2, or any detectable PARP activity, in the eWAT of LPS-treated mice, whereas basal PARylation was noticeably increased in Cd38 KO mice (Fig. 7i and Extended Data Fig. 7e,f).
To determine whether Cd38 KO mice were protected from LPS-induced NAD decline, we measured NAD and related metabolites in the eWAT of LPS and control Cd38 KO and WT mice. As expected, based on the increased expression of CD38 in the eWAT of WT mice, an acute dose of LPS reduced NAD and NADP levels by 50% in WT mice, whereas Cd38 KO mice were protected from major NAD and NADP declines (Fig. 7j). Further analysis revealed robust change on exposure to LPS in many NAD metabolites (Fig. 7j and Extended Data Fig. 7g). Similar to that in eWAT, Cd38 expression significantly increased in the livers of WT mice treated with LPS, but not PBS (Fig. 7k). Notably, LPS significantly decreased NAD and NADP in the livers of WT mice (Fig. 7l). Cd38 KO mice treated with LPS were protected from a decline in NADP (Fig. 7l). However, Cd38 KO mice treated with LPS had significantly less NAD than PBS-treated Cd38 KO mice, although they still had signficantly higher NAD levels than LPS-treated WT mice (Fig. 7l). Furthermore, NA increased and nicotinic acid dinucleotide declined in both the eWAT and liver of WT and Cd38 KO mice treated with LPS (Extended Data Fig. 7g,h), indicative of decreased NAD biosynthesis by the Preiss–Handler pathway and NAD de novo pathway upon LPS exposure.
Enhanced NAD turnover by CD38 or decreased NAD biosynthesis does not fully explain why NAD levels decline in response to LPS, because the NAM-salvage pathway should be able to convert excess NAM to NAD (Fig. 7m). Thus, we asked whether CD38-dependent turnover of NAD in response to LPS was coupled to the activity of nicotinamide-N-methyltransferase (NNMT), an enzyme that uses S-methyl-adenosine as a methyl donor to methylate the ring nitrogen of NAM, converting it to N1-methylnicotinamide (MeNAM). NNMT conversion of NAM to MeNAM effectively diverts NAM from recycling back to NAD by the NAM-salvage pathway and thus affects global NAD+ levels (Fig. 7m). In support of our hypothesis, LPS exposure increased the expression of Nnmt in the liver, but not eWAT, in both WT and Cd38 KO mice (Fig. 7k and Extended Data Fig. 7e). However, only WT LPS-treated mice showed increased production of MeNAM in both eWAT and liver; WT PBS-treated mice and Cd38 KO mice did not (Fig. 7j,l). Further, MeNAM can be further oxidized via aldehyde oxidase (AOX) to produce N1-methyl-pyridone-5-carboxamide (2py) and N1-methyl-4-pyridone-carboxamide (4py), which, along with MeNAM, are secreted in the urine (Fig. 7m). Both 2py and 4py metabolites increased in the livers of WT LPS-treated mice, but not in Cd38 KO mice. These data suggest that, in inflammatory conditions, excess NAM, produced by consumption of NAD by CD38, is methylated by NNMT in the liver and shunted away from the NAM-salvage pathway (Fig. 7m). Additionally, excess NAM was reported to be oxidized by the cytochrome P450 enzyme CYP2E, converting NAM to NAM-N-oxide (oxNAM), which is also secreted in urine60 (Fig. 7m). However, we did not detect a significant increase in oxNAM in the eWAT of WT LPS-treated mice compared with WT mice treated with PBS (Fig. 7j). Furthermore, the overall abundance of oxNAM was tenfold less than that of MeNAM in the eWAT (Fig. 7j), and we were unable to detect oxNAM in the liver. Thus, our data suggest that clearance of excess NAM downstream of CD38 is primarily via its methylation by NNMT.
Lastly, a recent study using stable-isotope tracing has demonstrated that, outside the liver, NAM is the major NAD precursor used by peripheral cells and tissues to maintain NAD levels6. Thus, the methylation and clearance of excess NAM, derived from CD38 in the liver, may also influence NAD levels in peripheral tissues under inflammatory conditions. Understanding the conditions and disease states that influence this connection between CD38 and NNMT will be the focus of future studies, and will have implications for restoring NAD levels during ageing and diseases associated with chronic and acute inflammation.
Discussion
In this study, we show that ageing-related inflammation drives the polarization of tissue-resident macrophages to an M1-like state characterized by increased expression of CD38 and enhanced NAD consumption (Figs. 1 and 3). As organisms age, genotoxic and other stressors cause some cells to undergo senescence. Although senescent cells can be beneficial during wound healing and embryogenesis, they also secrete inflammatory and other factors that can disrupt normal cellular functions, damage tissues and promote carcinogenesis61,62. Here, we provide evidence that the SASP is necessary and sufficient to increase CD38 expression in macrophages and promote macrophage proliferation, leading to an increase in CD38+ macrophages in aged mice and young mice with an increased senescent-cell burden due to treatment with the genotoxic agent doxo (Figs. 5 and 6). These results link the accumulation of senescent cells with age to the age-related NAD decline. Our study also raises new questions regarding how senescent cells and macrophages communicate and how their interactions influence each other’s function. Although we show that the SASP, in particular the immunomodulatory cytokines, such as IL-6, TNF-α and IL-10, can promote CD38 expression in macrophages (Extended Data Fig. 5f), it will be important to investigate other SASP components that can activate CD38 expression in macrophages.
Previous studies have reported that CD38 regulates immune-cell homing, innate immune responses to pathogens and antitumour immunity63-65. However, Cd38 remains a largely uncharacterized gene, despite its widespread use as an activation marker. Although the teleological purpose of increased NAD consumption by CD38+ M1 macrophages is not obvious, it is tempting to speculate that NAD degradation by CD38 may influence NAD levels to control other NAD-consuming enzymes, such as PARPs and sirtuins. In support of competition among different NAD-consuming enzymes for NAD, one of the most prominent phenotypes in Cd38 KO tissue is the enhanced basal PARylation in Cd38 KO BMDMs (Extended Data Fig. 3g) and eWAT (Extended Data Fig. 7f), similar to the enhanced SIRT3 activity that was reported in Cd38 KO mice7. Thus, CD38 may be expressed by inflammatory M1 macrophages to modulate cell-autonomous NAD-dependent processes to control macrophage activation and gene expression (Fig. 2), or metabolic pathways in neighbouring cells. Indeed, a recent study has shown that metabolic rate is substantially decreased in LPS-treated mice, as a means to conserve energy for the innate immune system66. Given the role of NAD as critical coenzyme in many metabolic pathways, future studies to investigate how NAD levels and CD38 are connected to this reduction in the metabolic rate will be important to better understand how inflammation, immune responses and metabolism are interconnected. Another mechanism by which CD38 may influence cellular processes and immune responses is by the production of secondary messenger NAD metabolites, including cADPR and NaADP, to control Ca2+ mobilization and cell migration67. In addition, NAM has immunomodulatory properties, and high NAM levels can reduce inflammatory cytokine production in macrophages68, suggesting that CD38 may act as a feedback mechanism to initiate the resolution of acute inflammation.
In conclusion, our study demonstrates that increased expression of CD38 in macrophages is an important source of increased NADase activity during ageing. Notably, in a study published in this issue of Nature Metabolism, Chini and colleagues have reported similar findings, which are consistent with the role of inflammaging as a driver of NAD decline during ageing69. The data from both articles provide insight into how the immune system and NAD metabolism are integrated. Although our study focuses on metabolic tissues, including visceral white adipose tissue and the liver, we speculate that a similar mechanism linking the SASP to macrophage CD38 NADase activity may contribute to NAD declines in other tissues, particularly those in which tissue-resident macrophages reside in high abundance, such as the brain. This newly discovered cross-talk between senescent cells and macrophages, which modulates tissue NAD levels, may provide a therapeutic avenue to target age-related inflammation and help to restore NAD levels and metabolic health in ageing individuals.
Methods
Mice.
Male and female C57BL/6J and Cd38 KO mice on a C57BL/6J background were used for in vivo studies. Six-to 12-week-old C57BL/6J and Cd38 KO male mice were used as a source of BMDMs. Mice were maintained at the Buck Institute vivarium (although initially at UCSF-Gladstone Institute) on a standard chow diet; the light cycle was 12-h light–12-h dark (6:00–18:00, light), temperature throughout facility was set at 20–22.2 °C, and E.V.’s vivarium room stays at 21.1–22.2 °C. The vivarium records daily humidity to make sure it stays within the allowable range of 30–70%—over the year it averages approximately 30–40% in the room used to house mice. All procedures were performed in accordance with federal guidelines and those set forth by the Buck Institute and the UCSF Gladstone Institutional Animal Care and Use Committees (IACUC).
Cd38 KO mice on a C57BL/6J background were from The Jackson Laboratory. NamptFl/Fl mice were developed by the laboratory of O. Leo (Université Libre de Bruxelles), and shared by the laboratory of S.-I. Imai (Washington University).
Bone marrows used in experiments comparing WT, Cd157 (Bst1) KO, Cd38 KO and Cd38/Cd157 DKO mice were generously provided by the laboratory of K. Ishihara (Kawasaki Medical School).
In vivo experiments.
Male and female C57BL/6J mice were obtained from the National Institute on Aging colony and The Jackson Laboratory. Colonies of old and young male and female Cd38 KO mice were and maintained at the Buck Institute. Male and female mice aged 2 to 30 months were used for natural ageing studies. As designated in the figure legends the following age, sex and number of animals were used in the following in vivo experiments: Fig. 4a, Extended Data Fig. 4a: WT young male mice (6 months old) n = 7 mice per group, Cd38 KO young male mice (3 months old) n = 5 mice per group, WT old male mice (25 months old) n = 10 mice per group, and Cd38 KO old male mice (26 months old) n = 5 mice per group. Fig. 4b, Extended Data Fig. 4b: 6- and 25-month-old WT male mice; WT young male mice (6 months old) n = 7 mice per group, WT old male mice (25 months old) n = 9 mice per group). Fig. 4c,d: young (3 months old) and old (30 months old) WT male mice. Each lane represents 1 mouse (young n = 7 mice per group, old n = 4 mice per group). Extended Data Fig. 4c: young (3 months old) and old (19 months old) WT male mice, each lane represents 1 mouse (young n = 4 mice per group, old n = 4 mice per group). Extended Data Fig. 4d: young (3 months old) and old (19 months old) WT male mice (young n = 4 mice per group, old n = 4 mice per group). Fig. 4e, Extended Data Fig. 4f: WT male mice for the indicated ages. (2 months old n = 6 mice per group, 6 months old n = 5 mice per group, 12 months old n = 5 mice per group, 18 months old n = 5 mice per group, 25+ months old n = 7 mice per group). Fig. 4f: representative image from young (4 months old) and old (26 months old) male and female WT mice. Images are representative of 4–5 mice per group. Fig. 5a: young (3 months old) and old (19 months old) eWAT from WT male mice. Fig. 5b: young (4 months old) and old (26 months old) WT male mice. Fig. 5c, Extended Data Fig. 5a: 6-month-old WT male mice (PBS n = 8 mice per group, doxo n = 7 mice per group). Fig. 5d, Extended Data Fig. 5b: 6-month-old WT male mice (PBS n = 8 mice per group, doxo n = 8 mice per group). Fig. 6a: young (4 months old) and old (26 months old) WT male and WT female mice (old n = 6 male and 6 female mice per group, young n = 6 male and 6 female mice per group), and Cd38 KO young (3 months old) and old (26 months old) male and female mice (old Cd38 KO n = 5 male and 5 female mice per group, young Cd38 KO n = 5 male and 5 female mice per group). Fig. 6b: Representative image from WT young (4 months old) and WT old (26 months old) male mice, and a young (3 months old) Cd38 KO male mouse. Representative of 7–8 mice per group. Fig. 6c: analysis of WT male old and young liver slides from Fig. 6b (each dot represents 1 slide), n = 20 slides per mouse (young n = 8 mice per group, old n = 7 mice per group). Fig. 6j: young (3 months old) and old (19 months old) WT male mice. Representative images of two out of four mice per group. Fig. 6k: 4- to 6-month-old p16-3MR male mice treated with PBS (vehicle) n = 7 mice per group, doxo (vehicle) n = 4 mice per group and doxo (GCV) n = 4 mice per group. Fig. 7c,d and Extended Data Fig. 7a,b: 2-month-old WT male mice treated with LPS or PBS (PBS n = 10 mice per group, LPS n = 9 mice per group). Extended Data Fig. 7c: 3-month-old WT male mice injected with PBS or LPS for 4 weeks (PBS n = 4 mice per group, LPS n = 5 mice per group). Fig. 7e, Extended Data Fig. 7d: 3-month-old WT male mice injected with PBS or LPS for 4 weeks. PBS n = 4 mice per group, LPS n = 5 mice per group); Fig. 7f: NAD and NADP concentrations are shown in 3-month-old WT male mice i.p. injected with PBS or LPS for 4 weeks (PBS n = 5 mice per group, LPS n = 5 mice per group). Fig. 7g: WT male mice (4 months old) treated with 1 mg per kg (body weight) LPS or PBS over a 24-h period (PBS n = 3 mice per group, LPS n = 3 mice per group). Fig. 7i-l and Extended Data Fig. 7e,g,h: 4-month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 h. n = 10 mice per group. Extended Data Fig. 7f: 4-month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 h (n = 3 mice per group), each lane represents 1 mouse.
In doxo experiments, 4- to 6-month-old male mice were injected i.p. with a single dose of 10 mg doxo (Sigma Aldrich) per kg (body weight) or vehicle (PBS) and euthanized 6 weeks later. To eliminate p16+ cells, p16-3MR male mice were i.p. injected with 25 mg per kg (body weight) GCV for 5 consecutive days before euthanasia. For chronic LPS experiments, 2-month-old male C57BL/6J mice from The Jackson Laboratory were injected with 0.25 mg per kg (body weight) LPS or vehicle (PBS) twice weekly for 4 weeks (LPS EK-Ultrapure, Invivogen). For acute LPS experiments, 3- to 4-month-old male Cd38 KO mice bred in-house, and aged-matched C57BL/6J control male mice from The Jackson Laboratory were injected with a single dose of 1 mg per kg (body weight) LPS or vehicle (PBS) (LPS EK-Ultrapure, Invivogen). All animals were euthanized with CO2, followed by cervical dislocation. Epididymal fat and/or liver tissues were excised, and portions of these were fixed for microscopy, flash frozen in liquid nitrogen for quantitative PCR and western blot analysis or processed for flow cytometry. Littermates were not used to compare WT and Cd38 KO mice. All procedures were approved by the Buck Institute IACUC.
BMDM culture and experiments.
BMDMs were derived from bone marrow extracted from the femurs of euthanized mice (6- to 12-week-old male C57BL/6J or Cd38 KO mice). Briefly, femurs were placed in a mortar and washed with 70% ethanol, followed by 2 washes with complete RPMI (cRPMI; standard RPMI 1640 (Corning) supplemented with 10% foetal calf serum, penicillin–streptomycin (Corning), 1 mM sodium pyruvate (Corning), 2 mM l-glutamine (Corning), 10 mM HEPES buffer (Corning) and 50 μM 2-mercaptoethanol). After washing, 10 ml of cRPMI was added to the mortar, and the femurs were gently crushed. The suspension was collected and filtered through a 70-μm filter, placed in a conical tube, and centrifuged at 1,200 r.p.m. (150 RCF) for 5 min. Cells were resuspended, counted and plated at 3 × 106 cells per 10-cm dish in 10 ml of macrophage growth medium (cRPMI containing 25% M-CSF containing L929-conditioned medium). Cells were cultured for 7 d to differentiate and were supplemented with 5 ml of macrophage growth medium on day 5. On day 7, BMDMs (10–12 × 106 cells per 10-cm dish) were lifted off the plate using cold PBS containing 5 mM EDTA. BMDMs were counted and replated in macrophage growth medium overnight prior to experiments. On the day of experiments, the medium was replaced with cRPMI 6 h prior to stimulation. M2 polarization was performed by stimulating macrophages with 10 ng ml−1 recombinant mouse IL-4 (Peprotech). For M1 polarization, macrophages were stimulated with 100 ng ml−1 LPS from Escherichia coli K12 (LPS EK-Ultrapure, Invivogen). To test different TLR ligands for CD38 activation, we used a Mouse TLR Agonist kit (Invivogen), and BMDMs were treated with each ligand for 16 h. The NAMPT inhibitor FK866 (Sigma Aldrich) was used at a final concentration of 50 nM, and sirtuin inhibitors Agk2 and EX527 were from Sigma Aldrich and used at the indicated concentrations.
Measurement of gene expression.
For cell-culture experiments, RNA was isolated using RNA STAT 60 (Amsbio) as per the manufacturer’s protocol.
One microgramme of RNA was converted to complementary DNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems), then diluted with 280 μl H2O. Gene expression was measured using a CFX384 Real Time System (Bio-Rad) and Thermo Scientific Maxima 2× SYBR Green. Data were analysed using the Bio-Rad CFX Manager 3.1 software and normalized using the 2−ΔΔCt method. BMDMs were normalized to hypoxanthine phosphoribosyltransferase (Hprt). For in vivo experiments, RNA was extracted from flash-frozen epididymal fat using TRIzol (Thermo Fisher) in a TissueLyser system (Qiagen). cDNA was generated as described above, and gene expression was measured in a LightCycler480 II using the Universal Probe Library system (Roche). Expression was normalized to actin and Hprt using the 2−ΔΔCt method.
NAD metabolome mass spectrometry.
NAD, NADP and other metabolite levels were measured by shipping tissue samples in dry ice from E.V.’s laboratory to C.B.’s laboratory, and stored at −80 °C until further processing. Samples were pulverized prior to extraction by freezing in LN2 and being added to a Bessman pulverizer cooled to LN2 temperature. Samples were weighed frozen in microcentrifuge tubes cooled to LN2 temperature. The appropriate internal standard solution was added, and the sample was extracted with hot 75% ethanol/25% HEPES (pH 7.1), as previously described70. After centrifugation, the aqueous layer between the pellet and lipid layer was transferred to a clean tube; the solvent was evaporated to dryness using a vacuum centrifuge. Standards and controls were prepared by adding internal standard, the appropriate amount of standard working solution and ethanol/HEPES buffer; the standards were dried along with the samples, and stored at −20 °C until reconstitution immediately prior to analysis. For analysis, samples were reconstituted in 70 μl of 10 mM ammonium acetate and analysed using 2 separations70,71, and tissue NAD and NADP levels were normalized to tissue weight. For cells, 7 × 106 BMDMs were cultured in 10-cm dishes overnight in macrophage growth medium to ~90% confluence. The following day, media were replaced with cRPMI 6 h prior to stimulating with IL-4 (10 ng ml−1) or LPS (100 ng ml−1) for the indicated timepoints. BMDMs were washed twice with cold PBS, gently scraped off the plates, counted, pelleted, flash frozen and shipped on dry ice to C.B.’s laboratory. The cell pellets were stored at −80 °C. To extract metabolites, the appropriate standard was added along with 400 μl of hot ethanol/HEPES buffer70,71. Calibrators and controls were prepared and solvent removed in a vacuum centrifuge. The analytical runs were conducted as above. BMDM metabolite levels were normalized to cell number (pmol per 1 × 106 cells). Internal standards were stable-isotope analogues of nucleotides and nucleoside from yeast grown in broth with universally labelled 13C-labelled glucose, resulting in all ribose rings being fully labelled. [13C10]NAD was used as an internal standard. For the second analysis, a mix of [18O]NR, [18O]Nam, d4-NA, d3-MeNam and d3-methyl-4-pyridone-3-carboxamide was used as a standard.
Quantification of tryptophan and metabolites.
We cultured 7 × 106 BMDMs in 10-cm dishes overnight in macrophage growth medium to ~90% confluence. The following day, the medium was replaced with cRPMI 6 h prior to stimulation with IL-4 (10 ng ml−1) or LPS (100 ng ml−1) for 24 h. Cells were washed twice with cold PBS, and gently scraped off, pelleted, flash frozen and shipped to Y.-M.L.’s laboratory on dry ice. The extracted metabolites were analysed by LC–electrospray ionization–tandem MS (LC–ESI–MS/MS) (API 5000 QTRAP mass, AB/SCIEX) by MRM mode. Tryptophan and metabolite MS/MS transitions (m/z) were 205 → 188 for tryptophan, 209 → 192 for kynurenine, 225 → 208 for 3-hydroxy-l-kynurenine, 154 → 136 for 3-hydroxyanthranilic acid, 206 → 160 for kynurenic acid, 190 → 144 for xanthurenic acid, 138 → 120 for anthranilic acid, 124 → 106 for 2-picolinic acid, 168 → 150 for quinolinic acid and 199 → 112 for l-mimosine as an internal standard. The metabolites were separated on a Luna C18 column (2.1 × 150 mm, 5.0 μm) with an injection volume of 5 μl and flow rate of 0.3 ml min−1 using 0.1% trifluoroacetic acid for mobile phase A and acetonitrile for mobile phase B. The gradient was as follows: 0 min, 2% B; 12 min, 60% B; 13 min, 60% B; 14 min, 2% B; 20 min, 2% B. Data were acquired using Analyst 1.5.1 software (Applied Biosystems), and are presented as pmol per mg protein.
Extraction of nicotinamide riboside and LC–MS/MS analysis.
The relative abundance of NR was assessed by LC–MS/MS. Briefly, 7 × 106 BMDMs were cultured to ~90% confluence, as outlined above. The following day, the medium was replaced with cRPMI 6 h prior to stimulation with LPS (100 ng ml−1) for the indicated timepoints. Cells were washed twice with cold PBS, and scraped from the plates, pelleted, flash frozen and shipped overnight to I.B.-S.’s laboratory on dry ice. Metabolites were extracted on ice with 250 μl of a 40:40:20 mixture of acetonitrile/methanol/(10 mM Tris, pH 9.2, 200 mM NaCl). Supernatants were stored at −80 °C (for 1 week or less), and 25 μl was injected into a Thermo Q-Exactive (LC–MS) in line with an electrospray source using an Ultimate3000 (Thermo) series high-performance LC (HPLC) consisting of a binary pump, degasser and auto-sampler outfitted with a Xbridge Amide column (Waters; dimensions of 4.6 mm × 100 mm; 3.5-μm particle size). The mobile phase A contained 95% (vol/vol) water, 5% (vol/vol) acetonitrile, 20 mM ammonium hydroxide, 20 mM ammonium acetate pH 9.0; B was 100% acetonitrile. The gradient was as follows: 0 min, 15% A; 2.5 min, 30% A; 7 min, 43% A; 16 min, 62% A; 16–18 min, 75% A; 18–25 min, 15% A with a flow rate of 400 μl min−1. The ESI source capillary was set to 275 °C, with sheath gas at 45 arbitrary units, auxiliary gas at 5 arbitrary units and spray voltage at 4.0 kV. In positive/negative polarity switching mode, an m/z scan range from 70 to 850 was chosen, and MS1 data collected at a resolution of 70,000. The automatic gain control (AGC) target was set at 1 × 106, and the maximum injection time was 200 ms. The top 5 precursor ions were fragmented, in a data-dependent manner, using the higher energy collisional dissociation cell set to 30% normalized collision energy in MS2 at a resolution power of 17,500. Samples (25 μl) were injected. Data acquisition and analysis were carried out by Xcalibur 4.0 and Tracefinder 2.1 software, respectively (Thermo Fisher Scientific). NR peak area was normalized to protein concentration for each sample and graphed as normalized peak area. Metabolomics was performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Adipose-tissue digestion.
Briefly, the tissue was minced, prior to digestion with 2 mg ml−1 collagenase (Sigma) diluted with KRBH buffer containing 2% FBS for 30 min at 37 °C. The suspension was filtered through a 70-μm mesh and centrifuged for 5 min at 2000 r.p.m. (425 RCF). The fat layer and supernatant were discarded, and the cell pellet of stromal vascular cells (SVF) was resuspended, and washed twice with PBS containing 1 mM EDTA and 2% FBS prior to lysing red blood cells with ACK lysis buffer and staining cells with antibodies for flow cytometry.
Flow cytometry.
Adipose-tissue SVF cells (isolated as described above), were counted and blocked with anti-mouse FC-block (BD Biosciences) per the manufacturer’s protocol for 15 min on ice. Cells were labelled on ice for 30 min with the following antibodies diluted 1:200: CD38–FITC (Biolegend, clone: 90, cat. no. 102705), CD11c–APC (Biolegend clone: N418, cat. no. 117309), CD11b–PECy7 (eBioscience, clone: M1/70, cat. no. 25-0112-82), CD45.2 PE (eBioscience, clone: 104, cat. no. 12-0454-82), F4/80-BV510 (Biolegend, clone: BM8, cat. no. 123135). Cells were immediately analysed using the BD FACSAria flow cytometer and the software BD FACSDiva 8.0.2 to acquire the data. Analysis of the data was performed using the FlowJo 10.5.2 software. For cell-culture experiments, activated BMDMs were lifted from non-TC-treated plates after activation using cold PBS with 5 mM EDTA for 15 min prior to blocking with Fc block and staining with antibodies as described above. Click-iT EdU 488 Flow Cytometry Kit (Thermo Fisher) was used to measure macrophage proliferation as per manufacturers protocol. Leukocyte suspensions were prepared from mouse spleens by crushing, dissociation of cells by pipetting, straining through 40-μm nylon mesh and ACK lysis. After a 15-min treatment with Fc block, surface staining with fluorochrome-conjugated antibodies was performed for 30 min on ice, at 1 μg ml−1 for each antibody, in PBS with 2% FCS and 2 mM EDTA, followed by fixation in PBS/1% PFA and analysis on a BD LSR II flow cytometer using the BD FACSDiva 8.0.2 Software. Compensation and gating were performed using FlowJo. All antibodies were obtained from Biolegend and diluted 1:200: CD4-BV650 (clone: GK1.5, cat. no. 100469); CD8α–Alexa 488 (clone: 53-6.7, cat. no. 100726) TCRβ–PerCP–Cy5.5 (clone: H57-597, cat. no. 109227) or TCRβ–PE (clone: H57-597, cat. no. 109207); Ly-6G–FITC (clone: 1A8, cat. no. 127605); CD11b–PerCP–Cy5.5 (clone: M1/70, cat. no. 101227); CD62L–APC (clone: MEL-14, cat. no. 104411); CD44–APC–Cy7 (clone: IM7, cat. no. 103027); CD25–Pacific Blue (clone: PC61, cat. no. 102021); CD69–BV711 (clone: H1.2F3, cat. no. 104537); F4/80-BV510 (clone: BM8, cat. no. 123135); CD11c–Pacific Blue (clone: N418, cat. no. 117321); Class II MHC (I-AI/E)–APC–Cy7 (clone: M5/114.15.2, cat. no. 107627); NK1.1-BV650 (clone: PK136, cat. no. 108735); CD38–PE–Cy7 (clone: 90, cat. no. 102717).
The gating strategy for Figs. 4e and 5d and for Extended Data Figs. 4f and 5b was as follows. A flow-cytometry gating strategy (Extended Data Fig. 4e) was used to identify CD45+ immune cells from the stromal vascular fraction of eWAT. Cells were first gated on forward scatter (FSCA) versus side scatter (SSCA) to discard cell debris and dead or dying cells. Next, FSCH (height) versus FSCA (area) was used to select single cells. Single cells were then gated for auto-fluorescence using the Empty(E) BV421 versus BV711 channels (not used as antibody fluorophores) to discard cells that showed auto-fluorescence in these channels. Then, CD45+ cells were selected and analysed for CD38 and macrophage markers. Flow cytometry gating strategy to identify resident and non-resident macrophages from the stromal vascular fraction of eWAT, showing representative flow plots and histograms for the indicated ages of mice is described in Extended Data Fig. 4e.
Representative gating was used for the splenic leukocyte populations quantified in Fig. 7c,d and Extended Data Fig. 7b. The left six panels (Extended Data Fig. 7a) show gating for identification of B cells and different myeloid cells, as indicated, as well as gating for CD38+ cells in all populations. The right six panels (Extended Data Fig. 7a) show gating for T-cell subsets, as indicated. Red arrows indicate sequential gating, pointing from parent plots towards child plots. For both the myeloid/B-cell and T-cell analyses, cells were first gated as shown on FSC area (FSCA) versus SSC area (SSCA) to identify intact cells larger than erythrocytes, which was followed by gating on FSC area versus FSC height (FSCH) to identify single cells. CD38 expression was gated for all populations as shown in the lower-leftmost panel. For the myeloid/B-cell analysis, single cells were gated on CD19 (B cells) versus CD11b (myeloid/NK cells), and CD11b+ cells were further gated on Ly-6G (granulocytes) versus F4/80 (macrophages). Cells expressing neither of these population markers were then gated on NK1.1 (NK cells) versus IaE (Class II MHC, a marker of myeloid cell activation). For the T cells, single cells were gated on TCR-β versus SSCA to identify αβ T cells, followed by CD4 versus CD8. Each of these T-cell subsets was then gated on CD44 versus CD62L, to identify naive and memory subsets as indicated.
Immunofluorescence staining and confocal microscopy.
Liver and eWAT tissue were dissected and fixed in 4% paraformaldehyde for 24 h at room temperature, followed by paraffin embedding. Paraffin sections (5 μm) were prepared for staining. After deparaffinization and rehydration, antigen retrieval was performed by boiling slides in 1× citrate buffer (pH 6.0). The slides were blocked in 10% normal serum from the host species of the secondary antibody and 5% BSA for 1 h in a humidified chamber. Primary-antibody incubation was at 4 °C overnight. The sheep anti-CD38 polyclonal antibody (R&D Systems, cat. no. AF4947, diluted 1:100) and rabbit anti-F4/80 monoclonal antibody (Cell Signaling, cat. no. 70076, diluted 1:100) were used for both single and costaining. For p21 and F4/80+ costaining the antibodies were: rat anti-F4/80 monoclonal antibody (Abcam, cat. no. ab6640, diluted 1:200) and rabbit anti-p21 polyclonal antibody (Invitrogen, cat. no. PA1-30399, diluted 1:200). The slides were washed 3 times with 1× PBS, followed by incubation with fluorescent-dye-conjugated secondary antibodies (Jackson ImmunoResearch) at room temperature for 1 h. Following 3 washes with 1× PBS, slides were mounted with ProLong Gold antifade reagent with DAPI (Thermo Fisher Scientific, cat. no. P36935). Images were acquired on a Zeiss LSM700.
Image analysis.
Immunofluorescence images of liver tissue and eWAT were analysed using a deep convolutional neural network with 23 layers (based on U-Net), trained to detect macrophages. For each model, sample images were selected and marked up to identify macrophages (16 samples for Kupffer cells, 12 for macrophages in eWAT and 16 for macrophages in eWAT for the Cd38 KO experiment). All training sets were extended with data augmentation, where the set of images was automatically and randomly manipulated during training (by adjusting size, brightness and rotation) to prevent overfitting and improve robustness. For each model, the neural network was trained for 200 epochs. After training on Kupffer cells, the neural network identified 6,019 cells on the basis of F4/80 and DAPI staining in 300 slides for 15 mice (8 young at 4 months and 7 old at 26 months). In each region, the intensities of CD38 or F4/80 were analysed, and the Pearson correlation coefficient was used to measure colocalization. Trained on eWAT, a second neural network detected 457 macrophage regions in 44 slides for 5 young (3 males and 2 females) and 4 old (2 males and 2 females) mice. To compare young and old tissue, the average number of macrophages and the area covered by the macrophages were calculated per slide. A third model was used to analyse p21+ macrophages in Cd38 KO and WT, identifying 1052 macrophage regions in 119 slides (all male) for counting The code is available on request.
Human PBMC macrophage experiment.
PBMCs were purified from healthy donor blood (Blood Centers of the Pacific) using density centrifugation on a Ficoll–Paque gradient (GE Healthcare Life Sciences). Monocyte were isolated by adherence: 3×106 PBMCs were seeded into 10-cm culture dishes and allowed to adhere in a 5% CO2 incubator at 37 °C for 2–3 h in cRPMI. Non-adherent cells were removed and adherent cells were washed twice with PBS. To generate human macrophages, monocytes isolated by adherence were cultured in complete RPMI supplemented with 100 ng ml−1 recombinant human M-CSF (Peprotech) in a 5% CO2 at 37 °C for 7 d. Cells were then stimulated with 100 ng ml−1 LPS or 10 ng ml−1 recombinant human IL-4 for 18 h.
ELISA.
TNF-α and IL-6 were measured in the medium of activated BMDMs using ELISA kits from Biolegend per the manufacturer’s protocol. Supernatants were cleared of debris or dead cells by centrifuging and were diluted 1:150 with the ELISA dilution buffer recommended in the kit.
Western blot.
Cell lysates were prepared by lysing cells in RIPA Buffer containing Halt protease and phosphatase inhibitor cocktail (Thermo Fisher). Protein concentrations were determined using the BCA Assay kit (Thermo Fisher). Approximately 15 μg of protein was loaded onto 8–15% polyacrylamide gels and transferred to PVDF membranes. The following primary antibodies from Cell Signaling were used: tubulin (clone: 9F3, cat. no., diluted 1:3,000), PARP1 (clone: 46D11, cat. no. 9532, diluted 1:1,000), SIRT1 (clone: D1D7, cat. no. 9475, diluted 1:1,000), SIRT2 (clone: D4O5O, cat. no. 12650, diluted 1:1,000), SIRT3 (clone: D22A3, cat. no. 5490, diluted 1:1,000), SIRT5 (clone: D8C3, cat. no. 8782, diluted 1:1,000), SIRT6 (clone: D8D12, cat. no. 1248, diluted 1:1,000), SIRT7 (clone: D3K5A, cat. no. 5360, diluted 1:1,000), SOD2 (clone: D9V9C, cat. no. 13194, diluted 1:1,000), Poly/Mono ADP ribosylated proteins, PARylation (clone: E6F6A, cat. no. 83732, diluted 1:1,000) and γH2AX (clone: 20E3, cat. no. 9718, diluted 1:1,000). CD38 (cat. no. AF4947, diluted 1:1,000) and CD157 (cat. no. AF4710, diluted 1:1,000) from R&D Systems. Actin (cat. no. A5441, diluted 1:3,000) was from Sigma and NAMPT (cat. no. A700-058-T, diluted 1:1,000) from Bethyl Laboratories.
Arginase assay.
The arginase assay was performed as follows. Briefly, 0.5×106 stimulated BMDMs were lysed with 75 μl 0.1% Triton X-100 buffer with Halt protease inhibitor cocktail (Thermo Fisher). Fifty microlitres of 25 mM Tris-HCL and 10 μl of 2 mM MnCl2 was added to each sample with heating at 56 °C for 10 min; 100 μl of 500 mM l-arginine (pH 9.7) was added incubated for 45 min at 37 °C; 800 μl H2SO4:H3PO4:H2O (1:3:7)) was used to stop each reaction. Urea production was measured by adding 40 μl of 9% α-isonitrosopropiophenone (in 100% ethanol) and heating to 100 °C for 15 min. A standard curve was run in parallel, and standards/samples were measured at 540 nm using a plate reader.
NADase assay.
NADase activity was measured using a fluorescence-based assay. Briefly, 0.5×106 BMDMs per sample were lysed with 0.1% Triton X-100 Sucrose (0.25 M sucrose, 40 mM Tris pH 7.4) buffer with protease and phosphatase inhibitors (Thermo Fisher). For NADase in intact macrophages, 2.5×106 BMDMs per sample were lifted with the cell scraper, centrifuged and resuspended in 100 μl PBS per well. The reaction was started by adding 80 μM nicotinamide 1,N6-etheno-adenine dinucleotide (Sigma Aldrich). The samples were excited at 340 nm, and fluorescence was measured at 460 nm at 37 °C every min for 1 hr in a PHERAstar FS microplate reader (BMG LABTECH). Data acquisition and analysis were carried out by PHERAstar 5.41 and MARS 3.32 software, respectively. NADase activity was calculated as the slope of the linear portion of the fluorescence–time curve, corrected for the amount of protein in each sample. Protein concentrations were determined using the BCA Assay Kit (Thermo Fisher).
Senescence and senescent-cell-conditioned media.
MDFs were exposed to 10-Gy X-rays, and medium was changed every 2 d. Control cells were mock irradiated. Primary preadipocytes (PAs) were collected from eWAT of 8-week-old C57BL/6J. Briefly, eWAT was digested as described above, and the SVF resuspended in complete MEM (Sigma Aldrich). Cells were allowed to adhere to culture dishes overnight. The next day, attached cells were washed with PBS and trypsinized. The trypsinized cells (~ 85% PAs) were confirmed by flow cytometry, replated, passaged and exposed to 10-Gy X-rays. Medium was changed every 2 d. Control cells were mock irradiated and passaged in parallel. At day 10 postradiation, irradiated and mock PA-conditioned medium and cells were collected. Alternatively, MDFs were treated with doxorubicin at 250 nM in DMSO for 24 h. The medium was replaced and refreshed every 2 d. Control cells were treated with DMSO. After day 10, conditioned media were prepared by washing of cells 3 times in PBS, and then incubation in serum-free medium for 24 h. BMDMs were incubated in CM for 24 h and collected for RNA isolation. For coculture experiments, 1×106 WT or Cd38 KO BMDMs were counted and added to 6-well tissue culture plates containing 1×104 senescent or control MDFs at 10:1 in 3.5 ml DMEM supplemented with 10% FBS in a 6-well plate for 24 h. Coculture cells were collected with RNA Stat 60 as described above.
Proteomics.
Chemicals.
Acetonitrile (no. AH015) and water (no. AH365) were from Burdick & Jackson. Iodoacetamide (IAA, no. I1149), dithiothreitol (DTT, no. D9779), formic acid (FA, no. 94318-50ML-F) and triethylammonium bicarbonate buffer 1.0 M, pH 8.5 ± 0.1 (no. T7408) were from Sigma Aldrich. Urea (no. 29700) was from Thermo Scientific. Sequencing-grade trypsin (no. V5113) was from Promega. HLB Oasis SPE cartridges (no. 186003908) were from Waters
X-ray irradiation.
Induction of senescence by ionizing radiation (IR) was performed on MDFs as described above. Quiescent control cells were mock irradiated. Senescent cells were cultured for 10 d, and quiescent cells were cultured in 0.2% serum for 3 d. Subsequently, cells were washed with PBS (Gibco no. 10010023) and placed in serum-free and phenol-red-free DMEM (Gibco no. 21063-029), and conditioned media were collected after 24 h.
Isolation, concentration and quantification of secreted proteins.
Conditioned media were collected, concentrated using Amicon Ultra-15 Centrifugal Filter Units with 3-kDa molecular-weight cut-off (MilliporeSigma no. UFC900324) as per the manufacturer’s instructions and transferred to a solution of 8 M urea/50 mM triethylammonium bicarbonate buffer at pH 8. Protein quantitation was performed using the BCA Protein Assay Kit (Pierce, no. 23225).
Digestion.
For digestion, 25–100 μg of each sample was brought to equal volume with 50 mM triethylammonium bicarbonate buffer in 8 M urea at pH 8. The protein mixtures were reduced with 20 mM DTT (37 °C for 1 h), and subsequently alkylated with 40 mM iodoacetamide (30 min at RT in the dark). Samples were diluted tenfold with 50 mM triethylammonium bicarbonate at pH 8 and incubated overnight at 37 °C with sequencing-grade trypsin (Promega) at a 1:50 enzyme/substrate ratio (wt/wt).
Desalting.
Peptide supernatants were collected and desalted with Oasis HLB Sorbent Cartridges (Waters no. 186003908), concentrated and resuspended in a solution containing mass spectrometric ‘Hyper Reaction Monitoring’ peptide standards (HRM, Biognosys no. Kit-3003)) and 0.2% formic acid in water.
Mass-spectrometry analysis.
Samples were analysed by reverse-phase HPLC–ESI–MS/MS using the Eksigent Ultra Plus nano-LC 2D HPLC system combined with a cHiPLC System connected to a quadrupole time-of-flight SCIEX TripleTOF 6600 or a TripleTOF 5600 mass spectrometer (SCIEX). Typically, mass resolution in precursor scans was ~45,000 (TripleTOF 6600), while fragment-ion resolution was ~15,000 in ‘high sensitivity’ product-ion-scan mode. After injection, peptide mixtures were transferred onto a C18 precolumn chip (200 μm × 6 mm ChromXP C18-CL chip, 3 μm, 300 Å, SCIEX) and washed at 2 μl min−1 for 10 min with the loading solvent (H2O/0.1% formic acid) for desalting. Subsequently, peptides were transferred to the 75 μm × 15 cm ChromXP C18-CL chip, 3 μm, 300 Å (SCIEX), and eluted at a flow rate of 300 nl min−1 with a 3-h gradient using aqueous and acetonitrile solvent buffers.
For label-free relative quantification, samples were analysed by data-independent acquisitions, or specifically variable window SWATH acquisitions. In these SWATH acquisitions, instead of the Q1 quadrupole transmitting a narrow mass range through to the collision cell, windows of variable width (m/z 5–90) are passed in incremental steps over the full mass range (m/z 400–1,250). The cycle time of 3.2 s includes a 250-ms precursor ion scan followed by 45 ms if accumulation time for each of the 64 SWATH segments. The variable windows were determined according to the complexity of the typical MS1 ion current observed within a certain m/z range using a SCIEX ‘variable window calculator’ algorithm (https://sciex.com/support/knowledge-base-articles/how-to-use-the-swath-variable-calculator-excel-sheet)72. SWATH MS2 produces complex MS/MS spectra composed of all analytes within each selected Q1 m/z window. Data-dependent acquisitions were carried out on 8 samples containing secreted proteins from MDFs to obtain MS/MS spectra for the 30 most abundant precursor ions (100 ms per MS/MS) following each survey MS1 scan (250 ms), yielding a total cycle time of 3.3 s. For collision-induced dissociation MS/MS, the mass window for precursor ion selection of the quadrupole mass analyser was set to ±1 m/z using the Analyst 1.7 software.
Quantification and statistical analysis.
Raw files from DDA analyses of eight secreted samples were searched using ProteinPilot 5.0 software (Revision 4769, Paragon Algorithm 5.0.0.0, 4767 (SCIEX)), and results were imported into Spectronaut (Biognosys) to create a spectral library. SWATH acquisitions were quantitatively processed using the proprietary Spectronaut v12 (12.020491.3.1543) software73. Quantitative SWATH MS2 analysis was based on ion chromatograms (XICs) of 6–10 of the most abundant fragment ions in the identified spectra. Relative quantification was done by comparing different conditions (senescent versus control) and assessing fold-changes in proteins. The number of biological replicates analysed were five senescent secretome samples and six quiescent (control) samples. Significance was assessed using false-discovery-rate-corrected q values < 0.05.
Glycolysis and mitochondrial respiration in BMDMs.
Glycolysis and mitochondrial respiration of BMDMs were assessed by a Seahorse XFe96 Analyzer. Briefly, BMDMs were seeded at 50,000 cells per well in Seahorse XF96 microplates in RPMIc and allowed to adhere for 6 h prior to treatment with 50 nM FK866 and NMN. At 6 h post-treatment, BMDMs were treated with IL-4 (10 ng ml−1) and LPS (100 ng ml−1) for 16 and 6 h, respectively. Cells were then washed twice with XF-RPMI plus 2 mM glutamine followed by a 30-min incubation with glutamine-supplemented XF-RPMI in an air incubator at 37 °C. The rates of glycolysis and mitochondrial respiration were examined using Seahorse XF Glycolysis and Mito Stress Test Kits (cat. nos. 103020-100 and 103015-100 from Agilent), according to the manufacturer’s protocols. To measure mitochondrial function, maximum mitochondrial respiration capacity was defined as the maximum rate induced by FCCP (0.5 μM). Rates were normalized to cell number and are expressed as mpH per minute for glycolysis and pmol O2 per minute for mitochondrial respiration.
SA-Bgal assay.
Cells and tissues were fixed and processed with the Biovision Senescence Detection Kit per the manufacturer’s instructions (Biovision Senescence Detection Kit, no. K320-250). Liver tissues, frozen in OCT, were cut (~10-μm sections) and processed for SA-Bgal and nuclear Fast Red (Fisher Scientific, cat. no. R5463200-500A) staining74,75.
Statistical analysis.
Statistical analysis was performed using GraphPad Prism 8.4.3 software and Microsoft Excel. Student’s t-tests (one-sided and two-sided) were applied to comparisons between two conditions, as indicated below each figure, with mean ± s.e.m. shown. For statistical analysis of proteomic data, see above.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
For proteomics data, all files are uploaded to the Center for Computational Mass Spectrometry, and can be downloaded using the following link ftp://massive.ucsd.edu/MSV000083726 (MassIVE ID number: MSV000083726). Data uploads include protein identification and quantification details, spectral library and FASTA file used for analysis. Tabula Muris Senis database is accessible at this link: https://tabula-muris-senis.ds.czbiohub.org. All other data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.
Code availability
Coding used in image analysis is available upon request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 ∣. CD38 expression in human M1 macrophages and analysis of the de novo NAD pathway in BMDMs.
a, mRNA levels of CD38 in human peripheral blood monocytes (PBMC)-derived macrophages treated with recombinant human IL-4 (M2) or LPS (M1) for 18 hours. Representative data from one of three patient samples. (n = 4 independent biological experiments) b, Immunofluorescence of human PBMC derived macrophages stimulated as described above using an anti-human CD38 antibody (Green) and nuclei with DAPI (Blue). Scale bars represents 10μm. Analyzed in PBMCs derived from one patient. c, NADase activity in human PBMC-derived macrophages treated with recombinant human IL-4 (M2) or LPS (M1) for 18 hours. Shown is the mean of two separate experiments from different donors with 2 replicates each. d, Schematic of the de novo NAD synthesis pathway. e, mRNA levels of de novo NAD synthesis pathway enzymes. f, Quantification of tryptophan metabolites measured by LC-MS in M0, M2 and M1 mouse BMDMs activated for 24 hours. ND=not detected. Data shows the mean ± SEM n=3 independent experiments except in A and B. Statistical significance indicated as *P<0.05, **P<0.01, and ***P<0.001; two-sided Student’s t-test.
Extended Data Fig. 2 ∣. Analysis of the role of the NAM-salvage pathway and sirtuins in macrophage activation and polarization.
a, NAD levels quantified by LC-MS in M0, M1 and M2 BMDMs pre-treated or not with 50 nM FK866 and NR for 6 hours prior to stimulation with LPS for an additional 6 hours or IL-4 for 16 hours. b, mRNA levels of M2 genes in BMDMs pre-treated or not with FK866 and NR for 6 hours prior to stimulation with IL-4 for 16 hours. All statistical comparisons are relative to M2 + FK866. c, mRNA levels of M1 genes in BMDMs pre-treated or not with FK866 and NR for 6 hours prior to stimulation with LPS for 6 hours. All statistical comparisons are relative to M1 + FK866. d, Western analysis of Nampt Fl/Fl CreER and Nampt Fl/Fl BMDMs treated with 1 ug/ml tamoxifen. e, mRNA levels of M2 genes in Nampt Fl/Fl CreER and Nampt Fl/Fl BMDMs treated with IL-4 for 16 hours. f, mRNA levels of M1 genes in Nampt Fl/Fl CreER and Nampt Fl/Fl BMDMs treated with LPS for 6 hours. g, mRNA levels of M2 genes in WT BMDMs pretreated with the indicated concentration of the sirtuin inhibitors Ex527 and AGK2 for 30 minutes prior to stimulation with IL-4 for 16 hours. All statistical comparisons are relative to M2. h, mRNA levels of M1 genes in WT BMDMs pretreated with the indicated concentration of the sirtuin inhibitors Ex527 and AGK2 for 30 minutes prior to stimulation with LPS for 6 hours. All statistical comparisons are relative to M1. Data show the mean ± SEM. (n=3 independent experiments). Statistical significance defined as *P<0.05, **P<0.01, and ***P<0.001; two-sided Student’s t-test.
Extended Data Fig. 3 ∣. Heightended NADase activity in M1 macrophages is CD38 dependent and PARP1 independent.
a, Flow cytometry results comparing CD38 surface staining in naive (M0) WT and Cd38 KO BMDMs or BMDMs treated with IL-4 (M2) and LPS (M1) for 16 hours. b-c, NADase activity measured with non-cell permeable εNAD in intact M0, M2, and M1 WT and Cd38 KO BMDMs activated for 16 hours relative to cell number (B) and protein content (C). d, mRNA levels of Cd157 in M0 and M1 WT and Cd38 KO BMDMs treated for 16 hours. e, LC-MS quantification of NR in M0 and M1 WT and Cd38 KO BMDMs treated for 16 hours. f, mRNA levels of anti-oxidant genes in WT and Cd38 KO BMDMs treated with IL-4 (M2) and LPS (M1) for the indicated intervals. g, Western analysis of PARP activity (PARylation) and DNA damage (γH2AX) in WT and Cd38 KO BMDMs treated with IL-4 (M2) and LPS (M1) for the indicated intervals compared to WT MO macrophage treated with 1 mM H2O2 for 10 minutes. h, Western analysis of PARP activity (PARylation) and DNA damage (γH2AX) in WT and Cd38 KO BMDMs treated with IL-4 (M2) and LPS (M1) for 8 hours prior to treatment with 1 mM H2O2 for 10 minutes. Data show the mean ± SEM. (n= at least 3 independent experiments). Statistical significance indicated as *P<0.05, **P<0.01, and ***P<0.001; two-sided Student’s t-test. Unless noted with a bar, statistical comparisons are relative to the appropriate MO WT or Cd38 KO sample of the same genotype.
Extended Data Fig. 4 ∣. CD38+ resident macrophages accumulate in ageing adipose tissue.
a, LC-MS quantification of NAD in eWAT from WT young male mice (6 month) n=7 mice/group, Cd38 KO young male mice (3 month) n=5 mice/group, WT old male mice (25 month) n=10 mice/group, and Cd38 KO old male mice (26 month) n=5 mice/group. NAD concentrations are shown as pmol/mg of tissue. (same WT data from Fig. 4a). b, mRNA levels of Il-1α and IL-10 in eWAT from 6 and 25 month-old WT male mice. (WT young male mice (6 month) n=7 mice/group, WT old male mice (25 month) n=9 mice/group) c, Western analysis of adipose tissue from young (3 Month) and old (19 month) WT male mice to detect PARP activity (PARylation) and DNA damage (γH2AX). Each lane represents one mouse (young n=4 mice/group, old n=4 mice/group). d, mRNA levels of Cd38 in visceral adipose tissue, the stromal vascular fraction, and adipocyte fraction from young (3 month) and old (19 month) WT male mice. (young n=4 mice/group, old n=4 mice/group). e, Flow cytometry gating strategy to identify CD45+ immune cells from the stromal vascular fraction of eWAT. Cells were first gated on forward scatter (FSCA) vs side scatter (SSCA) to discard cell debris and dead or dying cells. Next FSCH (height) vs FSCA (Area) was used to select single cells. Single cells were then gated for auto-fluorescent using the Empty(E) BV421 vs BV711 channels (not used as antibody fluorophores) to discard cells that showed auto-fluorescence in these channels. Then CD45+ cells were selected and analyzed for CD38 and macrophage markers. Flow cytometry gating strategy to identify resident and non-resident macrophages from the stromal vascular fraction of eWAT, showing representative flow plots and histograms for the indicated ages of mice. f, Flow cytometry quantification of CD38- (low) resident macrophages, CD38- non-resident macrophages, and CD38+ (high) non-macrophage immune cells from eWAT of WT male mice for the ages shown. (2 months n=6 mice/group, 6 months n=5 mice/group, 12 months n=5 mice/group, 18 months n=5 mice/group, 25+ months n=7 mice/group) For in vivo experiments, data from individual mice are shown. Statistical significance indicated as *P<0.05, **P<0.01, and ***P<0.001; two-sided Student’s t-test.
Extended Data Fig. 5 ∣. Senescent cell burden is causally linked to increased macrophage CD38 expression.
a, mRNA levels of Il-1α, Cxcl1, and IL-10 in eWAT from 6 month-old WT male mice i.p. injected with Doxo or PBS. (PBS n=8 mice/group, Doxo n=7 mice/group) b, Quantification of CD38-low resident macrophages, and CD38-low non-resident macrophages from eWAT of 6 month-old WT male mice injected with Doxo or PBS. (PBS n=8 mice/group, Doxo n=8 mice/group). c, CD38 mRNA levels in WT and Cd38 KO BMDMs co-cultured (10:1) with non-senescent control mouse dermal fibroblasts (CTRL-MDF) or irradiated senescent MDF (Sen(IR)-MDF) for 24 hours. (n=4 independent biological experiments per condition) d, mRNA levels of Cd38 in WT BMDMs treated with the indicated DAMPs for 16 hours. (n=3 independent biological experiments per condition) e, mRNA levels of inflammatory genes in CTRL-MDF and Sen(IR)-MDF. (n=4 independent biological experiments per condition) f, mRNA levels of Cd38 in BMDMs treated with the indicated concentrations (ng/ml) of recombinant mouse cytokines for 24 hours. (n=3 independent biological experiments per condition) g, Heatmap of significantly upregulated proteins identified by mass spectrometry in conditioned media from CTRL-MDF and Sen(IR)-MDF. (n=4-6 independent biological experiments per condition). For in vivo experiments, data from individual mice are shown. Data show the mean ± SEM. (n= at least 3 independent experiments). Statistical significance indicated as *P<0.05, **P<0.01, and ***P<0.001; two-sided Student’s t-test.
Extended Data Fig. 6 ∣. Single-cell RNA sequencing analysis of inflammatory, NAD consuming, and biosynthetic genes in ageing hepatocytes and liver endothelial cells.
a, Dot-plot and heatmap of the indicated genes in liver hepatocytes based on age. Logarithmic axes, base-10. b, Dot-plot and heatmap of the indicated genes in liver endothelial cells based on age. Logarithmic axes, base-10.
Extended Data Fig. 7 ∣. LPS promotes tissue NAD decline via CD38.
a, Representative gating for the splenic leukocyte populations quantified in Fig. 7c, d and Extended Data Fig. 7a. Left six panels show gating for identification of B cells and different myeloid cells, as indicated, as well as gating for CD38-positive cells in all populations. Right six panels show gating for T cell subsets, as indicated. Red arrows indicate sequential gating, pointing from parent plots towards child plots. b, Quantification of immune cell populations and CD38+ immune cells in the spleen of 3 month-old WT male mice i.p. injected with PBS or LPS for 4 weeks, and analyzed by flow cytometry. (PBS n=10 mice/group, LPS n=9 mice/group) c, Western analysis of CD38, CD157, CD68, and NAMPT in eWAT of 3 month-old WT male mice injected with PBS or LPS for 4 weeks and Image J quantification of CD38 protein levels relative to NAMPT. Each lane represents one mouse (PBS n=4 mice/group, LPS n=5 mice/group) d, mRNA levels of NAD consuming enzymes in eWAT from 3 month-old WT male mice injected with PBS or LPS for 4 weeks. (PBS n=4 mice/group, LPS n=5 mice/group) e, mRNA levels of the indicated genes in whole eWAT from 4 month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 hours. (n=10 mice/group) f, Western analysis of eWAT from 4 month-old WT and Cd38 KO male mice injected with PBS or LPS for 12 hours (n=3 mice/group). g, LC-MS quantification of NAD-related metabolites in eWAT from 4 month-old WT and Cd38 KO male mice IP injected with PBS or LPS for 12 hours. (n=10 mice/group) h, LC-MS quantification of NAD-related metabolites in liver from 4 month-old WT and Cd38 KO male mice IP injected with PBS or LPS for 12 hours. (n=10 mice/group) Data from individual mice are shown for in vivo experiments. Data show the mean ± SEM. Statistical significance indicated as *P<0.05, **P<0.01, and ***P<0.001; two-sided Student’s t-test except for 7 g and 7 h one-tailed t-test was used.
Acknowledgements
This project was supported by NIH grant R24DK085610 (E.V.), Gladstone Institute intramural funds (E.V.) and Buck Institute intramural funds (E.V. and J.C.). A.J.C. is a recipient of the UC President’s Postdoctoral Fellowship at UCSF and was supported by NIH training grant T32AG000266 (Buck Institute). A.K. was supported by the SENS Research Foundation and NIH grant R01AG051729 (J.C.). B.S. and N.B. were supported by NIH grant U01AG060906 (B.S., principal investigator) and NIH Shared Instrumentation Grant 1S10OD016281 (Buck Institute). M.S.S. and C.B. were supported by NIH grant R01HL147545 and the Roy J. Carver Trust to C.B. We thank M. Walter for help with imaging cells, and P. Dighe for help optimizing Seahorse assay conditions. We thank R. Camarda, V. Byles and D. Powell for reviewing the manuscript and helpful discussions.
Footnotes
Competing interests
All authors have reviewed and approved the manuscript. C.B. is the inventor of intellectual property on the nutritional and therapeutic uses of NR, serves as chief scientific advisor of and holds stock in ChromaDex. E.V. is a scientific cofounder of NAPA Therapeutics. A.J.C., R.P., Q.W., E.S. and K.V. received partial salary support from NAPA Therapeutics. J.C. is a scientific cofounder of Unity Biotechnology. The other authors declare no competing interests.
Supplementary information is available for this paper at https://doi.org/10.1038/s42255-020-00305-3.
Peer review information Primary Handling Editor: Pooja Jha.
Reprints and permissions information is available at www.nature.com/reprints.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Canto C, Menzies KJ & Auwerx J NAD+ metabolism and the control of energy homeostasis: a balancing act between mitochondria and the nucleus. Cell Metab. 22, 31–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshino J, Baur JA & Imai SI NAD+ intermediates: the biology and therapeutic potential of NMN and NR. Cell Metab. 27, 513–528 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitchell SJ et al. Nicotinamide improves aspects of healthspan, but not lifespan, in mice. Cell Mefab. 27, 667–676 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verdin E NAD+ in aging, metabolism, and neurodegeneration. Science 350, 1208–1213 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Bogan KL & Brenner C Nicotinic acid, nicotinamide and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Ann. Rev. Nutr 28, 115–130 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Liu L et al. Quantitative analysis of NAD synthesis-breakdown fluxes. Cell Metab. 27, 1067–1080 e1065 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho-Pereira J et al. CD38 dictates age-related NAD decline and mitochondrial dysfunction through an SIRT3-dependent mechanism. Cell Mefab. 23, 1127–1139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson DG & Bell JI Isolation of a cDNA encoding the human CD38 (T10) molecule, a cell surface glycoprotein with an unusual discontinuous pattern of expression during lymphocyte differentiation. J. Immunol 144, 2811–2815 (1990). [PubMed] [Google Scholar]
- 9.Schneider M et al. CD38 is expressed on inflammatory cells of the intestine and promotes intestinal inflammation. PLoS ONE 10, e0126007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savarino A, Bottarel F, Malavasi F & Dianzani U Role of CD38 in HIV-1 infection: an epiphenomenon of T-cell activation or an active player in virus/host interactions? AIDS 14, 1079–1089 (2000). [DOI] [PubMed] [Google Scholar]
- 11.Franceschi C et al. Inflamm-aging. An evolutionary perspective on immunosenescence. Ann. N. Y. Acad. Sci 908, 244–254 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Franceschi C & Campisi J Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. A Biol. Sci. Med Sci 69, S4–S9 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Ganeshan K & Chawla A Metabolic regulation of immune responses. Annu. Rev. Immunol 32, 609–634 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coppe JP et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray PJ et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity 41, 14–20 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bieganowski P & Brenner C Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss–Handler independent route to NAD+ in fungi and humans. Cell 117, 495–502 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Ratajczak J et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun 7, 13103 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cameron AM et al. Inflammatory macrophage dependence on NAD+ salvage is a consequence of reactive oxygen species-mediated DNA damage. Nat. Immunol 20, 420–432 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Langston PK, Shibata M & Horng T Metabolism supports macrophage activation. Front. Immunol 8, 61 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Van Gool F et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat. Med 15, 206–210 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amano SU et al. Local proliferation of macrophages contributes to obesity-associated adipose tissue inflammation. Cell Metab. 19, 162–171 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Z et al. Mouse macrophage specific knockout of SIRT1 influences macrophage polarization and promotes angiotensin II-induced abdominal aortic aneurysm formation. J. Genet Genomics 45, 25–32 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Cockayne DA et al. Mice deficient for the ecto-nicotinamide adenine dinucleotide glycohydrolase CD38 exhibit altered humoral immune responses. Blood 92, 1324–1333 (1998). [PubMed] [Google Scholar]
- 24.Jablonski KA et al. Novel markers to delineate murine M1 and M2 macrophages. PLoS ONE 10, e0145342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shrimp JH et al. Revealing CD38 cellular localization using a cell permeable, mechanism-based fluorescent small-molecule probe. J. Am. Chem. Soc 136, 5656–5663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao YJ, Lam CM & Lee HC The membrane-bound enzyme CD38 exists in two opposing orientations. Sci. Signal 5, ra67 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Liu J et al. Cytosolic interaction of type III human CD38 with CIB1 modulates cellular cyclic ADP-ribose levels. Proc. Natl Acad. Sci. USA 114, 8283–8288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preugschat F et al. A pre-steady state and steady state kinetic analysis of the N-ribosyl hydrolase activity of hCD157. Arch. Biochem. Biophys 564, 156–163 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Tarrago MG et al. A potent and specific CD38 inhibitor ameliorates age-related metabolic dysfunction by reversing tissue NAD+ decline. Cell Metab. 27, 1081–1095 e1010 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Regdon Z et al. LPS protects macrophages from AIF-independent parthanatos by downregulation of PARP1 expression, induction of SOD2 expression, and a metabolic shift to aerobic glycolysis. Free Radic. Biol. Med 131, 184–196 (2019). [DOI] [PubMed] [Google Scholar]
- 31.Virag L, Jaen RI, Regdon Z, Bosca L & Prieto P Self-defense of macrophages against oxidative injury: fighting for their own survival. Redox Biol. 26, 101261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshino J, Mills KF, Yoon MJ & Imai S Nicotinamide mononucleotide, a key NAD+ intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab. 14, 528–536 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rabinowitz SS & Gordon S Macrosialin, a macrophage-restricted membrane sialoprotein differentially glycosylated in response to inflammatory stimuli. J. Exp. Med 174, 827–836 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu X et al. Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 18, 816–830 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho KW, Morris DL & Lumeng CN Flow cytometry analyses of adipose tissue macrophages. Methods Enzymol. 537, 297–314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Consortium TTM Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chakarov S et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363, eaau0964 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Mrdjen D et al. High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 380–395 e386 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Dimri GP et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coppe JP, Desprez PY, Krtolica A & Campisi J The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol 5, 99–118 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tchkonia T et al. Fat tissue, aging, and cellular senescence. Aging Cell 9, 667–684 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu M et al. Senolytics improve physical function and increase lifespan in old age. Nat. Med 24, 1246–1256 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Demaria M et al. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 7, 165–176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pommier Y, Leo E, Zhang H & Marchand C DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol 17, 421–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baar MP et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell 169, 132–147 e116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaefer L Complexity of danger: the diverse nature of damage-associated molecular patterns. J. Biol. Chem. 289, 35237–35245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kratz M et al. Metabolic dysfunction drives a mechanistically distinct proinflammatory phenotype in adipose tissue macrophages. Cell Metab. 20, 614–625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iqbal J & Zaidi M TNF regulates cellular NAD+ metabolism in primary macrophages. Biochem. Biophys. Res. Commun 342, 1312–1318 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Jenkins SJ et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science 332, 1284–1288 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hashimoto D et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity 38, 792–804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davies LC et al. Distinct bone marrow-derived and tissue-resident macrophage lineages proliferate at key stages during inflammation. Nat. Commun 4, 1886 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ginhoux F et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jager J, Aparicio-Vergara M & Aouadi M Liver innate immune cells and insulin resistance: the multiple facets of Kupffer cells. J. Intern. Med 280, 209–220 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Stahl EC, Haschak MJ, Popovic B & Brown BN Macrophages in the aging liver and age-related liver disease. Front. Immunol 9, 2795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.The Tabula Muris Consortium. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogrodnik M et al. Cellular senescence drives age-dependent hepatic steatosis. Nat. Commun 8, 15691 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demaria M et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buford TW (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome 5, 80 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thevaranjan N et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe 21, 455–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Real AM, Hong S & Pissios P Nicotinamide N-oxidation by CYP2E1 in human liver microsomes. Drug Metab. Dispos 41, 550–553 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Campisi J & Robert L Cell senescence: role in aging and age-related diseases. Interdiscip. Top. Gerontol 39, 45–61 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campisi J Aging, cellular senescence, and cancer. Annu Rev. Physiol 75, 685–705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatterjee S et al. CD38–NAD+ axis regulates immunotherapeutic anti-tumor T cell response.Cell Metab. 27, 85–100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lischke T et al. CD38 controls the innate immune response against Listeria monocytogenes. Infect. Immun. 81, 4091–4099 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Partida-Sanchez S et al. Chemotaxis of mouse bone marrow neutrophils and dendritic cells is controlled by ADP-ribose, the major product generated by the CD38 enzyme reaction. J. Immunol 179, 7827–7839 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Ganeshan K et al. Energetic trade-offs and hypometabolic states promote disease tolerance. Cell 177, 399–413 e312 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei W, Graeff R & Yue J Roles and mechanisms of the CD38/cyclic adenosine diphosphate ribose/Ca2+ signaling pathway. World J. Biol. Chem 5, 58–67 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weiss R et al. Nicotinamide: a vitamin able to shift macrophage differentiation toward macrophages with restricted inflammatory features. Innate Immun. 21, 813–826 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Chini CCS et al. CD38 ecto-enzyme in immune cells is induced during aging and regulates NAD+ and NMN levels. Nat. Metab 2, 1284–1304 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Trammell SA & Brenner C Targeted, LCMS-based metabolomics for quantitative measurement of NAD+ metabolites. Comput. Struct. Biotechnol. J 4, e201301012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trammell SA et al. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun 7, 12948 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schilling B, Gibson BW & Hunter CL Generation of high-quality SWATH® acquisition data for label-free quantitative proteomics studies using tripleTOF® mass spectrometers. Methods Mol. Biol 1550, 223–233 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bruderer R et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen- treated three-dimensional liver microtissues. Mol. Cell Proteom 14, 1400–1410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Velarde MC, Demaria M, Melov S & Campisi J Pleiotropic age-dependent effects of mitochondrial dysfunction on epidermal stem cells. Proc. Natl Acad. Sci. USA 112, 10407–10412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiley CD et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metab. 23, 303–314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
For proteomics data, all files are uploaded to the Center for Computational Mass Spectrometry, and can be downloaded using the following link ftp://massive.ucsd.edu/MSV000083726 (MassIVE ID number: MSV000083726). Data uploads include protein identification and quantification details, spectral library and FASTA file used for analysis. Tabula Muris Senis database is accessible at this link: https://tabula-muris-senis.ds.czbiohub.org. All other data that support the findings of this study are available from the corresponding author upon request. Source data are provided with this paper.