Abstract
Background
Although imbalanced intestinal flora contributes to the pathogenesis of nonalcoholic fatty liver disease (NAFLD), conflicting results have been obtained for patient-derived microbiome composition analyses. A meta-analysis was performed to summarize the characteristics of intestinal microbiota at the species level in NAFLD patients.
Methods
Following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement, a completed search (last update: December 30, 2020) of databases was performed to identify eligible case-control studies detecting gut microbiota in NAFLD patients. The meta-analysis results are presented as the standard mean difference (SMD) and 95% confidence interval (CI). Bias controls were evaluated with the Newcastle-Ottawa Scale (NOS), funnel plot analysis, and Egger’s and Begg’s tests.
Results
Fifteen studies (NOS score range: 6–8) that detected the gut microbiota in the stools of 1265 individuals (577 NAFLD patients and 688 controls) were included. It was found that Escherichia, Prevotella and Streptococcus (SMD = 1.55 [95% CI: 0.57, 2.54], 1.89 [95% CI: 0.02, 3.76] and 1.33 [95% CI: 0.62, 2.05], respectively) exhibited increased abundance while Coprococcus, Faecalibacterium and Ruminococcus (SMD = − 1.75 [95% CI: − 3.13, − 0.37], − 9.84 [95% CI: − 13.21, − 6.47] and − 1.84 [95% CI, − 2.41, − 1.27], respectively) exhibited decreased abundance in the NAFLD patients compared with healthy controls. No differences in the abundance of Bacteroides, Bifidobacterium, Blautia, Clostridium, Dorea, Lactobacillus, Parabacteroides or Roseburia were confirmed between the NAFLD patients and healthy controls.
Conclusions
This meta-analysis revealed that changes in the abundance of Escherichia, Prevotella, Streptococcus, Coprococcus, Faecalibacterium and Ruminococcus were the universal intestinal bacterial signature of NAFLD.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12944-021-01440-w.
Keywords: Nonalcoholic fatty liver disease, Nonalcoholic steatohepatitis, Gut, Microbiome, Bacterial composition, Microbiota
Background
Nonalcoholic fatty liver disease (NAFLD) is characterized by excessive intrahepatic lipid accumulation and consequent necroinflammation and fibrosis under conditions of metabolic disturbance, and it represents the leading cause of chronic liver disease worldwide [1]. The global prevalence of NAFLD is greater than 25%, and these levels are dramatically increased in areas with previously low incidence rates, especially in China [2]. Although the pathogenesis of NAFLD remains unclear, interactions between the environment and individual genetic backgrounds promote disease susceptibility. Moreover, the gut microbiota, which are shaped by the host immune response, and environmental factors, such as a high caloric diet, dietary fiber deficiency and a sedentary lifestyle, contribute greatly to the predisposition to NAFLD [3].
In clinical studies, ample evidence suggests that aberrations in the gut microbiota may play a key role in the development of NAFLD. Small intestinal bacterial overgrowth is more prevalent in NAFLD patients than in healthy individuals [4, 5]. High levels of lipopolysaccharide (LPS), the major constituent of the cytoplasm of gram-negative bacteria, were identified as an independent risk factor for NAFLD incidence [6]. Escherichia enriched in NAFLD patients is capable of ethanol synthesis [7], inducing oxidative stress that is involved in NAFLD progression. Changes in the community structure of intestinal microbes are also associated with inflammatory activity and the fibrosis stage of NAFLD [8]. Moreover, modulation of the gut microbiota by probiotic treatment has been demonstrated to improve liver injuries, metabolic abnormalities, and inflammatory chemokine levels in NAFLD patients [9]. Therefore, identifying the microflora signature in NAFLD is of great value because imbalanced microbiota may serve as a novel target for establishing diagnostic or treatment methods.
Although the gut microbiota is profoundly important in the pathogenesis of NAFLD, the distinct composition of the gut microbiota in NAFLD patients remains under debate. A limited number of studies on stool microbiota differences between NAFLD patients and controls have reported inconsistent or even opposing results. These discrepancies may be associated with geographic origins and dietary habits [10]. Therefore, we performed a meta-analysis to explore the profiles of intestinal dysbiosis in NAFLD patients in regions around the world.
Methods
Data sources
This systematic review and meta-analysis is reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) Statement [11] and the PRISMA 2009 checklist, which has been with a registration code in PROSPERO registry (registration number is: CRD42020220632, https://www.crd.york.ac.uk/PROSPERO). A complete and computerized search (last update: December 30, 2020) was performed without restriction based on region, language or publication type in the following electronic databases: PubMed, EMBASE and Cochrane Library. The Medical Subject Heading (MeSH) database was used to acquire MeSH terms and their relevant entry terms based on the following formatted terms: (NAFLD OR NASH OR NAFL OR “nonalcoholic fatty liver disease” OR “nonalcoholic steatohepatitis” OR “nonalcoholic fatty liver disease” OR “nonalcoholic steatohepatitis” OR “fatty liver” OR “steatohepatitis” OR “steatosis”) AND (microbiota OR microbes OR microbiome OR microflora OR flora OR bacteria). The query was adapted to different databases with minimal differences as required. The literature search was also supplemented with a manual search using the Related Articles Function and the reference lists of all retrieved articles.
Study selection
Any study that provides necessary data on detecting gut microbiota for NAFLD patients and comparable controls was eligible for this systematic review and meta-analysis. The inclusion criteria for this study were as follows: 1) original full-text publications, 2) NAFLD diagnosed with sonographic or histologic evidence, 3) healthy controls with comparable age and sex proportions as the case group, 4) studies comparing the gut microbiota between NAFLD patients and healthy controls, and 5) available and sufficient data (sample size, mean and standard deviation or any data that can be converted to) to calculate the standardized mean difference (SMD) with 95% confidence interval (CI) in these two groups.
The exclusion criteria were as follows: 1) editorials, letters to the editor, reviews, case reports, hypotheses, book chapters, conference abstracts or studies on animals or cell lines; 2) studies that included NAFLD patients with other liver diseases, such as alcoholic fatty liver disease, autoimmune liver disease and viral hepatitis; and 3) studies that performed any intervention on participants at baseline.
Data extraction
Two investigators (FX L and JZ Y) were responsible for selecting studies and extracting data from the included studies independently according to predesigned inclusion and exclusion criteria. Any disagreement was discussed and finally resolved by the adjudicating senior author (BH Z). If additional data or data transformations were required for analysis, the corresponding or first authors were contacted by email for assistance. When the authors did not reply, standard statistical formulas were applied for data transformation [12, 13]. If important additional data were not available from the corresponding or first author, the study was excluded.
A full-text review of the included studies was performed independently by the two aforementioned investigators, and the following variables were collected: 1) first author; 2) year of publication; 3) location; 4) study design; 5) number, age and sex of NAFLD (or NAFL or NASH) patients and healthy controls; 6) methods for diagnosing NAFLD; 7) methods for detecting and analyzing gut microbiota; and 8) bacterial counts and units for expressing the values.
Quality assessment
To evaluate the risk of bias of individual studies, the methodological quality of the included studies was assessed using the Newcastle-Ottawa Scale (NOS) by two investigators (FX L and CX S). A senior author (BH Z) who was not involved in the initial assessment was responsible for adjudication on any disagreement between the two investigators. The NOS is a widely and frequently used tool for assessing the quality of nonrandomized studies included in systematic reviews and/or meta-analyses. Using this tool, study quality is assessed based on eight items that are categorized into three groups: study group selection, group comparability (a maximum of two stars can be given for comparability), and exposure or outcome of interest determination for case-controlled or cohort studies. A star rating system was used to rate the quality of the included studies based on a scale ranging from 0 (low quality) to 9 (high quality).
Statistical analysis
In this study, statistical analyses were performed using R language Version 3.4.3 (The package of meta and metafor) and STATA/SE 12.0 (StataCorp, Texas, USA) with a Meta module for Windows. After extraction and transformation, all bacterial count data are presented as the mean (), standard deviation (SD) and total sample size (n). The standardized mean difference (SMD) with 95% confidence interval (CI) was adopted to evaluate the effect size when a bacterial genus was detected by different techniques in the included studies [14]. The α criterion was set at 0.05. Accordingly, when the total SMD of a bacterial genus was more than 0 (favors NAFLD) with a 95% CI not including the value 0, we assumed that this bacterial genus exhibited overgrowth in NAFLD patients. Otherwise, the bacterial genus was assumed to be deficient in NAFLD patients. Additionally, subgroup analysis was performed based on the areas where these studies were conducted (Western and Eastern). Meta regression analysis was also performed to evaluate the effect of alanine aminotransferase (ALT) and body mass index (BMI) levels on mean different abundance of bacterial genus for NAFLD versus control patients. Those univariate significant variances would be entered in the multivariate models.
Heterogeneity across the included studies was evaluated using χ2 and I2 statistics. Higher χ2 and I2 statistics indicated greater heterogeneity, and an I2 value greater than 50% with a P-value less than 0.1 represented substantial heterogeneity in this analysis. A random-effects model was reported when substantial heterogeneity existed. The 95% prediction intervals which reflected the true effects of future similar studies with 95% certainties were calculated based on the formulas reported by Joanna IntHout et al. [15].
To assess the risk of publication bias, funnel plot analyses, Egger’s test and Begg’s test were conducted for genus-specific gut microbiota. Publication bias can cause asymmetry in funnel plots, which can be quantified with Egger’s and Begg’s tests. When P-values derived from Egger’s or Begg’s regressions were greater than 0.05, we assumed that no publication bias was present. Moreover, a sensitivity analysis was performed to evaluate the impact of each study on the overall results. In addition, considering the observational design limitations of each included study despite of its large sample size and quality, there would be the maximum certainty of an effect of case-control comparison, which was defined as credibility ceiling. Credibility ceilings tests setting different degrees of credibility to further adjust the included study in a meta-analysis would favor lessening the reporting of exaggerated associations with statistic significant results, therefore decrease the unmeasured bias. A sensitivity analysis based on ceiling value(s) of 0, 20 and 40% were conducted for ccredibility ceilings tests as the literature reported [16].
Results
Study selection
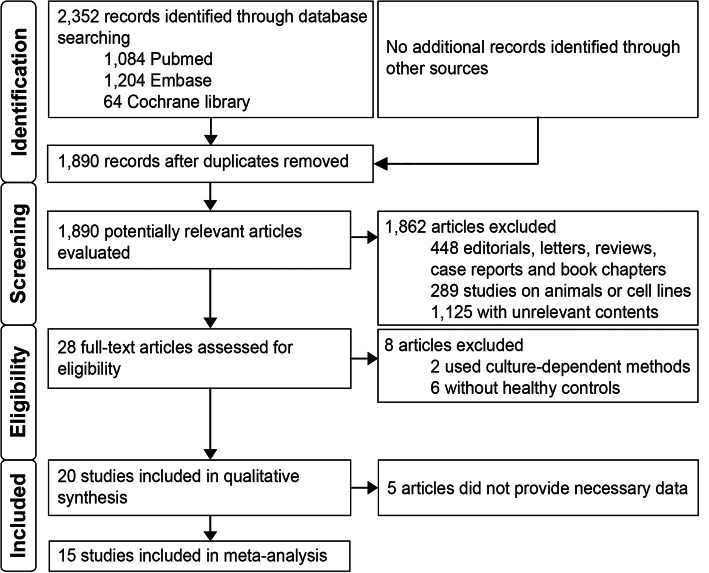
Using the predefined search strategy, 2352 citations were identified, as shown in Fig. 1. No additional records were identified through other sources. After removing duplicates, 1890 citations were screened by reviewing titles and abstracts. Of the remaining citations, 1862 articles were excluded because of unsuitable article type or studies on animals or cell lines or irrelevance. Then, the full texts of the remaining 28 articles were retrieved and assessed for eligibility. Of these full-text studies, 2 were excluded for using culture-dependent methods for detecting gut microbiota, 6 were excluded for not including a control group, and 5 were excluded because the authors did not provide necessary data. Finally, 15 studies published from 2012 to 2020 were included in the meta-analysis [7, 17–30]. Additional examination of the references listed in these studies did not provide any additional eligible studies. Agreement between the two investigators on study selection was 93.3%.
Fig. 1.
PRISMA flow diagram of studies identified in the meta-analysis. Abbreviations: PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses
Characteristics of the included studies
The main characteristics of the 15 included studies are listed in Table 1. Among the 15 studies consisting of 1265 individuals (577 NAFLD patients and 688 controls), six studies were performed in China, including one in Taiwan, one in Hong Kong, two in the USA, two in Canada, one in Thailand, one in Turkey, one in Spain, one in Italy and one in Korea. Nine of the 15 included studies adopted liver biopsy to diagnose NAFLD. All of the studies collected stools of the patients and healthy controls as samples for gut microbiota detection and utilized 16S rRNA sequencing.
Table 1.
Characteristics of the included studies in the meta-analysis
| First Author | Year | Location | Diagnostic methods | Numbers of NAFLD/Control | Age, years | Sex (Males, %) | BMI, kg/m2 | ALT, U/L | Sample | Microbiota detection method | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NAFLD Control | NAFLD | Control | NAFLD | Control | NAFLD | Control | ||||||||
| Caussy [17] | 2019 | USA | Histology | 42/51 | NAFL54.0 ± 14.9NASH65.1 ± 9.8 | 45.9 ± 19.9 | 13 (31.0%) | 15 (29.4%) | NAFL31.1 ± 6.6NASH31.3 ± 6.1 | 26.2 ± 6.8 | NAFL23.2 ± 11.2NASH45.0 ± 37.4 | 18.8 ± 8.8 | Stool | Sequencing |
| Chierico [18] | 2016 | Italy | Not mentioned | 53/62 | NAFL 12.04 ± 2.8NASH 12.27 ± 2.5 | Healthy 10.24 ± 2.5Obese 11.25 ± 2.7 | 32 (60.4%) | 26 (41.9%) | NAFL26.46 ± 4.43NASH27.42 ± 6.45 | Healthy 17.59 ± 1.79Obese26.15 ± 4.38 | NAFL32.3 ± 22.74NASH44.46 ± 16.73 | HealthyNAObese 41.50 ± 47.70 | Stool | Sequencing |
| Jiang [19] | 2015 | China | Histology or ultrasound | 53/32 | 48.00 | 41.00 | 26 (49.1%) | 5 (15.6%) | 26.4 (21.5–33.3) | 22.5 (18.2–33.5) | 42.7 (11–145) | 21 (6–29) | Stool | Sequencing |
| Li [20] | 2018 | China | Ultrasound | 30/37 | 47.53 ± 8.5 | 44.24 ± 9.2 | 15 (50%) | 11 (29.7%) | 27.19 ± 2.56 | 23.37 ± 2.21 | 27.0 ± 17.63 | 16.7 ± 8.51 | Stool | Sequencing |
| Nistal [21] | 2019 | Spain | Ultrasound | 36/17 | 49.70 (31–64) | 40.12 (25–56) | 14 (38.8%) | 5 (29.4%) | 45.64 (38.9–61.1) | 46.9 (39–63) | 34.22 (15–91) | 27.12 (10–52) | Stool | Sequencing |
| Özkul [22] | 2017 | Turkey | Histology | 46/38 | 48.0 ± 12.0 | 36.0 ± 10.0 | 22 (47.8%) | 12 (31.5%) | 29.0 ± 4.0 | 22.0 ± 2.0 | 50.0 ± 41.0 | 20.0 ± 11.0 | Stool | Sequencing |
| Raman [23] | 2013 | Canada | Ultrasound | 30/30 | 49 (34–57) | 51 (57–56) | 13 (43.3%) | 13 (43.3%) | 33 (29–35) | 22 (21–24) | 51 (31–82) | 18 (14–23) | Stool | Sequencing |
| Shen [24] | 2017 | China | Histology | 25/22 | 45.5 ± 10.1 | 50.5 ± 9.5 | 19 (76.0%) | 17 (73.7%) | 28.6 ± 3.5 | 21.6 ± 1.7 | 51.6 ± 34.5 | 17.7 ± 5.3 | Stool | Sequencing |
| Silva [25] | 2018 | Canada | Histology | 39/28 | NAFL48 (33–61)NASH46.5 (29–68) | 36.5 (21–58) | 20 (51.3%) | 15 (53.6%) | NAFL27.4 (23.5–44.2)NASH32.1 (24.17–49.53) | 26.6 (19.5–35.3) | NAFL45(14–116)NASH70(22–168) | 17.5 (7–41) | Stool | Sequencing |
| Sobhonslidsuk [26] | 2018 | Thailand | Histology | 16/8 | 59.8 ± 9.6 | 43.4 ± 6.8 | 3 (18.8%) | 0 (0%) | 27.7 ± 4.8 | 21.3 ± 1.2 | 59 ± 30 | 17 ± 6 | Stool | Sequencing |
| Tsai [27] | 2020 | China (Taiwan) | Histology | 50/25 | 51.2 ± 15.0 | 36.7 ± 15.0 | 24 (48.0%) | 12 (48.0%) | 31.3 ± 8.9 | 24.8 ± 5.2 | 50.2 ± 43.1 | 22.5 ± 15.7 | Stool | Sequencing |
| Wang [28] | 2016 | China | Ultrasound | 43/83 | 47.0 ± 12.0 | 40.5 ± 16.0 | 36 (83.4%) | 70 (84.3%) | 23.19 (22.19–24.22) | 21.77 (20.7–23.38) | 29 (20.5–39.5) | 14.5 (12–20.75) | Stool | Sequencing |
| Wong [29] | 2013 | China (HongKong) | Histology | 16/22 | 51 ± 9 | 44 ± 10 | 9 (56.3%) | 9 (40.9%) | 29.1 ± 5.6 | 22.2 ± 2.7 | 80 (44–94) | 22 (17–30) | Stool | Sequencing |
| Yun [30] | 2019 | Korea | Histology | 76/192 | 45.3 ± 8.2 | 42.9 ± 8.2 | 55 (72.4%) | 83 (43.2%) | 25.7 ± 2.6 | 22.2 ± 2.4 | 24.5 ± 12.9 | 20.9 ± 17.9 | Stool | Sequencing |
| Zhu [7] | 2012 | USA | Histology | 22/41 | 13.6 ± 3.5 | Healthy14.4 ± 1.8Obese12.7 ± 3.2 | 12 (54.5%) | 23 (56.1%) | 34.0 ± 0.4 | Healthy20.4 ± 0.1Obese33.4 ± 0.3 | 66.9 ± 1.9 | HelathyNAObese 27.7 ± 0.6 | Stool | Sequencing |
Rank by the beginning letter of the first authors. Abbreviations: NAFLD nonalcoholic fatty liver disease, NAFL nonalcoholic fatty liver, NASH nonalcoholic steatohepatitis, BMI body mass index, ALT alanine aminotransferase
Quality assessment of the included studies
As shown in Table 2, quality assessment of the included studies was carefully performed using the NOS, and the general quality was moderate (six studies scored 8 points, seven scored 7 points, and two scored 6 points, mean ± SD: 7.27 ± 0.70). Agreement between the two investigators for quality assessment was 93.3%. No study was excluded due to poor NOS score.
Table 2.
Quality assessment of included studies using the Newcastle-Ottawa Scale (Case-Control Studies)
| First Author | Selectiona | Comparabilityb | Exposurec | Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | A | B | C | ||||
| Caussy [17] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Chierico [18] | – | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 6 |
| Jiang [19] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Li [20] | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 8 |
| Nistal [21] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Özkul [22] | ☆ | ☆ | – | ☆ | – | ☆ | ☆ | ☆ | – | 6 |
| Raman [23] | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 8 |
| Shen [24] | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 8 |
| Silva [25] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Sobhonslidsuk [26] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Tsai [27] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Wang [28] | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 8 |
| Wong [29] | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 8 |
| Yun [30] | ☆ | ☆ | ☆ | ☆ | – | ☆ | ☆ | ☆ | – | 7 |
| Zhu [7] | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | ☆ | – | 8 |
Rank by the beginning letter of the first authors
a1: Is the case definition adequate? 2: Representativeness of the cases. 3: Selection of Controls. 4: Definition of Controls
bComparability: Comparability of cases and controls on the basis of the design or analysis. A maximum of two stars can be given for Comparability
cA: Ascertainment of exposure. B: Same method of ascertainment for cases and controls. C: Nonresponse rate
Meta-analysis of gut microbiota
The included studies analyzed different taxa of gut microbiota in NAFLD patients at different levels. Based on available data, this meta-analysis primarily focused on alterations at the genus level, including Escherichia, Prevotella, Streptococcus, Coprococcus, Faecalibacterium, Ruminococcus, Bacteroides, Bifidobacterium, Blautia, Clostridium, Dorea, Lactobacillus, Parabacteroides and Roseburia.
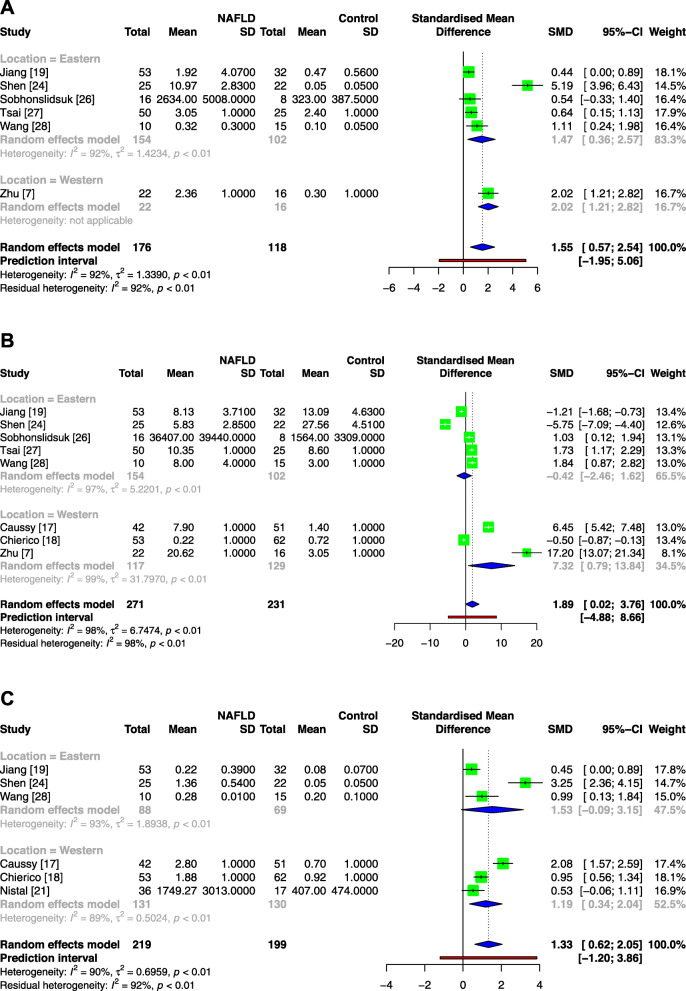
As noted in Fig. 2, the total SMDs of the genera Escherichia, Prevotella and Streptococcus were 1.55 [95% CI: 0.57, 2.54], 1.89 [95% CI: 0.02, 3.76] and 1.33 [95% CI: 0.62, 2.05], respectively, revealing that all these genera exhibited increased abundance in NAFLD. Significant heterogeneities were noted in the included studies of the three genera with I2 values greater than 50% and p values less than 0.1. In the subgroup analysis, the results of the two subgroups of the genus Escherichia were consistent with the overall results, while in the analysis of the genera Prevotella and Streptococcus, the results of the Eastern subgroups were not significantly different.
Fig. 2.
Meta-analysis comparing the abundance of Escherichia (a), Prevotella (b) and Streptococcus (c) between NAFLD patients and healthy controls. Abbreviations: NAFLD, nonalcoholic fatty liver disease; Tau2, tau-squared; Chi2, chi-square test; df, degrees of freedom; I2, I-squared
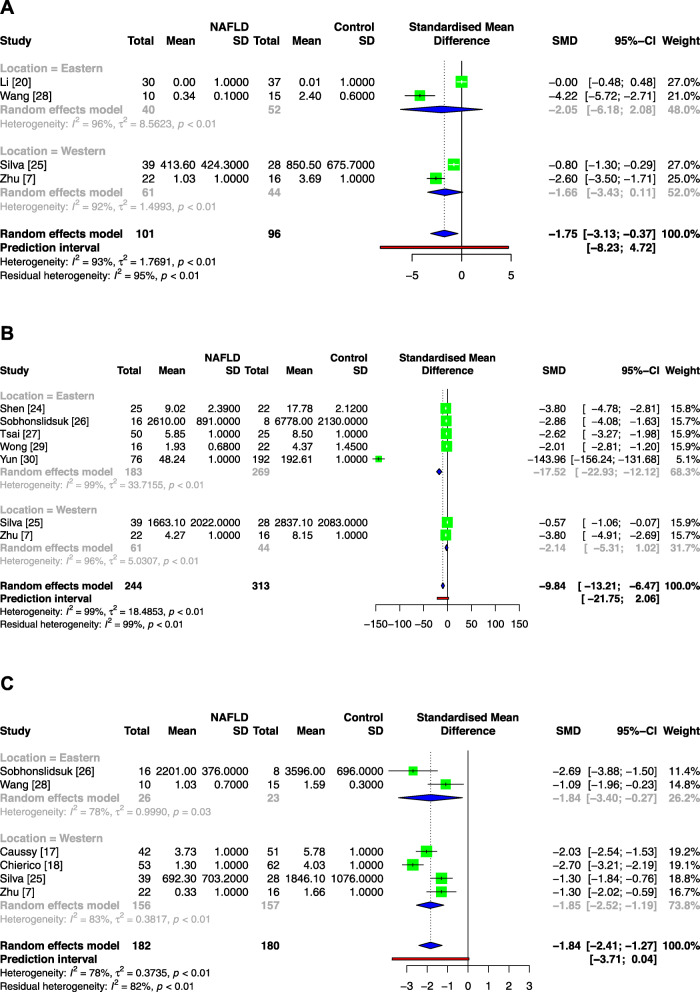
In Fig. 3, the total SMDs of the genera Coprococcus, Faecalibacterium and Ruminococcus were − 1.75 [95% CI: − 3.13, − 0.37], − 13.23 [95% CI: − 17.59, − 8.87] and − 1.84 [95% CI: − 2.41, − 1.27], respectively, revealing that these genera were deficient in NAFLD patients. Significant heterogeneities were also noted. In the subgroup analysis, neither subgroup of the genus Coprococcus showed significant differences, the Western subgroup of the genus Faecalibacterium did not reach a significant difference either, and two subgroups of the genus Ruminococcus were consistent with the overall results.
Fig. 3.
Meta-analysis comparing the abundance of Coprococcus (a), Faecalibacterium (b) and Ruminococcus (c) between NAFLD patients and healthy controls. Abbreviations: NAFLD, nonalcoholic fatty liver disease; Tau2, tau-squared; Chi2, chi-square test; df, degrees of freedom; I2, I-squared
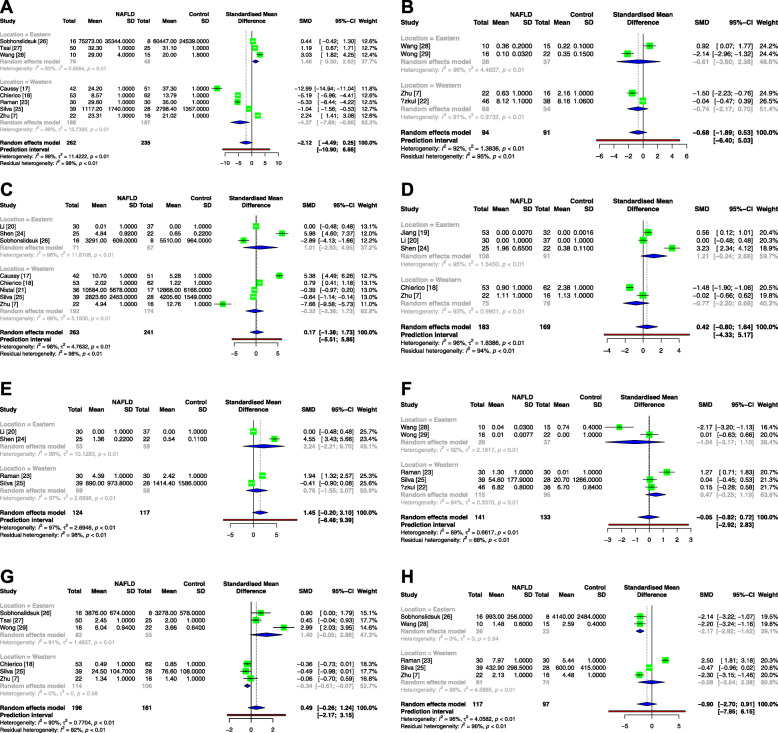
Regarding the genera Bacteroides, Bifidobacterium, Blautia, Clostridium, Dorea, Lactobacillus, Parabacteroides and Roseburia (Fig. 4), no significant differences were observed between the NAFLD patients and healthy controls in the overall analysis. Interestingly, in the subgroup analysis, the abundance of the genus Bacteroides in the NAFLD patients was decreased in the Western population but increased in the Eastern population. Moreover, the genera Parabacteroides and Roseburia in the NAFLD patients presented decreased abundance in the Western and Eastern subgroups, respectively.
Fig. 4.
Meta-analysis comparing the abundance of Bacteroides (a), Bifidobacterium (b), Blautia (c), Clostridium (d), Dorea (e), Lactobacillus (f), Parabacteroides (g) and Roseburia (h) between NAFLD patients and healthy controls. Abbreviations: NAFLD, nonalcoholic fatty liver disease; Tau2, tau-squared; Chi2, chi-square test; df, degrees of freedom; I2, I-square
Analysis of publication bias
Supplemental Figure 1 and Supplemental Table 1 provide an analysis of the funnel plots and Egger’s and Begg’s tests for publication bias. According to Egger’s and Begg’s tests, none of the analyses presented publication bias.
Sensitivity analysis
The sensitivity analysis (Supplemental Figure 2) showed that omitting each single study did not significantly change the results in the analysis for the genera Escherichia, Streptococcus, Faecalibacterium and Ruminococcus. However, after removing some of the included studies, the results of the genera Prevotella and Coprococcus did not reach significant differences. The results of the credibility ceiling test for ceilings of 0–40% are shown in Supplemental Table 2. All 3 meta-analyses that retained statistical significance with c = 20% or 40% for abundance changes of genera [Coprococcus, Faecalibacterium, Ruminococcus] in NAFLD. The meta-analysis on Escherichia, Prevotella and NAFLD none survived a 20% ceiling, whereas the Coprococcus remained statistically significant even with 20% ceiling but not 40% ceiling.
Meta regression analysis of BMI and ALT for the significant genera
On univariate meta-regression model (Supplemental Figure 3), when plotting log odds ratio of mean different abundance of bacterial genus for NAFLD versus control patients (y-axis) against alanine aminotransferase (ALT) or body mass index (BMI) levels age (x-axis), significant association was found between BMI and Faecalibacterium (log regression coefficient β = 2.38, P = 0.00001), and Prevotella (regression coefficient β = 1.0554, P = 0.0005). While ALT levels was associated with different abundance of Faecalibacterium (log regression coefficient β = 1.11, P = 0.001), Prevotella (log regression coefficient β = 0.2018, P = 0.031), Streptococcus (log regression coefficient β = 0.09, P = 0.041). After multivariate adjustments, BMI remained significant for Prevotella (log regression coefficient β = 1.20, P = 0.019), while both BMI and ALT were significant for Faecalibacterium (log regression coefficient β = − 6.2, P = 0.00001 and β = 2.51, respectively, both P < 0.00001).
Discussion
To our knowledge, this study is the first meta-analysis to investigate microbial shifts in NAFLD patients. This meta-analysis included 15 studies that assessed alterations in gut microbiota between 577 NAFLD patients and 688 healthy controls from eight different countries. Although the bacterial composition of the gut microbiota varies among individuals, the current study provides evidence for the existence of an overall profile of gut microbiota in NAFLD. Specifically, Escherichia, Prevotella and Streptococcus levels were increased in NAFLD patients (with 95% prediction interval of − 1.95 to 5.06, − 4.88 to 8.66 and − 1.2 to 3.86, respectively), while Coprococcus, Faecalibacterium and Ruminococcus were decreased (with 95% prediction interval of − 8.23 to 4.72, − 21.75 to 2.06 and − 3.71 to 0.04, respectively). However, no differences in the abundance of Bacteroides, Bifidobacterium, Blautia, Clostridium, Dorea, Lactobacillus, Parabacteroides or Roseburia were confirmed between the NAFLD patients and healthy controls.
Patients with NAFLD exhibited an increased proportion of gut Enterobacter (Escherichia) and Streptococcus in this study. Five studies [7, 19, 22, 24, 28] detected overgrowth of potentially antigenic bacteria, including Enterobacter (Escherichia) and Streptococcus, which is consistent with this study. Overgrowth of the Proteobacteria phylum (especially Escherichia coli and Enterobacteriaceae families) might increase intestinal permeability and portal LPS levels, which could trigger inflammasome activation and contribute to liver injury [31]. Moreover, Escherichia coli and other Enterobacteriaceae families produce ethanol [32], which might be responsible for the overproduction of endogenous ethanol that is involved in the development of NASH.
Prevotella has been identified as a fruit- and vegetable-rich diet-associated species that is also linked with short-chain fatty acid (SCFA) production [33]. The overall microbial differences across the studies revealed an increase in Prevotella in NAFLD. The reasons for the different conclusions across studies are not clear. One possible explanation might be that dietary habits, inflammatory conditions, and age confounded the relationship between Prevotella and NAFLD susceptibility [34]. Another probable explanation might be the different roles of the varied Prevotella copri strains in different genetic backgrounds [33]. Further studies analyzing Prevotella abundance at the strain level in NAFLD patients remain necessary.
It was also found that the abundance of Coprococcus, Faecalibacterium and Ruminococcus was reduced in NAFLD patients. Ruminococcaceae and Faecalibacterium have been well demonstrated to produce SCFAs via fermentation of dietary soluble fibers, which can activate their free fatty acid receptors (FFARs), including G-protein coupled receptor 43 and 41 (GPR43, GPR41) [35]. These pathways inhibit proinflammatory functions on neutrophils, monocytes and macrophages, thereby reducing the generation of tumor necrosis factor (TNF)-α and monocyte chemotactic protein-1 [36]. As SCFA-rich diets showed efficacy in alleviating insulin resistance and inflammation in both experimental mouse models and clinical trials [36], decreased numbers of Ruminococcaceae and Faecalibacterium may lower the SCFA levels in the gut, thereby escalating gut inflammation involved in the pathogens of NAFLD.
In this study, significant heterogeneities were present among the evaluated studies, which might be attributed to population characteristics, diet, obesity degree, NAFLD severity, associated comorbidities (i.e., metabolic syndrome), detection methods and other factors. Although subgroup analysis stratified with Eastern and Western to further demonstrated the potential effect of different regions with varied lifestyles, another important subgroup analysis of disease related features of NAFLD including steatosis degree, inflammation severity or fibrosis stages were not conducted due to the lack of these detailed information in the included studies.
Study strength and limitations
This study is the first to identify microbial signatures in NAFLD patients via a meta-analysis. A subgroup analysis was also conducted by classifying the research locations to validate the generalities of the results in the overall generation. Certain limitations were observed in this meta-analysis. First, the included studies only analyzed gut microbiomes recovered in the stool; however, several studies have indicated that mucosa-associated bacteria might greatly differ from those recovered in stool and could play a more important role in the pathogenesis of associated diseases [31, 32]. In addition, most of the analyses presented significant heterogeneities, which can only be minimized but not eliminated using a random-effects model. Third, several eligible studies [37–41] were excluded because of lack of necessary data. Fourth, the sensitivity of Egger’s and Begg’s test is low in the context of small sample size (most of which does not exceed 100) of the included studies. Last but not least, meta-regression was performed for BMI and ALT values based on aggregate data; therefore, the risk of ecollogic fallacy exists.
Conclusion
In conclusion, this study sought to assess gut microbial signatures in NAFLD patients via a meta-analysis. This study confirmed increases in the genera Escherichia, Prevotella and Streptococcus and decreases in the genera Coprococcus, Faecalibacterium and Ruminococcus as compositional patterns of fecal microbiota in NAFLD patients. Furthermore, BMI may contribute to the abundance change of Faecalibacterium and Prevotella in NAFLD relative to the control, whereas inflammation markers of ALT was associated with the abundance change of Streptococcus and Faecalibacterium in the meta regression analysis. This work will help identify specific differences in the proportion of several bacterial taxa in stool samples correlated with the presence of NAFLD, which will provide evidence for the design of probiotic and antibiotic treatments for NAFLD patients. This study suggested that targeting these gut microbiota in NAFLD patients may be another approach for treatment in the future.
Supplementary Information
Additional file 1. Supplemental Material.
Acknowledgements
We are grateful to Dr. Xinxiang Fan in Sun Yat-sen Memorial Hospital, Sun Yat-sen University for his assistance in statistical analysis of this study.
Abbreviations
- CI
Confidence interval
- FFARs
Free fatty acid receptors
- GPR
G-protein coupled receptor
- LPS
Lipopolysaccharide
- MeSH
Medical subject heading
- NAFL
Non-alcoholic fatty liver
- NAFLD
Nonalcoholic fatty liver disease
- NASH
Nonalcoholic steatohepatitis
- NOS
Newcastle Ottawa scale
- PRISMA
Preferred reporting items for systematic reviews and meta-analyses
- SCFAs
Short-chain fatty acids
- SD
Standard deviation
- SMD
Standard mean difference
- TNF
Tumor necrosis factor
Authors’ contributions
Bihui Zhong: conceive, design, critical revision of the manuscript for important intellectual content; Fuxi Li, and Junzhao Ye contributed equally to this work: data collection, analysis, manuscript drafting; Congxiang Shao: quality assessment of the included studies. All authors have read and approved the final manuscript.
Funding
This study was supported by National Natural Science Foundation of China (81,870,404, 81,670,518, 81,170,392), Medical Scientific Research Foundation of Guangdong Province (A2019496), and Science and Technology Program of Guangdong province, China (2013B021800290, 2014A020212118, 2017A020215015).
Availability of data and materials
All data generated or analysed during the current study are included in this published article and its supplementary information files.
Competing interests
none.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fuxi Li and Junzhao Ye contributed equally to this work.
References
- 1.Friedman SL, Neuschwander-Tetri BA, Rinella M, et al. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24(7):908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Younossi ZM, Koenig AB, Abdelatif D, et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84. doi: 10.1002/hep.28431. [DOI] [PubMed] [Google Scholar]
- 3.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 4.Kapil S, Duseja A, Sharma BK, et al. Small intestinal bacterial overgrowth and toll-like receptor signaling in patients with non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2016;31(1):213–221. doi: 10.1111/jgh.13058. [DOI] [PubMed] [Google Scholar]
- 5.Belei O, Olariu L, Dobrescu A, et al. The relationship between non-alcoholic fatty liver disease and small intestinal bacterial overgrowth among overweight and obese children and adolescents. J Pediatr Endocrinol Metab. 2017;30(11):1161–1168. doi: 10.1515/jpem-2017-0252. [DOI] [PubMed] [Google Scholar]
- 6.Wong VW, Wong GL, Chan HY, et al. Bacterial endotoxin and non-alcoholic fatty liver disease in the general population: a prospective cohort study. Aliment Pharmacol Ther. 2015;42(6):731–740. doi: 10.1111/apt.13327. [DOI] [PubMed] [Google Scholar]
- 7.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology. 2013;57(2):601–609. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 8.Boursier J, Mueller O, Barret M, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology. 2016;63(3):764–775. doi: 10.1002/hep.28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma YY, Li L, Yu CH, et al. Effects of probiotics on nonalcoholic fatty liver disease: a meta-analysis. World J Gastroenterol. 2013;19(40):6911–6918. doi: 10.3748/wjg.v19.i40.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandl K, Schnabl B. Intestinal microbiota and nonalcoholic steatohepatitis. Curr Opin Gastroenterol. 2017;33(3):128–133. doi: 10.1097/MOG.0000000000000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moher D, Liberati A, Tetzlaff J, et al. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. doi: 10.1136/bmj.b2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J.P. Higgins, J.J. Deeks, Selecting studies and collecting data, in Cochrane handbook for systematic reviews of interventions, J.P. Higgins and S. Green, J.P. Higgins and S. Green^Editors. 2011.
- 13.Hozo SP, Djulbegovic B, Hozo I. Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol. 2005;5:13. doi: 10.1186/1471-2288-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhuang X, Xiong L, Li L, et al. Alterations of gut microbiota in patients with irritable bowel syndrome: a systematic review and meta-analysis. J Gastroenterol Hepatol. 2017;32(1):28–38. doi: 10.1111/jgh.13471. [DOI] [PubMed] [Google Scholar]
- 15.IntHout J, Ioannidis JPA, Rovers MM, et al. Plea for routinely presenting prediction intervals in meta-analysis. BMJ Open. 2016;6(7):e10247. doi: 10.1136/bmjopen-2015-010247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salanti G, Ioannidis JP. Synthesis of observational studies should consider credibility ceilings. J Clin Epidemiol. 2009;62(2):115–122. doi: 10.1016/j.jclinepi.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Caussy C, Tripathi A, Humphrey G, et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat Commun. 2019;10(1):1406. [DOI] [PMC free article] [PubMed]
- 18.Del CF, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric nonalcoholic fatty liver disease and obese patients unveiled by an integrated meta-omics-based approach. Hepatology. 2017;65(2):451–464. doi: 10.1002/hep.28572. [DOI] [PubMed] [Google Scholar]
- 19.Jiang W, Wu N, Wang X, et al. Dysbiosis gut microbiota associated with inflammation and impaired mucosal immune function in intestine of humans with non-alcoholic fatty liver disease. Sci Rep. 2015;5:8096. doi: 10.1038/srep08096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li F, Sun G, Wang Z, et al. Characteristics of fecal microbiota in non-alcoholic fatty liver disease patients. Sci China Life Sci. 2018;61(7):770–778. doi: 10.1007/s11427-017-9303-9. [DOI] [PubMed] [Google Scholar]
- 21.Nistal E, Sáenz De Miera LE, Ballesteros Pomar M, et al. An altered fecal microbiota profile in patients with non-alcoholic fatty liver disease (NAFLD) associated with obesity. Rev Esp Enferm Dig. 2019;111(4):275–82. [DOI] [PubMed]
- 22.Ozkul C, Yalinay M, Karakan T, et al. Determination of certain bacterial groups in gut microbiota and endotoxin levels in patients with nonalcoholic steatohepatitis. Turk J Gastroenterol. 2017;28(5):361–9. [DOI] [PubMed]
- 23.Raman M, Ahmed I, Gillevet PM, et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013;11(7):868–875. doi: 10.1016/j.cgh.2013.02.015. [DOI] [PubMed] [Google Scholar]
- 24.Shen F, Zheng RD, Sun XQ, et al. Gut microbiota dysbiosis in patients with non-alcoholic fatty liver disease. Hepatobiliary Pancreat Dis Int. 2017;16(4):375–381. doi: 10.1016/S1499-3872(17)60019-5. [DOI] [PubMed] [Google Scholar]
- 25.Silva HE, Teterina A, Comelli EM, et al. Nonalcoholic fatty liver disease is associated with dysbiosis independent of body mass index and insulin resistance. Sci Rep. 2018;8(8):1466. doi: 10.1038/s41598-018-19753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sobhonslidsuk A, Chanprasertyothin S, Pongrujikorn T, et al. The Association of gut Microbiota with nonalcoholic steatohepatitis in Thais. Biomed Res Int. 2018;2018:1–08. doi: 10.1155/2018/9340316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai MC, Liu YY, Lin CC, et al. Gut microbiota Dysbiosis in patients with biopsy-proven nonalcoholic fatty liver disease: a cross-sectional study in Taiwan. Nutrients. 2020;12(3):820. [DOI] [PMC free article] [PubMed]
- 28.Wang B, Jiang X, Cao M, et al. Altered fecal microbiota correlates with liver biochemistry in nonobese patients with non-alcoholic fatty liver disease. Sci Rep. 2016;6:32002. doi: 10.1038/srep32002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong VW, Tse CH, Lam TT, et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis--a longitudinal study. PLoS One. 2013;8(4):e62885. doi: 10.1371/journal.pone.0062885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yun Y, Kim H, Lee E, et al. Fecal and blood microbiota profiles and presence of nonalcoholic fatty liver disease in obese versus lean subjects. Plos One. 2019;14(3):e213692. doi: 10.1371/journal.pone.0213692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DuPont AW, DuPont HL. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8(9):523–531. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- 32.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Filippis F, Pasolli E, Tett A, et al. Distinct genetic and functional traits of human intestinal Prevotella copri strains are associated with different habitual diets. Cell Host Microbe. 2019;25(3):444–453. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Jazayeri O, Daghighi SM, Rezaee F. Lifestyle alters GUT-bacteria function: linking immune response and host. Best Pract Res Clin Gastroenterol. 2017;31(6):625–635. doi: 10.1016/j.bpg.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Guo Z, Xue Z, et al. A phylo-functional core of gut microbiota in healthy young Chinese cohorts across lifestyles, geography and ethnicities. ISME J. 2015;9(9):1979–1990. doi: 10.1038/ismej.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kimura I, Ichimura A, Ohue-Kitano R, et al. Free fatty acid receptors in health and disease. Physiol Rev. 2020;100(1):171–210. doi: 10.1152/physrev.00041.2018. [DOI] [PubMed] [Google Scholar]
- 37.Mouzaki M, Comelli EM, Arendt BM, et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology. 2013;58(1):120–127. doi: 10.1002/hep.26319. [DOI] [PubMed] [Google Scholar]
- 38.Michail S, Lin M, Frey MR, et al. Altered gut microbial energy and metabolism in children with non-alcoholic fatty liver disease. FEMS Microbiol Ecol. 2015;91(2):1–09. doi: 10.1093/femsec/fiu002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duarte S, Stefano JT, Miele L, et al. Gut microbiome composition in lean patients with NASH is associated with liver damage independent of caloric intake: A prospective pilot study. 2017. [DOI] [PubMed] [Google Scholar]
- 40.Vernekar M, Singhal R, Joshi K, et al. Variation in the plasma levels of polyunsaturated fatty acids in control Vis-à-Vis nonalcoholic fatty liver disease subjects and its possible association with gut microbiome. Metab Syndr Relat Disord. 2018;16(7):329–335. doi: 10.1089/met.2018.0008. [DOI] [PubMed] [Google Scholar]
- 41.Yuan J, Baker SS, Liu W, et al. Endotoxemia unrequired in the pathogenesis of pediatric nonalcoholic steatohepatitis. J Gastroenterol Hepatol. 2014;29(6):1292–1298. doi: 10.1111/jgh.12510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplemental Material.
Data Availability Statement
All data generated or analysed during the current study are included in this published article and its supplementary information files.