Abstract
Background
Malaria is one of the most serious infectious diseases in the world. The malaria burden is greatly affected by human immunity, and immune responses vary between populations. Genetic diversity in KIR and HLA-C genes, which are important in immunity to infectious diseases, is likely to play a role in this heterogeneity. Several studies have shown that KIR and HLA-C genes influence the immune response to viral infections, but few studies have examined the role of KIR and HLA-C in malaria infection, and these have used low-resolution genotyping. The aim of this study was to determine whether genetic variation in KIR and their HLA-C ligands differ in Ugandan populations with historically varied malaria transmission intensity using more comprehensive genotyping approaches.
Methods
High throughput multiplex quantitative real-time PCR method was used to genotype KIR genetic variants and copy number variation and a high-throughput real-time PCR method was developed to genotype HLA-C1 and C2 allotypes for 1344 participants, aged 6 months to 10 years, enrolled from Ugandan populations with historically high (Tororo District), medium (Jinja District) and low (Kanungu District) malaria transmission intensity.
Results
The prevalence of KIR3DS1, KIR2DL5, KIR2DS5, and KIR2DS1 genes was significantly lower in populations from Kanungu compared to Tororo (7.6 vs 13.2%: p = 0.006, 57.2 vs 66.4%: p = 0.005, 33.2 vs 46.6%: p < 0.001, and 19.7 vs 26.7%: p = 0.014, respectively) or Jinja (7.6 vs 18.1%: p < 0.001, 57.2 vs 63.8%: p = 0.048, 33.2 vs 43.5%: p = 0.002, and 19.7 vs 30.4%: p < 0.001, respectively). The prevalence of homozygous HLA-C2 was significantly higher in populations from Kanungu (31.6%) compared to Jinja (21.4%), p = 0.043, with no significant difference between Kanungu and Tororo (26.7%), p = 0.296.
Conclusions
The KIR3DS1, KIR2DL5, KIR2DS5 and KIR2DS1 genes may partly explain differences in transmission intensity of malaria since these genes have been positively selected for in places with historically high malaria transmission intensity. The high-throughput, multiplex, real-time HLA-C genotyping PCR method developed will be useful in disease-association studies involving large cohorts.
Keywords: Genetic diversity, Human leukocyte antigen, Killer-cell immunoglobulin-like receptor, Malaria, Uganda
Background
Malaria is estimated to cause nearly half a million deaths each year worldwide [1]. Malaria is a known evolutionary driving force in the selection of several human genetic polymorphisms that protect against malaria. Red blood cell alterations are the most studied genetic abnormalities that impact on malaria [2]. These include mutations in the alpha- and beta-globin genes that lead to sickle cell anaemia or thalassemias, glucose-6-phosphate dehydrogenase (G6PD) deficiency and the Duffy antigen protein [3]. It has been suggested that many of these polymorphisms were selected in human populations due to their role in protection from the detrimental effects of Plasmodium falciparum infection [4]. It has been demonstrated that different populations have developed independent evolutionary responses to malaria [5]. For example, 3 haemoglobin variants (HbS, HbC, and HbE) appear to confer protection against malaria in different parts of the world [6]. The HbS allele is common in Africa, but rare in Southeast Asia, and the opposite is true for the HbE allele [7, 8].
A recent genome wide association study of 17,000 individuals in Africa reported that known genetic variants account for only 11% of the total genetic influence of malaria on the human genome [9]. Among other genes potentially influencing malaria responses are those mediating innate immunity, which is important in protection from P. falciparum infection. Natural killer (NK) cells play an important role in the innate immune response to malaria infection [10, 11]. NK cells are the first cells in peripheral blood to produce interferon gamma (IFN- γ) in response to P. falciparum infection [11], and they have also been shown to participate in adaptive immunity. Recent evidence indicates a role for NK cells in malaria infection in humans and in mouse models [10, 12]. It has been shown that copy number variation (CNV) in KIR genes influences immunity to infections [13] and plays an important role in NK cell education [14] through interactions with their HLA class I ligands. Hence, the expression of multiple copies of KIR genes could potentially lead to enhanced NK cell education, thereby strengthening immunity to pathogens. This has been well studied in viral infections, but not in malaria.
Some studies have demonstrated that individuals may vary in their ability to elicit an innate immune response to malaria infection, with clear implications for disease manifestations [15]. Heterogeneity in response could arise from variations in KIR and their major ligands, HLA-C molecules, that have a direct impact on NK cell functions [11, 16]. The frequencies of different KIR and HLA-C genes vary remarkably across world populations, which might reflect differential selection pressures as well as persistence of ancestral genotypes [17]. The KIR and HLA loci have been suggested to be fast evolving and under positive selection, with pathogen pressure as the driving force [18, 19]. Genetic variation of KIR and their HLA-C ligands across the African continent is not well documented. Several studies have linked high KIR and HLA genetic diversity in Africa to malaria pressure [20–22]. However, there is limited data regarding the distribution of KIR and HLA variants in populations with varied malaria transmission intensity. Since interactions between the genetically diverse KIR and HLA molecules modulate the functionality of the NK cell response to malaria infections, a better understanding of the distribution of KIR and HLA genes in populations with varied malaria transmission intensity will be important in appreciating the impact of P. falciparum malaria on the evolution of KIR and HLA genes.
To date, there is limited data on the distribution of KIR and their HLA-C ligands in populations with varied malaria transmission intensity. This is partly due to the genotyping approaches that only reveal information about presence or absence of KIR and HLA genes. Furthermore, the few studies that have been carried out are case–control comparisons of severe versus uncomplicated malaria, with limited genetic information about KIR and HLA genes. As an alternative approach, more comprehensive genotyping techniques that provide additional information like copy number variation in these genes were used to evaluate the diversity of KIR genes and their HLA-C ligands in humans living in populations with historically varied malaria transmission intensity.
Methods
Study samples and populations
Samples from cohorts enrolled at 3 sites in Uganda were utilized, that is, Nagongera Sub-county in Tororo District, a rural area in southeastern Uganda with historically high malaria transmission intensity; Walukuba Sub-county in Jinja District, a peri-urban area near the city of Jinja in south-central Uganda with historically moderate malaria transmission intensity; and Kihihi Sub-county in Kanungu District, a rural area in southwestern Uganda with historically low malaria transmission intensity.
The total population of Nagongera sub-county, Tororo District was 37,500. All participants at this site were recruited within Nagongera Health Center IV, which is the largest public health facility in the sub-county treating an average of 2044 patients per month. Walukuba sub-county, Jinja District has an estimated population of 31,900. Study participants from Walukuba sub-county were recruited within Walukuba health centre IV, the largest public health facility in the sub-county and treating an average of 3198 patients per month. Kihihi is a rural sub-county in Kanungu District with an estimated population of 55,700. Study participants were recruited within Kihihi health centre IV, which is the largest healthcare facility in Kihihi sub-county, a public health facility that treated an average of 1945 patients per month.
To establish these cohorts, all households within the 3 sites were enumerated and mapped, and randomly selected 100 households, that included at least one resident 6 months to 10 years of age, and which were enrolled and followed from August 2011 to September 2013, as previously described [23]. All the participants enrolled in these cohorts provided thick blood smears and a blood sample for genetic analysis. For this study, all participants whose parents consented to future use of their samples were considered. No a priori power calculation was performed.
Sample collection and DNA purification
Blood samples from previous studies [23] were collected into EDTA tubes, and human genomic DNA was purified from buffy coats using QIAamp DNA Mini Kits (Qiagen), following manufacturer’s instructions using kit inserts with minor modifications. For each sample, 300 μl of buffy coat was mixed with 20 μl of kit protease enzyme solution and then 200 μl of lysis buffer; the mixture was vortexed for 15 s and incubated at 56 °C for 10 min, and then 200 μl of absolute ethanol was added. The mixture was vortexed briefly and transferred to a QIAamp column, and the column was spun for 1 min at 8000 rpm. The column was then washed twice with kit wash buffer, and DNA was eluted by incubating with 80 μl of kit elution buffer at room temperature for 5 min followed by centrifugation at 8000 rpm for 5 min. The DNA concentration was determined using a Qubit fluorimeter (Life Technologies, Carlsbad, CA, USA), and the isolated DNA was stored at − 20 °C.
Preparation of DNA for multiplex qPCR
To prepare human genomic DNA for KIR and HLA-C genotyping as well as KIR copy number identification, 10 ng samples of genomic DNA (2.5 µl of 4 ng/µl) were aliquoted into 384-well plates using the Hydra 96 micro dispenser (Art Robbins, San Jose, CA, USA) [24]. The DNA was air dried in the plates for subsequent multiplex quantitative PCR assays. Molecular grade water was used in all reactions.
KIR genotyping by high-throughput, multiplex, real-time qPCR
Two pairs of primers were used for each gene, as previously described [25]. Additional KIR primers were designed using sequence information from the immuno polymorphism database-KIR (IPD-KIR) database (release 2.4.0) to detect rare alleles of KIR2DS5 and KIR2DL3 (KIR2DS5, 2DS5rev2: TCC AGA GGG TCA CTG GGA and KIR2DL3, 2DL3rev3: AGA CTC TTG GTC CAT TAC CG) [26]. Samples were genotyped for copy number by multiplex quantitative PCR for all the KIR genes (KIR2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 2DP1, 3DP1, 3DL1, 3DL2, 3DL3, 3DS1) [24]. Reactions were carried out in quadruplicate to ensure accuracy of the copy number scoring. Two controls with known copy number and one non-template control were included in each run. Two assays for both 3DL1 and 3DL2 genes that target different exons of the genes were included to identify known fusion genes [27], which are carried on a truncated haplotype (with 2DS4 completely deleted) seen in individuals of African descent. There is a drop in copy number for exon 9 of 3DL1 and exon 4 of 3DL2 (i.e., discordance between the exon 4 and exon 9 copy numbers in the same gene) when the fusion gene is present. Assays for 2DS4 variants, 2DS4DEL (a 22-bp deletion in exon 5 that causes a frameshift mutation) and 2DS4WT (full-length gene) were also included.
HLA-C genotyping by high-throughput, multiplex, real-time qPCR
A high-throughput real-time qPCR for genotyping HLA-C allotypes was developed. For every reaction, KAPA SYBR buffer (5 µl), forward primer (1 µl), reverse primer (1 µl), and water (4 µl) were added to dried DNA in the 384-well plates. HLA-C1 PCR conditions were: denaturation at 95 °C for 3 min, 5 cycles of 95 °C for 3 s and 72 °C for 30 s, followed by 35 cycles of 95 °C for 3 s and 70 °C for 30 s, dissociation at 60 °C for 1 s, and finally 95 °C. HLA-C2 PCR conditions were: denaturation at 95 °C for 3 min, 5 cycles of 95 °C for 3 s and 72 °C for 45 s, followed by 40 cycles of 95 °C for 3 s and 70 °C for 45 s, dissociation at 60 °C for 1 s, and finally 95 °C. In each HLA-C allotype, primers were used at 5 µM concentrations. Primer combinations for C1 and C2 were: C1 = C1Fa and C1Fb with C1R, and C2 = C2F with C2R, respectively (Table 1). This method was validated against a large range of samples, the HLA Reference Panel from Coriell with known HLA-C allotypes, and in families. The sequences for each of the primers used are shown in Table 1.
Table 1.
HLA-C primers for high-throughput qPCR
| Primer name | Sequence |
|---|---|
| C1Fa | GCCGCGAGTCCAAGAGG |
| C1Fb | GCCGCGAGTCCGAGAGG |
| C2F | CTGACCGAGTGAACCTGCGGAAA |
| C2R | GGAGATGGGGAAGGCTCCCCAC |
| C1R | GCGCAGGTTCCGCAGGC |
KIR and HLA-C genotypes analysis
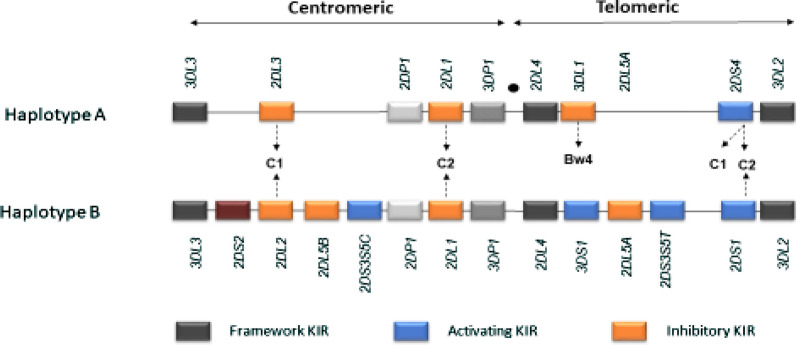
KIR genotypes were defined following the recommendations from the 2011 KIR workshop that was held at Tammsvik, Stockholm, Sweden [28]. Briefly, the centromeric A region (cenA) was defined by the presence of KIR2DL3 and KIR2DL1 and absence of any A haplotype gene; the centromeric B (cenB) region was defined by presence of any centromeric B haplotype gene (KIR2DS2 and/or KIR2DL2, and/or 2DL5B and/or centromeric 2DS3/5). The telomeric A (telA) region was defined by KIR3DL1 and KIR2DS4 and absence of any A haplotype gene, and the telomeric B (telB) region was defined by presence of any centromeric B haplotype gene (KIR3DS1 and/or KIR2DS1 and or 2DL5B and/or telomeric 2DS5) (Fig. 1). The KIR and HLA-C genotypes were ascertained according to the Allele Frequency Net Database (http://www.allele.frequencies.net).
Fig. 1.
KIR haplotypes. KIR A haplotypes (a, b) are present in all populations worldwide. KIR A haplotype is composed of mainly inhibitory KIR except KIR2DS4. Allelic polymorphism is very high in the KIR A haplotype (KIR3DL1, 3DL2 and 3DL3 exhibit > 100 alleles, and 2DL1 and 2DL3 exhibit ~ 50 alleles). Haplotypes (b) has several activating receptors, with variable number of genes and fewer allelic polymorphisms. Some KIR B haplotypes are composed of combinations of haplotypes (a, b) (CenA-TelB, CenB-TelA). The HLA epitopes bound by some KIRs are known and are indicated as C1, C2 or Bw4
KIR Copy number determination by multiplex quantitative PCR
Copy numbers for all KIR genes (KIR2DL1–5, 2DS1–5, 2DS4 (separate assays for the gene, wild-type variant [2DS4WT], and deletion variant [2DS4DEL]), 2DP1, 3DP1, 3DL1-3 and 3DS1) were determined using a Roche Light Cycler 480. Copy numbers were measured by relative quantification analysis of the target KIR gene and reference gene (signal transducer and activator of transcription 6; STAT 6) using the comparative Cq method [24, 29]. Cq value is the qPCR cycle at which fluorescence from amplification exceeds the background fluorescence (also referred to as threshold cycle, Ct). The ΔΔCq was used to calculate KIR copy number. The first ΔCq was calculated by the cycle threshold difference between the target and reference assay of the same sample. The second ΔCq was calculated by the difference of ΔCq values from a test sample and a calibrator sample with known copy number of the target. Two controls with known copy number and one non-template control were included in each run. COPYCALLER software from Applied Biosystems (Foster City, CA, USA) was used to score KIR copy numbers. When the Cq of the reference gene was greater than 32 or a data point was more than 4 SD from the mean ΔCq of four replicates, the reaction was not analysed. KIR copy number frequencies were calculated for all the samples.
Statistical methods
Data across the 3 sites was described using frequencies and percentages for categorical variables. Frequencies of KIR genes, KIR genotypes and HLA-C allotypes were calculated by direct counting. Differences in the distribution of KIR and HLA-C genetic variants within the three populations were compared by Chi-square and Mid-P exact tests. A p-value < 0.05 was considered significant.
Results
Characteristics of study participants and populations
Among the 1,344 subjects in the 3 cohorts, 44% were under 5 years of age, and 56% between 5 and 10 years. A recent report defined malaria transmission, prevalence, and incidence in the 3 cohorts (Table 2) [23]. The 3 sites differed markedly, with very high transmission intensity, parasite prevalence and malaria incidence in Tororo District, lower levels of all of these parameters in Kanungu District, and the lowest levels in Jinja District [23, 30]. Of note, malaria transmission was considerably greater in earlier surveys in Jinja District [31], with decreasing transmission likely due to the peri-urban nature of the study area. In Tororo District, transmission has subsequently decreased greatly, after annual indoor residual spraying of insecticides was launched in 2014 [32]. Historically, malaria transmission intensity followed the rank order Tororo > Jinja > Kanungu [31]. The aim of this study was to compare KIR and HLA-C genetic variants that may have been selected due to differential malaria selection pressures at these sites.
Table 2.
Characteristics of study participants and populations
| Study sites | |||
|---|---|---|---|
| Tororo | Jinja | Kanungu | |
| Characteristics of sites | |||
| Location | South-eastern | South-central | South-western |
| Setting | Rural | Peri-urban | Rural |
| Altitude | 695–1443 m | 1102–1500 m | 886–1329 m |
| Number of study subjects | |||
| Children below 5 years | 340 | 321 | 365 |
| Children 5–10 years | 106 | 114 | 98 |
| Total | 446 | 435 | 463 |
| Malaria indicators (children)a | |||
| Entomological inoculation rate per year | 310 | 2.8 | 32.0 |
| Parasite prevalence | 28.7% | 7.4% | 9.3% |
| Malaria incidence per year | 2.81 | 0.43 | 1.43 |
aDetermined August 2011-September 2013 [23]
Comparative prevalence of KIR and HLA-C genetic variants at 3 sites in Uganda
Analysis for differential prevalence of KIR genes, KIR genotypes, HLA-C allotypes (HLA-C1C1, C1C2, C2C2), centromeric and telomeric KIR motifs and KIR/HLA-C combinations across the 3 populations was carried out (Table 3). More than 90% of samples from all the study populations were successfully analysed for KIR and HLA-C genetic variants. The prevalence of the inhibitory KIR genes KIR2DL1 and KIR3DL1 and the activating gene KIR2DS4 was very high (> 95%). The prevalence of KIR3DS1 was generally low across the 3 populations, with the lowest prevalence in Kanungu (7.6%) compared to Jinja (18.1%) and Tororo (13.2%). The prevalence of KIR2DS5 was lower in Kanungu (33.2%) compared to Jinja (43.5%) and Tororo (46.6%). The prevalence of HLA-C1C2 heterozygotes was higher (53.4%) in all 3 populations compared to homozygous HLA-C1 (20.1%) or homozygous HLA-C2 (26.5%).
Table 3.
Distribution of KIR and HLA-C genetic variants at the 3 sites in Uganda
| Genetic variants | Study sites | ||
|---|---|---|---|
| Tororo, N = 438 n (%) | Jinja, N = 414 n (%) | Kanungu, N = 446 n (%) | |
| KIR genes | |||
| 2DS2 | 224 (51.1) | 218 (52.7) | 228 (51.1) |
| 2DL2 | 254 (58.0) | 240 (58.0) | 254 (57.0) |
| 2DL3 | 366 (83.6) | 352 (85.0) | 380 (85.2) |
| 2DP1 | 432 (98.6) | 409 (98.8) | 442 (99.1) |
| 2DL1 | 432 (98.6) | 409 (98.8) | 441 (98.9) |
| 3DL1 | 421 (96.1) | 394 (95.2) | 439 (98.4) |
| 3DS1 | 58 (13.2) | 75 (18.1) | 34 (7.6) |
| 2DL5 | 291 (66.4) | 264 (63.8) | 255 (57.2) |
| 2DS5 | 204 (46.6) | 180 (43.5) | 148 (33.2) |
| 2DS3 | 92 (21.0) | 84 (20.3) | 111 (24.9) |
| 2DS1 | 117 (26.7) | 126 (30.4) | 88 (19.7) |
| 2DS4 | 424 (96.8) | 392 (94.7) | 438 (98.2) |
| Tororo, N = 385 n (%) | Jinja, N = 392 n (%) | Kanungu, N = 433 n (%) | |
|---|---|---|---|
| KIR genotypes | |||
| AA | 150 (39.0) | 133 (33.9) | 171 (39.5) |
| BX | 235 (61.0) | 259 (66.1) | 262 (60.5) |
| Tororo, N = 356 n (%) | Jinja, N = 168 n (%) | Kanungu, N = 405 n (%) | |
|---|---|---|---|
| HLA-C allotypes | |||
| C1C1 | 66 (18.5) | 39 (23.2) | 75 (18.5) |
| C1C2 | 195 (54.8) | 93 (55.4) | 202 (49.9) |
| C2C2 | 95 (26.7) | 36 (21.4) | 128 (31.6) |
| Tororo, N = 438 n (%) | Jinja, N = 414 n (%) | Kanungu, N = 446 n (%) | |
|---|---|---|---|
| Centromeric KIR region | |||
| CenAA | 191 (43.6) | 171 (41.3) | 190 (42.6) |
| CenAB | 175 (40.0) | 182 (44.0) | 192 (43.1) |
| CenBB | 72 (16.4) | 61 (14.7) | 64 (14.3) |
| Tororo, N = 438 n (%) | Jinja, N = 413 n (%) | Kanungu, N = 446 n (%) | |
|---|---|---|---|
| Telomeric KIR region | |||
| TelAA | 314 (71.7) | 283 (68.5) | 356 (79.8) |
| TelAB | 115 (26.3) | 112 (27.1) | 83 (18.6) |
| TelBB | 9 (2.0) | 18 (4.4) | 7 (1.6) |
| Tororo, N = 313 n (%) | Jinja, N = 152 n (%) | Kanungu, N = 393 n (%) | |
|---|---|---|---|
| Combinations of KIR haplotypes/HLA-C | |||
| AA/C1C1 | 21 (6.7) | 11 (7.3) | 22 (5.6) |
| AA/C1C2 | 62 (19.8) | 37 (24.3) | 93 (23.7) |
| AA/C2C2 | 37 (11.8) | 11 (7.2) | 46 (11.7) |
| BX/C1C1 | 36 (11.5) | 23 (15.1) | 50 (12.7) |
| BX/C1C2 | 109 (34.8) | 46 (30.3) | 105 (26.7) |
| BX/C2C2 | 48 (15.4) | 24 (15.8) | 77 (19.6) |
The prevalence of KIR3DS1, 2DL5, 2DS5, and 2DS1 genes was significantly lower in Kanungu compared to both Tororo (7.6 vs 13.2%: p = 0.006, 57.2 vs 66.4%: p = 0.005, 33.2 vs 46.6%: p < 0.001, and 19.7% vs 26.7%: p = 0.014, respectively) and Jinja (7.6 vs 18.1%: p < 0.001, 57.2 vs 63.8%: p = 0.048, 33.2 vs 43.5%: p = 0.002 and 19.7 vs 30.4%: p < 0.001, respectively). There was no significant difference in the prevalence of inhibitory KIR2DL1, 2DL2, 2DL3, and 3DL1 and activating KIR2DS2, 2DS3 and 2DS4 (Table 4). The prevalence of homozygous HLA-C2 was significantly higher in Kanungu (31.6%) compared to Jinja (21.4%), p = 0.043. No significant difference was observed between the prevalence of HLA-C2 in Tororo (26.7%) and Kanungu (31.6%), p = 0.296. There was no significant difference in the prevalence of KIR AA and KIR BX genotypes in Tororo and Jinja (p = 0.145), Tororo and Kanungu (p = 0.877), or Jinja and Kanungu (p = 0.098). Combinations of KIR genotypes with HLA-C ligands did not differ in the 3 populations. There was no significant difference in the KIR centromeric or telomeric motifs across the 3 populations (Table 5).
Table 4.
Comparative prevalence of KIR genes from 3 sites of Uganda with historically varied malaria transmission intensity
| Study sites | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genes | Tororo, N = 438 | Jinja, N = 414 | Kanungu, N = 446 | Mid-P p-values | |||||
| n | % | n | % | n | % | T vs J | T vs K | J vs K | |
| 2DS2 | 224 | 51.1% | 218 | 52.7% | 228 | 51.1% | 0.658 | 0.995 | 0.652 |
| 2DL2 | 254 | 58.0% | 240 | 58..0% | 254 | 57.0% | 0.995 | 0.754 | 0.762 |
| 2DL3 | 366 | 83.6% | 352 | 85.0% | 380 | 85.2% | 0.558 | 0.502 | 0.942 |
| 2DP1 | 432 | 98.6% | 409 | 98.8% | 442 | 99.1% | 0.834 | 0.506 | 0.654 |
| 2DL1 | 432 | 98.6% | 409 | 98.8% | 441 | 98.9% | 0.834 | 0.739 | 0.906 |
| 3DL1 | 421 | 96.1% | 394 | 95.2% | 439 | 98.4% | 0.497 | 0.064 | 0.061 |
| 3DS1 | 58 | 13.2% | 75 | 18.1% | 34 | 7.6% | 0.052 | 0.006 | < 0.001 |
| 2DL5 | 291 | 66.4% | 264 | 63.8% | 255 | 57.2% | 0.414 | 0.005 | 0.048 |
| 2DS5 | 204 | 46.6% | 180 | 43.5% | 148 | 33.2% | 0.364 | < 0.001 | 0.002 |
| 2DS3 | 92 | 21.0% | 84 | 20.3% | 111 | 24.9% | 0.797 | 0.170 | 0.108 |
| 2DS1 | 117 | 26.7% | 126 | 30.4% | 88 | 19.7% | 0.229 | 0.014 | < 0.001 |
| 2DS4 | 424 | 96.8% | 392 | 94.7% | 438 | 98.2% | 0.125 | 0.181 | 0.059 |
Mid-P exact p-values for comparisons of KIR genes in Tororo vs Jinja, Tororo vs Kanungu and Jinja vs Kanungu
Table 5.
Comparative prevalence of KIR and HLA-C genetic variants from 3 sites of Uganda with historically varied malaria transmission intensity
| Study sites | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Tororo, N = 385 | Jinja, N = 392 | Kanungu, N = 433 | p-values | ||||||
| n | (%) | n | (%) | n | (%) | T vs J | T vs K | J vs K | |
| KIR genotypes | |||||||||
| AA | 150 | 39.0% | 133 | 33.9% | 171 | 39.5% | 0.145 | 0.877 | 0.098 |
| BX | 235 | 61.0% | 259 | 66.1% | 262 | 60.5% | |||
| Tororo, N = 356 | Jinja, N = 168 | Kanungu, N = 405 | p-values | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | T vs J | T vs K | J vs K | |
| HLA-C | |||||||||
| C1C1 | 66 | 18.5% | 39 | 23.2% | 75 | 18.5% | 0.285 | 0.296 | 0.043 |
| C1C2 | 195 | 54.8% | 93 | 55.4% | 202 | 49.9% | |||
| C2C2 | 95 | 26.7% | 36 | 21.4% | 128 | 31.6% | |||
| Tororo, N = 120 | Jinja, N = 59 | Kanungu, N = 161 | p-values | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | T vs J | T vs K | J vs K | |
| KIRAA/HLA-C | |||||||||
| AA/C1C1 | 21 | 17.5% | 11 | 18.6% | 22 | 13.7% | 0.213 | 0.537 | 0.282 |
| AA/C1C2 | 62 | 51.7% | 37 | 62.8% | 93 | 57.8% | |||
| AA/C2C2 | 37 | 30.8% | 11 | 18.6% | 46 | 28.5% | |||
| Tororo, N = 193 | Jinja, N = 93 | Kanungu, N = 232 | p-values | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | T vs J | T vs K | J vs K | |
| KIRBX/HLA-C | |||||||||
| BX/C1C1 | 36 | 18.6% | 23 | 24.7% | 50 | 21.5% | 0.424 | 0.062 | 0.424 |
| BX/C1C2 | 109 | 56.5% | 46 | 49.5% | 105 | 45.3% | |||
| BX/C2C2 | 48 | 24.9% | 24 | 25.8% | 77 | 33.2% | |||
| Tororo, N = 438 | Jinja, N = 414 | Kanungu, N = 446 | p-values | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | T vs J | T vs K | J vs K | |
| Centromeric KIR motif | |||||||||
| CenAA | 191 | 43.6% | 171 | 41.3% | 190 | 42.6% | 0.478 | 0.552 | 0.928 |
| CenAB | 175 | 40.0% | 182 | 44.0% | 192 | 43.0% | |||
| CenBB | 72 | 16.4% | 61 | 14.7% | 64 | 14.4% | |||
| Tororo, N = 438 | Jinja, N = 413 | Kanungu, N = 446 | p-values | ||||||
|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | n | (%) | T vs J | T vs K | J vs K | |
| Telomeric KIR Motif | |||||||||
| TelAA | 314 | 71.7% | 283 | 68.5% | 326 | 73.1% | 0.141 | 0.088 | 0.061 |
| TelAB | 115 | 26.3% | 112 | 27.1% | 113 | 25.3% | |||
| TelBB | 9 | 2.0% | 18 | 4.4% | 7 | 1.6% | |||
p-values for comparisons of the prevalence of KIR AA vs BX genotypes, HLA (C1C1) vs C1C2 vs C2C2, KIR/HLA (AA/C1C1) vs AA/C1C2 vs AA/C2C2, KIR/HLA (BX/C1C1) vs BX/C1C2 vs BX/C2C2, centromeric (CenAA) vs CenAB vs CenBB, telomeric (TelAA) vs TelAB vs TelBB in Tororo (T), Jinja (J) and Kanungu (K) districts were determined using Fisher’s exact test.
Copy number variation in KIR genes and malaria transmission intensity
Determination of whether CNV in KIR genes is influenced by malaria transmission intensity was done by comparing KIR CNV in children from the 3 populations. Comparisons were done for inhibitory KIR2DL1, 2DL2, 2DL3, and 2DL5, and the activating KIR2DS2 and 2DS5. All the KIR genes, including framework genes, were subject to CNV. The majority of study participants (over 90%) had 0–2 copies. However, there was no significant difference in KIR CNV across the 3 study populations (Table 6), suggesting that CNV in KIR genes may not be influenced by P. falciparum pathogen pressure.
Table 6.
Comparison of CNV in KIR genes in populations with historically varied malaria transmission intensity
| Tororo | Jinja | Kanungu | p-value | ||||
|---|---|---|---|---|---|---|---|
| N = 438 | F (%) | N = 414 | F (%) | N = 446 | F (%) | ||
| KIR2DL1 CNV | |||||||
| 0 | 6 | 1.4 | 5 | 1.2 | 5 | 1.2 | 0.985 |
| 1 | 104 | 23.7 | 99 | 23.9 | 101 | 22.6 | |
| 2 | 328 | 74.9 | 310 | 74.9 | 340 | 76.2 | |
| KIR2DL2 CNV | |||||||
| 0 | 184 | 42 | 174 | 42 | 192 | 43.1 | 0.834 |
| 1 | 220 | 50.2 | 199 | 48.1 | 216 | 48.4 | |
| 2 | 34 | 7.8 | 41 | 9.9 | 38 | 8.5 | |
| KIR2DL3 CNV | |||||||
| 0 | 72 | 16.4 | 62 | 15 | 66 | 14.8 | 0.195 |
| 1 | 154 | 35.2 | 171 | 41.3 | 191 | 42.8 | |
| 2 | 212 | 48.4 | 181 | 43.7 | 189 | 42.4 | |
| KIR2DS2 CNV | |||||||
| 0 | 214 | 48.8 | 196 | 47.3 | 218 | 48.9 | 0.387 |
| 1 | 207 | 47.3 | 189 | 45.7 | 204 | 45.7 | |
| 2 | 17 | 3.9 | 29 | 7 | 24 | 5.4 | |
| KIR2DS5 CNV | |||||||
| 0 | 336 | 76.7 | 324 | 78.3 | 372 | 83.4 | 0.144 |
| 1 | 98 | 22.3 | 86 | 20.7 | 71 | 16 | |
| 2 | 4 | 1 | 4 | 1 | 3 | 0.6 | |
| KIR2DL5 CNV | |||||||
| 0 | 292 | 66.8 | 282 | 68.2 | 318 | 71.4 | 0.650 |
| 1 | 138 | 31.4 | 126 | 30.5 | 121 | 27 | |
| 2 | 8 | 1.8 | 6 | 1.3 | 7 | 1.6 | |
CNV is copy number variation of KIR genes. The value can be 0, 1 or 2 in these populations, F is the frequency of participants with the different copies of KIR genes
Discussion
Most studies about genetic variation in KIR and HLA class I molecules and malaria have focused mainly on protection from severe malaria [33]. The aim of this study was to determine whether KIR and HLA-C genetic variants and CNV in KIR genes from 3 populations of Uganda with historically varied malaria transmission intensity have been shaped by selection pressure from falciparum malaria. Appreciation of malaria transmission prior to recent intensive control efforts and urbanization suggests a rank order for historical transmission intensity of Tororo > Jinja > Kanungu [31]. Thus, the measured prevalence of KIR genes and their HLA-C ligands in populations with historically varied malaria transmission was aimed at understanding impacts of malaria evolutionary pressure on KIR and HLA genes.
There was high KIR diversity in the 3 studied populations, as has been seen in previous studies in Uganda [20] and in other African populations [34]. Generally, the frequency of KIR3DS1 was low across the 3 populations, similar to that reported in previous studies from other African populations [35]. The frequency of KIR3DS1 was significantly lower in Kanungu compared to Tororo and Jinja, implying that KIR3DS1 could have been positively selected for in Tororo and Jinja to offer some advantage against malaria. The prevalence of KIR2DS5 and KIR2DL5 genes was significantly lower in Kanungu. Interestingly, results from a previous study in Nigeria demonstrated that KIR2DS5 and KIR2DL5 genes were associated with reduced parasitaemia [36]. The KIR3DS1, KIR2DL5, KIR2DS5 and KIR2DS1 genes can be present together on a particular haplotype in sub-Saharan Africans [37]. Differences in the prevalence of this haplotype across the three sites could potentially be explained by the selective pressure imposed by malaria. If so, the responsible gene or genes on the haplotype are not known, but KIR3DS1 has a low frequency and is present on few other haplotypes in Ugandans [38]. This gene is more prevalent in other populations, including Europeans [39], suggesting that it is selected against in Uganda or it evolved outside Africa [35]. The observed differences may be due, in part, to genetic differences between the ethnic groups principally inhabiting these populations. Indeed, in the previous study from these cohorts, it was observed that the populations of Tororo and Kanungu were homogeneous, based on language groups, but the Jinja population had ethnic groups from all over Uganda [40]. Although the specific ligands and expression details for KIR2DS3 and KIR2DS5 are yet to be defined, it is speculated that under functionally relevant combinations these activating genes, in conjunction with their putative ligands, may increase the threshold of NK cell activation and subsequent recruitment of other immune factors that mediate protection against malaria.
Although there was no significant difference in KIR/HLA-C combinations across the three sites in Uganda, it should be noted that, interactions between KIR and their HLA-C ligands within an individual play a key role in modulating the activity of NK cells [41]. For instance, the presence of particular HLA-C allotypes and inhibitory KIR2DL1, KIR2DL2 and KIR2DL3 genes determines the strength of NK cell inhibition during malaria infection [33]. The best characterized KIR-HLA ligand interactions are KIR2DL1 with the HLA-C2 subgroup and KIR2DL2/L3 with the HLA-C1 subgroup. Generally, KIR2DL1/HLA-C2 provides the strongest inhibition, followed by KIR2DL2/HLAC1, and KIR2DL3/HLA-C1 [42, 43]. HLA-C1/C1 individuals are only able to receive inhibitory signals via KIR2DL2 and KIR2DL3, whereas HLA-C2/C2 individuals receive inhibitory signals predominantly via KIR2DL1, and heterozygous individuals have the ligand for all three of these KIR genes [44]. Lower KIR inhibition may allow unrestrained NK cell activation that could contribute to immune-mediated pathology. This would be consistent with the theory that mechanisms that prevent malaria infection and those that prevent severe disease are distinct and may have a balancing effect on the maintenance of different KIR and their HLA ligands in malaria-endemic populations.
The role of KIR/HLA compound genotypes during falciparum malaria requires more attention given that malaria parasites spend most of the life cycle outside of HLA-expressing cells. Sporozoites infect hepatocytes after injection by mosquitoes. This is the only stage in the parasite replicative life cycle which is within an HLA-expressing host cell (39). Because erythrocyte membranes contain little to no HLA (42), it is postulated that the influence of KIR on cell-mediated anti-parasite immunity may occur primarily during the liver stage. This implies that cellular immune responses play an important role in restricting P. falciparum infection. During the blood stage, KIR-expressing effector cells may respond more strongly to an HLA-devoid cell due to the loss of inhibitory signalling via inhibitory KIR (43). KIR inhibition may also influence the clearance of parasites through antibody-dependent cellular cytotoxity (44, 45).
Previous studies have indicated that variation in KIR copy number, which leads to expression differences [14], may be important for susceptibility to some diseases. For example, CNV of KIR3DL1/S1 influences HIV control [45] and expression differences of KIR2DL3, interacting with HLA-C, may have a profound effect on resolution of hepatitis C virus infection [46]. However, there was no significant difference between KIR CNV across the three populations of Uganda.
Although different KIR and HLA variants may have been selected in different populations primarily due to differential risk of malaria, the role of other infectious pathogens that are prevalent in these malaria-endemic populations should not be overlooked, as they may also have exerted selective pressure on the evolution of KIR and HLA. Therefore, the role of other co-infections should be considered in future studies involving KIR and malaria, especially in populations affected by many infectious pathogens.
This study had some limitations. First, the genotyping technique for both KIR and HLA-C could not give detailed information up to the allele level. Second, other HLA class I genes, for instance HLA-B (e.g., HLA Bw4 and HLA Bw6 allotypes), which may play a role in malaria risk were not looked at. Nevertheless, analysis for HLA-C allotypes, which are the major ligands for KIR genes, was done. Third, the status of KIR genes and their HLA-C ligands remain unknown in a larger part of Uganda that was not covered in this study given its limited coverage. Despite these limitations, description of the genetic diversity of KIR and their HLA-C ligands in populations with historically varied malaria transmission intensity offered an opportunity to identify KIR and HLA-C genetic variants that are under positive selection and are potentially important in protection against malaria.
Conclusions
This study has provided baseline information about differences in the prevalence KIR genes and their HLA-C ligands in populations of Uganda with historically varied malaria transmission intensity. The KIR3DS1, KIR2DL5, KIR2DS5, and KIR2DS1 genes may partly explain differences in transmission intensity of malaria since these genes have been positively selected for in places with historically high malaria transmission intensity. This is the largest cohort ever studied investigating KIR, HLA-C and malaria risk. A new high-throughput real-time PCR assay for HLA-C genotyping has been developed from this study. This will be useful in disease association studies that involve larger cohorts.
Acknowledgements
We thank cohort study participants and their parents and guardians; the cohort study team; and the staffs of the Infectious Diseases Research Collaboration, the MUII-Plus Monitoring and Evaluation team and the Makerere/UVRI infection and immunity program for administrative and technical support.
Authors’ contributions
JIN, JR, EA, PJ, MRK, and GD directed the clinical study that provided samples for analysis. ST, SLN, AN, AM, SC, FC, JT and PJR conceived the study design. ST, JT, OC, AM, WJ and JJ designed and carried out the reported laboratory studies. ST, FM, GA and OC performed the data analysis. All authors contributed to the preparation of this manuscript and approval of its content. All authors read and approved the final manuscript.
Funding
This work was supported through the DELTAS Africa Initiative (Grant no. 107743), that funded Stephen Tukwasibwe through a PhD fellowship award, and Annettee Nakimuli through a group leader award. The DELTAS Africa Initiative is an independent funding scheme of the African Academy of Science (AAS), Alliance for Accelerating Excellence in Science in Africa (AESA), and supported by the New Partnership for Africa’s Development Planning and Coordinating Agency (NEPAD Agency) with funding from the Wellcome Trust (Grant no. 107743) and the UK government. Francesco Colucci is funded by Wellcome Trust grant 200841/Z/16/Z. The project received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grant agreement No. 695551) for John Trowsdale and James Traherne. Jyothi Jayaraman is a recipient of a fellowship from the Centre for Trophoblast Research. JIN is supported by the Fogarty International Center (Emerging Global Leader Award grant number K43TW010365). EA is supported by the Fogarty International Center of the National Institutes of Health under Award Number D43TW010526. This study was partly supported by funding from the National Institutes of Health (AI075045, AI089674, TW009343, and TW007375). The views expressed in this publication are those of the authors and not necessarily those of the funding bodies.
Availability of data and materials
The datasets utilized for this study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The cohort study that supplied samples for analysis and this specific study were approved by the Makerere University School of Medicine Research and Ethics Committee, the Uganda National Council for Science and Technology, the University of California, San Francisco Committee on Human Research, and the University of Cambridge, UK Committee on Human Research. Written informed consent was obtained from study participants.
Consent for publication
Not applicable.
Competing interests
OC had started in a role as an employee of AstraZeneca, UK, at the time of manuscript preparation. Other authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Stephen Tukwasibwe, Email: stephentukwasibwe@yahoo.com.
James A. Traherne, Email: jat51@cam.ac.uk
Olympe Chazara, Email: olympechazara@gmail.com.
Jyothi Jayaraman, Email: jj329@cam.ac.uk.
John Trowsdale, Email: jt233@cam.ac.uk.
Ashley Moffett, Email: am485@cam.ac.uk.
Wei Jiang, Email: wj224@cam.ac.uk.
Joaniter I. Nankabirwa, Email: jnankabirwa@idrc-uganda.org
John Rek, Email: jrek@idrc-uganda.org.
Emmanuel Arinaitwe, Email: earinaitwe@idrc-uganda.org.
Samuel L. Nsobya, Email: samnsobya@yahoo.co.uk
Maxine Atuheirwe, Email: maxinejasmine194@gmail.com.
Mubiru Frank, Email: mubiruf09@gmail.com.
Anguzu Godwin, Email: tukumbogodwin@gmail.com.
Prasanna Jagannathan, Email: prasj@stanford.edu.
Stephen Cose, Email: stephen.cose@lshtm.ac.uk.
Moses R. Kamya, Email: mkamya@infocom.co.ug
Grant Dorsey, Email: grant.dorsey@ucsf.edu.
Philip J. Rosenthal, Email: philip.rosenthal@ucsf.edu
Francesco Colucci, Email: fc287@medschl.cam.ac.uk.
Annettee Nakimuli, Email: annettee.nakimuli@gmail.com.
References
- 1.WHO . World Malaria Report 2019. Geneva: World Health Organization; 2019. [Google Scholar]
- 2.Weatherall DJ. Genetic variation and susceptibility to infection: the red cell and malaria. Br J Haematol. 2008;141:276–286. doi: 10.1111/j.1365-2141.2008.07085.x. [DOI] [PubMed] [Google Scholar]
- 3.Kwiatkowski DP. How malaria has affected the human genome and what human genetics can teach us about malaria. Am J Hum Genet. 2005;77:171–192. doi: 10.1086/432519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hedrick PW. Population genetics of malaria resistance in humans. Heredity. 2011;107:283–304. doi: 10.1038/hdy.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter R, Mendis KN. Evolutionary and historical aspects of the burden of malaria. Clin Microbiol Rev. 2002;15:564. doi: 10.1128/CMR.15.4.564-594.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López C, Saravia C, Gómez Camacho A, Hoebeke J, Patarroyo M. Mechanisms of genetically-based resistance to malaria. Gene. 2010;467:1–12. doi: 10.1016/j.gene.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Manjurano A, Clark TG, Nadjm B, Mtove G, Wangai H, Sepulveda N, et al. Candidate human genetic polymorphisms and severe malaria in a Tanzanian population. PLoS ONE. 2012;7:e47463. doi: 10.1371/journal.pone.0047463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toure O, Konate S, Sissoko S, Niangaly A, Barry A, Sall AH, et al. Candidate polymorphisms and severe malaria in a Malian population. PLoS ONE. 2012;7:e43987. doi: 10.1371/journal.pone.0043987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Band G, Le QS, Clarke GM, Kivinen K, Hubbart C, Jeffreys AE, et al. Insights into malaria susceptibility using genome-wide data on 17,000 individuals from Africa Asia and Oceania. Nat Commun. 2019;10:5732. doi: 10.1038/s41467-019-13480-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burrack KS, Hart GT, Hamilton SE. Contributions of natural killer cells to the immune response against Plasmodium. Malar J. 2019;18:321. doi: 10.1186/s12936-019-2953-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf A-S, Sherratt S, Riley EM. NK Cells: uncertain allies against malaria. Front Immunol. 2017;8:212. doi: 10.3389/fimmu.2017.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khakoo S, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 13.Hellmann I, Lim SY, Gelman RS, Letvin NL. Association of activating KIR copy number variation of NK cells with containment of SIV replication in rhesus monkeys. PLoS Pathog. 2011;7:e1002436. doi: 10.1371/journal.ppat.1002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Béziat V, Traherne JA, Liu LL, Jayaraman J, Enqvist M, Larsson S, et al. Influence of KIR gene copy number on natural killer cell education. Blood. 2013;121:4703–4707. doi: 10.1182/blood-2012-10-461442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doolan DL, Dobaño C, Baird JK. Acquired immunity to malaria. Clin Microbiol Rev. 2009;22:13. doi: 10.1128/CMR.00025-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tukwasibwe S, Nakimuli A, Traherne J, Chazara O, Jayaraman J, Trowsdale J, et al. Variations in killer-cell immunoglobulin-like receptor and human leukocyte antigen genes and immunity to malaria. Cell Mol Immunol. 2020;17:799–806. doi: 10.1038/s41423-020-0482-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kulkarni S, Martin MP, Carrington M. The Yin and Yang of HLA and KIR in human disease. Semin Immunol. 2008;20:343–352. doi: 10.1016/j.smim.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Radwan J, Babik W, Kaufman J, Lenz TL, Winternitz J. Advances in the Evolutionary Understanding of MHC Polymorphism. Trends Genet. 2020;36:298–311. doi: 10.1016/j.tig.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Meyer D, Aguiar CVR, Bitarello BD, Brandt CDY, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. doi: 10.1007/s00251-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakimuli A, Chazara O, Farrell L, Hiby SE, Tukwasibwe S, Knee O, et al. Killer cell immunoglobulin-like receptor (KIR) genes and their HLA-C ligands in a Ugandan population. Immunogenetics. 2013;65:765–775. doi: 10.1007/s00251-013-0724-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chazara O, Xiong S, Moffett A. Maternal KIR and fetal HLA-C: a fine balance. J Leukoc Biol. 2011;90:703–716. doi: 10.1189/jlb.0511227. [DOI] [PubMed] [Google Scholar]
- 22.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 23.Kamya MR, Arinaitwe E, Wanzira H, Katureebe A, Barusya C, Kigozi SP, et al. Malaria transmission, infection, and disease at three sites with varied transmission intensity in Uganda: implications for malaria control. Am J Trop Med Hyg. 2015;92:903–912. doi: 10.4269/ajtmh.14-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang W, Johnson C, Simecek N, López-Álvarez MR, Di D, Trowsdale J, et al. qKAT: a high-throughput qPCR method for KIR gene copy number and haplotype determination. Genome Med. 2016;8:99. doi: 10.1186/s13073-016-0358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiby SE, Walker JJ, O'Shaughnessy KM, Redman CW, Carrington M, Trowsdale J, et al. Combinations of maternal KIR and fetal HLA-C genes influence the risk of preeclampsia and reproductive success. J Exp Med. 2004;200:957–965. doi: 10.1084/jem.20041214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19:757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malmberg K-J, Michaëlsson J, Parham P, Ljunggren H-G. Killer cell immunoglobulin-like receptor workshop: Insights into evolution, genetics, function, and translation. Immunity. 2011;35:653–657. doi: 10.1016/j.immuni.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Jayaraman J, Kirgizova V, Di D, Johnson C, Jiang W, Traherne JA. KAT: quantitative semi-automated typing of killer-cell immunoglobulin-like receptor genes. J Vis Exp. 2019 doi: 10.3791/58646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rek J, Katrak S, Obasi H, Nayebare P, Katureebe A, Kakande E, et al. Characterizing microscopic and submicroscopic malaria parasitaemia at three sites with varied transmission intensity in Uganda. Malar J. 2016;15:470. doi: 10.1186/s12936-016-1519-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeka A, Gasasira A, Mpimbaza A, Achan J, Nankabirwa J, Nsobya S, et al. Malaria in Uganda: challenges to control on the long road to elimination: I. Epidemiology and current control efforts. Acta Trop. 2012;121:184–195. doi: 10.1016/j.actatropica.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katureebe A, Zinszer K, Arinaitwe E, Rek J, Kakande E, Charland K, et al. Measures of malaria burden after long-lasting insecticidal net distribution and indoor residual spraying at three sites in Uganda: a prospective observational study. PLoS Med. 2016;13:e1002167. doi: 10.1371/journal.pmed.1002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirayasu K, Ohashi J, Kashiwase K, Hananantachai H, Naka I, Ogawa A, et al. Significant association of KIR2DL3-HLA-C1 combination with cerebral malaria and implications for co-evolution of KIR and HLA. PLoS Pathog. 2012;8:e1002565. doi: 10.1371/journal.ppat.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, et al. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLoS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 36.Ademola S, Amodu O, Yindom L-M, Conway D, Aka P, Bakare A, et al. Killer-cell immunoglobulin-like receptors and falciparum malaria in southwest Nigeria. Hum Immunol. 2014;75:816–821. doi: 10.1016/j.humimm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Nemat-Gorgani N, Guethlein LA. Diversity of KIR, HLA Class I, and their interactions in seven populations of sub-Saharan Africans. J Immunol. 2019;202:2636–2647. doi: 10.4049/jimmunol.1801586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, et al. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci USA. 2015;112:845–850. doi: 10.1073/pnas.1413453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Körner C, Altfeld M. Role of KIR3DS1 in human diseases. Front Immunol. 2012;3:326. doi: 10.3389/fimmu.2012.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Walakira A, Tukwasibwe S, Kiggundu M, Verra F, Kakeeto P, Ruhamyankaka E, et al. Marked variation in prevalence of malaria-protective human genetic polymorphisms across Uganda. Infect Genet Evol. 2017;55:281–287. doi: 10.1016/j.meegid.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pende D, Falco M, Vitale M, Cantoni C, Vitale C, Munari E, et al. Killer Ig-like receptors (KIRs): their role in nk cell modulation and developments leading to their clinical exploitation. Front Immunol. 2019;10:1179. doi: 10.3389/fimmu.2019.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180:3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 43.Carrington M, Wang S, Martin MP, Gao X, Schiffman M, Cheng J, et al. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. doi: 10.1084/jem.20042158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by "group 2" or "group 1" NK clones. J Exp Med. 1995;182:605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, et al. Copy number variation of KIR genes influences HIV-1 control. PLoS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets utilized for this study are available from the corresponding author on reasonable request.