Abstract
Defects in the motor domain of kinesin family member 1A (KIF1A), a neuron-specific ATP-dependent anterograde axonal transporter of synaptic cargo, are well-recognized to cause a spectrum of neurological conditions, commonly known as KIF1A-associated neurological disorders (KAND). Here we report one mutation-negative female with classic Rett syndrome (RTT) harboring a de novo heterozygous novel variant [NP_001230937.1:p.(Asp248Glu)] in the highly-conserved motor domain of KIF1A. In addition, three individuals with severe neurodevelopmental disorder along with clinical features overlapping with KAND are also reported carrying de novo heterozygous novel [NP_001230937.1:p.(Cys92Arg) & p.(Pro305Leu)] or previously reported [NP_001230937.1:p.(Thr99Met)] variants in KIF1A. In silico tools predicted these variants to be likely pathogenic, and 3D molecular modelling predicted defective ATP hydrolysis and/or microtubule binding. Using the neurite tip accumulation assay, we demonstrated that all novel KIF1A variants significantly reduced the ability of the motor domain of KIF1A to accumulate along neurite lengths of differentiated SH-SY5Y cells. In vitro microtubule gliding assays showed significantly reduced velocities for the variant p.(Asp248Glu) and reduced microtubule binding for the p.(Cys92Arg) and p.(Pro305Leu) variants, suggesting decreased ability of KIF1A to move along microtubules. Thus, this study further expanded the phenotypic characteristics of KAND individuals with pathogenic variants in KIF1A motor domain to include clinical features commonly seen in RTT individuals.
Keywords: KAND, KIF1A, neurite tip accumulation, microtubule, kinesin, Rett syndrome, MECP2
1. INTRODUCTION
Pathogenic variants in KIF1A were initially associated with autosomal recessive hereditary sensory neuropathy type IIC (HSNIIC; MIM# 614213), autosomal recessive hereditary spastic paraplegia 30 (SPG30; MIM# 610357) and autosomal dominant non-syndromic intellectual disability 9 (MRD9; MIM# 614255) (Erlich et al., 2011; Esmaeeli Nieh et al., 2015; Hotchkiss et al., 2016; Klebe et al., 2012; Krenn et al., 2017; Riviere et al., 2011). HSNIIC-affected individuals have progressive degeneration of sensory neurons, resulting in loss of feeling in extremities, ulceration and ultimately amputation of fingers and toes (Riviere et al., 2011). SPG30 is a slowly progressive spastic paraplegia with unsteady gait, hyperreflexia of the lower limbs (Cheon et al., 2017; Yoshikawa et al., 2019) and the autosomal dominant form also features intellectual disability, speech delay, hypotonia, evolving into hyperreflexia and hypertonia with age. More severely affected individuals have acquired microcephaly, cerebellar atrophy, seizures, scoliosis, contractures, optic nerve atrophy, peripheral neuropathy, ataxia, strabismus, nystagmus, ptosis, facial diplegia and gastroesophageal reflux. In addition, some male patients have been found to have small testes/ penis or cryptorchidism (Boyle & Chung, 2017). More recently, pathogenic variations in KIF1A have been recognized to be responsible for a broad spectrum of neurological disorders, broadly termed KIF1A-associated neurological disorders (KAND), in which the severity of clinical symptoms largely depends upon the variation introduced and its location in KIF1A (Boyle & Chung, 2017). Thus, the phenotypic spectrum of KAND individuals is continuously expanding.
Rett syndrome (RTT; MIM# 312750) is a severe X-linked neurodevelopmental disorder that predominantly affects females and is notable for its progressive nature (Hagberg, Aicardi, Dias, & Ramos, 1983; Rett, 1966). As per the Neul revised diagnostic criteria, RTT patients can be classified broadly into two major categories; classic (or typical RTT) and variant (or atypical RTT) (Neul et al., 2010). Classic RTT patients have normal development until 6 −18 months of age, after which developmental regression occurs, along with the onset of characteristic abnormal hand wringing movements, a hallmark of classic RTT patients. Using the Neul classification, atypical RTT patients do not exhibit all four of the required criteria for classic RTT and must have at least 5 of the 11 supportive criteria. Moreover, there is a further group of RTT-like patients who exhibit some clinical features of RTT, but not enough in order to be classified as either classic or atypical RTT (Ip, Mellios, & Sur, 2018). Up to 97% of classic RTT and 86% of atypical patients have pathogenic variants in Methyl CpG Protein 2 (MECP2), and a proportion of individuals with clinical features that overlap with RTT may have variants in cyclin-dependent kinase-like 5 (CDKL5), or Forkhead box protein G1 (FOXG1) (Mitter et al., 2018; Neul et al., 2014; Schonewolf-Greulich et al., 2019). Due to considerable overlap of clinical symptoms of RTT patients, especially those who are MECP2-mutation negative, with other neurodevelopment disorders, it can be challenging to establish a precise genetic diagnosis. However, recent advances in next generation sequencing (NGS) have identified pathogenic variants in a growing list of genes known to cause intellectual disability, severe epilepsy and/or autistic behaviors where some individuals appear to have a RTT-like clinical picture, thus providing a definitive genetic diagnosis for patients and ‘closure’ for affected families (Cogliati et al., 2019; Henriksen, Ravn, Paus, von Tetzchner, & Skjeldal, 2018; Iwama et al., 2019; Schonewolf-Greulich et al., 2019; Vidal et al., 2019; Yoo et al., 2017).
There is clear evidence that the RTT-associated genes MECP2 and CDKL5 regulate microtubule stability and neuronal transport, and that pathogenic mutations in those genes disrupt microtubule dynamics (Barbiero, De Rosa, & Kilstrup-Nielsen, 2019; Delepine, Nectoux, Bahi-Buisson, Chelly, & Bienvenu, 2013; Gold, Lacina, Cantrill, & Christodoulou, 2015). Microtubules are highly dynamic structures that are critical for maintaining correct neuronal architecture and for the transport of cargos via motor proteins such as kinesins (KIFs) and dynein (Kevenaar & Hoogenraad, 2015). KIFs have been implicated in the pathogenesis of RTT suggesting an important role in this disorder (Gibson et al., 2010; Pelka et al., 2006). KIF’s are ATP-dependent molecular motors critical for cytoskeleton organization (Hirokawa, Noda, Tanaka, & Niwa, 2009; Stanhope & Ross, 2015) and in neurons, the kinesin-3 family member KIF1A is involved in the anterograde transport of synaptic vesicle precursors containing brain-derived neurotrophic factor (BDNF; MIM# 113505) (Lo, Kuzmin, Unger, Petersen, & Silverman, 2011; Stucchi et al., 2018). BDNF is critical for neuronal differentiation and maintaining synaptic plasticity (Ben Zeev et al., 2009; Li, Calfa, Larimore, & Pozzo-Miller, 2012; Zhou et al., 2006). Interestingly, environmental enrichment in the Kif1a+/− mouse induces BDNF-dependent upregulation of kinesin family member 1A (KIF1A; MIM# 601255) and its cargo synaptophysin in the hippocampus, suggesting that BDNF is both a cargo and an upstream regulator of KIF1A (Kondo, Takei, & Hirokawa, 2012). KIF1A is also involved in axonal transport of cargo containing synaptophysin, synaptotagmin and Ras-associated protein Rab-3A (RAB3A; MIM# 179490). The N-terminal highly conserved motor domain of KIF1A has ATP hydrolysis functionality and microtubule binding capacity, which are critical for KIF1A-driven trafficking of specific cargo bound to the C-terminal Pleckstrin-homology (PH) tail domain. KIF1A dimerizes via its neck coil region (Figure 1) (Klopfenstein, Tomishige, Stuurman, & Vale, 2002; Soppina et al., 2014; Tomishige, Klopfenstein, & Vale, 2002).
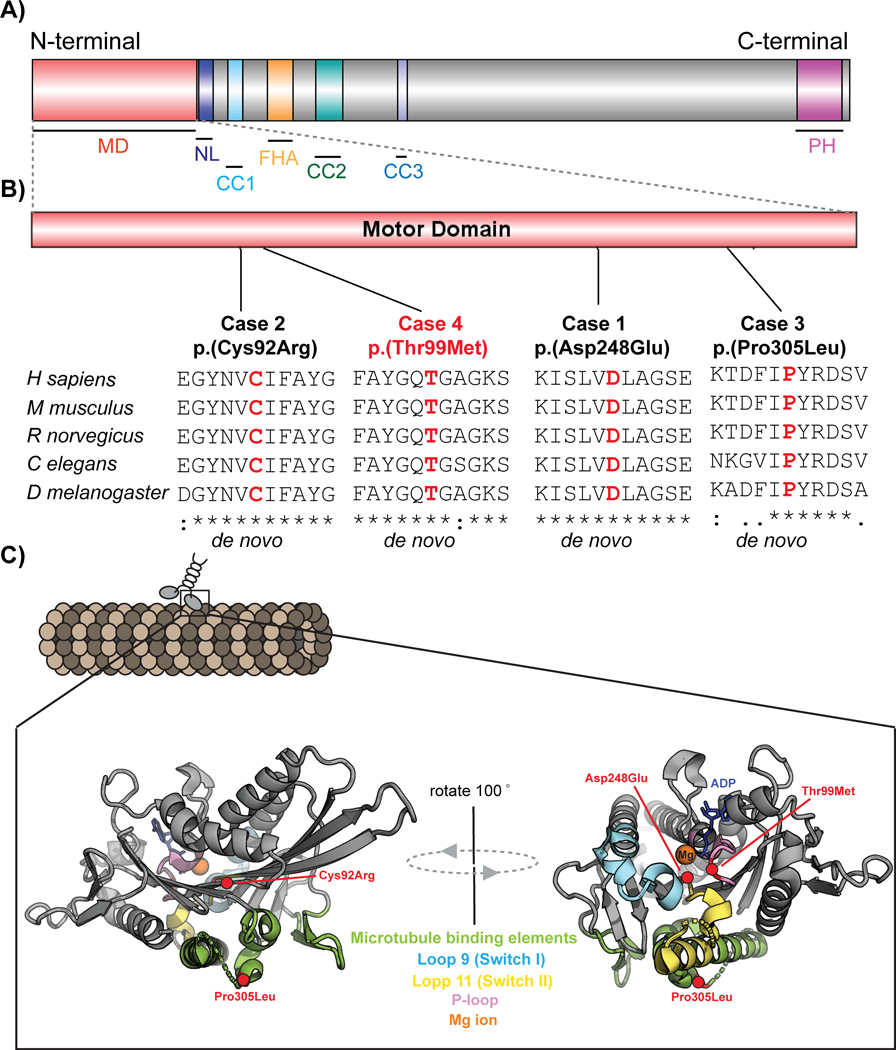
Figure 1: Schematic representing KIF1A domain structure, evolutionary conservation at variant site and 3D molecular modelling.
A) Domain structure of full length KIF1A containing N-terminal motor domain (MD: 1–361 aa), neck coil (NC: 365–397 aa), coiled coil domains [(CC1: 429–462 aa), (CC2; 622–681 aa) and (CC3: 801–822 aa)], forkhead-associated domain (FH: 516–572 aa), and pleckstrin homology domain (PH: 1676–1774 aa)]. B) Representation showing location of four patient variants in four reported cases in motor domain of KIF1A. The variants NP_001230937.1:p.(Asp248Glu), NP_001230937.1:p.(Cys92Arg) and NP_001230937.1:p.(Pro305Leu) are novel (in black), whereas NP_001230937.1:p.(Thr99Met) is previously reported (in red). Multiple sequence alignment underneath each variant shows the sequence conservation at each variant site. Sequences were aligned using Clustal Omega (URL: https://www.ebi.ac.uk/Tools/msa/clustalo/) using the RefSeqs of H sapiens (NP_001230937.1), M musculus (NP_032466.2), R norvegicus (XP_017452404.1), C elegans (NP_001022041.2) and D melanogaster (NP_001246373.1). C) Schematic structures representing location of variants on crystal structure of KIF1A motor domain before Mg ion release (PDB: 2ZFI) using PyMOL software. The P-loop, switch I and switch II, P-loop are indicated in light magenta, aquamarine and yellow-orange respectively and rest of motor domain is represented in grey. The ligand Adenosine-5’-diphosphate (ADP) is shown by a stick model in blue and magnesium ion is represented as orange sphere. The residue affected in reported cases are represented as red circles and red labels.
A subset of KIF1A patients share some clinical features with RTT including gait abnormalities, hypotonia, scoliosis, seizures and intellectual disability. Recently a de novo heterozygous truncating variant [NM_001244008.1: c.275_276insAA; NP_001230937.1: p.(Cys92*)] in the motor domain of KIF1A has been reported in a single female case presenting with classic RTT including developmental delay, microcephaly, lack of independent ambulation and speech, loss of hand skills, hand clapping and mouthing, bruxism, breathing and sleep disturbances (Wang et al., 2019). Interestingly, the well-conserved KIF1A motor domain has been identified as a mutation hotspot (with over 80 reported variants) resulting in a spectrum of phenotypes that overlap with other neurological disorders including RTT. In this study, we report one classic RTT and three individuals with a severe neurodevelopmental disorder carrying variants in motor domain of KIF1A. We have confirmed the pathogenicity of the variants using a range of in silico analyses and in vitro functional studies.
2. METHODS
2.1. Subjects and genetic analysis
All methods followed in this study were approved by the human research ethics committee of the participating institutes with written consents obtained from the legal guardians of each participant. The affected participants with variants in the motor domain of KIF1A were recruited from Australia (case 1), Italy (case 2 and 3) and USA (case 4; deceased). Next generation sequencing (NGS) was performed on genomic DNA isolated from patient blood, and variant filtering was performed as described in the Supplementary Material. The segregation of identified KIF1A variants was also confirmed by Sanger sequencing of the proband and the parents.
2.2. In silico analysis, variant classification and structural modelling
The pathogenicity of KIF1A variants, denoted as per RefSeq NM_001244008.1, was determined using in silico prediction tools including PolyPhen-2 (v2.2.2r398; http://genetics.bwh.harvard.edu/pph2/) (Adzhubei et al., 2010), SIFT (v6.2.1; http://sift.jcvi.org/) (Kumar, Henikoff, & Ng, 2009) and MutationTaster (v2.0; www.mutationtaster.org/) (Schwarz, Cooper, Schuelke, & Seelow, 2014). The population frequency was determined using the Genome Aggregation Database (gnomAD) (v2.1.1;http://gnomad.broadinstitute.org/) (Karczewski et al., 2019). All variants were classified according to the guidelines of the American College of Medical Genetics and Genomics (ACMG) (Richards et al., 2015). Amino acid substitutions were assigned a Grantham score (range 0 to 215), with values above 100 generally considered significant (Grantham, 1974). The presence of all these variants was checked in the ClinVar public archive of human genetic variants (https://www.ncbi.nlm.nih.gov/clinvar/; accessed on 16 September 2019) (Landrum et al., 2018).
Since the full length human KIF1A crystal structure is yet to be resolved, the motor domain of Mus musculus KIF1A (PDB ID: 2ZFI) was used for in silico modelling using HOPE (URL: http://www.cmbi.ru.nl/hope/) (Venselaar, Te Beek, Kuipers, Hekkelman, & Vriend, 2010). The position of residue affected in reported cases is shown on 3D structure images generated using PyMOL (http://www.pymol.org/) molecular visualization software (Schrödinger, 2010). A BLAST search showed 99% identity of the Mus musculus KIF1A motor domain (NP_032466.2) and the motor domain of human KIF1A (NP_001230937.1) (Supplementary Material).
2.3. Constructs and mutagenesis
We used a constitutively-active dimeric KIF1A motor (amino acids 1 – 393) containing the motor (amino acid 1—361), neck linker, and neck coil domains and dimerized via a GCN4 leucine zipper (LZ) (Hammond et al., 2009). KIF1A(1–393)-LZ was tagged at the C-terminus with three tandem mCitrine (mCit) fluorescent proteins [KIF1A(1–393)-3xmCit] for the neurite tip accumulation assay and with mNeonGreen (mNG; Allele Biotech) and AviTag (amino acid sequence GLNDIFEAQKIEWHE) [KIF1A(1–393)-LZ-mNG-AviTag] for the microtubule gliding assay. KIF1A(1–393)-LZ-mNG-AviTag was biotinylated by co-expression with HA-tag labeled bacterial biotin ligase BirA as described (Yue et al., 2018).
Site-directed mutagenesis was used to introduce patient-specific variants into the KIF1A constructs (QuikChange lightning mutagenesis kit, Agilent technologies) according to the manufacturer’s instructions and using their software for mutagenesis primer design (QuikChange Primer Design, www.agilent.com/genomics/qcpd). Primers are listed in Supp. Table S1.
2.4. Cell culture, transfection, immunostaining and fluorescence microscopy
COS-7 (African green monkey kidney fibroblast, American Type Culture Collection) cells were grown in DMEM media (DMEM with 3.7g/L NaHCO3, 10% (vol/vol) fetal bovine serum (FBS), 1X Pen-Strep) at 37°C in a humidified incubator and 5% CO2. SH-SY5Y (human neuroblastoma) cells were grown in RPMI media (RPMI with 3.7g/L NaHCO3, 10% (vol/vol) fetal bovine serum (FBS), 1X Pen-Strep) at 37 °C in a humidified incubator and 5% CO2.
For the neurite tip accumulation assay, 50,000 SH-SY5Y cells were plated on sterile glass coverslips coated with rat collagen (3.3%) and matrigel (2%) (recipe optimized by Dr. Wendy Gold) in RPMI media without serum and antibiotics in a 12 well plate. The following day, cells were transfected with 1μg of KIF1A(1–393)-3xmCit DNA using 5μl of Lipofectamine 2000 (Life Technologies; 11668019) according to the manufacturer’s instructions. After 24 hours the media was refreshed to RPMI with 1% serum supplemented with 10μM retinoic acid (RA; Sigma; R2625) to induce neuronal differentiation and the media was refreshed daily for 4 days. The expression of KIF1A motor protein in SH-SY5Y cells did not appear to cause any qualitative alteration in neurite outgrowth during RA-induced differentiation (data not shown). On day 3, cells were washed with 1X PBS, fixed in 4% (vol/vol) paraformaldehyde (PFA) in PBS for 20 minutes at room temperature before mounting on glass slides using Prolong Gold with DAPI (Life Technologies). Images were acquired using Zeiss AxioVision fluorescence microscope with 40X objective (zoom factor =2). Quantification of the mean florescence intensity (MFI) along the length of the neurites and within the cell bodies of SH-SY5Y cells was measured manually using ImageJ. Mann Whitney test (n=20) was performed on the ratio of average MFI at the neurite length versus the cell body using Prism software (GraphPad). The length of the neurites was calculated using Simple Neurite Tracer plugin in ImageJ and statistical analysis was performed using Unpaired t-test. All experiments were performed in triplicate and repeated three times prior to data analysis.
2.5. In-vitro microtubule gliding assay
Cell lysates containing biotinylated KIF1A motor proteins were prepared by co-expressing wild type or variant KIF1A(1–393)-LZ-mNG-AviTag constructs together with HA-BirA in COS-7 cells as previously described (Yue et al., 2018). Briefly, COS-7 cells were plated at a density of 200,000 per well in a 6-well plate and transfected with 1 μg of KIF1A(1–393)-LZ-mNG-AviTag and 1 μg HA-BirA DNA using 5 μl of Lipofectamine 2000. Cells were washed, trypsinized and harvested (3000g) at 4⁰C 16 hours after transfection. The pellet was washed with PBS and resuspended in ice cold lysis buffer (25 mM HEPES/KOH, pH 7.4, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, and 1% [vol/vol] Triton X-100) supplemented with 1 mM ATP, 1 mM phenylmethylsulphonyl fluoride (PMSF; Sigma; 10837091001), and protease inhibitor cocktail (Thermo Fisher Scientific; A32965) and incubated at 4⁰C for 30 minutes. The soluble fraction was collected after centrifugation (20,000g) for 15 mins at 4⁰C, snap frozen in single use aliquots, and stored at −80⁰C. Biotinylation of KIF1A(1–393)-LZ-mNG-AviTag was confirmed by staining transfected cells with Alexa Fluor® 555 streptavidin (S21381; Thermo Fisher Scientific) and determining coincident staining with green (mNG) fluorescent KIF1A(1–393).
To polymerize microtubules, lyophilized tubulin (Cytoskeleton Inc (USA); T240-A) and rhodamine-labeled tubulin (TL590M-A) were reconstituted to a final concentration of 10 mg/ml using ice-cold general tubulin buffer (80mM PIPES, 2mM MgCl2, 2.5mM EGTA pH 7.0) supplemented with fresh 1mM guanosine triphosphate (GTP; Cytoskeleton; BST06), snap frozen in single use aliquots, and stored at −80⁰C. Unlabeled tubulin and labeled tubulin were mixed together in the molar ratio of 10:1, respectively, and incubated at 37⁰C for 30 minutes to induce tubulin polymerization. To maintain the polymerized state, paclitaxel (Cytoskeleton Inc; TXD01) was then added stepwise in increasing concentrations (2μM, 20μM and 200μM) at room temperature with incubation for 10 minutes at each step. The polymerized microtubules were then pelleted through a 40% glycerol cushion at 300,000g for 10 minutes at 25⁰C. The pellet was then resuspended using warm general tubulin buffer supplemented with 5% glycerol, 1mM GTP and 20μM paclitaxel.
A 10 μl flow chamber was prepared by attaching two stripes of double-sided tape spaced 5 mm apart to a glass slide and mounting a clean #1.5 coverslip (Thermo Fisher Scientific) over the top. Biotinylated KIF1A proteins were immobilized on the attached coverslip by sequential addition of different solutions for 5 minutes each (Yue et al., 2018). Briefly, the flow chamber was first incubated with 1 mg/ml BSA-biotin (Sigma; A8549) followed by blocking solution containing 0.5mg/ml casein (Sigma; C7078) in BRB80 buffer (80mM PIPES, 1mM MgCl2 and 1mM EGTA) supplemented with 10μM paclitaxel. NeutrAvidin (0.5 mg/ml; Thermo Fischer Scientific; 31000) was then added to the chamber followed by blocking solution before adding 30 μl of COS-7 cell lysates supplemented with 2mM ATP. Then 10μl of motility mix containing taxol-stabilized rhodamine-labeled microtubules, 2mM ATP, 2mM MgCl2 and enzymatic oxygen scavenging system (10mM glucose, 200μg/ml glucose oxidase, 1mM dithiothreitol, 100 μg/ml catalase), to avoid microtubule breaking due to oxidation, was added into the chamber. Finally, the ends of the flow cell were sealed with varnish and microtubule movement was captured by Leica DMIB6000 total internal reflection microscopy (TIRF) microscope fitted with a 100X U PlaApo 1.49 NA oil immersion objective and solid state 488, 561 nm laser line) at room temperature. Signals were collected using EMCCD camera (Andor iXon 897 Ultra). Images were acquired continuously at 50 msec per frame for 4000 frames. Kymographs (width = 3 pixels) were generated by using the multi-kymograph plugin in ImageJ (National Institute of Health) and manually drawing tracks generated from maximum intensity projections as previously published (Yue et al., 2018). The velocity (μm/sec) of microtubule movement was calculated by dividing distance (x-axis) to time (y-axis). Measurements were taken from three independent cell lysate preparations. Plots and statistical analysis using ANOVA and Mann-Whitney test was carried out using Prism software (GraphPad). All data are presented as mean ± standard deviation (SD).
3. RESULTS
In this study we present four unrelated individuals harboring different de novo heterozygous missense variants restricted to the motor domain of KIF1A. For these four cases, the clinical features are captured in Table 1.
Table 1:
Evaluation of KIF1A associated features in our cases.
| Category | Case 1 | Case 2 | Case 3 | Case 4 (deceased) |
|---|---|---|---|---|
| Neurological features | ||||
| Hypotonia | No | Yes (Severe) | Yes (first 2 years of life) | Yes (since birth) |
| Hypertonia | Yes (lower limbs) | No | Yes (since birth) | |
| Microcephaly | No | Yes | Yes | No |
| Global developmental delay | Yes | Yes | Yes† | Yes |
| Intellectual disability | Severe | Yes | Severe‡ | Yes |
| Ataxia | Yes | Yes | No | NA |
| Spastic paraplegia | Yes (lower limbs) | Yes | No | NA |
| Peripheral neuropathy | No | Not evaluated | No | Yes |
| Nerve conduction studies | Not performed | Not performed | No | Yes; EMG at 10m$ |
| Seizures | Yes (past history) | Yes | No¶ | Yes |
| Neuromuscular features | ||||
| Upper limb spasticity | No | No | No | NA |
| lower limb spasticity | Yes (Progressive) | Yes | No | NA |
| Muscle hypertrophy | No | No | No | No |
| Muscle weakness | No | Yes | Yes | Yes |
| Gait abnormalities | Yes | Yes | Yes | Yes (non-ambulatory) |
| Ophthalmological abnormalities | ||||
| Cataracts | No | No | No | Yes |
| Cortical visual impairment | No | Yes | No | Yes |
| Optic nerve abnormalities | Yes (mild optic nerve atrophy) | Yes | No | Yes |
| Ptosis | No | No | No | No |
| Strabismus | No | Yes | No | No |
| Other features | ||||
| Gastroesophageal reflux disease | No | No | No | Yes |
| Eczema | No | Skin xerosis on hands | No | No |
| Genital abnormalities | No | No | No | No |
| MRI changes | ||||
| Year (Age) when test was performed | 2002 (2 year) |
2013 (4 year) |
2016 (3 year) |
7 month |
| Cerebral atrophy | Yes | Yes | Yes! | No |
| Cerebellar atrophy | No | Yes | Yes | No |
| Reduced cerebral white matter | No | Yes | No | Reduced white matter: unclear if cerebellar |
| Thin or absent corpus callosum | No | Yes | No | No |
| Cerebellar abnormalities | No | Yes | No | No |
| Other abnormalities | small pineal cyst, tortuosity of the vessels (internal cerebral veins) |
Yes | No | No |
: treated with growth hormone;
: global development score 45 (scale Bayley III), language 42, gross motor and fine 43;
: slow paroxysmal bilateral posterior activity in sleep;
periventricular white matter (temporal and occipital) alterations of signal;
: EMG at 10m showed ‘anomaly in axon’ and as per the mother of case 4: muscle biopsy at 14m showed “light axonal neuropathy with mitochondrial anomalies in the nerve”
3.1. Variant curation
The four missense variants identified in KIF1A are described in Table 2. All the variants were confirmed to be de novo heterozygous (Supp. Figure S1) and are located at evolutionarily-conserved sites in the motor domain of KIF1A (Figure 1 and Supp. Figure S2), which is a mutation hot-spot region in KIF1A-related disorders (Gabrych, Lau, Niwa, & Silverman, 2019). Several in silico tools (MutationTaster, PolyPhen-2 and SIFT) predicted these variants to be disease causing and/or damaging for protein function. Using the Grantham scoring system, the variant c.274T>C; p.(Cys92Arg) was predicted to harbor a major amino acid change with scores of 180, whereas the amino acid changes in c.744C>A; p.(Asp248Glu), c.914C>T; p.(Pro305Leu) and c.296C>T; p.(Thr99Met) were predicted to be minor due to their low Grantham scores of 45, 98 and 81 respectively (Table 2).
Table 2:
Variant analysis for our cases
| Category | Case 1 | Case 2 | Case 3 | Case 4 (deceased) | |
|---|---|---|---|---|---|
| Genetic screening method | trio; WGS |
trio; WES |
trio; targeted sequencing |
Singleton; targeted sequencing |
|
| Variant | chr2:241723210G>T; c.744C>A; p.(Asp248Glu) |
chr2:241727557A>G; c.274T>C; p.(Cys92Arg) |
chr2:241715312G>A; c.914C>T; p.(Pro305Leu) |
chr2:241727535G>A; c.296C>T; (Thr99Met) |
|
| Zygosity | Heterozygous | Heterozygous | Heterozygous | Heterozygous | |
| Segregation | de novo | de novo | de novo | de novo | |
| Conserved site | Yes | Yes | Yes | Yes | |
| Novel or reported | Novel | Novel | Reported | Reported | |
| Affected exon | Exon 8 | Exon 4 | Exon 11 | Exon 4 | |
| Amino acid change (Grantham distance) | Minor (score: 45) | Major (score: 180) | Minor (score: 98) | Minor (score: 81) | |
| population database (gnomAD) | No | No | No | No | |
| Location of variant | Motor domain | Motor domain | Motor domain | Motor domain | |
| in silico tools | Mutation Taster# | Disease causing (p=0.999) | Disease causing (0.999) | Disease causing (0.999) | Disease causing (0.999) |
| PolyPhen-2## | Probably damaging (p=0.998) | Probably damaging (p=1.0) | Probably damaging (p=1.0) | Probably damaging (p=1.0) | |
| SIFT### | Damaging (score = 0.001) | Damaging (score = 0.000) | Damaging (score = 0.001) | Damaging (score = 0.000) | |
| ClinVar | No | No | Yes | Yes | |
| ACMG classification | Likely pathogenic | Likely pathogenic | Likely pathogenic | Likely pathogenic | |
RefSeq ID: NM_001244008.1 (longest isoform): Position: GRCh37 chr2:241,653,181–241,759,725, size: 106,545 and total exon count: 49; NCBI Protein ID: NP_001230937.1 (longest isoform); Ensembl transcript ID: ENST00000320389. WGS = whole genome sequencing, WES = whole exome sequencing, ACMG= American College of Medical Genetics, gnomAD = Genome Aggregation Database.
: v2.0. URL = www.mutationtaster.org/
: v2.2.2r398. URL = http://genetics.bwh.harvard.edu/pph2/. Score range: 0.0 (tolerated) to 1.0 (deleterious)
: v6.2.1. URL = http://sift.jcvi.org/. Score range: 0.0 (deleterious) to 1.0 (benign). Prediction cut-off = 0.05.
All variants were absent in gnomAD (accessed on 16 September 2019) and classified as likely pathogenic using ACMG guidelines. Of the four identified variants, two variants [p.(Asp248Glu) and p.(Cys92Arg)] were absent from ClinVar. For the p.(Pro305Leu) variant, three cases with developmental delay, cerebellar atrophy, ataxia, and eye movement abnormalities were reported in ClinVar. The p.(Thr99Met) is a common recurrent variant and has been reported as a de novo heterozygous variant in six unrelated individuals with complex neurological phenotype characterized by moderate to severe development delay and/or intellectual disability, spastic paraparesis, cerebellar atrophy, axonal neuropathy, visual disturbances and/or progressive neurodegeneration (Esmaeeli Nieh et al., 2015; Hamdan et al., 2011; Lee et al., 2015; Okamoto et al., 2014). Moreover, a recent report also found this variant in a female diagnosed with Progressive encephalopathy with Edema, Hypsarrhythmia and Optic atrophy syndrome (PEHO; MIM# 260565) (Langlois et al., 2016).
3.2. Patient variants are located in sequences critical for kinesin enzymatic activity
All four de novo variants are located within the kinesin motor domain of KIF1A which harbors the sequences required for ATP-dependent microtubule-based transport. ATP hydrolysis in the motor domain facilitates a step-wise movement of KIF1A along the microtubule network (Atherton et al., 2014; Kikkawa & Hirokawa, 2006; Kikkawa et al., 2001). Three motifs of the kinesin motor domain are involved in coordinating ATP binding and hydrolysis: the phosphate-binding loop (P-loop) with a Walker A fold (GXXXXGK[T/S]), Switch I (loop L9), and Switch II (loop L11) (Kull & Endow, 2013). The variant in case 1 [p.(Asp248Glu)] is present in Switch II and thus, it is predicted to affect nucleotide binding. The wild type (Asp) amino acid coordinates the metal ion (Mg2+) critical for ATP binding, and the increased bulk of the side chain of the introduced amino acid (Glu) may preclude effective nucleotide binding. This is predicted to affect the velocity with which the variant KIF1A would move along the microtubules (Figure 1).
The variant in case 2 [p.(Cys92Arg)] is located in β3 immediately preceding the P-loop. The substitution of the neutral wild-type amino acid (Cys), which is buried in the protein core, with the larger and positively charged variant residue (Arg) is predicted to interfere with KIF1A protein folding and therefore with nucleotide or microtubule binding or both.
Three motifs of the kinesin motor domain have been implicated in microtubule binding: β5-L8, L11-β7, and α4-L12-α5. The variant in case 3 [p.(Pro305Leu)] alters a motif in loop L12 that is evolutionarily-conserved across kinesins for microtubule binding (FIPYRD →FILYRD) (Figure 1). We thus predicted that this variant motor would be ineffective at microtubule binding and motility. The variant in cases 4 [p.(Thr99Met)] affects wild type (Thr) amino acid in the P-loop (nucleotide binding pocket) and hence is predicted to affect the ATP binding to motor domain, thereby altering ATP hydrolysis rate. Structural models published in previous reports suggest that this variant specifically affects interaction between KIF1A motor domain and the γ -phosphate of ATP (Lee et al., 2015).
3.2.1. Patient variants result in defective motility of KIF1A in neuronal SH-SY5Y cells
To examine the effect of patient variants on the efficiency of KIF1A’s microtubule-based motility, we utilized the neurite tip accumulation assay. In this assay, kinesin motors that are capable of processive microtubule-based motility accumulate at the tips of neurites in differentiated neuronal cells (Yang, Bentley, Huang, & Banker, 2016). For this assay, we used a truncated and constitutively active version of KIF1A containing amino acids 1–393 tagged with the fluorescent protein monomeric Citrine (mCit) and we expressed wild-type and mutant versions in differentiated neuronal SH-SY5Y cells.
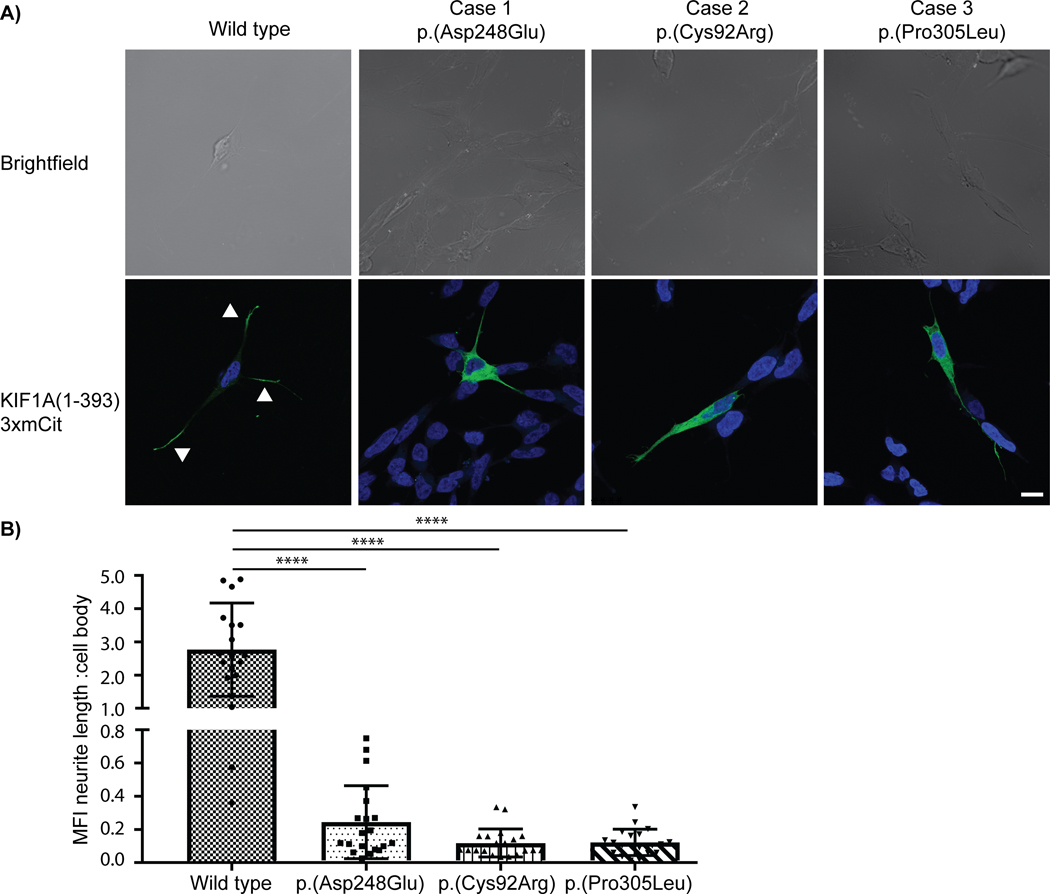
Consistent with previous work (Hammond et al., 2009; Huang & Banker, 2012), wild-type KIF1A(1–393) accumulated at the end of the growing neurites in differentiated SH-SY5Y cells, indicative of directed movement of KIF1A protein towards the plus ends of the microtubules. However, the KIF1A variant [p.(Asp248Glu), p.Cys92Arg) and p.(Pro305Leu)] proteins did not preferentially accumulate at neurite tips, rather, a significant fraction of the expressed mutant protein was found to be localized in the cell body. The mean fluorescence intensity (MFI) ratio of motor fluorescence in the neurite length compared to the cell body was significantly higher for the wild-type motor protein (2.8±1.4) compared to the patient variants [p.(Asp248Glu) 0.244±0.219; p<0.0001], [p.Cys92Arg) 0.118±0.084; p<0.0001] and [p.(Pro305Leu) 0.122±0.079; p<0.0001] (Figure 2; Supp. Figure S3). However, no significant difference was observed in neurite length of SH-SY5Y cells transfected with wild-type or patient specific KIF1A(1–393) variant construct (Supp. Figure S4). Previously published reports have already shown the reduced accumulation of KIF1A motor domain harboring the case 4 variant p.(Thr99Met) in the distal neurites of rat hippocampal neurons (Hamdan et al., 2011; Lee et al., 2015) These results demonstrate that the variants in KIF1A found in patients reported in this study have a severe negative impact on the capacity of KIF1A to undergo processive, long-distance transport along the cellular microtubule network.
Figure 2: Neurite tip accumulation in differentiated SH-SY5Y cells.
A) Representative images showing the localization of rat KIF1A(1–393)-3xmCit motors (green) in differentiated SH-SY5Y cells (Nucleus/ DAPI = blue). Whereas the wild-type motor accumulated in neurite tips (white arrows), the variant proteins remained near the cell body. B) Quantification of the mean florescence intensity (MFI) at the neurite length versus the cell body for the wild-type and three pathogenic KIF1A patient variants. Data are from three independent experiments (n=20 cells for each experiment) and are plotted as mean ± standard deviation. **** represents p<0.0001, Mann-Whitney test. Scale bar (white line, bottom right) = 10μm.
3.2.2. In-vitro microtubule gliding assay reveals impaired microtubule binding
To directly examine the ability of patient variant KIF1A motor proteins to undergo microtubule-based motility, we performed in-vitro TIRF-based microtubule gliding assays. In this assay, truncated KIF1A(1–393) motors are immobilized on a slide and their ability to work in concert to “glide” microtubules along the surface is examined by fluorescence microscopy. A unique property of the TIRF microscopy is that it captures the signal from fluorescent microtubules in a limited specimen region immediately adjacent to the adsorbed KIF1A motor protein and the glass coverslip. This exclusive property minimizes any noise coming from the floating microtubules that were not able to bind to the KIF1A motor protein.
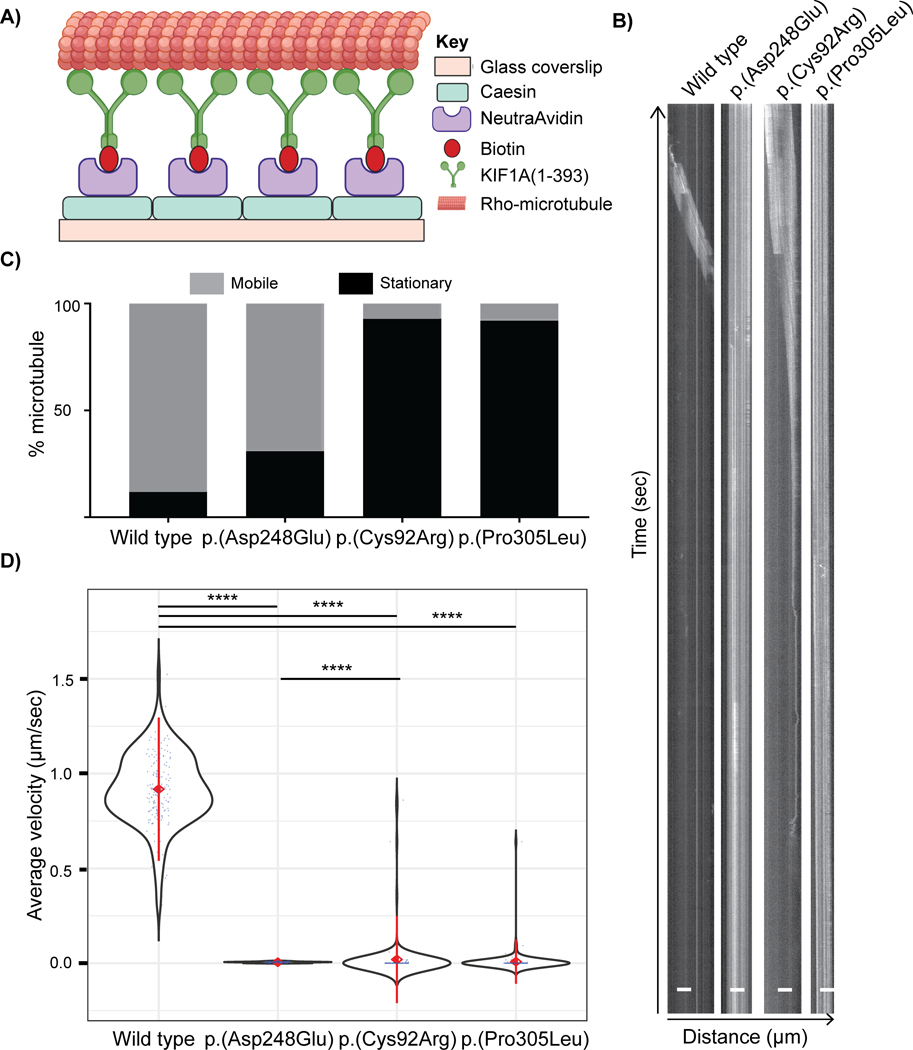
For the wild-type KIF1A(1–393) motors, the majority of microtubules (~85–90%) introduced into the flow chamber bound to the immobilized KIF1A and moved along the surface with an average velocity of 0.91 ± 0.09 μm (Figure 3, Supp. Figure S5 and Supp. Video 1).
Figure 3: in vitro microtubule gliding driven by wild-type and variant KIF1A motors.
A) Schematic of the microtubule gliding assay in a flow chamber modified from (Yue et al., 2018). The biotinylated KIF1A(1–393)-LZ-mNG-AviTag motor proteins expressed in COS-7 cells were immobilized on a NeutrAvidin-coated glass coverslip. The motility mix containing rhodamine-labelled microtubules, ATP, and oxygen-scavenging system was then added into the flow chamber. Time lapse images were captured using the TIRF microscope. B) Representative kymographs (distance versus time graphs) for rhodamine-labeled microtubule movement over “lawns” of wild-type or variant KIF1A proteins. Time (seconds) is on y-axis and distance travelled (μm) is on x-axis. The angled line represents movement and straight line signifies stationary microtubules. C) Quantification of the percentage of microtubules that were stationary versus motile. D) Violin plot representing average velocity (μm/sec) of microtubules moved by KIF1A(1–393) proteins. Black = violin plot, blue = each data point, red diamond with center = mean, red line = +/− standard deviation. The average velocity for every variant KIF1A motor domain protein was significantly reduced compared to the wild type protein. Data are analyzed from three independent experiments (n=50 microtubules each) and is plotted as mean ± standard deviation. **** represents p<0.0001, Mann-Whitney test.
For the case 1 variant [p.(Asp248Glu)] in switch II, which is predicted to impact the coordination of the divalent cation and ATP, the majority of microtubules introduced into the chamber (~70%) were able to bind to the variant KIF1A motors but moved at a significantly lower velocity (0.005 ± 0.002 μm/s) compared to the wild-type motor protein (Figure 3, Supp. Figure S5 and Supp. Video 2). For the case 2 [p.(Cys92Arg)] and case 3 [p.(Pro305Leu)] variants, very few of the microtubules introduced to the chamber were able to bind to the variant KIF1A motors (~10%) (Figure 3, Supp. Figure S5 and Supp. Video 3, 4). The lack of the microtubule binding was mainly assessed qualitatively by the observation that a significantly smaller proportion of fluorescently labelled microtubules were found to bind to the adsorbed variant as compared to the wild type KIF1A motor protein at the same TIRF interface optimized using wild type protein. Moreover, in the raw videos, the ends of the partially bound microtubules for the p.(Cys92Arg) and p.(Pro305Leu) variants were observed to be flickering before dissociating from the adsorbed protein after a short period of time and moving out of the pre-set interface for TIRF. This phenomenon during the gliding assay suggests that there was impaired microtubule binding, consistent with the hypothesis that these variants alter protein folding and/or microtubule binding. We analyzed the movement of the bound fraction of microtubules and found that these microtubules moved at a much slower velocity [0.014 ± 0.06 μm/s [p.(Cys92Arg); Video 3] and 0.009 ± 0.05 μm/s [p.(Pro305Leu); Video 4] compared to the wild-type motor protein (Figure 3, Supp. Figure S5). Nieh et al observed no microtubule motility in case of KIF1A motor protein with p.(Thr99Met) variant which was predicted to affect the ATP binding to the KIF1A motor domain (Esmaeeli Nieh et al., 2015). Overall, these results suggest that patient KIF1A variants negatively affect the ability of KIF1A to bind and move along microtubules.
4. DISCUSSION
In this report we describe one classic RTT individual and three individuals with a severe neurodevelopmental disorder carrying de novo variants in the motor domain of the kinesin-3 motor protein KIF1A. The variants are predicted to be pathogenic by in silico and functional analyses. Heterozygous de novo variants in KIF1A have previously been associated with HSNIIC, SPG30 and MRD9 as well as a much broader spectrum of KIF1A-related disorders (Gabrych et al., 2019).
Here we report a MECP2-negative classic RTT female (case 1) with a novel de novo heterozygous p.(Asp248Glu) variant in the motor domain of KIF1A. Characteristic of SPG30, she has progressive lower limb spasticity as well as mild optic nerve atrophy consistent with observations in KIF1A-related cases. However, in contrast to KIF1A-related disorders, her brain MRI at 12 years of age was normal (Lee et al., 2015; Ohba et al., 2015; Raffa et al., 2017). Recently a classic RTT female patient was reported carrying a de novo heterozygous nonsense variant p.(Cys92*) in KIF1A with a normal brain MRI (age not specified) (Wang et al., 2019). Our in vitro analysis indicates that the p.(Asp248Glu) variant results in a marked decrease in the ability of KIF1A to undergo processive motility to the plus ends of microtubules in the neurite tips of SH-SY5Y cells and a significantly lower velocity of microtubule movement compared to wild-type KIF1A. Together these results suggest that change of Asp248 in switch II of KIF1A results in compromised ATP hydrolysis/cycling rates.
Two cases [case 2 with p.(Cys92Arg) and case 3 with p.(Pro305Leu)] also had features overlapping with the other KIF1A-related phenotypes. Of interest, they had a number of features seen in RTT including microcephaly, global developmental delay, seizures, stereotypic hand movements, gait abnormalities, abnormal muscle tone and scoliosis. Our functional assays provide strong evidence that these variants negatively impact KIF1A function. The variant KIF1A proteins, p.(Cys92Arg) and p.(Pro305Leu) were unable to accumulate at the neurite tips in differentiated SH-SY5Y cells, indicating a lack of processive motility along microtubules for these KIF1A variant proteins. While the p.(Cys92Arg) variant was able to bind to microtubules in the gliding assay, the velocity of movement was dramatically decreased. The p.(Pro305Leu) variant in L12 was predicated to impact the ability of KIF1A to bind to microtubules and indeed, the variant KIF1A protein showed a significant reduction in its ability to bind to microtubules in the in vitro microtubule gliding assay.
Case 4 with p.(Thr99Met) variant in KIF1A shared common clinical features with the previously reported patients with this variant such as abnormal muscle tone, seizures, intellectual disability, gait abnormalities, optic nerve abnormalities (Hamdan et al., 2011; Langlois et al., 2016; Lee et al., 2015). In addition, she exhibited features described in RTT such as period of regression followed by stabilization as well as characteristic stereotypic hand movements. Already published functional data showed a significant decrease in the accumulation of variant KIF1A protein at the distal ends of primary rat hippocampal neurons as well as no microtubule motility using the in-vitro microtubule gliding assay. This is consistent with molecular modelling results predicting loss in interaction between the KIF1A motor domain and ATP, which is crucial for the movement of kinesin head on the microtubule structure (Esmaeeli Nieh et al., 2015; Hamdan et al., 2011; Lee et al., 2015). Together these findings imply that the KIF1A variant proteins are defective for their ability to bind to and move along microtubules, and support the conclusion that they are pathogenic.
Of interest, MeCP2 and KIF1A are both regulated by BDNF which is a well-known target of MeCP2 (Li & Pozzo-Miller, 2014). Several reports have uncovered BDNF defects in RTT mouse models and patients at the genetic as well as functional levels (Ben Zeev et al., 2009; Li et al., 2012; Sampathkumar et al., 2016; Zhou et al., 2006). BDNF has also been shown to be an upstream regulator of KIF1A and its specific cargo such as synaptophysin (Kondo et al., 2012). Importantly, variants in KIF1A have been previously described to cause intellectual disability, and we suggest that defects in KIF1A could contribute towards the development of features overlapping with RTT, but the exact molecular mechanism still needs to be explored.
In conclusion, we have identified four de novo heterozygous variants in KIF1A in a classic RTT patient and three other individuals who share common clinical features with the other KIF1A-related disorders and appear to have expanded phenotype including features seen in RTT. We recommend that genetic testing for KIF1A should be considered in MECP2 negative RTT patients.
Supplementary Material
7. ACKNOWLEDGMENTS
We thank all the family members for taking part in this study. The research conducted at the Murdoch Children’s Research Institute was supported by the Victorian Government’s Operational Infrastructure Support Program. SK was the recipient of Research Training Program scholarship. Work in the laboratory of KJV was supported by grants (R01GM070862 and R35GM131744) from the United States’ National Institutes of Health (NIH). BB was supported by a Graduate Research Fellowship from the National Science Foundation under grant no. DGE 1256260, the Rackham Pre-doctoral Fellowship, and the Endowment for the Development of Graduate Education Student Fellowship. The research work performed in WKC’s lab was supported by KIF1A.org and grant (R01NS114636) from NIH. LB was supported by the grant TL1TR001875.
GRANT NUMBERS
R01GM070862 (KJV), R35GM131744 (KJV), DGE 1256260 (BB), R01NS114636 (WKC) and TL1TR001875 (LB).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
8 REFERENCES
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, . . . Sunyaev SR (2010). A method and server for predicting damaging missense mutations. Nat Methods, 7(4), 248–249. doi: 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atherton J, Farabella I, Yu IM, Rosenfeld SS, Houdusse A, Topf M, & Moores CA (2014). Conserved mechanisms of microtubule-stimulated ADP release, ATP binding, and force generation in transport kinesins. Elife, 3, e03680. doi: 10.7554/eLife.03680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbiero I, De Rosa R, & Kilstrup-Nielsen C (2019). Microtubules: A Key to Understand and Correct Neuronal Defects in CDKL5 Deficiency Disorder? Int J Mol Sci, 20(17), E4075. doi: 10.3390/ijms20174075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Zeev B, Bebbington A, Ho G, Leonard H, de Klerk N, Gak E, . . . Christodoulou J (2009). The common BDNF polymorphism may be a modifier of disease severity in Rett syndrome. Neurology, 72(14), 1242–1247. doi: 10.1212/01.wnl.0000345664.72220.6a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle L, & Chung W (2017). Expanding the natural history of KIF1A associated neurological disorders (KAND). [Google Scholar]
- Cheon CK, Lim SH, Kim YM, Kim D, Lee NY, Yoon TS, . . . Lee JR (2017). Autosomal dominant transmission of complicated hereditary spastic paraplegia due to a dominant negative mutation of KIF1A, SPG30 gene. Sci Rep, 7(1), 12527. doi: 10.1038/s41598-017-12999-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati F, Giorgini V, Masciadri M, Bonati MT, Marchi M, Cracco I, . . . Russo S (2019). Pathogenic Variants in STXBP1 and in Genes for GABAa Receptor Subunities Cause Atypical Rett/Rett-like Phenotypes. Int J Mol Sci, 20(15), E3621. doi: 10.3390/ijms20153621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delepine C, Nectoux J, Bahi-Buisson N, Chelly J, & Bienvenu T (2013). MeCP2 deficiency is associated with impaired microtubule stability. FEBS Lett, 587(2), 245–253. doi: 10.1016/j.febslet.2012.11.033 [DOI] [PubMed] [Google Scholar]
- Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, Shaag A, . . . Elpeleg O (2011). Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res, 21(5), 658–664. doi: 10.1101/gr.117143.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeeli Nieh S, Madou MR, Sirajuddin M, Fregeau B, McKnight D, Lexa K, . . . Sherr EH (2015). De novo mutations in KIF1A cause progressive encephalopathy and brain atrophy. Ann Clin Transl Neurol, 2(6), 623–635. doi: 10.1002/acn3.198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrych DR, Lau VZ, Niwa S, & Silverman MA (2019). Going Too Far Is the Same as Falling Short(dagger): Kinesin-3 Family Members in Hereditary Spastic Paraplegia. Front Cell Neurosci, 13, 419. doi: 10.3389/fncel.2019.00419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JH, Slobedman BK,NH, Williamson SL, Minchenko D, El-Osta A, . . . Christodoulou J (2010). Downstream targets of methyl CpG binding protein 2 and their abnormal expression in the frontal cortex of the human Rett syndrome brain. BMC Neurosci, 11, 53. doi: 10.1186/1471-2202-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold WA, Lacina TA, Cantrill LC, & Christodoulou J (2015). MeCP2 deficiency is associated with reduced levels of tubulin acetylation and can be restored using HDAC6 inhibitors. J Mol Med (Berl), 93(1), 63–72. doi: 10.1007/s00109-014-1202-x [DOI] [PubMed] [Google Scholar]
- Grantham R (1974). Amino acid difference formula to help explain protein evolution. Science, 185(4154), 862–864. doi: 10.1126/science.185.4154.862 [DOI] [PubMed] [Google Scholar]
- Hagberg B, Aicardi J, Dias K, & Ramos O (1983). A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann Neurol, 14(4), 471–479. doi: 10.1002/ana.410140412 [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, . . . Michaud JL (2011). Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet, 88(3), 306–316. doi: 10.1016/j.ajhg.2011.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond JW, Cai D, Blasius TL, Li Z, Jiang Y, Jih GT, . . . Verhey KJ (2009). Mammalian Kinesin-3 motors are dimeric in vivo and move by processive motility upon release of autoinhibition. PLoS Biol, 7(3), e72. doi: 10.1371/journal.pbio.1000072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksen MW, Ravn K, Paus B, von Tetzchner S, & Skjeldal OH (2018). De novo mutations in SCN1A are associated with classic Rett syndrome: a case report. BMC Med Genet, 19(1), 184. doi: 10.1186/s12881-018-0700-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, & Niwa S (2009). Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol, 10(10), 682–696. doi: 10.1038/nrm2774 [DOI] [PubMed] [Google Scholar]
- Hotchkiss L, Donkervoort S, Leach ME, Mohassel P, Bharucha-Goebel DX, Bradley N, . . . Bonnemann CG (2016). Novel De Novo Mutations in KIF1A as a Cause of Hereditary Spastic Paraplegia With Progressive Central Nervous System Involvement. J Child Neurol, 31(9), 1114–1119. doi: 10.1177/0883073816639718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CF, & Banker G (2012). The translocation selectivity of the kinesins that mediate neuronal organelle transport. Traffic, 13(4), 549–564. doi: 10.1111/j.1600-0854.2011.01325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ip JPK, Mellios N, & Sur M (2018). Rett syndrome: insights into genetic, molecular and circuit mechanisms. Nat Rev Neurosci, 19(6), 368–382. doi: 10.1038/s41583-018-0006-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama K, Mizuguchi T, Takeshita E, Nakagawa E, Okazaki T, Nomura Y, . . . Matsumoto N (2019). Genetic landscape of Rett syndrome-like phenotypes revealed by whole exome sequencing. J Med Genet, 56(6), 396–407. doi: 10.1136/jmedgenet-2018-105775 [DOI] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, . . . MacArthur DG (2019). Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. 531210. doi: 10.1101/531210%JbioRxiv [DOI] [Google Scholar]
- Kevenaar JT, & Hoogenraad CC (2015). The axonal cytoskeleton: from organization to function. Front Mol Neurosci, 8, 44. doi: 10.3389/fnmol.2015.00044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, & Hirokawa N (2006). High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J, 25(18), 4187–4194. doi: 10.1038/sj.emboj.7601299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, & Hirokawa N (2001). Switch-based mechanism of kinesin motors. Nature, 411(6836), 439–445. doi: 10.1038/35078000 [DOI] [PubMed] [Google Scholar]
- Klebe S, Lossos A, Azzedine H, Mundwiller E, Sheffer R, Gaussen M, . . . Stevanin G (2012). KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet, 20(6), 645–649. doi: 10.1038/ejhg.2011.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DR, Tomishige M, Stuurman N, & Vale RD (2002). Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell, 109(3), 347–358. doi: 10.1016/s0092-8674(02)00708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Takei Y, & Hirokawa N (2012). Motor protein KIF1A is essential for hippocampal synaptogenesis and learning enhancement in an enriched environment. Neuron, 73(4), 743–757. doi: 10.1016/j.neuron.2011.12.020 [DOI] [PubMed] [Google Scholar]
- Krenn M, Zulehner G, Hotzy C, Rath J, Stogmann E, Wagner M, . . . Zimprich F (2017). Hereditary spastic paraplegia caused by compound heterozygous mutations outside the motor domain of the KIF1A gene. Eur J Neurol, 24(5), 741–747. doi: 10.1111/ene.13279 [DOI] [PubMed] [Google Scholar]
- Kull FJ, & Endow SA (2013). Force generation by kinesin and myosin cytoskeletal motor proteins. J Cell Sci, 126(Pt 1), 9–19. doi: 10.1242/jcs.103911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, & Ng PC (2009). Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc, 4(7), 1073–1081. doi: 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown GR, Chao C, Chitipiralla S, . . . Maglott DR (2018). ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res, 46(D1), D1062-D1067. doi: 10.1093/nar/gkx1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois S, Tarailo-Graovac M, Sayson B, Drogemoller B, Swenerton A, Ross CJ, . . . van Karnebeek CD (2016). De novo dominant variants affecting the motor domain of KIF1A are a cause of PEHO syndrome. Eur J Hum Genet, 24(6), 949–953. doi: 10.1038/ejhg.2015.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Srour M, Kim D, Hamdan FF, Lim SH, Brunel-Guitton C, . . . Michaud JL (2015). De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum Mutat, 36(1), 69–78. doi: 10.1002/humu.22709 [DOI] [PubMed] [Google Scholar]
- Li W, Calfa G, Larimore J, & Pozzo-Miller L (2012). Activity-dependent BDNF release and TRPC signaling is impaired in hippocampal neurons of Mecp2 mutant mice. Proc Natl Acad Sci U S A, 109(42), 17087–17092. doi: 10.1073/pnas.1205271109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, & Pozzo-Miller L (2014). BDNF deregulation in Rett syndrome. Neuropharmacology, 76 Pt C, 737–746. doi: 10.1016/j.neuropharm.2013.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo KY, Kuzmin A, Unger SM, Petersen JD, & Silverman MA (2011). KIF1A is the primary anterograde motor protein required for the axonal transport of dense-core vesicles in cultured hippocampal neurons. Neurosci Lett, 491(3), 168–173. doi: 10.1016/j.neulet.2011.01.018 [DOI] [PubMed] [Google Scholar]
- Mitter D, Pringsheim M, Kaulisch M, Plumacher KS, Schroder S, Warthemann R, . . . Brockmann K (2018). FOXG1 syndrome: genotype-phenotype association in 83 patients with FOXG1 variants. Genet Med, 20(1), 98–108. doi: 10.1038/gim.2017.75 [DOI] [PubMed] [Google Scholar]
- Neul JL, Kaufmann WE, Glaze DG, Christodoulou J, Clarke AJ, Bahi-Buisson N, . . . RettSearch C (2010). Rett syndrome: revised diagnostic criteria and nomenclature. Ann Neurol, 68(6), 944–950. doi: 10.1002/ana.22124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neul JL, Lane JB, Lee HS, Geerts S, Barrish JO, Annese F, . . . Percy AK (2014). Developmental delay in Rett syndrome: data from the natural history study. J Neurodev Disord, 6(1), 20. doi: 10.1186/1866-1955-6-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, Haginoya K, Osaka H, Kubota K, Ishiyama A, Hiraide T, . . . Matsumoto N (2015). De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J Hum Genet, 60(12), 739–742. doi: 10.1038/jhg.2015.108 [DOI] [PubMed] [Google Scholar]
- Okamoto N, Miya F, Tsunoda T, Yanagihara K, Kato M, Saitoh S, . . . Kosaki K (2014). KIF1A mutation in a patient with progressive neurodegeneration. J Hum Genet, 59(11), 639–641. doi: 10.1038/jhg.2014.80 [DOI] [PubMed] [Google Scholar]
- Pelka GJ, Watson CM, Radziewic T, Hayward M, Lahooti H, Christodoulou J, & Tam PP (2006). Mecp2 deficiency is associated with learning and cognitive deficits and altered gene activity in the hippocampal region of mice. Brain, 129(Pt 4), 887–898. doi: 10.1093/brain/awl022 [DOI] [PubMed] [Google Scholar]
- Raffa L, Matton MP, Michaud J, Rossignol E, Decarie JC, & Ospina LH (2017). Optic nerve hypoplasia in a patient with a de novo KIF1A heterozygous mutation. Can J Ophthalmol, 52(5), e169–e171. doi: 10.1016/j.jcjo.2017.02.021 [DOI] [PubMed] [Google Scholar]
- Rett A (1966). [On a unusual brain atrophy syndrome in hyperammonemia in childhood]. Wien Med Wochenschr, 116(37), 723–726. [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, . . . Committee ALQA (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere JB, Ramalingam S, Lavastre V, Shekarabi M, Holbert S, Lafontaine J, . . . Rouleau GA (2011). KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet, 89(2), 219–230. doi: 10.1016/j.ajhg.2011.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar C, Wu YJ, Vadhvani M, Trimbuch T, Eickholt B, & Rosenmund C (2016). Loss of MeCP2 disrupts cell autonomous and autocrine BDNF signaling in mouse glutamatergic neurons. Elife, 5. doi: 10.7554/eLife.19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonewolf-Greulich B, Bisgaard AM, Moller RS, Duno M, Brondum-Nielsen K, Kaur S, . . . Tumer Z (2019). Clinician’s guide to genes associated with Rett-like phenotypes-Investigation of a Danish cohort and review of the literature. Clin Genet, 95(2), 221–230. doi: 10.1111/cge.13153 [DOI] [PubMed] [Google Scholar]
- Schrödinger. (2010). The PyMOL Molecular Graphics System. [Google Scholar]
- Schwarz JM, Cooper DN, Schuelke M, & Seelow D (2014). MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods, 11(4), 361–362. doi: 10.1038/nmeth.2890 [DOI] [PubMed] [Google Scholar]
- Soppina V, Norris SR, Dizaji AS, Kortus M, Veatch S, Peckham M, & Verhey KJ (2014). Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc Natl Acad Sci U S A, 111(15), 5562–5567. doi: 10.1073/pnas.1400759111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhope K, & Ross J (2015). Microtubules, MAPs, and Motor Proteins. In Ross J & Marshall W (Eds.), Methods in Cell Biology (Vol. 128, pp. 23–37 ). [DOI] [PubMed] [Google Scholar]
- Stucchi R, Plucinska G, Hummel JJA, Zahavi EE, Guerra San Juan I, Klykov O, . . . Hoogenraad CC (2018). Regulation of KIF1A-Driven Dense Core Vesicle Transport: Ca(2+)/CaM Controls DCV Binding and Liprin-alpha/TANC2 Recruits DCVs to Postsynaptic Sites. Cell Rep, 24(3), 685–700. doi: 10.1016/j.celrep.2018.06.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M, Klopfenstein DR, & Vale RD (2002). Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science, 297(5590), 2263–2267. doi: 10.1126/science.1073386 [DOI] [PubMed] [Google Scholar]
- Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, & Vriend G (2010). Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinformatics, 11, 548. doi: 10.1186/1471-2105-11-548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal S, Brandi N, Pacheco P, Maynou J, Fernandez G, Xiol C, . . . Armstrong J (2019). The most recurrent monogenic disorders that overlap with the phenotype of Rett syndrome. Eur J Paediatr Neurol, 23(4), 609–620. doi: 10.1016/j.ejpn.2019.04.006 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Q, Chen Y, Yu S, Wu X, & Bao X (2019). Rett and Rett-like syndrome: Expanding the genetic spectrum to KIF1A and GRIN1 gene. Mol Genet Genomic Med, e968. doi: 10.1002/mgg3.968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Bentley M, Huang CF, & Banker G (2016). Analyzing kinesin motor domain translocation in cultured hippocampal neurons. Methods Cell Biol, 131, 217–232. doi: 10.1016/bs.mcb.2015.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo Y, Jung J, Lee YN, Lee Y, Cho H, Na E, . . . Choi, M. (2017). GABBR2 mutations determine phenotype in rett syndrome and epileptic encephalopathy. Ann Neurol, 82(3), 466–478. doi: 10.1002/ana.25032 [DOI] [PubMed] [Google Scholar]
- Yoshikawa K, Kuwahara M, Saigoh K, Ishiura H, Yamagishi Y, Hamano Y, . . . Kusunoki S (2019). The novel de novo mutation of KIF1A gene as the cause for Spastic paraplegia 30 in a Japanese case. eNeurologicalSci, 14, 34–37. doi: 10.1016/j.ensci.2018.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Blasius TL, Zhang S, Jariwala S, Walker B, Grant BJ, . . . Verhey KJ (2018). Altered chemomechanical coupling causes impaired motility of the kinesin-4 motors KIF27 and KIF7. J Cell Biol, 217(4), 1319–1334. doi: 10.1083/jcb.201708179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, . . . Greenberg ME (2006). Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron, 52(2), 255–269. doi: 10.1016/j.neuron.2006.09.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.