Abstract
Significant barriers to the diagnosis of latent and acute SARS-CoV-2 infection continue to hamper population-based screening efforts required to contain the COVID-19 pandemic in the absence of widely available antiviral therapeutics or vaccines. We report an aptamer-based SARS-CoV-2 salivary antigen assay employing only low-cost reagents ($3.20/test) and an off-the-shelf glucometer. The test was engineered around a glucometer as it is quantitative, easy to use, and the most prevalent piece of diagnostic equipment globally, making the test highly scalable with an infrastructure that is already in place. Furthermore, many glucometers connect to smartphones, providing an opportunity to integrate with contact tracing apps, medical providers, and electronic health records. In clinical testing, the developed assay detected SARS-CoV-2 infection in patient saliva across a range of viral loads - as benchmarked by RT-qPCR - within 1 h, with 100% sensitivity (positive percent agreement) and distinguished infected specimens from off-target antigens in uninfected controls with 100% specificity (negative percent agreement). We propose that this approach provides an inexpensive, rapid, and accurate diagnostic for distributed screening of SARS-CoV-2 infection at scale.
Keywords: Aptamer, Glucometer, SARS-CoV-2, COVID-19, Point-of-care, Population screening
1. Introduction
Since the first reports of a deadly respiratory illness from Wuhan, China in the winter of 2019, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, has spread globally and resulted in the most impactful pandemic in more than a century. This pathogen joins a growing number of emerging infectious diseases, including Avian and H1N1 “swine” influenza, human immunodeficiency virus (HIV), Ebola virus, Zika virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV), which are increasing in frequency and severity as a consequence of human-related activities such as globalization, societal unrest, and changes in environmental conditions (Morens and Fauci, 2020). To a greater or lesser extent, these viruses share important features, including passage through intermediate non-human hosts leading to genetic alterations that result in increased infectivity and virulence in human populations, asymptomatic carriage in a proportion of infected individuals allowing for extensive undetected spread, and inadequate availability of accurate diagnostic tests. Even as COVID-19 has reached over 214 countries, true prevalence remains difficult to estimate due to continued limitations in the capacity and performance of existing molecular diagnostics (CSSEGISandData, 2020; Dong et al., 2020; Kaplan and Forman, 2020). In this context, the number of confirmed cases exceeds 104 million, and there have been over 2.27 million attributed deaths to date. The United States has the greatest number of cases of any country in the world (over 27 million as of March 2, 2021), nearly 25% of global incidence (CDC, 2020a), and has conducted 92 million tests or ~22% of the more than 2 billion viral tests that have been performed globally (CDC, 2020b).
There are currently three accepted methods for the diagnosis of SARS-CoV-2 infection: 1) Viral RNA detection; 2) Viral protein detection, typically against the nucleocapsid (N) protein or spike (S) surface glycoprotein; and 3) Measurement of specific antibodies directed towards viral proteins. While the initial antibody response may be detected within a week of symptoms (IgM as early as day 7 and IgG >14-days post-infection) (CDC, 2020c), direct viral testing has been the preferred screening method in asymptomatic populations and acute presentations. For this, reverse transcription polymerase chain reaction (RT-PCR) remains the gold standard nucleic acid amplification test (NAAT), with samples collected from nasopharyngeal, mid-turbinate, oropharyngeal, saliva, and bronchoscopy specimens. While sensitivity and specificity approach 99% in ideal settings, reported real-world sensitivities are estimated to be as low as 70% (Arevalo-Rodriguez et al., 2020; Yang et al., 2020), likely due to variation in sample collection methods and differences in viral shedding across the oropharynx and respiratory tract (Afzal, 2020). Importantly, it is now recognized that the presence of RNA detected by PCR may not reflect infectivity (Tahamtan and Ardebili, 2020), potentially making viral antigen detection a more appropriate biomarker of transmissible disease (Kubina and Dziedzic, 2020).
Tromberg and others have discussed the continued challenges in SARS-CoV-2 testing (Tromberg et al., 2020). Current efforts are hampered by limited capacity, cost, and deployment logistics, leading to prioritized testing of specific high-risk groups and leaving many populations without the level of screening necessary to control the spread of infection (Tan et al., 2020). Moreover, we still lack a “perfect test” with high sensitivity to rule in, high specificity to rule out, the ability to discern active and past infection, rapid turn-around-time, and a price-point to allow testing at scale. Ideally, such a test could be performed by an inexperienced user (e.g., at-home or in the community), be able to reliably detect early (asymptomatic or acute) infection with a low false positive rate, and have results that can be objectively read and easily transmitted to patients’ medical providers and public health personnel. Lateral flow immunoassays (LFIA) are a promising point-of-care (POC) solution but are associated with significant limitations, including qualitative readout and reportedly low sensitivity with high false positive rates (Pallett et al., 2020; Scohy et al., 2020; Wu et al., 2020). Hence, there remains an urgent need for accurate and cost-effective diagnostic tests that can be broadly deployed.
A potential solution to this problem is to develop quantitative, rapid tests around infrastructure that has already been adopted in the market at-scale. It has been previously shown that commercial glucometers can be repurposed to detect a variety of non-glucose-based targets, quantitatively measuring cocaine, Ebola, hepatitis B, foodborne pathogens, and interferon-gamma (Chen et al., 2020; Du et al., 2015; Guo et al., 2020; Lan et al, 2015, 2016; Taebi et al., 2018; Xiang and Lu, 2011; Zhang et al, 2019, 2020). There are currently 422 million people worldwide who rely on these devices daily to manage their blood sugar, making the glucometer the most prevalent piece of diagnostic equipment globally (CDC, 2020d; “Diabetes Statistics). These meters are small, inexpensive ($20–50 USD), user-friendly, highly accurate (Klonoff et al., 2018), and many connect to smartphones through Bluetooth, providing an opportunity to integrate SARS-CoV-2 detection results with contact tracing apps and electronic health records. The major hurdle in repurposing a glucometer for direct detection of SARS-CoV-2 is that the target biomarkers (e.g., protein N and S) are present at low concentrations in biological samples. The average SARS-CoV-2 viral load in nasal/throat, sputum, and saliva samples is 3×106, 7.50×105, and 3.5×107 copies/mL (To et al., 2020; Zou et al., 2020), respectively, necessitating signal amplification to generate product (i.e. glucose) in quantities similar to physiological levels in human blood (i.e. 10–600 mg/dL or 0.6–33 mM) (Diabetes Statistics; Montagnana et al., 2009).
Aiming to hit a SARS-CoV-2 diagnostic sweet-spot, we report a point-of-care saliva-based test that can measure viral antigen with a glucometer. To transduce antigen binding events into glucose signal production, we exploit the native catalytic property of invertase and an aptamer-based competitive assay. The proposed workflow is illustrated in Fig. 1 , where aptamers directed at the viral S or N protein are pre-conjugated to invertase through a small antisense oligonucleotide strand complementary to a portion of the aptamer's binding domain (aptatope). The biotinylated aptamer-oligo-invertase complex is pre-assembled on magnetic beads. In the presence of the cognate antigen, the aptamer undergoes a conformational change, displacing the lower affinity antisense strand, thus creating an antigen-sensitive switch. After magnetic separation, the released enzyme hydrolyzes sucrose into glucose with a turnover rate of 5×103 glucose mol/sec (Xiang and Lu, 2011), enabling many orders of magnitude signal enhancement. This amplification allows readout with an off-the-shelf glucometer where the signal is proportional to the viral antigen concentration. In this work, we designed and optimized the system for saliva given the simplicity of sample collection (Ceron et al., 2020). However, this approach would work equally well with other sample types. Through testing, we demonstrate that the assay has minimal cross-reactivity to proteins from other respiratory viruses, recognizes native antigens in conditioned media of cells infected with SARS-CoV-2, and clinically discriminates infected and non-infected individuals with an unmodified $29 glucometer.
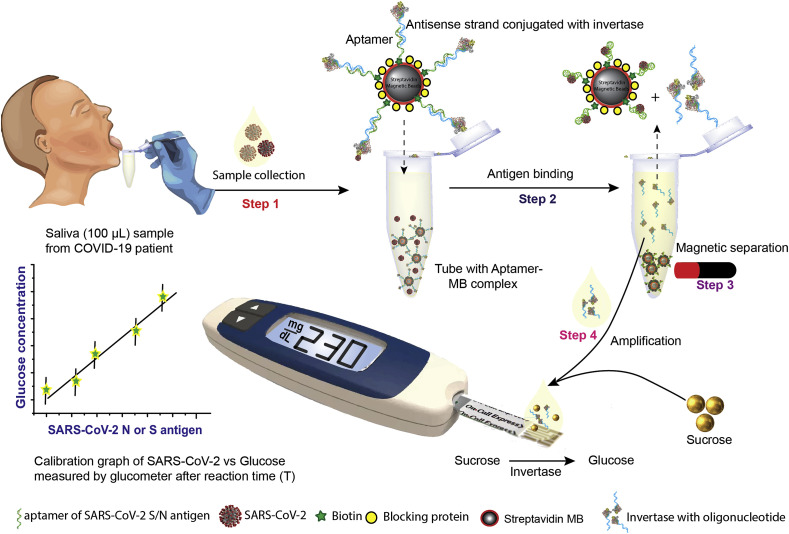
Fig. 1.
Overview of proposed point-of-care, aptamer-based COVID-19 assay. SARS-CoV-2 N or S protein-specific biotinylated aptamer is conjugated to a streptavidin-coated magnetic bead (MB) and pre-hybridized with a complementary antisense oligonucleotide strand that is covalently attached to an invertase enzyme. The saliva sample is added to this cocktail (Step 1). Upon binding to the viral antigen or the SARS-CoV-2 virion, the invertase-antisense oligo is released (Step 2). A magnet is used to remove the MB conjugated to the aptamer-antigen complex and the remaining aptamer-antisense-invertase complex (Step 3). The solution containing the released antisense-invertase is then collected and incubated with sucrose (Step 4). Invertase converts sucrose to glucose that is directly readout using a glucometer. The glucose concentration is correlated with the SARS-CoV-2 N or S protein concentration. (Glucometer image adopted with permission from Hangzhou Sejoy Electronics & Instruments ltd, China.)
2. Materials and Methods
Reagents. Biotin-tagged, HPLC-grade purified aptamers against SARS-CoV-2 N (Chen et al., 2020) and S (Song et al., 2020) antigen and complementary thiolated antisense DNA oligonucleotides were designed and ordered from Integrated DNA Technology (IDT). The aptamer and antisense sequences used in this work are listed in Supplemental Table S1. Streptavidin-coated Dynabeads M-280 (2.8 μm), 10% bovine serum albumin (BSA), dithiobis (succinimidyl propionate) (DSP), and tris(2-carboxyethyl) phosphine (TCEP) were purchased from Thermo Fisher. Dulbecco's potassium phosphate buffer (DPBS) with calcium and magnesium, citrate buffer, calcium chloride (CaCl2), magnesium chloride (MgCl2), ethylenediaminetetraacetic acid (EDTA), sodium borohydride (NaBH4), sucrose, glucose, 4-(N-Maleimidomethyl)cyclohexane-1-carboxylic acid 3-sulfo-N-hydroxysuccinimide ester sodium salt (sulfo-SMCC), glucose oxidase type-VII from Aspergillus niger, and invertase (Grade VII) from Saccharomyces cerevisiae were purchased from Sigma Aldrich. All reagents were analytical grade and used without further processing. Buffer compositions are described in Supplemental Table S2. SARS-CoV-2 N and S, Influenza A (H1N1) hemagglutinin and neuraminidase, and MERS nucleocapsid and spike RBD fragment proteins were purchased from Sino Biological. Amicon filters (3, 10, and 100 kDa cutoffs) were purchased from Millipore. Dulbecco's Modified Eagle's Medium (DMEM) culture media and Penicillin-Streptomycin (10,000 U/mL) antibiotics were obtained from Corning and Gibco, respectively. An “Accu-Chek GuideMe” glucometer was used for all assays. The reagents with catalog numbers are listed in Supplemental Table S3.
Conjugation of invertase with the antisense oligomer strand. Invertase was covalently linked with a thiolated antisense oligomer strand (specific to the N or S aptamer) using a modified version of a previously reported protocol (Xiang and Lu, 2011). Briefly, 30 μL of 1 mM thiolated antisense oligomer was mixed with 6 μL of 0.5 M TCEP and stirred at room temperature (RT) for 2 h. After incubation, the antisense strand was purified through centrifugation with a 3 kDa cutoff filter. This was repeated 8× in DPBS buffer. Next, 400 μL of invertase was mixed with 1 mg of water-soluble sulfo-SMCC by gentle pipetting for 5 min. The mixture was then placed on a shaker for 2 h at RT. After incubation, unbound sulfo-SMCC was removed by centrifugation with a 10 kDa cutoff filter. This process was repeated 8× in DPBS buffer. The purified, sulfo-SMCC linked invertase (sulfo-SMCC-invertase) was mixed with the purified, reduced, thiolated antisense strand and kept on a shaker for 48 h at RT. Unreacted, free antisense oligomers were removed by centrifugation with a 10 kDa cutoff filter. This was repeated 8× in DPBS. The purified antisense-invertase conjugate was stored at 4 °C for downstream application.
Hybridization of aptamer with antisense-invertase conjugate. The biotinylated (N or S) aptamer was refolded by heat treatment at 80 °C for 3 min, followed by gentle cooling at RT for 5 min. Similarly, the antisense-invertase conjugate was heat-treated at 40 °C for 10 min before hybridization. 10 μL of heat-treated N or S aptamer (0.5 mM) was mixed with 20 μL of heat-treated antisense-invertase conjugate and 170 μL of DPBS and placed on a shaker for 2 h at RT. The unhybridized, free aptamer was removed by centrifugation with a 100 kDa cutoff filter. This was repeated 8× washing with DPBS. The purified, hybridized aptamer/antisense-invertase complex (~200 μL) was stored at 4 °C.
Conjugation of aptamer/antisense-invertase complex and magnetic beads. 200 μL of streptavidin-coated magnetic beads (MBs) was placed near a rare earth magnet. The supernatant was discarded and replaced with 600 μL of washing and binding buffer (see Supplemental Table S2). This process was repeated 3×. The MBs were then equilibrated with DPBS buffer for 10 min, the incubation buffer discarded and resuspended in 200 μL of the biotinylated aptamer/antisense-invertase complex. This was kept on a shaker for 1 h at RT. Excess unbound aptamer/antisense-invertase complex was washed 3-5× with buffer. The resulting aptamer/antisense-invertase magnetic bead complex (MBC) was treated with 1% BSA in DPBS for 30 min. After incubation, the BSA solution was discarded, and the MBC was resuspended in 400 μL of DPBS. 50 μL of the MBC (~200 μg) was aliquoted in test tubes and stored at 4 °C.
Fabrication of the custom electrochemical glucose sensor. A glass slide with an evaporated gold electrode (5 nm Ti/50 nm Au) was chemically cleaned in piranha solution (3:1 of H2SO4:H2O2) for 1 min followed by washing with ultrapure (Milli-Q) water. The electrode was then sonicated in acetone and isopropanol sequentially for 5 min followed by washing with ultrapure water. The electrode was then electrochemically cleaned in 0.5 M H2SO4 by sweeping the potential from −0.5 to +1.2 V vs. Ag/AgCl electrode, washed with water, and air-dried. A surface assembled monolayer (SAM) was formed by incubating the electrode in 1 mL DSP (2 mg/mL in Dimethyl sulfoxide) reduced with 5 μL of NaBH4 (10 mg/mL in ultrapure water) for 2 h at RT. The electrode was then washed with acetone, methanol, isopropanol, and ultrapure water, followed by air drying. The DSP-modified electrode was incubated with 5 μM of glucose oxidase in PBS overnight at 4 °C to covalently link to the DSP-modified surface. The unbound GOx was washed off with PBS, and the sensor was incubated in 1% ethanolamine for 15 min to block any remaining active succinimidyl and then 1% BSA for 10 min. The electrode was stored at 4 °C when not in use. Layer by layer assembly was monitored by cyclic voltammetry (CV) with a CHI-760 E electrochemical workstation in a three-electrode configuration (BASi Ag/AgCl reference electrode and a platinum wire counter electrode). Voltammograms were measured from −0.5 to +0.8 V at a scan rate of 50 mV/s with 1 mM ferrocene in 1× PBS and 0.25 M KCl.
Custom-made glucose sensor SARS-CoV-2 assay. 100 μL of DPBS buffer spiked with SARS-CoV-2 N or S protein was incubated with 200 μg of MBC (N or S) with gentle shaking for 30 min at RT in a 1.5 mL centrifuge tube. The MBC was pulled down using a rare earth magnet. 90 μL of the supernatant was transferred to another centrifuge tube prefilled with 100 μL of 2× Measurement buffer (Supplemental Table S2). After incubating for 30 min, 200 μL was placed on the glucose sensor, and readout using a CHI-760 E electrochemical workstation with the three-electrode configuration described previously. Voltammograms were measured from −0.5 to +0.8 V at a scan rate of 50 mV/s.
Collection of nasopharyngeal swab and saliva samples. We collected matched nasopharyngeal swabs (NPS) in RNA Shield DNA/RNA storage medium (Zymo) or viral transport medium and saliva samples (no additive) from symptomatic and asymptomatic study subjects with a prior positive clinical COVID-19 RT-qPCR result under institutional review board (IRB) approval (UCSD protocol #200477). Informed consent was provided from all subjects. These samples were subjected to viral RNA extraction using the MagMax Viral/Pathogen Nucleic Acid Isolation Kit (Thermo), and the TaqPath COVID-19 multiplex RT-qPCR assay was performed on the resulting RNA samples. Saliva samples in 300 μL aliquots were provided as blinded specimens for testing in the Biosafety Level 3 (BSL-3) lab. All experiments were performed in accordance with the relevant guidelines and regulations designated by the UCSD Human Research Protections Program (HRPP). Demographic information about the cohort is listed in Supplemental Table S4.
Glucometer-based SARS-CoV-2 assay. Saliva samples (~300–500 μL) were collected in sterile tubes from volunteers using the standard passive drooling technique (Granger et al., 2012). To test N protein, saliva was mixed with 1% Triton, whereas no detergent was added to samples where S protein was to be detected. 100 μL of sample (contrived, conditioned media, or saliva) was then diluted two-fold in DPBS buffer and assayed without further processing or treatment. Half of the diluted sample was then incubated with 200 μg of MBC (N or S) with gentle shaking for 30 min at RT in a 1.5 mL centrifuge tube. The MBC was pulled down using a rare earth magnet. 90 μL of supernatant was transferred to a centrifuge tube prefilled with the Sucrose buffer (Supplemental Table S2). The other half of the diluted sample was placed in a separate centrifuge tube prefilled with the Sucrose buffer as a background control. After mixing, the tubes were incubated at 60 °C in a water bath for 30 min. Finally, 10 μL of each reaction solution was placed on a glucometer test strip and read out using a glucometer. The difference between the two readings was recorded. All measurements were repeated in triplicates.
Authentic SARS-CoV-2 production and quantification. Vero E6 cells were obtained from ATCC and grown in DMEM with 10% fetal bovine serum (FBS) and Penicillin-Streptomycin. SARS-CoV-2 isolate USA-WA1/2020 (BEI Resources) was propagated, and aliquots of the secreted virus in culture media were stored at −80 °C. RNA copies were quantified by digital droplet PCR (ddPCR) and ininfectious units (IU) using Vero E6 cells. For ddPCR, viral stock media was added to TRIzol LS (ThermoFisher), and RNA was extracted using a Directzol RNA miniprep kit (Zymo Research). The ddPCR quantified SARS-CoV-2 ORF1a and was performed by the UCSD Center for Aids Research (CFAR) Genomics and Sequencing Core. For plaque assay quantification, viral supernatants were 10-fold serially diluted in DMEM without serum. Vero E6 cells in 12-well plates were washed with PBS, and 200 μL of virus dilution was added per well and incubated for 1 h at 37 °C with rocking every 10–15 min. The inoculum was removed, and 1 mL of overlay (0.6% agarose in MEM with 4% FBS) was added to each well. Overlays were prepared by mixing equal volumes of 1.2% agarose and 2× MEM supplemented with 8% FBS, 2× L-glutamine, 2× non-essential amino acids, and 2× sodium bicarbonate. Assays were incubated for 48 h at 37 °C and fixed by adding 2 mL of 10% formaldehyde for at least 24 h. Overlays were removed, and monolayers were stained with 0.025% crystal violet in 2% EtOH and plaques counted.
Safety. Piranha solution is highly corrosive and extreme precaution is needed in the handling. All work involving infectious SARS-CoV-2 samples was undertaken in the UC San Diego Division of Infectious Diseases BSL-3 laboratory with oversight from the UC San Diego Institutional Biosafety Committee (IBC).
Statistical analysis. All data was from a minimum of three independent experiments. Error bars represent one standard deviation. Statistical analysis was performed with Origin 9.0 and/or MATLAB. The limit of detection (LOD) was calculated using the slope method where LOD = 3×standard deviation (SD) of blank/slope (Shrivastava and Gupta, 2011).
3. Results
Antigen-sensitive aptamer switch validation. We selected aptamers reported in the literature to have high a affinity towards SARS-CoV-2 N and S antigen. We then analyzed their secondary structures using Mfold (Zuker, 2003) and designed complementary antisense oligonucleotide strands to overlap the predicted secondary stem-loop structures. The 5′ end of the aptamers and antisense strands were extended with a linker (6 and 12 thymine oligomers, respectively) to increase the distance between the aptamer and the magnetic beads, allowing the aptamer room to properly fold and reduce steric hindrance. To test the release of the antisense strand from the aptamer upon ligand binding, we designed a PCR-based assay where protein binding induces a conformation change in the aptamer, releasing the antisense oligo, as shown in Supplemental Fig. S1. The free antisense oligo was then collected from the supernatant and used as the reverse primer in a PCR reaction with the S and N aptamer as the template and the corresponding forward primers (Supplemental Table S5). We confirmed PCR amplification through agarose gel electrophoresis. These data demonstrated specific, antigen-mediated release of the antisense oligonucleotide from the aptamer and established baseline conditions for the subsequent assay development.
Tuning assay conditions for glucometer read out. We carried out the antisense oligo conjugation to invertase via amine functionalization to preserve the aspartic and glutamic acids in the enzyme's active site (Reddy and Maley, 1996). Crosslinking the antisense oligonucleotide with invertase was performed with sulfo-SMCC and evaluated by an electrophoretic mobility shift assay (EMSA). The results, shown in Supplemental Fig. S2, indicate successful crosslinking between the DNA and invertase, as visible from the gel image. We verified enzyme activity in the presence of the substrate (sucrose) with a redox mediator and quantified the amount of glucose using custom-made glucose sensors that have higher sensitivity and dynamic range than glucometers (Supplemental Fig. S3). As enzyme activity is dependent on various factors, we assessed different incubation temperatures, substrate concentrations, buffers, pH, and salts to optimize for efficiency and linearity (Supplemental Figs. S4 and S5). Based on our findings, the optimum conditions for the amplification phase are 1 M sucrose, 5 mM CaCl2, 1 mM MgCl2, and 0.5 mM EDTA in 0.1 M citrate buffer at pH 5.0. Maximum invertase activity was observed at 60 °C, roughly 6× higher than at room temperature in PBS buffer. We then conjugated the aptamer to magnetic beads using streptavidin-biotin chemistry and optimized the magnetic bead to aptamer ratio to avoid overcrowding, which causes steric hindrance reducing antigen binding efficiency. We found that magnetic beads saturated with a 1:5 ratio for both the N and S directed aptamers (Supplemental Fig. S6). We performed studies to identify the optimum aptamer-target interaction binding time and signal amplification (invertase) time (Supplemental Fig. S7). We selected 30 min for both. These optimized conditions were used for all subsequent assays.
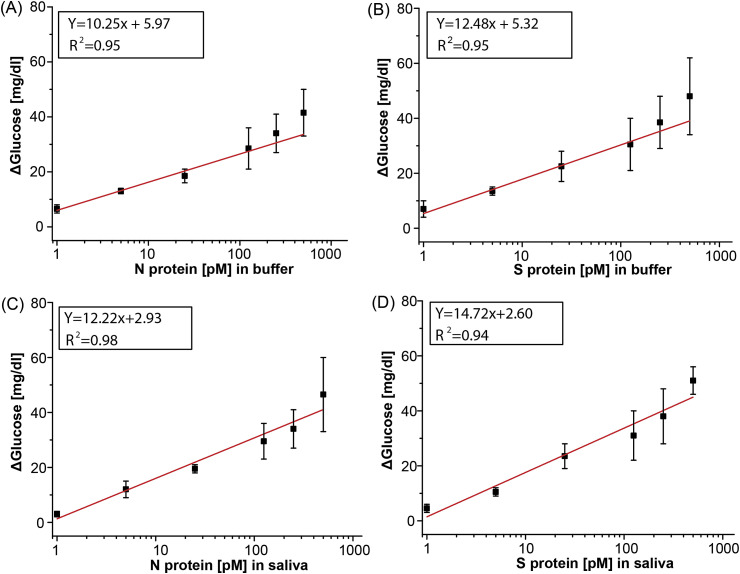
SARS-CoV-2 protein detection in buffer and saliva. We measured the correlation between glucose readout and antigen concentration by spiking SARS-CoV-2 N and S antigen in buffer and healthy saliva (Fig. 2 ). A calibration plot was generated by varying the antigen concentration across the same range as commercially available SARS-CoV-2 ELISA kits (”2019-nCoV Coronavirus Nucleocapsid ELISA Kit, KIT40588 | Sino Biological,” n. d.; “2019-nCoV Coronavirus spike ELISA Kit, KIT40592 | Sino Biological,” n. d.). Using off-the-shelf glucose test strips and a glucometer, we observed a broad linear dynamic range of 1–500 pM for all combinations of sample matrix and antigen. The LODs in buffer were 1.50 pM and 1.31 pM for protein N and S, respectively. In saliva, the LODs increased to 5.27 pM and 6.31 pM for protein N and S, respectively. This indicates a small but consistent matrix effect. We further evaluated the assay performance with custom-made glucose sensors (Supplemental Fig. S8). The custom electrochemical sensor performed similarly but achieved lower LODs (0.71 pM and 0.34 pM for protein N and S, respectively) due to the use of a high-performance, benchtop potentiostat. This assay showed similar performance to ELISA kits for both N (3.5–226 pM) and S (2–128 pM) proteins despite using a glucometer.
Fig. 2.
Calibration curves for SARS-CoV-2 N and S protein in buffer and saliva measured with a glucometer. Linear calibration curve with subtracted background signal of protein N spiked into (A) buffer and (C) saliva and protein S spiked in (B) buffer and (D) saliva. All measurements taken with n = 3 and error bars represent ±1σ.
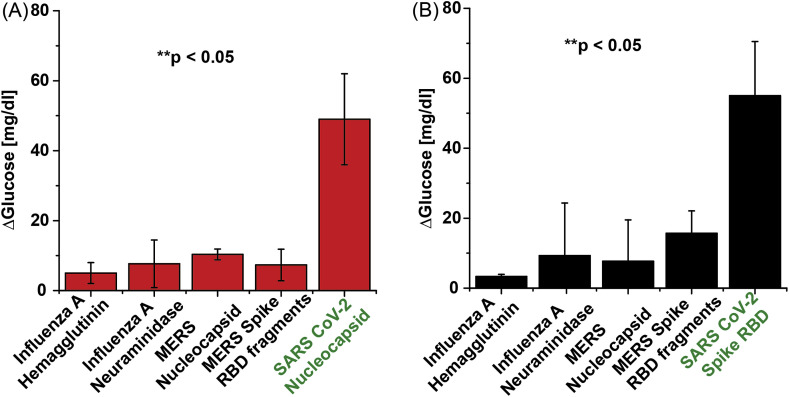
Determination of assay specificity. As a preliminary assessment of assay specificity, we measured signal generation in the presence of antigen from non-SARS-CoV-2 respiratory viruses, namely: Influenza A (H1N1) and MERS-CoV. Protein N and S specific aptamer complexes were assayed with the off-target antigens at a fixed concentration of 500 pM with the conditions described above. This assay was performed in buffer rather than saliva to isolate the source of the non-specific binding. The results, shown in Fig. 3 , indicate minimal cross-reactivity in the assay, even when the off-target antigens are present in high concentration. Unsurprisingly, the SARS-CoV-2 N aptamer displayed the highest signal with the MERS nucleocapsid antigen and the S aptamer with the MERS-CoV RBD antigen, consistent with reported homology between the two coronavirus genomes. However, in all cases, the corresponding SARS-CoV-2 signal is >3× higher than the off-target recording (p < 0.05).
Fig. 3.
Cross-reactivity study. Assays using analogous proteins with (A) N aptamer complex and (B) S aptamer complex. All proteins were spiked into DPBS at 500 pM. All measurements taken with n = 3 and error bars represent ±1σ.
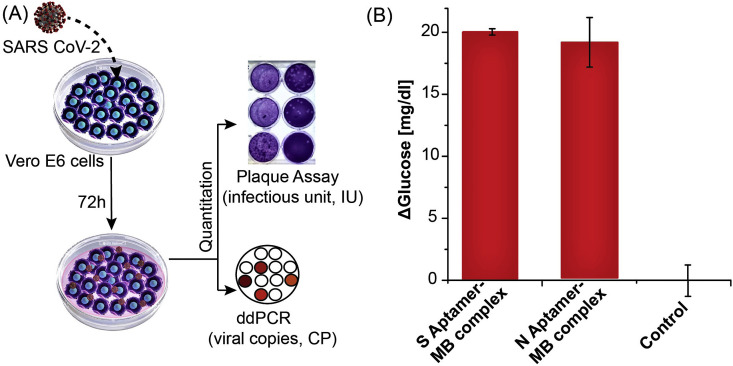
Detection of authentic SARS-CoV-2 N and S protein in cultured media. Previous studies of protein S and N directed aptamers validated binding with only recombinant purified proteins (Chen et al., 2020; Song et al., 2020). To determine if our S and N aptamer/antisense-invertase system could recognize authentic virus and native proteins produced during SARS-CoV-2 infection, we created and quantified viral stocks of SARS-CoV-2 in a biosafety level 3 (BSL-3) laboratory. We inoculated authentic SARS-CoV-2 isolate USA-WA1/2020 onto Vero E6 cells and allowed these to propagate and secrete virus into the supernatant (Fig. 4 A). The supernatant was collected, aliquoted, and frozen. The quantity of SARS-CoV-2 in these preparations was determined by two methods: 1) we measured the amount of SARS-CoV-2 ORF1a RNA using ddPCR, and 2) we determined the number of infectious virions by plaque assay. SARS-CoV-2 supernatants were diluted 1:10 with DPBS to 52×106 copies and 12.5×103 IU and assayed with the S and N aptamer complex. The cell media used to propagate the virus has a high glucose level (450 mg/dL). To nullify the effect of background glucose in the conditioned media, we also conducted glucometer readings with control diluted media in a similar manner but in the absence of the aptamer complexes. Both the N and S aptamers showed significant increases in glucometer readings compared to control samples (Fig. 4B). This demonstrates that the aptamer/antisense-invertase systems recognize their native targets when produced by replicating authentic SARS-CoV-2.
Fig. 4.
Detection of N and S protein using authentic SARS-CoV-2. (A) Schematic of authentic SARS-CoV-2 virus preparation and quantification of viral RNA by ddPCR and infectious units by plaque assay. (B) N and S aptamer/antisense MB complex detection of the SARS-CoV-2 native protein in 1:10 diluted virus culture media. Background media (control) values were subtracted from the measurement results. All measurements taken with n = 3 and error bars represent ±1σ.
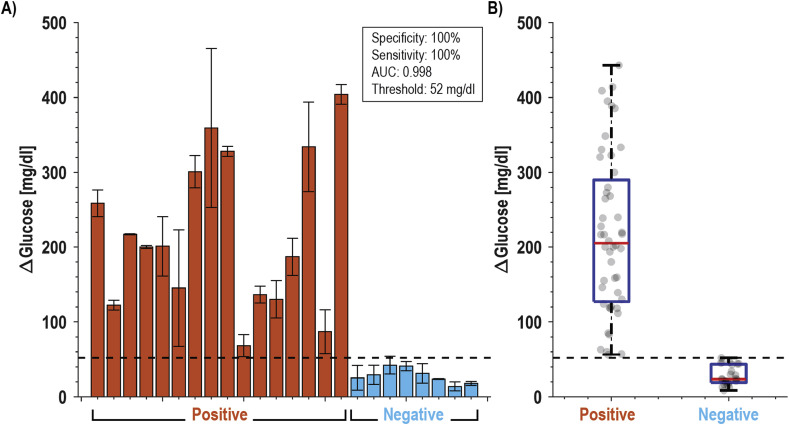
Detection of SARS-CoV-2 in clinical specimens. Finally, we evaluated whether the developed assays could discriminate between SARS-CoV-2 infected and healthy individuals with validated saliva samples (see Materials and Methods). We started with a small cohort of 3 infected persons (confirmed positive with RT-qPCR) and 4 healthy controls tested for protein S and N binding. The results, shown in Supplemental Fig. S9, demonstrate the ability of both assays to correctly differentiate between infected and non-infected individuals; however, the protein S assay showed significantly higher signal-to-control than the protein N assay and was selected as the focus of the larger study. In this cohort of 24 individuals, 42% were female, and the average age was 31 years. Of the 16 infected individuals, the average time between symptom onset and testing was 7 days, 63% had a fever, and 50% had a cough. Two subjects self-reported having asthma, one was pregnant, and none were diabetic. Fig. 5 shows the results that were presented as a blind panel run under BSL-3 conditions over two days using the same glucometer and a single lot of commercial test strips. All SARS-CoV-2 confirmed positive samples demonstrated higher glucose production (μ = 218 mg/dL, range = 68–404 mg/dL) than healthy individuals (μ = 24 mg/dL, range = 14–37 mg/dL). Receiver operator characteristic (ROC) analysis yielded an ideal cutoff of 52 mg/dL, which classified positive and negative samples with a sensitivity and specificity of 100% (AUC = 0.998), as shown in Supplemental Fig. S10. These data have 100% positive percent agreement (PPA) and 100% negative percent agreement (NPA) with the RT-qPCR data performed on the same samples (Supplemental Table S5).
Fig. 5.
Clinical performance of saliva samples from COVID-19 patients (n=16) and healthy volunteers (n=8). (A) Bar plot of all subjects using S aptamer magnetic bead complex and (B) box and whisker plot of all data points. With a cut-off threshold of 52 mg/dL, this assay has 100% PPA and 100% NPA with RT-qPCR data.
4. Discussion
Based on the need for rapid, accurate, and easily scalable tests that can detect acute SARS-CoV-2 infection in large populations (Paltiel et al., 2020), we developed and validated a novel aptamer-based sensor (an aptasensor) capable of sensitive detection of virus in human saliva using only low-cost reagents and “detectors” that are inexpensive and already ubiquitous worldwide – glucometers. The assay design required integration and optimization of three novel components: to bind antigen in human samples, to transduce binding into signal, and to detect that signal. There were several considerations that determined the selection of an aptamer affinity reagent over more commonly used protein-based molecules such as classical, single-chain, or camelid antibodies (Chen and Yang, 2015). Aptamers are oligonucleotide (DNA or RNA) ligands selected through an iterative process known as SELEX (Ellington and Szostak, 1990; Tuerk and Gold, 1990). They share similar affinity and specificity to monoclonal antibodies, yet can be mass-produced at low cost, are stable at ambient temperatures for long-term storage, and can be chemically modified and engineered to produce conformational switches (Baker et al., 2006; Banerjee et al., 2020; Huang et al., 2020). Specifically, the amount of overlap between the aptatope and the antisense strand in this assay allows the affinity to be engineered where it is intentionally designed to have low affinity for facile release when the viral protein is present. Notably, the sequence-based design of antigen-dependent aptamer switches cannot be easily recapitulated with antibodies.
To develop a test that could be fielded for clinical use in months rather than years, we selected aptamers in the literature that had been previously validated for SARS-CoV-2 affinity. This allowed us to employ the second key element, a signal transduction and amplification step that converts viral antigen binding to signal production. Here the aptamer switch is coupled to invertase, a high-efficiency sucrose-converting enzyme that produces signal, glucose, in physiologic ranges. In this assay, we chose an invertase from S. cerevisiae that has a high turnover rate to amplify biomarkers from low concentrations to those that could then be read by a glucometer. However, this enzyme has a peak efficiency near 60 °C, which requires a heating element. While other POC tests have also used higher temperature to increase the assay kinetics (Broughton et al., 2020), there is potential to remove the heating element and to shorten time-to-answer with a room-temperature invertase from Bacillus sp. (Zhou et al., 2016) or other glucose converting enzymes.
The choice of saliva as a target specimen for testing presents challenges and opportunities. Saliva is the most readily obtained human specimen; however, it is also a complex and viscous sample consisting of various electrolytes, enzymes, and antibodies (Chauncey et al., 1954; Jasim et al., 2016) posing hurdles for specific and interference-free biomarker detection (Paltiel et al., 2020). This is reflected in our data, where we found a higher LOD for protein spiked in saliva as compared to buffer, likely a consequence of the viscosity and proteases found in saliva. While invertase is not present in saliva, glucose is, especially after eating and/or drinking, interfering with the proposed readout method. Even with fasting, there will be some background glucose in saliva which could confound the measurement. To mitigate this, we dilute the saliva sample two-fold to reduce the effect from possible interferents and take a differential measurement to account for the initial (background) glucose and “signal leakage” from the low-affinity antisense strand. This differential sampling would also account for persons with diabetic ketoacidosis (DKA). If using nasal swabs (nasopharyngeal or mid-turbinate), differential sampling would likely not be necessary.
Next, we tested the ability of the assay to detect native viral antigens during authentic cellular infection. Here we found significant differences between measurements of infected and uninfected viral media. These tests confirmed that findings determined using free antigen in buffer and saliva could be replicated with native antigens, setting the stage for clinical testing. We then tested a blinded clinical panel comprising 24 saliva samples in a BSL-3 facility. With a cutoff of 52 mg/dL determined by ROC analysis, all positive samples were discriminated from negative samples, giving a 100% level of agreement (LOA) with PCR testing and sensitivities and specificities of 100%. For each clinical sample, we correlated the mean glucose signal to patient and sample characteristics. Contrary to prior reports referencing better performance of antigen tests in specimens with high viral loads, we found no correlation (R2 < 0.2) between the Ct value obtained from PCR and the reported test, nor did we find any relationship between days since onset and testing or clinical symptoms.
While at first glance, this would seem to indicate no quantitative relationship between infectious viral load and signal level, there are key pieces of missing data. First, PCR detects circulating nucleic acid, which may be a poor proxy for infectiousness. Second, there is evidence of wide variation between nucleic acid copy number and viral antigen levels (Gry et al., 2009; Taniguchi et al., 2010), which may complicate the comparison of detected antigen and nucleic acid levels. Likewise, in this study, we explored the S protein, rather than the more commonly reported nucleocapsid protein. Each of these may have different measured correlations to viral load, and depending on site and state of infection, each may be accessible at different levels due to the presence of blocking antibodies. Even as it misses some resolved infections, salivary antigen detection has thus been suggested as more appropriate for population testing than NAATs as it may be more likely to diagnose truly infectious individuals (Ceron et al., 2020; Xu et al., 2020). A definitive study, which is beyond the scope of this work, would be to use quantitative methods to measure antigen concentration in the samples and measure intact viral genomes concomitantly with in vitro models to measure intact, infectious virions. As a proxy, we estimated the concentration of N protein in clinical samples from our controlled titrations (Supplemental Fig. S11). We found values of approximately 250–1000 pM for N protein, which are ~10–100× higher than other reports (Li et al., 2020; Torrente-Rodríguez et al., 2020); however, these had no addition of Triton or other form of sample treatment. Thus, one would expect a lower amount of free protein in non-lysed/unprocessed samples. We have found no published literature reporting protein S concentrations in saliva.
Since our test consists of readily available reagents and low-cost glucose test strips, each test has a modest $3.20 USD production cost with the current low volume production (see Supplemental Table 2). It is worth noting that nearly 70% of the cost is the magnetic beads, which could be replaced with polystyrene beads and separated with a size selection filter, possibly in a syringe, rather than magnetic separation to reduce cost. Assuming that it is viable to reduce this to below $1 USD, our approach could provide a price point needed for population screening and repeated testing in both well- and under-resourced settings. Moreover, the numeric readout can be transmitted electronically, allowing test reporting and tracking. With minor workflow improvements, a test such as this could be conducted at home, in dormitories, nursing homes, or other ambulatory settings with support for remotely-observed-testing, digital reporting, and results notification via telemedicine or a smartphone app.
To situate our assay in the landscape of available SARS-CoV-2 diagnostics, Table 1 shows a summary of reported point-of-care diagnostic tests. Here we see a diversity of approaches across the spectrum of nucleic acid and antigen modalities. However, there are notable gaps, including a paucity of saliva-based antigen tests and a complete lack of tests that provide a quantitative readout. The reported test fills this gap without sacrificing the sensitivity or specificity, using devices that already exist at scale with an easily acquired sample.
Table 1.
Comparison of point-of-care SARS-CoV-2 diagnostic tests. Unless otherwise noted, data compiled from the FDA Emergency Use Authorization (EUA) submission.
| Test | Target | Sample | Method | Device (readout) | Time (min) | LOD | Cost/Test (USD) | Complexity (Labor) | PPA (n) | NPA (n) |
|---|---|---|---|---|---|---|---|---|---|---|
| Nucleic Acid Amplification Tests (NAAT) | ||||||||||
| ID Now (Abbott) | ORF1ab gene | NP, NS | RT-NEAR | ID Now | 13 | 125 copies/mL | 40–60 | Moderate (6 steps) | 100% (30) | 100% (30) |
| Xpert Xpress (Cepheid) | E, N genes | NP, NS, MT, OP | RT-PCR | GeneXpert Dx | 30 | 250 copies/mL | 20 | Moderate (6 steps) | 97.8% (46) | 95.6% (44) |
| Cue (Cue Health) | N gene | NS | Isothermal PCR | Cue Health Monitoring System | 25 | 1300 copies/mL | 10 | Moderate (5 steps) | 98.8% (81) | 95.6% (68) |
| DETECTR (Mammoth) Broughton et al. (2020) |
E, N genes | NP, OP | LFIA w\ RT-LAMP and CAS-12 | None – visual | 45 | 10,000 copies/mL | 5 | High (multistep) | 95% (36) | 100% (42) |
| SHERLOCK (Sherlock Biosciences) Patchsung et al., (2020) |
E, N, ORF1ab genes | LFIA w\ RT-RPA and CAS-13a | None – visual | 70 | 210 copies/mL | 30 | High (multistep) | 100% (81) | 97% (73) | |
| Rapid Antigen Tests | ||||||||||
| Veritor (BD) | N protein | NS | Chromatographic immunoassay | Veritor Plus Analyzer | 15 | 140 TCID50/swab | 35 | Moderate (5 steps) | 83.9% (31) | 100% (195) |
| LumiraDx (LumiraDx) | N protein | NS | Fluorescence immunoassay | LumiraDx Instrument | 12 | 32 TCID50/swab | 160 | Moderate (5 steps) | 97.6% (83) | 96.6% (174) |
| Sofia 2 (Quidel) | N protein | NP, NS | LFIA | Sofia 2 Fluorescent Analyzer | 15 | 113 TCID50/swab | 20 | Moderate (6 steps) | 96.7% (30) | 100% (179) |
| BinaxNow (Abbott) | N protein | NS | LFIA | None – visual | 15 | 22.5 TCID50/swab | 25 | Low (3 steps) | 97.1% (35) | 98.5% (67) |
| Biocredit (Rapigen) Weitzel et al., (2020) |
N protein | NP | LFIA | None – visual | 8 | N/A | 20 | Low (3 steps) | 62% (79) | 100% (30) |
| Respi-Strip (Coris Bio) Mertens et al., (2020) |
N protein | NP | Dipstick LFIA | None – visual | 15–30 | 5.4 pM | 5 | Low (3 steps) | 87.9% (99) | 77.3% (132) |
| This Work | S and N protein | OP | Aptamer-based competitive assay | Glucometer | 60 | 6.31 pM [S] 5.27 pM [N] |
3.2* | Low (4 steps) | 100% (16) | 100% (8) |
NS = Nasal sample, NP = Nasopharyngeal swab, MT = mid-turbinate swab, OP = oropharyngeal swab, * Cost of goods.
5. Study limitations
Our study has limitations. First, we selected aptamers based on the literature rather than using SELEX to develop bespoke aptamers. Even so, the aptamers chosen in this work demonstrated high affinity and specificity for SARS-CoV-2 antigens and have the advantage of prior validation and peer-reviewed reporting. Second, for testing of binding during authentic viral transmission, it should be noted that we did not attempt to lyse the virions (i.e. no detergent was added), potentially leaving a complex mixture of intact virus, partially assembled virion, and protein which may or may not be recognized due to association with neutralizing antibodies. Third, our clinical dataset was relatively small and was conducted retrospectively. Larger prospective studies are needed to establish the true LOD, ROC characteristics, and correlation to clinical characteristics such as symptom status, viral load, and infectivity. Fourth, at higher antigen concentrations this assay showed increased variability. Likewise, we noted more than expected variability in clinical saliva specimens. We attribute this to the heterogeneity of saliva, which could cause differences in the amount of functional sample assayed, as well as potential carryover and signal leakage occurring from magnetic bead transfer or non-specific enzyme release. Finally, our assay was performed with only minimal sample preparation and only preliminary optimizations in assay conditions and workflow. Finding optimal antigen-aptamer binding conditions, including the addition of detergents, optimization of enzyme selection and reaction conditions to accommodate faster, low-temperature testing, and lyophilization to increase shelf life, would each need to be addressed prior to testing at scale or approval for clinical use.
6. Outlook
We developed, validated, and tested a novel SARS-CoV-2 biosensor that can sensitively and specifically detect acute viral infection from human saliva using low-cost reagents and widespread commercially available glucometers. The detection method could be automated by performing the assay in a syringe or multi-compartment tube with a size exclusion-based approach. Additionally, the assay could be integrated with Bluetooth wireless-enabled software technology for easy “sample to answer” reporting of SARS-CoV-2 infection. Our preliminary results suggest such an approach could be used at scale for repeated population screening and diagnosis, however, prospective clinical trials are needed to determine assay performance across a range of clinical contexts. As discussed by Paltiel and Walensky (“Screening To Prevent SARS-CoV-2 Outbreaks,” n. d.), we propose that such rapid, saliva-based antigen testing can be the “essential weapon in the fight to resume many of the activities and reopen many of the venues that comprise what we used to call normal life.”
CRediT authorship contribution statement
Naveen K. Singh: Conceptualization, Investigation, Formal analysis. Partha Ray: Conceptualization, Investigation, Formal analysis. Aaron F. Carlin: Investigation. Celestine Magallanes: Resources. Sydney C. Morgan: Resources. Louise C. Laurent: Resources, Supervision. Eliah S. Aronoff-Spencer: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition. Drew A. Hall: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported in part by the National Institutes of Health (NIH) Rapid Acceleration of Diagnostics (RADx) program (3U54EB027690-03S1), a Career Award for Medical Scientists from the Burroughs Wellcome Fund to A.F.C., and a National Science Foundation (NSF) CAREER Award to D.A.H. (ECCS-1454608). We thank the UCSD Center for AIDS Research Genomics and Sequencing Core and support from the John and Mary Tu Foundation for ddPCR SARS-CoV-2 quantification. We thank Efren Sandoval, Aakash Amin, and David Becker at Helix for performing the SARS-CoV-2 RNA quantification on the clinical samples. The following reagent was deposited by the Centers for Disease Control and Prevention (CDC) and obtained through BEI Resources, NIAID, NIH: SARS-related Coronavirus 2, Isolate USA-WA1/2020, NR-52281. We thank Dr. Robert Schooley for valuable feedback and the volunteers who provided samples.
References
- 2019-nCoV Coronavirus Nucleocapsid Elisa Kit KIT40588 | Sino biological. https://www.sinobiological.com/elisa-kits/cov-nucleocapsid-kit40588 [WWW Document], n.d. URL. (accessed 7.12.20)
- 2019-nCoV Coronavirus spike Elisa Kit KIT40592 | Sino biological. https://www.sinobiological.com/elisa-kits/cov-spike-kit40592 [WWW Document], n.d. URL. (accessed 7.12.20)
- Afzal A. J. Adv. Res. S2090123220301788. 2020 [Google Scholar]
- Arevalo-Rodriguez I., Buitrago-Garcia D., Simancas-Racines D., Zambrano-Achig P., del Campo R., Ciapponi A., Sued O., Martinez-Garcia L., Rutjes A., Low N., Bossuyt P.M., Perez-Molina J.A., Zamora J. FALSE-NEGATIVE results OF initial rt-pcr assays for COVID-19: a SYSTEMATIC REVIEW (preprint) Infectious Diseases (except HIV/AIDS) 2020 doi: 10.1371/journal.pone.0242958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B.R., Lai R.Y., Wood M.S., Doctor E.H., Heeger A.J., Plaxco K.W. J. Am. Chem. Soc. 2006;128:3138–3139. doi: 10.1021/ja056957p. [DOI] [PubMed] [Google Scholar]
- Banerjee S., Yoon H., Yebra M., Tang C.-M., Gilardi M., Narayanan J.S.S., White R.R., Sicklick J.K., Ray P. Mol. Canc. Therapeut. 2020;19:1173–1182. doi: 10.1158/1535-7163.MCT-19-0959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.-Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cdc International locations with confirmed COVID-19 cases. Cent. Dis. Control Prev. 2020 https://www.cdc.gov/coronavirus/2019-ncov/global-covid-19/world-map.html URL. (accessed 4.30.20) [Google Scholar]
- Cdc Coronavirus disease 2019 (COVID-19) 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/testing-in-us.html [WWW Document]. Cent. Dis. Control Prev. URL. (accessed 5.17.20)
- Cdc Information for laboratories about coronavirus (COVID-19) Cent. Dis. Control Prev. 2020 https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html URL. (accessed 9.14.20) [Google Scholar]
- Cdc . 2020. National Diabetes Statistics Report.https://www.cdc.gov/diabetes/library/features/diabetes-stat-report.html 2020 [WWW Document]. Cent. Dis. Control Prev. URL. (accessed 12.21.20) [Google Scholar]
- Ceron J., Lamy E., Martinez-Subiela S., Lopez-Jornet P., Capela-Silva F., Eckersall P., Tvarijonaviciute A. J. Clin. Med. 2020;9:1491. doi: 10.3390/jcm9051491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauncey H.H., Lionetti F., Winer R.A., Lisanti V.F. J. Dent. Res. 1954;33:321–334. doi: 10.1177/00220345540330030501. [DOI] [PubMed] [Google Scholar]
- Chen A., Yang S. Biosens. Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- Chen Z., Wu Q., Chen J., Ni X., Dai J. Virol. Sin. 2020;1–4 doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CSSEGISandData . 2020. CSSEGISandData/COVID-19. [Google Scholar]
- Diabetes Statistics https://www.diabetesresearch.org/diabetes-statistics [WWW Document], n.d. URL. (accessed 5.17.20)
- Dong E., Du H., Gardner L. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y., Hughes R.A., Bhadra S., Jiang Y.S., Ellington A.D., Li B. Sci. Rep. 2015;5:11039. doi: 10.1038/srep11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Granger D.A., Johnson S.B., Szanton S.L., Out D., Schumann L.L. Biol. Res. Nurs. 2012;14:347–356. doi: 10.1177/1099800412443892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gry M., Rimini R., Strömberg S., Asplund A., Pontén F., Uhlén M., Nilsson P. BMC Genom. 2009;10:365. doi: 10.1186/1471-2164-10-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Lu B., Dong Q., Tang Y., Du Y., Li B. Anal. Chim. Acta. 2020;1106:191–198. doi: 10.1016/j.aca.2020.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.-C., Ray P., Chan M., Zhou X., Hall D.A. Bioengineering; 2020. An Aptamer-Based Magnetic Flow Cytometer Using Matched Filtering (Preprint) [DOI] [PubMed] [Google Scholar]
- Jasim H., Olausson P., Hedenberg-Magnusson B., Ernberg M., Ghafouri B. Sci. Rep. 2016;6:39073. doi: 10.1038/srep39073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E.H., Forman H.P. JAMA Health Forum. 2020;1 doi: 10.1001/jamahealthforum.2020.0565. [DOI] [PubMed] [Google Scholar]
- Klonoff D.C., Parkes J.L., Kovatchev B.P., Kerr D., Bevier W.C., Brazg R.L., Christiansen M., Bailey T.S., Nichols J.H., Kohn M.A. Diabetes Care. 2018;41:1681–1688. doi: 10.2337/dc17-1960. [DOI] [PubMed] [Google Scholar]
- Kubina R., Dziedzic A. Diagnostics. 2020;10:434. doi: 10.3390/diagnostics10060434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T., Xiang Y., Lu Y. Methods Mol. Biol. Clifton NJ. 2015;1256:99–109. doi: 10.1007/978-1-4939-2172-0_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan T., Zhang J., Lu Y. Biotechnol. Adv., Trends in In Vitro Diagnostics and Mobile Healthcare. 2016;34:331–341. [Google Scholar]
- Li T., Wang L., Wang H., Li X., Zhang S., Xu Y., Wei W. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens P., De Vos N., Martiny D., Jassoy C., Mirazimi A., Cuypers L., Van den Wijngaert S., Monteil V., Melin P., Stoffels K., Yin N., Mileto D., Delaunoy S., Magein H., Lagrou K., Bouzet J., Serrano G., Wautier M., Leclipteux T., Van Ranst M., Vandenberg O. Front. Med. 2020;7 doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnana M., Caputo M., Giavarina D., Lippi G. Clin. Chim. Acta. 2009;402:7–13. doi: 10.1016/j.cca.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Morens D.M., Fauci A.S. Cell. 2020;182:1077–1092. doi: 10.1016/j.cell.2020.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallett S.J.C., Rayment M., Patel A., Fitzgerald-Smith S.A.M., Denny S.J., Charani E., Mai A.L., Gilmour K.C., Hatcher J., Scott C., Randell P., Mughal N., Jones R., Moore L.S.P., Davies G.W. Lancet Respir. Med. 2020;8:885–894. doi: 10.1016/S2213-2600(20)30315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltiel A.D., Zheng A., Walensky R.P. medRxiv. 2020 2020.07.06.20147702. [Google Scholar]
- Patchsung M., Jantarug K., Pattama A., Aphicho K., Suraritdechachai S., Meesawat P., Sappakhaw K., Leelahakorn N., Ruenkam T., Wongsatit T., Athipanyasilp N., Eiamthong B., Lakkanasirorat B., Phoodokmai T., Niljianskul N., Pakotiprapha D., Chanarat S., Homchan A., Tinikul R., Kamutira P., Phiwkaow K., Soithongcharoen S., Kantiwiriyawanitch C., Pongsupasa V., Trisrivirat D., Jaroensuk J., Wongnate T., Maenpuen S., Chaiyen P., Kamnerdnakta S., Swangsri J., Chuthapisith S., Sirivatanauksorn Y., Chaimayo C., Sutthent R., Kantakamalakul W., Joung J., Ladha A., Jin X., Gootenberg J.S., Abudayyeh O.O., Zhang F., Horthongkham N., Uttamapinant C. Nat. Biomed. Eng. 2020:1–10. doi: 10.1038/s41551-020-00603-x. [DOI] [PubMed] [Google Scholar]
- Reddy A., Maley F. J. Biol. Chem. 1996;271:13953–13958. doi: 10.1074/jbc.271.24.13953. [DOI] [PubMed] [Google Scholar]
- Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. J. Clin. Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swab [WWW Document], n.d. URL/do/10.1377/hblog20200909.430047/full (accessed 9.19.20).
- Shrivastava A., Gupta V. Chronicles Young Sci. 2011;2:21. [Google Scholar]
- Song, Y., Song, J., Wei, X., Huang, M., Sun, M., Zhu, L., Lin, B., Shen, H., Zhu, Z., Yang, C., 2020.
- Taebi S., Keyhanfar M., Noorbakhsh A. J. Immunol. Methods. 2018;458:26–32. doi: 10.1016/j.jim.2018.04.001. [DOI] [PubMed] [Google Scholar]
- Tahamtan A., Ardebili A. Expert Rev. Mol. Diagn. 2020;20:453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Torres Edejer T., Hanssen O., Mirelman A., Verboom P., Lolong G., Watson O.J., Boulanger L.L., Soucat A. 2020. Lancet Glob. Health S2214109X20303831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, Y., Choi, P.J., Li, G.-W., Chen, H., Babu, M., Hearn, J., Emili, A., Xie, X.S., 2010. 329, 8. [DOI] [PMC free article] [PubMed]
- To K.K.-W., Tsang O.T.-Y., Leung W.-S., Tam A.R., Wu T.-C., Lung D.C., Yip C.C.-Y., Cai J.-P., Chan J.M.-C., Chik T.S.-H., Lau D.P.-L., Choi C.Y.-C., Chen L.-L., Chan W.-M., Chan K.-H., Ip J.D., Ng A.C.-K., Poon R.W.-S., Luo C.-T., Cheng V.C.-C., Chan J.F.-W., Hung I.F.-N., Chen Z., Chen H., Yuen K.-Y. Lancet Infect. Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromberg B.J., Schwetz T.A., Pérez-Stable E.J., Hodes R.J., Woychik R.P., Bright R.A., Fleurence R.L., Collins F.S. N. Engl. J. Med. 2020;383:1071–1077. doi: 10.1056/NEJMsr2022263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Weitzel T., Legarraga P., Iruretagoyena M., Pizarro G., Vollrath V., Araos R., Munita J.M., Porte L. bioRxiv. 2020 doi: 10.7717/peerj.10801. 2020.05.27.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-L., Tseng W.-P., Lin C.-H., Lee T.-F., Chung M.-Y., Huang C.-H., Chen S.-Y., Hsueh P.-R., Chen S.-C. J. Infect. 2020;81:435–442. doi: 10.1016/j.jinf.2020.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Lu Y. Nat. Chem. 2011;3:697–703. doi: 10.1038/nchem.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Cui B., Duan X., Zhang P., Zhou X., Yuan Q. Int. J. Oral Sci. 2020;12:1–6. doi: 10.1038/s41368-020-0080-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Yang M., Shen C., Wang F., Yuan J., Li Jinxiu, Zhang M., Wang Z., Xing L., Wei J., Peng L., Wong G., Zheng H., Liao M., Feng K., Li Jianming, Yang Q., Zhao J., Zhang Z., Liu L., Liu Y. medRxiv. 2020 2020.02.11.20021493. [Google Scholar]
- Zhang L., Gu C., Ma H., Zhu L., Wen J., Xu H., Liu H., Li L. Anal. Bioanal. Chem. 2019;411:21–36. doi: 10.1007/s00216-018-1361-7. [DOI] [PubMed] [Google Scholar]
- Zhang J., Lan T., Lu Y. Trends Anal. Chem. TRAC. 2020;124 doi: 10.1016/j.trac.2019.115782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., He L., Gao Y., Han N., Zhang R., Wu Q., Li J., Tang X., Xu B., Ding J., Huang Z. Sci. Rep. 2016;6:32081. doi: 10.1038/srep32081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., Guo Q., Song T., He J., Yen H.-L., Peiris M., Wu J. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]