Abstract
Objectives
To compare the Lumipulse® SARS-CoV-2 antigen test with the gold standard real-time reverse transcription-polymerase chain reaction (RT-PCR) for diagnosis of SARS-CoV-2 infection and to evaluate its role in screening programs.
Methods
Lumipulse® SARS-CoV-2 antigen assay was compared with the gold standard RT-PCR test in a selected cohort of 226 subjects with suspected SARS-CoV-2 infection, and its accuracy was evaluated. Subsequently, the test was administered to a real-life screening cohort of 1738 cases. ROC analysis was performed to explore test features and cutoffs. All tests were performed in the regional reference laboratory in Umbria, Italy.
Results
A 42.0% positive result at RT-PCR was observed in the selected cohort. The Lumipulse® system showed 92.6% sensitivity (95% CI 85.4–97.0%) and 90.8% specificity (95% CI 84.5–95.2%) at 1.24 pg/mL optimal cutoff. In the screening cohort, characterized by 5.2% prevalence of infection, Lumipulse® assay showed 100% sensitivity (95% CI 96.0–100.0%) and 94.8% specificity (95% CI 93.6–95.8%) at 1.645 pg/mL optimal cutoff; the AUC was 97.4%, NPV was 100% (95% CI 99.8–100.0%) and PPV was 51.1% (95% CI 43.5–58.7%).
Conclusions
The Lumipulse® SARS-CoV-2 antigen assay can be safely employed in the screening strategies in small and large communities and in the general population.
Keywords: Lumipulse®, Antigen NP, COVID-19, SARS-CoV-2, Diagnosis, Screening
Introduction
In December 2019, several cases of unknown-etiology pneumonia in Wuhan City, Hubei Province, were reported by the Chinese Health Authority (Lu et al., 2020). Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) was identified on 07 January 2020 (Hui et al., 2020) as the etiologic agent of this disease. On 30 January, the World Health Organization declared SARS-CoV-2 outbreaks a Public Health Emergency of International Concern (Burki, 2020a) and on 11 March declared the Coronavirus Disease 2019 (COVID-19) pandemic. To date, more than 107 million people and more than 2.4 million deaths have been reported globally (WHO, 2020b).
The current gold standard for COVID-19 microbiological diagnosis is the detection of SARS-CoV-2 genetic targets in respiratory samples using molecular real-time reverse transcription-polymerase chain reaction (RT-PCR) tests (WHO, 2020a). The same test is used for epidemiological surveillance of the infection, aiming to contain virus spread in the community. As individuals before symptom onset and those who never develop symptoms can be highly contagious (Huff and Singh, 2020), extensive community testing is considered to be one of the cornerstones of control strategies of the spread of the infection. Despite the high sensitivity and specificity, RT-PCR suffers from a series of limitations, such as: the long time it takes to be performed, the need for dedicated equipment and highly specialized laboratory technicians, and high costs. Hence, there is a pressing need to introduce new diagnostic technologies that are equally reliable, but at the same time rapid, easily suitable for laboratory work-flow, and economically advantageous (ECDC, 2020a) for screening strategies based on extensive community testing.
The Lumipulse® SARS-CoV-2 antigen assay (Fujirebio, Inc., Tokyo, Japan) automated test was recently introduced into the market. The system, capable of processing up to 120 samples per hour, is widely used in Japan for COVID-19 surveillance; it obtained the European CE-IVD mark for in vitro diagnostic use in August 2020. The system is based on chemiluminescence enzyme immunoassay (CLEIA) technology, capable of detecting and quantitatively estimating the presence of SARS-CoV-2 nucleocapsid protein (NP) in nasopharyngeal swabs or saliva. Hirotsu et al. recently demonstrated that it can successfully identify SARS-CoV-2-infected patients with moderate-to-high viral load (Hirotsu et al., 2020b).
The present study evaluated the possible role of the Lumipulse® SARS-CoV-2 antigen assay in selected communities (e.g. health professionals, schools, residential and nursing home for the elderly) or population-wide screening for SARS-CoV-2 infection. To this aim, the antigen assay was first evaluated on a small high-prevalence selected series of 226 nasopharyngeal swabs (selected cohort), sent to the reference laboratory of the Umbria Italian Region (coverage about 870,000 people) at the start of the second epidemic wave and analyzed by RT-PCR. Subsequently, the antigen test was administered to a second unselected cohort (screening cohort) comprising 1738 swabs from real-life screening scenarios (e.g. schools, hospital healthcare workers, and other communities), and the results were compared with those of RT-PCR.
Materials and methods
Samples
The Lumipulse® assay was first tested on a selected series of 226 nasopharyngeal swabs (selected cohort) analyzed 10–15 September 2020 at the Microbiology Unit of the Santa Maria della Misericordia Perugia General Hospital, which is the Umbria Regional Reference Laboratory for SARS-CoV-2 diagnosis. Selection of the swabs to be evaluated with the antigen assay was performed in order to include a large number of samples in the study – with one or more target genes detected, with high variability of Ct, as a proxy of viral load – to be able to evaluate the test in terms of sensitivity and specificity with adequate numbers of RT-PCR-positive and RT-PCR-negative cases. Samples from both symptomatic and close contact individuals were included, while samples from patients already diagnosed with SARS-CoV-2 infection were excluded. Subsequently, over the period 1–26 January 2021, the Lumipulse® assay was employed in real-life screening strategies in a second unselected cohort (screening cohort) of 1738 swabs collected in schools, prisons, elderly care homes, and from hospital healthcare worker surveillance programs. All swabs were also analyzed by RT-PCR test.
Testing
In the selected cohort, all samples were first tested for SARS-CoV-2 by RT-PCR, stored at 4 °C, and then analyzed with the Lumipulse® system within 24 h. In the screening cohort, samples were analyzed with the antigen test, and soon after by RT-PCR. Swabs were collected in Universal Transport Medium (UTM, Copan, Brescia, Italy). For RT-PCR, samples were analyzed by the Allplex™ SARS-CoV-2 assay (Seegene, Seoul, South Korea). The test was performed according to the manufacturer’s instructions, using 300 μL of UTM and 10 μL of the provided internal control (IC). The envelope (E) gene (specific of the subgenus Sarbecovirus), the nucleocapsid gene (N), and the RNA-dependent-RNA-polymerase (RdRP) gene (both specific of the SARS-CoV-2) were the target genes. The assay was considered valid if the Ct value of the IC was ≤40.
For the antigen test, after removing the swabs, UTM tubes were centrifuged at 1400 × g for 10 min, and loaded on the Lumipulse® G1200 automated immunoassay analyzer (Fujirebio) to measure the NP antigen level with the Lumipulse® SARS-CoV-2 antigen kit (Fujirebio) following the manufacturer’s instruction. Briefly, the treatment solution and the sample were mixed and the mixture was dispensed into the anti-SARS-CoV-2 antigen monoclonal antibody-coated magnetic particle solution and then incubated for 10 min at 37 °C. After the first wash step, alkaline phosphatase-conjugated anti-SARS-CoV-2 antigen monoclonal antibody was added and incubated again for 10 min at 37 °C. After another wash step, the substrate solution was added, incubated for 5 min at 37 °C, and the amount of NP antigen (pg/mL) in the samples was determined in relation to the obtained luminescent signal.
Statistical analysis
The results obtained were analyzed using Stata Statistical Software (Release 16.1, College Station, Houston, TX: StataCorp LLC). Regarding the molecular test, analysis was performed using the cutoff of 35 Ct to discriminate samples positive for SARS-CoV-2 (≤35 for at least one of the target genes detected) from negative (>35 for all target genes detected), based on the accepted notion that subjects with SARS-CoV-2-positive samples with Ct >35 were not contagious (Binnicker, 2020, Bullard et al., 2020, Gupta, 2020, Singanayagam et al., 2020, Tom and Mina, 2020). Moreover, the cutoff was inferred by the Italian Health Ministerial Circular no. 9774 of 20 March 2020, stating that “confirmation tests should be performed only for samples in which the result is difficult to interpret or the Ct in RT-PCR is greater than 35. In these cases it is recommended to repeat the test on a new sample" (Ministero della Salute, 2020).
The following cutoff values were considered for the antigen assay: 1.340 pg/mL, suggested by the manufacturer to discriminate samples positive for SARS-CoV-2 antigen NP from negative samples, and 1.240 pg/mL and 1.645 pg/mL to optimize sensitivity and specificity of the test in the selected and screening cohort, respectively. Optimal cutoffs were obtained by the Youden approach (area under ROC curve at cutoff 0.92 in selected cohort and 0.97 in screening cohort) and were also confirmed by the Liu approach (Fluss et al., 2005, Liu, 2012, Youden, 1950). The 1.645 pg/mL screening cohort cutoff was bootstrapped to estimate 95% confidence intervals (100 replications).
Test sensitivity, specificity, ROC area, positive likelihood ratio (LR+) and negative likelihood ratio (LR-), odds ratio (LR+/LR-), positive and negative predicted value (PPV and NPV), prior and posterior probability (Odds) with 95% CI were calculated. To calculate PPV and NPV in the 2.5% low-prevalence scenario, estimated by an Italian National seroprevalence study performed in June 2020 (ISTAT (National Institute of Statistics, Italy), 2021)), values of sensitivity and specificity of the screening cohort that were unselected and larger than the selected cohort were used.
Results
Evaluation of Lumipulse® antigen assay in the selected and real-life screening cohort
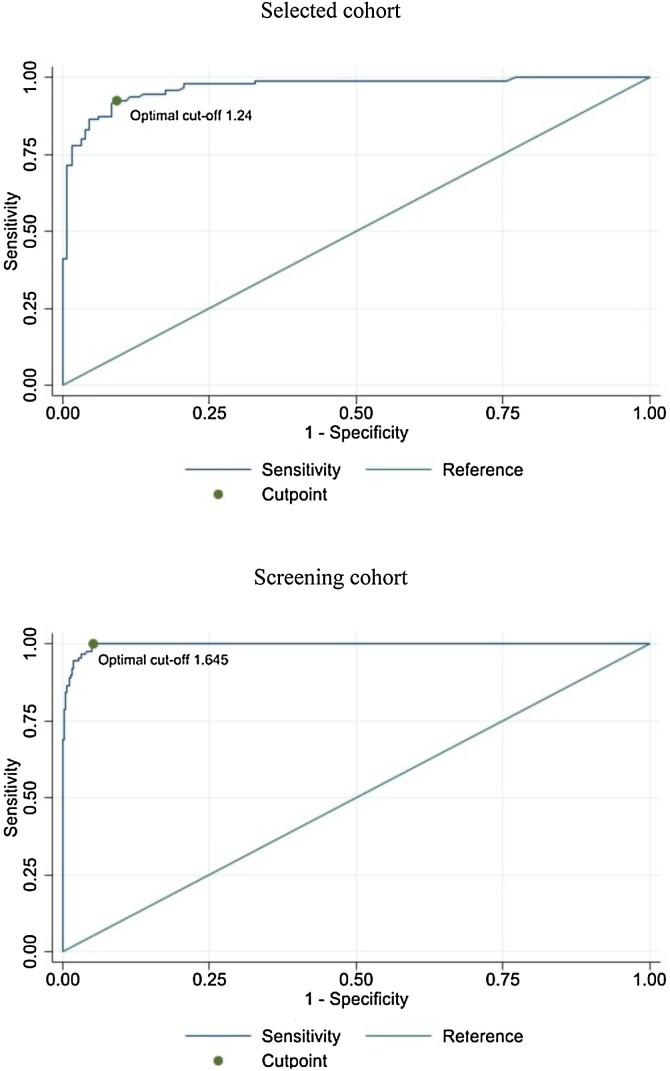
In the selected cohort, among 226 nasopharyngeal swabs, 116 (51.3%) were negative for all target genes, 95 (42.0%) were positive for 1, 2, or 3 target genes, with Ct ≤5. The other 15 (6.7%) samples were positive for one or more target genes with Ct >35 and were considered as negative. The median Ct for positive samples was 29. Table 1 compares RT-PCR results with those obtained by the Lumipulse® assay. Positive or negative samples for the NP antigen were discriminated according to the manufacturer cutoff of 1.340 pg/mL or the optimal cutoff of 1.240 pg/mL, obtained as described in the Material and Methods section. At a 1.340 pg/mL cutoff value, RT-PCR and antigen assay overall agreement was 91.2% (206/226 samples), and area under the curve (AUC) was 91.1%. Sensitivity and specificity were 90.5% and 91.6%, respectively, and LR + reached 10.8. NPV and PPV were 93.0% and 88.7%, respectively. At the optimal cutoff of 1.240 pg/mL, sensitivity and NPV increased to 92.6% and 94.4%, respectively (Table 2 ). Overall agreement raised to 91.6% (207/226 samples) and AUC to 91.7% (Figure 1 ). Specificity, PPV, and LR + decreased to 90.8%, 88.0%, and 10.1, respectively (Table 1 and Figure 2 ).
Table 1.
Comparison of RT-PCR and antigen assay results in the selected cohort of 226 nasopharyngeal swabs (A) and in the real-time screening cohort of 1738 swabs (B), according to different cutoff values to discriminate positive and negative samples.
| RT-PCR (35 Ct cutoff) |
|||||
|---|---|---|---|---|---|
| A | Antigen assay cutoff (ng/mL) | Positive (%) | Negative (%) | Total (%) | |
| Selected cohort | 1.340 | Positive | 86 (38.0) | 11 (5.0) | 97 (43.0) |
| Negative | 9 (4.0) | 120 (53.0) | 129 (57.0) | ||
| Total | 95 (42.0) | 131 (58.0) | 226 (100.0) | ||
| 1.240 | Positive | 88 (38.9) | 12 (5.3) | 100 (44.2) | |
| Negative | 7 (3.1) | 119 (52.7) | 126 (55.8) | ||
| Total | 95 (42.0) | 131 (58.0) | 226 (100.0) | ||
| RT-PCR (35 Ct cutoff) |
|||||
|---|---|---|---|---|---|
| B | Antigen assay cutoff (ng/mL) | Positive (%) | Negative (%) | Total (%) | |
| Screening cohort | 1.340 | Positive | 90 (5.2) | 130 (7.5) | 220 (12.7) |
| Negative | 0 (0.0) | 1518 (87.3) | 1518 (87.3) | ||
| Total | 90 (5.2) | 1648 (94.8) | 1738 (100.0) | ||
| 1.645 | Positive | 90 (5.2) | 86 (4.9) | 176 (10.1) | |
| Negative | 0 (0.0) | 1562 (89.9) | 1562 (89.9) | ||
| Total | 90 (5.2) | 1648 (94.8) | 1738 (100.0) | ||
Table 2.
Evaluation of Lumipulse® antigen assay with 1.240 pg/mL and 1.645 pg/mL optimal cutoffs on selected cohort and screening cohort, respectively.
| Selected cohort | Screening cohort | |
|---|---|---|
| Prevalence | 42% | 5.2% |
| ROC curve’s AUC at cutoff | 91.7% (88.1–95.4) | 97.4% (96.9–97.9) |
| Sensitivity | 92.6% (85.4–97.0) | 100% (96–100) |
| Specificity | 90.8% (84.5–95.2) | 94.8% (93.6–95.8) |
| LR+ | 10.1 (5.88–17.4) | 19.2 (15.6–23.5) |
| LR- | 0.0811 (0.0397–0.166) | 0 |
| Odds Ratio | 125 (47.7–325) | n. a.a |
| PPV | 88.0% (80.0–83.6) | 51.1% (43.5–58.7) |
| NPV | 94.4% (88.9–97.7) | 100% (99.8–100) |
Values in brackets refer to 95% CI, when applicable. AUC: area under the curve; LR-: negative likelihood ratio; LR+: positive likelihood ratio; NPV: negative predictive value; PPV: positive predictive value; ROC: receiver operating characteristic
n. a.: not applicable.
Figure 1.
Estimated optimal cutoff and ROC curve’s AUC for selected cohort and screening cohort.
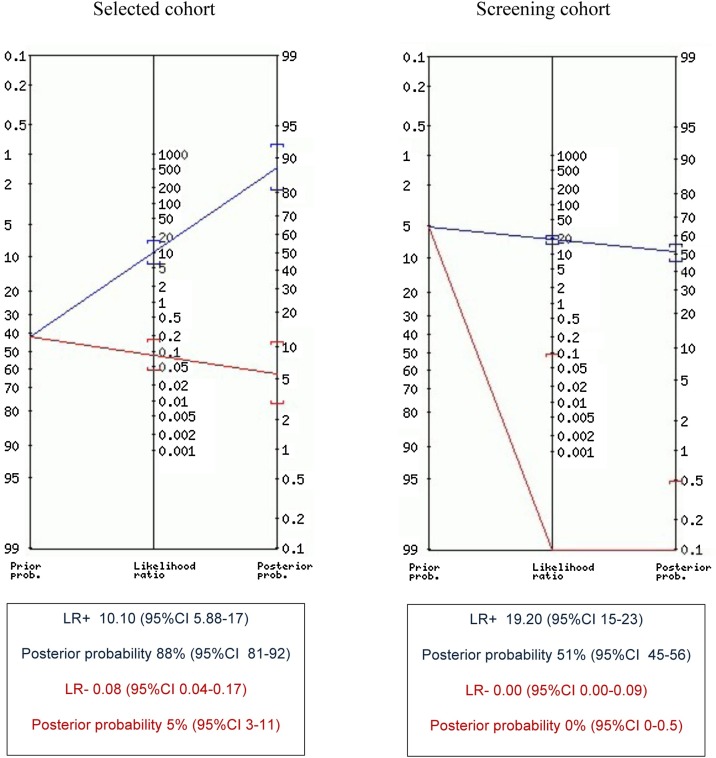
Figure 2.
Positive (blue) and negative (red) likelihood ratio, prior and posterior probability positive (blue) and negative (red), calculated at 1.240 pg/mL optimal cutoff for selected cohort and at 1.645 pg/mL optimal cutoff for screening cohort.
Among 1738 nasopharyngeal swabs, 1644 (94.6%) were negative for all target genes, 90 (5.2%) were positive for one, two, or three target genes with Ct ≤35. The remaining four (0.2%) samples were positive for one or more target genes with Ct >35 and were considered as negative. The median Ct for positive samples was 22. Table 1 compares RT-PCR and antigen assay results, according to the manufacturer cutoff of 1.340 pg/mL or the optimal cutoff of 1.645 pg/mL, obtained for this specific cohort, as described above (Figure 1). This optimal cutoff was not statistically different from that calculated for the selected cohort (95% CI 0.69–2.59 pg/mL). At the 1.340 pg/mL cutoff value, overall agreement was 92.5% (1608/1738), AUC was 96.1%; sensitivity and specificity were 100% and 92.1%, respectively; LR + was 12.7; and NPV and PPV were 100% and 40.9%, respectively. At 1.645 pg/mL optimal cutoff, overall agreement reached 95.1% (1652/1738), AUC was 97.4% (Figure 1); sensitivity and NPV were 100%; specificity, PPV, and LR + increased to 94.8%, 51.1%, and 19.2, respectively (Table 2 and Figure 2).
Performance of Lumipulse® antigen assay in hypothetical and real-life low-prevalence scenarios
The performance of the test in a 2.5% prevalence scenario, calculated on sensitivity and specificity (100% and 94.8%, respectively) of the screening cohort that was unselected and larger than the selected cohort, was estimated to be 100% NPV (95% CI 99.8–100%) and 32.9% PPV (95% CI 28.6–37.6%).
Assuming to test all samples with antigen concentrations >1.645 pg/mL with RT-PCR to confirm infection, in a scenario with a prevalence of 2.5%, the estimated posterior positive probability was 33%, indicating that eight out of 100 samples tested for the NP antigen should have been evaluated by RT-PCR. Of the eight samples, two would be positive for SARS-CoV-2 at the molecular test and six would be negative.
By the same assumption, 176 samples with antigen concentrations >1.645 pg/mL would have been evaluated by RT-PCR and 86/176 (48.9%) would be negative for SARS-CoV-2 infection in the screening cohort. To optimize the use of the Lumipulse® antigen test and reduce the number of samples to be confirmed by RT-PCR in routine laboratory practice, the cutoff value was searched for optimal specificity and LR + of the assay based on the screening cohort (i.e. a cutoff associated with a very high probability of a true positive result). The best LR+ (320.5) was found at the cutoff value of 10.4 pg/mL, with a specificity of 99.8%. According to this cutoff, the RT-PCR test could have been avoided for 74/176 samples in this screening cohort, with a reduction of 42% RT-PCR tests.
Discussion and conclusions
The main result of this study is that Lumipulse® SARS-CoV-2 antigen assay, compared with RT-PCR from nasopharyngeal swabs, showed an excellent NPV for the presence of SARS-CoV-2 infection both in high- and low-prevalence scenarios. This result supports the use of this assay in selected high-risk communities and for community and population screening purposes. In the low-prevalence screening cohort, concordance between Lumipulse® and RT-PCR was ≥94.8% at the 1.645 optimal cutoff and LR + was 19.2, a figure which is excellent according to the Deeks and Altman classification (Deeks and Altman, 2004). A sensitivity of ≥92.5% was found in both the high-prevalence selected cohort and low-prevalence screening cohort, which was much higher than the 55.2% found by Hirotsu et al. (Hirotsu et al., 2020b). The difference could be explained by the fact that the study by Hirotsu et al. was performed in a population of hospitalized patients, mostly with low viral load already known to be infected with SARS-CoV-2. Moreover, many patients studied by Hirotsu et al. were probably in the late phase of the infection. Since the current study was aimed at evaluating the performance of the test for screening purposes, it considered a population of non-hospitalized subjects with no laboratory evidence of previous SARS-CoV-2 infection. It found a median Ct value of 29 in the selected cohort and 22 in the screening cohort, which suggests a viral load higher than that of Hirotsu et al. (Hirotsu et al., 2020b). The current results are in accordance with the optimal correlation between RT-PCR and the Lumipulse® antigen test found in the early phase of the infection, characterized by high viral load (Hirotsu et al., 2020a, Hirotsu et al., 2020b). The performance of the antigen test was better in the screening cohort than in the selected one, which may reflect a high prevalence of early cases with high viral load diagnosed at screening during a phase of rapid increase of SARS-CoV-2 spread in this regional population. In addition, results from a semi-quantitative RT-PCR method, like the one used in this study, cannot be directly compared with those obtained with the quantitative method used by Hirotsu et al., especially in cases of low viral load in the late phase of infection (Yu et al., 2020).
As expected, specificity was lower than that found in the validation study of 99.6% (Hirotsu et al., 2020b). Test specificity of 90.8% and 94.8%, according to the study best cutoffs, was found for the selected and screening study cohorts, respectively (Figure 1). When the test was used in the low-prevalence population, many cases with NP antigen >1.645 pg/mL had a false positive antigen test result (48.9% of all positive tests), as expected. However, two strategies can be adopted to reduce RT-PCR confirmation in cases of limited availability of this resource: a) assume that the antigen test positivity is diagnostic of SARS-CoV-2 infection for high antigen concentrations, and b) isolate all people with a positive antigen test results (this choice may be costly in cases of population screening).
This study found that 74/176 (42%) RT-PCR tests performed to confirm a positive Lumipulse® antigen test could have been omitted based on the 10.4 ng/mL cutoff, which could safely be assumed as diagnostic of SARS-CoV-2 infection at a 99.8% specificity level. This cutoff was very close to that of 10.0 pg/mL proposed by the manufacturer to establish sample positivity for SARS-CoV-2 infection.
Both at the observed prevalence in this screening cohort (5.2%) and at the hypothetical 2.5% prevalence scenario, NPV remained >99.0%, which is an excellent value to be used in community or population screening. A screening based on this test could aim to greatly reduce or even eliminate transmission in the target communities as schools, retirement homes, clinics, prisons, and others in which it is difficult to guarantee strict observance of containment measures.
The gold standard RT-PCR test takes a minimum of 3–4 h for results, needs specialized laboratory equipment and technicians, and has higher costs. On the other hand, the Lumipulse® antigen assay is completely automated, can easily be introduced into routine laboratory workflow, and is capable of processing up to 120 samples per hour with a time-to-report of about 1 h. As accessibility, frequency, and sample to-answer time are crucial for effective surveillance of COVID-19 (Larremore et al., 2020), the current findings suggest that this antigen assay can be optimally employed to control the spread of the virus in this pandemic under conditions in which molecular testing is not widely feasible.
Novel SARS-CoV-2 virus variants of potential concern with different mutations of the Spike protein have recently emerged (ECDC, 2021). Detection ability of the NP antigen by the Lumipulse® assay is not theoretically impacted by these variants. Indeed, preliminary results of genome sequencing of some positive samples included in this screening cohort showed that the antigen assay detected both P1 Brazilian and VOC 202012/01 UK variants actively spreading in the Perugia region in January 2021 (data not shown).
Diagnostic tests for SARS-CoV-2, including antigenic tests, are increasingly being used for mass screening purposes in the different epidemic phases. In Wuhan city, at the end of the epidemic, a very large screening program using nucleic acid testing on about 10 million people found 300 asymptomatic cases (Cao et al., 2020). Similar findings have been made in Liverpool, UK (ECDC 2020b ) and Alto Adige, Italy (Euronews, 2020). In Europe, lateral flow antigenic tests were recently introduced during an active epidemic phase, with the purpose of reducing the effective reproduction number (Rt) and improve transmission control. However, mass screening based on such low sensitivity tests, although sustainable and affordable, may be ineffective because of false reassurance of infectious people testing negative (Iacobucci, 2020). Indeed, solid evidence of the effectiveness of these screenings is still lacking; Slovakia was recently forced to adopt new lockdown measures shortly after mass screening based on a lateral flow rapid test with a declared 30% false negative rate (Burki, 2020b). To overcome the influence of low sensitivity on screening performance it is necessary to repeat individual testing many times in close screening rounds (Mina et al., 2020, Larremore et al., 2020). Thus, the automated, fast and cheap Lumipulse® assay could be a good alternative to lateral flow tests because of its higher sensitivity. Studies have reported saliva as a reliable sample for COVID-19 molecular diagnosis (Azzi et al., 2020, Yu et al., 2020). It would be interesting to explore the performance of the Lumipulse® antigen test on this non-invasive specimen.
Finally, as viral RNA load can be swinging in the late phase of the infection, while antigen NP does not seem to be affected by significant variability (Hirotsu et al., 2020a, Hirotsu et al., 2020b), further studies are needed to explore the role of the antigen test to discriminate early and late phases of the infection, to monitor its course, and, eventually, to predict its outcome.
In conclusion, this study found that the Lumipulse® antigen test can be used for screening purposes in communities with high and low prevalence of SARS-CoV-2 infection. In particular, when adopting the 1.645 pg/mL cutoff, the test showed high sensitivity and excellent NPV, which are desirable features in a mass or large community screening.
Ethical approval
This study was reviewed and approved by Bioethics Review Board of University of Perugia (no. 36371).
Conflict of interest
None.
Financial support
This paper and the research behind it received a financial contribution from Brunello Cucinelli S.p.A. (Italy) without involvement at any stage.
Author contributions
Study concept and design: Alessio Gili, Riccardo Paggi, Fabrizio Stracci, Antonella Mencacci; Drafting manuscript: Alessio Gili, Riccardo Paggi, Fabrizio Stracci, Antonella Mencacci;
Critical revision of manuscript: Carla Russo, Elio Cenci, Donatella Pietrella, Alessandro Graziani
Statistical analysis: Alessio Gili, Riccardo Paggi, Fabrizio Stracci, Antonella Mencacci Experiments: Alessandro Graziani, Carla Russo
Approval of manuscript: Alessio Gili, Riccardo Paggi, Fabrizio Stracci, Antonella Mencacci, Carla Russo, Elio Cenci, Donatella Pietrella, Alessandro Graziani.
References
- Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81(1):e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnicker M.J. Can the SARS-CoV-2 PCR cycle threshold value and time from symptom onset to testing predict infectivity? Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa735. Epub ahead of print. PMID: 32504529; PMCID: PMC7314221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. Epub ahead of print. PMID: 32442256; PMCID: PMC7314198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki T.K. Coronavirus in China. Lancet Respir Med. 2020;8(3):238. doi: 10.1016/S2213-2600(20)30056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki T.K. Mass testing for COVID-19. Lancet Microbe. 2020;1(8):e317. doi: 10.1016/S2666-5247(20)30205-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Gan Y., Wang C., Bachmann M., Wei S., Gong J. Post-lockdown SARS-CoV-2 nucleic acid screening in nearly ten million residents of Wuhan, China. Nat Commun. 2020;11(1):5917. doi: 10.1038/s41467-020-19802-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks Jonathan J., Altman Douglas G. Diagnostic tests 4: likelihood ratios. BMJ. 2004;329:168. doi: 10.1136/bmj.329.7458.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ECDC . 2020. Diagnostic testing and screening for SARS-CoV-2. ECDC.https://www.ecdc.europa.eu/en/covid-19/latest-evidence/diagnostic-testing [Google Scholar]
- ECDC . 2020. Population-wide testing of SARS-CoV-2: country experiences and potential approaches in the EU/EEA and the UK Stockholm. ECDC.https://www.ecdc.europa.eu/en/publications-data/population-wide-testing-sars-cov-2-country-experiences-and-potential-approaches [Google Scholar]
- ECDC. Risk Assessment: Risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA – first update. 21 January 2021. [DOI] [PMC free article] [PubMed]
- Euronews . 2020. Covid-19, screening di massa in Alto Adige: testate oltre 320mila persone in 72 ore.https://it.euronews.com/2020/11/22/covid-19-screening-di-massa-in-alto-adige-testate-oltre-320mila-persone-in-72-ore [Google Scholar]
- Fluss R., Faraggi D., Reiser B. Estimation of the Youden Index and its associated cutoff point. Biom J. 2005;47(4):458–472. doi: 10.1002/bimj.200410135. [DOI] [PubMed] [Google Scholar]
- Gupta S. 2020. Strong inverse correlation between SARS-CoV-2 infectivity and cycle threshold value.https://www.infectiousdiseaseadvisor.com/home/topics/covid19/ct-value-may-inform-when-patients-with-covid-19-can-be-safely-discharged/ [Google Scholar]
- Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K. Analysis of a persistent viral shedding patient infected with SARS-CoV-2 by RT-qPCR, FilmArray Respiratory Panel v2.1, and antigen detection. J Infect Chemother. 2020;27(2):406–409. doi: 10.1016/j.jiac.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsu Y., Maejima M., Shaibusawa M., Nagakubo Y., Hosaka K., Amemiya K. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int J Infect Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff H.V., Singh A. Asymptomatic transmission during the COVID-19 pandemic and implications for public health strategies. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa654. Epub ahead of print. PMID: 32463076; PMCID: PMC7314132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health – the latest 2019 novel coronavirus outbreak in Wuhan, China. Int J Infect Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobucci G. Covid 19: Mass population testing is rolled out in Liverpool. BMJ. 2020;371:m4268. doi: 10.1136/bmj.m4268. [DOI] [PubMed] [Google Scholar]
- ISTAT (National Institute of Statistics, Italy) First reports of SARS-CoV-2 Seroprevalence. https://www.istat.it/it/files/2020/08/ReportPrimiRisultatiIndagineSiero.pdf
- Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A. Test sensitivity is secondary to frequency and turnaround time for COVID-19 surveillance. Sci Adv. 2021;7(1):eabd5393. doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. Classification accuracy and cut point selection. Stat Med. 2012;31(23):2676–2686. doi: 10.1002/sim.4509. [DOI] [PubMed] [Google Scholar]
- Lu H., Stratton C.W., Tang Y.W. Outbreak of pneumonia of unknown etiology in Wuhan China: the mystery and the miracle. J Med Virol. 2020;92(4):401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina J.M., Parker R., Larremore B.D. Rethinking Covid-19 test sensitivity – a strategy for containment. N Engl J Med. 2020;383(22):e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- Ministero della Salute (Health Ministry, Italy) 2020. Update of contact tracing and laboratory diagnosis of SARS-CoV-2 infection. 19 March.https://www.trovanorme.salute.gov.it/norme/renderNormsanPdf?anno=2020&codLeg=73714&parte=1%20&serie=null [Google Scholar]
- Singanayagam A., Patel M., Charlett A., Bernal J.L., Saliba V., Ellis J. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom M.R., Mina M.J. To Interpret the SARS-CoV-2 Test, consider the cycle threshold value. Clin Infect Dis. 2020;71(16):2252–2254. doi: 10.1093/cid/ciaa619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . WHO; 2020. Laboratory testing for coronavirus disease (COVID-19) in suspected human cases: interim guidance, 19 March 2020.https://apps.who.int/iris/handle/10665/331501 [Google Scholar]
- WHO . WHO; 2020. World Health Organization Coronavirus disease 2019 (COVID-19) Situation Report – 20/12/2020.https://covid19.who.int/ [Google Scholar]
- Youden W.J. Index for rating diagnostic tests. Cancer. 1950;3(1):32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71(15):793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]