Abstract
Background
While SARS-CoV-2 similarly infects men and women, COVID-19 outcome is less favorable in men. Variability in COVID-19 severity may be explained by differences in the host genome.
Methods
We compared poly-amino acids variability from WES data in severely affected COVID-19 patients versus SARS-CoV-2 PCR-positive oligo-asymptomatic subjects.
Findings
Shorter polyQ alleles (≤22) in the androgen receptor (AR) conferred protection against severe outcome in COVID-19 in the first tested cohort (both males and females) of 638 Italian subjects. The association between long polyQ alleles (≥23) and severe clinical outcome (p = 0.024) was also validated in an independent cohort of Spanish men <60 years of age (p = 0.014). Testosterone was higher in subjects with AR long-polyQ, possibly indicating receptor resistance (p = 0.042 Mann-Whitney U test). Inappropriately low serum testosterone level among carriers of the long-polyQ alleles (p = 0.0004 Mann-Whitney U test) predicted the need for intensive care in COVID-19 infected men. In agreement with the known anti-inflammatory action of testosterone, patients with long-polyQ and age ≥60 years had increased levels of CRP (p = 0.018, not accounting for multiple testing).
Interpretation
We identify the first genetic polymorphism that appears to predispose some men to develop more severe disease. Failure of the endocrine feedback to overcome AR signaling defects by increasing testosterone levels during the infection leads to the polyQ tract becoming dominant to serum testosterone levels for the clinical outcome. These results may contribute to designing reliable clinical and public health measures and provide a rationale to test testosterone as adjuvant therapy in men with COVID-19 expressing long AR polyQ repeats.
Funding
MIUR project “Dipartimenti di Eccellenza 2018-2020” to Department of Medical Biotechnologies University of Siena, Italy (Italian D.L. n.18 March 17, 2020) and “Bando Ricerca COVID-19 Toscana” project to Azienda Ospedaliero-Universitaria Senese. Private donors for COVID-19 research and charity funds from Intesa San Paolo.
Keywords: Androgen receptor gene, Testosterone, COVID-19, LASSO logistic regression, WES, Viral infection and host genome
Research in context.
Evidence before this study
We searched on Medline, EMBASE, and Pubmed for articles published from January 2020 to August 2020 using various combinations of the search terms “sex-difference”, “gender” AND SARS-Cov-2, or COVID. Epidemiological studies indicate that men and women are similarly infected by COVID-19, but the outcome is less favorable in men, independently of age. Several studies also showed that patients with hypogonadism tend to be more severely affected. A prompt intervention directed toward the most fragile subjects with SARS-Cov-2 infection is currently the only strategy to reduce mortality. Glucocorticoid treatment is a cost-effective measure to improve the outcome of severe cases. Clinical algorithms have been proposed, but little is known on the ability of genetic profiling to predict outcome and disclose novel therapeutic strategies.
Added-value of this study
In a cohort of 1178 men and women with COVID-19, we used a supervised Machine Learning approach on a synthetic representation of genetic variability due to poly-amino acid repeats. Comparing the genotype of patients with extreme manifestations (severe vs. asymptomatic), we found an association between the poly-glutamine repeat number of the androgen receptor (AR) gene, serum testosterone concentrations, and COVID-19 outcome in male patients. Failure of the endocrine feedback to overcome AR signaling defects by increasing testosterone levels during the infection leads to the fact that polyQ ≥ 23 becomes dominant to testosterone levels for the clinical outcome.
Implications of all the available evidence
We identify the first genetic polymorphism predisposing some men to develop a more severe disease irrespectively of age. Based on this, we suggest that sizing the AR poly-glutamine repeat has important implications in the diagnostic pipeline of patients affected by life-threatening COVID-19 infection. Most importantly, our studies open to the potential of using testosterone as adjuvant therapy for patients with severe COVID-19 having defective androgen signaling, defined by this study as ≥23 PolyQ repeats, and inappropriately low levels of circulating androgens.
Alt-text: Unlabelled box
1. Introduction
Alongside the mode of transmission, viral load, comorbidities, and demographic factors (such as age and sex), the host genetic background appears to play an important role in COVID-19 severity and progression [1], [2], [3], [4], [5], [6], [7], [8]. We hypothesized that common polymorphisms may contribute to COVID-19 severity, including poly-amino acids repeat polymorphisms, such as the polyQ tract of the Androgen Receptor (AR). AR contains in its N-terminus domain a polymorphic polyQ tract, ranging between 9 and 36 repeated CAG units in the normal population [9]. In vitro and in vivo studies have demonstrated that the transactivation potential of AR is inversely correlated to repeat length, and Q-tract size can significantly influence androgen-dependent physiological functions [9], [10], [11], [12].
Several lines of evidence lead to the concept that androgens are relevant to both SARS-CoV-2 infection and COVID-19 disease presentation; however, they seem to have a Janus bifacial way of action [13,14]. On one side, androgens promote the transcription of the TMPRSS2 gene that encodes a serine protease known to prime the spike (S) protein of coronaviruses, facilitating viral entry into the cells [15]. On the other hand, hypogonadism is known to correlate with severe COVID-19 [16] and other chronic conditions, partly due to the loss of attenuation of the inflammatory immune response exerted by testosterone (T) [17], [18], [19].
2. Methods
2.1. Patients
We performed a nested case-control study (NCC). Cases and controls were drawn from the Italian GEN-COVID cohort of 1178 subjects infected with SARS-CoV-2 diagnosed by RT-PCR on nasopharyngeal swab [2]. Demographic characteristics of patients enrolled in the cohort are summarized in Table 1 according to their clinical status. In the current NCC study, cases were selected according to the following inclusion criteria: i. CPAP/biPAP ventilation (230 subjects); ii. endotracheal intubation (108 subjects). As controls, 300 subjects were selected using the sole criterion of not requiring hospitalization. Exclusion criteria for both cases and controls were i. SARS-CoV-2 infection not confirmed by PCR; ii. non-caucasian ethnicity. Demographic characteristics of the subjects in the NCC study are summarized in Table 1. A similar Spanish cohort, composed of male COVID-19 patients (117 cases and 41 controls) was used to validate the results in another representative European population highly impacted by COVID-19. All subjects were white European. The Spanish Covid HGE cohort is under IRB approval PR127/20 from Bellvitge University Hospital, Barcelona, Spain.
Table 1.
Demographics characteristics of the Italian GEN-COVID Cohort and NCC study.
| Intubation | CPAP/BiPAP Ventilation | Oxygen Therapy | Hospitalized w/o respiratory support | Oligo^-asymptomatics w/o hospitalization | ||
|---|---|---|---|---|---|---|
| GEN-COVID | Number of Sybjects | 108 | 230 | 352 | 188 | 300 |
| Male/Female | 80/28 | 157/73 | 208/144 | 104/84 | 116/184 | |
| Age males (years) | 61,52±11,43 | 62,75±13,48 | 63,41±14,53 | 55,99±15,44 | 47,40±13,23 | |
| Age females (years) | 63,71±13,96 | 66,23±15,25 | 68,40±14,74 | 52,88±16,39 | 48,61±11,06 | |
| Cases | Controls | |||||
| NCC study | Number of Subjects | 338 | 300 | |||
| Male/Female | 237/101 | 116/184 | ||||
| Age males (years) | 62,34±12,84 | 47,40±13,23 | ||||
| Age females (years) | 65,53±14,94 | 48,61±11,06 | ||||
Oligosymtpomatic: individuals with minor symptoms of COVID-19 (mild fever, cough, sore throat, etc.)
2.2. Ethics
The GEN-COVID study was approved by the University Hospital of Siena Ethics Review Board (Protocol n. 16917, dated March 16, 2020). This observational study has been inserted in www.clinicaltrial.org (NCT04549831). The Spanish Covid HGE cohort is under IRB approval PR127/20 from Bellvitge University Hospital, Barcelona Spain. Written informed consent was obtained from all individuals who contributed samples and data.
2.3. Analysis of triplets size in the AR locus
To establish allele sizes of the polymorphic triplet in the AR locus, we used the HUMARA assay with minor modifications [20]. Specifically, we performed a fluorescent PCR followed by capillary electrophoresis on an ABI3130 sequencer. Allele size was established using the Genescan Analysis software.
2.4. Binary representation of WES data
Variants calling was performed according to the GATK4 best practice guidelines, using BWA for mapping, and ANNOVAR for annotating. WES data were represented in a binary mode on a gene-by-gene basis. Poly-amino acids triplet repeats were represented in a binary mode: long and short repeats in respect to the reference sequence on the genome. A total of 40 genes with 43 triplet repeat regions were taken from UniProtKB (Supplementary Table S1). In the boolean representation of poly-amino acids triplet repeats, for each of these 40 genes two features were defined, Dij and Iij, with Dij being equal to 1 if gene i in sample j has a repeated region shorter than the reference, 0 otherwise, and Iij being equal to 1 if gene i in sample j has a repeated region longer than the reference, 0 otherwise.
2.5. LASSO logistic regression
We adopted the LASSO logistic regression that provides a feature selection method within the classification tasks able to enforce both the sparsity and the interpretability of the results. The weights of the logistic regression algorithm can be interpreted as the importance of the subset of the most relevant features for the task [21].
The input features of the LASSO logistic regression are the poly-amino acids triplet repeats as well as gender, comorbidity (1 if there is at least one comorbidity) and age, the latter as a continuous variable normalized between 0 and 1. Comorbidities were defined as the presence of one or more clinical conditions (i.e. cardiac, endocrine, neurological, neoplastic diseases) at the time of infection. During the fitting procedure, the class slight unbalancing is tackled by penalizing the misclassification of the minority class with a multiplicative factor inversely proportional to the class frequencies. The data pre-processing was coded in Python, whereas for the logistic regression model we used the scikit-learn module with the liblinear coordinate descent optimization algorithm.
2.6. Total T measurement
Blood samples were collected after an overnight fast, immediately centrifuged at 4 °C and stored at -20 °C until assayed. Serum and plasma total T (TT), SHBG levels in plasma and serum LH were measured following standard procedures.
Serum TT was measured using the Access testosterone assay (Beckman Coulter Inc., Fullerton, CA, USA) with a minimum detection limit of 0.35 nmol/L. Reference range for this assay was 6.07–27.1 nmol/L and liquid chromatography - tandem mass spectrometry (LC-MS/MS) according to a previously validated method provided with reference values between 9.8–28.4 nmol/L [22]. Thawed plasma underwent 15 min incubation at 56 °C for virus inactivation, and TT measured in 100 µl of plasma, with sensitivity limit being 0.270 nmol/L, imprecision ranging 9.8 to 0.7% and accuracy 90.6 to 101.5% at concentration levels between 1.12 and 39.2 nmol/L. A stability test under viral inactivation conditions was performed in 6 samples, revealing a T mean (min-max) % loss of 9.7% (4.6–16.7%).
SHBG levels were measured in plasma samples using Quantikine ELISA Kit (DSHB G0B, R&D Systems, Minneapolis, MN, USA) according to the manufacturers' instructions. Serum LH was measured using “Access LH assay“ a chemiluminescenSert, two-step enzyme immunoassay (Beckman Coulter Inc., Fullerton, CA, USA). Sensitivity for the LH determination is 0.2 mIU/mL. Reference range in adult males for this assay is 1.2–8.6 mIU/mL.
2.7. Statistical analysis
Since serum and plasma T values were not normally distributed, the statistical analyses were performed using non-parametric tests. When appropriate, transformation was used for skewed data in regression models. We used the Mann-Whitney U test to compare T levels in males with AR long-polyQ (≥23) versus males with short-polyQ repeat (≤22). Logistic regression analysis was performed to test the contribution of age, T, and the number of polyglutamine repetitions on COVID-19 outcome. The only prespecified interaction tested was the T by polyQ (categorical). Box-Tidwell procedure was used to assess linearity and the Hosmer and Lemeshow to assess goodness of fit test. Multicollinearity was assessed by variance inflation factor, and dealt with by dropping the offending variables from the analysis on the basis of clinical grounds.
2.8. Role of funders
The work was financially supported by MIUR project “Dipartimenti di Eccellenza 2018-2020” to Department of Medical Biotechnologies University of Siena, Italy (Italian D.L. n.18 March 17, 2020) and by “Bando Ricerca COVID-19 Toscana” project to Azienda Ospedaliero-Universitaria Senese. It was also funded by private donors for COVID-19 research and charity funds from Intesa San Paolo “Fondo di Beneficenza n. b/2020/0119”. The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the manuscript. The authors collected the data, and had full access to all of the data in the study. They also had the final decision and responsibility to submit the study results for publication.
3. Results
3.1. Testing the role of common poly-amino acid repeat polymorphisms in COVID-19 outcome
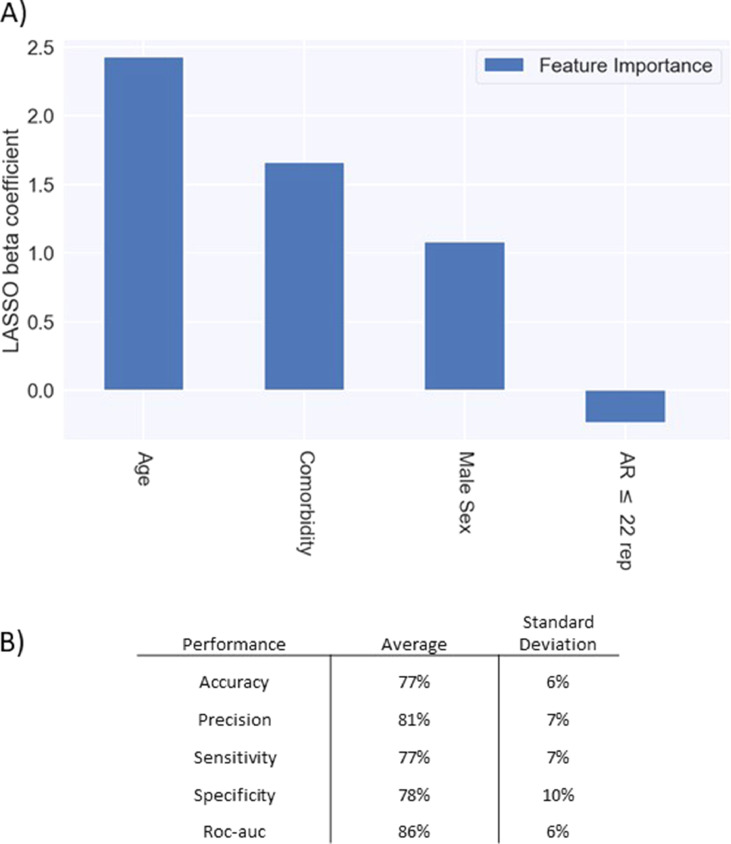
In order to test the role of common poly-amino acid repeat polymorphisms in determining COVID-19 clinical severity, we performed a NCC, selecting the extreme phenotypic ends of our entire GEN-COVID cohort (Table 1 and Fig. 1). Among 18,439 annotated genes, we selected those with amino acid repeats, namely 40 genes, and represented them as a boolean variable. Logistic regression with LASSO regularization analysis identified AR as the only protective gene (Fig. 1, panel a). The 10-fold cross-validation provides good performances in terms of accuracy (77%), precision (81%), sensitivity (77%), specificity (78%) and Area Under the Curve (AUC) score (86%) (Fig. 1, panel b). The performances of the logistic regression without LASSO regularization for the selected set of features (age, gender, comorbidity and AR gene) are 79% accuracy, 81% precision, 81% sensitivity, 78% specificity, 88% roc-auc. The model shows a slight decrease of almost all the performance measures when the AR gene is removed from the set (accuracy -1.2%, precision -1.3%, sensitivity -1.4%, specificity -1.2%, roc-auc +0.3%). Finally, the logistic regression on the male cohort with the AR gene alone provides results quite higher than the random guess (accuracy 58%, precision 71%, sensitivity 64%, specificity 55%, roc-auc 55%).
Fig. 1.
LASSO logistic regression. The bar of the LASSO logistic regression beta coefficients represents the importance of each feature for the classification task (Fig. 1) (Panel a). The positive beta coefficients of the LASSO (upward bars) reflect a susceptible behaviour of the features to the target COVID-19 disease, whereas the negative coefficients (downward bars) a protective action. The calculated odd ratio of AR short repeats (≤22) is 0.79 i.e. protective. Therefore, the odd ratio of long repeats (≥23) is 1/0.79 = 1.27 i.e. severity. Panel b: Table reporting the averages and the standard deviations of accuracy, precision, sensitivity, specificity, and ROC-AUC scores for the 10-folds of the cross-validation.
3.2. Validation of polyQ polymorphism by sizing the PolyQ repeat of the AR gene
In order to validate the results on AR obtained by LASSO logistic regression, we sized the number of triplets in the male subset (351 subjects) using the gold standard technique that uses a fluorescent PCR reaction followed by the use of GeneScan Analysis software® (Applied Biosystems) [20]. We identified a 98% concordance between the results of the two techniques in measuring the polyQ repeats. Based on the AR polyQ length, male patients were subdivided into two categories, those having a number of PolyQ repeats less than or equal to 22 repeats, and those having a number of PolyQ repeats greater than or equal to 23 repeats, being 23 repeats the reference sequence on genome browsers and the reported cut-off value [23], [24]. We found that PolyQ repeats below 22 are enriched in the asymptomatic cohort of males. The difference was statistically significant in the group of males younger than 60 years of age in which genetic factors are expected to have a major impact (p-value 0.024 by χ2 test) (Table 2; Supplementary Table S2).
Table 2.
PolyQ alleles correlation with COVID-19 outcome - males with age <60.
| Males <60 | |||
|---|---|---|---|
| <22 | >23 | Marginal Row Totals | |
| Cases | 52 (59,1%) | 36 (40,9%) | 88 (48,1%) |
| Controls | 71 (74,7%)* | 24 (25,3%) | 95 (51,9%) |
| Marginal Column Totals | 123 (67,2%) | 60 (32,8%) | 183 (Grand Total) |
p-value (cases vs controls) =0.024
3.3. Validation of polyQ polymorphism in the Spanish Cohort
We then sized the polyQ repeat in an independent cohort consisting of 158 <60 years old Spanish males without known comorbidities (117 cases and 41 controls). The association with shorter repeats (≤ 22) and protection was confirmed (p-value 0.014 by χ2 test) (Table 3).
Table 3.
Validation in Spanish cohort
| Spanish validation (χ2) Males global | |||
|---|---|---|---|
| ≤22 | ≥23 | Marginal Row Totals | |
| Cases | 51 (43,6%) | 66 (56,4%) | 117 (74,1%) |
| Controls | 27 (65,9%)* | 14 (34,1%) | 41 (25,9%) |
| Marginal Column Totals | 78 (49,4%) | 80 (50,6%) | 158 (Grand Total) |
p-value (cases vs controls)=0.014 (Significant at p<0.05)
3.4. Males with longer polyQ have receptor resistance
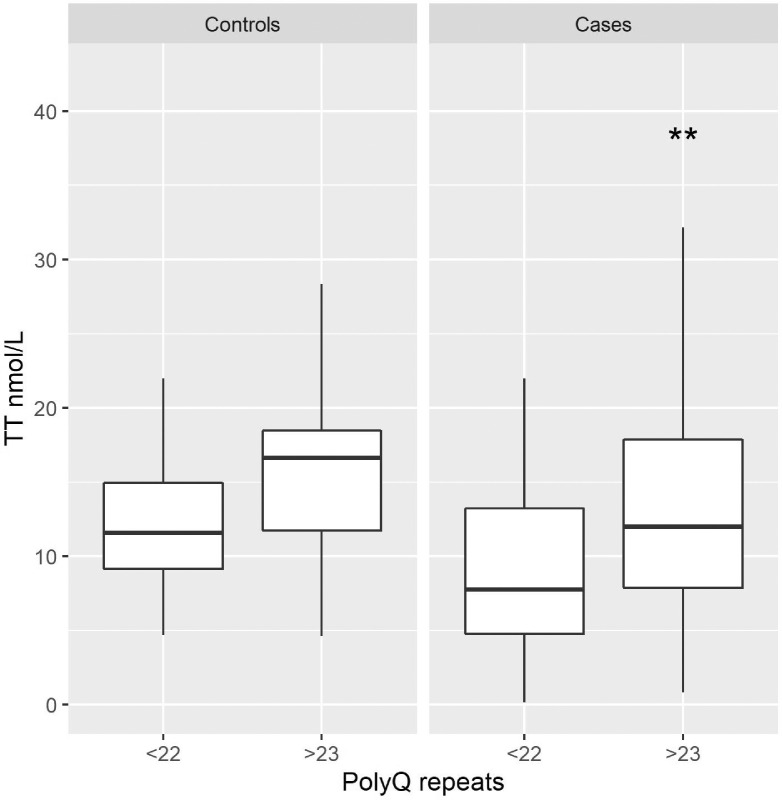
To functionally link the length of the PolyQ repeats to AR functionality, we measured TT in 183 men using LCMS/MS (Supplementary Table S2). TT was higher in patients carrying ≥23 vs ≤22 glutamines (13.45 vs 11.23 nmol/L, p-value 0.042), reflecting reduced negative feedback from the less active receptors present in patients carrying a PolyQ repeat of ≥23. This difference was evident also comparing the TT value and polyQ repeats in the case and the control group (Fig. 2).
Fig. 2.
Relationship between Total Testosterone and polyQ repeats in the case and the control group. Box-plot showing values of Total Testosterone (TT), expressed in nmol/L, in subjects with shorter (≤22) and longer (≥23) polyQ repeats in AR gene grouped between controls (left panel) and cases (right panel). The TT median value, represented by the black horizontal line, is higher in patients with ≥23 polyQ repeats in the case group, (**p-value = 0.023; Mann-Whitney U test). No statistically significant difference was present in the control group (p-value = 0.088; Mann-Whitney U test).
3.5. Unbalanced T-AR axis in males with longer polyQ repeats
The hormonal status of the entire male cohort revealed lower TT and calculated free T levels and higher SHBG levels with increasing age (Supplementary Table S3).
To evaluate whether the AR receptor reduced activity resulted in signs and symptoms of hypogonadism, subjects were interviewed, post-infection, using a modified version of the Androstest® [25]. Interviews were available for 61 subjects (43 short and 18 long) representative of the extremes genotypes (≤19 and ≥25 repeats) of the cohort. An Androtest score ≥8 was found in 38% of men with longer repeats as compared to 16% of those with ≤19 glutamines (likelihood ratio, p = 0.046). Similarly, cryptorchidism (11% in long repeats vs. 2% in short repeats), and anemia (11% in long repeats vs. 2% in short repeats), two powerful sings of low androgenicity, and severe erectile dysfunction (22% in long repeats vs. 9% in short repeats) were more frequently reported in subjects with longer repeats, but not osteopenia/osteoporosis (6% in long repeats vs. 7% in short repeats) (Supplementary Table S4). These results indicate a trend toward clinical hypogonadism for those with longer repeats. Conversely, in the entire male dataset, 6 cases of prostate cancer were found annotated in the past-medical history, all in the ≤22 glutamines group, suggesting an increased prostate sensitivity to androgens in this group. No difference was found in the prevalence of BPH or 5-alpha-reductase inhibitors use.
As the reduced signal transduction of AR might be partially compensated by higher T levels, we tested whether the decreased AR negative feedback was sufficient to overcome larger polyQ repeats size (Fig. 2). Logistic regression was performed to investigate the joint effect of T level and polyglutamine receptor length on the likelihood that subjects require intensive care during COVID infections, adjusting for age in the model. The logistic regression model was highly significant (χ2 (3) = 18,881, p < 0.0001), with the model explaining 7.5% (Nagelkerke /R2) of the variance in COVID-19 outcome (Supplementary Table S5). To test whether the association between T and the outcome changes when the polyQ is short (≤ 22) or long (≥23), an interaction term was included in the model. A significant interaction was found (p-value 0.018), suggesting impaired feedback as a predictor of the worst outcome, namely intubation or CPAP/BiPAP versus hospitalization not requiring respiratory assistance. To provide an intuitive graphical representation, we plotted the ratio between TT serum concentrations and polyQ number vs. clinical outcome (Supplementary Figure 1). Results show a decreased mean ratio, a sign of an inappropriate rise of TT for increasing polyglutamine repeats, and association with a worse outcome (p = 0.0004).
3.6. Inflammatory phenotype in males with longer polyQ repeats
Finally, we tested the relationship between the AR polyQ repeat size and 5 laboratory markers of immunity/inflammation, including CRP, Fibrinogen, IL6, CD4 and NK count. We found that older (≥60) males with AR polyQ tract ≥23 have a higher (55.92 versus 48.21 mg/dl) mean value of CRP (p-value 0.018, not accounting for multiple testing) and lower mean value of Fibrinogen and a trend of higher IL6 (Table 4).
Table 4.
Correlation between polyQ repeats in AR gene and laboratory values
| CRP M≥60y cases |
CRP M<60y cases |
||||
|---|---|---|---|---|---|
| Triplets | Mean | Count | Triplets | Mean | Count |
| ≤22 | 48,21 | 78 | ≤22 | 54,5 | 43 |
| ≥23 | 55,92 | 38 | ≥23 | 26,41 | 29 |
| p-value = 0.018 (Significant at p<0,05) | p-value = 0.2 | ||||
| Fibrinogen M≥60y cases |
Fibrinogen M<60y cases |
||||
|---|---|---|---|---|---|
| Triplets | Mean | Count | Triplets | Mean | Count |
| ≤22 | 401,33 | 57 | ≤22 | 316,93 | 22 |
| ≥23 | 320,34 | 27 | ≥23 | 356,91 | 19 |
| p-value = 0.093 | p-value = 0.53 | ||||
| IL6 times the upper limit of normal M≥60y cases |
IL6 times the upper limit of normal M<60y cases |
||||
|---|---|---|---|---|---|
| Triplets | Mean | Count | Triplets | Mean | Count |
| ≤22 | 54,56 | 40 | ≤22 | 40,43 | 17 |
| ≥23 | 75,78 | 16 | ≥23 | 31,8 | 14 |
| p-value = 0,249 | p-value = 0,81 | ||||
| CD4 Lymphocytes M≥60y cases |
CD4 Lymphocytes M<60 cases |
||||
|---|---|---|---|---|---|
| Triplets | Mean | Count | Triplets | Mean | Count |
| ≤22 | 264,06 | 32 | ≤22 | 503,68 | 16 |
| ≥23 | 357,52 | 21 | ≥23 | 396,13 | 15 |
| p-value = 0.22 | p-value = 0.45 | ||||
| NK Cells M≥60y cases |
NK Cells M<60y cases |
||||
|---|---|---|---|---|---|
| Triplets | Mean | Count | Triplets | Mean | Count |
| ≤22 | 70,71 | 28 | ≤22 | 147,3 | 13 |
| ≥23 | 102,25 | 16 | ≥23 | 107,14 | 14 |
| p-value = 0.179 | p-value = 0.098 | ||||
4. Discussion
We employed machine learning methodologies to identify a set of genes involved in the severity of COVID-19. In the presence of very high dimensionality, as for instance in a WES study, it is crucial to select the most predictive genes representing patterns of variation (mutations or variants) in subjects with different classes of response (i.e., disease state: from asymptomatic to severe cases). This problem is even more complex in diseases where multiple genes are involved in determining the severity and clinical variability of the pathology. Here, we wanted to represent poly-amino acids repeat polymorphisms that are typically missed in classical GWAS analysis, which concentrates on bi-allelic polymorphisms.
We used a machine learning approach and logistic regression with a LASSO regularization to test if using such a simplified representation could lead to a reliable prediction of extreme clinical outcomes (asymptomatic versus severely affected). This approach enabled us to predict such clinical outcomes with 77% sensitivity.
AR contains a highly variable polyglutamine repeat (poly-Q) located in the N-terminal domain of the protein, spanning from 9 to 36 glutamine residues in the normal population [5]. AR polyQ length correlates with receptor functionality, with shorter polymorphic glutamine repeats typically associated with higher and longer PolyQ tracts with lower receptor activity [5]. AR is expressed in both males and females, but the bioavailability of its ligands T and dihydroT (DHT) differs significantly, being much higher in males. As previous studies linked male hypogonadism to a poorer outcome in COVID-19 patients we decided to focus on male patients and demonstrated that shorter polymorphic glutamine repeats (≤22) confer protection against life-threatening COVID-19 in a subpopulation of individuals with age <60 years.
We also confirmed the association between polyQ size and receptor activity. Specifically, we showed that longer polyQ size (≥23) is associated with higher serum T levels, suggestive of impaired negative feedback (p=0.004 at Mann-Whitney U test) at the level of the hypothalamus and pituitary gland. While this is compensated in healthy subjects [26], during non-gonadal illnesses (NGI) such as COVID-19, some patients are unable to compensate for the reduced AR activity with higher T levels [27]. The result is a status of reduced androgenicity even in the presence of apparently normal T values [27].
As T is known to have an immunomodulatory activity attenuating inflammatory immune responses [26], [27], [28], [29], [30], [31], [32], we hypothesized that a long PolyQ repeat would lead to a pro-inflammatory status heralded by increased proinflammatory markers [19,33] by conferring decreased AR transcriptional activity. Conversely, men with a more active receptor (short PolyQ tract) would be protected because they can tame the inflammatory response and increase survival regardless of serum T levels. We found that -CRP-, one of the main inflammatory markers, was higher in subjects with a long AR PolyQ tract. This observation not only is in line with the known anti-inflammatory function of T, but also reinforces the functional importance of the AR PolyQ tract and its association with COVID-19 clinical outcome. Furthermore, this observation suggests that CRP is hierarchically more relevant than serum T level, which can be inappropriately normal and mask a status of low androgenicity in men with a long PolyQ repeat.
The allele distribution of the PolyQ repeat length varies among different populations, with the shortest in Africans, medium in Caucasians, and longest in Asians [34]. Interestingly, WHO data on mortality rates during the first pandemic wave indicated a higher fatality rate in China and Italy (https://covid19.who.int/) [35] with respect to African. Hence, AR polyQ length variability could represent an explanation for the observed differences in death rate. Moreover, Africans seem to be more prone to infection [36]. This observation could be due to a more active AR receptor, leading to a higher expression of TMPRSS2, a protease essential for SARS-CoV-2 spread [15].
Different studies have shown an association between hypogonadism or long polyQ repeats and severe COVID-19 [16,37] and other chronic obstructive pulmonary diseases [17,18]. Our results are in line with these initial observations and provide a possible mechanism explaining these associations. The present study brings these observations to the next level, revealing that is the overall androgenic effect -resulting from the interaction of polyQ polymorphism and circulating T levels- that predicts the need for intensive care. In infected men, we observed impaired feedback no longer sufficient to compensate for the reduced AR transcriptional activity, leading to the conclusion that polyQ tract length is hierarchically more important than serum T levels. This concept helps to solve some inconsistencies, including the early reports of a slightly better outcome in prostate cancer patients -who tend to have smaller polyQ repeats, as in our cohort - when compared to other cancers. Interestingly, previous studies failed to link polyQ with mortality, in healthy subjects [26] or individuals with chronic diseases such as diabetes mellitus [38]. Thus, the observed association between low androgenicity and outcome seems related to the hyperinflammatory state present in severe COVID-19.
An improvement in peak oxygen saturation in men receiving T replacement therapy has been demonstrated in a randomized controlled trial [39] and could be one of the mechanisms responsible for the observed protective effect of AR's with shorter polyQ tract in COVID-19 patients. The observations reported in this study prompt organizing a clinical trial where patients selected based on their serum T concentration and polyQ repeat size are randomized to receive T vs. placebo. Such study could introduce the concept that a simple genetic test measuring the AR polyQ repeat can be used in male patients to screen for those who are more likely to benefit from T therapy.
Variants of another X-linked gene, TLR7, have been associated with severe COVID-19 outcomes in young men [6]. In the 2 reported families, the rare TLR7 mutations segregated as a highly penetrant monogenic X-linked recessive trait. While variants in TLR7 gene are expected to account for a small number of severely affected cases, our findings involve a much larger number of subjects, as long polyQ alleles are relatively common (27%) [40]. Overall, X-linked genetic variants keep coming up as important for defining severe COVID-19 cases in males.
In conclusion, we present a method that can predict if subjects infected by SARS-CoV-2 are at risk for life-threatening complications. This approach has 77% accuracy, 81% precision, 77% sensitivity, and 78% specificity. Furthermore, we present evidence suggesting that a more active AR has the potential to confer protection against COVID-19 severity. If confirmed, these observations should be followed by properly conducted clinical trials exploring if T replacement may decrease morbidity and mortality in patients affected by the most severe forms of the disease. Finally, as shown by regression analysis, ORs ranges between 1.26 and 1.45, therefore the risk of carrying a longer AR is much smaller than other already known strong predictors such as age and sex, but still is highly significant, relatively common, and among the very few known genetic predictors of COVID-19 outcome.
Declaration of Competing Interest
The authors declare no competing interests.
Acknowledgments
Additional information
GEN-COVID Multicenter Study (https://sites.google.com/dbm.unisi.it/gen-covid)
Francesca Montagnani2,22, Laura Di Sarno1,2, Andrea Tommasi1,2,5, Maria Palmieri1,2, Massimiliano Fabbiani22, Barbara Rossetti22, Giacomo Zanelli2,22, Fausta Sestini20, Laura Bergantini23, Miriana D'Alessandro23, Paolo Cameli23, David Bennet23, Federico Anedda24, Simona Marcantonio24, Sabino Scolletta24, Federico Franchi24, Maria Antonietta Mazzei25, Susanna Guerrini25, Edoardo Conticini26, Luca Cantarini26, Bruno Frediani26, Danilo Tacconi27, Chiara Spertilli27, Marco Feri28, Alice Donati28, Raffaele Scala29, Luca Guidelli29, Genni Spargi30, Marta Corridi30, Cesira Nencioni31, Leonardo Croci31, Gian Piero Caldarelli32, Maurizio Spagnesi33, Paolo Piacentini33, Maria Bandini33, Elena Desanctis33, Silvia Cappelli33, Anna Canaccini34, Agnese Verzuri34, Valentina Anemoli34, Agostino Ognibene35, Massimo Vaghi36, Antonella D'Arminio Monforte37, Esther Merlini37, Federica Gaia Miraglia37, Mario U. Mondelli38,39, Stefania Mantovani38, Serena Ludovisi38,39, Massimo Girardis40, Sophie Venturelli40, Marco Sita40, Andrea Cossarizza41, Andrea Antinori42, Alessandra Vergori42, Arianna Emiliozzi2,22,42, Stefano Rusconi43,44, Matteo Siano44, Arianna Gabrieli44, Agostino Riva43,44, Daniela Francisci45,46, Elisabetta Schiaroli45, Pier Giorgio Scotton47, Francesca Andretta47, Sandro Panese48, Stefano Baratti48 Renzo Scaggiante49, Francesca Gatti49, Saverio Giuseppe Parisi50, Francesco Castelli51, Eugenia Quiros-Roldan51, Melania Degli Antoni51, Isabella Zanella52, Matteo Della Monica53, Carmelo Piscopo53, Mario Capasso54,55,56, Roberta Russo54,55, Immacolata Andolfo54,55, Achille Iolascon54,55, Giuseppe Fiorentino57, Massimo Carella58, Marco Castori58, Giuseppe Merla58, Filippo Aucella59, Pamela Raggi60, Carmen Marciano60, Rita Perna60, Matteo Bassetti61,62, Antonio Di Biagio62, Maurizio Sanguinetti63,64, Luca Masucci63,64, Serafina Valente65, Maria Antonietta Mencarelli5, Caterina Lo Rizzo5, Elena Bargagli23, Marco Mandalà66, Alessia Giorli66, Lorenzo Salerni66, Patrizia Zucchi67, Pierpaolo Parravicini67, Elisabetta Menatti68, Tullio Trotta69, Ferdinando Giannattasio69, Gabriella Coiro69, Fabio Lena70, Domenico A. Coviello71, Cristina Mussini72, Giancarlo Bosio73, Enrico Martinelli73, Sandro Mancarella74, Luisa Tavecchia74, Lia Crotti75,76,77,78,79
22Dept of Specialized and Internal Medicine, Tropical and Infectious Diseases Unit, Azienda Ospedaliera Universitaria Senese, Siena, Italy
23Unit of Respiratory Diseases and Lung Transplantation, Department of Internal and Specialist Medicine, University of Siena, Italy
24Dept of Emergency and Urgency, Medicine, Surgery and Neurosciences, Unit of Intensive Care Medicine, Siena University Hospital, Italy
25Department of Medical, Surgical and Neuro Sciences and Radiological Sciences, Unit of Diagnostic Imaging, University of Siena, Italy
26Rheumatology Unit, Department of Medicine, Surgery and Neurosciences, University of Siena, Policlinico Le Scotte, Italy
27Department of Specialized and Internal Medicine, Infectious Diseases Unit, San Donato Hospital Arezzo, Italy
28Dept of Emergency, Anesthesia Unit, San Donato Hospital, Arezzo, Italy
29Department of Specialized and Internal Medicine, Pneumology Unit and UTIP, San Donato Hospital, Arezzo, Italy
30Department of Emergency, Anesthesia Unit, Misericordia Hospital, Grosseto, Italy
31Department of Specialized and Internal Medicine, Infectious Diseases Unit, Misericordia Hospital, Grosseto, Italy
32Clinical Chemical Analysis Laboratory, Misericordia Hospital, Grosseto, Italy
33Department of Preventive Medicine, Azienda USL Toscana Sud Est, Italy
34Territorial Scientific Technician Department, Azienda USL Toscana Sud Est, Italy
35Clinical Chemical Analysis Laboratory, San Donato Hospital, Arezzo, Italy
36Chirurgia Vascolare, Ospedale Maggiore di Crema, Italy
37Department of Health Sciences, Clinic of Infectious Diseases, ASST Santi Paolo e Carlo, University of Milan, Italy
38Division of Infectious Diseases and Immunology, Fondazione IRCCS Policlinico San Matteo, Pavia, Italy
39Department of Internal Medicine and Therapeutics, University of Pavia, Italy
40Department of Anesthesia and Intensive Care, University of Modena and Reggio Emilia, Modena, Italy
41Department of Medical and Surgical Sciences for Children and Adults, University of Modena and Reggio Emilia, Modena, Italy
42HIV/AIDS Department, National Institute for Infectious Diseases, IRCCS, Lazzaro Spallanzani, Rome, Italy
43III Infectious Diseases Unit, ASST-FBF-Sacco, Milan, Italy
44Department of Biomedical and Clinical Sciences Luigi Sacco, University of Milan, Milan, Italy
45Infectious Diseases Clinic, Department of Medicine 2, Azienda Ospedaliera di Perugia and University of Perugia, Santa Maria Hospital, Perugia, Italy
46Infectious Diseases Clinic, "Santa Maria" Hospital, University of Perugia, Perugia, Italy
47Department of Infectious Diseases, Treviso Hospital, Local Health Unit 2 Marca Trevigiana, Treviso, Italy
48Clinical Infectious Diseases, Mestre Hospital, Venezia, Italy.
49Infectious Diseases Clinic, ULSS1, Belluno, Italy
50Department of Molecular Medicine, University of Padova, Italy
51Department of Infectious and Tropical Diseases, University of Brescia and ASST Spedali Civili Hospital, Brescia, Italy
52Department of Molecular and Translational Medicine, University of Brescia, Italy; Clinical Chemistry Laboratory, Cytogenetics and Molecular Genetics Section, Diagnostic Department, ASST Spedali Civili di Brescia, Italy
53Medical Genetics and Laboratory of Medical Genetics Unit, A.O.R.N. "Antonio Cardarelli", Naples, Italy
54Department of Molecular Medicine and Medical Biotechnology, University of Naples Federico II, Naples, Italy
55CEINGE Biotecnologie Avanzate, Naples, Italy
56IRCCS SDN, Naples, Italy
57Unit of Respiratory Physiopathology, AORN dei Colli, Monaldi Hospital, Naples, Italy
58Division of Medical Genetics, Fondazione IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, Italy
59Department of Medical Sciences, Fondazione IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, Italy
60Clinical Trial Office, Fondazione IRCCS Casa Sollievo della Sofferenza Hospital, San Giovanni Rotondo, Italy
61Department of Health Sciences, University of Genova, Genova, Italy
62Infectious Diseases Clinic, Policlinico San Martino Hospital, IRCCS for Cancer Research Genova, Italy
63Microbiology, Fondazione Policlinico Universitario Agostino Gemelli IRCCS, Catholic University of Medicine, Rome, Italy
64Department of Laboratory Sciences and Infectious Diseases, Fondazione Policlinico Universitario A. Gemelli IRCCS, Rome, Italy
65Department of Cardiovascular Diseases, University of Siena, Siena, Italy
66Otolaryngology Unit, University of Siena, Italy
67Department of Internal Medicine, ASST Valtellina e Alto Lario, Sondrio, Italy
68Study Coordinator Oncologia Medica e Ufficio Flussi Sondrio, Italy
69First Aid Department, Luigi Curto Hospital, Polla, Salerno, Italy
70Local Health Unit-Pharmaceutical Department of Grosseto, Toscana Sud Est Local Health Unit, Grosseto, Italy
71U.O.C. Laboratorio di Genetica Umana, IRCCS Istituto G. Gaslini, Genova, Italy.
72Infectious Diseases Clinics, University of Modena and Reggio Emilia, Modena, Italy.
73Department of Respiratory Diseases, Azienda Ospedaliera di Cremona, Cremona, Italy
74U.O.C. Medicina, ASST Nord Milano, Ospedale Bassini, Cinisello Balsamo (MI), Italy
75Istituto Auxologico Italiano, IRCCS, Department of Cardiovascular, Neural and Metabolic Sciences, San Luca Hospital, Milan, Italy.
76Department of Medicine and Surgery, University of Milano-Bicocca, Milan, Italy
77Istituto Auxologico Italiano, IRCCS, Center for Cardiac Arrhythmias of Genetic Origin, Milan, Italy.
78Istituto Auxologico Italiano, IRCCS, Laboratory of Cardiovascular Genetics, Milan, Italy
79Member of the European Reference Network for Rare, Low Prevalence and Complex Diseases of the Heart-ERN GUARD-Heart
Spanish COVID HGE
Sergio Aguilera-Albesa80, Sergiu Albu81, Carlos Casasnovas82,13, Valentina Vélez-Santamaria82,13, Juan Pablo Horcajada83, Judit Villar83, Agustí Rodríguez-Palmero84,13,14, Montserrat Ruiz13,14, Luis M Seijo85, Jesús Troya86, Juan Valencia-Ramos87, Marta Gut88
80Navarra Health Service Hospital, Pamplona, Spain
81Institut Guttmann Foundation, Badalona, Barcelona, Spain
82Bellvitge University Hospital, L'Hospitalet de Llobregat, Barcelona, Spain
83Hospital del Mar, Parc de Salut Mar, Barcelona, Spain
84University Hospital Germans Trias i Pujol, Badalona, Barcelona, Spain
85Clínica Universitaria de Navarra, Madrid, Spain
86Infanta Leonor University Hospital, Madrid, Spain
87University Hospital of Burgos, Burgos, Spain
88CNAG-CRG, Centre for Genomic Regulation (CRG), Barcelona Institute of Science and Technology (BIST), Carrer Baldiri i Reixac 4, 08028, Barcelona, Spain
Contributors
EF, FM, AR designed the study. CF and IM, were in charge of biological samples’ collection and biobanking. MB, FF were in charge of clinical data collection. MB, FF, AR, and FM performed analysis/interpretation of clinical data. UP, FF and MM performed T measurement by LC-MS/MS. EDG, AS, AP and LPS performed the validation of association between shorter repeats and protection in a Spanish cohort. MM and AI critically reviewed the manuscript and interpreted clinical data/androgen physiopathological processes. SA and MB were in charge of DNA isolations from peripheral blood samples. FV, GD, AG, RT carried the sequencing experiments. EB, NP, SF, CG, MG and MS, performed bioinformatics and statistical analyses. EB, NP, SD, CF, and SC prepared Figures and Tables. EB, NP, AMP, FPC, AR, EF and FM wrote the manuscript. All authors have reviewed and approved the manuscript.
Acknowledgements
This study is part of the GEN-COVID Multicenter Study, https://sites.google.com/dbm.unisi.it/gen-covid, the Italian multicenter study aimed at identifying the COVID-19 host genetic bases. Specimens were provided by the COVID-19 Biobank of Siena, which is part of the Genetic Biobank of Siena, member of BBMRI-IT, of Telethon Network of Genetic Biobanks (project no. GTB18001), of EuroBioBank, and of RD-Connect. We thank the CINECA consortium for providing computational resources and the Network for Italian Genomes (NIG) http://www.nig.cineca.it for its support. We thank private donors for the support provided to A.R. (Department of Medical Biotechnologies, University of Siena) for the COVID-19 host genetics research project (D.L n.18 of March 17, 2020). We also thank the COVID-19 Host Genetics Initiative (https://www.covid19hg.org/), MIUR project “Dipartimenti di Eccellenza 2018-2020” to the Department of Medical Biotechnologies University of Siena, Italy and “Bando Ricerca COVID-19 Toscana” project to Azienda Ospedaliero-Universitaria Senese. We also thank Intesa San Paolo for the 2020 charity fund dedicated to the project N. B/2020/0119 “Identificazione delle basi genetiche determinanti la variabilità clinica della risposta a COVID-19 nella popolazione italiana”.
Data availability and data sharing statement
The samples referenced here are housed in the GEN-COVID Patient Registry and the GEN-COVID Biobank and are available for sharing. The sequencing data are deposited in http://www.nig.cineca.it/, specifically, http://nigdb.cineca.it) and available for consultation. For further information, you may contact the corresponding author, Prof. Alessandra Renieri (e-mail: alessandra.renieri@unisi.it).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103246.
Appendix. Supplementary materials
References
- 1.Benetti E., Giliberti A., Emiliozzi A. Clinical and molecular characterization of COVID-19 hospitalized patients. PLoS One. 2020;15(11) doi: 10.1371/journal.pone.0242534. Published 2020 Nov 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daga S, Fallerini C, Baldassarri M, et al. Employing a systematic approach to biobanking and analyzing clinical and genetic data for advancing COVID-19 research [published online ahead of print, 2021 Jan 17]. Eur J Hum Genet. 2021;1-15. doi: 10.1038/s41431-020-00793-7. [DOI] [PMC free article] [PubMed]
- 3.Zhang X., Tan Y., Ling Y. Viral and host factors related to the clinical outcome of COVID-19. Nature. 2020;583(7816):437–440. doi: 10.1038/s41586-020-2355-0. [DOI] [PubMed] [Google Scholar]
- 4.Ellinghaus D., Degenhardt F., Bujanda L. Genomewide association study of severe Covid-19 with respiratory failure [published online ahead of print, 2020 Jun 17] N Engl J Med. 2020 doi: 10.1056/NEJMoa2020283. NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q., Bastard P., Liu Z. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020:eabd4570. doi: 10.1126/science.abd4570. Epub ahead of print. PMID: 32972995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Made C.I., Simons A., Schuurs-Hoeijmakers J. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324(7):1–11. doi: 10.1001/jama.2020.13719. Epub ahead of print. PMID: 32706371; PMCID: PMC7382021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bastard P., Rosen L.B., Zhang Q. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370(6515):eabd4585. doi: 10.1126/science.abd4585. Epub 2020 Sep 24. PMID: 32972996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pivonello R., Auriemma R.S., Pivonello C. Sex disparities in COVID-19 severity and outcome: are men weaker or women stronger? [published online ahead of print, 2020 Nov 26] Neuroendocrinology. 2020 doi: 10.1159/000513346. 10.1159/000513346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callewaert L., Christiaens V., Haelens A., Verrijdt G., Verhoeven G., Claessens F. Implications of a polyglutamine tract in the function of the human androgen receptor. Biochem Biophys Res Commun. 2003;306(1):46–52. doi: 10.1016/s0006-291x(03)00902-1. [DOI] [PubMed] [Google Scholar]
- 10.Simanainen U., Brogley M., Gao Y.R. Length of the human androgen receptor glutamine tract determines androgen sensitivity in vivo. Mol Cell Endocrinol. 2011;342:81–86. doi: 10.1016/j.mce.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirabassi G., Cignarelli A., Perrini S. Influence of CAG repeat polymorphism on the targets of testosterone action. Int J Endocrinol. 2015;2015 doi: 10.1155/2015/298107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindström S., Ma J., Altshuler D. A large study of androgen receptor germline variants and their relation to sex hormone levels and prostate cancer risk. Results from the national cancer institute breast and prostate cancer cohort consortium. J Clin Endocrinol Metab. 2010;95(9):E121–E127. doi: 10.1210/jc.2009-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wambier C.G., Goren A. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is likely to be androgen mediated. J Am Acad Dermatol. 2020;83(1):308–309. doi: 10.1016/j.jaad.2020.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pozzilli P., Lenzi A. Commentary: testosterone, a key hormone in the context of COVID-19 pandemic. Metabolism. 2020;108 doi: 10.1016/j.metabol.2020.154252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann M., Kleine-Weber H., Schroeder S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. .e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rastrelli G., Di Stasi V., Inglese F. Low testosterone levels predict clinical adverse outcomes in SARS-CoV-2 pneumonia patients [published online ahead of print, 2020 May 20] Andrology. 2020 doi: 10.1111/andr.12821. 10.1111/andr.12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Vliet M., Spruit M.A., Verleden G. Hypogonadism, quadriceps weakness, and exercise intolerance in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2005;172(9):1105–1111. doi: 10.1164/rccm.200501-114OC. [DOI] [PubMed] [Google Scholar]
- 18.Mohan S.S., Knuiman M.W., Divitini M.L. Higher serum testosterone and dihydrotestosterone, but not oestradiol, are independently associated with favourable indices of lung function in community-dwelling men. Clin Endocrinol (Oxf) 2015;83(2):268–276. doi: 10.1111/cen.12738. [DOI] [PubMed] [Google Scholar]
- 19.Mohamad N.V., Wong S.K., Wan Hasan W.N. The relationship between circulating testosterone and inflammatory cytokines in men. Aging Male. 2019;22(2):129–140. doi: 10.1080/13685538.2018.1482487V. [DOI] [PubMed] [Google Scholar]
- 20.Allen R.C., Zoghbi H.Y., Moseley A.B. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51(6):1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 21.Tibshirani R. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological) 1996;58(21):267–288. .36. [Google Scholar]
- 22.Fanelli F., Belluomo I., Di Lallo V.D. Serum steroid profiling by isotopic dilution-liquid chromatography-mass spectrometry: comparison with current immunoassays and reference intervals in healthy adults. Steroids. 2011 Feb;76(3):244–253. doi: 10.1016/j.steroids.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Mengual L., Oriola J., Ascaso C., Ballescà J.L., Oliva R. An increased CAG repeat length in the androgen receptor gene in azoospermic ICSI candidates. J Androl. 2003;24(2):279–284. doi: 10.1002/j.1939-4640.2003.tb02673.x. [DOI] [PubMed] [Google Scholar]
- 24.Möhlig M., Arafat A.M., Osterhoff M.A. Androgen receptor CAG repeat length polymorphism modifies the impact of testosterone on insulin sensitivity in men. Eur J Endocrinol. 2011 Jun;164(6):1013–1018. doi: 10.1530/EJE-10-1022. [DOI] [PubMed] [Google Scholar]
- 25.Millar A.C., Lau A.N.C., Tomlinson G., Kraguljac A., Simel D.L., Detsky A.S., Lipscombe L.L. Predicting low testosterone in aging men: a systematic review. CMAJ. 2016 Sep 20;188(13):E321–E330. doi: 10.1503/cmaj.150262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eendebak R.J., Huhtaniemi I.T., Pye S.R. The androgen receptor gene CAG repeat in relation to 4-year changes in androgen-sensitive endpoints in community-dwelling older European men. Eur J Endocrinol. 2016;175(6):583–593. doi: 10.1530/EJE-16-0447. [DOI] [PubMed] [Google Scholar]
- 27.Zitzmann M., Nieschlag E. The CAG repeat polymorphism within the androgen receptor gene and maleness. Int J Androl. 2003;26(2):76–83. doi: 10.1046/j.1365-2605.2003.00393.x. [DOI] [PubMed] [Google Scholar]
- 28.Gubbels Bupp M.R., Jorgensen T.N. Androgen-induced immunosuppression. Front Immunol. 2018;9:794. doi: 10.3389/fimmu.2018.00794. Published 2018 Apr 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoebe K., Janssen E., Beutler B. The interface between innate and adaptive immunity. Nat Immunol. 2004;5(10):971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 30.Janeway C.A., Jr., Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 31.Lai J.J., Lai K.P., Zeng W., Chuang K.H., Altuwaijri S., Chang C. Androgen receptor influences on body defense system via modulation of innate and adaptive immune systems: lessons from conditional AR knockout mice. Am J Pathol. 2012;181(5):1504–1512. doi: 10.1016/j.ajpath.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medzhitov R., Janeway C., Jr. Innate immunity. N Engl J Med. 2000;343(5):338–344. doi: 10.1056/NEJM200008033430506. [DOI] [PubMed] [Google Scholar]
- 33.Pierotti S., Lolli F., Lauretta R. Androgen modulation of pro- inflammatory and anti-inflammatory cytokines during preadipocyte differentiation. Horm Mol Biol Clin Investig. 2010;4(1):483–488. doi: 10.1515/HMBCI.2010.059. [DOI] [PubMed] [Google Scholar]
- 34.Ackerman C.M., Lowe L.P., Lee H. Ethnic variation in allele distribution of the androgen receptor (AR) (CAG)n repeat. J Androl. 2012;33(2):210–215. doi: 10.2164/jandrol.111.013391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porcheddu R., Serra C., Kelvin D., Kelvin N., Rubino S. Similarity in case fatality rates (CFR) of COVID-19/SARS-COV-2 in Italy and China. J Infect Dev Ctries. 2020;14(2):125–128. doi: 10.3855/jidc.12600. Published 2020 Feb 29. [DOI] [PubMed] [Google Scholar]
- 36.de Lusignan S., Dorward J., Correa A. Risk factors for SARS-CoV-2 among patients in the oxford royal college of general practitioners research and surveillance centre primary care network: a cross-sectional study. Lancet Infect Dis. 2020;20(9):1034–1042. doi: 10.1016/S1473-3099(20)30371-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCoy J., Wambier C.G., Herrera S. Androgen receptor genetic variant predicts COVID-19 disease severity: a prospective longitudinal study of hospitalized COVID-19 male patients [published online ahead of print, 2020 Sep 25] J Eur Acad Dermatol Venereol. 2020 doi: 10.1111/jdv.16956. 10.1111/jdv.16956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong P.W., Lee H.M., Lau E.S.H. Interactive effects of testosterone and the androgen receptor CAG repeat length polymorphism on cardiovascular- renal events and mortality in men with diabetes. Diabetes Metab Res Rev. 2019;35(1):e3081. doi: 10.1002/dmrr.3081. [DOI] [PubMed] [Google Scholar]
- 39.Caminiti G., Volterrani M., Iellamo F. Effect of long-acting testosterone treatment on functional exercise capacity, skeletal muscle performance, insulin resistance, and baroreflex sensitivity in elderly patients with chronic heart failure a double-blind, placebo-controlled, randomized study. J Am Coll Cardiol. 2009;54(10):919–927. doi: 10.1016/j.jacc.2009.04.078. [DOI] [PubMed] [Google Scholar]
- 40.Canale D., Caglieresi C., Moschini C. Androgen receptor polymorphism (CAG repeats) and androgenicity [published correction appears in Clin Endocrinol (Oxf). 2005 Oct; 63(4):482] Clin Endocrinol (Oxf) 2005;63(3):356–361. doi: 10.1111/j.1365-2265.2005.02354.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The samples referenced here are housed in the GEN-COVID Patient Registry and the GEN-COVID Biobank and are available for sharing. The sequencing data are deposited in http://www.nig.cineca.it/, specifically, http://nigdb.cineca.it) and available for consultation. For further information, you may contact the corresponding author, Prof. Alessandra Renieri (e-mail: alessandra.renieri@unisi.it).