Key Points
Markers of neutrophil activation (RETN, LCN2, HGF, IL-8, G-CSF) are among the most potent discriminators of critical illness in COVID-19.
Evidence of neutrophil activation precedes the onset of critical illness and predicts mortality in COVID-19.
Abstract
Pathologic immune hyperactivation is emerging as a key feature of critical illness in COVID-19, but the mechanisms involved remain poorly understood. We carried out proteomic profiling of plasma from cross-sectional and longitudinal cohorts of hospitalized patients with COVID-19 and analyzed clinical data from our health system database of more than 3300 patients. Using a machine learning algorithm, we identified a prominent signature of neutrophil activation, including resistin, lipocalin-2, hepatocyte growth factor, interleukin-8, and granulocyte colony-stimulating factor, which were the strongest predictors of critical illness. Evidence of neutrophil activation was present on the first day of hospitalization in patients who would only later require transfer to the intensive care unit, thus preceding the onset of critical illness and predicting increased mortality. In the health system database, early elevations in developing and mature neutrophil counts also predicted higher mortality rates. Altogether, these data suggest a central role for neutrophil activation in the pathogenesis of severe COVID-19 and identify molecular markers that distinguish patients at risk of future clinical decompensation.
Visual Abstract
Introduction
As the death toll from COVID-19 exceeds 2.1 million worldwide, it remains a pressing concern to understand how the disease causes such a wide spectrum of clinical outcomes. For most patients, COVID-19 manifests as an upper respiratory tract infection that is self-limited. However, the progression of COVID-19 in a large subset of patients to respiratory distress, multiorgan failure, and death has resulted in an enormous global impact. A number of studies have highlighted important roles for monocytes and macrophages in severe COVID-19,1,2 but our knowledge of the immunologic drivers of critical illness is otherwise limited.
To achieve a deeper understanding of the immunologic phenotypes of COVID-19 across the spectrum of disease severity, we performed proteomic profiling of blood obtained from hospitalized patients with COVID-19 and used a machine learning algorithm to define the biomarkers that best discriminate between critically ill patients and those with milder disease. We discovered a unique neutrophil activation signature composed of neutrophil activators (granulocyte colony-stimulating factor [G-CSF] and interleukin-8 [IL-8]) and neutrophil-derived effectors (resistin [RETN], lipocalin-2 [LCN2], and hepatocyte growth factor [HGF]), which had the greatest power of all measured biomarkers to identify critically ill patients. These effector proteins were highly transcriptionally enriched in a developing neutrophil population that was recently identified specifically in critically ill COVID-19 patients and were strongly correlated with absolute neutrophil count (ANC) in our patient cohorts.3,4 This signature of neutrophil activation was predictive of in-hospital mortality and, most compellingly, was elevated at the time of hospital admission in patients who only later progressed to critical illness, thus preceding and predicting the onset of critical illness. We identify neutrophil activation as a defining feature of severe COVID-19 that occurs before the onset of critical illness, implicating neutrophils as a central player in the pathogenesis of severe COVID-19 and highlighting opportunities for clinical prediction and therapeutic intervention.
Methods
Study design and participants
We first conducted a study of 49 adult patients admitted to Yale-New Haven Hospital between 13 and 24 April 2020 with a confirmed diagnosis of COVID-19 via polymerase chain reaction (cross-sectional cohort). The protocol was approved by the Yale University Institutional Review Board (IRB; IRB 2000027792). Plasma was analyzed from 40 patients treated in a medical intensive care unit (ICU) and 9 patients treated on standard medical floors (non-ICU) in our hospital. Admission to the ICU was based on established ICU admission guidelines (supplemental Table 1). For this cohort, blood was collected at a single time point during each patient’s hospitalization (supplemental Figure 1). Blood samples from 13 additional asymptomatic, nonhospitalized controls were also analyzed after signed consents were obtained for a separate IRB-approved protocol (IRB 1401013259).
We also analyzed blood samples obtained longitudinally on days 1 (within 24 hours), 4, and 7 of hospitalization from a separate cohort of 23 consecutive adult patients admitted for treatment of laboratory-confirmed COVID-19 between 23 and 28 May 2020 who remained hospitalized until at least day 4 (longitudinal cohort). These patients were identified prospectively based on real-time electronic chart review of hospital admissions, and analysis was performed for consecutive patients who met the following criteria: (1) COVID-19–positive polymerase chain reaction test; (2) admission for the purpose of COVID-19 treatment; (3) patient identification in a time frame allowing for sample collection within 24 hours of admission; and (4) collection of at least 2 longitudinal samples (resulting in the exclusion of 8 patients discharged before day 4 sample collection).
Last, we used the Yale Department of Medicine Covid Explorer (DOM-CovX) database to evaluate laboratory results from 3325 deidentified COVID-19–positive patients admitted to the 6 hospitals within the Yale New Haven Health System (DOM-CovX cohort; IRB 2000028509). Patients with a confirmed positive COVID-19 test within 14 days preceding hospitalization were included in the cohort. This dataset combined all clinical variables extracted from the electronic medical record (Epic, Verona, WI) including demographics, comorbidities, procedures, and all laboratory values recorded during the hospitalization.
Procedures
Blood was collected in 3.2% sodium citrate tubes and centrifuged at 2000g for 20 minutes at room temperature; the resulting plasma supernatant was frozen at −80°C and used for further testing. Biomarker profiling analyses were conducted at Eve Technologies (Calgary, Alberta, Canada). For the cross-sectional cohort, the following assays were performed: Human Cytokine 71-Plex, Human Complement Panels 1 and 2, Human SAA & ADAMTS13, and Human Adipokine 5-Plex. For the longitudinal cohort, the following assays were performed: Human Cytokine 48-Plex, Human Complement Panel 1, Human Adipokine 5-Plex, and Human MMP 9-Plex and TIMP 4-Plex. Five control samples were evaluated concurrently with the longitudinal cohort samples. Heatmaps were generated using concentrations of circulating biomarkers obtained from biomarker profiling analyses using Heatmapper as described.5 For confirmation, RETN levels were also measured by enzyme-linked immunosorbent assay (ELISA) (category no. DRSN00; R&D Systems) following a fivefold sample dilution per the manufacturer’s protocol. Linear regression analysis was performed to assess the relationship between ELISA-based concentrations and values reported by Eve Technologies based on multiplex biomarker profiling.
Principal component analysis and random forest classifier
Values for biomarker profiles in the cross-sectional cohort were log-transformed. A pseudocount of half the minimum observed nonzero value per biomarker was added to each observed zero value before log 10 transformation. IL-6 was excluded from modeling because 38 of 40 patients in the ICU subset of the cross-sectional cohort had received IL-6 receptor blockade before blood collection. The remaining biomarker values were used in a principal component analysis (PCA) and a random forest classifier using the scikit-learn Python package.6 Data were partitioned into 66-33 train-validation split (training cohort: 26 ICU patients, 7 non-ICU patients, 3 controls; validation cohort: 14 ICU patients, 2 non-ICU patients, 5 controls), and all biomarkers were then used to predict ICU status. A maximal tree depth of 10 was used with the minimal cost-complexity set to 0.02; all other parameters were set to their defaults. Feature importance was assessed using mean decrease in impurity. Data restricted to 5 biomarkers of interest with high feature importance were then used in a separate random forest classifier, using the same methods.
Single cell analyses
Processed count matrices with deidentified metadata and embeddings from a single-cell RNA sequencing (scRNAseq) dataset published by Wilk et al3 were downloaded from the COVID-19 Cell Atlas (https://www.covid19cellatlas.org/#2iki20) hosted by the Wellcome Sanger Institute. This dataset was further analyzed using Seurat (version 3.0, https://satijalab.org/seurat). Clusters in this object were renamed based on cell type, using UMAPs and provided heatmaps. A new metadata column, ARDSstatus, was added to the object, which grouped samples into Control, COVID-19–non-acute respiratory distress syndrome (ARDS), and COVID-19–ARDS categories. Violin and feature plots were generated to examine specific genes of interest. Cells identified as developing neutrophil were made a subset from the overall object to examine this population more closely.
Statistical analyses
To evaluate differences in mean age across groups, we used 1-way analysis of variance with post hoc Tukey's multiple comparisons tests in the cross-sectional cohort and Kruskal-Wallis test in the longitudinal cohort, because the samples in the former cohort but not the latter conformed to the normal distribution. For comparisons of sex proportions, we used χ2 tests in the cross-sectional cohort and Fisher’s exact test in the longitudinal cohort. In both cross-sectional and longitudinal cohorts, we examined differences in proportions of patients with comorbidities using Fisher’s exact test.
To compare biomarker values, we used 2-sample tests. Because most sample distributions did not satisfy Anderson-Darling and D'Agostino-Pearson tests of normality, we used the unpaired 2-tailed Mann-Whitney U test. For samples that conformed to the normal distribution, we used unpaired 2-sided Student t tests with Welch correction for unequal variances, where applicable; where appropriate, P values were corrected for multiple comparisons using the false discovery rate procedure with the discovery rate Q of 5%. P < .05 was considered significant.
Correlation coefficients between ANC and each biomarker were computed using Spearman’s rank method. For Kaplan-Meier survival analyses, we used median values of evaluated variables as cutpoints to classify patients into high and low groups, based on whether values for a particular biomarker or cell count were above or below the cutpoint. We then used log-rank test to compare the 2 groups for each evaluated variable. To determine P values for cell type–specific enrichment of genes in scRNAseq data, we used the Wilcoxon rank sum test. All statistical analyses were carried out using GraphPad Prism (v8.4.3; GraphPad Software, San Diego, CA), Stata (v16; StataCorp. College Station, TX), and R (v4; R Core Team, 2020).
Results
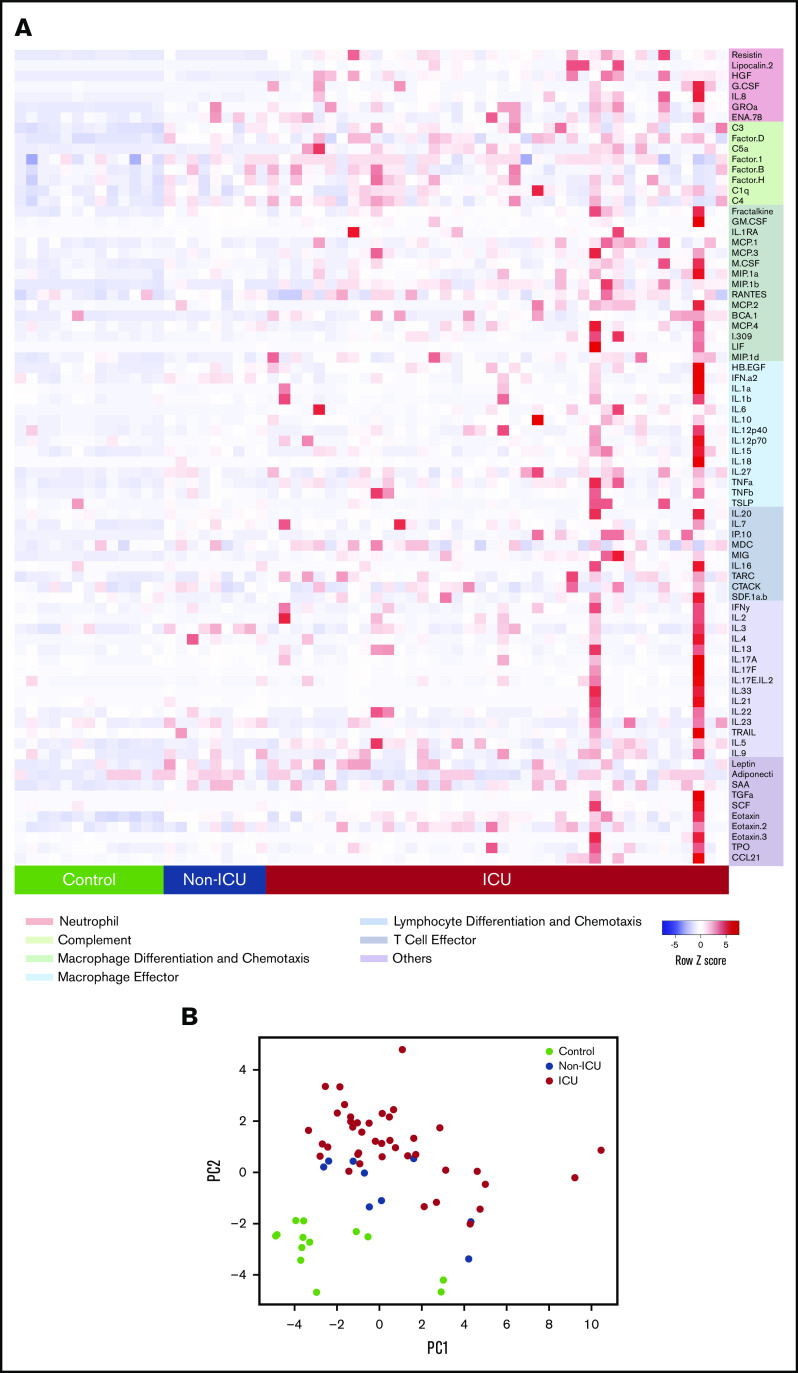
Forty-nine adult COVID-19 patients (40 in the medical ICU and 9 in non-ICU units) and 13 nonhospitalized controls were included in our initial cross-sectional cohort (Table 1). We performed multiplexed biomarker profiling of plasma to measure circulating concentrations of 78 proteins with immunologic functions (Figure 1A; supplemental Figure 2). PCA showed separation of the control, non-ICU COVID-19, and ICU COVID-19 samples, indicating that these circulating markers capture a spectrum of illness severity (Figure 1B).
Table 1.
Demographics of patients in the cross-sectional cohort
| Subjects | ICU(N = 40) | Non-ICU (N = 9) | Controls (N = 13) | P | ||
|---|---|---|---|---|---|---|
| Mean age (SD), y | 62 (16) | 69 (21) | 48 (10) | 0.0062* | ||
| ICU vs non-ICU, 0.4559† | ||||||
| ICU vs controls, 0.0196† | ||||||
| Non-ICU vs controls, 0.0089† | ||||||
| Sex, n (%) | Male | 30 (75) | 3 (33) | 5 (39) | 0.0112‡ | |
| Female | 10 (25) | 6 (67) | 8 (61) | ICU vs non-ICU, 0.043§ | ||
| ICU vs controls, 0.022§ | ||||||
| Non-ICU vs controls, 0.806§ | ||||||
| Comorbidities, n (%) | Obesity|| | 23 (58) | 3 (33) | 0.27¶ | ||
| CHF | 4 (10) | 0 (0) | 1.0¶ | |||
| Hyperlipidemia | 11 (28) | 1 (11) | 0.42¶ | |||
| Hypertension | 24 (60) | 6 (67) | 1.0¶ | |||
| Diabetes | 12 (30) | 1 (11) | 0.41¶ | |||
| CAD, MI, or heart disease | 6 (15) | 0 (0) | 0.58¶ | |||
| Atrial fibrillation | 3 (8) | 0 (0) | 1.0¶ | |||
| Stroke or TIA | 3 (8) | 2 (22) | 0.22¶ | |||
| CKD | 6 (15) | 0 (0) | 0.58¶ | |||
| Active malignancy | 3 (8) | 0 (0) | 1.0¶ | |||
CAD, coronary artery disease; CHF, congestive heart failure; CKD, chronic kidney disease; MI, myocardial infarction; SD, standard deviation; TIA, transient ischemic attack.
One-way analysis of variance
Post hoc Tukey's multiple comparisons tests
Group-wise χ2 test.
Individual χ2 tests.
Obesity is defined as body mass index >30 kg/m2.
Fisher's exact test.
Figure 1.
Circulating biomarkers separate COVID-19 patients according to disease severity. (A) Heatmap of proteomic data from the cross-sectional cohort, indicating relative protein levels detected in each subject (columns) for all biomarkers tested (rows). Proteins are categorized by biological function. (B) Visualization of the first 2 principal components (PCs) of a PCA of all biomarker data for each subject.
To assess whether this profile of plasma biomarkers could distinguish between critically ill and noncritically ill patients, we applied a random forest machine learning prediction model. The model was trained on data from two-thirds of the patients, and its ability to predict ICU status was tested on the remaining one-third. The model accurately identified 14 of 14 patients in the ICU and 7 of 7 subjects not in the ICU (Figure 2A). To avoid introducing bias based on treatment, the random forest model and PCA analyses excluded IL-6 because most (38 of 40) ICU patients received tocilizumab, which is known to increase IL-6 levels.2 These results demonstrate that our proteomic profile provides a reliable circulating signature of critical illness in COVID-19.
Figure 2.
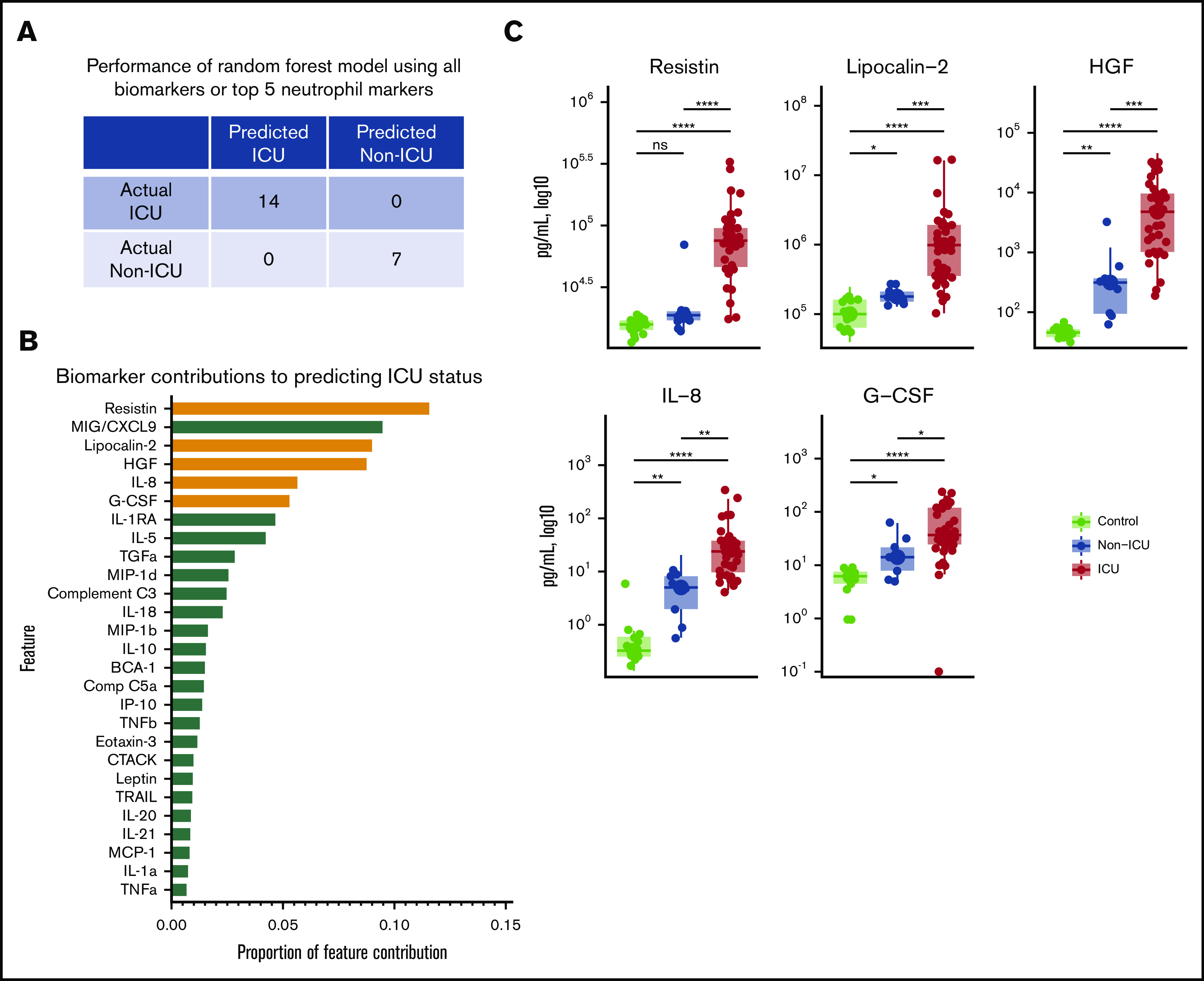
Markers of neutrophil activation accurately identify patients with critical illness. (A) Performance of a random forest (RF) model trained on data from two-thirds of the study subjects in the cross-sectional cohort, predicting ICU status for the remaining one-third of subjects not included in the training set. Perfect classification as depicted is achieved when using data from all biomarkers or from only the top 5 neutrophil markers (highlighted in orange in panel B). (B) Feature importance ranked by proportion of feature contribution to the RF model. Members of the neutrophil activation signature are highlighted in orange. (C) Comparisons of circulating levels of neutrophil markers in controls, non-ICU COVID-19, and ICU COVID-19 patients in the cross-sectional cohort. Asterisks denote statistically significant differences between groups (*P < .05, **P < .01, ***P < .001, ****P < .0001).
To gain insight into the specific plasma proteins that may affect disease severity, we next examined the importance of each feature in the random forest prediction model. Five of the top 6 features contributing to the model were proteins related to neutrophil activation (Figure 2B-C). Three of the top 4 features were RETN, LCN2, and HGF, each produced by neutrophils, stored in neutrophil secondary granules, and released on neutrophil activation.7-12 The next 2 highest ranking features were IL-8 and G-CSF, which stimulate neutrophil chemotaxis and development, respectively.13,14
Next, we assessed whether these 5 proteins alone could discriminate between critically ill (ICU) and noncritically ill (non-ICU) patients. We repeated random forest modeling using only the markers related to neutrophil activation (RETN, LCN2, HGF, IL-8, and G-CSF) and found that circulating levels of this selective panel also accurately classified all test subjects into ICU and non-ICU categories (Figure 2A). Interestingly, each of the neutrophil activation markers showed greater discriminatory power for critical illness than previously described monocyte/macrophage markers (with the exception of MIG/CXCL9) that were also included in our proteomic panel (Figure 1A; Figure 2B; supplemental Figure 2),2,15-17 suggesting that neutrophils may play a central role in critical illness associated with COVID-19.
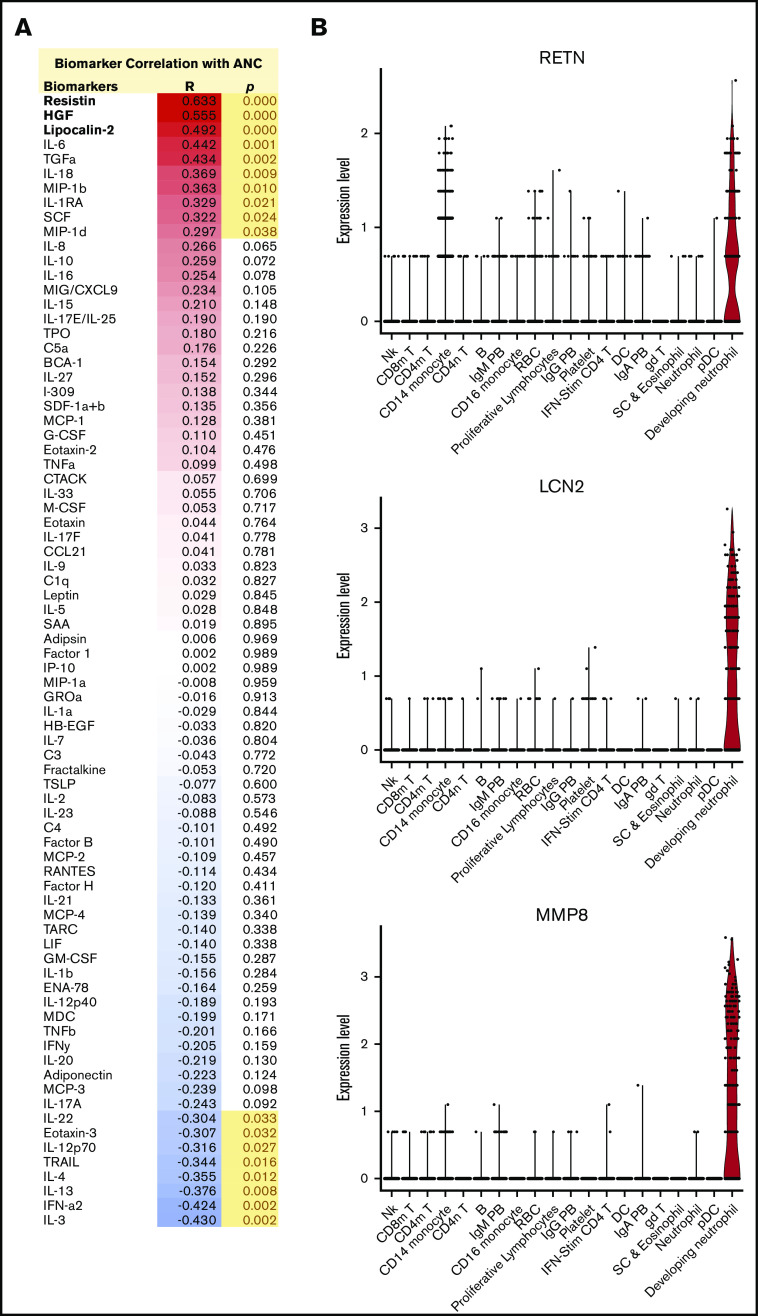
Although the neutrophil granule proteins RETN, LCN2, and HGF are produced in large quantities by neutrophils, they can also be secreted by other cell types. To determine whether increases in these proteins were associated with other indicators of neutrophil activity, we analyzed the correlation of the ANC at the time of blood draw with all circulating markers measured. We found that RETN, HGF, and LCN2 were the 3 proteins that most strongly correlated with ANC (Figure 3A), further suggesting that neutrophils are the likely common source of these proteins. We then analyzed data from a scRNAseq study of peripheral blood mononuclear cells (PBMCs) from patients with COVID-19.3 We found that RETN and LCN2 transcripts were highly enriched in a circulating cell population designated as developing neutrophils (P < 10e−300 for each), which was detected almost exclusively in severely ill patients with ARDS (Figure 3B; supplemental Figure 3).3 Another highly enriched marker (P < 10e−300) for this population was matrix metallopeptidase 8 (MMP8) or neutrophil collagenase (Figure 3B). Notably, constituents of neutrophil granules, like RETN, LCN2, and MMP8, are transcribed at earlier stages of neutrophil development, packaged into granules, and later released from mature neutrophils on degranulation.18 This set of observations strongly supports the conclusion that neutrophils are the primary source of these circulating markers of critical illness in COVID-19.
Figure 3.
Circulating neutrophil granule proteins are likely derived from neutrophilic source in COVID-19 patients. (A) Correlations of circulating biomarkers with ANC in the cross-sectional cohort. Neutrophil granule proteins, which show the highest correlation with ANC, are highlighted. R, Spearman’s rank correlation coefficient; P, P value. (B) Violin plots of RETN, LCN2, and MMP-8 mRNA, showing enrichment in a developing neutrophil population, based on reanalysis of single-cell RNAseq data published by Wilk et al3 of PBMCs from patients with COVID-19.
In our initial cross-sectional cohort, blood was collected during the course of hospitalization, limiting our capacity to determine whether the observed neutrophil signature was a consequence of critical illness or whether it preceded its onset. To address this, we established a second longitudinal cohort of patients. Proteomic plasma profiling was conducted on blood samples collected serially, starting on day 1 (within 24 hours of hospital admission), from 23 consecutive patients admitted for treatment of confirmed COVID-19 who remained hospitalized for ≥4 days (Figure 4A; supplemental Figures 4 and 5). To further test the role of neutrophil activation, we included a panel of matrix metalloproteinases (MMPs) and tissue inhibitor of metalloproteinases including MMP-8, a marker of the developing neutrophil population described above (Figure 3B).19 Consistent with our findings in the cross-sectional cohort, RETN, HGF, and LCN2 in the day 1 samples of the longitudinal cohort showed strong correlations with ANC, and MMP-8 was the protein most highly correlated with ANC, further supporting a neutrophilic source of these proteins in COVID-19 (supplemental Figure 6).
Figure 4.
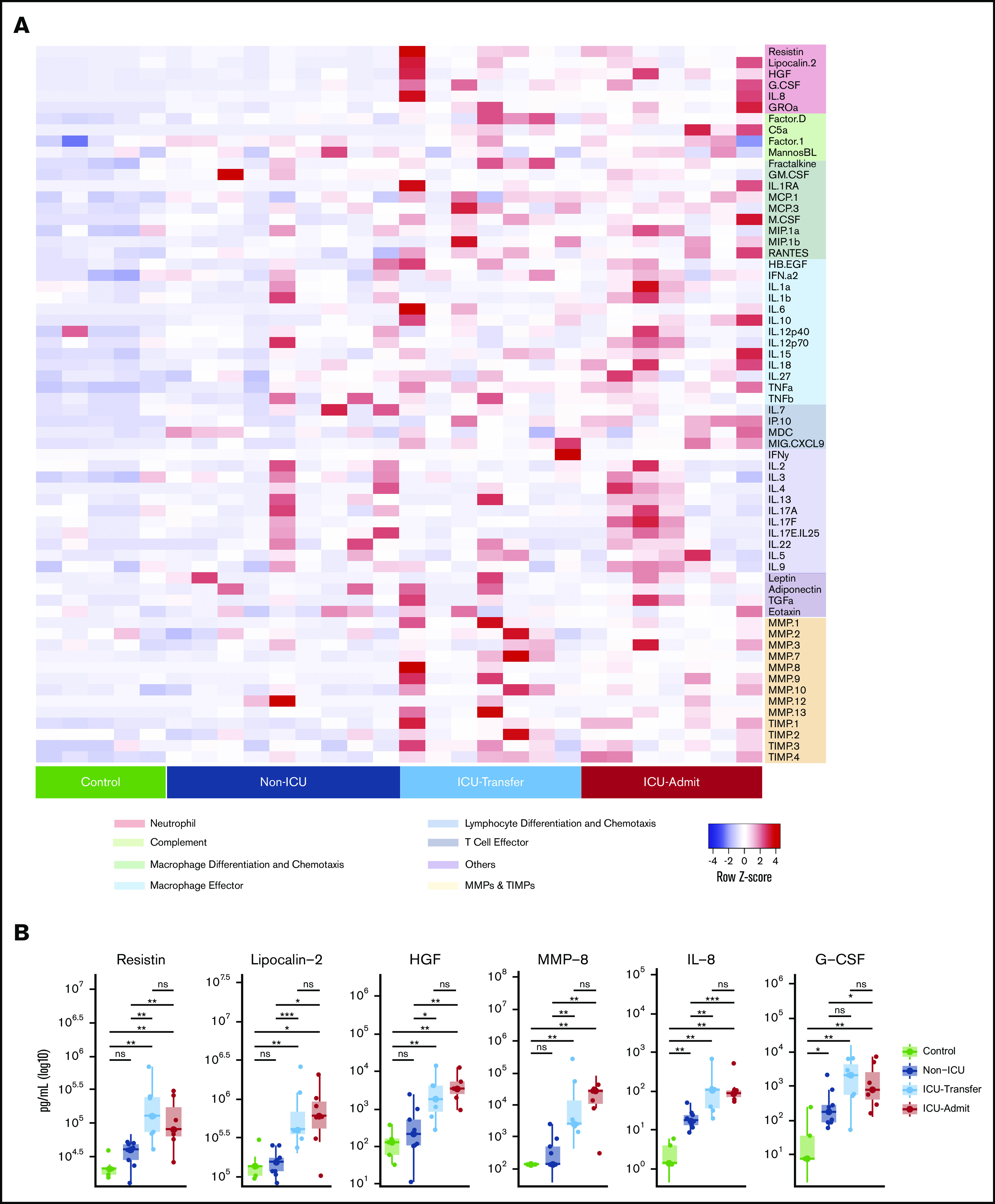
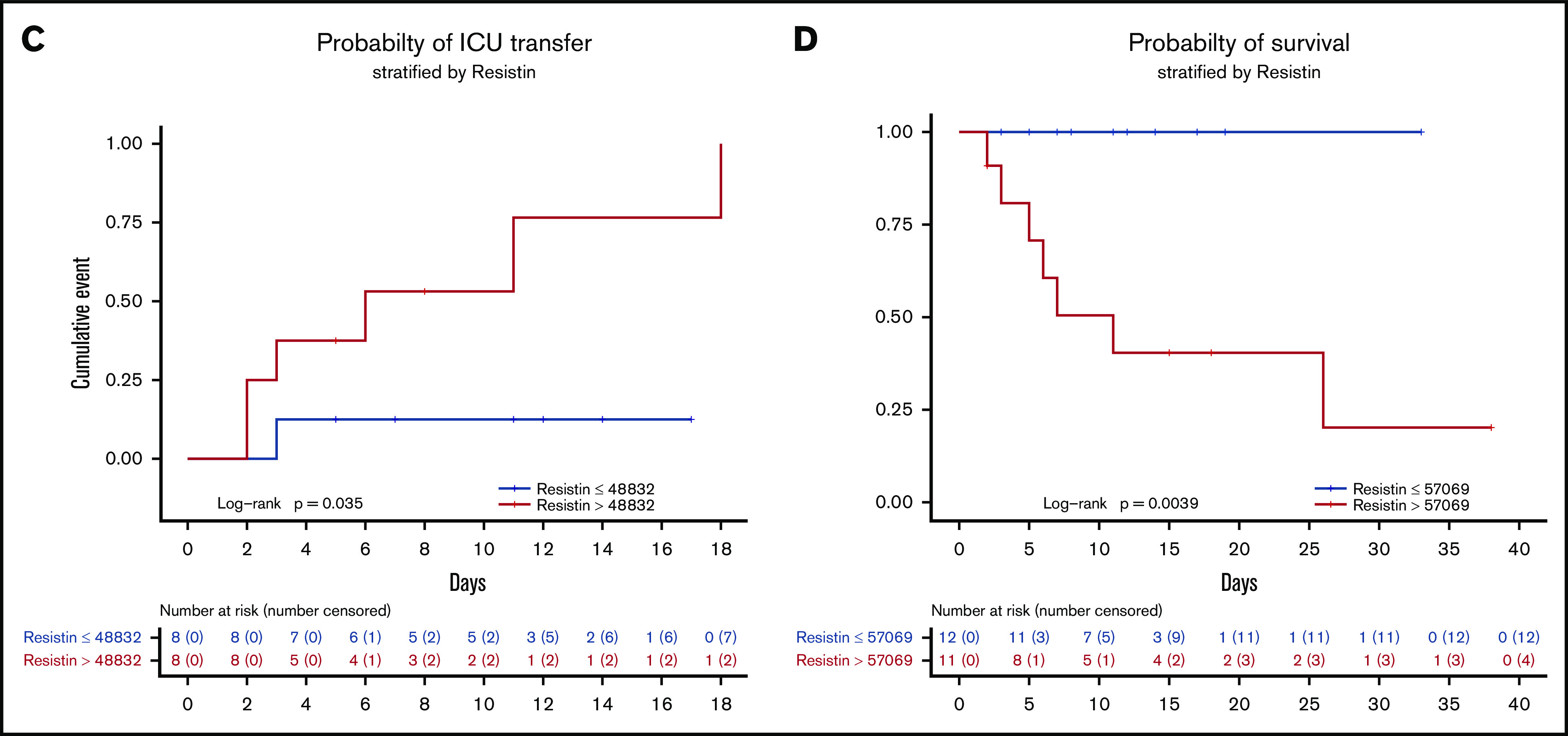
Elevation of the neutrophil activation signature precedes the onset of critical illness. (A) Heatmap indicating relative protein levels detected on day 1 in each subject in the longitudinal cohort (columns) for all biomarkers tested (rows). Proteins are categorized by biological function. (B) Comparisons of circulating levels of neutrophil markers in subjects categorized as controls, non-ICU, ICU-Transfer, and ICU-Admit. Non-ICU indicates patients who remained in a non-ICU unit until discharge; ICU-Transfer indicates patients who were admitted to a non-ICU unit and were transferred to an ICU unit during hospitalization; ICU-Admit indicates patients who were admitted directly to an ICU unit. Asterisks denote statistically significant differences between groups (*P < .05, **P < .01, ***P < .001, ****P < 0.0001). (C) Kaplan-Meier curve depicting the likelihood of ICU admission depending on resistin (RETN) levels on day 1, using the median of the group (non-ICU and ICU-Transfer) as the cutoff value (pg/mL). (D) Kaplan-Meier curve depicting the likelihood of survival depending on resistin levels on day 1, using the median of the group (entire longitudinal cohort) as the cutoff value (pg/mL). ns, not significant.
Sixteen patients in the longitudinal cohort were initially admitted to lower-acuity non-ICU units, and 7 were admitted directly to the ICU (ICU-Admit) (Table 2). Of the patients admitted to non-ICU units, 9 remained in those units until discharge (non-ICU), whereas 7 were later transferred to the ICU (ICU-Transfer). Consistent with our observations in the cross-sectional cohort, ICU-Admit patients had significantly higher levels of all neutrophil activation markers than non-ICU patients on day 1 of hospitalization (Figure 4B; supplemental Figure 4b). As predicted, the same pattern was observed for MMP-8, which, like RETN, HGF, and LCN2, is known to be stored and released from neutrophil secondary granules and is transcriptionally enriched in developing neutrophils in COVID-19 patients (Figure 3B).20,21
Table 2.
Demographics of patients in the longitudinal cohort
| Subjects | Controls (N = 5) | Non-ICU (N = 9) | ICU-Transfer (N = 7) | ICU-Admit (N = 7) | P | |
|---|---|---|---|---|---|---|
| Mean age (SD), y | 48 (11.5) | 63.6 (10.6) | 70.1 (17.5) | 62.6 (9.8) | 0.09* | |
| Sex, n (%) | Male | 2 (40) | 5 (56) | 3 (43) | 6 (86) | 0.403† |
| Female | 3 (60) | 4 (44) | 4 (57) | 1 (14) | ||
| Comorbidities, n (%) | Obesity‡ | 4 (44) | 4 (57) | 3 (43) | 1.0† | |
| CHF | 2 (22) | 1 (14) | 1 (14) | 1.0† | ||
| COPD or asthma | 2 (22) | 3 (43) | 2 (29) | 0.84† | ||
| Hyperlipidemia | 4 (44) | 5 (71) | 3 (43) | 0.58† | ||
| Hypertension | 6 (67) | 4 (57) | 3 (43) | 0.86† | ||
| Diabetes | 3 (33) | 4 (57) | 2 (29) | 0.64† | ||
| CAD, MI, or heart disease | 2 (22) | 2 (29) | 2 (29) | 1.0† | ||
| Stroke or TIA | 1 (11) | 0 | 2 (29) | 0.46† | ||
| CKD | 0 | 3 (43) | 1 (14) | 0.07† | ||
| Active malignancy | 0 | 1 (14) | 1 (14) | 0.50† | ||
COPD, chronic obstructive pulmonary disease.
Kruskal-Wallis test.
Fisher's exact test.
Obesity is defined as body mass index >30 kg/m2.
Remarkably, the levels of the neutrophil activation markers on day 1 of hospitalization were also significantly elevated in ICU-Transfer patients (who were at that time in non-ICU units), at levels comparable to ICU-Admit patients and well above levels in non-ICU patients (Figure 4B). In both ICU-Transfer and ICU-Admit patients, the neutrophil activation markers remained elevated from the day of admission to day 7 and did not change appreciably over time, whereas these levels in non-ICU patients remained stably low (supplemental Figure 4b). Thus, despite the fact that patients in the ICU-Transfer group were not critically ill at the time of the day 1 blood draw, evidence of neutrophil activation was already present. The neutrophil activation signature identified patients who were primed for eventual transfer to the ICU before the onset of critical illness.
Previously reported markers of macrophage activation, including IL-6, IL-10, tumor necrosis factor-α (TNF-α), and MIG/CXCL9, were also elevated in ICU-Admit and ICU-Transfer patients compared with non-ICU patients on day 1 of hospitalization (supplemental Figure 5). Notably, most of these markers (as well as IL-8 and G-CSF) were also significantly elevated in non-ICU patients compared with controls, whereas the neutrophil granule proteins (RETN, LCN2, HGF, MMP-8) that may more directly reflect neutrophil activation were not elevated in non-ICU patients compared with controls and were only significantly different in patients who later became critically ill (Figure 4B; supplemental Figure 5). This suggests that proteins released by neutrophil degranulation may be more specific to severe COVID-19 than elevations in traditional cytokines. Altogether, these longitudinal data offer the first evidence, to our knowledge, that neutrophil activation and monocyte/macrophage activation precede the onset of critical illness.
To further validate our findings, we conducted additional analyses using RETN, the factor most important in distinguishing critical illness in our random forest prediction model. Among patients initially admitted to non-ICU units (non-ICU, ICU-Transfer), those with day 1 RETN levels above the median value were much more likely to later require ICU transfer (Figure 4C). Moreover, patients with day 1 RETN levels above the median were significantly less likely to survive (Figure 4D). To confirm these results, we also measured RETN levels in the longitudinal cohort by a separate ELISA assay and found that concentrations measured by ELISA showed a strong linear relationship with those measured by multiplex profiling (R2 = 0.9863). In addition to RETN, we found that higher levels on hospital day 1 of LCN2, G-CSF, IL-8, and MMP-8, as well as IL-6, IL-10, TNF-α, IL-1RA, and M-CSF, were significantly associated with mortality (supplemental Figure 7). Meanwhile, we found that the neutrophil to lymphocyte ratio (NLR), previously described as a prognostic indicator in COVID-19,16 was not significantly different between non-ICU and ICU-Transfer patients on day 1 of hospitalization (supplemental Figure 8a), suggesting that NLR does not distinguish critical illness as clearly as the molecular markers, which may more directly reflect cellular activation. Among established clinical markers of severe COVID-19, C-reactive protein and D-dimer were elevated in ICU-Transfer and ICU-Admit patients compared with non-ICU patients (supplemental Figure 8a), but none of these markers were significantly different between patients who did and did not survive their hospitalization (supplemental Figure 8b), unlike the neutrophil and macrophage activation markers (supplemental Figure 7).
Last, we explored whether these mechanistic insights regarding the role of neutrophils in severe COVID-19 could be validated in a large patient population hospitalized with COVID-19. Using a database of 3325 patients admitted to the Yale-New Haven Health System who tested positive for SARS-CoV-2 (DOM-CovX cohort; Table 3), we examined the first recorded values of immature granulocyte (IG) and neutrophil counts during the course of hospitalization. We used these early values to avoid potential confounding effects of hospitalization, such as therapeutic interventions and secondary infections. We found that in-hospital mortality was significantly higher among patients with elevated initial IG absolute count (Figure 5A), IG percent (Figure 5B), and ANC (Figure 5C). Interestingly, we did not find that initial absolute monocyte count was associated with increased mortality (Figure 5D). Although our molecular markers that would allow us to assess neutrophil activation are not available in this large cohort, these findings confirm that early signs of neutrophil development are associated with future mortality in one of the largest COVID-19 patient cohorts assessed to date.
Table 3.
Demographics of patients in the DOM-CovX cohort
| Subjects | N = 3325 | |
|---|---|---|
| Mean age (SD), y | 63.2 (19) | |
| Sex, n (%) | Male | 1665 (50) |
| Female | 1660 (49) | |
| Comorbidities, n (%) | Obesity* | 1096 (33) |
| CHF | 803 (24) | |
| Chronic pulmonary disease | 1112 (33) | |
| Hypertension | 2199 (66) | |
| Diabetes | 1360 (41) | |
| CKD | 803 (24) | |
| Active malignancy | 382 (11) | |
Obesity is defined as body mass index >30 kg/m2.
Figure 5.
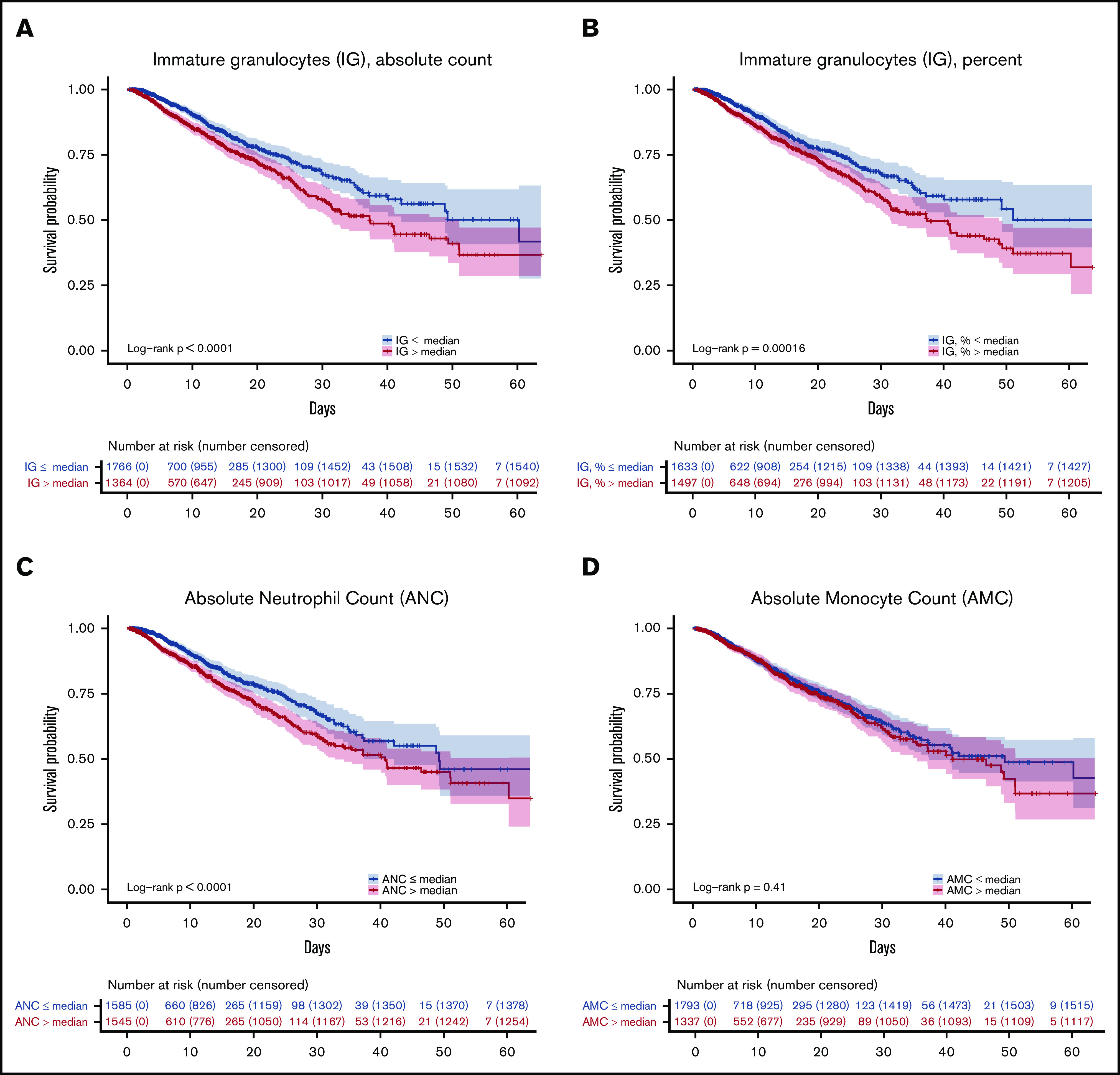
Early elevations in developing and mature neutrophil counts predict increased mortality. Kaplan-Meier curves depicting likelihood of survival based on patients’ first recorded values in the DOM-CovX cohort of absolute immature granulocyte count (A), immature granulocyte percent (B), ANC (C), and absolute monocyte count (D), using the median value as the cutoff.
Discussion
Our analyses of multiple hospitalized patient cohorts with COVID-19 reveal a prominent signature of neutrophil development and activation in critically ill patients with COVID-19. Moreover, we demonstrate for the first time that elevations in circulating markers of neutrophil and macrophage activation precede the onset of critical illness, identifying noncritically ill patients who are at risk of becoming critically ill.
The host immune response is essential for pathogen resistance, but it can also cause costly tissue injury.22 Neutrophils are often the first responders to infection and injury yet can also cause significant collateral damage.23 Although important roles for monocytes and macrophages in severe COVID-19 have been well described,15 a potential role for neutrophils has not received the same attention. However, suggestive evidence has begun to emerge, including the prognostic value of NLR,24 the identification of a population of immature neutrophils in the blood of critically ill COVID-19 patients, and the observation of increased neutrophils in bronchoalveolar lavage.1,3,4,25 It has remained unclear, however, whether there is heightened neutrophil activation in severe COVID-19 and whether the observed innate immune response is a consequence or potential cause of critical illness.
Using proteomic profiling and a machine learning prediction algorithm, we determined that several markers of neutrophil activation had the greatest discriminatory power for detecting critical illness, outperforming many other cytokines previously associated with severe COVID-19.16,17 This neutrophil signature includes G-CSF and IL-8, which can drive neutrophil activation and have been detected by other groups, with a recent study reporting an association of IL-8 levels with survival.2,26
Unlike in prior studies, we identify a group of proteins that are released from neutrophils themselves. RETN, LCN2, and MMP8 are all novel markers in COVID-19 (whereas HGF has been reported),26 and the fact that they are among the strongest predictors of critical illness offers perhaps the most direct evidence to date that neutrophil activation is a hallmark of severe disease. These signs of conspicuous neutrophil activation may not have been detected in other analyses because neutrophil granule proteins are not included in standard cytokine panels.
One limitation of this study is that we did not directly demonstrate that neutrophils are the source of these proteins in COVID-19. However, multiple lines of evidence support this conclusion. First, RETN, LCN2, HGF, and MMP8 are all well-established products of neutrophils and are each known to be stored in the same subcellular compartment, from which they would be released together on neutrophil activation.8-11,27,28 Second, consistent with this, we found that each of these proteins was highly correlated with neutrophil counts and with one another. Third, by interrogating published scRNAseq data from blood samples of COVID-19 patients, where a developing neutrophil population was recently identified specifically in critically ill patients,3 we found that RETN, LCN2, and MMP-8 were highly enriched in these neutrophil precursors and were not expressed at high levels in any other circulating cell population. Fourth, our findings are supported by other studies that have observed shifts in neutrophil populations in both the blood and lungs of patients with severe COVID-19.1,3,4,25 Importantly, we have not relied on the specificity of any single biomarker but rather the combination of all of these proteins (including MMP-8, which we measured as an additional test of our hypothesis) to draw the inference that their high levels are likely attributable to neutrophil activation. Nonetheless, we cannot exclude contributions from other cellular sources.
The temporal relationship between markers of immune activation and the onset of critical illness in COVID-19 has not been previously established. We reasoned that if neutrophils play a causal role in the development of critical illness, then evidence of neutrophil activation should be present before its onset. Remarkably, in our longitudinal cohort of patients, we found that the neutrophil activation signature was already elevated on day 1 of hospitalization in patients who were not critically ill at that time but later developed critical illness and required transfer to the ICU. Among all patients, day 1 elevations in neutrophil activation markers also predicted increased mortality. These observations additionally held for a few cytokines associated with monocyte/macrophage activation (IL-6, IL-10, TNF-α), although these proteins were also markedly increased in non-ICU patients compared with controls, indicating that they are less specific to critical illness than the neutrophil signature. Altogether, these findings indicate that neutrophil activation specifically, and innate immune activation more broadly, precede the onset of critical illness and may play a key role in its pathogenesis.
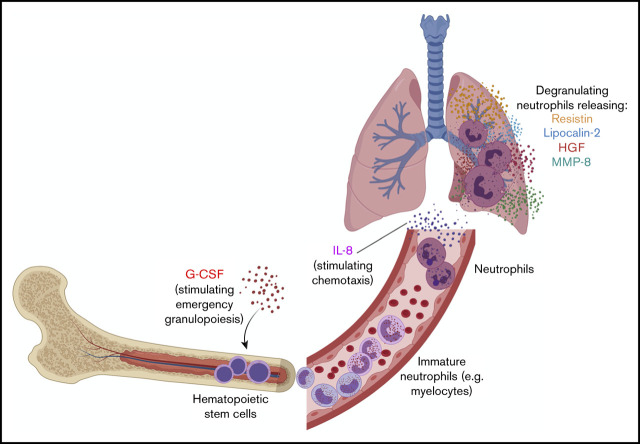
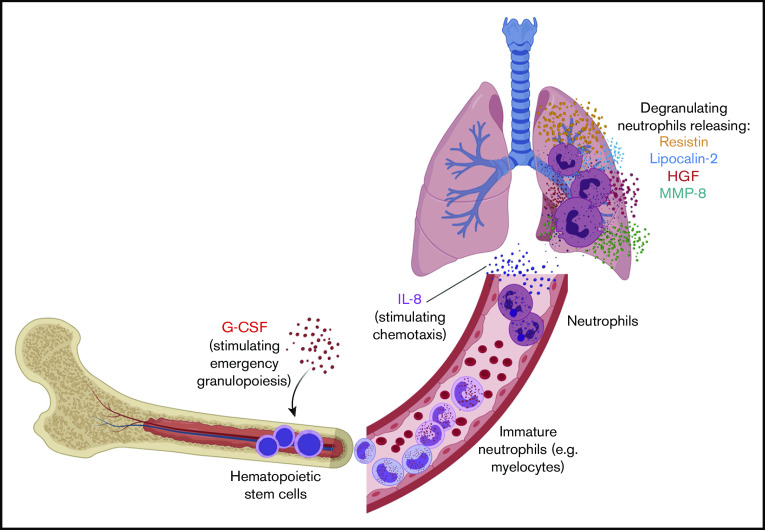
Our findings suggest a model of neutrophil development and activation in the pathogenesis of severe COVID-19 (Figure 6). We hypothesize that high levels of G-CSF stimulate emergency granulopoiesis to increase neutrophil production and that IL-8 (CXCL8) drives neutrophil migration into the lungs and perhaps other tissues.13,14,29 There, neutrophils are activated and release RETN, LCN2, HGF, MMP8, and other proteins with antimicrobial and other inflammatory functions that also cause significant collateral damage to the lungs, vasculature, and other organs.
Figure 6.
Model of neutrophil development and activation driving pathogenesis of severe COVID-19. We hypothesize that high levels of G-CSF drive emergency granulopoiesis, stimulating rapid neutrophil development and egress of immature neutrophils into the bloodstream. These cells then differentiate into neutrophils and are attracted to the lung, and perhaps other tissues, by the chemokine IL-8 (CXCL8). When activated, these mature neutrophils degranulate, releasing granule proteins that include resistin, lipocalin-2, HGF, and MMP-8. Neutrophil activation causes significant collateral damage that may contribute to severe COVID-19 pathology and clinical decompensation. Created with BioRender.com.
The occurrence of emergency granulopoiesis in COVID-19, which we propose in our model, is suggested by the presence of a developing neutrophil population specifically in severely ill patients.3,4 We find that these developing neutrophils transcribe the genes encoding the circulating proteins (RETN, LCN2, MMP8) that we detect in the blood of critically ill patients. This is consistent with the sequence of events described in the neutrophil literature: neutrophil secondary granule contents are transcribed by the transcription factor C/EBPε during the myelocyte stage of development, are packaged into granules, and are later released from mature neutrophils.9,18,28,30,31 Typically, this neutrophil development takes place in the bone marrow, but during emergency granulopoiesis (driven by G-CSF, which we detect at elevated levels in patients with severe disease), immature granulocytes can be detected in the circulation (Figure 6).13 Our model accounts for the detection of developing neutrophils in the blood of only severely ill COVID-19 patients and is supported by our analysis of a hospital-wide dataset, where we find that the earliest counts of immature granulocytes predict mortality.
The neutrophil granule proteins that we detected may simply be markers of neutrophil activation or may themselves be detrimental to patients with COVID-19. RETN regulates production of multiple cytokines that are elevated in severely ill COVID-19 patients, including IL-6, IL-8, and TNF-α,32 and is associated with increased endothelial permeability and thrombosis risk, another feature of severe COVID-19.33 LCN2 is upregulated in multiple autoimmune diseases.34,35 Elevated levels of MMP-8 are associated with worsening clinical outcomes in pediatric patients with ARDS.35
Neutrophils are known to contribute to acute lung injury in other contexts, including viral lung infections, and their role in COVID-19 may represent an exaggerated version of that pathophysiology.36,37 Interestingly, a recent study demonstrated a higher degree of neutrophil activation in severe COVID-19 than in influenza pneumonia.38 Our findings raise essential questions. What are the initial triggers for neutrophil development, recruitment, and activation in COVID-19? Is there a threshold of injury to the lungs or vascular beds that stimulates neutrophil activation,39,40 setting in motion the cascade of events that propagate critical illness in COVID-19? Directly testing the causal role of neutrophil activation in COVID-19 will require either animal models that recapitulate neutrophil activation or clinical trials of targeted therapeutic interventions. Therapeutic strategies such as inhibition of G-CSF or IL-8 should be considered, but the potential benefits of these approaches will need to be weighed carefully against the risks of immunosuppression. Although multiple trials are underway for granulocyte-macrophage colony-stimulating factor (GM-CSF) inhibitors, this growth factor has a distinct functional role from G-CSF, and we observe an association of only G-CSF levels with disease severity in this study.41,42 To our knowledge, there are no ongoing clinical trials of G-CSF inhibitors in COVID-19. Overall, our study highlights a central role for neutrophil activation in the pathogenesis of severe COVID-19, which may help guide the development of new therapeutic strategies and more accurate predictive markers of severe disease.
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors thank all front-line staff taking care of our community of patients affected by COVID-19 and are grateful for the multidisciplinary collaboration at the Yale School of Medicine and Yale-New Haven Health System in the care of our community.
This work was supported by a gift donation from Jack Levin to the Benign Hematology program at Yale, the National Institutes of Health, National Heart, Lung, and Blood Institute (HL142818 [H.J.C.] and HL139116 [M.L.M.]), National Institute of Diabetes and Digestive and Kidney Diseases (DK079310 and DK113191) (F.P.W.), and National Institute of General Medical Sciences (GM136651) (M.L.M.), the American Heart Association, Transformational Project Award (H.J.C.), and the DeLuca Center for Innovation in Hematology Research at Yale Cancer Center.
Footnotes
For data sharing, please contact the corresponding authors at hyung.chun@yale.edu or alfred.lee@yale.edu.
Authorship
Contribution: M.L.M., A.B.P., J.D.B., G.G., E.R.N., M. Simonov, D.v.D., A.I.L., and H.J.C. designed and executed the project; C-H.C. and H.Z. performed experiments related to the project; P.B. and H.M.R. oversaw sample collection; M. Simonov created the DOM-CovX database; all authors provided valuable ideas and contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hyung J. Chun, Yale Cardiovascular Research Center, 300 George St, Room 770H, New Haven, CT 06511; e-mail: hyung.chun@yale.edu; and Alfred I. Lee, Yale Hematology, 333 Cedar St, PO Box 208028, New Haven, CT 06520-8028; e-mail: alfred.lee@yale.edu.
References
- 1.Liao M, Liu Y, Yuan J, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat Med. 2020;26(6):842-844. [DOI] [PubMed] [Google Scholar]
- 2.Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26(10):1636-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilk AJ, Rustagi A, Zhao NQ, et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat Med. 2020;26(7):1070-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schulte-Schrepping J, Reusch N, Paclik D, et al. ; Deutsche COVID-19 OMICS Initiative (DeCOI) . Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182(6):1419-1440.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babicki S, Arndt D, Marcu A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44(W1):W147-W153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazan H, Saunders DJ, Khan H, et al. BindsNET: a machine learning-oriented spiking neural networks library in Python. Front Neuroinform. 2018;12:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang S, Park DW, Tadie JM, et al. Human resistin promotes neutrophil proinflammatory activation and neutrophil extracellular trap formation and increases severity of acute lung injury. J Immunol. 2014;192(10):4795-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boström EA, Tarkowski A, Bokarewa M. Resistin is stored in neutrophil granules being released upon challenge with inflammatory stimuli. Biochim Biophys Acta. 2009;1793(12):1894-1900. [DOI] [PubMed] [Google Scholar]
- 9.Chumakov AM, Kubota T, Walter S, Koeffler HP. Identification of murine and human XCP1 genes as C/EBP-epsilon-dependent members of FIZZ/Resistin gene family. Oncogene. 2004;23(19):3414-3425. [DOI] [PubMed] [Google Scholar]
- 10.Crestani B, Dehoux M, Hayem G, et al. Differential role of neutrophils and alveolar macrophages in hepatocyte growth factor production in pulmonary fibrosis. Lab Invest. 2002;82(8):1015-1022. [DOI] [PubMed] [Google Scholar]
- 11.Grenier A, Chollet-Martin S, Crestani B, et al. Presence of a mobilizable intracellular pool of hepatocyte growth factor in human polymorphonuclear neutrophils. Blood. 2002;99(8):2997-3004. [DOI] [PubMed] [Google Scholar]
- 12.Bundgaard JR, Sengeløv H, Borregaard N, Kjeldsen L. Molecular cloning and expression of a cDNA encoding NGAL: a lipocalin expressed in human neutrophils. Biochem Biophys Res Commun. 1994;202(3):1468-1475. [DOI] [PubMed] [Google Scholar]
- 13.Manz MG, Boettcher S. Emergency granulopoiesis. Nat Rev Immunol. 2014;14(5):302-314. [DOI] [PubMed] [Google Scholar]
- 14.Kolaczkowska E, Kubes P. Neutrophil recruitment and function in health and inflammation. Nat Rev Immunol. 2013;13(3):159-175. [DOI] [PubMed] [Google Scholar]
- 15.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages [correction published in Nat Rev Immunol. 2020;20:448]. Nat Rev Immunol. 2020;20(6):355-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vabret N, Britton GJ, Gruber C, et al. ; Sinai Immunology Review Project . Immunology of COVID-19: current state of the science. Immunity. 2020;52(6):910-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence SM, Corriden R, Nizet V. The ontogeny of a neutrophil: mechanisms of granulopoiesis and homeostasis. Microbiol Mol Biol Rev. 2018;82(1):e00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasty KA, Pourmotabbed TF, Goldberg GI, et al. Human neutrophil collagenase. A distinct gene product with homology to other matrix metalloproteinases. J Biol Chem. 1990;265(20):11421-11424. [PubMed] [Google Scholar]
- 20.Dorweiler B, Torzewski M, Dahm M, Kirkpatrick CJ, Lackner KJ, Vahl CF. Subendothelial infiltration of neutrophil granulocytes and liberation of matrix-destabilizing enzymes in an experimental model of human neo-intima. Thromb Haemost. 2008;99(2):373-381. [DOI] [PubMed] [Google Scholar]
- 21.Khanna-Gupta A, Zibello T, Idone V, Sun H, Lekstrom-Himes J, Berliner N. Human neutrophil collagenase expression is C/EBP-dependent during myeloid development. Exp Hematol. 2005;33(1):42-52. [DOI] [PubMed] [Google Scholar]
- 22.Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science. 2012;335(6071):936-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardoel BW, Kenny EF, Sollberger G, Zychlinsky A. The balancing act of neutrophils. Cell Host Microbe. 2014;15(5):526-536. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Du X, Chen J, et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J Infect. 2020;81(1):e6-e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Ren L, Zhang L, et al. heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27(6):883-890.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Zhang C, Huang F, et al. Elevated plasma levels of selective cytokines in COVID-19 patients reflect viral load and lung injury. Natl Sci Rev. 2020;7(6):1003-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cramer EP, Dahl SL, Rozell B, et al. Lipocalin-2 from both myeloid cells and the epithelium combats Klebsiella pneumoniae lung infection in mice. Blood. 2017;129(20):2813-2817. [DOI] [PubMed] [Google Scholar]
- 28.Serwas NK, Huemer J, Dieckmann R, et al. CEBPE-mutant specific granule deficiency correlates with aberrant granule organization and substantial proteome alterations in neutrophils. Front Immunol. 2018;9(588):588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacy P. Mechanisms of degranulation in neutrophils. Allergy Asthma Clin Immunol. 2006;2(3):98-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cowland JB, Borregaard N. Granulopoiesis and granules of human neutrophils. Immunol Rev. 2016;273(1):11-28. [DOI] [PubMed] [Google Scholar]
- 31.Gombart AF, Kwok SH, Anderson KL, Yamaguchi Y, Torbett BE, Koeffler HP. Regulation of neutrophil and eosinophil secondary granule gene expression by transcription factors C/EBP ε and PU.1. Blood. 2003;101(8):3265-3273. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z, Xing X, Hensley G, et al. Resistin induces expression of proinflammatory cytokines and chemokines in human articular chondrocytes via transcription and messenger RNA stabilization. Arthritis Rheum. 2010;62(7):1993-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamaluddin MS, Weakley SM, Yao Q, Chen C. Resistin: functional roles and therapeutic considerations for cardiovascular disease. Br J Pharmacol. 2012;165(3):622-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rubinstein T, Pitashny M, Putterman C. The novel role of neutrophil gelatinase-B associated lipocalin (NGAL)/Lipocalin-2 as a biomarker for lupus nephritis. Autoimmun Rev. 2008;7(3):229-234. [DOI] [PubMed] [Google Scholar]
- 35.Kong MY, Li Y, Oster R, Gaggar A, Clancy JP. Early elevation of matrix metalloproteinase-8 and -9 in pediatric ARDS is associated with an increased risk of prolonged mechanical ventilation. PLoS One. 2011;6(8):e22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narasaraju T, Yang E, Samy RP, et al. Excessive neutrophils and neutrophil extracellular traps contribute to acute lung injury of influenza pneumonitis. Am J Pathol. 2011;179(1):199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grommes J, Soehnlein O. Contribution of neutrophils to acute lung injury. Mol Med. 2011;17(3-4):293-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicolai L, Leunig A, Brambs S, et al. Vascular neutrophilic inflammation and immunothrombosis distinguish severe COVID-19 from influenza pneumonia [published online ahead of print 20 November 2020]. J Thromb Haemostasis., doi:10.1111/jth.15179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goshua G, Pine AB, Meizlish ML, et al. Endotheliopathy in COVID-19-associated coagulopathy: evidence from a single-centre, cross-sectional study. Lancet Haematol. 2020;7(8):e575-e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varga Z, Flammer AJ, Steiger P, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395(10234):1417-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lang FM, Lee KMC, Teijaro JR, Becher B, Hamilton JA. GM-CSF-based treatments in COVID-19: reconciling opposing therapeutic approaches. Nat Rev Immunol. 2020;20(8):507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanley E, Lieschke GJ, Grail D, et al. Granulocyte/macrophage colony-stimulating factor-deficient mice show no major perturbation of hematopoiesis but develop a characteristic pulmonary pathology. Proc Natl Acad Sci USA. 1994;91(12):5592-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.