Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged into the human population in late 2019 and caused the global COVID-19 pandemic. SARS-CoV-2 has spread to more than 215 countries and infected many millions of people. Despite the introduction of numerous governmental and public health measures to control disease spread, infections continue at an unabated pace, suggesting that effective vaccines and antiviral drugs will be required to curtail disease, end the pandemic, and restore societal norms. Here, we review the current developments in antibody and vaccine countermeasures to limit or prevent disease.
Keywords: SARS-CoV-2, COVID-19, Pathogenesis, Immunity, Vaccines, Antibody, Therapy, Protection
1. Introduction
In late 2019, for the third time this century, a highly pathogenic betacoronavirus crossed into the human population (Zhou et al., 2020; Zhu et al., 2020c). Unlike the first two coronavirus outbreaks, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of Coronavirus Disease 2019 (COVID-19), caused a global pandemic resulting in immense loss of life and economic hardship. Although the case-fatality rates for SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) are higher, SARS-CoV-2 is considerably more transmissible, most likely through sustained community and asymptomatic human-to-human spread by direct contact, respiratory droplets, or airborne transmission (Petersen et al., 2020; Day, 2020; Li et al., 2020b). At present, more than 108 million confirmed cases and over 2.3 million deaths have been reported globally (WHO, 2020a).
Coronaviruses are enveloped viruses encoded by an extraordinarily large, single-stranded, positive-sense RNA genome. For all human coronaviruses, the first two-thirds of the genome encodes nonstructural proteins that primarily contribute to viral replication and RNA synthesis. The remaining one-third of the genome encodes structural proteins (spike (S), envelope (E), membrane (M), and nucleocapsid (N)) that comprise the roughly spherical virion as well as accessory open reading frame (ORF) proteins, which often mediate immune antagonism. The SARS-CoV-2 S protein mediates cellular attachment and entry. Most SARS-CoV-2 S proteins contain 27 amino acid differences compared to SARS-CoV, including 6 substitutions in the receptor binding domain (RBD) (Wu et al., 2020a). Despite these differences, both SARS-CoV and SARS-CoV-2 utilize angiotensin converting enzyme 2 (ACE2) as a dominant receptor for cell entry (Hoffmann et al., 2020; Letko et al., 2020; Wan et al., 2020). After receptor engagement by the S1 subunit of the trimeric SARS-CoV-2 S protein (Fig. 1 A), cleavage occurs by a plasma membrane-associated serine protease, TMPRSS2, which facilitates membrane fusion by the S2 subunit and release of the viral genome into the host cytoplasm (Hoffmann et al., 2020; Matsuyama et al., 2020). The SARS-CoV-2 S protein, because of its key role in target cell entry, is the primary target of antibody and vaccine development. This review highlights some of the key countermeasures currently in development to control or prevent COVID-19.
Fig. 1.
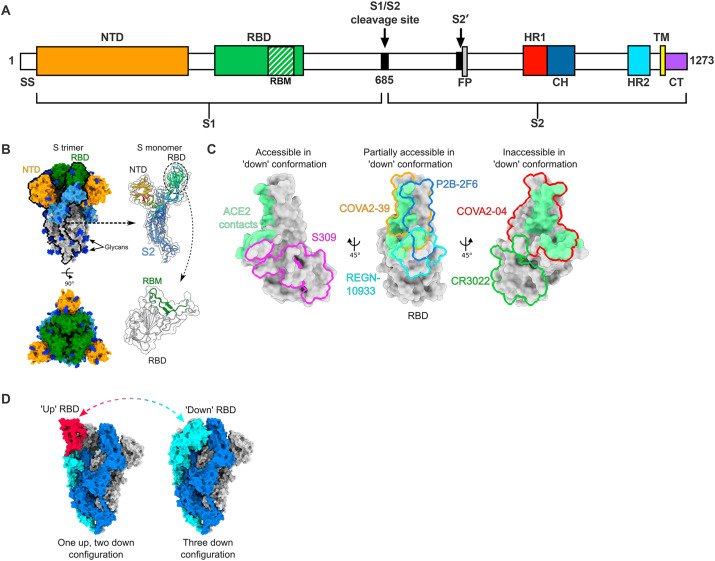
SARS-CoV-2 S protein organization and structural targets of neutralizing mAbs. (A) SARS-CoV-2 S protein schematic with key domains. SS; signal sequence, NTD; N-terminal domain, RBD; receptor-binding domain, RBM; receptor-binding motif, FP; fusion peptide, HR; heptad repeat, CH; central helix, TM; transmembrane domain, CT; cytoplasmic tail. (B) Structural model of the SARS-CoV-2 S protein. Left panels show trimeric spike in the three ‘down’ conformation (PDB: 6VXX), with N-terminal domain (NTD) colored yellow, receptor-binding domain (RBD) colored green, the rest of S1 colored light blue, S2 colored silver, and glycans colored as blue. Right panels show a single S monomer with S1 colored as a rainbow from N to C-terminus, and S2 displayed in light blue. For the closeup of the RBD, the RBM is shown in green. (C) Epitopes of select antibodies on the SARS-CoV-2 RBD. ACE2 contacts are shaded green at the top of the RBD. Interfaces were calculated based on buried surface area using UCSF ChimeraX. PDB codes used in visualization are as follows: S309; 6WPS, COVA2-39; 7JMP, REGN-10933; 6XDG, P2B-2F6; 7BWJ, CR3022; 6W41, COVA2-04; 7JMO. (D) Depiction of the structural transition of a single RBD on the trimeric spike moving between the ‘up’ and ‘down’ conformations (PDB 6VXX for three down conformation, PDB 6VYB for one up two down conformation). The N-terminal domain of the blue S monomer has been removed to aid visualization of the RBD.
2. Animal models for testing SARS-CoV-2 countermeasures
Under standard development timelines, animal models of disease are generated and refined in preparation for determining vaccine or antibody efficacy. However, out of necessity, as the virus was unknown prior to 2019, animal models for SARS-CoV-2 were established concurrently with countermeasure development. Indeed, several of the lead candidate vaccines progressed to human trials with minimal animal or efficacy data. A few of the most promising animal models that are used to test SARS-CoV-2 countermeasures are briefly summarized below and have been reviewed recently in greater detail (Muñoz-Fontela et al., 2020).
Mice. A major hurdle for evaluating antiviral treatments and vaccine efficacy in mice is the inability of SARS-CoV-2 to infect common laboratory mouse strains (Hassan et al., 2020). Because SARS-CoV-2 utilizes human but not mouse ACE2 as a receptor for entry, many of the reagents and approaches developed for SARS-CoV research were repurposed for SARS-CoV-2 such as mice with Krt1810, chicken β-actin, HFHF/FOXJ1, or endogenous mouse ACE2 promoters that drive transgenic expression of human ACE2 (Bao et al., 2020; Jiang et al., 2020; Tseng et al., 2007; Winkler et al., 2020). In addition, viral vector gene delivery systems, which were initially developed to introduce DPP4, a MERS-CoV receptor, and sensitize mice to infection by this virus, have been utilized to transduce human ACE2 expression in mice (Hassan et al., 2020; Sun et al., 2020; Zhao et al., 2014). The adenovirus-based hACE2 delivery method has the added benefit that it can be used in pre-existing, genetically modified mouse strains without the need for months of backcrossing. In many of these models, mice inoculated with SARS-CoV-2 develop appreciable lung viral infection and an interstitial pneumonia similar to that seen in COVID-19 patients (Hassan et al., 2020; Jiang et al., 2020; Winkler et al., 2020). SARS-CoV-2 also has been adapted by mutagenesis and passage, so that it can bind to and utilize murine ACE2 and productively replicate in the upper and lower respiratory tracts of non-transgenic young and aged BALB/c mice (Dinnon et al., 2020; Gu et al., 2020).
Hamsters. Syrian hamsters have been used as an animal model to study several respiratory viruses, including influenza A virus and adenovirus (Iwatsuki-Horimoto et al., 2018; Miao et al., 2019) and are known to support SARS-CoV infection (Schaecher et al., 2008). Multiple groups have reported that SARS-CoV-2 infection of hamsters results in labored breathing, high levels of viral RNA in the upper and lower respiratory tracts, immune cell infiltration, and extensive lung pathology (Chan et al., 2020; Imai et al., 2020). However, weight loss in hamster models can be variable, and furin cleavage site mutations in the SARS-CoV-2 S protein, which are readily generated when viral stocks are produced in vitro, greatly alter pathogenesis (Johnson et al., 2020).
Ferrets. Ferrets have been used to study respiratory virus infection and transmission of orthomyxoviruses and paramyxoviruses (Enkirch et al., 2015). In comparison, SARS-CoV-2 pathogenesis in ferrets is relatively benign, consisting of slightly elevated body temperatures, viral RNA predominantly in the upper respiratory tract, and mild alveolar and bronchoalveolar inflammation (Kim et al., 2020; Shi et al., 2020a). Transmission of SARS-CoV-2 within cages has been described for both hamsters and ferrets (Chan et al., 2020; Richard et al., 2020), suggesting that these animal models have utility for interventions that disrupt transmission.
Nonhuman primates. While expensive and in limited supply, nonhuman primates (NHPs) are important for establishing treatment and vaccine efficacy and providing rationale for initiating phase I clinical trials in humans. African green monkeys, rhesus macaques, and cynomolgus macaques have been the primary NHPs used for SARS-CoV-2 studies to date. Mild to moderate disease, including viral replication in the upper and lower respiratory tract and lung pathology consistent with pneumonia, has been described for NHPs (Munster et al., 2020; Rockx et al., 2020). Humoral and cellular immune responses as well as upper and lower respiratory tract protection have been observed in several vaccine studies (Corbett et al., 2020; Mercado et al., 2020; van Doremalen et al., 2020; Wang et al., 2020a). However, NHP protection data across studies should be interpreted carefully, as the challenge doses, virus stocks, and routes of inoculation vary between groups. Furthermore, the efficacy of intranasal treatments may be underestimated because most NHP models require intratracheal or intrabronchial virus challenge, which bypasses mucosal immunity that may be present in the upper respiratory tract.
3. Antibody therapy against SARS-CoV-2
Convalescent plasma therapy. The induction of a humoral response against SARS-CoV-2 during natural infection is well-documented (Long et al., 2020), and consistent with similar observations following SARS-CoV or MERS-CoV infection (Ko et al., 2017; Shi et al., 2004). The majority of patients infected with SARS-CoV-2 mount robust B cell responses and generate neutralizing S-specific antibodies (Atyeo et al., 2020; Long et al., 2020; Ni et al., 2020) of the IgM, IgA, and IgG subtypes (Sterlin et al., 2021; Wang et al., 2021a). A broader examination of the humoral response landscape has shown production of antibodies against other structural (e.g., N and M proteins) and non-structural (e.g., Orf3b and Orf8) proteins (Hachim et al., 2020), although the role these antibodies have in protective immunity remains to be established. The initial description of protective anti-S neutralizing antibody responses prompted the development, evaluation, and ultimately emergency use authorization (EUA) of convalescent plasma therapy for the treatment of SARS-CoV-2.
Convalescent immune plasma therapy was first used more than a century ago for other infectious diseases including rabies (Aoki et al., 1989). The evidence supporting plasma therapy for COVID-19 is limited, and because of the clinical urgency, the majority of initial studies were not powered adequately or designed in a randomized, controlled manner (Duan et al., 2020; Liu et al., 2020c). An additional challenge has been the lack of unifying metrics in the selection of donor plasma, as different studies have used anti-SARS-CoV-2 IgG titers, anti-S or anti-RBD IgG titers, neutralization titers, or a combination of multiple parameters. The most recent findings from the few randomized, controlled clinical studies suggests that the convalescent plasma has no or marginal benefit in moderate or severe COVID-19 (Agarwal et al., 2020; Li et al., 2020a; Salazar et al., 2020; Simonovich et al., 2020). However, one limitation of these studies was the late timing of initiated therapy after patient hospitalization. Indeed, in a subsequent multicenter, randomized, controlled trial, when high-titer convalescent plasma was administered within 72 h of symptom onset, the incidence of severe COVD-19 in older adults was reduced (Libster et al., 2021). Thus, the use of pre-selected convalescent plasma coupled with earlier intervention might still limit disease progression.
Monoclonal antibody development. As convalescent plasma has several limitations including the requirement for appropriate blood type matching, a dependence on donors to maintain sufficient supplies, and inherent donor-to-donor variability in the quantity and quality of the antibodies, the development of monoclonal antibodies (mAbs) against SARS-CoV-2 is an attractive therapeutic alternative. Historically, the use of mAbs in patients has been dominated by oncology and autoimmune but not infectious disease targets, with the exception of respiratory syncytial (Pavilizumab [Synagis®]) and Ebola (REGN-EB3 [Inmazebo®]) viruses. However, advances in mAb manufacturing as well as the discovery of antibodies with robust neutralization breadth and potency for other viral infections (i.e., HIV and Ebola) (Flyak et al., 2018; Halper-Stromberg and Nussenzweig, 2016) suggest that optimized single or cocktails of anti-SARS-CoV-2 mAbs could be a treatment option in certain clinical contexts.
Multiple approaches have been used to generate neutralizing anti-SARS-CoV-2 mAbs including classical animal immunization strategies and combinatorial protein-display libraries (Table 1 ) (Alsoussi et al., 2020; Hansen et al., 2020; Noy-Porat et al., 2020). The most common method has been the isolation of B cells from SARS-CoV-2 convalescent donors (Barnes et al., 2020b; Brouwer et al., 2020; Hansen et al., 2020; Ju et al., 2020; Kreye et al., 2020; Liu et al., 2020b; Robbiani et al., 2020; Rogers et al., 2020; Shi et al., 2020b; Wu et al., 2020d; Zost et al., 2020). In the majority of these studies, convalescent serum responses are screened for antibody binding to S or RBD in conjunction with neutralization tests against fully infectious or pseudotyped SARS-CoV-2. Sorting or recovery of single antigen-specific memory B cells from donors and subsequent sequencing and cloning of the immunoglobulin heavy and light chain genes has enabled rapid antibody expression and characterization.
Table 1.
SARS-CoV-2 mAb candidates in phase II/III clinical trials.
| Antibody candidate | Developer | Clinical Stage | Antibody(s) | Is IsIsotype/Fc Modifications | Animal Model | Trial number/References |
|---|---|---|---|---|---|---|
| REGN-COV2 | Regeneron/NIAID | EUA (11/20/20) | REGN109333(Casirivimab)+ REGN10987(imdevimab) | hIgG1 | Hamster, Rhesus Macaques (Baum et al., 2020a) | NCT04452318 NCT04426695 NCT04425629 |
| LY-CoV555 | AbCellera/Eli Lilly/NIH | EUA (11/09/20) | LY3819253 (Bamlanivimab) | hIgG1 | Rhesus Macaques (Jones et al., 2020) |
NCT04634409 NCT04518410 NCT04501978 NCT04497987 |
| LY-CoV016 | AbCellera/Eli Lilly/NIH | EUA (2/10/21) | LY3819253 (Etesevimab) | hIgG1 | NR |
NCT04441931 NCT04427501 |
| VIR-7831/GSK4182136 | Vir Biotechnology/GlaxoSmithKline | Phase III | VIR-7831, GSK4182136 (Sotrovimab) Based on mAb S309 |
hIgG1 (extended half-life, enhanced Fc effector functions) | NR | NCT04545060 |
| AZD7442 | AstraZeneca, Vanderbilt University Medical Center | Phase III | AZD8895 + AZD1061 (Sotrovimab) | hIgG1 (extended half-life, abrogated Fc effector functions) | NR |
NCT04507256 NCT04625972 NCT04625725 |
| CT-P59 | Celltrion | Phase II-III | CT-P59 (Regdanvimab) | hIgG1 | Hamster Ferret Rhesus Macaques (Lee et al., 2020) |
NCT04525079 |
| JS016, LYCoV016, LY3832479, CB6-LALA | Shanghai Junshi Biosciences Co., / Eli Lilly and Company; Institute of Microbiology, Chinese Academy of Sciences | Phase II | JS016, LYCoV016, LY3832479, CB6-LALA (Etesevimab) | hIgG1 (abrogated Fc effector functions) | Rhesus Macaques (CB6-LALA) (Shi et al., 2020b) |
NCT04634409 NCT04611789 |
| TY027 | Tychan | Phase III Pending | TY027 | NR | NR |
NCT04649515 NCT04429529 |
| SCTA01 | Sinocelltech Ltd. | Phase II-III Pending | SCTA01, H014 (assumed) |
NR | NR |
NCT04644185 NCT04483375 |
| BGB DXP-593 | BeiGene, Ltd. and Singlomics | Phase II Pending | DXP-593 | NR | NR |
NCT04551898 NCT04532294 |
NR; not reported.
EUA; emergency use authorization.
Structural targets of neutralizing mAbs to SARS-CoV-2. To date, all neutralizing mAbs to SARS-CoV-2 target the S protein (1273 residues), which is displayed on the surface of the virus as 15 to 30 trimers (Ke et al., 2020). The receptor-binding S1 subunit (residues 14-685) consists of the N-terminal domain (NTD) and the RBD, whereas the S2 subunit (residues 685-1273) contains the fusion peptide and transmembrane domain (Fig. 1A-B). The majority of potently neutralizing mAbs bind epitopes in the RBD (residues 319-541) (Fig. 1C) (Alsoussi et al., 2020; Barnes et al., 2020b; Brouwer et al., 2020; Hansen et al., 2020; Ju et al., 2020; Kreye et al., 2020; Liu et al., 2020b; Robbiani et al., 2020; Rogers et al., 2020; Shi et al., 2020b; Wu et al., 2020d; Zost et al., 2020) with a smaller number engaging the NTD (residues 14-305) (Chi et al., 2020; Liu et al., 2020b). The description of two neutralizing mAbs that bind a cryptic epitope on the S protein, but not the RBD or NTD also was recently reported (Liu et al., 2020b), whereas neutralizing antibodies to the S2 stalk region of the S trimer have not been described. The limited diversity of neutralizing epitopes reported could be biased by use of RBD to sort virus-specific B cells rather than trimeric S, virus-like particles, or infectious virus. Additionally, most of the B cells obtained are from donors who experienced relatively mild COVD-19, rather than severely ill subjects who have higher neutralization titers likely due to persistent viral antigen, prolonged germinal center reactions, and more extensive somatic hypermutation (Ju et al., 2020; Robbiani et al., 2020; Wu et al., 2020b).
Although many neutralizing mAbs bind the RBD, distinct, non-overlapping epitopes have been identified. X-ray crystallographic or cryo-electron microscopy structures of Fab fragments of mAbs in complex with either the RBD or trimeric spike have been solved (Yuan et al., 2020a). Each RBD in the S trimer can exist in an ‘up’ or ‘down’ conformation (Fig. 1D), with binding to human ACE2 occurring only in the ‘up’ conformation, leading to four possible conformational configurations per spike trimer (Ke et al., 2020; Walls et al., 2020; Wrapp et al., 2020). Some RBD-binding mAbs target only ‘up’ conformation RBDs, whereas others preferentially bind the ‘down’ conformation; a third group can bind both conformations (Barnes et al., 2020a).
The majority of neutralizing anti-RBD mAbs are believed to function by directly blocking ACE2 through engagement with the receptor-bind motif (RBM) of the S protein. As more RBM-specific antibodies have been characterized, several trends have been observed. For example, in one study, a large number of the neutralizing RBM antibodies utilized a restricted set of germline heavy chain genes, most commonly IGHV3-53, with many of their paratope contact residues retaining their germline identities (Yuan et al., 2020a). Additionally, these antibodies tended to encode shorter CDRH3 loops, although others described mAbs (e.g., COVA2-39, another IGHV3-53 antibody) that adopt a different binding orientation on the RBM, which accommodates a longer CDRH3 (Wu et al., 2020c). Antibodies utilizing alternative germline variants also have been reported, but still tend to display relatively low levels of somatic hypermutation (Barnes et al., 2020a; Ju et al., 2020).
A smaller subset of mAbs target epitopes outside the human ACE2 binding site, and thus may neutralize through alternative mechanisms of action including blockade of one or more other cellular attachment factors (heparan sulfate (Clausen et al., 2020), HDL scavenger receptor B type 1 (Wei et al., 2020)) or possibly alternative receptors (neuropilin (Cantuti-Castelvetri et al., 2020)), inhibition of conformational changes necessary for fusion, or allosteric blockade of ACE2 interactions. Recently, the structure of potently neutralizing antibodies C144 and S2M11 showed that both target a similar quaternary epitope, binding adjacent RBD subunits, and locking them into an ACE2-inaccessible ‘down’ conformation (Barnes et al., 2020a; Tortorici et al., 2020). However, it is unclear how important this mechanism is for neutralization, since both mAbs also directly block ACE2 binding (Barnes et al., 2020a; Tortorici et al., 2020). Among RBD antibodies, binding affinity to recombinant protein does not necessarily correlate with neutralization potency (Liu et al., 2020b). This could be due to differences in how epitopes on the trimeric S are displayed or glycan shielding of epitopes present on the native virion (Watanabe et al., 2020; Zhao et al., 2020).
A third subset of antibodies targets epitopes further away from the RBM and typically display low to no neutralizing potency. Some of these antibodies, such as the SARS-CoV derived CR3022, engage cryptic epitopes. Notably, despite its ability to bind SARS-CoV-2 RBD and cause S trimer dissociation, CR3022 does not neutralize SARS-CoV-2 (Huo et al., 2020; Wrobel et al., 2020; Yuan et al., 2020b). In contrast, the recently characterized mAb COVA1-16 targets an epitope outside the RBM that overlaps with CR3022, but still blocks ACE2 binding as a result of steric hindrance with its light chain, indicating that direct overlap with binding residues is not necessarily a requirement for ACE2 blockade (Liu et al., 2020a). Although non-RBM antibodies tend to be less potently neutralizing, several have been shown to cross-react and neutralize SARS-CoV due to the increased conservation in the RBD outside of the RBM, highlighting their potential use against future betacoronaviruses (Pinto et al., 2020; Yuan et al., 2020b).
Combatting viral resistance in mAb therapy design. Although coronaviruses have a slower mutation rate relative to other RNA viruses because of their proofreading 3′-to-5′ exoribonuclease (nsp14), escape from antibody neutralization is still a major concern. Multiple groups have described viral escape under the selective pressure of single mAb treatments and identified viral escape mutations in the RBD (Baum et al., 2020b, Greaney et al., 2021a; Liu et al., 2020d; Weisblum et al., 2020). The most concerning viral escape mutations are those that do not cause a loss-of-fitness to the virus and can occur through single nucleotide changes. Two approaches have been employed to prevent viral resistance against mAb therapies: (1) the selection of antibodies that bind highly conserved residues and epitopes that do not tolerate mutations without marked loss-of-function phenotypes and (2) the development of antibody cocktails targeting distinct, non-overlapping regions of the RBD or S, which would require the less likely occurrence of simultaneous mutations at two separate genetic sites.
The emergence of rapidly-spreading SARS-CoV-2 variants in the United Kingdom (B.1.1.7), South Africa (B.1.351), and Brazil (B.1.1.248) as well as enzootic crossover events on mink farms (Oude Munnink et al., 2021) has highlighted the need to monitor frequently whether viral evolution could compromise mAb therapy or even vaccine efficacy. Of particular concern, the South Africa and Brazil variants contain an amino acid substitution at position 484 in the RBM, which previously was identified as a key monoclonal and polyclonal antibody escape mutation in the context of yeast and chimeric vesicular stomatitis virus screening campaigns (Greaney et al., 2021a, 2021b; Starr et al., 2020; Weisblum et al., 2020). Structural and computational approaches outlining the footprints of different classes of neutralizing antibodies that synergize within the RBD (Barnes et al., 2020b; Greaney et al., 2020; Hansen et al., 2020) and the discovery of additional classes of neutralizing antibodies, such as the NTD mAbs (Suryadevara et al., 2021) that bind epitopes outside the RBM and RBD, may be important tools in designing mAb cocktails against COVID-19 and minimizing the potential for emergence of resistance.
Testing of mAbs in animal models. In the majority of humans, viral replication is followed by clearance and rapid clinical resolution. However, a small, yet important, fraction of patients develop severe COVID-19 requiring hospitalization and intensive care. Severe COVD-19 is thought to be driven by an over-exuberant immune response at a time in which viral replication is waning. Prophylactic and therapeutic efficacy of neutralizing anti-RBD mAbs has been demonstrated in vivo in murine, hamster, and NHP models of SARS-CoV-2 pathogenesis (Table 2 ) (Alsoussi et al., 2020; Baum et al., 2020a; Hassan et al., 2020; Kreye et al., 2020; Rogers et al., 2020; Shi et al., 2020b; Wu et al., 2020d; Zost et al., 2020). Treatment benefit is typically assessed by measuring reductions in viral loads, clinical signs of disease (weight loss and body temperature), and lung inflammation (cytokine levels and histopathology). Pre-exposure administration of neutralizing mAbs prior to SARS-CoV-2 infection reduces viral titers in the upper respiratory tract and can completely prevent viral replication in the lower respiratory tract of the lungs (Baum et al., 2020a). However, post-exposure mAb therapy demonstrates more modest reductions in viral titers (especially when measuring viral RNA levels in the lung), although improvements in clinical disease and pulmonary pathology still are demonstrated. Of note, in all therapeutic studies in animals published to date, mAbs have been administered within 24 h of viral infection (often earlier), highlighting that treatment success may require an early window of intervention. Although mAb treatment might have a role later in the course of COVID-19 disease, the pathophysiology is complicated by immune dysregulation and immunopathology in both pre-clinical animal models and human disease.
Table 2.
SARS-CoV-2 mAb Tested in Animal Models.
| Antibody | Animal Model | Prophylactic | Therapeutic | Virus Dose | Outcome compared to control |
|---|---|---|---|---|---|
| 2B04 (Alsoussi et al., 2020) |
murine BALB/C+ hACE2-AdV (Hassan et al., 2020) |
1 day prior to infection (IP) (~10 mg/kg) |
4 x 105 PFU Intranasal |
|
|
| 2H04 (Alsoussi et al., 2020) |
murine BALB/C+ hACE2-AdV (Hassan et al., 2020) |
1 day prior to infection (IP) (~10 mg/kg) |
4 x 105 PFU Intranasal |
|
|
| COV2-2196 COV2-2130 (Cocktail) COV2-2196+ COV2-2130 (Zost et al., 2020) |
BALB/C + Mouse-Adapted SARS-CoV-2 (Dinnon et al., 2020) | 1 day prior to infection (IP) (~10 mg/kg) |
105 PFU Intranasal |
|
|
| murine BALB/C+ hACE2-AdV (Hassan et al., 2020) |
1 day prior to infection (IP) (~10 mg/kg) |
4 x 105 PFU Intranasal |
|
||
| COV2-2196 COV2-2381 |
Rhesus Macaques | 1 day prior to infection (IV) (50 mg/kg) |
105 PFU Intranasal and Intratracheal |
|
|
| COV2-2196 COV2-2130 (Cocktail) COV2-2196+ COV2-2130 |
BALB/C + Mouse-Adapted SARS-CoV-2 (Hassan et al., 2020) |
12 h post-infection (~20 mg/kg) | 106 PFU Intranasal |
For 2196 and Cocktail:
|
|
| (Cocktail) COV2-2196+ COV2-2130 |
BALB/C+ hACE2-AdV (Hassan et al., 2020) |
12 h post-infection (~20 mg/kg) | 4 x 105 PFU Intranasal |
|
|
| REGN-COV2 (REGN10987 + REGN10933) (Baum et al., 2020a) |
Syrian Hamsters | 2 day prior to infection (IP) (50, 5, and 0.5 mg/kg) |
2.3 x 104 PFU Intranasal |
All doses:
|
|
| Syrian Hamsters | 1 day post infection (IP) (50, 5, and 0.5 mg/kg) |
2.3 x 104 PFU Intranasal |
For 50 and 5 mg/kg doses:
|
||
| Rhesus Macaques | 3 day prior to infection (IV) (50 mg/kg) |
105 PFU Intranasal and Intratracheal |
For 50 mg/kg:
|
||
| Rhesus Macaques | 3 day prior to infection (IV) (0.3 and 50 mg/kg) |
106PFU Intranasal and Intratracheal |
For 50 mg/kg:
|
||
| Rhesus Macaques | 1 day post infection (IP) (150 and 25 mg/kg) |
105 PFU Intranasal and Intratracheal |
For both 150 mg/kg and 25 mg/kg:
|
||
| CCL12.1 (Rogers et al., 2020) |
Syrian Hamsters | 12 h pre-infection (IP) (16.5, 4.125, 1.03, 0.25, 0.06 mg/kg) |
106PFU Intranasal |
For 16.5, 4.125, and 1.03 mg doses:
|
|
| CB6 LALA (Shi et al., 2020b) |
Rhesus Macaques | 1 day prior to infection (IV) 50 mg/kg |
105 TCID50 Intratracheal |
|
|
| CB6 LALA | Rhesus Macaques | 1 day and 3 post infection (IV) 50 mg/kg |
105 TCID50 Intratracheal |
|
|
| B38 (Wu et al., 2020d) |
Murine hACE2 Transgenic | 12 h post-infection (25 mg/kg) | NR |
|
|
| H4 (Wu et al., 2020d) |
Murine hACE2 Transgenic | 12 h post-infection (25 mg/kg) | NR |
|
|
| CV07-209 (Kreye et al., 2020) |
Syrian Hamsters | 24 h prior (18 mg/kg) | 105PFU Intranasal |
|
|
| Syrian Hamsters | 2 h post-infection (18 mg/kg) | 105PFU Intranasal |
|
||
| S2M11 S2E12 Cocktail (S2M11 + S2E12) (Tortorici et al., 2020) |
Syrian Hamsters | 48 h prior (1 or 0.5 mg/kg) | 2x105 TCID50 Intranasal |
All doses:
|
|
| LY-CoV555 (Jones et al., 2020) |
Rhesus Macaques | 1 day prior to infection (IV) (50, 15, 2.5, and 1 mg/kg) |
1.1x105 PFU Intranasal and Intratracheal |
For all doses:
|
|
| ADG-2 (Rappazzo et al., 2021) |
BALB/c + Mouse-Adapted SARS-CoV-2 | 12 h prior (10mg/kg) | 103 PFU Intranasal |
|
|
| BALB/c + Mouse-Adapted SARS-CoV-2 | 12 h post-infection (10mg/kg) | 103 PFU Intranasal |
|
Fc region considerations in mAb development. Mechanisms of antibody protection in vivo can be due to multiple factors including direct Fab-dependent virus neutralization and Fc-dependent engagement of complement or Fc-gamma receptors (FcγRs) on leukocytes. The vast majority of currently described anti-S mAbs are of the human IgG1 subclass that can engage FcγRs on a variety of immune cells. By binding individual FcγRs on specific hematopoietic cells, antibodies and their Fc effector functions can promote immune-mediated clearance by antibody-dependent cellular cytotoxicity, antibody-dependent cellular phagocytosis, and complement activation, enhanced antigen presentation and CD8+ T cell responses, and reshaping of inflammation (Bournazos et al., 2020; Lu et al., 2018). The importance of Fc effector functions for antibody efficacy in vivo has been illustrated in many virus models, including HIV, Ebola, West Nile, hepatitis B, chikungunya, and influenza viruses (Bournazos et al, 2014, 2019; 2014; DiLillo et al, 2014, 2016; Fox et al., 2019; Hessell et al., 2007; Li et al., 2017; Liu et al., 2017; Vogt et al., 2011). While preliminary in vitro (Tortorici et al., 2020) and in vivo studies highlight a possible beneficial role of Fc receptor engagement by mAb therapies against SARS-CoV-2 (Schäfer et al., 2020), further work is needed to identify which FcγRs, Fc effector functions, and cell subsets mediate protection.
Notwithstanding these possible beneficial functions, under certain circumstances, Fc-FcγR interactions can contribute to pathological outcomes. As one example, antibody-dependent enhancement of virus infection (ADE) (Morens, 1994) can occur when sub-neutralizing concentrations or non-neutralizing antibodies promote virus uptake and infection of FcγR-expressing immune cells. Although well-documented in dengue virus infection, ADE is at least a theoretical concern of antibody-based therapies and vaccines against SARS-CoV-2 (Diamond and Pierson, 2020). Another potential concern is the generation of non-productive Fc-mediated immune responses, such as the pathological TH2 immune skewing seen with SARS-CoV in NHPs in the setting of poorly neutralizing antibodies (Bolles et al., 2011). However, in pre-clinical animal models and COVID-19 patients treated with convalescent plasma or anti-SARS-CoV-2 mAbs, there has been no reported evidence for ADE, immune skewing, or enhancement of disease.
4. Vaccines against SARS-CoV-2
A race for the most sought-after vaccine in history. Vaccination is one of the most effective means by which to prevent infectious diseases (WHO, 2019); with a goal of inducing immune responses that prevent or limit disease upon subsequent natural infection. Smallpox, a lethal disease caused by variola virus, was officially eradicated in 1980 due to the global implementation of a highly effective vaccine (Belongia and Naleway, 2003). After smallpox vaccine immunization, approximately 95% of individuals developed neutralizing antibodies that endured for 5–10 years or longer (Belongia and Naleway, 2003). The measles, mumps, and rubella vaccines are other examples of highly effective vaccines that prevented what once were devastating childhood diseases (Amanna and Slifka, 2018). Historically, developing a safe, efficacious, and cost-effective human vaccine took between 4 to 15 years, if not longer. In comparison, COVID-19 vaccine development has proceeded at an unprecedented pace. The complete viral genome sequence of SARS-CoV-2 from patient isolates was reported on January 11, 2020 – less than a month after the first cases were documented in Wuhan, China (Zhou et al., 2020). With this information, academic research laboratories and biotechnology companies globally embarked on a race to generate an effective SARS-CoV-2 vaccine. Due to knowledge gained during previous vaccine development efforts for MERS-CoV and SARS-CoV, the initial step of target identification was essentially skipped without loss. For both SARS-CoV and MERS-CoV, the S protein had been demonstrated as the dominant antigen responsible for inducing neutralizing antibodies and an important target of protective T cell responses (Graham et al., 2013; Yong et al., 2019). A plethora of vaccine candidates for SARS-CoV-2 were designed rapidly using the SARS-CoV-2 S protein as the primary immunogen (Fig. 2 and Table 3 ).
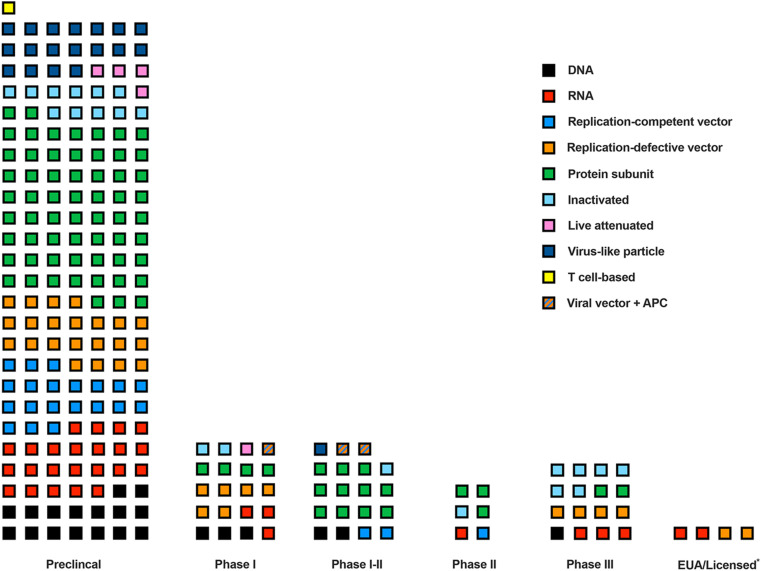
Fig. 2.
SARS-CoV-2 vaccine candidate number and platform by developmental status. Each square represents a SARS-CoV-2 vaccine candidate officially recognized by the World Health Organization (WHO, 2020b). The color of each square indicates the vaccine platform utilized. EUA indicates emergency use authorization of vaccines that are still in phase III trials. *Licensed only for select groups or in a single country without completing clinical trials.
Table 3.
SARS-CoV-2 vaccine candidates in phase III clinical trialsa.
| Vaccine candidate | Developer | Platform | Dose rangeb | Route | Peak neut titerb | T cell responsesb | Trial number/References |
|---|---|---|---|---|---|---|---|
| BNT162b2 | Pfizer/BioNTech | RNA | 10–100 μg; 2 doses | IM | 1:363 (30 μg)c | NR | NCT04368728 (Walsh et al., 2020) |
| mRNA-1273 | Moderna/NIAID | RNA | 25–250 μg; 2 doses | IM | 1:654.3 (100 μg)d | CD4+; low CD8+ | NCT04470427 (Jackson et al., 2020) |
| ChAdOx1 nCoV19 | AstraZeneca/ University of Oxford | Non-replicating viral vector | 5 x 1010 VPs; 1 or 2 doses | IM | 1:218 (2 doses)e | Yes; bulk PBMCs |
NCT04516746 NCT04540393 (Folegatti et al., 2020) |
| Ad5-nCOV | CanSino Biologics Inc./ Beijing Institute of Biotechnology | Non-replicating viral vector | 5 x 1010 VPs; 1 dose | IM | 1:19.5 (1 dose)f | Yes; bulk PBMCs |
NCT04526990 NCT04540419 (Zhu et al., 2020a, 2020b) |
| Ad26.COV2.S | Janssen Pharmaceutical Companies | Non-replicating viral vector | 5 x 1010 VPs; 1 dose | IM | NR | NR | NCT04505722 |
| Gam-COVID-Vac Lyo | Gamaleya Research Institute | Non-replicating viral vector | NR; 2 heterologous doses | IM | 1:45.95g | CD4+ and CD8+ |
NCT04530396 NCT04564716 NCT04642339 (Logunov et al., 2020) |
| NVX-nCoV2373 | Novavax | Protein subunit | 2.5–25 μg; 2 doses | IM | 1:8344g | CD4+ | NCT04611802 (Keech et al., 2020) |
| COVID-19 vaccine | Anhui Zhifei Longcom Biopharmaceutical/Institute of Microbiology, Chinese Academy of Sciences | Protein subunit | NR | IM | NR | NR | ChiCTR2000040153 |
| CoronaVac | Sinovac | Inactivated | 3–6 μg; 2 doses |
IM | 1:65 (6 μg dose)h | NR |
NCT04456595 669/UN6.KEP/EC/2020 NCT04582344 NCT04617483 (Zhang et al., 2020) |
| BBIBP-CorV | Sinopharm/ Beijing Institute of Biological Products | Inactivated | 2–8 μg; 2–3 doses |
IM | 1:282.7 (4 μg dose)i | NR | ChiCTR2000034780 (Xia et al., 2020b) |
| COVID-19 vaccine | Sinopharm/ Wuhan Institute of Biolocial Products | Inactivated | 2.5–10 μg; 2–3 doses | IM | 1:316 (2.5 μg dose)j | NR | ChiCTR2000034780 ChiCTR2000039000 (Xia et al., 2020a) |
| BBV152 | Bharat Biotech | Inactivated | NR | IM | NR | NR |
NCT04641481 CTRI/2020/11/028976 |
| CoVLP | Medicago | VLP | NR | IM | NR | NR | NCT04636697 |
NR; not reported.
IM, intramuscular; VP, virus particles; VLP, virus-like particle.
Vaccine candidates in phase III clinical trials according to the World Health Organization (WHO, 2020b).
These data are from published phase I-II or phase III trials, when available.
SARS-CoV-2 – mNeonGreen reporter virus neutralization assay.
PRNT80 assay with authentic SARS-CoV-2.
Public Health England PRNT50 assay.
Neutralization assay with authentic SARS-CoV-2.
Microneutralization assay with authentic SARS-CoV-2.
Microcytopathic effect assay with authentic SARS-CoV-2.
Neutralization assay with infectious SARS-CoV-2.
PRNT50 assay with authentic SARS-CoV-2.
5. SARS-CoV-2 vaccine platforms
Nucleic acid vaccines. At present, six DNA-based vaccines and six mRNA-based vaccines against SARS-CoV-2 are in clinical trials (Fig. 2) (WHO, 2020b). Although DNA vaccines are currently used in veterinary medicine, due to their relatively modest immunogenicity as well as delivery hurdles, they have not been widely utilized in humans (Suschak et al., 2017). mRNA vaccines are a new platform commonly consisting of an mRNA, which typically includes a 5′ cap, regulatory elements in the 5′ and/or 3′ untranslated regions (UTRs), a poly(A) tail, and modified nucleosides to increase RNA stability and decrease innate immune activation, that is engineered to encode specific viral antigens and delivered by polymer-based or lipid nanoparticles (Pardi et al., 2018). Upon entry into the cell cytoplasm, the mRNA is translated by host cell ribosomes similarly to endogenous mRNA, resulting in antigen expression for immune system recognition and response. Two mRNA vaccines, Pfizer/BioNtech's BNT162b2 and Moderna/NIAID's mRNA-1273, are currently in phase III clinical trials and recently received EUA status from the U.S. Food and Drug Administration and other countries (Cohen, 2020; WHO, 2020b). BNT162b2 is an mRNA-lipid nanoparticle-formulated vaccine that encodes a membrane-bound, prefusion stabilized form of the full-length SARS-CoV-2 S protein (Walsh et al., 2020). In a phase I, placebo-controlled, dose escalation trial, BNT162b2 and a similar vaccine candidate, BNT162b1, which encodes a secreted trimerized form of the SARS-CoV-2 RBD, were tested for safety and immunogenicity. Both vaccine candidates were evaluated at four doses (10, 20, 30, or 100 μg) in a prime-boost strategy with three weeks between immunizations. Whereas serum neutralizing activity was dose-dependent for both vaccine candidates and peaked at levels higher than convalescent serum from COVID-19 infected subjects, BNT162b2 had fewer adverse reactions and was selected for clinical development. A recently published safety and efficacy trial of a 30 μg dose of BNT162b2 reported 95% protective efficacy against symptomatic COVID-19 (Polack et al., 2020).
mRNA-1273 encodes a prefusion stabilized form of the SARS-CoV-2 S protein and is delivered into cells by lipid-encapsulated nanoparticles (Jackson et al., 2020). In a phase I, dose-escalation (25, 100, or 250 μg), open-label clinical trial in 45 healthy adults using a 28-day prime-boost intramuscularly administered regimen, mRNA-1273 induced serum neutralizing activity in all participants. Participants receiving the 25 μg or 100 μg doses also elicited strong TH1-polarized CD4+ T cell responses. In a promising unpublished report from a 30,000-person efficacy trial, 185 placebo group participants developed COVID-19 symptoms over the course of the trial compared to 11 such individuals in the mRNA-1273 vaccinated group, suggesting a 94% efficacy rate (Cohen, 2020). Moreover, of the 11 mRNA-1273 vaccinated individuals that contracted SARS-CoV-2, none progressed to severe disease compared to 30 of the 185 placebo group participants.
Other mRNA vaccines in clinical trials include those developed by Curevac, Arcturus-/Duke-NUS, Imperial College of London, and the People's Liberation Army (WHO, 2020b). Although prior to COVID-19, an mRNA vaccine had never been licensed for use in humans, the recent deployment of mRNA-1273 and BN162b2 confirms the promising status of this platform.
Viral-vectored vaccines. Viral-vectored vaccines utilize the genetic backbone of another virus to deliver and express antigens of the targeted pathogen. Two forms of viral vectors exist – replication-deficient or replication-competent. Currently, 14 viral-vectored vaccines (9 replication-defective and 5 replication-competent) are in clinical trials (Fig. 2) (WHO, 2020b). Replication-defective viral vectors are engineered such that components of the viral genome essential for replication have been deleted and a transgene encoding the target antigen is inserted. Hence, in a self-limiting fashion, the targeted antigen (S protein in the case of SARS-CoV-2) is delivered and expressed in cells of the vaccine recipient, provoking an immune response. In contrast, replication-competent viral vectors are derived from attenuated or vaccine strains of viruses that have been engineered to encode a target pathogen antigen but still are capable of one or more rounds of replication. Numerous SARS-CoV-2 vaccine candidates are in preclinical or early stages of clinical development that utilize vesicular stomatitis virus, influenza virus, adenovirus, and modified vaccinia virus viral vectors (Case et al., 2020; Chiuppesi et al., 2020; Logunov et al., 2020; WHO, 2020b).
Currently, four viral-vectored vaccines against SARS-CoV-2 are in phase III clinical trials, all of which utilize replication-defective adenovirus strains in prime-only or prime-boost regimens (Table 3) (WHO, 2020b). ChAdOx1 nCoV-19, which is jointly developed by the University of Oxford and AstraZeneca, uses a genetically modified Y25 simian (chimpanzee) adenovirus strain to deliver and express a full-length, codon-optimized form of the SARS-CoV-2 S protein (van Doremalen et al., 2020). Preclinical studies showed strong cellular and humoral responses in ChAdOx1 nCoV-19 vaccinated mice and NHPs (van Doremalen et al., 2020). Phase I-II and II-III safety and immunogenicity trials in humans reported promising S-specific T cell responses and seroconversion rates of >99% in all ChAdOX1 nCoV-19 prime-boost vaccination groups (Folegatti et al., 2020; Ramasamy et al., 2020). Adverse events attributed to the vaccine were not detected, and immunogenicity data were similar across the multiple age groups (18-55, 56-69, and ≥70 years). In efficacy analyses, vaccination with two standard doses or a low dose followed by a standard dose resulted in 62% and 90% protection against symptomatic disease, respectively (Voysey et al., 2020).
Ad5-nCOV, a replication-defective viral vector vaccine developed by CanSino Biologics, uses a human adenovirus serotype-5 vector expressing a full-length, codon-optimized form of the SARS-CoV-2 S protein (Zhu et al., 2020b). Phase I and II clinical trials were conducted at a single center in Wuhan, China between March and April, 2020 (Zhu et al., 2020a, 2020b). The highest dose (1.5 x 1011 viral particles) caused considerable adverse reactions and was excluded from further development. T cell and neutralizing antibody responses peaked at 14- and 28-days post-vaccination, respectively, but were generally modest for the low (5 x 1010 viral particles) and middle doses (1 x 1011 viral particles). Indeed, neutralizing antibody responses were observed in only 50% of participants receiving the low or middle doses. In contrast to most viral-vectored vaccines in development, Ad5-nCOV is administered as a single intramuscular injection. While an Ad5-nCOV booster dose could theoretically improve the elicited antibody and T cell responses, this approach might be hampered by immunity generated by the first dose or even pre-existing natural immunity to the Ad5 serotype. Ad5-nCOV is currently in phase III clinical trials in Russia, Pakistan, and several Latin American countries (NCT04526990).
Other replication-defective human adenoviral vectors, such as those based on adenovirus serotype 26 (Ad26), which there is little pre-existing immunity in humans, also are under clinical evaluation. Janssen developed an Ad26 vector-based vaccine (Ad26.COV2.S) that encodes for a full-length SARS-CoV-2 S protein containing a mutated furin cleavage site between S1 and S2 (Fig. 1A) and two proline stabilizing mutations in S2 (Mercado et al., 2020). Ad26.COV2.S was selected as the best of seven constructs initially tested in NHPs, with each candidate differing in the leader sequences used, status of the furin cleavage site, introduction of proline stabilizing mutations, and/or truncation of C-terminal components of the spike protein. At 28-days post-vaccination, Ad26.COV2.S induced the highest neutralizing antibody titers of the different Ad26-vaccine formulations. T cell responses were notably poor, especially for Ad26.COV2.S. Despite this limitation, in challenged animals, viral RNA was absent in all bronchoalveolar lavage fluid and all but one nasal wash samples obtained from Ad26.COV2.S vaccinated animals. Currently, Ad26.COV2.S is under evaluation in phase III clinical trials as a single intramuscular dose of 5 x 1010 virus particles (NCT04505722).
Viral-vectored vaccines often can induce robust B and T cell responses, and some are suitable for mucosal administration, a key consideration for SARS-CoV-2 immunity (Mudgal et al., 2020). However, a number of issues such as safety in immunocompromised individuals as well as pre-existing vector immunity (against Ad5 and Ad26) may need to be overcome.
Recombinant protein vaccines. Subunit vaccines incorporating only a specific component(s) of SARS-CoV-2 (e.g., S protein) are also under evaluation. With 16 in clinical trials and 56 in preclinical development against SARS-CoV-2, subunit vaccines are the most abundant platform (Fig. 2) (WHO, 2020b). This is due in part to the relative ease in generating recombinant proteins and the number of variations possible. Novavax's NVX-CoV2373 vaccine is a recombinant, prefusion stabilized S protein adjuvanted with Matrix-M1 (Keech et al., 2020). A 3-week prime-boost regimen of different doses (2.5, 5, or 25 μg) given by intramuscular injection to cynomolgus macaques induced high levels of neutralizing antibody titers. Upon challenge of NVX-CoV2373 immunized macaques, viral RNA was undetectable in all but one bronchoalveolar lavage fluid and all nasal swab samples isolated. In a phase I-II clinical trial, NVX-CoV2373 appeared safe without serious adverse events, and participants receiving the vaccine developed neutralizing antibody responses that exceeded convalescent patient-derived serum controls (Keech et al., 2020). In addition, CD4+ T cell responses were observed and skewed toward a TH1 phenotype. NVX-CoV2373 is currently being evaluated in phase III clinical trials (NCT04611802).
Another adjuvanted recombinant protein vaccine (RBD-dimer), which is being developed by Anhui Zhifei Longcom Biopharmaceuticals in collaboration with the Chinese Academy of Sciences’ Institute of Microbiology, is currently in phase III clinical trials (WHO, 2020b). However, no published immunogenicity or efficacy data are available for this vaccine candidate.
Inactivated virus vaccines. Inactivation of a virus using chemical or physical methods is another proven vaccine strategy, as many antiviral vaccines of this type are licensed for use in humans including against polio, influenza, rabies, and hepatitis A viruses (Amanna and Slifka, 2018; Murdin et al., 1996; Vellozzi et al., 2009). Seven inactivated SARS-CoV-2 vaccines are in clinical trials (Fig. 2) (WHO, 2020b). Inactivated vaccines are relatively easy to produce, most often by generating virus in cell culture on a massive scale, followed by inactivation. In the case of SARS-CoV-2, production is more complex due to the need for BSL3 production facilities prior to inactivation. Since the entire SARS-CoV-2 virion is present in the vaccine, the resulting immunity may be more complete and include responses against S as well as the other structural proteins present in the virion (E, M, and N). The SARS-CoV-2 vaccine candidate PiCoVacc (now known as CoronaVac), an alum-adjuvanted vaccine developed by Sinovac Biotech Ltd., elicited potent neutralizing antibodies in mice, rats, and NHPs after two or three immunizations (Table 3) (Gao et al., 2020). NHPs receiving the highest vaccine dose were protected from severe interstitial pneumonia, and viral RNA was absent in the lungs. In another study, the inactivated vaccine candidate BBIBP-CorV, developed by Sinopharm, induced neutralizing antibody responses in a dose-dependent manner in mice, rats, guinea pigs, and rabbits after one, two, or three immunizations (Wang et al., 2020a). However, CD4+ and CD8+ T cell responses were not observed with either inactivated vaccine (Gao et al., 2020; Wang et al., 2020a). While inactivated vaccines are safe when given intramuscularly, they poorly induce mucosal immunity or secretory IgA in the upper respiratory tract.
Live-attenuated vaccines. Live-attenuated vaccines are derived from a pathogenic virus that has been weakened to the point where it no longer causes disease despite replicating in an individual for a few days. Serial passage of the virus in vitro, rational deletion of virulence genes, chimerizaton, and codon de-optimization techniques are strategies to reduce viral pathogenicity (Barrett, 1997; Coleman et al., 2008; Talon et al., 2000; Theiler and Smith, 1937). Live-attenuated vaccines often generate strong humoral and cellular immune responses at the site of infection. While several live-attenuated antiviral vaccines are approved for use in humans (Amanna and Slifka, 2018), only three are in development against SARS-CoV-2, and only one, COVI-VAC, has advanced beyond preclinical stages (WHO, 2020b). These vaccine candidates all utilize codon-deoptimization strategies for attenuation. Codon-deoptimization introduces silent mutations into the nucleic acid sequences to yield a virus that uses codons that are relatively rare in human host cells, which attenuates viral infection due to poor genome translation efficiency. The primary concern of live-attenuated vaccines is safety due to possible reversion to virulence by mutation or recombination with a circulating strain. Since coronaviruses are known to recombine in nature (Su et al., 2016), this issue may be challenging to overcome.
6. Conclusions and SARS-CoV-2 countermeasures outlook
The deployment of effective vaccines against SARS-CoV-2 could prevent disease and transmission, ultimately resulting in community protection. However, large-scale manufacturing of EUA vaccines with a requirement of two doses (for most vaccines) to all countries of the world may take many months if not longer, especially in places where cold-chain distribution infrastructure is lacking. Another consideration is whether the current vaccines against SARS-CoV-2 will prevent transmission. While phase III clinical trial data suggest that Pfizer's BNT162b2 and Moderna's mRNA-1273 are effective against symptomatic disease and prevent progression to severe disease, more data over a longer period of time is needed to determine their durability and ability to prevent or mitigate upper airway infection and transmission.
Prophylactic and/or therapeutic administration of mAbs could have utility in populations that respond poorly to vaccination or remain unvaccinated. Currently two antibody drugs, Lilly's LY-CoV555 (Chen et al., 2020; Jones et al., 2020) and Regeneron's REGN-COV2 (Baum et al., 2020a) have been granted EUA status with at least 11 others undergoing advanced human testing in phase II or III clinical trials. Whereas both REGN-COV2 and LY-CoV555 are potently neutralizing anti-RBD human IgG1s, LY-CoV555 is a monotherapy and REGN-COV2 is a cocktail of two mAbs. Preliminary results with REGN-COV2 and LY-CoV555 demonstrated reduced hospitalization rates in mAb-treated subjects with mild to-moderate COVID-19. However, clinical trials showed no benefit of LY-CoV55 in hospitalized patients (ACTIV-3/TICO LY-CoV555 Study Group, 2020) highlighting that current mAb treatments may have their greatest effect early in the course of COVID-19. As more mAbs are developed and tested, several factors may be considered for progression to human evaluation including production capacity, stability, possible synergy in antibody combination, prevention of resistance, the role and optimization of Fc effector functions, and mechanism of neutralization. Furthermore, as SARS-CoV-2 viral evolution continues, as seen with the emergence of novel strains in Great Britan, South Africa, and Brazil, existing antibody therapies and vaccines may need to be reformulated to remain effective. In the case of vaccine-induced immunity, one might predict that a polyclonal response against the SARS-CoV-2 S would not result in a complete loss of vaccine efficacy due to the small number of point mutations present in the spike of the circulating variants and the likelihood of generating multiple antibodies against different regions. However, preliminary data demonstrating reduced neutralizing activity of plasma from vaccinated individuals against psuedoviruses containing E484K, N501Y or K417N/E484K/N501Y substitutions has raised concern (Wang et al., 2021b, 2021c; Wibmer et al., 2021). These data contrast with a study utilizing infectious SARS-CoV-2 virus with similar mutations that showed only modest reductions in neutralization potency by mRNA vaccine-elicited sera (Xie et al., 2021). At present, there is insufficient data to conclude what level of efficacy will be achieved by vaccine immunity against the emerging variants. Updated mAb cocktails and adjustments to the spike sequences of vaccines may be needed to prevent loss of protection.
In summary, while the current pandemic continues to rage, due to the remarkable efforts of numerous groups and partnerships across the world, SARS-CoV-2 countermeasures are advancing at record speed and now being deployed. In the months to come, the ongoing need for public health practices such as universal mask-wearing and social distancing will continue to be particularly important until a sufficient proportion of the population is immunized or achieve herd immunity, and emerging variants are shown to be controlled by natural or vaccine-induced immunity. With four new countermeasures granted EUA status and others on the horizon, hopefully, by spring 2021, the COVID-19 pandemic will reach a turning point, as long as viral evolution does not overcome our immune-based interventions.
CRediT authorship contribution statement
James Brett Case: Conceptualization, Writing, Figure and Table preparation, Reviewing, and Editing. Emma S. Winkler: Conceptualization, Writing, Figure and Table preparation, Reviewing, and Editing. John M. Errico: Conceptualization, Writing, Figure and Table preparation, Reviewing, and Editing. Michael S. Diamond: Conceptualization, Writing, Figure and Table preparation, Reviewing, and Editing.
Declaration of competing interest
M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, Carnival Corporation and on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
Acknowledgements
This work was supported by R01 AI157155 and the Defense Advanced Research Project Agency (HR001117S0019). J.B.C. is supported by a Helen Hay Whitney Foundation postdoctoral fellowship. E.S.W. is supported by F30 AI152327-01. We thank Daved Fremont for critical comments on the manuscript. We apologize to our colleagues for omissions due to space constraints or the fluid nature of advances. An up-to-date list of SARS-CoV-2 vaccines and therapeutic mAbs can be found at https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines and https://www.antibodysociety.org/covid-19-biologics-tracker/.
References
- ACTIV-3/TICO LY-CoV555 Study Group A neutralizing monoclonal antibody for hospitalized patients with COVID-19. N Engl J Med NEJMoa2033130. 2020 doi: 10.1056/NEJMoa2033130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., Mukherjee A., Kumar G., Chatterjee P., Bhatnagar T., Malhotra P. Convalescent plasma in the management of moderate covid-19 in adults in India: open label phase II multicentre randomised controlled trial (PLACID Trial) BMJ. 2020;371:m3939. doi: 10.1136/bmj.m3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., Winkler E.S., Fox J.M., Kafai N.M., Thackray L.B., Hassan A.O., Amanat F., Krammer F., Watson C.T., Kleinstein S.H., Fremont D.H., Diamond M.S., Ellebedy A.H. A potently neutralizing antibody protects mice against SARS-CoV-2 infection. J. Immunol. 2020;205:915. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amanna I.J., Slifka M.K. Vaccination Strategies against Highly Variable Pathogens, Current Topics in Microbiology and Immunology. Springer International Publishing; Cham: 2018. Successful vaccines; pp. 1–30. [DOI] [Google Scholar]
- Aoki F.Y., Rubin M.E., Friesen A.D., Bowman J.M., Saunders J.R. Intravenous human rabies immunoglobulin for post-exposure prophylaxis: serum rabies neutralizing antibody concentrations and side-effects. J. Biol. Stand. 1989;17:91–104. doi: 10.1016/0092-1157(89)90032-2. [DOI] [PubMed] [Google Scholar]
- Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J., Shuey K., Feldman J., Hauser B.M., Caradonna T., Schmidt A.G., Suscovich T.J., Linde C., Cai Y., Barouch D., Ryan E.T., Charles R.C., Lauffenburger D., Chu H., Alter G. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532. doi: 10.1016/j.immuni.2020.07.020. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Deng W., Huang B., Gao H., Liu J., Ren L., Wei Q., Yu P., Xu Y., Qi F., Qu Y., Li F., Lv Q., Wang W., Xue J., Gong S., Liu M., Wang G., Wang S., Song Z., Zhao L., Liu P., Zhao L., Ye F., Wang H., Zhou W., Zhu N., Zhen W., Yu H., Zhang X., Guo L., Chen L., Wang C., Wang Y., Wang X., Xiao Y., Sun Q., Liu H., Zhu F., Ma C., Yan L., Yang M., Han J., Xu W., Tan W., Peng X., Jin Q., Wu G., Qin C. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.-M.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., Robbiani D.F., Nussenzweig M.C., West A.P., Bjorkman P.J. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., Lorenzi J.C.C., Finkin S., Hägglöf T., Hurley A., Millard K.G., Weisblum Y., Schmidt F., Hatziioannou T., Bieniasz P.D., Caskey M., Robbiani D.F., Nussenzweig M.C., Bjorkman P.J. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett A.D.T. Yellow fever vaccines. Biologicals. 1997;25:17–25. doi: 10.1006/biol.1997.0056. [DOI] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., Atwal G.S., Oyejide A., Goez-Gazi Y., Dutton J., Clemmons E., Staples H.M., Bartley C., Klaffke B., Alfson K., Gazi M., Gonzalez O., Dick E., Carrion R., Pessaint L., Porto M., Cook A., Brown R., Ali V., Greenhouse J., Taylor T., Andersen H., Lewis M.G., Stahl N., Murphy A.J., Yancopoulos G.D., Kyratsous C.A. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belongia E.A., Naleway A.L. Smallpox vaccine: the good, the bad, and the ugly. Clin. Med. Res. 2003;1:87–92. doi: 10.3121/cmr.1.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Corti D., Virgin H.W., Ravetch J.V. Fc-optimized antibodies elicit CD8 immunity to viral respiratory infection. Nature. 2020;588:485–490. doi: 10.1038/s41586-020-2838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., DiLillo D.J., Goff A.J., Glass P.J., Ravetch J.V. Differential requirements for FcγR engagement by protective antibodies against Ebola virus. Proc. Natl. Acad. Sci. U.S.A. 2019;116:20054. doi: 10.1073/pnas.1911842116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bournazos S., Klein F., Pietzsch J., Seaman M.S., Nussenzweig M.C., Ravetch J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell. 2014;158:1243–1253. doi: 10.1016/j.cell.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., Bentlage A.E.H., van Haaren M.M., Guerra D., Burger J.A., Schermer E.E., Verheul K.D., van der Velde N., van der Kooi A., van Schooten J., van Breemen M.J., Bijl T.P.L., Sliepen K., Aartse A., Derking R., Bontjer I., Kootstra N.A., Wiersinga W.J., Vidarsson G., Haagmans B.L., Ward A.B., de Bree G.J., Sanders R.W., van Gils M.J. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Österlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Kafai N.M., Fox J.M., Smith B.K., Shrihari S., McCune B.T., Harvey I.B., Keeler S.P., Bloyet L.-M., Zhao H., Ma M., Adams L.J., Winkler E.S., Holtzman M.J., Fremont D.H., Whelan S.P.J., Diamond M.S. Replication-competent vesicular stomatitis virus vaccine vector protects against SARS-CoV-2-mediated pathogenesis in mice. Cell Host Microbe. 2020;28:465–474. doi: 10.1016/j.chom.2020.07.018. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.-W., Zhang A.J., Yuan S., Poon V.K.-M., Chan C.C.-S., Lee A.C.-Y., Chan W.-M., Fan Z., Tsoi H.-W., Wen L., Liang R., Cao J., Chen Y., Tang K., Luo C., Cai J.-P., Kok K.-H., Chu H., Chan K.-H., Sridhar S., Chen Z., Chen H., To K.K.-W., Yuen K.-Y. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin. Infect. Dis. 2020;71:2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., Shawa I., Adams A.C., Van Naarden J., Custer K.L., Shen L., Durante M., Oakley G., Schade A.E., Sabo J., Patel D.R., Klekotka P., Skovronsky D.M. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N Engl J Med NEJMoa2029849. 2020 doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F., Salazar M.D., Contreras H., Nguyen V.H., Martinez J., Park Y., Nguyen J., Kha M., Iniguez A., Zhou Q., Kaltcheva T., Levytskyy R., Ebelt N.D., Kang T.H., Wu X., Rogers T.F., Manuel E.R., Shostak Y., Diamond D.J., Wussow F. Development of a multi-antigenic SARS-CoV-2 vaccine candidate using a synthetic poxvirus platform. Nat. Commun. 2020;11:6121. doi: 10.1038/s41467-020-19819-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P.L.S.M., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057. doi: 10.1016/j.cell.2020.09.033. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen J. “Absolutely remarkable”: No one who got Moderna's vaccine in trial developed severe COVID-19. Science. 2020 doi: 10.1126/science.abf9360. [DOI] [Google Scholar]
- Coleman J.R., Papamichail D., Skiena S., Futcher B., Wimmer E., Mueller S. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., Flach B., O'Connell S., Bock K.W., Minai M., Nagata B.M., Andersen H., Martinez D.R., Noe A.T., Douek N., Donaldson M.M., Nji N.N., Alvarado G.S., Edwards D.K., Flebbe D.R., Lamb E., Doria-Rose N.A., Lin B.C., Louder M.K., O'Dell S., Schmidt S.D., Phung E., Chang L.A., Yap C., Todd J.-P.M., Pessaint L., Van Ry A., Browne S., Greenhouse J., Putman-Taylor T., Strasbaugh A., Campbell T.-A., Cook A., Dodson A., Steingrebe K., Shi W., Zhang Y., Abiona O.M., Wang L., Pegu A., Yang E.S., Leung K., Zhou T., Teng I.-T., Widge A., Gordon I., Novik L., Gillespie R.A., Loomis R.J., Moliva J.I., Stewart-Jones G., Himansu S., Kong W.-P., Nason M.C., Morabito K.M., Ruckwardt T.J., Ledgerwood J.E., Gaudinski M.R., Kwong P.D., Mascola J.R., Carfi A., Lewis M.G., Baric R.S., McDermott A., Moore I.N., Sullivan N.J., Roederer M., Seder R.A., Graham B.S. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N. Engl. J. Med. 2020;383:1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M. Covid-19: identifying and isolating asymptomatic people helped eliminate virus in Italian village. BMJ. 2020;368:m1165. doi: 10.1136/bmj.m1165. [DOI] [PubMed] [Google Scholar]
- Diamond M.S., Pierson T.C. The challenges of vaccine development against a new virus during a pandemic. Cell Host Microbe. 2020;27:699–703. doi: 10.1016/j.chom.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Palese P., Wilson P.C., Ravetch J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Invest. 2016;126:605–610. doi: 10.1172/JCI84428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiLillo D.J., Tan G.S., Palese P., Ravetch J.V. Broadly neutralizing hemagglutinin stalk–specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat. Med. 2014;20:143–151. doi: 10.1038/nm.3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinnon K.H., Leist S.R., Schäfer A., Edwards C.E., Martinez D.R., Montgomery S.A., West A., Yount B.L., Hou Y.J., Adams L.E., Gully K.L., Brown A.J., Huang E., Bryant M.D., Choong I.C., Glenn J.S., Gralinski L.E., Sheahan T.P., Baric R.S. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. U.S.A. 2020;117:9490. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enkirch T., Messling von V. Ferret models of viral pathogenesis. Virology. 2015;479–480:259–270. doi: 10.1016/j.virol.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak A.I., Kuzmina N., Murin C.D., Bryan C., Davidson E., Gilchuk P., Gulka C.P., Ilinykh P.A., Shen X., Huang K., Ramanathan P., Turner H., Fusco M.L., Lampley R., Kose N., King H., Sapparapu G., Doranz B.J., Ksiazek T.G., Wright D.W., Saphire E.O., Ward A.B., Bukreyev A., Crowe J.E. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2–MPER region. Nat. Microbiol. 2018;3:670–677. doi: 10.1038/s41564-018-0157-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aboagye J., Adams K., Ali A., Allen E., Allison J.L., Anslow R., Arbe-Barnes E.H., Babbage G., Baillie K., Baker M., Baker N., Baker P., Baleanu I., Ballaminut J., Barnes E., Barrett J., Bates L., Batten A., Beadon K., Beckley R., Berrie E., Berry L., Beveridge A., Bewley K.R., Bijker E.M., Bingham T., Blackwell L., Blundell C.L., Bolam E., Boland E., Borthwick N., Bower T., Boyd A., Brenner T., Bright P.D., Brown-O'Sullivan C., Brunt E., Burbage J., Burge S., Buttigieg K.R., Byard N., Cabera Puig I., Calvert A., Camara S., Cao M., Cappuccini F., Carr M., Carroll M.W., Carter V., Cathie K., Challis R.J., Charlton S., Chelysheva I., Cho J.-S., Cicconi P., Cifuentes L., Clark H., Clark E., Cole T., Colin-Jones R., Conlon C.P., Cook A., Coombes N.S., Cooper R., Cosgrove C.A., Coy K., Crocker W.E.M., Cunningham C.J., Damratoski B.E., Dando L., Datoo M.S., Davies H., De Graaf H., Demissie T., Di Maso C., Dietrich I., Dong T., Donnellan F.R., Douglas N., Downing C., Drake J., Drake-Brockman R., Drury R.E., Dunachie S.J., Edwards N.J., Edwards F.D.L., Edwards C.J., Elias S.C., Elmore M.J., Emary K.R.W., English M.R., Fagerbrink S., Felle S., Feng S., Field S., Fixmer C., Fletcher C., Ford K.J., Fowler J., Fox P., Francis E., Frater J., Furze J., Fuskova M., Galiza E., Gbesemete D., Gilbride C., Godwin K., Gorini G., Goulston L., Grabau C., Gracie L., Gray Z., Guthrie L.B., Hackett M., Halwe S., Hamilton E., Hamlyn J., Hanumunthadu B., Harding I., Harris S.A., Harris A., Harrison D., Harrison C., Hart T.C., Haskell L., Hawkins S., Head I., Henry J.A., Hill J., Hodgson S.H.C., Hou M.M., Howe E., Howell N., Hutlin C., Ikram S., Isitt C., Iveson P., Jackson S., Jackson F., James S.W., Jenkins M., Jones E., Jones K., Jones C.E., Jones B., Kailath R., Karampatsas K., Keen J., Kelly S., Kelly D., Kerr D., Kerridge S., Khan L., Khan U., Killen A., Kinch J., King T.B., King L., King J., Kingham-Page L., Klenerman P., Knapper F., Knight J.C., Knott D., Koleva S., Kupke A., Larkworthy C.W., Larwood J.P.J., Laskey A., Lawrie A.M., Lee A., Ngan Lee K.Y., Lees E.A., Legge H., Lelliott A., Lemm N.-M., Lias A.M., Linder A., Lipworth S., Liu X., Liu S., Lopez Ramon R., Lwin M., Mabesa F., Madhavan M., Mallett G., Mansatta K., Marcal I., Marinou S., Marlow E., Marshall J.L., Martin J., McEwan J., McInroy L., Meddaugh G., Mentzer A.J., Mirtorabi N., Moore M., Moran E., Morey E., Morgan V., Morris S.J., Morrison H., Morshead G., Morter R., Mujadidi Y.F., Muller J., Munera-Huertas T., Munro C., Munro A., Murphy S., Munster V.J., Mweu P., Noé A., Nugent F.L., Nuthall E., O'Brien K., O'Connor D., Oguti B., Oliver J.L., Oliveira C., O'Reilly P.J., Osborn M., Osborne P., Owen C., Owens D., Owino N., Pacurar M., Parker K., Parracho H., Patrick-Smith M., Payne V., Pearce J., Peng Y., Peralta Alvarez M.P., Perring J., Pfafferott K., Pipini D., Plested E., Pluess-Hall H., Pollock K., Poulton I., Presland L., Provstgaard-Morys S., Pulido D., Radia K., Ramos Lopez F., Rand J., Ratcliffe H., Rawlinson T., Rhead S., Riddell A., Ritchie A.J., Roberts H., Robson J., Roche S., Rohde C., Rollier C.S., Romani R., Rudiansyah I., Saich S., Sajjad S., Salvador S., Sanchez Riera L., Sanders H., Sanders K., Sapaun S., Sayce C., Schofield E., Screaton G., Selby B., Semple C., Sharpe H.R., Shaik I., Shea A., Shelton H., Silk S., Silva-Reyes L., Skelly D.T., Smee H., Smith C.C., Smith D.J., Song R., Spencer A.J., Stafford E., Steele A., Stefanova E., Stockdale L., Szigeti A., Tahiri-Alaoui A., Tait M., Talbot H., Tanner R., Taylor I.J., Taylor V., Water Naude Te R., Thakur N., Themistocleous Y., Themistocleous A., Thomas M., Thomas T.M., Thompson A., Thomson-Hill S., Tomlins J., Tonks S., Towner J., Tran N., Tree J.A., Truby A., Turkentine K., Turner C., Turner N., Turner S., Tuthill T., Ulaszewska M., Varughese R., van Doremalen N., Veighey K., Verheul M.K., Vichos I., Vitale E., Walker L., Watson M.E.E., Welham B., Wheat J., White C., White R., Worth A.T., Wright D., Wright S., Yao X.L., Yau Y. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J.M., Roy V., Gunn B.M., Huang L., Edeling M.A., Mack M., Fremont D.H., Doranz B.J., Johnson S., Alter G., Diamond M.S. Optimal therapeutic activity of monoclonal antibodies against chikungunya virus requires Fc-FcγR interaction on monocytes. Sci. Immunol. 2019;4 doi: 10.1126/sciimmunol.aav5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., Li Y., Zhu L., Wang N., Lv Z., Gao H., Ge X., Kan B., Hu Y., Liu J., Cai F., Jiang D., Yin Y., Qin C., Li J., Gong X., Lou X., Shi W., Wu D., Zhang H., Zhu L., Deng W., Li Y., Lu J., Li C., Wang X., Yin W., Zhang Y., Qin C. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369:77. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H., Chen Q., Yang G., He L., Fan H., Deng Y.-Q., Wang Y., Teng Y., Zhao Z., Cui Y., Li Y., Li X.-F., Li J., Zhang N.-N., Yang X., Chen S., Guo Y., Zhao G., Wang X., Luo D.-Y., Wang H., Yang X., Li Y., Han G., He Y., Zhou X., Geng S., Sheng X., Jiang S., Sun S., Qin C.-F., Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R.L., Donaldson E.F., Baric R.S. A decade after SARS: strategies for controlling emerging coronaviruses. Nat. Rev. Microbiol. 2013;11:836–848. doi: 10.1038/nrmicro3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57. doi: 10.1016/j.chom.2020.11.007. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. 2021. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., Tsang O.T.Y., Yeung Y.C., Perera R.A.P.M., Poon L.L.M., Peiris J.S.M., Valkenburg S.A. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat. Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- Halper-Stromberg A., Nussenzweig M.C. Towards HIV-1 remission: potential roles for broadly neutralizing antibodies. J. Clin. Invest. 2016;126:415–423. doi: 10.1172/JCI80561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., Chung K.M., Hermann A., Ullman E., Cruz J., Rafique A., Huang T., Fairhurst J., Libertiny C., Malbec M., Lee W.-Y., Welsh R., Farr G., Pennington S., Deshpande D., Cheng J., Watty A., Bouffard P., Babb R., Levenkova N., Chen C., Zhang B., Romero Hernandez A., Saotome K., Zhou Y., Franklin M., Sivapalasingam S., Lye D.C., Weston S., Logue J., Haupt R., Frieman M., Chen G., Olson W., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., Turner J.S., Schmitz A.J., Lei T., Shrihari S., Keeler S.P., Fremont D.H., Greco S., McCray P.B., Jr., Perlman S., Holtzman M.J., Ellebedy A.H., Diamond M.S. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hessell A.J., Hangartner L., Hunter M., Havenith C.E.G., Beurskens F.J., Bakker J.M., Lanigan C.M.S., Landucci G., Forthal D.N., Parren P.W.H.I., Marx P.A., Burton D.R. Fc receptor but not complement binding is important in antibody protection against HIV. Nature. 2007;449:101–104. doi: 10.1038/nature06106. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., Tan T.K., Rijal P., Coombes N., Bewley K.R., Tree J.A., Radecke J., Paterson N.G., Supasa P., Mongkolsapaya J., Screaton G.R., Carroll M., Townsend A., Fry E.E., Owens R.J., Stuart D.I. Neutralization of SARS-CoV-2 by destruction of the prefusion spike. Cell Host Microbe. 2020;28:445–454. doi: 10.1016/j.chom.2020.06.010. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M., Iwatsuki-Horimoto K., Hatta M., Loeber S., Halfmann P.J., Nakajima N., Watanabe T., Ujie M., Takahashi K., Ito M., Yamada S., Fan S., Chiba S., Kuroda M., Guan L., Takada K., Armbrust T., Balogh A., Furusawa Y., Okuda M., Ueki H., Yasuhara A., Sakai-Tagawa Y., Lopes T.J.S., Kiso M., Yamayoshi S., Kinoshita N., Ohmagari N., Hattori S.-I., Takeda M., Mitsuya H., Krammer F., Suzuki T., Kawaoka Y. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:16587–16595. doi: 10.1073/pnas.2009799117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatsuki-Horimoto K., Nakajima N., Ichiko Y., Sakai-Tagawa Y., Noda T., Hasegawa H., Kawaoka Y., Dermody T.S. Syrian hamster as an animal model for the study of human influenza virus infection. J. Virol. 2018;92(JVI) doi: 10.1128/JVI.01693-17. 01693–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J., McDermott A., Flach B., Doria-Rose N.A., Corbett K.S., Morabito K.M., O'Dell S., Schmidt S.D., Swanson P.A., II, Padilla M., Mascola J.R., Neuzil K.M., Bennett H., Sun W., Peters E., Makowski M., Albert J., Cross K., Buchanan W., Pikaart-Tautges R., Ledgerwood J.E., Graham B.S., Beigel J.H. An mRNA vaccine against SARS-CoV-2 — preliminary report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R.-D., Liu M.-Q., Chen Y., Shan C., Zhou Y.-W., Shen X.-R., Li Q., Zhang L., Zhu Y., Si H.-R., Wang Q., Min J., Wang X., Zhang W., Li B., Zhang H.-J., Baric R.S., Zhou P., Yang X.-L., Shi Z.-L. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B.A., Xie X., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., Zhang L., Bopp N., Schindewolf C., Vu M., Vanderheiden A., Swetnam D., Plante J.A., Aguilar P., Plante K.S., Lee B., Weaver S.C., Suthar M.S., Routh A.L., Ren P., Ku Z., An Z., Debbink K., Shi P.-Y., Freiberg A.N., Menachery V.D. Furin cleavage site is key to SARS-CoV-2 pathogenesis. bioRxiv. 2020;581 doi: 10.1101/2020.08.26.268854. 2020.08.26.268854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., Higgs R.E., Balasubramaniam D., Wang L., Bidshahri R., Kraft L., Hwang Y., Žentelis S., Jepson K.R., Goya R., Smith M.A., Collins D.W., Hinshaw S.J., Tycho S.A., Pellacani D., Xiang P., Muthuraman K., Sobhanifar S., Piper M.H., Triana F.J., Hendle J., Pustilnik A., Adams A.C., Berens S.J., Baric R.S., Martinez D.R., Cross R.W., Geisbert T.W., Borisevich V., Abiona O., Belli H.M., de Vries M., Mohamed A., Dittmann M., Samanovic M., Mulligan M.J., Goldsmith J.A., Hsieh C.-L., Johnson N.V., Wrapp D., McLellan J.S., Barnhart B.C., Graham B.S., Mascola J.R., Hansen C.L., Falconer E. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv. 2020 2020.09.30.318972. [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., Tang X., Yu J., Lan J., Yuan J., Wang H., Zhao J., Zhang S., Wang Y., Shi X., Liu L., Zhao J., Wang X., Zhang Z., Zhang L. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., Lu J.M., Peukes J., Xiong X., Kräusslich H.-G., Scheres S.H.W., Bartenschlager R., Briggs J.A.G. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., Plested J.S., Zhu M., Cloney-Clark S., Zhou H., Smith G., Patel N., Frieman M.B., Haupt R.E., Logue J., McGrath M., Weston S., Piedra P.A., Desai C., Callahan K., Lewis M., Price-Abbott P., Formica N., Shinde V., Fries L., Lickliter J.D., Griffin P., Wilkinson B., Glenn G.M. Phase 1–2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383:2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.-I., Kim S.-G., Kim S.-M., Kim E.-H., Park S.-J., Yu K.-M., Chang J.-H., Kim E.J., Lee S., Casel M.A.B., Um J., Song M.-S., Jeong H.W., Lai V.D., Kim Y., Chin B.S., Park J.-S., Chung K.-H., Foo S.-S., Poo H., Mo I.-P., Lee O.-J., Webby R.J., Jung J.U., Choi Y.K. Infection and rapid transmission of SARS-CoV-2 in ferrets. Cell Host Microbe. 2020;27:704–709. doi: 10.1016/j.chom.2020.03.023. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J.-H., Müller M.A., Seok H., Park G.E., Lee J.Y., Cho S.Y., Ha Y.E., Baek J.Y., Kim S.H., Kang J.-M., Kim Y.-J., Jo I.J., Chung C.R., Hahn M.-J., Drosten C., Kang C.-I., Chung D.R., Song J.-H., Kang E.-S., Peck K.R. Serologic responses of 42 MERS-coronavirus-infected patients according to the disease severity. Diagn. Microbiol. Infect. Dis. 2017;89:106–111. doi: 10.1016/j.diagmicrobio.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye J., Reincke S.M., Kornau H.-C., Sánchez-Sendin E., Corman V.M., Liu H., Yuan M., Wu N.C., Zhu X., Lee C.-C.D., Trimpert J., Höltje M., Dietert K., Stöffler L., Wardenburg von N., van Hoof S., Homeyer M.A., Hoffmann J., Abdelgawad A., Gruber A.D., Bertzbach L.D., Vladimirova D., Li L.Y., Barthel P.C., Skriner K., Hocke A.C., Hippenstiel S., Witzenrath M., Suttorp N., Kurth F., Franke C., Endres M., Schmitz D., Jeworowski L.M., Richter A., Schmidt M.L., Schwarz T., Müller M.A., Drosten C., Wendisch D., Sander L.E., Osterrieder N., Wilson I.A., Prüss H. A therapeutic non-self-reactive SARS-CoV-2 antibody protects from lung pathology in a COVID-19 hamster model. Cell. 2020;183:1058–1069. doi: 10.1016/j.cell.2020.09.049. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-Y., Kim C., Ryu D.-K., Lee J., Kim Y.-I., Seo J.-M., kim Y.-G., Jeong J.-H., Kim M., Kim J.-I., Kim P., Bae J.S., Shim E.Y., Lee M.S., Kim M.S., Noh H., Park G.-S., Park J.S., Son D., An Y., Lee J.N., Kwon K.-S., Lee J.-Y., Lee H., Yang J.-S., Kim K.-C., Kim S.S., Woo H.-M., Kim J.-W., Park M.-S., Yu K.-M., Kim S.-M., Kim E.-H., Park S.-J., Jeong S.T., Yu C.H., Song Y., Gu S.H., Oh H., Koo B.-S., Hong J.J., Ryu C.-M., Park W.B., Oh M.-D., Choi Y.K. Research Square; 2020. A Novel Neutralizing Antibody Targeting Receptor-Binding Domain of SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., He W., Liu X., Zheng S., Qi Y., Li H., Mao F., Liu J., Sun Y., Pan L., Du K., Ye K., Li W., Sui J. A potent human neutralizing antibody Fc-dependently reduces established HBV infections. Elife. 2017;6 doi: 10.7554/eLife.26738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhang W., Hu Y., Tong X., Zheng S., Yang J., Kong Y., Ren L., Wei Q., Mei H., Hu C., Tao C., Yang R., Wang J., Yu Y., Guo Y., Wu X., Xu Z., Zeng L., Xiong N., Chen L., Wang J., Man N., Liu Y., Xu H., Deng E., Zhang X., Li C., Wang C., Su S., Zhang L., Wang J., Wu Y., Liu Z. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. J. Am. Med. Assoc. 2020;324:460–470. doi: 10.1001/jama.2020.10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei Sen, Chen Bin, Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libster R., Pérez Marc G., Wappner D., Coviello S., Bianchi A., Braem V., Esteban I., Caballero M.T., Wood C., Berrueta M., Rondan A., Lescano G., Cruz P., Ritou Y., Fernández Viña V., Álvarez Paggi D., Esperante S., Ferreti A., Ofman G., Ciganda Á., Rodriguez R., Lantos J., Valentini R., Itcovici N., Hintze A., Oyarvide M.L., Etchegaray C., Neira A., Name I., Alfonso J., López Castelo R., Caruso G., Rapelius S., Alvez F., Etchenique F., Dimase F., Alvarez D., Aranda S.S., Sánchez Yanotti C., De Luca J., Jares Baglivo S., Laudanno S., Nowogrodzki F., Larrea R., Silveyra M., Leberzstein G., Debonis A., Molinos J., González M., Perez E., Kreplak N., Pastor Argüello S., Gibbons L., Althabe F., Bergel E., Polack F.P. Early high-titer plasma therapy to prevent severe covid-19 in older adults. N. Engl. J. Med. 2021 doi: 10.1056/nejmoa2033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., Van Schooten J., Zhu X., Lee C.-C.D., Brouwer P.J.M., Van Gils M.J., Sanders R.W., Ward A.B., Wilson I.A. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020;53:1272–1280. doi: 10.1016/j.immuni.2020.10.023. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.W., Sahi V., Figueroa A., Guo X.V., Cerutti G., Bimela J., Gorman J., Zhou T., Chen Z., Yuen K.-Y., Kwong P.D., Sodroski J.G., Yin M.T., Sheng Z., Huang Y., Shapiro L., Ho D.D. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Q., Fan C., Li Q., Zhou S., Huang W., Wang L., Sun C., Wang M., Wu X., Ma J., Li B., Xie L., Wang Y. Antibody-dependent-cellular-cytotoxicity-inducing antibodies significantly affect the post-exposure treatment of Ebola virus infection. Sci. Rep. 2017;7:45552. doi: 10.1038/srep45552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Lin H.-M., Baine I., Wajnberg A., Gumprecht J.P., Rahman F., Rodriguez D., Tandon P., Bassily-Marcus A., Bander J., Sanky C., Dupper A., Zheng A., Nguyen F.T., Amanat F., Stadlbauer D., Altman D.R., Chen B.K., Krammer F., Mendu D.R., Firpo-Betancourt A., Levin M.A., Bagiella E., Casadevall A., Cordon-Cardo C., Jhang J.S., Arinsburg S.A., Reich D.L., Aberg J.A., Bouvier N.M. Convalescent plasma treatment of severe COVID-19: a propensity score–matched control study. Nat. Med. 2020;26:1708–1713. doi: 10.1038/s41591-020-1088-9. [DOI] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Rothlauf P.W., Bloyet L.-M., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. bioRxiv. 2020 doi: 10.1016/j.chom.2021.01.014. 2020.11.06.372037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., Grousova D.M., Erokhova A.S., Kovyrshina A.V., Botikov A.G., Izhaeva F.M., Popova O., Ozharovskaya T.A., Esmagambetov I.B., Favorskaya I.A., Zrelkin D.I., Voronina D.V., Shcherbinin D.N., Semikhin A.S., Simakova Y.V., Tokarskaya E.A., Lubenets N.L., Egorova D.A., Shmarov M.M., Nikitenko N.A., Morozova L.F., Smolyarchuk E.A., Kryukov E.V., Babira V.F., Borisevich S.V., Naroditsky B.S., Gintsburg A.L. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396:887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., Wang D.-Q., Hu Y., Ren J.-H., Tang N., Xu Y.-Y., Yu L.-H., Mo Z., Gong F., Zhang X.-L., Tian W.-G., Hu L., Zhang X.-X., Xiang J.-L., Du H.-X., Liu H.-W., Lang C.-H., Luo X.-H., Wu S.-B., Cui X.-P., Zhou Z., Zhu M.-M., Wang J., Xue C.-J., Li X.-F., Wang L., Li Z.-J., Wang K., Niu C.-C., Yang Q.-J., Tang X.-J., Zhang Y., Liu X.-M., Li J.-J., Zhang D.-C., Zhang F., Liu P., Yuan J., Li Q., Hu J.-L., Chen J., Huang A.-L. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lu L.L., Suscovich T.J., Fortune S.M., Alter G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 2018;18:46–61. doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., Sakata M., Tahara M., Kutsuna S., Ohmagari N., Kuroda M., Suzuki T., Kageyama T., Takeda M. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. Unit. States Am. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., Liu J., Peter L., McMahan K., Tostanoski L.H., He X., Martinez D.R., Rutten L., Bos R., van Manen D., Vellinga J., Custers J., Langedijk J.P., Kwaks T., Bakkers M.J.G., Zuijdgeest D., Rosendahl Huber S.K., Atyeo C., Fischinger S., Burke J.S., Feldman J., Hauser B.M., Caradonna T.M., Bondzie E.A., Dagotto G., Gebre M.S., Hoffman E., Jacob-Dolan C., Kirilova M., Li Z., Lin Z., Mahrokhian S.H., Maxfield L.F., Nampanya F., Nityanandam R., Nkolola J.P., Patel S., Ventura J.D., Verrington K., Wan H., Pessaint L., Van Ry A., Blade K., Strasbaugh A., Cabus M., Brown R., Cook A., Zouantchangadou S., Teow E., Andersen H., Lewis M.G., Cai Y., Chen B., Schmidt A.G., Reeves R.K., Baric R.S., Lauffenburger D.A., Alter G., Stoffels P., Mammen M., Van Hoof J., Schuitemaker H., Barouch D.H. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586:583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao J., Chard L.S., Wang Z., Wang Y. Syrian hamster as an animal model for the study on infectious diseases. Front. Immunol. 2019;10:2704. doi: 10.3389/fimmu.2019.02329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D.M. Antibody-dependent enhancement of infection and the pathogenesis of viral disease. Clin. Infect. Dis. 1994;19:500–512. doi: 10.1093/clinids/19.3.500. [DOI] [PubMed] [Google Scholar]
- Mudgal R., Nehul S., Tomar S. Prospects for mucosal vaccine: shutting the door on SARS-CoV-2. Hum. Vaccines Immunother. 2020;59:1–11. doi: 10.1080/21645515.2020.1805992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Feldmann F., Williamson B.N., van Doremalen N., Pérez-Pérez L., Schulz J., Meade-White K., Okumura A., Callison J., Brumbaugh B., Avanzato V.A., Rosenke R., Hanley P.W., Saturday G., Scott D., Fischer E.R., de Wit E. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature. 2020;585:268–272. doi: 10.1038/s41586-020-2324-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela C., Dowling W.E., Funnell S.G.P., Gsell P.-S., Balta X.R., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., Qin C., Crozier I., Dallmeier K., de Waal L., de Wit E., Delang L., Dohm E., Duprex W.P., Falzarano D., Finch C.L., Frieman M.B., Graham B.S., Gralinski L., Guilfoyle K., Haagmans B.L., Hamilton G.A., Hartman A.L., Herfst S., Kaptein S.J.F., Klimstra W., Knezevic I., Krause P.R., Kuhn J.H., Le Grand R., Lewis M., Liu W.-C., Maisonnasse P., McElroy A.K., Munster V., Oreshkova N., Rasmussen A.L., Rocha-Pereira J., Rockx B., Rodríguez E., Rogers T., Salguero F.J., Schotsaert M., Stittelaar K., Jan Thibaut H., Tseng C.-T., Vergara-Alert J., Beer M., Brasel T., Chan J.F.W., García-Sastre A., Neyts J., Perlman S., Reed D.S., Richt J.A., Roy C.J., Segalés J., Vasan S.S., Henao-Restrepo A.M., Barouch D.H. Animal models for COVID-19. Nature. 2020;20:669. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdin A.D., Barreto L., Plotkin S. Inactivated poliovirus vaccine: past and present experience. Vaccine. 1996;14:735–746. doi: 10.1016/0264-410x(95)00211-i. [DOI] [PubMed] [Google Scholar]
- Ni L., Ye F., Cheng M.-L., Feng Y., Deng Y.-Q., Zhao H., Wei P., Ge J., Gou M., Li X., Sun L., Cao T., Wang P., Zhou C., Zhang R., Liang P., Guo H., Wang X., Qin C.-F., Chen F., Dong C. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity. 2020;52:971–977. doi: 10.1016/j.immuni.2020.04.023. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., Epstein E., Achdout H., Melamed S., Chitlaru T., Weiss S., Peretz E., Rosen O., Paran N., Yitzhaki S., Shapira S.C., Israely T., Mazor O., Rosenfeld R. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oude Munnink B.B., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., van der Spek A., Tolsma P., Rietveld A., Brouwer M., Bouwmeester-Vincken N., Harders F., Hakze-van der Honing R., Wegdam-Blans M.C.A., Bouwstra R.J., GeurtsvanKessel C., van der Eijk A.A., Velkers F.C., Smit L.A.M., Stegeman A., van der Poel W.H.M., Koopmans M.P.G. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021;371:172–177. doi: 10.1126/science.abe5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines — a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen E., Koopmans M., Go U., Hamer D.H., Petrosillo N., Castelli F., Storgaard M., Al Khalili S., Simonsen L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020;20:e238–e244. doi: 10.1016/S1473-3099(20)30484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Jr., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl. J. Med. NEJMoa2034577. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy M.N., Minassian A.M., Ewer K.J., Flaxman A.L., Folegatti P.M., Owens D.R., Voysey M., Aley P.K., Angus B., Babbage G., Belij-Rammerstorfer S., Berry L., Bibi S., Bittaye M., Cathie K., Chappell H., Charlton S., Cicconi P., Clutterbuck E.A., Colin-Jones R., Dold C., Emary K.R.W., Fedosyuk S., Fuskova M., Gbesemete D., Green C., Hallis B., Hou M.M., Jenkin D., Joe C.C.D., Kelly E.J., Kerridge S., Lawrie A.M., Lelliott A., Lwin M.N., Makinson R., Marchevsky N.G., Mujadidi Y., Munro A.P.S., Pacurar M., Plested E., Rand J., Rawlinson T., Rhead S., Robinson H., Ritchie A.J., Ross-Russell A.L., Saich S., Singh N., Smith C.C., Snape M.D., Song R., Tarrant R., Themistocleous Y., Thomas K.M., Villafana T.L., Warren S.C., Watson M.E.E., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Faust S.N., Pollard A.J., Aboagye J., Adams K., Ali A., Allen E.R., Allen L., Allison J.L., Andritsou F., Anslow R., Arbe-Barnes E.H., Baker M., Baker N., Baker P., Baleanu I., Barker D., Barnes E., Barrett J.R., Barrett K., Bates L., Batten A., Beadon K., Beckley R., Bellamy D., Berg A., Bermejo L., Berrie E., Beveridge A., Bewley K., Bijker E.M., Birch G., Blackwell L., Bletchly H., Blundell C.L., Blundell S.R., Bolam E., Boland E., Bormans D., Borthwick N., Boukas K., Bower T., Bowring F., Boyd A., Brenner T., Brown P., Brown-O'Sullivan C., Bruce S., Brunt E., Burbage J., Burgoyne J., Buttigieg K.R., Byard N., Cabera Puig I., Camara S., Cao M., Cappuccini F., Carr M., Carroll M.W., Cashen P., Cavey A., Chadwick J., Challis R., Chapman D., Charles D., Chelysheva I., Cho J.-S., Cifuentes L., Clark E., Collins S., Conlon C.P., Coombes N.S., Cooper R., Cooper C., Crocker W.E.M., Crosbie S., Cullen D., Cunningham C., Cuthbertson F., Datoo B.E., Dando L., Datoo M.S., Datta C., Davies H., Davies S., Davis E.J., Davis J., Dearlove D., Demissie T., Di Marco S., Di Maso C., DiTirro D., Docksey C., Dong T., Donnellan F.R., Douglas N., Downing C., Drake J., Drake-Brockman R., Drury R.E., Dunachie S.J., Edwards C.J., Edwards N.J., Muhanna El O., Elias S.C., Elliott R.S., Elmore M.J., English M.R., Felle S., Feng S., Ferreira Da Silva C., Field S., Fisher R., Fixmer C., Ford K.J., Fowler J., Francis E., Frater J., Furze J., Galian-Rubio P., Galloway C., Garlant H., Gavrila M., Gibbons F., Gibbons K., Gilbride C., Gill H., Godwin K., Gordon-Quayle K., Gorini G., Goulston L., Grabau C., Gracie L., Graham N., Greenwood N., Griffiths O., Gupta G., Hamilton E., Hanumunthadu B., Harris S.A., Harris T., Harrison D., Hart T.C., Hartnell B., Haskell L., Hawkins S., Henry J.A., Hermosin Herrera M., Hill D., Hill J., Hodges G., Hodgson S.H.C., Horton K.L., Howe E., Howell N., Howes J., Huang B., Humphreys J., Humphries H.E., Iveson P., Jackson F., Jackson S., Jauregui S., Jeffers H., Jones B., Jones C.E., Jones E., Jones K., Joshi A., Kailath R., Keen J., Kelly D.M., Kelly S., Kelly D., Kerr D., Khan L., Khozoee B., Killen A., Kinch J., King L.D.W., King T.B., Kingham L., Klenerman P., Knight J.C., Knott D., Koleva S., Lang G., Larkworthy C.W., Larwood J.P.J., Law R., Lee A., Lee K.Y.N., Lees E.A., Leung S., Li Y., Lias A.M., Linder A., Lipworth S., Liu S., Liu X., Lloyd S., Loew L., Lopez Ramon R., Madhavan M., Mainwaring D.O., Mallett G., Mansatta K., Marinou S., Marius P., Marlow E., Marriott P., Marshall J.L., Martin J., Masters S., McEwan J., McGlashan J.L., McInroy L., McRobert N., Megson C., Mentzer A.J., Mirtorabi N., Mitton C., Moore M., Moran M., Morey E., Morgans R., Morris S.J., Morrison H.M., Morshead G., Morter R., Moya N.A., Mukhopadhyay E., Muller J., Munro C., Murphy S., Mweu P., Noé A., Nugent F.L., O'Brien K., O'Connor D., Oguti B., Olchawski V., Oliveira C., O'Reilly P.J., Osborne P., Owen L., Owino N., Papageorgiou P., Parracho H., Parsons K., Patel B., Patrick-Smith M., Peng Y., Penn E.J., Peralta Alvarez M.P., Perring J., Petropoulos C., Phillips D.J., Pipini D., Pollard S., Poulton I., Pratt D., Presland L., Proud P.C., Provstgaard-Morys S., Pueschel S., Pulido D., Rabara R., Radia K., Rajapaska D., Ramos Lopez F., Ratcliffe H., Rayhan S., Rees B., Reyes Pabon E., Roberts H., Robertson I., Roche S., Rollier C.S., Romani R., Rose Z., Rudiansyah I., Sabheha S., Salvador S., Sanders H., Sanders K., Satti I., Sayce C., Schmid A.B., Schofield E., Screaton G., Sedik C., Seddiqi S., Segireddy R.R., Selby B., Shaik I., Sharpe H.R., Shaw R., Shea A., Silk S., Silva-Reyes L., Skelly D.T., Smith D.J., Smith D.C., Smith N., Spencer A.J., Spoors L., Stafford E., Stamford I., Stockdale L., Stockley D., Stockwell L.V., Stokes M., Strickland L.H., Stuart A., Sulaiman S., Summerton E., Swash Z., Szigeti A., Tahiri-Alaoui A., Tanner R., Taylor I., Taylor K., Taylor U., Water Naude Te R., Themistocleous A., Thomas M., Thomas T.M., Thompson A., Thompson K., Thornton-Jones V., Tinh L., Tomic A., Tonks S., Towner J., Tran N., Tree J.A., Truby A., Turner C., Turner R., Ulaszewska M., Varughese R., Verbart D., Verheul M.K., Vichos I., Walker L., Wand M.E., Watkins B., Welch J., West A.J., White C., White R., Williams P., Woodyer M., Worth A.T., Wright D., Wrin T., Yao X.L., Zbarcea D.-A., Zizi D. Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial. Lancet. 2020;396:1979–1993. doi: 10.1016/S0140-6736(20)32466-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappazzo C.G., Tse L.V., Kaku C.I., Wrapp D., Sakharkar M., Huang D., Deveau L.M., Yockachonis T.J., Herbert A.S., Battles M.B., O'Brien C.M., Brown M.E., Geoghegan J.C., Belk J., Peng L., Yang L., Hou Y., Scobey T.D., Burton D.R., Nemazee D., Dye J.M., Voss J.E., Gunn B.M., Mclellan J.S., Baric R.S., Gralinski L.E., Walker L.M. Broad and potent activity against SARS-like viruses by an engineered human monoclonal antibody. Sci. eabf4830. 2021 doi: 10.1126/science.abf4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M., Kok A., de Meulder D., Bestebroer T.M., Lamers M.M., Okba N.M.A., Fentener van Vlissingen M., Rockx B., Haagmans B.L., Koopmans M.P.G., Fouchier R.A.M., Herfst S. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020;11:3496. doi: 10.1038/s41467-020-17367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hägglöf T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.-H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Oude Munnink B.B., de Meulder D., van Amerongen G., van den Brand J., Okba N.M.A., Schipper D., van Run P., Leijten L., Sikkema R., Verschoor E., Verstrepen B., Bogers W., Langermans J., Drosten C., Fentener van Vlissingen M., Fouchier R., de Swart R., Koopmans M., Haagmans B.L. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.-T., Limbo O., Smith C., Song G., Woehl J., Yang L., Abbott R.K., Callaghan S., Garcia E., Hurtado J., Parren M., Peng L., Ramirez S., Ricketts J., Ricciardi M.J., Rawlings S.A., Wu N.C., Yuan M., Smith D.M., Nemazee D., Teijaro J.R., Voss J.E., Wilson I.A., Andrabi R., Briney B., Landais E., Sok D., Jardine J.G., Burton D.R. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar E., Christensen P.A., Graviss E.A., Nguyen D.T., Castillo B., Chen J., Lopez B.V., Eagar T.N., Yi X., Zhao P., Rogers J., Shehabeldin A., Joseph D., Leveque C., Olsen R.J., Bernard D.W., Gollihar J., Musser J.M. Treatment of coronavirus disease 2019 patients with convalescent plasma reveals a signal of significantly decreased mortality. Am. J. Pathol. 2020;190:2290–2303. doi: 10.1016/j.ajpath.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaecher S.R., Stabenow J., Oberle C., Schriewer J., Buller R.M., Sagartz J.E., Pekosz A. An immunosuppressed Syrian golden hamster model for SARS-CoV infection. Virology. 2008;380:312–321. doi: 10.1016/j.virol.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R., Baric R.S., Robbiani D.F., Hatziioannou T., Ravetch J.V., Bieniasz P.D., Bowen R.A., Nussenzweig M.C., Sheahan T.P. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2020;218 doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020;368:1016–1020. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.-S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shi Y., Wan Z., Li L., Li P., Li C., Ma Q., Cao C. Antibody responses against SARS-coronavirus and its nucleocaspid in SARS patients. J. Clin. Virol. 2004;31:66–68. doi: 10.1016/j.jcv.2004.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonovich V.A., Burgos Pratx L.D., Scibona P., Beruto M.V., Vallone M.G., Vázquez C., Savoy N., Giunta D.H., Pérez L.G., Sánchez M.D.L., Gamarnik A.V., Ojeda D.S., Santoro D.M., Camino P.J., Antelo S., Rainero K., Vidiella G.P., Miyazaki E.A., Cornistein W., Trabadelo O.A., Ross F.M., Spotti M., Funtowicz G., Scordo W.E., Losso M.H., Ferniot I., Pardo P.E., Rodriguez E., Rucci P., Pasquali J., Fuentes N.A., Esperatti M., Speroni G.A., Nannini E.C., Matteaccio A., Michelangelo H.G., Follmann D., Lane H.C., Belloso W.H. A randomized trial of convalescent plasma in covid-19 severe pneumonia. N. Engl. J. Med. NEJMoa2031304. 2020 doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. 2020. Prospective Mapping of Viral Mutations that Escape Antibodies Used to Treat COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlin D., Mathian A., Miyara M., Mohr A., Anna F., Claër L., Quentric P., Fadlallah J., Devilliers H., Ghillani P., Gunn C., Hockett R., Mudumba S., Guihot A., Luyt C.-E., Mayaux J., Beurton A., Fourati S., Bruel T., Schwartz O., Lacorte J.-M., Yssel H., Parizot C., Dorgham K., Charneau P., Amoura Z., Gorochov G. IgA dominates the early neutralizing antibody response to SARS-CoV-2. Sci. Transl. Med. eabd2223. 2021;13 doi: 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., Gao G.F. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24:490–502. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.-Y., Liu D., Huang J., He J., Zhu A., Zhao J., Li X., Xi Y., Chen R., Alshukairi A.N., Chen Z., Zhang Z., Chen C., Huang X., Li F., Lai X., Chen D., Wen L., Zhuo J., Zhang Y., Wang Y., Huang S., Dai J., Shi Y., Zheng K., Leidinger M.R., Chen J., Li Y., Zhong N., Meyerholz D.K., McCray P.B., Jr., Perlman S., Zhao J. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743. doi: 10.1016/j.cell.2020.06.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., Vanblargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C., Fouch M.E., Davidson E., Doranz B.J., Carnahan R.H., Thackray L.B., Diamond M.S., Crowe J.E. 2021. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suschak J.J., Williams J.A., Schmaljohn C.S. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum. Vaccines Immunother. 2017;13:2837–2848. doi: 10.1080/21645515.2017.1330236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon J., Salvatore M., Neill R.E., Nakaya Y., Zheng H., Muster T., García-Sastre A., Palese P. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4309. doi: 10.1073/pnas.070525997. O 039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theiler M., Smith H.H. The effect OF prolonged cultivation IN vitro UPON the pathogenicity OF yellow fever virus. J. Exp. Med. 1937;65:767–786. doi: 10.1084/jem.65.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S., Zatta F., De Marco A., Guarino B., Bianchi S., Lauron E.J., Tucker H., Zhou J., Peter A., Havenar-Daughton C., Wojcechowskyj J.A., Case J.B., Chen R.E., Kaiser H., Montiel-Ruiz M., Meury M., Czudnochowski N., Spreafico R., Dillen J., Ng C., Sprugasci N., Culap K., Benigni F., Abdelnabi R., Foo S.-Y.C., Schmid M.A., Cameroni E., Riva A., Gabrieli A., Galli M., Pizzuto M.S., Neyts J., Diamond M.S., Virgin H.W., Snell G., Corti D., Fink K., Veesler D. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng C.-T.K., Huang C., Newman P., Wang N., Narayanan K., Watts D.M., Makino S., Packard M.M., Zaki S.R., Chan T.-S., Peters C.J. Severe acute respiratory syndrome coronavirus infection of mice transgenic for the human angiotensin-converting enzyme 2 virus receptor. J. Virol. 2007;81:1162–1173. doi: 10.1128/JVI.01702-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., Avanzato V.A., Bushmaker T., Flaxman A., Ulaszewska M., Feldmann F., Allen E.R., Sharpe H., Schulz J., Holbrook M., Okumura A., Meade-White K., Pérez-Pérez L., Edwards N.J., Wright D., Bissett C., Gilbride C., Williamson B.N., Rosenke R., Long D., Ishwarbhai A., Kailath R., Rose L., Morris S., Powers C., Lovaglio J., Hanley P.W., Scott D., Saturday G., de Wit E., Gilbert S.C., Munster V.J. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellozzi C., Burwen D.R., Dobardzic A., Ball R., Walton K., Haber P. Safety of trivalent inactivated influenza vaccines in adults: background for pandemic influenza vaccine safety monitoring. Vaccine. 2009;27:2114–2120. doi: 10.1016/j.vaccine.2009.01.125. [DOI] [PubMed] [Google Scholar]
- Vogt M.R., Dowd K.A., Engle M., Tesh R.B., Johnson S., Pierson T.C., Diamond M.S. Poorly Neutralizing Cross-Reactive Antibodies against the Fusion Loop of West Nile Virus Envelope Protein Protect <em>In Vivo</em> via Fcγ Receptor and Complement-Dependent Effector Mechanisms. J. Virol. 2011;85:11567. doi: 10.1128/JVI.05859-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Bibi S., Briner C., Cicconi P., Collins A.M., Colin-Jones R., Cutland C.L., Darton T.C., Dheda K., Duncan C.J.A., Emary K.R.W., Ewer K.J., Fairlie L., Faust S.N., Ferreira D.M., Finn A., Goodman A.L., Green C.M., Green C.A., Heath P.T., Hill C., Hill H., Hirsch I., Hodgson S.H.C., Izu A., Jackson S., Jenkin D., Joe C.C.D., Kerridge S., Koen A., Kwatra G., Lazarus R., Lawrie A.M., Lelliott A., Libri V., Lillie P.J., Mallory R., Mendes A.V.A., Milan E.P., Minassian A.M., McGregor A., Morrison H., Mujadidi Y.F., Nana A., O'Reilly P.J., Padayachee S.D., Pittella A., Plested E., Pollock K.M., Ramasamy M.N., Rhead S., Schwarzbold A.V., Singh N., Smith A., Song R., Snape M.D., Sprinz E., Sutherland R.K., Tarrant R., Thomson E.C., Török M.E., Toshner M., Turner D.P.J., Vekemans J., Villafana T.L., Watson M.E.E., Williams C.J., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aban M., Abayomi F., Abeyskera K., Aboagye J., Adam M., Adams K., Adamson J., Adelaja Y.A., Adlou S., Ahmed K., Akhalwaya Y., Akhalwaya S., Alcock A., Ali A., Allen E.R., Allen L., Almeida T.C.D.S.C., Alves M.P.S., Amorim F., Andritsou F., Anslow R., Appleby M., Arbe-Barnes E.H., Ariaans M.P., Arns B., Arruda L., Awedetan G., Azi P., Azi L., Babbage G., Bailey C., Baker K.F., Baker M., Baker N., Baker P., Baldwin L., Baleanu I., Bandeira D., Bara A., Barbosa M.A.S., Barker D., Barlow G.D., Barnes E., Barr A.S., Barrett J.R., Barrett J., Bates L., Batten A., Beadon K., Beales E., Beckley R., Belij-Rammerstorfer S., Bell J., Bellamy D., Bellei N., Belton S., Berg A., Bermejo L., Berrie E., Berry L., Berzenyi D., Beveridge A., Bewley K.R., Bexhell H., Bhikha S., Bhorat A.E., Bhorat Z.E., Bijker E., Birch G., Birch S., Bird A., Bird O., Bisnauthsing K., Bittaye M., Blackstone K., Blackwell L., Bletchly H., Blundell C.L., Blundell S.R., Bodalia P., Boettger B.C., Bolam E., Boland E., Bormans D., Borthwick N., Bowring F., Boyd A., Bradley P., Brenner T., Brown P., Brown C., Brown-O'Sullivan C., Bruce S., Brunt E., Buchan R., Budd W., Bulbulia Y.A., Bull M., Burbage J., Burhan H., Burn A., Buttigieg K.R., Byard N., Cabera Puig I., Calderon G., Calvert A., Camara S., Cao M., Cappuccini F., Cardoso J.R., Carr M., Carroll M.W., Carson-Stevens A., Carvalho Y. de M., Carvalho J.A.M., Casey H.R., Cashen P., Castro T., Castro L.C., Cathie K., Cavey A., Cerbino-Neto J., Chadwick J., Chapman D., Charlton S., Chelysheva I., Chester O., Chita S., Cho J.-S., Cifuentes L., Clark E., Clark M., Clarke A., Clutterbuck E.A., Collins S.L.K., Conlon C.P., Connarty S., Coombes N., Cooper C., Cooper R., Cornelissen L., Corrah T., Cosgrove C., Cox T., Crocker W.E.M., Crosbie S., Cullen L., Cullen D., Cunha D.R.M.F., Cunningham C., Cuthbertson F.C., Da Guarda S.N.F., da Silva L.P., Damratoski B.E., Danos Z., Dantas M.T.D.C., Darroch P., Datoo M.S., Datta C., Davids M., Davies S.L., Davies H., Davis E., Davis J., Davis J., De Nobrega M.M.D., De Oliveira Kalid L.M., Dearlove D., Demissie T., Desai A., Di Marco S., Di Maso C., Dinelli M.I.S., Dinesh T., Docksey C., Dold C., Dong T., Donnellan F.R., Santos Dos T., Santos dos T.G., Santos Dos E.P., Douglas N., Downing C., Drake J., Drake-Brockman R., Driver K., Drury R., Dunachie S.J., Durham B.S., Dutra L., Easom N.J.W., van Eck S., Edwards M., Edwards N.J., Muhanna El O.M., Elias S.C., Elmore M., English M., Esmail A., Essack Y.M., Farmer E., Farooq M., Farrar M., Farrugia L., Faulkner B., Fedosyuk S., Felle S., Feng S., Ferreira Da Silva C., Field S., Fisher R., Flaxman A., Fletcher J., Fofie H., Fok H., Ford K.J., Fowler J., Fraiman P.H.A., Francis E., Franco M.M., Frater J., Freire M.S.M., Fry S.H., Fudge S., Furze J., Fuskova M., Galian-Rubio P., Galiza E., Garlant H., Gavrila M., Geddes A., Gibbons K.A., Gilbride C., Gill H., Glynn S., Godwin K., Gokani K., Goldoni U.C., Goncalves M., Gonzalez I.G.S., Goodwin J., Goondiwala A., Gordon-Quayle K., Gorini G., Grab J., Gracie L., Greenland M., Greenwood N., Greffrath J., Groenewald M.M., Grossi L., Gupta G., Hackett M., Hallis B., Hamaluba M., Hamilton E., Hammersley D., Hanrath A.T., Hanumunthadu B., Harris S.A., Harris C., Harris T., Harrison T.D., Harrison D., Hart T.C., Hartnell B., Hassan S., Haughney J., Hawkins S., Hay J., Head I., Henry J., Hermosin Herrera M., Hettle D.B., Hill J., Hodges G., Horne E., Hou M.M., Houlihan C., Howe E., Howell N., Humphreys J., Humphries H.E., Hurley K., Huson C., Hyder-Wright A., Hyamns C., Ikram S., Ishwarbhai A., Ivan M., Iveson P., Iyer V., Jackson F., De Jager J., Jaumdally S., Jeffers H., Jesudason N., Jones B., Jones K., Jones E., Jones C., Jorge M.R., Jose A., Joshi A., Júnior E.A.M.S., Kadziola J., Kailath R., Kana F., Karampatsas K., Kasanyinga M., Keen J., Kelly E.J., Kelly D.M., Kelly D., Kelly S., Kerr D., Kfouri R. de Á., Khan L., Khozoee B., Kidd S., Killen A., Kinch J., Kinch P., King L.D.W., King T.B., Kingham L., Klenerman P., Knapper F., Knight J.C., Knott D., Koleva S., Lang M., Lang G., Larkworthy C.W., Larwood J.P.J., Law R., Lazarus E.M., Leach A., Lees E.A., Lemm N.-M., Lessa A., Leung S., Li Y., Lias A.M., Liatsikos K., Linder A., Lipworth S., Liu S., Liu X., Lloyd A., Lloyd S., Loew L., Lopez Ramon R., Lora L., Lowthorpe V., Luz K., MacDonald J.C., MacGregor G., Madhavan M., Mainwaring D.O., Makambwa E., Makinson R., Malahleha M., Malamatsho R., Mallett G., Mansatta K., Maoko T., Mapetla K., Marchevsky N.G., Marinou S., Marlow E., Marques G.N., Marriott P., Marshall R.P., Marshall J.L., Martins F.J., Masenya M., Masilela M., Masters S.K., Mathew M., Matlebjane H., Matshidiso K., Mazur O., Mazzella A., McCaughan H., McEwan J., McGlashan J., McInroy L., McIntyre Z., McLenaghan D., McRobert N., McSwiggan S., Megson C., Mehdipour S., Meijs W., Mendonça R.N.Á., Mentzer A.J., Mirtorabi N., Mitton C., Mnyakeni S., Moghaddas F., Molapo K., Moloi M., Moore M., Moraes-Pinto M.I., Moran M., Morey E., Morgans R., Morris S., Morris S., Morris H.C., Morselli F., Morshead G., Morter R., Mottal L., Moultrie A., Moya N., Mpelembue M., Msomi S., Mugodi Y., Mukhopadhyay E., Muller J., Munro A., Munro C., Murphy S., Mweu P., Myasaki C.H., Naik G., Naker K., Nastouli E., Nazir A., Ndlovu B., Neffa F., Njenga C., Noal H., Noé A., Novaes G., Nugent F.L., Nunes G., O'Brien K., O'Connor D., Odam M., Oelofse S., Oguti B., Olchawski V., Oldfield N.J., Oliveira M.G., Oliveira C., Oosthuizen A., O'Reilly P., Osborne P., Owen D.R.J., Owen L., Owens D., Owino N., Pacurar M., Paiva B.V.B., Palhares E.M.F., Palmer S., Parkinson S., Parracho H.M.R.T., Parsons K., Patel D., Patel B., Patel F., Patel K., Patrick-Smith M., Payne R.O., Peng Y., Penn E.J., Pennington A., Peralta Alvarez M.P., Perring J., Perry N., Perumal R., Petkar S., Philip T., Phillips D.J., Phillips J., Phohu M.K., Pickup L., Pieterse S., Piper J., Pipini D., Plank M., Plessis Du J., Pollard S., Pooley J., Pooran A., Poulton I., Powers C., Presa F.B., Price D.A., Price V., Primeira M., Proud P.C., Provstgaard-Morys S., Pueschel S., Pulido D., Quaid S., Rabara R., Radford A., Radia K., Rajapaska D., Rajeswaran T., Ramos A.S.F., Ramos Lopez F., Rampling T., Rand J., Ratcliffe H., Rawlinson T., Rea D., Rees B., Reiné J., Resuello-Dauti M., Reyes Pabon E., Ribiero C.M., Ricamara M., Richter A., Ritchie N., Ritchie A.J., Robbins A.J., Roberts H., Robinson R.E., Robinson H., Rocchetti T.T., Rocha B.P., Roche S., Rollier C., Rose L., Ross-Russell A.L., Rossouw L., Royal S., Rudiansyah I., Ruiz S., Saich S., Sala C., Sale J., Salman A.M., Salvador N., Salvador S., Sampaio M., Samson A.D., Sanchez-Gonzalez A., Sanders H., Sanders K., Santos E., Santos Guerra M.F.S., Satti I., Saunders J.E., Saunders C., Sayed A., Schim van der Loeff I., Schmid A.B., Schofield E., Screaton G., Seddiqi S., Segireddy R.R., Senger R., Serrano S., Shah R., Shaik I., Sharpe H.E., Sharrocks K., Shaw R., Shea A., Shepherd A., Shepherd J.G., Shiham F., Sidhom E., Silk S.E., da Silva Moraes A.C., Silva-Junior G., Silva-Reyes L., Silveira A.D., Silveira M.B.V., Sinha J., Skelly D.T., Smith D.C., Smith N., Smith H.E., Smith D.J., Smith C.C., Soares A., Soares T., Solórzano C., Sorio G.L., Sorley K., Sosa-Rodriguez T., Souza C.M.C.D.L., Souza B.S.D.F., Souza A.R., Spencer A.J., Spina F., Spoors L., Stafford L., Stamford I., Starinskij I., Stein R., Steven J., Stockdale L., Stockwell L.V., Strickland L.H., Stuart A.C., Sturdy A., Sutton N., Szigeti A., Tahiri-Alaoui A., Tanner R., Taoushanis C., Tarr A.W., Taylor K., Taylor U., Taylor I.J., Taylor J., Water Naude Te R., Themistocleous Y., Themistocleous A., Thomas M., Thomas K., Thomas T.M., Thombrayil A., Thompson F., Thompson A., Thompson K., Thompson A., Thomson J., Thornton-Jones V., Tighe P.J., Tinoco L.A., Tiongson G., Tladinyane B., Tomasicchio M., Tomic A., Tonks S., Tran N., Tree J., Trillana G., Trinham C., Trivett R., Truby A., Tsheko B.L., Turabi A., Turner R., Turner C., Ulaszewska M., Underwood B.R., Varughese R., Verbart D., Verheul M., Vichos I., Vieira T., Waddington C.S., Walker L., Wallis E., Wand M., Warbick D., Wardell T., Warimwe G., Warren S.C., Watkins B., Watson E., Webb S., Webb-Bridges A., Webster A., Welch J., Wells J., West A., White C., White R., Williams P., Williams R.L., Winslow R., Woodyer M., Worth A.T., Wright D., Wroblewska M., Yao A., Zimmer R., Zizi D., Zuidewind P. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2020 doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.-Y., Türeci Ö., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Şahin U., Gruber W.C. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N. Engl. J. Med. NEJMoa2027906. 2020;383:2439–2450. doi: 10.1056/nejmoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F., Gallagher T. Receptor recognition by the novel coronavirus from wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:1986. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., Xu W., Zhao Y., Li N., Zhang J., Liang H., Bao L., Xu Y., Ding L., Zhou W., Gao H., Liu J., Niu P., Zhao L., Zhen W., Fu H., Yu S., Zhang Z., Xu G., Li C., Lou Z., Xu M., Qin C., Wu G., Gao G.F., Tan W., Yang X. Development of an inactivated vaccine candidate, BBIBP-CorV, with potent protection against SARS-CoV-2. Cell. 2020;182:713–721. doi: 10.1016/j.cell.2020.06.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Viant C., Gaebler C., Cipolla M., Hoffmann H.-H., Oliveira T.Y., Oren D.A., Ramos V., Nogueira L., Michailidis E., Robbiani D.F., Gazumyan A., Rice C.M., Hatziioannou T., Bieniasz P.D., Caskey M., Nussenzweig M.C. Science Translational Medicine 13; 2021. Enhanced SARS-CoV-2 neutralization by dimeric IgA. eabf1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Nair M.S., Huang Y., Ho D.D. 2021. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., Oliveira T.Y., Yang Z., Abernathy M.E., Huey-Tubman K.E., Hurley A., Turroja M., West K.A., Gordon K., Millard K.G., Ramos V., Da Silva J., Xu J., Colbert R.A., Patel R., Dizon J., Unson-O’Brien C., Shimeliovich I., Gazumyan A., Caskey M., Bjorkman P.J., Casellas R., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C. 2021. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., Sun J., Gong J., Yang X., Wang Y., Wang X., Li J., Yang H., Li H., Zhang Z., Wang R., Du P., Zong Y., Yin F., Zhang W., Wang N., Peng Y., Lin H., Feng J., Qin C., Chen W., Gao Q., Zhang R., Cao Y., Zhong H. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metabol. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.-H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO coronavirus disease (COVID-19) dashboard. 2020. http://scholar.google.comjavascript:void(0 [WWW Document]. httpcovid.who.int. URL. accessed 10.24.20a.
- WHO . 2020. DRAFT Landscape of COVID-19.https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines candidate vaccines [WWW Document]. URL. accessed 12.8.20b. [Google Scholar]
- WHO Immunization. 2019. https://www.who.int/news-room/facts-in-pictures/detail/immunization#:~:text=Immunization%20prevents%20deaths%20every%20year,cost%2Deffective%20public%20health%20interventions [WWW Document]. URL. accessed 11.12.20.
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Lambson B.E., Vermeulen M., Van Den Berg K., Rossouw T., Boswell M., Ueckermann V., Meiring S., Von Gottberg A., Cohen C., Morris L., Bhiman J.N., Moore P.L. 2021. SARS-CoV-2 501Y.V2 Escapes Neutralization by South African COVID-19 Donor Plasma. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P., Ritter J.H., Kang L.-I., Dort S., Robichaud A., Head R., Holtzman M.J., Diamond M.S. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;181:1–9. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel A.G., Benton D.J., Hussain S., Harvey R., Martin S.R., Roustan C., Rosenthal P.B., Skehel J.J., Gamblin S.J. Antibody-mediated disruption of the SARS-CoV-2 spike glycoprotein. Nat. Commun. 2020;11:5337. doi: 10.1038/s41467-020-19146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J., Sheng J., Quan L., Xia Z., Tan W., Cheng G., Jiang T. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27:325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Liu M., Wang A., Lu L., Wang Q., Gu C., Chen J., Wu Y., Xia S., Ling Y., Zhang Y., Xun J., Zhang R., Xie Y., Jiang S., Zhu T., Lu H., Wen Y., Huang J. Evaluating the association of clinical characteristics with neutralizing antibody levels in patients who have recovered from mild COVID-19 in shanghai, China. JAMA Intern. Med. 2020;180:1356–1362. doi: 10.1001/jamainternmed.2020.4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.-C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., Van Gils M.J., Sanders R.W., Ward A.B., Wilson I.A. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Reports. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., Gong Y., Xiao H., Fan Z., Tan S., Wu G., Tan W., Lu X., Fan C., Wang Q., Liu Y., Zhang C., Qi J., Gao G.F., Gao F., Liu L. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Duan K., Zhang Y., Zhao D., Zhang H., Xie Z., Li X., Peng C., Zhang Y., Zhang W., Yang Y., Chen W., Gao X., You W., Wang X., Wang Z., Shi Z., Wang Y., Yang X., Zhang L., Huang L., Wang Q., Lu J., Yang Y., Guo J., Zhou W., Wan X., Wu C., Wang W., Huang S., Du J., Meng Z., Pan A., Yuan Z., Shen S., Guo W., Yang X. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. J. Am. Med. Assoc. 2020;324:951–960. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Zhang Y., Wang Y., Wang H., Yang Y., Gao G.F., Tan W., Wu G., Xu M., Lou Z., Huang W., Xu W., Huang B., Wang H., Wang W., Zhang W., Li N., Xie Z., Ding L., You W., Zhao Y., Yang X., Liu Y., Wang Q., Huang L., Yang Y., Xu G., Luo B., Wang W., Liu P., Guo W., Yang X. Safety and immunogenicity of an inactivated SARS-CoV-2 vaccine, BBIBP-CorV: a randomised, double-blind, placebo-controlled, phase 1/2 trial. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30831-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Liu Y., Liu J., Zhang X., Zou J., Fontes-Garfias C.R., Xia H., Swanson K.A., Cutler M., Cooper D., Menachery V.D., Weaver S., Dormitzer P.R., Shi P.-Y. 2021. Neutralization of SARS-CoV-2 Spike 69/70 Deletion, E484K, and N501Y Variants by BNT162b2 Vaccine-Elicited Sera. [DOI] [PubMed] [Google Scholar]
- Yong C.Y., Ong H.K., Yeap S.K., Ho K.L., Tan W.S. Recent advances in the vaccine development against Middle East respiratory syndrome-coronavirus. Front. Microbiol. 2019;10:1999. doi: 10.3389/fmicb.2019.01781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.-C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., Rogers T.F., Landais E., Sok D., Jardine J.G., Burton D.R., Wilson I.A. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.-C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zeng G., Pan H., Li C., Hu Y., Chu K., Han W., Chen Z., Tang R., Yin W., Chen X., Hu Y., Liu X., Jiang C., Li J., Yang M., Song Y., Wang X., Gao Q., Zhu F. Safety, tolerability, and immunogenicity of an inactivated SARS-CoV-2 vaccine in healthy adults aged 18–59 years: a randomised, double-blind, placebo-controlled, phase 1/2 clinical trial. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30843-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Li K., Wohlford-Lenane C., Agnihothram S.S., Fett C., Zhao J., Gale M.J., Baric R.S., Enjuanes L., Gallagher T., McCray P.B., Perlman S. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., Brindley M.A., Lewis N.E., Tiemeyer M., Chen B., Woods R.J., Wells L. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601. doi: 10.1016/j.chom.2020.08.004. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu Ben, Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., Li J.-X., Yang B.-F., Wang L., Wang W.-J., Wu S.-P., Wang Z., Wu X.-H., Xu J.-J., Zhang Z., Jia S.-Y., Wang B.-S., Hu Y., Liu J.-J., Zhang J., Qian X.-A., Li Q., Pan H.-X., Jiang H.-D., Deng P., Gou J.-B., Wang X.-W., Wang X.-H., Chen W. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396:479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., Wu S.-P., Wang B.-S., Wang Z., Wang L., Jia S.-Y., Jiang H.-D., Wang L., Jiang T., Hu Y., Gou J.-B., Xu S.-B., Xu J.-J., Wang X.-W., Wang W., Chen W. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Martinez D.R., Williamson L.E., Chen E.C., Jones T., Day S., Myers L., Hassan A.O., Kafai N.M., Winkler E.S., Fox J.M., Shrihari S., Mueller B.K., Meiler J., Chandrashekar A., Mercado N.B., Steinhardt J.J., Ren K., Loo Y.-M., Kallewaard N.L., McCune B.T., Keeler S.P., Holtzman M.J., Barouch D.H., Gralinski L.E., Baric R.S., Thackray L.B., Diamond M.S., Carnahan R.H., Crowe J.E. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]