Escherichia coli strain FEX669 was isolated from retail ground chicken and shown to contain the extraintestinal pathogenic E. coli (ExPEC) virulence genes sfaD/focC and iutA. Because this presumptive ExPEC strain was isolated from a retail food item and it was a weak biofilm former, it was characterized using whole-genome sequencing using the PacBio RS II platform.
ABSTRACT
Escherichia coli strain FEX669 was isolated from retail ground chicken and shown to contain the extraintestinal pathogenic E. coli (ExPEC) virulence genes sfaD, focC, and iutA. Because this presumptive ExPEC strain was isolated from a retail food item and it was a weak biofilm former, it was characterized using whole-genome sequencing using the PacBio RS II platform. Genomic analysis showed that the FEX669 chromosome is 4,973,943 bp long, with a GC content of 50.47%, and is accompanied by a ColV plasmid that is 237,102 bp long, with a GC content of 50.49%.
ANNOUNCEMENT
Foodborne extraintestinal pathogenic Escherichia coli (ExPEC) represents an underappreciated public health threat. Millions of ExPEC infections occur annually in the United States, producing diseases ranging from urinary tract infections (UTIs) to acute sepsis (1). It is well established that ExPEC strains colonize livestock and contaminate food products. ExPEC strains isolated from food and human patient samples were screened for biofilm production via a microtiter plate assay (2). Here, the E. coli strain FEX669, an isolate with scant (8 times less than the positive control) biofilm production, was selected for whole-genome sequencing (Fig. 1). This strain was originally isolated from ground chicken in Minneapolis, Minnesota, in May 2002 through the survey of 1,648 diverse food items from 10 retail markets. In addition, its ExPEC-associated features were identified through PCR-based assays and O serotyping (3).
FIG 1.
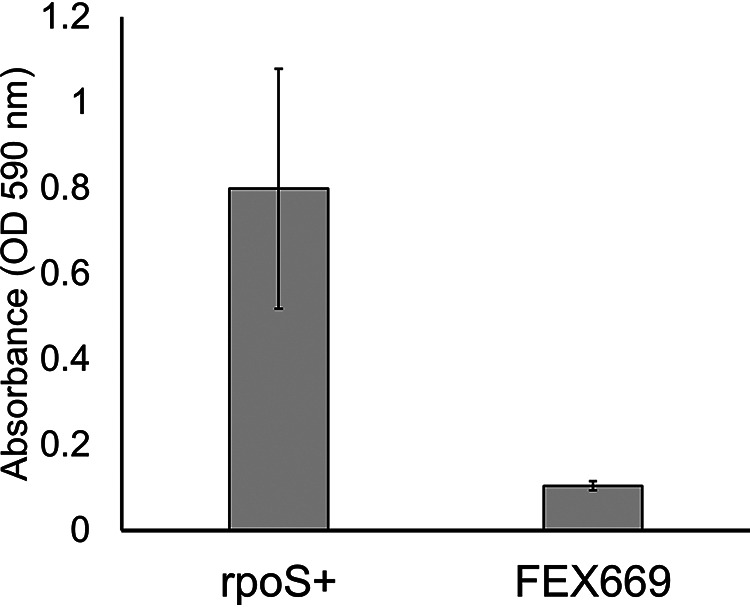
Biofilm production by Escherichia coli strain FEX669. Strain 43894OR (OR), a constitutive rpoS-producing (rpoS+) strain, was used as a positive control for biofilm (10). Biofilms were stained with crystal violet as previously described (2). Bars represent the mean crystal violet absorbance measured at an optical density at 590 nm (OD590) ± standard deviation (SD) of three independent samples.
Genomic DNA (gDNA) was isolated from overnight cultures grown in LB broth at 37°C, with shaking at 180 rpm, by using the Genomic-tip 500/G kit (Qiagen, Valencia, CA). The gDNA concentration was determined using the Qubit 2.0 instrument (Invitrogen, Carlsbad, CA), was sheared with a g-TUBE (Covaris, Inc., Woburn, MA), and underwent size selection for 20-kb fragments using the BluePippin kit (Sage Science, Inc., Beverly, MA). The sequencing library was prepared using the SMRTbell library preparation kit (Pacific Biosciences, Menlo Park, CA) and was sequenced on an RS II machine with P6C4 chemistry at the University of Delaware Sequencing Center (Newark, DE). The sequencing generated 150,292 reads, with 1,519,409,874 bases. For all software analyses, default parameters were used except where otherwise noted. SMRT Portal v2.3.0 of SMRT Link v7.0 was used for quality control and filtering, which resulted in 75,495 reads, 1,316,564,288 bases, a read N50 value of 24,304 bases, and a mean read length of 17,439 bases. The postfiltered reads were used for genome de novo assembly using Hierarchical Genome Assembly version 3 (HGAP3) (Pacific Biosciences). The genome was polished using Quiver software (4). Benchmarking universal single-copy orthologues (BUSCO) and blast analysis were used to assess the quality and completeness of the assembled genome. The final assembly included one circular, 4,973,943-bp chromosome and one circular, 237,102-bp plasmid.
We annotated the genome using NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (5) and identified 5,013 genes in the genome and 117 RNA sequences, including 85 tRNA genes. PHASTER (6) identified four phages on the chromosome and one on the plasmid. The SerotypeFinder-2.0 (7) and MLST-2.0 databases from the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/) assigned FEX669 to serotype O150:H6, sequence type 6570 (ST6570), and phylogroup B2. ResFinder-3.2 (8) found tetracycline and ampicillin resistance genes on the plasmid and no resistance genes on the chromosome.
A total of 27 virulence genes were identified on the FEX669 chromosome and plasmid using VirulenceFinder-2.0 software (with 90% threshold and 60% minimum length parameters) (9) (accessed 2 November 2020). They included genes for iron acquisition (chuA, fyuA, iroN, sitA, iucC, irp2, and iutA), attachment (hra, focC, sfaD, and yfcV), and toxin production (hlyF, vat, and usp). According to BLAST analysis, the plasmid was highly similar (83% coverage; 99.90% identity) to a ColV plasmid from E. coli strain PU-1, a blood isolate from a piglet with acute sepsis (GenBank accession number CP042245.1).
The food product origin of strain FEX669 makes its possession of virulence genes that could facilitate extraintestinal and even systemic infection a cause for concern. Genomic sequencing and characterization of potentially pathogenic E. coli are important tools for identifying hazards in the food supply and preventing human illness outbreaks.
Data availability.
This whole-genome project has been deposited under BioProject number PRJNA642167, Sequence Read Archive (SRA) number SRP293948, and BioSample number SAMN15822731 with the DDBJ/ENA/GenBank accession numbers CP065152.1 for the FEX669 chromosome and CP065153 for the plasmid.
ACKNOWLEDGMENTS
We thank Amy Ream for performing biofilm assays and Pina Fratamico for critical reading of the manuscript.
This research was supported in part by an appointment of J.R.E. to the Agricultural Research Service (ARS) Research Participation Program, as administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the U.S. Department of Energy (DOE) and the U.S. Department of Agriculture (USDA). ORISE is managed by Oak Ridge Associated Universities (ORAU) under DOE contract number DE-AC05-06OR23100. This work was also supported in part by Office of Research and Development, Department of Veterans Affairs (J.R.J.). This research used resources provided by the SciNet project of the USDA Agricultural Research Service, ARS project number 0500-00093-001-00-D.
Mention of trade names or commercial products is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
REFERENCES
- 1.Russo TA, Johnson JR. 2003. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes Infect 5:449–456. doi: 10.1016/S1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 2.Uhlich GA, Chen CY, Cottrell BJ, Hofmann CS, Dudley EG, Strobaugh TP, Nguyen L-H. 2013. Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157: H7. Microbiology (Reading) 159:1586–1596. doi: 10.1099/mic.0.066118-0. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JR, Kuskowski MA, Smith K, O’Bryan TT, Tatini S. 2005. Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J Infect Dis 191:1040–1049. doi: 10.1086/428451. [DOI] [PubMed] [Google Scholar]
- 4.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 5.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI Prokaryotic Genome Annotation Pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. 2015. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol 53:2410–2426. doi: 10.1128/JCM.00008-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tetzschner AMM, Johnson JR, Johnson BD, Lund O, Scheutz F. 2020. In silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by use of whole-genome sequencing data. J Clin Microbiol 58:e0126920. doi: 10.1128/JCM.01269-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uhlich GA, Keen JE, Elder RO. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl Environ Microbiol 67:2367–2370. doi: 10.1128/AEM.67.5.2367-2370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This whole-genome project has been deposited under BioProject number PRJNA642167, Sequence Read Archive (SRA) number SRP293948, and BioSample number SAMN15822731 with the DDBJ/ENA/GenBank accession numbers CP065152.1 for the FEX669 chromosome and CP065153 for the plasmid.


