Abstract
Cryptosporidiumfelis is an important cause of feline and human cryptosporidiosis. However, the transmission of this pathogen between humans and cats remains controversial, partially due to a lack of genetic characterization of isolates from cats. The present study was conducted to examine the genetic diversity of C. felis in cats in China and to assess their potential zoonotic transmission. A newly developed subtyping tool based on a sequence analysis of the 60-kDa glycoprotein (gp60) gene was employed to identify the subtypes of 30 cat-derived C. felis isolates from Guangdong and Shanghai. Altogether, 20 C. felis isolates were successfully subtyped. The results of the sequence alignment showed a high genetic diversity, with 13 novel subtypes and 2 known subtypes of the XIXa subtype family being identified. The known subtypes were previously detected in humans, while some of the subtypes formed well-supported subclusters with human-derived subtypes from other countries in a phylogenetic analysis of the gp60 sequences. The results of this study confirmed the high genetic diversity of the XIXa subtype family of C. felis. The common occurrence of this subtype family in both humans and cats suggests that there could be cross-species transmission of C. felis.
Keywords: Cryptosporidium felis, 60-kDa glycoprotein, subtypes, zoonotic transmission
1. Introduction
Cryptosporidium spp. are important apicomplexan parasites inhabiting the gastrointestinal tract of humans and other vertebrates, causing severe diarrhea [1]. Human cryptosporidiosis has been associated with over 20 Cryptosporidium species, but C. hominis, C. parvum, C. meleagridis, C. felis, and C. canis are the most common ones [2]. Among them, C. felis mainly infects cats and is therefore considered a host-adapted species [3]. Human C. felis infections, however, are common in developing countries [4,5,6,7], and at least one possible zoonotic transmission of C. felis between a household cat and the owner has been reported [8]. Nevertheless, the limited number of reports of zoonotic infections with this species has raised questions on the importance of zoonotic transmission in the epidemiology of human C. felis infections [9].
Subtyping tools based on sequence analysis of the 60-kDa glycoprotein (gp60) gene have been developed for human-pathogenic Cryptosporidium spp. to track infection sources [10]. Currently, the gp60-based subtyping tools are available for C. hominis, C. parvum, C. meleagridis, C. ubiquitum, C. viatorum, Cryptosporidium skunk genotype, and Cryptosporidium chipmunk genotype I [11,12,13,14,15,16]. These subtyping methods have been used in characterizing the transmission of these Cryptosporidium spp. in humans and animals [3].
A gp60 subtyping tool has been developed recently for genetic characterizations of C. felis [17]. Thus far, nearly 200 C. felis isolates have been examined, which has led to the identification of approximately 100 subtypes in five subtype families (XIXa, XIXb, XIXc, XIXd, and XIXe) worldwide [17,18]. Most of the isolates, however, were from humans, and only two isolates from a human and a rhesus macaque have been characterized from China [18]. As C. felis has been identified in children and immunocompromised patients in China [19,20,21,22], we sought to examine the subtype identity of cat-derived C. felis isolates from Guangdong and Shanghai for assessment of their zoonotic potential.
2. Results
2.1. Amplification of the gp60 Gene
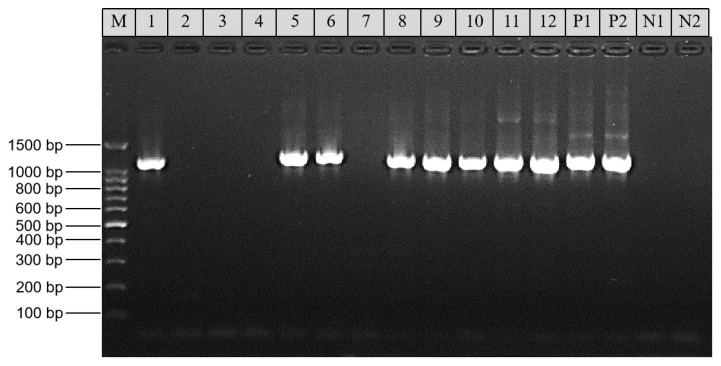
Among the 30 DNA preparations that were positive for C. felis based on nested PCR analysis of the small subunit (SSU) rRNA gene, 20 (66.7%) generated the expected products in the gp60 PCR. Among them, 9 were from cats in pet shelters, 5 were from cats visiting animal hospitals, 4 were from cats in pet shops, and 2 were from stray cats (Table 1). These PCR products differed slightly in size (Figure 1) and were all sequenced successfully.
Table 1.
Sources of Cryptosporidium felis samples used in the study and their gp60 subtype identity.
| Sample ID | Host | Region | Sample Source | Subtype |
|---|---|---|---|---|
| SCAU320 | Cat | Guangzhou | Animal hospital | XIXa-90 |
| SCAU1149 | Cat | Guangzhou | Animal hospital | XIXa-89 |
| SCAU1850 | Cat | Guangzhou | Animal hospital | XIXa-87 |
| SCAU1851 | Cat | Guangzhou | Animal hospital | XIXa-87 |
| SCAU1857 | Cat | Guangzhou | Animal hospital | XIXa-88 |
| SCAU2396 | Cat | Shenzhen | Animal hospital | N/A |
| SCAU252 | Cat | Guangzhou | Pet shop | XIXa-81 |
| SCAU343 | Cat | Guangzhou | Pet shelter | XIXa-88 |
| SCAU356 | Cat | Guangzhou | Pet shelter | N/A |
| SCAU392 | Cat | Guangzhou | Pet shelter | XIXa-39 |
| SCAU405 | Cat | Guangzhou | Pet shelter | XIXa-89 |
| SCAU1309 | Cat | Shantou | Pet shelter | XIXa-86 |
| SCAU4854 | Cat | Guangzhou | Pet shelter | N/A |
| SCAU4905 | Cat | Guangzhou | Pet shelter | N/A |
| SCAU143 | Cat | Guangzhou | Stray animal | XIXa-84 |
| SCAU731 | Cat | Guangzhou | Stray animal | N/A |
| SCAU732 | Cat | Guangzhou | Stray animal | N/A |
| SCAU754 | Cat | Guangzhou | Stray animal | XIXa-40 |
| ECUST19937 | Cat | Shanghai | Pet shop | XIXa-81 |
| ECUST26245 | Cat | Shanghai | Pet shop | N/A |
| ECUST26246 | Cat | Shanghai | Pet shop | XIXa-82 |
| ECUST26248 | Cat | Shanghai | Pet shop | XIXa-81 |
| ECUST19997 | Cat | Shanghai | Pet shelter | N/A |
| ECUST20283 | Cat | Shanghai | Pet shelter | N/A |
| ECUST20286 | Cat | Shanghai | Pet shelter | XIXa-91 |
| ECUST20309 | Cat | Shanghai | Pet shelter | N/A |
| ECUST26244 | Cat | Shanghai | Pet shelter | XIXa-92 |
| ECUST26249 | Cat | Shanghai | Pet shelter | XIXa-93 |
| ECUST26250 | Cat | Shanghai | Pet shelter | XIXa-85 |
| ECUST26251 | Cat | Shanghai | Pet shelter | XIXa-83 |
N/A: PCR negative.
Figure 1.
Analysis of the 60-kDa glycoprotein (gp60) gene in Cryptosporidium felis by nested PCR: lanes 1 and 2, SCAU320; lanes 3 and 4, SCAU2396; lanes 5 and 6, SCAU1149; lanes 7 and 8, SCAU1850; lanes 9 and 10, SCAU1851; lanes 11 and 12, SCAU1857; lane M, 100-bp molecular marker; P1 and P2, positive control (C. felis DNA); and N1 and N2, negative control (reagent-grade water).
2.2. Nucleotide Sequence Variations in the gp60 Gene of C. felis
A comparison of the nucleotide sequences generated led to the identification of 15 sequence types. They differed from each other by both nucleotide substitutions and copy numbers of repetitive sequences. The sequence alignments showed the presence of numerous single nucleotide substitutions (SNPs) over the partial gp60 gene among the 20 C. felis isolates. Three types of simple tandem repeats were detected in the gp60 genes (Table 2). Among them, 2–5 copies of the 33-bp repeat sequence 5′-CCACCTAGTGGCGGTAGTGGCGTGTCCCCTGCT-3′ and 2–4 copies of the 39-bp repeat sequence 5′-CCACCTAGTGGCGGTAGTGGCGTGTCCCCTGCT-3′ were observed at nucleotides 460–577 and 778–911 of the sequence alignment, respectively. In both repeat types, the last copy had only half of the usual length. In addition, 5 copies of the trinucleotide repeat GTT were found in the gp60 sequences at nucleotides 1143-1154.
Table 2.
Tandem repeats in nucleotide sequences of the gp60 gene of Cryptosporidium felis.
| Sample ID a | GenBank Accession no. | Subtype | 33-bp Repeat (No.) at 460–577 bp | 39-bp Repeat (No.) at 778–911 bp | GGT Repeat (No.) at 1143–1154 |
|---|---|---|---|---|---|
| SCAU392 | MH240852 | XIXa-39 | R1 b (2) | R2 c (3) | 4 |
| SCAU754 | MH240853 | XIXa-40 | R1 (3) | R2 (3) | 4 |
| SCAU252 | MW351820 | XIXa-81 | R1 (2) | R2 (4) | 4 |
| ECUST19937 | MW351820 | XIXa-81 | R1 (2) | R2 (4) | 4 |
| ECUST26248 | MW351820 | XIXa-81 | R1 (2) | R2 (4) | 4 |
| ECUST26246 | MW351821 | XIXa-82 | - | R2 (2) | 4 |
| ECUST26251 | MW351822 | XIXa-83 | - | R2 (3) | 4 |
| SCAU143 | MW351823 | XIXa-84 | R1 (5) | R2 (3) | 4 |
| ECUST26250 | MW351824 | XIXa-85 | - | R2 (3) | 4 |
| SCAU1309 | MW351825 | XIXa-86 | - | R2 (3) | 4 |
| SCAU1850 | MW351826 | XIXa-87 | R1 (2) | R2 (2) | 4 |
| SCAU1851 | MW351826 | XIXa-87 | R1 (2) | R2 (2) | 4 |
| SCAU343 | MW351827 | XIXa-88 | R1 (2) | R2 (2) | 4 |
| SCAU1857 | MW351827 | XIXa-88 | R1 (2) | R2 (2) | 4 |
| SCAU405 | MW351828 | XIXa-89 | R1 (2) | R2 (3) | 4 |
| SCAU1149 | MW351828 | XIXa-89 | R1 (2) | R2 (3) | 4 |
| SCAU320 | MW351829 | XIXa-90 | R1 (2) | R2 (4) | 4 |
| ECUST20286 | MW351830 | XIXa-91 | R1 (2) | R2 (3) | 4 |
| ECUST26244 | MW351831 | XIXa-92 | R1 (2) | R2 (3) | 4 |
| ECUST26249 | MW351832 | XIXa-93 | R1 (2) | R2 (3) | 4 |
a Sample IDs labelled with ECUST were from cats in Shanghai, while those with SCAU were from cats in Guangdong; b 33-bp tandem repeat (5′-CCACCTAGTGGCGGTAGTGGCGTGTCCCCTGCT-3′) with a partial copy at the end; c 39-bp tandem repeat (5′-AGCACAACTGCGGCTACAGCGAGCACTGCGAGTTCGACA-3′) with a partial copy at the end and 0–2 nucleotide differences.
2.3. Cryptosporidium felis Subtypes Identified
Altogether, 15 subtypes were identified, with two subtypes in samples SCAU392 and SCAU754 having nucleotide sequences identical to the XIXa-39 and XIXa-40 subtypes (GenBank reference sequences MH240852 and MH240853) from humans in the United Kingdom, respectively (Table 1). According to the nomenclature system of the XIXa subtypes [18], the 13 new subtypes were named XIXa-81 (3), XIXa-82 (1), XIXa-83 (1), XIXa-84 (1), XIXa-85 (1), XIXa-86 (1), XIXa-87 (2), XIXa-88 (2), XIXa-89 (2), XIXa-90 (1), XIXa-91 (1), XIXa-92 (1), and XIXa-93 (1).
2.4. Phylogenetic Analysis of C. felis Subtypes
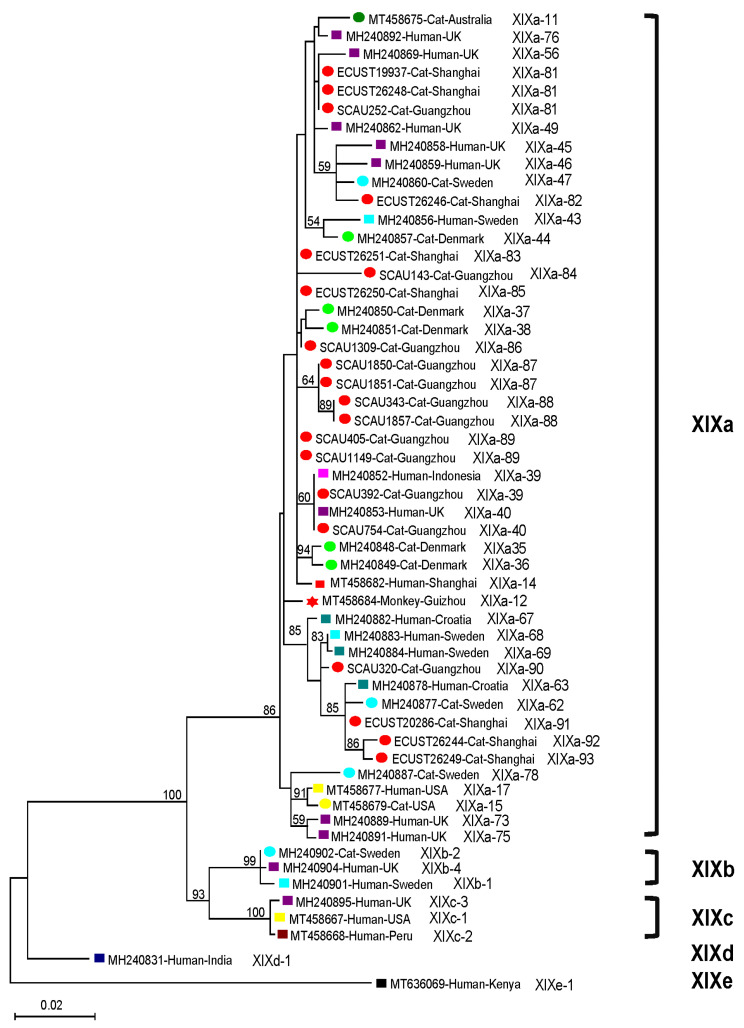
The gp60 sequences from the 20 C. felis isolates were all placed in the XIXa subtype family in the phylogenetic tree (Figure 2). They formed several subclusters with strong bootstrap support with C. felis subtypes from humans. Among the novel subtypes identified in the study, 4 subtypes (XIXa-90, XIXa-91, XIXa-92, and XIXa-93) formed a subcluster with several subtypes identified from humans and cats in several European countries and another subtype (XIXa-82) formed a subcluster with several subtypes from humans and cats in the United Kingdom and Sweden. In addition, two cat-derived subtypes (XIXa-87 and XIXa-88) in 4 isolates from this study formed their own subcluster. As expected, the known subtypes (XIXa-39 and XIXa-40) identified in the present study and previously in humans from the United Kingdom and Indonesia formed a subcluster (Figure 2).
Figure 2.
Phylogenetic relationship among XIXa subtypes of Cryptosporidium felis identified in this study and references from GenBank based on maximum likelihood analysis of the sequences of partial gp60 gene: bootstrap values over 50 percent are shown on the branches of the phylogenetic tree. The human-, monkey-, and cat-derived isolates are indicated by rhombus, star, and round labels, respectively. Sequences from different countries are shown in different colors. The names of known subtype families of C. felis are shown on the right side of the phylogenetic tree. The subtype name of each isolate is labeled at the end of the sequence.
3. Discussion
The gp60 subtyping tools have been widely used in assessing the intra-species diversity and zoonotic transmission of human-pathogenic Cryptosporidium spp. Several genes including gp60, other genetic loci with simple tandem repeats, and double-stranded viral RNA have been used to further differentiate the Cryptosporidium species into different subtypes [10]. Among them, the gp60 gene is highly polymorphic in all Cryptosporidium spp. examined thus far and therefore can be categorized into multiple subtype families by nucleotide sequence differences [4]. Moreover, the gp60 is an invasion-related protein and the subtype families identified have been linked to differences in host ranges and with virulence in C. parvum and C. hominis [3,23,24]. In this study, the newly developed gp60 subtyping tool was employed to characterize the C. felis and to understand its zoonotic potential in China.
This represents the first subtyping study of C. felis in China. The previous two studies on C. felis subtypes mostly examined C. felis isolates from other countries, with five subtype families (XIXa, XIXb, XIXc, XIXd, and XIXe) being identified [17,18]. In the present study, 20 cat-derived isolates from China were subtyped and the results of the phylogenetic analysis showed that all of them belonged to the subtype family XIXa. The PCR amplification efficiency (66.7%) of this study was similar to that of a previous study (67.0%) [18]. The light infections of C. felis in healthy cats could be one of the reasons for the relatively low PCR amplification efficiency.
An analysis of the gp60 gene confirmed the high genetic diversity of C. felis isolates. Like observations in previous studies [17,18], 15 subtypes were seen in 20 C. felis isolates successfully analyzed from cats. However, all C. felis isolates in this study belonged to the subtype family XIXa. As 13 of the 15 XIXa subtypes identified were novel, there could be geographic isolation among some of the XIXa subtypes, as previously suggested by us [18].
The results of the present study suggest that cross-species transmission of C. felis could be possible. In this study, among the 15 XIXa subtypes, two were identical to subtypes previously found in humans, while others clustered with human-derived subtypes from other countries. Importantly, one of the subclusters formed included two sequences from an owner and the household cat (GenBank reference sequences MH240883 and MH240884) with possible zoonotic transmission [17]. Moreover, we previously subtyped two C. felis isolates from one child in Shanghai (XIXa-14) and one rhesus macaque (XIXa-12) in Guizhou, China [18], while the gp60 sequences were different with the 15 XIXa subtypes identified in the present study. They clustered together in a large clade within the XIXa subtype family in the phylogenetic tree (Figure 2). The subtype characteristics of C. felis identified in this suggest that the zoonotic XIXa subtype family could be the dominant subtypes in pet cats in China. As the sequences from C. felis in Guangzhou and Shanghai are dispersed throughout the large clade within the XIXa subtype family, they could be good representatives of C. felis in cats in China. Since C. felis infections have been reported in cats and humans in several locations in China [19,25,26,27,28], an analysis of the gp60 sequences of human- and cat-derived C. felis isolates from the same location is needed to understand the epidemiological importance of zoonotic transmission of C. felis.
Currently, the significance of the divergent C. felis subtypes in cats is not clear. As mentioned above, different C. parvum and C. hominis subtypes have been linked to differences in virulence. Therefore, the hyper-transmissible and virulent C. hominis IbA10G2 and C. parvum IIaA15G2R1 subtypes have caused numerous outbreaks of human cryptosporidiosis worldwide [10] and infections with the former have induced more clinical symptoms than other C. hominis subtypes [23]. In C. felis, although previous studies and results of the present study suggest that the XIXa is the dominant subtype family in humans and cats [18], the relationship between the pathogenicity and its high transmission is not yet clear. More subtyping studies at additional genetic loci are needed to understand the differences in pathogenicity among C. felis subtypes.
4. Materials and Methods
4.1. Ethics Statement
This research was reviewed and approved by the Ethics Committee of the South China Agricultural University. The fecal specimens were collected with the permission of the owners of the pets. Each sample was collected and placed into a 50-mL plastic centrifuge tube containing 2.5% potassium dichromate and was transferred to the laboratory for storage at 4 °C. The DNA extraction was completed within one week. The DNA was stored at −20 °C for less than three years before being used in the PCR analysis in the present study.
4.2. C. felis Isolates
The nested PCR targeting the small subunit (SSU) rRNA gene was used to detect Cryptosporidium spp. [29]. DNA preparations of 30 C. felis-positive fecal samples from China were used in the present study. Among them, 18 samples were obtained from cats in Guangdong province, including 7 from pet shelters, 6 from animal hospitals, 4 from stray cats, and 1 from a pet shop. The remaining 12 samples were obtained from cats in Shanghai, including 8 from pet shelters and 4 from pet shops. They were from two previous studies of molecular epidemiology of cryptosporidiosis in cats [27,28].
4.3. PCR Amplification
The newly developed nested PCR targeting the conserved region of the gp60 gene was employed to identify the subtypes of C. felis in this study [17]. Briefly, primers GP60-Felis-F1 (5′-TTT CCG TTA TTG TTG CAG TTG CA-3′) and GP60-Felis-R1 (5′-ATC GGA ATC CCA CCA TCG AAC-3′) were used in primary PCR, while GP60-Felis-F2 (5′-GGG CGT TCT GAA GGA TGT AA-3′) and GP60-Felis-R2 (5′-CGG TGG TCT CCT CAG TCT TC-3′) were used in secondary PCR. The sizes of primary and secondary PCR products were approximately 1200 and 900 bp, respectively. The PCR reaction and cycling program were described recently [18]. Each DNA preparation was analyzed in duplicate, with the inclusion of both positive (C. felis DNA) and negative (reagent-grade water) controls in each PCR run. The positive products from the secondary PCR were identified by 1.5% agarose electrophoresis.
4.4. DNA Sequence Analyses
All positive products of the expected size were sequenced bidirectionally on an ABI3730 autosequencer by the Sangon Biotech (Shanghai, China) using secondary PCR primers. The DNA sequences generated were assembled using ChromasPro 2.1.6 (http://technelysium.com.au/wp/) and edited using BioEdit 7.1.3.0 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). These and the reference sequences from GenBank were aligned with each other using the MUSCLE program implemented in MEGA 6 (https://www.megasoftware.net/). Tandem Repeats Finder 4.09 (http://tandem.bu.edu/trf/trf.html) was used to identify repetitive sequences within them. A maximum-likelihood tree was constructed using MEGA 6 based on substitution rates calculated using the general time reversible model and gamma distribution. The bootstrap method with 1000 replicates was used to assess the reliability of the phylogenetic clusters formed. Representative nucleotide sequences of the C. felis subtypes identified in the present study were deposited in the GenBank database under accession numbers MW351820-MW351832.
5. Conclusions
The present study reported the subtype characteristics of C. felis isolates from cats in China for the first time. The results of the phylogenetic analysis suggested the potential zoonotic transmission of this pathogen. More isolates from diverse areas and hosts should be analyzed to confirm this conclusion.
Author Contributions
Conceptualization, L.X. and Y.F.; data curation, J.L.; formal analysis, J.L.; funding acquisition, L.X. and Y.F.; investigation, J.L., F.Y., R.L. and S.G.; methodology, L.X. and Y.F.; project administration, N.L.; resources, Y.F.; software, L.X. and Y.F.; supervision, Y.G.; validation, L.X. and Y.F.; visualization, L.X. and Y.F.; writing—original draft, J.L.; writing—review and editing, L.X. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Guangdong Major Project of Basic and Applied Basic Research (2020B0301030007), by the National Natural Science Foundation of China (U1901208 and 31820103014), by the 111 Project (D20008), and by the Innovation Team Project of Guangdong University (2019KCXTD001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X.M., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L., et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahedi A., Ryan U. Cryptosporidium—An update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 2020;132:500–512. doi: 10.1016/j.rvsc.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Feng Y., Ryan U.M., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 4.Xiao L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 5.de Lucio A., Merino F.J., Martinez-Ruiz R., Bailo B., Aguilera M., Fuentes I., Carmena D. Molecular genotyping and sub-genotyping of Cryptosporidium spp. isolates from symptomatic individuals attending two major public hospitals in Madrid, Spain. Infect. Genet. Evol. 2016;37:49–56. doi: 10.1016/j.meegid.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Cieloszyk J., Goni P., Garcia A., Remacha M.A., Sanchez E., Clavel A. Two cases of zoonotic cryptosporidiosis in Spain by the unusual species Cryptosporidium ubiquitum and Cryptosporidium felis. Enferm. Infecc. Microbiol. Clin. 2012;30:549–551. doi: 10.1016/j.eimc.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 7.Caccio S., Pinter E., Fantini R., Mezzaroma I., Pozio E. Human infection with Cryptosporidium felis: Case report and literature review. Emerg. Infect. Dis. 2002;8:85–86. doi: 10.3201/eid0801.010269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beser J., Toresson L., Eitrem R., Troell K., Winiecka-Krusnell J., Lebbad M. Possible zoonotic transmission of Cryptosporidium felis in a household. Infect. Ecol. Epidemiol. 2015;5:28463. doi: 10.3402/iee.v5.28463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lucio-Forster A., Griffiths J.K., Cama V.A., Xiao L., Bowman D.D. Minimal zoonotic risk of cryptosporidiosis from pet dogs and cats. Trends Parasitol. 2010;26:174–179. doi: 10.1016/j.pt.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Xiao L., Feng Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017;8–9:14–32. doi: 10.1016/j.fawpar.2017.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan W., Alderisio K., Roellig D.M., Elwin K., Chalmers R.M., Yang F., Wang Y., Feng Y., Xiao L. Subtype analysis of zoonotic pathogen Cryptosporidium skunk genotype. Infect. Genet. Evol. 2017;55:20–25. doi: 10.1016/j.meegid.2017.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stensvold C.R., Elwin K., Winiecka-Krusnell J., Chalmers R.M., Xiao L., Lebbad M. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J. Clin. Microbiol. 2015;53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y., Cebelinski E., Matusevich C., Alderisio K.A., Lebbad M., McEvoy J., Roellig D.M., Yang C., Feng Y., Xiao L. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J. Clin. Microbiol. 2015;53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stensvold C.R., Beser J., Axen C., Lebbad M. High applicability of a novel method for gp60-based subtyping of Cryptosporidium meleagridis. J. Clin. Microbiol. 2014;52:2311–2319. doi: 10.1128/JCM.00598-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., Santin M., Fayer R., Kvac M., Ryan U., et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014;20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sulaiman I.M., Hira P.R., Zhou L., Al-Ali F.M., Al-Shelahi F.A., Shweiki H.M., Iqbal J., Khalid N., Xiao L. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 2005;43:2805–2809. doi: 10.1128/JCM.43.6.2805-2809.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rojas-Lopez L., Elwin K., Chalmers R.M., Enemark H.L., Beser J., Troell K. Development of a gp60-subtyping method for Cryptosporidium felis. Parasites Vectors. 2020;13:39. doi: 10.1186/s13071-020-3906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang W., Roellig D.M., Lebbad M., Beser J., Troell K., Guo Y., Li N., Xiao L., Feng Y. Subtype distribution of zoonotic pathogen Cryptosporidium felis in humans and animals in several countries. Emerg. Microbes Infect. 2020;9:2446–2454. doi: 10.1080/22221751.2020.1840312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y., Wang L., Duan L., Gomez-Puerta L.A., Zhang L., Zhao X., Hu J., Zhang N., Xiao L. Extended outbreak of cryptosporidiosis in a pediatric hospital, China. Emerg. Infect. Dis. 2012;18:312–314. doi: 10.3201/eid1802.110666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S., Ai L., Tian L., Zhang Y., Tong X., Li H., Chen J. Investigation and fecal specimens detection of cryptozoite and other protozoon infection from patients with diarrhea. Chin. J. Zoonoses. 2012;28:815–819. [Google Scholar]
- 21.Hung C.C., Tsaihong J.C., Lee Y.T., Deng H.Y., Hsiao W.H., Chang S.Y., Chang S.C., Su K.E. Prevalence of intestinal infection due to Cryptosporidium species among Taiwanese patients with human immunodeficiency virus infection. J. Formos. Med. Assoc. 2007;106:31–35. doi: 10.1016/S0929-6646(09)60213-8. [DOI] [PubMed] [Google Scholar]
- 22.Wang R., Zhang X., Zhu H., Zhang L., Feng Y., Jian F., Ning C., Qi M., Zhou Y., Fu K., et al. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp. Parasitol. 2011;127:42–45. doi: 10.1016/j.exppara.2010.06.034. [DOI] [PubMed] [Google Scholar]
- 23.Cama V.A., Bern C., Roberts J., Cabrera L., Sterling C.R., Ortega Y., Gilman R.H., Xiao L. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg. Infect. Dis. 2008;14:1567–1574. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cama V.A., Ross J.M., Crawford S., Kawai V., Chavez-Valdez R., Vargas D., Vivar A., Ticona E., Navincopa M., Williamson J., et al. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J. Infect. Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 25.Liu A., Gong B., Liu X., Shen Y., Wu Y., Zhang W., Cao J. A retrospective epidemiological analysis of human Cryptosporidium infection in China during the past three decades (1987–2018) PLoS Negl. Trop. Dis. 2020;14:e0008146. doi: 10.1371/journal.pntd.0008146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li W., Liu X., Gu Y., Liu J., Luo J. Prevalence of Cryptosporidium, Giardia, Blastocystis, and trichomonads in domestic cats in East China. J. Vet. Med. Sci. 2019;81:890–896. doi: 10.1292/jvms.19-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., Dan X., Zhu K., Li N., Guo Y., Zheng Z., Feng Y., Xiao L. Genetic characterization of Cryptosporidium spp. and Giardia duodenalis in dogs and cats in Guangdong, China. Parasites Vectors. 2019;12:571. doi: 10.1186/s13071-019-3822-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H., Jin Y., Wu W., Li P., Wang L., Li N., Feng Y., Xiao L. Genotypes of Cryptosporidium spp., Enterocytozoon bieneusi and Giardia duodenalis in dogs and cats in Shanghai, China. Parasites Vectors. 2016;9:121. doi: 10.1186/s13071-016-1409-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao L., Morgan U.M., Limor J., Escalante A., Arrowood M., Shulaw W., Thompson R.C., Fayer R., Lal A.A. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 1999;65:3386–3391. doi: 10.1128/AEM.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data is contained within the article.