Abstract
Human genome-wide association studies (GWASs) have identified more than 270 loci associated with pulmonary function; however, follow-up studies to determine causal genes at these loci are few. SNPs in low-density lipoprotein receptor–related protein 1 (LRP1) are associated with human pulmonary function in GWASs. Using murine models, we investigated the effect of genetic disruption of the Lrp1 gene in smooth muscle cells on pulmonary function in naive animals and after exposure to bacterial LPS or house dust mite extract. Disruption of Lrp1 in smooth muscle cells leads to an increase in tissue resistance, elastance, and tissue elastance at baseline. Furthermore, disruption of Lrp1 in smooth muscle increases airway responsiveness as measured by increased total lung resistance and airway resistance after methacholine. Immune cell counts in BAL fluid were increased in animals with Lrp1 disruption. The difference in airway responsiveness by genotype observed in naive animals was not observed after LPS or house dust mite extract exposure. To further explore the mechanisms contributing to changes in pulmonary function, we identified several ligands dysregulated with Lrp1 disruption in smooth muscle cells. These data suggest that dysregulation of LRP1 in smooth muscle cells affects baseline pulmonary function and airway responsiveness and helps establish LRP1 as the causal gene at this GWAS locus.
Keywords: genome-wide association studies, lung function, low-density lipoprotein receptor–related protein 1, genetic variation, chronic obstructive pulmonary disease
Clinical Relevance
Human genome-wide association studies (GWASs) have identified more than 270 loci associated with pulmonary function; however, follow-up studies to determine causal genes at these loci are few. SNPs in low-density lipoprotein receptor–related protein 1 (LRP1) are associated with the pulmonary function trait of the ratio of forced expiratory volume in 1 second to forced vital capacity. Although GWASs are very successful in identifying genetic loci associated with many phenotypes and diseases, identifying the causal genes or variants has proved more elusive. Experimental animal studies can establish causality but are costly and time consuming and thus have rarely been applied to follow-up GWAS associations. There are no studies following up the GWAS associations with pulmonary function for SNPs in LRP1. We provide updated GWAS results confirming the association between SNPs within LRP1 and the ratio of forced expiratory volume in 1 second to forced vital capacity in a multiethnic population of more than 90,000 individuals. Using animal models, we found that mice without Lrp1 in smooth muscle have altered baseline pulmonary function and exhibit increased airway responsiveness. These data recapitulate human findings from GWASs, lending evidence for a causal relationship between LRP1 and pulmonary function not possible to establish from association studies alone. Establishing causal genes underlying GWAS findings is an essential first step toward clinical translation.
Genome-wide association studies (GWASs) provide an unbiased, comprehensive approach to identifying genetic variants associated with disease relevant phenotypes. GWASs of pulmonary function have identified more than 270 genetic loci related to these traits (1, 2). Pulmonary function is a readily available and reliably measured index of the physiological state of the lungs that is used clinically to diagnose and assess the progression of chronic obstructive lung disease (COPD), asthma, and other lung conditions (3–5).
SNPs within the low-density lipoprotein receptor–related protein 1 (LRP1) gene associated with the pulmonary function trait of the ratio of forced expiratory volume in 1 second (FEV1) to forced vital capacity (FVC) have been identified in GWASs (6). Although GWASs have been very successful in identifying genetic loci associated with many phenotypes and diseases, identifying the causal genes or variants has proved more elusive. Experimental animal studies can establish causality but are costly and time consuming and thus have rarely been applied to follow-up GWAS associations. There are no studies following up the GWAS associations with pulmonary function for SNPs in LRP1.
LRP1 is a ubiquitously expressed, versatile scavenger receptor first characterized as mediating the uptake of extracellular or membrane-associated molecules (7). Further research into the function of LRP1 has described additional functions of the protein, which some suggest may be tissue specific (8). Deletion of Lrp1 in mice is embryonically lethal, outlining the importance of this receptor in embryogenesis and development (9). Within the immune system, this cell surface transmembrane scavenger receptor plays a significant role in regulation of cellular junctions and modulation of the inflammatory response (8). LRP1 in smooth muscle cells is known to mediate the endocytosis of numerous ligands that regulate the extracellular environment and promote survival of these cells (10, 11).
LRP1 has a wide range of functions in different tissues reflecting its diversity of interactions, activities, and expression. This receptor has been shown to interact with more than 100 proteins, including ∼40 extracellular ligands including lipoproteins, extracellular matrix proteins, proteases, protease-inhibitor complexes, and growth factors (12). These extracellular interactions coupled with unique, potentially tissue-specific intracellular interactions help to modulate cell migration, survival, proliferation, and differentiation. Although LRP1 is ubiquitously expressed, this protein is most abundant in the brain, lung, and muscle (7, 13). Recent data support a key role for LRP1 in lung immunity (14) and lung remodeling (15). No study has outlined the role of LRP1 in the modulation of pulmonary function.
In this study, we report the development of a novel murine model to facilitate studies on the function of LRP1 in the lung. Using a smooth muscle–specific Cre recombinase, we disrupted Lrp1 and assessed the impact of this disruption on pulmonary function. To identify the mechanisms contributing to the observed phenotypes, we used well-established pulmonary phenotyping techniques and proteomic assessment of BAL fluid to identify peptides altered by LRP1 disruption in the smooth muscle. This work in mice provides significant evidence that LRP1 is causal for the GWAS signal for modulation of pulmonary function identified in humans.
Methods
Human GWASs of LRP1 SNPs and Pulmonary Function Traits and Expression Quantitative Trait Analysis of Sentinel GWAS SNPs in LRP1
See data supplement.
Murine Studies
Lrp1flox (B6; 129S7-Lrp1tm2Her/J) mice were purchased from The Jackson Laboratory and maintained as Het x Het breeders to generate floxed mutant mice that were genotyped per the Jackson Laboratory protocol. These mice were bred to mice expressing Cre recombinase driven by the smooth muscle (SM) specific Tagln (Transgelin) promoter to generate experimental knockout (SM-Cre+-Lrp1flox/flox; represented as Lrp1−/−) and control (SM-Cre−-Lrp1flox/flox; represented as Lrp1+/+) animals. Given that SM-Cre+ and SM-Cre− did not significantly differ in pulmonary function (Figure E1 in the data supplement), SM-Cre−-Lrp1flox/flox animals were used as controls. Mice were fed NIH-31 rodent chow ad libitum and housed with alternating 12-hour light-dark cycles. All animal work described in this study was conducted according to National Institutes of Health guidelines and approved by the National Institute of Environmental Health Sciences Animal Care and Use Committee.
Immunofluorescence
Pulmonary tissue was sampled from control and knockout animals and assessed for LRP1 expression using immunofluorescence. See data supplement.
Pulmonary Function Assessment
Knockout and control animals between 9 and 12 weeks of age underwent pulmonary function analysis using the flexiVent Legacy system or flexiVent FX2 system (SCIREQ, Inc) as previously described (16). See data supplement.
BALF Cell Analyses
After exsanguination, BAL fluid (BALF) samples were collected with Hanks’ balanced salt solution (H6648; Sigma-Aldrich). BALF from each mouse was centrifuged to separate cells from supernatant. Cellular fractions were treated with ammonium–chloride–potassium buffer, centrifuged, resuspended in Hanks’ balanced salt solution, and quantified with a TC20 Automated Cell Counter (Bio-Rad), and cytospins were prepared for cell differential analysis.
BALF LRP1 Ligand Concentrations by ELISA or Bio-Plex
The concentrations of ligands of LRP1 were measured in the cell-free supernatant using Bio-Plex mouse assays and ELISAs. Matrix metalloproteinase 2 (MMP2) and matrix metalloproteinase 9 (MMP9) levels were assayed using the Bio-Plex mouse assay and Bio-Plex suspension array system (171 AM001M; Bio-Rad) according to the manufacturer’s instruction. ELISA kits were used according to manufacturer’s instruction to measure the levels of elastase (ab204730; Abcam), urokinase (ab198512; Abcam), and plasminogen activator inhibitor-1 (ab197752; Abcam) in the BALF.
BALF Proteomics
After samples were digested overnight with trypsin, the digests were analyzed by liquid chromatography/mass spectrometry (LC/MS) on a Q Exactive Plus mass spectrometer (ThermoFisher Scientific) interfaced with an M-Class nanoAcquity Ultra Performance LC (UPLC) system (Waters Corporation). Proteins from the LC/MS data were identified and quantified using Proteome Discoverer (ThermoFisher Scientific). Each sample was acquired in triplicate and the results searched against the RefSeq mouse protein database. See data supplement for details.
Statistical Analysis
For assessment of genotype differences in pulmonary function parameters at baseline and after methacholine administration, we used a Student’s t test. Values were reported as mean ± SEM and considered significant if the P value for difference by genotype was less than 0.05. See data supplement regarding proteomic analyses.
Results
Human GWASs
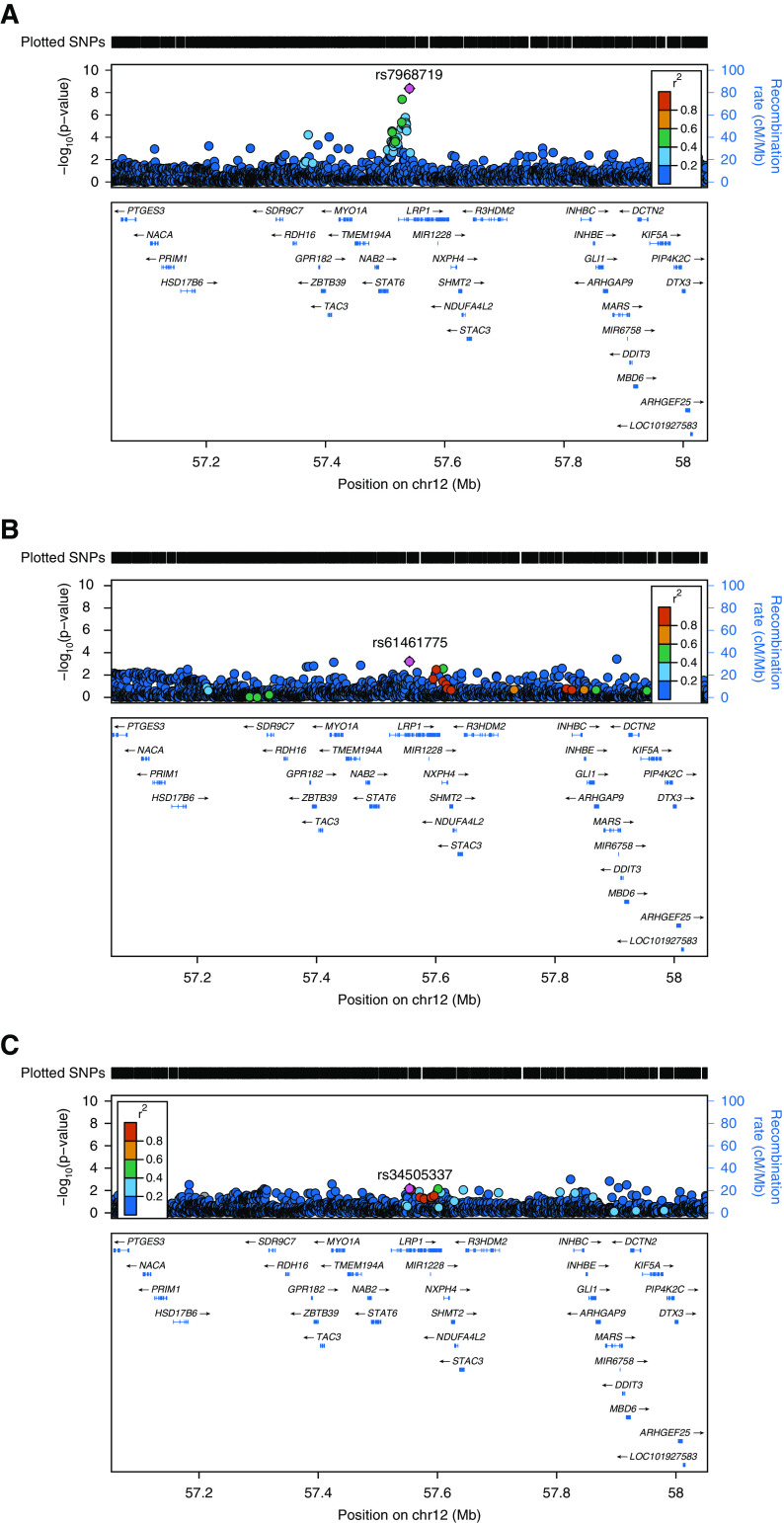
Using data from an updated large multiethnic GWAS meta-analysis (1), we confirmed earlier reported associations (6) from European ancestry individuals that SNPs within LRP1 are associated with FEV1/FVC (Figure 1A). Two common intronic SNPs in LRP1 were identified at genome wide significance (P < 5.00 × 10−8): rs7968719 (minor allele frequency 0.49, P = 4.34 × 10−9) and rs11172113 (minor allele frequency 0.43, P = 4.04 × 10−8). Genome-wide significant associations were not seen for LRP1 SNPs in relation to FEV1 (Figure 1B, smallest P value = 6.12 × 10−4 for SNP rs61461775) or FVC (Figure 1C, smallest P value = 6.9 × 10−3 for rs34505337).
Figure 1.
LRP1 (low-density lipoprotein receptor–related protein 1) is associated with pulmonary function parameters in a multiethnic meta-analysis. Locus zoom plots for lead variants in or near LRP1 identified in the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) consortium multiethnic meta-analysis of 1,000 genomes variants and the following pulmonary function measures: (A) The ratio of forced expiratory volume in 1 second (FEV1) to forced vital capacity (FVC), (B) FEV1, and (C) FVC. Insertion/deletion variants are omitted. Linkage disequilibrium is based on hg19/1000 Genomes Mar 2012 EUR (http://locuszoom.sph.umich.edu/). Nearby genes not shown on plots include SLC26A10 and B4GALNT1.
Human eQTL Analysis
Among the significant cis-expression quantitative trait locus (eQTL) results in Genotype-Tissue Expression (GTEx; obtained from https://www.gtexportal.org/home/ on January 3, 2019), rs7968719 and rs11172113, the genome-wide significant sentinel SNPs for LRP1 in the CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology) multiethnic meta-analysis of FEV1/FVC (1), indicated significant cis-eQTLs in three tissues: skin and artery (aorta and tibial). Because GTEx includes only a few hundred (or fewer) samples for each tissue (e.g., 383 lung tissue samples and 369 whole blood samples), we also looked up LRP1 variants in the following eQTL data sources: 5,311 blood samples in the Westra and colleagues study (17), and 2,116 blood samples in the BIOS (Biobank-based integrative omics study) Consortium (18). Both rs7968719 and rs11172113 indicated significant cis-eQTLs in blood in both studies.
Lrp1−/− Mouse Characterization
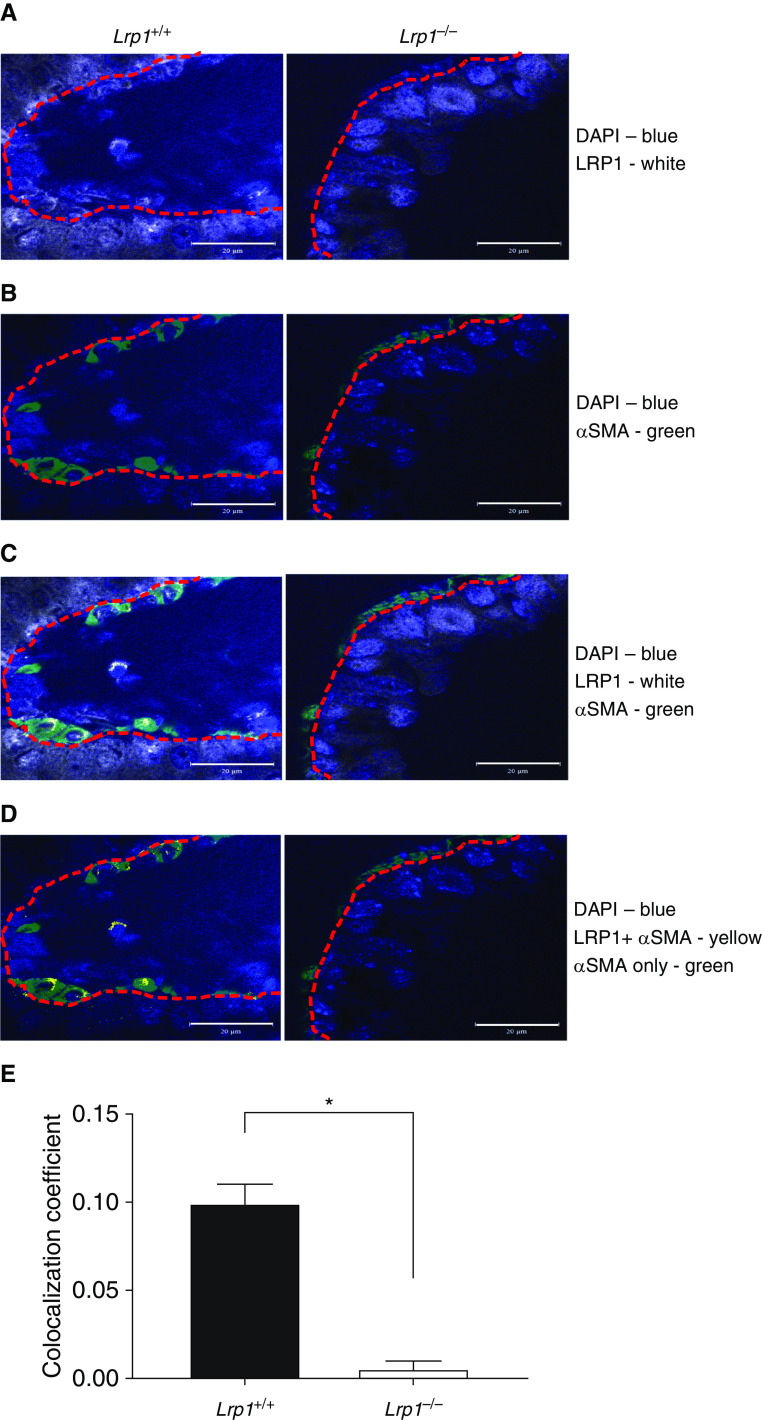
Using specific antibodies and immunofluorescence, we confirmed a significant decrease in LRP1 protein expression in smooth muscle cells of Lrp1−/− mice relative to Lrp1+/+ control mice (Figure 2). The changes in LRP1 expression were cell type specific as staining with markers for endothelial (CD31) and epithelial (EpCAM) cells did not yield significant differences in colocalization between Lrp1+/+ and Lrp1−/− mice (Figure E3). Examination of hematoxylin and eosin–stained sections revealed no obvious differences in lung histology or architecture with genetic disruption of Lrp1 (data not shown).
Figure 2.
Representative immunofluorescent images of LRP1 colocalization with αSMA (α-smooth muscle actin) in Lrp1+/+ and Lrp1−/− animals. (A) DAPI (blue) and anti-LRP1 (Cy5). (B) DAPI (blue) and anti-αSMA (Alexa488). (C) Merge of DAPI (blue), anti-αSMA (Alexa488), and anti-LRP1 (Cy5). (D) Areas of colocalization of anti-LRP1 and anti-αSMA with false color (yellow) and anti-LRP1 (Cy5) channel removed. Red dotted line is traced on the basement membrane separating smooth muscle from bronchial epithelium. Scale bars, 20 μm. (E) Quantification of colocalization signal in wild-type and Lrp1−/− animals. (n = 3/genotype, *P < 0.05).
Baseline Pulmonary Function Is Impacted by Lrp1 Disruption
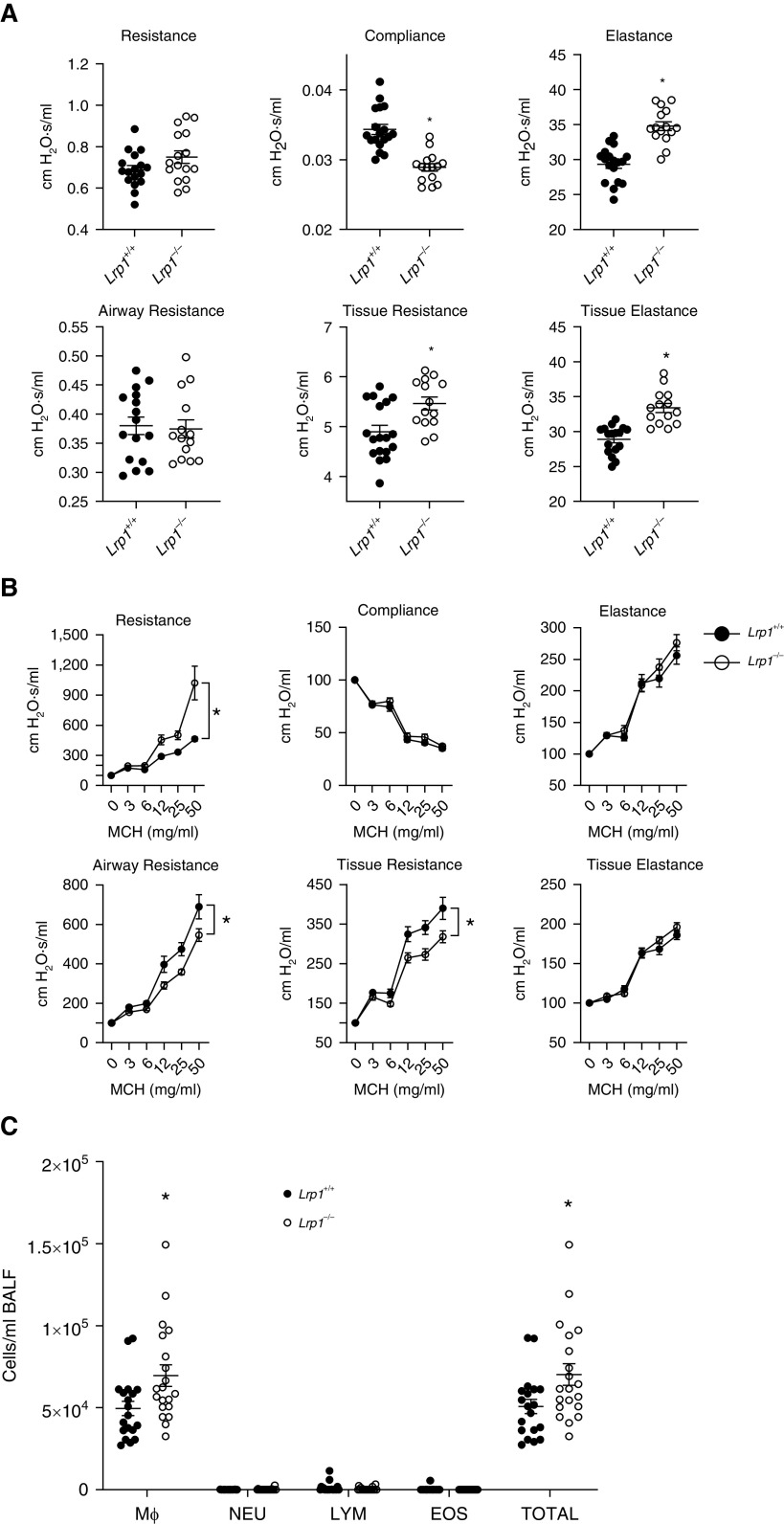
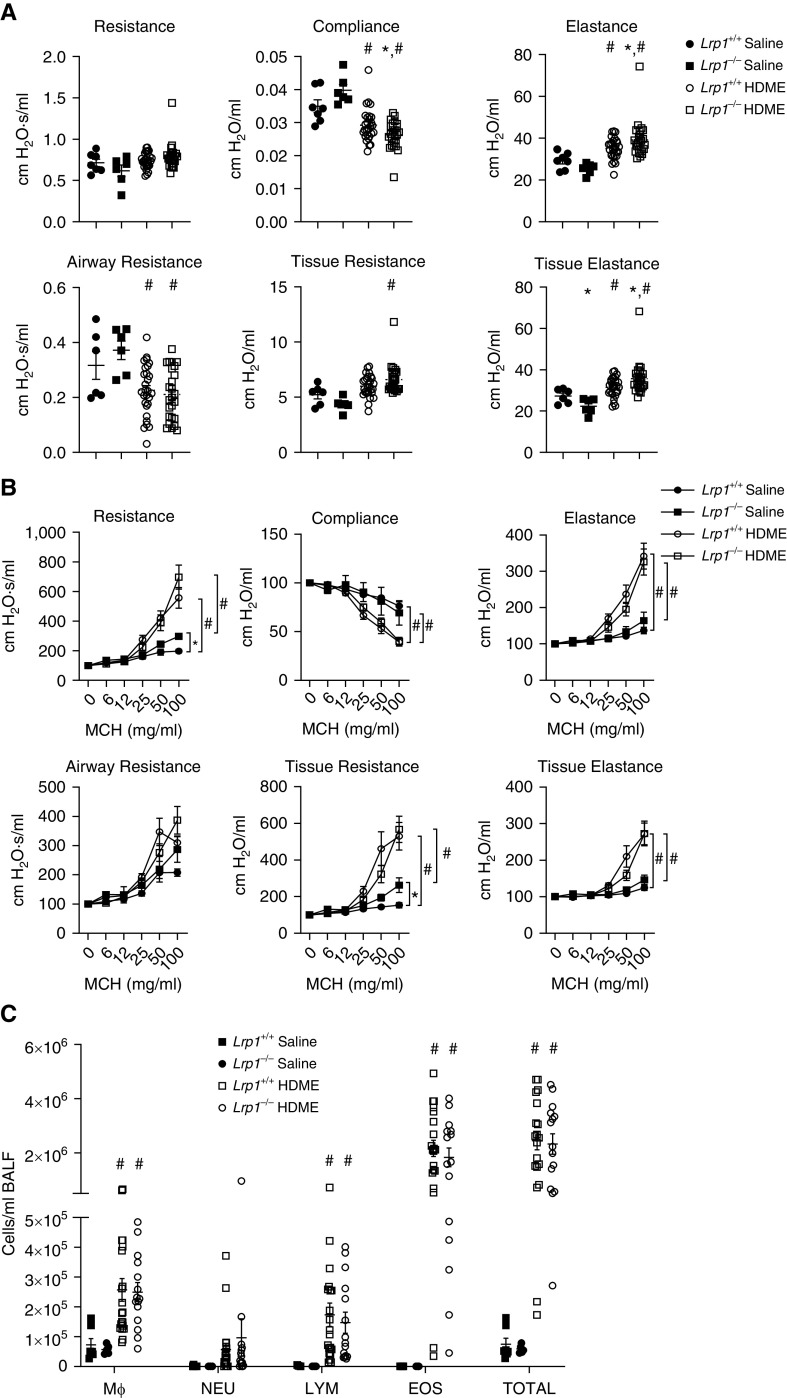
When analyzing the baseline pulmonary function in naive animals, we observed a significant increase in elastance, tissue resistance, and tissue elastance and a concomitant decrease in compliance in Lrp1−/− animals compared with Lrp1+/+ controls (Figure 3A). We also observed a significant increase in airway responsiveness to methacholine as assessed by resistance, airway resistance, and tissue resistance (Figure 3B and Table E1). We observed no changes in the numbers of neutrophils, eosinophils, or total lymphocytes in BAL fluid between Lrp1+/+ and Lrp1−/− animals, but did discern an increase in total cells and macrophages (Figure 3C).
Figure 3.
Smooth-muscle Lrp1−/− mice exhibit altered baseline pulmonary function, airway responsiveness, and BAL fluid (BALF) cell counts. (A) Baseline pulmonary function in Lrp1+/+ and Lrp1−/− mice; Means and SEMs are plotted (n > 18/genotype). (B) Airway responsiveness to methacholine (MCH) in Lrp1+/+ and Lrp1−/− mice; means and SEs are plotted (n > 18/genotype). (C) Cell differential counts between Lrp1+/+ and Lrp1−/− mice for macrophages (Mϕ), neutrophils (NEU), lymphocytes (LYM), and eosinophils (EOS). *P < 0.05.
Pulmonary Function in Lrp1−/− Animals after LPS Exposure
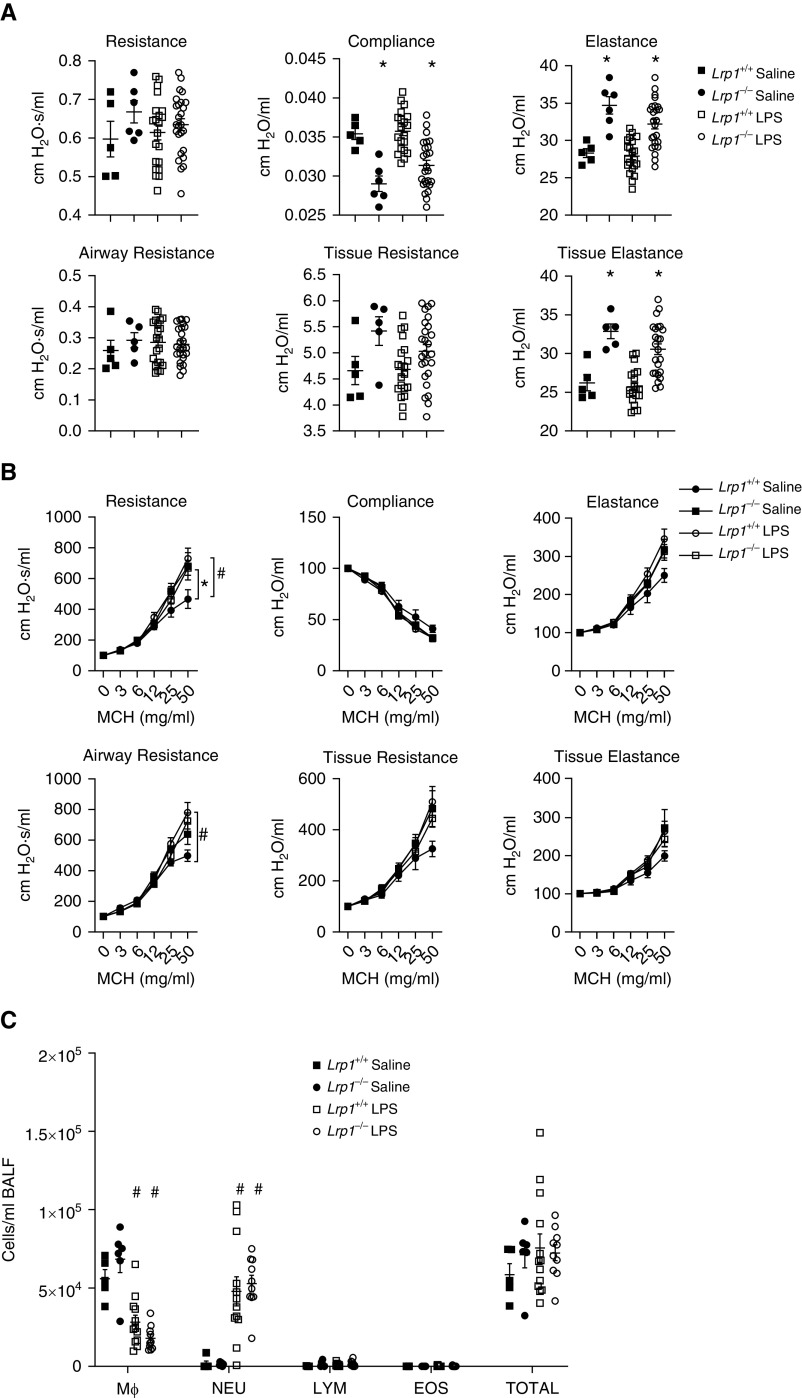
After exposure to LPS, there was a significant decrease in baseline compliance and significant increases in baseline elastance and tissue elastance in Lrp1−/− mice compared with Lrp1+/+ controls (Figure 4A). Moreover, after exposure to LPS, both Lrp1−/− and Lrp1+/+ animals exhibited significantly increased airway responsiveness to methacholine compared with saline (vehicle)-treated mice, with LPS-exposed Lrp1−/− animals responding similarly to their Lrp1+/+ counterparts (Figure 4B and Table E2). Thus, LPS exposure abolished genotype differences in airway responsiveness to methacholine. Furthermore, although there was an increase in the number of total cells and neutrophils BAL fluid in LPS-exposed Lrp1−/− and Lrp1+/+ animals compared with saline-exposed animals, there were no significant differences by genotype (Figure 4C). These data suggest that the innate immune response is unaffected by LRP1 deficiency.
Figure 4.
Smooth-muscle Lrp1−/− mice exhibit altered pulmonary function but not airway responsiveness or BALF cell counts after exposure to bacterial LPS. (A) Baseline pulmonary function in wild-type and Lrp1−/− mice 4 hours after exposure to bacterial LPS or saline. (B) Assessment of airway responsiveness to MCH in Lrp1+/+ and Lrp1−/− mice after exposure to bacterial LPS or saline. (C) Cell differential counts between Lrp1+/+ and Lrp1−/− mice for Mϕ, NEU, LYM, and EOS after exposure to bacterial LPS or saline. N > 5/genotype for saline exposure and N > 20/genotype for LPS exposure. *P < 0.05 for comparison between genotypes with the same exposure. #P < 0.05 for comparison between saline and LPS exposure of the same genotype.
Pulmonary Function in Lrp1−/− Animals after Exposure to House Dust Mite Extract
After house dust mite extract (HDME) exposure, baseline pulmonary function did not differ significantly between Lrp1−/− mice and Lrp1+/+ controls (Figure 5A). Moreover, after exposure to HDME, both Lrp1−/− and Lrp1+/+ animals had significantly increased airway responsiveness compared with their respective vehicle-exposed controls, without significant differences by genotype (Figure 5B and Table E3). Eosinophils and total cells were increased in the BAL fluid of HDME-exposed Lrp1−/− and Lrp1+/+ animals compared with their respective vehicle-exposed controls but there were no differences by genotype after exposure to HDME (Figure 5C). This suggests that the inflammation induced by HDME is similar in Lrp1−/− and Lrp1+/+ animals.
Figure 5.
Smooth-muscle Lrp1−/− mice exhibit few differences in pulmonary function, airway responsiveness, or BALF cell counts after house dust mite extract (HDME) exposure. (A) Baseline pulmonary function in Lrp1+/+ and Lrp1−/− mice after two sensitization exposures and three challenge exposures with HDME over 17 days. (B) Assessment of airway responsiveness to MCH in Lrp1+/+ and Lrp1−/− mice. (C) Cell differential counts between Lrp1+/+ and Lrp1−/− mice for Mϕ, NEU, LYM, and EOS. N > 6/genotype for saline exposure and N > 24/genotype for HDME exposure; *P < 0.05 for comparison between genotypes with the same exposure. #P < 0.05 for comparison between saline and HDME exposure of the same genotype.
LRP1 Ligands Are Dysregulated in BAL Fluid of Lrp1−/− Animals
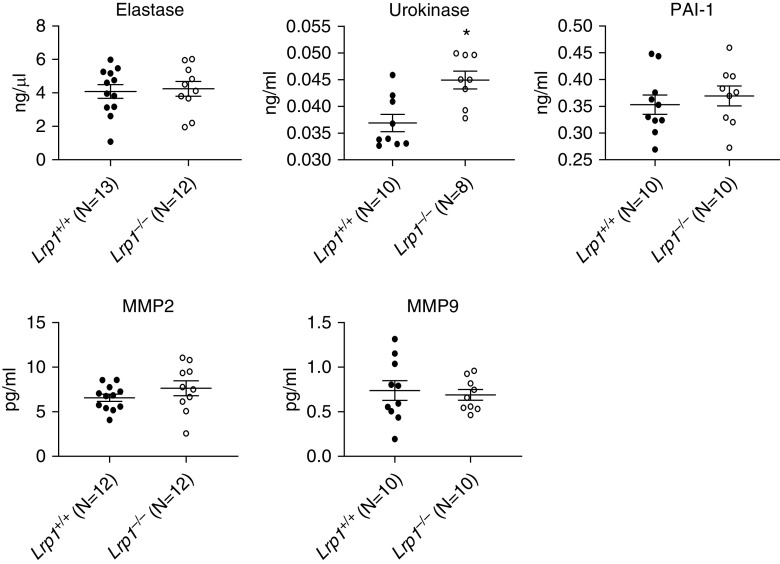
Given the observed differences in pulmonary function in naive Lrp1−/− mice relative to Lrp1+/+ controls and the absence of differences in immune cell response to LPS and HDME, we evaluated whether genetic disruption of Lrp1 in smooth muscle cells alters the levels of LRP1 ligands in the BAL fluid. We initially focused on LRP1 ligands previously shown to be associated with pulmonary disease including matrix metalloproteinase 9 (MMP9) (19), MMP2 (20), elastase (21), urokinase (22), and plasminogen activator inhibitor-1 (PAI-1) (23). We found that the concentration of urokinase was significantly increased in the BALF of Lrp1−/− animals compared with Lrp1+/+ controls. No significant differences in the concentrations of the other ligands were found between the genotypes (Figure 6).
Figure 6.
Smooth-muscle Lrp1−/− mice and LRP1-ligand concentrations in BALF. BALF was collected from Lrp1+/+ and Lrp1−/− mice and assessed via ELISA for concentrations of known LRP1 ligands: elastase, urokinase, PAI-1, MMP2, and MMP9. N ≥ 8/genotype. *P < 0.05. MMP2 = matrix metalloproteinase 2; MMP9 = matrix metalloproteinase 9; PAI-1 = plasminogen activator inhibitor-1.
BALF Proteomes Are Dysregulated in Lrp1−/− Animals
Because LRP1 has been reported to interact with more than 40 different proteins (24), we used a proteomic approach to further investigate differences in BALF proteins between Lrp1−/− and Lrp1+/+ animals using label-free mass spectrometry. A total of 1,850 proteins were identified after quality control and data processing. Of the proteins identified, 160 proteins were significantly dysregulated (adjusted P value < 0.05) in the BALF of Lrp1−/− compared with Lrp1+/+mice. Of the proteins identified, 21 are known to associate with LRP1 and 7 of these associated proteins were significantly dysregulated in Lrp1−/− animals (Table 1). Some proteins that were significantly upregulated include members of the serpin family (e.g., SERPINA1D, 21-fold increase). Apolipoprotein E (APOE) was significantly (28-fold) decreased in the BALF of Lrp1−/− animals relative to Lrp1+/+ controls. Some proteins altered by disruption of Lrp1 in smooth muscle are cytoskeletal proteins, for example, tubulin and microtubule-associated protein, RP/EB family, member 1. The presence of cytoskeletal proteins in the BALF has been reported in previous studies of mice under control conditions (25–29).
Table 1.
Proteins within the BAL Fluid Identified by Mass Spectrometry and Previously Associated with Low-Density Lipoprotein Receptor–related Protein 1
| Symbol | Protein Name | Adjusted P Value | Fold Change |
|---|---|---|---|
| APOE | Apolipoprotein E | 0.014 | −28.860 |
| CALR | Calreticulin | 0.064 | 23.348 |
| GRIN2A | Glutamate ionotropic receptor NMDA type subunit 2A | 0.641 | 1.404 |
| GLUL | Glutamate-ammonia ligase | 0.015 | −4.563 |
| HSP90AA1 | Heat shock protein HSP 90-α | 0.018 | −20.937 |
| HSP90B1 | Endoplasmin | 0.909 | −1.226 |
| HSPA5 | Heat shock protein family A member 5 | 0.010 | 9.401 |
| HPX | Hemopexin | 0.194 | 3.510 |
| IGFBP3 | Insulin-like growth factor–binding protein 3 | 0.386 | 1.694 |
| LPL | Lipoprotein lipase | 0.343 | −4.020 |
| MAPRE1 | Microtubule-associated protein RP/EB family member 1 | 0.131 | −2.652 |
| ROCK2 | Rho-associated coiled-coil–containing protein kinase 2 | 0.978 | 1.023 |
| SEPT7 | Septin-7 | 0.250 | −4.114 |
| SERPINA1D | Alpha-1-antitrypsin 1-4 | 0.003 | 21.471 |
| SERPINC1 | Serpin C1 | 0.333 | 2.226 |
| SERPIND1 | Serpin D1 | 0.070 | 3.295 |
| SERPING1 | Serpin G1 | 0.075 | 3.234 |
| TUBB4A | Tubulin β-4A chain | 0.130 | −4.047 |
| TUBB4B | Tubulin β-4B chain | 0.016 | −13.986 |
| YWHAG | 14-3-3 protein γ | 0.075 | 4.068 |
| VCL | Vinculin | 0.042 | 5.099 |
Discussion
In this study we used mouse models to follow up human GWAS–identified SNPs in LRP1 associated with pulmonary function to ascertain whether LRP1 is indeed the causal gene at this locus. We also provide updated GWAS results confirming the association between LRP1 SNPs and FEV1/FVC in a multiethnic population of more than 90,000 individuals (1). Furthermore, we investigate whether the sentinel LRP1 SNPs are expression quantitative trait loci that influence gene expression in humans. Using a murine model of Lrp1 disruption in smooth muscle cells we investigated the role of LRP1 in modulating pulmonary function and observed a unique pulmonary phenotype with Lrp1 knockout in smooth muscle showing a change in both baseline pulmonary function parameters and airway hyperresponsiveness compared with animals with intact Lrp1. We further investigated mechanisms that contribute to the unique pulmonary phenotype observed. Owing to the previously identified role of LRP1 in the modulation of the inflammatory response, we began by investigating the role of inflammation in the modulation of the inflammatory response in the lungs of Lrp1−/− animals and concluded that removal of LRP1 in the smooth muscle does not overtly impact the inflammatory response. Given our evidence that the inflammatory response modestly affects and is not responsible for the pulmonary phenotype, we investigated the dysregulation of ligands of LRP1 within the BALF and found that expression of some proteins known to be associated with pulmonary function and asthma are altered. Taken together, these data help to confirm that LRP1 is the gene at the locus identified in GWASs responsible for modulating pulmonary function in humans and establish that the pulmonary phenotype observed in animal models may in part be due to dysregulation of LRP1 ligands.
A previous GWAS meta-analysis had established the association of SNPs in LRP1 with FEV1/FVC in individuals of European descent (6). In the current study, we confirm this association in a much larger, multiethnic population. Of note, a recent large GWAS from the UK Biobank has implicated one of these two SNPs (rs11172113) in the additional spirometric trait of peak expiratory flow (2). However, simple association in GWASs does not establish LRP1 as causal at this locus. GWAS significant sentinel LRP1 variants identified for FEV1/FVC were significant cis-eQTLS in GTEx in skin (sun-exposed lower leg) and two arterial tissues (aorta and tibial artery) and in blood in two larger data sets. Interestingly, the two sentinel SNPs for lung function have also been associated in GWASs with aortic aneurysm (30) and arterial pulse pressure (31). Although we did not identify significant eQTLs in the lung, a heterogeneous tissue in GTEx samples (32), this result confirms that the sentinel LRP1 GWAS SNPs correlate with gene expression. However correlational analysis cannot establish a causal locus. Therefore, we used murine models to conduct follow-up analyses, to better establish the causal relationship between LRP1 and pulmonary function parameters.
Experimental follow-ups of GWAS findings are not a common undertaking as they are expensive and labor intensive and there are many loci identified in GWASs for a given trait. Previous experimental studies by us and others have followed up a handful of GWAS loci for pulmonary function traits and helped to clarify the causal gene at the locus and potential mechanisms for the GWAS associations (16, 33–35). Thus, this manuscript adds to this small body of work that highlights the value of experimental studies to better understand GWAS findings. Human GWASs of pulmonary function traits were done in population studies where it is not possible to administer different exposures. The baseline differences identified in this study by removal of LRP1 in smooth muscle cells in an animal model mimic the human phenotype. Because interventions are not part of the human population-based GWASs, we could build on the human data by exposing mice to methacholine and the environmentally relevant exposure models of LPS and HDME.
LRP1 has been demonstrated to have a wide variety of functions, but it is best known as a scavenger receptor. Furthermore, there is previous evidence for cell specificity of some of the functions of LRP1, prompting the need to investigate the impact of LRP1 in different cell types on pulmonary function. LRP1 has previously been shown to play a key role in vascular smooth muscle function (36, 37). In addition to documenting that Lrp1 disruption in smooth muscle alters pulmonary function in the mouse and establishing this locus as causal for the human association, we attempted to identify mechanistic insight for this observation. Lrp1 knockout induces rampant systemic inflammation suggesting that one canonical mechanism of LRP1 is to inhibit inflammatory reactions (38, 39). Thus, we began investigating this mechanism by treating Lrp1−/− animals with environmentally relevant exposures that are known to induce inflammatory reactions—LPS and HDME. After identifying that control and knockout animals responded similarly to these environmental exposures, we considered the other main function of LRP1: regulating the levels of its ligands within the extracellular space. Because LRP1 has more than 40 identified ligands, we used a combination of proteomics and well-established assays to gather information on the concentrations of these ligands within the bronchoalveolar fluid. Using these, we identified several ligands dysregulated by LRP1 removal that may contribute to the identified pulmonary phenotype. Although this study used proteomic approaches to identify some candidate proteins altered in the BALF of Lrp1−/− mice that could be involved in the pulmonary function phenotype, it does not establish a precise molecular mechanism. Future experiments to confirm the proteomic findings with another method and then to link dysregulation of these proteins to altered pulmonary function would be needed to establish whether any of the dysregulated ligands are responsible for the observed phenotype.
Deeper characterization of the cells in the BALF, as well as in vitro studies to understand how Lrp1 deletion in smooth muscle cells affects behavior of cytoskeletal proteins, would be of interest. Of direct relevance to this issue, a recent study examined the impact of Lrp1 deletion in smooth muscle in the mouse and also found various cytoskeletal proteins to be altered (40). In both their direct examination of vascular smooth muscle and ours of BALF, several cytoskeletal proteins were altered in the same direction with Lrp1 deletion in smooth muscle. At nominal P < 0.05 in both studies, ACTA2 and CLIC4 were decreased and levels of the following proteins were increased: ACTN4, MYH11, MYLK, and VCL. Both studies highlight the importance of LRP1 in the regulation of cytoskeletal proteins.
This is the first study to directly analyze the impact of a scavenger receptor on pulmonary function and to further investigate the mechanisms contributing to this phenotype. In the current study, urokinase and APOE were two ligands that were dysregulated in Lrp1−/− animals. Both of these ligands have been previously identified to be associated with pulmonary function and airway responsiveness (41, 42). In humans, urokinase is known to be increased in the BALF of humans with asthma (43), suggesting that in our study the increase in BALF urokinase levels may contribute to the increase in airway responsiveness. Decreased APOE levels in other animal models has been shown to increase susceptibility to cardiovascular and pulmonary injury (44, 45), especially after pulmonary environmental and occupational exposures (46–48). Furthermore, one study investigating the relationship between APOE and urokinase showed that decreased levels of urokinase lead to increased levels of ApoE, suggesting urokinase plays a role in regulating ApoE levels (49). The differential impact of ligands could be exacerbated with disruption of LRP1 in different cell types. Not only could this be due to the differential functions of LRP1 in different cell types, but further different ligands may differentially impact these different cell types.
In summary, following up human GWAS findings for LRP1 and pulmonary function, we found that Lrp1−/− mice have altered baseline pulmonary function (decreased compliance, increased elastance, tissue resistance, and tissue elastance) and exhibit increased airway responsiveness. These data recapitulate human findings from GWASs, lending evidence for a causal relationship between LRP1 and pulmonary function that was is not possible to establish from such association studies alone. We did not find that LRP1 deficiency leads to altered responses to either bacterial LPS or HDME. We found evidence that supported that ligand disruption contributes to the pulmonary phenotype observed in our animal model of Lrp1 disruption. In conclusion, we present here strong evidence for a causal relationship between LRP1 and pulmonary function in mice that supports human GWAS findings.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Katina Johnson of the Mass Spectrometry Research and Support Group and Charles J. Tucker, director of Fluorescence Microscopy and Imaging Center, both of the National Institute of Environmental Health Sciences, for their expert technical assistance.
Footnotes
Supported by the Intramural Research Program of the U.S. National Institutes of Health, the National Institute of Environmental Health Sciences (ZO1 ES025045 and ZO1 ES043012 [S.J.L.] and ES 025041 [D.C.Z.], and Contract No. HHSN2732016000031 [A.W.]), and National Heart, Lung, and Blood Institute grant R01HL105756.
Author Contributions: Conception and design: C.E.N., J.S.H., J.M.W., A.W., J.G.W., L.J.D., D.C.Z., and S.J.L. Analysis and interpretation: C.E.N., J.S.H., H.L., J.M.W., A.W., J.G.W., L.J.D., J.A.B., L.M., D.C.Z., and S.J.L. Drafting the manuscript for important intellectual content: C.E.N., J.M.W., A.W., L.J.D., D.C.Z., and S.J.L.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2019-0444OC on December 8, 2020
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Wyss AB, Sofer T, Lee MK, Terzikhan N, Nguyen JN, Lahousse L, et al. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nat Commun. 2018;9:2976. doi: 10.1038/s41467-018-05369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shrine N, Guyatt AL, Erzurumluoglu AM, Jackson VE, Hobbs BD, Melbourne CA, et al. Understanding Society Scientific Group. New genetic signals for lung function highlight pathways and chronic obstructive pulmonary disease associations across multiple ancestries. Nat Genet. 2019;51:481–493. doi: 10.1038/s41588-018-0321-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hole DJ, Watt GC, Davey-Smith G, Hart CL, Gillis CR, Hawthorne VM.Impaired lung function and mortality risk in men and women: findings from the Renfrew and Paisley prospective population study BMJ 1996313711–715.[Discussion, pp. 715–716.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Young RP, Hopkins R, Eaton TE. Forced expiratory volume in one second: not just a lung function test but a marker of premature death from all causes. Eur Respir J. 2007;30:616–622. doi: 10.1183/09031936.00021707. [DOI] [PubMed] [Google Scholar]
- 5.Schünemann HJ, Dorn J, Grant BJ, Winkelstein W, Jr, Trevisan M. Pulmonary function is a long-term predictor of mortality in the general population: 29-year follow-up of the Buffalo Health Study. Chest. 2000;118:656–664. doi: 10.1378/chest.118.3.656. [DOI] [PubMed] [Google Scholar]
- 6.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, et al. International Lung Cancer Consortium; GIANT consortium. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet. 2011;43:1082–1090. doi: 10.1038/ng.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herz J, Hamann U, Rogne S, Myklebost O, Gausepohl H, Stanley KK. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 1988;7:4119–4127. doi: 10.1002/j.1460-2075.1988.tb03306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lillis AP, Van Duyn LB, Murphy-Ullrich JE, Strickland DK. LDL receptor-related protein 1: unique tissue-specific functions revealed by selective gene knockout studies. Physiol Rev. 2008;88:887–918. doi: 10.1152/physrev.00033.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herz J, Clouthier DE, Hammer RE. LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell. 1992;71:411–421. doi: 10.1016/0092-8674(92)90511-a. [DOI] [PubMed] [Google Scholar]
- 10.Au DT, Arai AL, Fondrie WE, Muratoglu SC, Strickland DK. Role of the LDL receptor-related protein 1 in regulating protease activity and signaling pathways in the vasculature. Curr Drug Targets. 2018;19:1276–1288. doi: 10.2174/1389450119666180511162048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simone TM, Higgins SP, Archambeault J, Higgins CE, Ginnan RG, Singer H, et al. A small molecule PAI-1 functional inhibitor attenuates neointimal hyperplasia and vascular smooth muscle cell survival by promoting PAI-1 cleavage. Cell Signal. 2015;27:923–933. doi: 10.1016/j.cellsig.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wujak L, Markart P, Wygrecka M. The low density lipoprotein receptor-related protein (LRP) 1 and its function in lung diseases. Histol Histopathol. 2016;31:733–745. doi: 10.14670/HH-11-746. [DOI] [PubMed] [Google Scholar]
- 13.Strickland DK, Ashcom JD, Williams S, Burgess WH, Migliorini M, Argraves WS. Sequence identity between the alpha 2-macroglobulin receptor and low density lipoprotein receptor-related protein suggests that this molecule is a multifunctional receptor. J Biol Chem. 1990;265:17401–17404. [PubMed] [Google Scholar]
- 14.Mishra A, Yao X, Saxena A, Gordon EM, Kaler M, Cuento RA, et al. Low-density lipoprotein receptor-related protein 1 attenuates house dust mite-induced eosinophilic airway inflammation by suppressing dendritic cell-mediated adaptive immune responses. J Allergy Clin Immunol. 2018;142:1066–1079, e6. doi: 10.1016/j.jaci.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wujak L, Schnieder J, Schaefer L, Wygrecka M. LRP1: a chameleon receptor of lung inflammation and repair. Matrix Biol. 2018;68–69:366–381. doi: 10.1016/j.matbio.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 16.House JS, Li H, DeGraff LM, Flake G, Zeldin DC, London SJ. Genetic variation in HTR4 and lung function: GWAS follow-up in mouse. FASEB J. 2015;29:323–335. doi: 10.1096/fj.14-253898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westra HJ, Peters MJ, Esko T, Yaghootkar H, Schurmann C, Kettunen J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonder MJ, Luijk R, Zhernakova DV, Moed M, Deelen P, Vermaat M, et al. BIOS Consortium. Disease variants alter transcription factor levels and methylation of their binding sites. Nat Genet. 2017;49:131–138. doi: 10.1038/ng.3721. [DOI] [PubMed] [Google Scholar]
- 19.Barbaro MP, Spanevello A, Palladino GP, Salerno FG, Lacedonia D, Carpagnano GE. Exhaled matrix metalloproteinase-9 (MMP-9) in different biological phenotypes of asthma. Eur J Intern Med. 2014;25:92–96. doi: 10.1016/j.ejim.2013.08.705. [DOI] [PubMed] [Google Scholar]
- 20.Cataldo D, Munaut C, Noël A, Frankenne F, Bartsch P, Foidart JM, et al. MMP-2- and MMP-9-linked gelatinolytic activity in the sputum from patients with asthma and chronic obstructive pulmonary disease. Int Arch Allergy Immunol. 2000;123:259–267. doi: 10.1159/000024452. [DOI] [PubMed] [Google Scholar]
- 21.Polverino E, Rosales-Mayor E, Dale GE, Dembowsky K, Torres A. The role of neutrophil elastase inhibitors in lung diseases. Chest. 2017;152:249–262. doi: 10.1016/j.chest.2017.03.056. [DOI] [PubMed] [Google Scholar]
- 22.Bégin P, Tremblay K, Daley D, Lemire M, Claveau S, Salesse C, et al. Association of urokinase-type plasminogen activator with asthma and atopy. Am J Respir Crit Care Med. 2007;175:1109–1116. doi: 10.1164/rccm.200607-1012OC. [DOI] [PubMed] [Google Scholar]
- 23.Waschki B, Watz H, Holz O, Magnussen H, Olejnicka B, Welte T, et al. Plasminogen activator inhibitor-1 is elevated in patients with COPD independent of metabolic and cardiovascular function. Int J Chron Obstruct Pulmon Dis. 2017;12:981–987. doi: 10.2147/COPD.S128689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.May P, Herz J. LDL receptor-related proteins in neurodevelopment. Traffic. 2003;4:291–301. doi: 10.1034/j.1600-0854.2003.00086_4_5.x. [DOI] [PubMed] [Google Scholar]
- 25.Yue X, Guidry JJ. Differential protein expression profiles of bronchoalveolar lavage fluid following lipopolysaccharide-induced direct and indirect lung injury in mice. Int J Mol Sci. 2019;20:3401. doi: 10.3390/ijms20143401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo Y, Ma SF, Grigoryev D, Van Eyk J, Garcia JG. 1-DE MS and 2-D LC-MS analysis of the mouse bronchoalveolar lavage proteome. Proteomics. 2005;5:4608–4624. doi: 10.1002/pmic.200500052. [DOI] [PubMed] [Google Scholar]
- 27.Pounds JG, Flora JW, Adkins JN, Lee KM, Rana GS, Sengupta T, et al. Characterization of the mouse bronchoalveolar lavage proteome by micro-capillary LC-FTICR mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;864:95–101. doi: 10.1016/j.jchromb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 28.Schiller HB, Fernandez IE, Burgstaller G, Schaab C, Scheltema RA, Schwarzmayr T, et al. Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol Syst Biol. 2015;11:819. doi: 10.15252/msb.20156123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wattiez R, Falmagne P. Proteomics of bronchoalveolar lavage fluid. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:169–178. doi: 10.1016/j.jchromb.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 30.Bown MJ, Jones GT, Harrison SC, Wright BJ, Bumpstead S, Baas AF, et al. CARDIoGRAM Consortium; Global BPgen Consortium; DIAGRAM Consortium; VRCNZ Consortium. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011;89:619–627. doi: 10.1016/j.ajhg.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR, et al. Understanding Society Scientific Group; International Consortium for Blood Pressure; Blood Pressure-International Consortium of Exome Chip Studies; Million Veteran Program. Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Nat Genet. 2019;51:51–62. doi: 10.1038/s41588-018-0303-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCall MN, Illei PB, Halushka MK. Complex sources of variation in tissue expression data: analysis of the GTEx lung transcriptome. Am J Hum Genet. 2016;99:624–635. doi: 10.1016/j.ajhg.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.House JS, Nichols CE, Li H, Brandenberger C, Virgincar RS, DeGraff LM, et al. Vagal innervation is required for pulmonary function phenotype in Htr4-/- mice. Am J Physiol Lung Cell Mol Physiol. 2017;312:L520–L530. doi: 10.1152/ajplung.00495.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lao T, Jiang Z, Yun J, Qiu W, Guo F, Huang C, et al. Hhip haploinsufficiency sensitizes mice to age-related emphysema. Proc Natl Acad Sci USA. 2016;113:E4681–E4687. doi: 10.1073/pnas.1602342113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Z, Lao T, Qiu W, Polverino F, Gupta K, Guo F, et al. A chronic obstructive pulmonary disease susceptibility gene, FAM13A, regulates protein stability of β-Catenin. Am J Respir Crit Care Med. 2016;194:185–197. doi: 10.1164/rccm.201505-0999OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima C, Haffner P, Goerke SM, Zurhove K, Adelmann G, Frotscher M, et al. The lipoprotein receptor LRP1 modulates sphingosine-1-phosphate signaling and is essential for vascular development. Development. 2014;141:4513–4525. doi: 10.1242/dev.109124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J. LRP: role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–332. doi: 10.1126/science.1082095. [DOI] [PubMed] [Google Scholar]
- 38.Hu L, Boesten LS, May P, Herz J, Bovenschen N, Huisman MV, et al. Macrophage low-density lipoprotein receptor-related protein deficiency enhances atherosclerosis in ApoE/LDLR double knockout mice. Arterioscler Thromb Vasc Biol. 2006;26:2710–2715. doi: 10.1161/01.ATV.0000249641.96896.e6. [DOI] [PubMed] [Google Scholar]
- 39.Staudt ND, Jo M, Hu J, Bristow JM, Pizzo DP, Gaultier A, et al. Myeloid cell receptor LRP1/CD91 regulates monocyte recruitment and angiogenesis in tumors. Cancer Res. 2013;73:3902–3912. doi: 10.1158/0008-5472.CAN-12-4233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Au DT, Ying Z, Hernández-Ochoa EO, Fondrie WE, Hampton B, Migliorini M, et al. LRP1 (low-density lipoprotein receptor-related protein 1) regulates smooth muscle contractility by modulating Ca2+ signaling and expression of cytoskeleton-related proteins. Arterioscler Thromb Vasc Biol. 2018;38:2651–2664. doi: 10.1161/ATVBAHA.118.311197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Xiao W, Jiang Y, Wang H, Xu X, Ma D, et al. Levels of components of the urokinase-type plasminogen activator system are related to chronic obstructive pulmonary disease parenchymal destruction and airway remodelling. J Int Med Res. 2012;40:976–985. doi: 10.1177/147323001204000316. [DOI] [PubMed] [Google Scholar]
- 42.Yao X, Remaley AT, Levine SJ. New kids on the block: the emerging role of apolipoproteins in the pathogenesis and treatment of asthma. Chest. 2011;140:1048–1054. doi: 10.1378/chest.11-0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brooks AM, Bates ME, Vrtis RF, Jarjour NN, Bertics PJ, Sedgwick JB. Urokinase-type plasminogen activator modulates airway eosinophil adhesion in asthma. Am J Respir Cell Mol Biol. 2006;35:503–511. doi: 10.1165/rcmb.2006-0113OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamashita CM, Fessler MB, Vasanthamohan L, Lac J, Madenspacher J, McCaig L, et al. Apolipoprotein E-deficient mice are susceptible to the development of acute lung injury. Respiration. 2014;87:416–427. doi: 10.1159/000358438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damani SB, Topol EJ. Emerging genomic applications in coronary artery disease. JACC Cardiovasc Interv. 2011;4:473–482. doi: 10.1016/j.jcin.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Damiao Gouveia AC, Skovman A, Jensen A, Koponen IK, Loft S, Roursgaard M, et al. Telomere shortening and aortic plaque progression in Apoliprotein E knockout mice after pulmonary exposure to candle light combustion particles. Mutagenesis. 2018;33:253–261. doi: 10.1093/mutage/gey015. [DOI] [PubMed] [Google Scholar]
- 47.Christophersen DV, Jacobsen NR, Andersen MH, Connell SP, Barfod KK, Thomsen MB, et al. Cardiovascular health effects of oral and pulmonary exposure to multi-walled carbon nanotubes in ApoE-deficient mice. Toxicology. 2016;371:29–40. doi: 10.1016/j.tox.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Han SG, Howatt DA, Daugherty A, Gairola CG. Atherogenic and pulmonary responses of ApoE- and LDL receptor-deficient mice to sidestream cigarette smoke. Toxicology. 2012;299:133–138. doi: 10.1016/j.tox.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 49.Dellas C, Schremmer C, Hasenfuss G, Konstantinides SV, Schäfer K. Lack of urokinase plasminogen activator promotes progression and instability of atherosclerotic lesions in apolipoprotein E-knockout mice. Thromb Haemost. 2007;98:220–227. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.