Abstract
Immunoglobulins and T cell receptors (TCR) have obvious structural similarities as well as similar immunogenetic diversification and selection mechanisms. Nevertheless, the two receptor systems and the loci that encode them are distinct in humans and classical murine models, and the gene segments comprising each repertoire are mutually exclusive. Additionally, while both B and T cells employ recombination-activating genes (RAG) for primary diversification, immunoglobulins are afforded a supplementary set of activation-induced cytidine deaminase (AID)-mediated diversification tools. As the oldest-emerging vertebrates sharing the same adaptive B and T cell receptor systems as humans, extant cartilaginous fishes allow a potential view of the ancestral immune system. In this review, we discuss breakthroughs we have made in studies of nurse shark (Ginglymostoma cirratum) T cell receptors demonstrating substantial integration of loci and diversification mechanisms in primordial B and T cell repertoires. We survey these findings in this shark model where they were first described, while noting corroborating examples in other vertebrate groups. We also consider other examples where the gnathostome common ancestry of the B and T cell receptor systems have allowed dovetailing of genomic elements and AID-based diversification approaches for the TCR. The cartilaginous fish seem to have retained this T/B cell plasticity to a greater extent than more derived vertebrate groups, but representatives in all vertebrate taxa except bony fish and placental mammals show such plasticity.
Keywords: Immunoglobulins, T cell receptors, evolution: antigen receptor loci, activation induced cytidine deaminase, Shark, Vertebrate adaptive immune system, Bony fish, Placental mammals
Introduction
The vertebrate adaptive immune system (AIS) activates both humoral and cell-mediated responses against invading pathogens (Murphy and Weaver 2017), protecting the host from a multitude of potential pathogens over a lifetime. Pivotal to this function is the creation of a diverse repertoire of lymphocyte antigen receptors created by the assembly of gene segments into complete genes during lymphocyte development (Tonegawa 1983). In all jawed vertebrates including sharks, primary diversification of B and T lymphocyte receptors occurs during recombination-activating genes (RAG)-mediated somatic recombination of variable (V), diversifying (D), and joining (J) gene segments within primary lymphoid tissues. However, the V, D, and J gene segments themselves and the mechanisms of V(D)J recombination are nearly the same between B and T lymphocytes and demonstrate a shared origin in the primordial system (Ohta et al. 2019). In fact, herein, we review that T cells are adept at creating novel, diverse receptors by capitalizing on the accessibility of immunoglobulin heavy chain (IgH), IgH-like, and T cell receptor (TCR) V gene segments available to them. Chondrichthyes, which are the oldest evolutionary group of vertebrates with immunoglobulin superfamily-based lymphocyte antigen receptors, diversify their TCR repertoires by forming non-canonical TCR that incorporate both T and B cell receptor components. These unique TCRs are constructed by (1) recombining distinctly IgH V (and often one or two IgH D) gene segments with TCR D and TCR J gene segments (IgH-TCR trans-rearrangements), (2) incorporating unique IgH-like V gene segments from within the TCRαδ locus (TCR-associated Ig-like V, or TAILV), or (3) combining two V domains—one that includes IgH-like V, D, and J gene segments and the other that includes TCRδ V, D, and J gene segments (NAR-TCR). While most of our work has been done with the nurse shark, G. cirratum, wherever examined, these mechanisms have extended to all other cartilaginous fish, both Elasmobranchs and Holocephalans. While vestiges and sometimes convergence of these more cohesive systems of gene segment usage can be found in other classes of jawed vertebrates (e.g., VHδ gene segments; TCRμ locus), nurse sharks (and likely all cartilaginous fish) are unusual in that they also commandeer activation-induced cytidine deaminase (AID) to catalyze somatic hypermutation (SHM) of TCR α (and other TCR chains) to further diversify their developing TCR repertoire in the thymus.
Here, we begin with a brief overview of TCR and IgH locus organization, RAG-mediated somatic recombination, and thymic development of canonical αβTCR and γδTCR. We then discuss the unconventional (non-canonical) TCR observed specifically in nurse sharks and generally in other gnathostome vertebrates. Finally, we examine SHM as a TCR repertoire diversifying mechanism in nurse sharks and explore the use of SHM by T cells of other vertebrates (e.g., camelids). We end by proposing a model to explain how AID-mediated SHM is used to salvage TCR to facilitate selection, specifically by altering the TCR α chain.
Nurse sharks (like mammals) rearrange canonical TCR chains during thymocyte development
Most functional B cell receptors (BCR or immunoglobulin, Ig) are composed of a heterodimer of two protein chains—a heavy chain (IgH) and a light chain (IgL), and each IgH or IgL is composed of a variable (V) region that contains an antigen (Ag)-binding site and a constant (C) region that identifies the isotype. Additionally, all jawed vertebrates studied have four canonical T cell receptor (TCR) chains (α, β, γ, δ) and typically pair α chain with β chain to form αβ TCR and γ chain with δ chain to form γδ TCR. Both TCR types occur only as transmembrane proteins on the surface of T cells (Chien et al. 1987). A BCR isotype is defined by its H chain and can occur as either a membrane-bound receptor or a secreted antibody (Ab) protein. In humans, there are five IgH isotypes in mammals: Igμ (IgM), Igδ (IgD), Igγ (IgG), Igα (IgA), and Igε (IgE) (Murphy and Weaver 2017; Flajnik 2018). Only two of the conventional isotypes discovered in gnathostomes are found in sharks, IgM and an IgD-like isotype called IgW (Ohta and Flajnik 2006; Zhu et al. 2012).
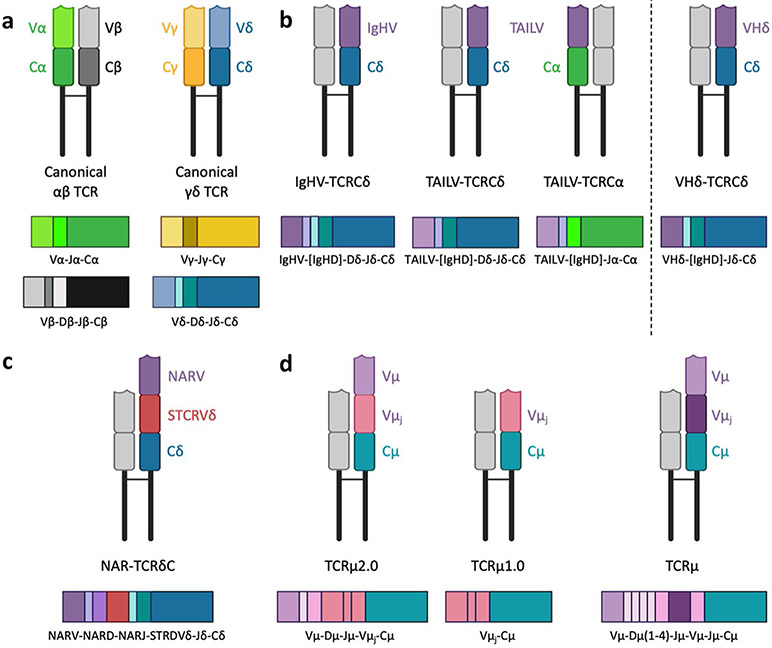
During lymphocyte development in primary lymphoid tissues, both B and T cells employ recombination activating genes (RAG1/RAG2) to assemble complete BCR and TCR variable region exons from V, (D), and J gene segments. Rearrangement is directed by recombination signal sequences (RSS) adjacent to each gene segment that guide RAG binding to the correct location and gene segment. B cells develop within bone marrow (or analogous primary tissue like epigonal or Leydig organ in sharks), while T cells develop within the thymus (Gellert 2002). Variable regions of IgH and TCR β and δ chains contain rearranged V, D, and J gene segments while those of Ig light chains (IgL) and TCR α and γ chains contain rearranged V and J gene segments only (Fig. 1a). The V gene segment encodes three of the four framework regions (FR) and the first two complementarity-determining regions (CDR) of the assembled chain. The V(D)J junction, located between the V and J segments of IgL, TCRα, and TCRγ chains or the V, D, and J segments of IgH, TCRβ, and TCRδ chains, encodes the third complementarity-determining region (CDR). The C-terminal part of the J gene segment forms the fourth FR (Tonegawa 1983; Gellert 2002; Lefranc et al. 2003; Lefranc 2014). Once assembled, each V gene encodes a domain that folds to form a nine β-strand support structure (composed of the FR) for the Ag-binding loops (CDR) at the membrane-distal end of the receptor (Kikutani et al. 1986). In a complete TCR, Ag specificity is determined by these six CDR loops (three from TCRβ or TCRδ and three from TCRα or TCRγ, respectively) that form a single paratope (Tonegawa 1983; Jack and Du Pasquier 2019). These same six CDR loops (three each from IgH and IgL) form the Ag-binding region in Igs, though the bivalent receptor can bind two antigens simultaneously. While γδ T cells generally bind free Ag in a manner similar to B cells (although there are many other types of binding (Hayday and Vantourout 2020), conventional αβ T cells typically are restricted to binding peptide Ag in complex with the major histocompatibility complex (MHC) (Jack and Du Pasquier 2019).
Fig. 1.
Cartoon depictions of putative assembled T cell receptors (TCR, top of each panel) and transcripts (bottom of each panel) illustrate how vertebrates refashion canonical TCR by incorporating immunoglobulin heavy chain (IgH) variable (V) gene segments. a Canonical αβTCR (alpha chain: α, green; beta chain: β, black) and γδTCR (gamma chain: γ, gold; delta chain: δ, blue) are composed of typical V, (D), and J gene segments; b non-canonical TCR replace Vδ (or Vα) with IgH or IgH-like V regions (purple) to form unique TCR chains [L to R: IgHV gene segments associate with nurse shark TCR Cδ (and rarely TCR Cα); TAILV gene segments, unique to nurse sharks, associate with both TCR Cδ and TCR Cα; and IgH-like Vδ (VHδ) gene segments are found in genomes of all gnathostome vertebrate groups except teleost fish and eutherian mammals (but not nurse sharks); c doubly rearranging NAR-TCR, also unique to cartilaginous fish, are composed of two variable domains that undergo separate RAG-mediated VDJ recombination events—a membrane-distal IgNAR-like V domain (NARV, purple) supported by a membrane-proximal TCR Vδ domain (STCRVδ, red)—associated with TCR Cδ; and d TCRμ, found in monotreme and marsupial mammals, combine IgH-like V gene segments (Vμ, light purple) with TCRδ-like C regions (Cμ, teal). Opossums express two isoforms of the receptor: a long form (TCRμ2.0) containing two variable domains—a membrane-distal domain formed by RAG-recombined Vμ, Dμ, and Jμ gene segments and an invariant, membrane-proximal Vμj domain that is pre-joined in the germline (dark pink); and a short form (TCRμ1.0) composed of a single invariant Vμj domain. Platypus express a single TCRμ isoform containing two variable domains that each undergoes a separate recombination event. Transcripts demonstrate the variable use of IgH and TCR D gene segments by non-canonical receptors [V: variable, D: diversifying, and J: joining gene segments; C: constant region; TAILV: TCR-associated Ig-like V; NAR: nurse shark (or new) antigen receptor; RAG: recombination activating genes]. All Ig and Ig-like V, D, and J gene segments are colored in shades of pink/purple. Figure created with BioRender.com
While ancestral, non-rearranging TCR and Ig genes likely occurred within a single locus linked to prototypic MHC genes within a pre-vertebrate primordial immune complex, one of these immune genes was invaded by the RAG transposon in an early gnathostome after the genome-wide duplications (Zhang et al. 2019). Our recent model suggests that this transposon-invaded immune gene underwent duplication, neofunctionalization, and translocation events to fashion the immune loci of extant vertebrates (Ohta et al. 2019). Presently, in the human example, genes and gene segments of IgH and IgL chains and TCR β and TCR γ chains each are encoded by separate loci, while TCR δ is embedded within the TCR α locus. As a result, rearrangement of TCR α chain deletes the embedded TCR δ locus. In most jawed vertebrates, loci are organized as discontiguous translocons, with numerous V, (D), and J gene segments preceding constant (C) region exons (Vn-Dn-Jn-C) that can stretch up to 3 Mbp in length (Tonegawa 1983; Flajnik and Rumfelt 2000; Gellert 2002; Schatz 2004; Criscitiello and Flajnik 2007; Hsu 2018). For rearrangement to occur, DNA must undergo conformational changes that permit chromatin to fold and bring segments together (Jhunjhunwala et al. 2009). However, the loci of some organisms (e.g., shark IgH and bony fish IgL) are organized as multiple clusters of V, (D), and J gene segments and C region exons (V-D-J-C)n, creating a different genomic environment for sequential rearrangement of loci and gene segments (Dooley and Flajnik 2006; Hsu and Criscitiello 2006; Hsu 2009, 2018).
Research in mouse and human models demonstrates that both B cells and αβ T cells rearrange and assemble Ag-binding receptors in similar ways during their development (though developing T cells in the subcapsular region of the thymus also rearrange γ and δ chains simultaneously with TCR β chain). Generally, IgH and TCRβ chain loci first combine D and J gene segments and then join V segments to the recombined DJ. The successful creation of a functional IgH or TCRβ chain halts RAG expression and gene rearrangement, and the cell undergoes a clonal expansion. Cells then express RAG again during rearrangement of V to J segments in IgL and TCRα (Bassing et al. 2002; Murphy and Weaver 2017).
Rearrangement of IgH, IgL, and TCR γ, δ, and β chains is regulated, in part, by allelic exclusion, which (by definition) permits only one allelic copy of a locus to be expressed at the surface of a cell, ensuring that each cell recognizes only a single ligand [reviewed in Brady et al. (2010)]. Locus rearrangement and expression of the first allele thus inhibit rearrangement of the second (Gascoigne and Alam 1999; Brady et al. 2010). The exception is TCRα and IgLκ, which can rearrange the loci of both alleles simultaneously. Developing B cells that produce autoreactive receptors, undergo unsuccessful IgL rearrangements, or produce an IgL that cannot associate with the IgH chain can undergo receptor editing, rearranging both alleles of the IgL chain locus multiple times until a productive arrangement is made or the cell undergoes apoptosis (McGargill et al. 2000; Schatz 2004; Kuklina 2006). Developing T cells that fail to produce a functional TCRα chain, or a TCRα chain that cannot associate with the TCRβ chain, or produce a TCRα chain that cannot be positively selected when associated with the TCRβ chain also can undergo receptor editing, rearranging at both TCR α alleles many times until a useful arrangement is made or the cell undergoes apoptosis (Borgulya et al. 1992; Livak and Schatz 1996; Kondo et al. 2019). We note that the very high numbers of J segments at the TCRα locus, which can allow for very high levels of receptor editing, is evolutionarily conserved and found in all vertebrates so far studied.
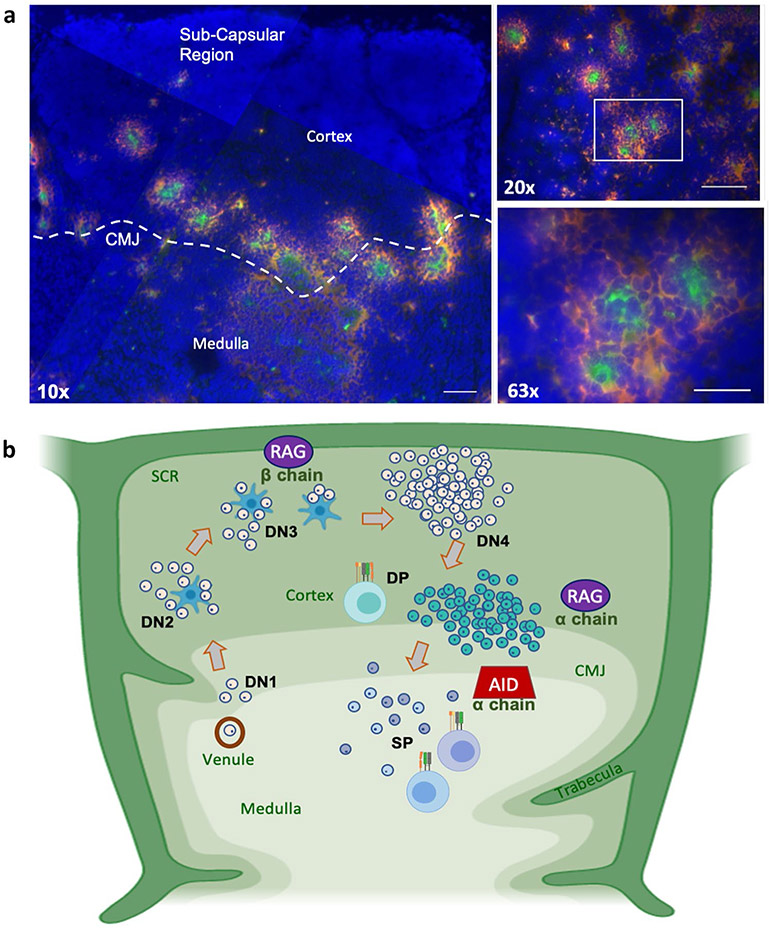
TCR gene rearrangement occurs as thymocytes develop within the thymus. In sharks, the thymus is bilaterally located dorsomedial to the gill arches and arranged as discrete lobules separated by trabeculae (Fig. 3). Similar to the architecture of human thymus, each lobule consists of a large outer cortical region containing densely packed immature thymocytes and branched cortical epithelial cells and a smaller interior medullary region of loosely packed mature thymocytes, medullary epithelial cells, macrophages, and dendritic cells (Luer et al. 1995). The junction between the cortex and medulla is called the cortico-medullary junction (CMJ) and the outer region of the cortex is called the subcapsular region (Luer et al. 1995; Criscitiello et al. 2010; Murphy and Weaver 2017). While the exact pathway that developing thymocytes take through the shark thymus is unknown, studies in mouse and human illustrate that stages of thymocyte development correlate with αβ TCR gene rearrangement and expression of key proteins on the T cell. In sharks, expressions of TCR αβ, MHC I/II, RAG, and TdT appear conserved with mammals where TCR γδ expression appears different (Criscitiello et al. 2010), though a detailed chronology of developmental checkpoints is still lacking in sharks (Germain 2002; Kuo and Schlissel 2009; Murphy and Weaver 2017).
Fig. 3.
In nurse sharks, AID is proposed to catalyze somatic hypermutation (SHM) of TCR alpha chain genes (α chain) during the late double-positive stage of thymocyte development, likely producing new TCR paratopes capable of passing thymic selection. a Single-molecule RNA fluorescence in situ hybridization (FISH) probing fixed shark thymus sections simultaneously for AID (probes labeled with Quasar 670; pseudo colored red) and TCR α chain (probes labeled with CalFluor Red 610; pseudo colored green) and counterstained with DAPI (blue). TCRα is highly expressed within thymic cortical regions near the cortico-medullary junction (CMJ), suggesting that late TCR α chain gene rearrangement is occurring here. Coincidently, cells expressing high levels of AID encircle groups of cells expressing high levels of the TCR α chain, suggesting that AID is involved in SHM of the α chain V region during late stages of positive selection or early stages of negative selection (Ott et al. 2018) (scale bars: 150 μm, 75 μm, and 30 μm at 10×, 20×, and 63× magnification, respectively. White box indicates the magnified regions of the 20× image shown in the 63× image). b Theoretical model illustrating putative rearrangement of TCR beta chain (β chain) and α chain in nurse shark thymus, based on what is known in mammals. CD4/CD8 double negative (DN) thymocytes utilize RAG to rearrange β chain in the sub-capsular region (SCR) and outer cortex. Cells with productive β chain arrangements then proliferate, expressing both CD4 and CD8 as double positive (DP) thymocytes. The strong distinction between small thymocytes in the cortex and larger cells in the medulla has been shown in sharks, as well as RAG and TCR β expression in the sub-capsular regions (Criscitiello et al. 2010). As DP thymocytes move toward the inner cortex and CMJ where RAG-mediated α chain re-rearrangement (editing) has been shown to occur in mice, shark (but not mouse) thymocytes begin to express AID, rescuing nonproductive receptor rearrangements from apoptosis through receptor editing and/or receptor salvaging via SHM. In the latter case, AID-catalyzed SHM can produce TCR with improved affinity to MHC: Ag complexes (to pass positive selection) or reduce recognition of selfpeptide, rescuing self-reactive thymocytes from apoptosis (to pass negative selection). Salvaged thymocytes then express either CD4 or CD8 on their surface as single-positive (SP) cells [AID: activation-induced cytidine deaminase; RAG: recombination activating genes]. Figure 3 b created with BioRender.com
In mammals, double positive (DP, expressing both CD8+ and CD4+ co-receptors) thymocytes migrate towards the inner cortex as a second wave of RAG activity rearranges the α chain V to J gene segments. RAG expression continues to mediate rearrangement of the α locus until an MHCcompatible receptor (i.e., a receptor poised to recognize antigen presented by MHC alleles of the individual) is rearranged or the cell dies, which happens to the vast majority of thymocytes. DP cells that successfully recognize self-MHC class I or class II “pass” positive selection and mature to express either CD8 or CD4, respectively, becoming CD8+ or CD4+ single positive (SP) thymocytes. TCRs also are tested for strong self-recognition (negative selection) during both DP and SP stages, eliminating cells that react to self Ag. In mouse thymus, only about 2% of thymocytes survive selection mechanisms in the cortex to become mature T cells that enter the medulla and exit the thymus to form the peripheral T cell repertoire. Thus, the cortex contains immature thymocytes actively rearranging and testing their receptor loci, and the medulla contains mature naíve CD8 or CD4 SP T cells postrecombination and selection poised to emigrate from the thymus. While a chronology of T cell movements through the shark thymus is unknown, we assume for this review that events occur in a similar fashion in sharks as it does for mammals (e.g., Fig. 8.21 of Janeway’s Immunobiology demonstrates the thymocyte developmental stages in mice and humans) (Murphy and Weaver 2017). In contrast to the MHC-restricted αβ T cells, both γ and δ chains of γδ T cells undergo receptor gene rearrangement simultaneously with β locus rearrangement during the double negative (DN, lacking both CD8+ and CD4+ co-receptors) stages 2 and 3 (DN2/DN3, respectively) of thymocyte development. Signal strength from the γδ receptor during the DN3 stage instructs αβ or γδ T cell lineage fate, with strong signaling promoting the γδ T cell line while weak TCR signaling favoring commitment to the αβ T cell line (Lafaille et al. 1990; Kreslavsky et al. 2010; Fahl et al. 2014). At the DN3 stage, three of the four T cell loci (β, γ, and δ) have undergone rearrangement. Cells that successfully express TCRβ and lack a strong γδ signal undergo proliferation, upregulate CD4 and CD8 co-receptors, cease TCRγ rearrangement, and ultimately rearrange the TCRα loci, resulting in the deletion of TCRδ genetic components from the locus. Progression to the CD4+/CD8+ DP stage commits cells to the αβ T cell lineage (Kreslavsky et al. 2010). However, cells that rearrange TCRγ and TCRδ loci successfully express γδ TCR at the surface, stimulating clonal proliferation but do not progress to the DP stage and thus emerge from the thymus committed to the γδ lineage (Kreslavsky et al. 2010). γδ T cells remain DN as mature thymocytes and express neither CD8 nor CD4 co-receptors. Further, because γδ T cells are not MHC-restricted, they likely do not undergo the same positive or negative selection processes during development as αβ T cells. The localization of shark γδ in the subcapsular region is not conserved with mammalian localization, but is consistent with the picture also emerging from bony fish (Criscitiello et al. 2010; Aghaallaei and Bajoghli 2018).
While RAG mediates receptor gene recombination, AID triggers BCR and antibody diversification through somatic hypermutation (SHM), class-switch recombination (CSR), and immunoglobulin gene conversion (IGC) events in humoral adaptive immunity (Muramatsu et al. 2000; Arakawa et al. 2002). AID is a member of the much larger AID/APOBEC (apolipoprotein B RNA-editing catalytic component) family of zinc-dependent deaminases (Liu et al. 2018). While zinc-dependent deaminases are found in nearly all life forms on Earth (including bacteria, archaea, yeast, plants, and animals), the ancestral APOBEC emerged at the beginning of the vertebrate radiation, with the appearance of AID coinciding with the evolution of RAG-mediated immunoglobulin-superfamily adaptive immunity and the divergence of cartilaginous fish (Flajnik 2002; Conticello et al. 2005). APOBEC2 genes, another ancestral member of the APOBEC family, are found in other vertebrates including bony fish but has not been found in sharks (Conticello et al. 2007). AID targets the ssDNA that is exposed during transcription of Ig loci in the nucleus, catalyzing the deamination of cytidine to uridine within the variable regions of lymphocyte antigen receptors. The presence of uridine in DNA creates a mismatch between guanidine and uridine, which activates DNA repair mechanisms (i.e., mismatch repair, base-excision repair) to correct the mismatch. B cells are capable of manipulating these pathways; so, the repair is less effective at Ig loci, resulting in the substitution of non-template bases at the affected site (Maul and Gearhart 2010; Álvarez-Prado et al. 2018).
In the T cell-dependent, antigen-driven immune responses of most jawed vertebrates, SHM is used to alter the affinity of BCR to Ag during affinity maturation. After a naive B cell is exposed to Ag, it is stimulated to proliferate within peripheral lymphoid tissues. In mammals and birds, activated B cells develop within germinal centers (GC) in B cell follicles within spleen, tonsils, Peyer’s patches, and (in mammals) lymph nodes [(Good and Finstad 1966); reviewed in (MacLennan 1994; Flajnik 2002)]. Affinity maturation occurs in a stepwise manner that repeatedly selects modified BCR with improved binding to the original Ag. Mutation is biased towards transitions and is targeted to particular motifs within variable region nucleotide sequences, focusing replacement mutation to particular hotspots of AID activity, particularly G and C residues within DGWY and WRCH motifs [where D is adenosine (A), guanosine (G), or thymidine (T); Y is cytosine (C) or T; W is A or T; and R is A or G]. There are intrinsic differences in codon use in CDR compared with FR, where CDR favor codons without wobble bases to favor amino acid replacement (Chang and Casali 1994). Further, an abundance of these motifs within CDR concentrates mutation within the Ag-binding regions of the structure, thereby improving humoral immunity (Muramatsu et al. 2000; Odegard and Schatz 2006; Saini and Hershberg 2015; Álvarez-Prado et al. 2018).
However, reptiles, amphibians, and fish do not form GC, and B cells develop within lymphocyte-rich follicles of splenic white pulp or (in teleost fish) melanomacrophage clusters of liver and kidney (Zapata et al. 1981; Rumfelt et al. 2002; Zimmerman et al. 2010; Magor 2015; Rios and Zimmerman 2015; Neely et al. 2018). They do, however, employ somatic hypermutation and a certain level of selection for higher affinity antibodies. SHM is well described in the nurse shark B cell receptors (Diaz and Flajnik 1998; Diaz et al. 2001; Lee et al. 2002) and affinity maturation has been confirmed at the sequence level, biochemically and structurally (Dooley and Flajnik 2005; Dooley et al. 2006b). Despite the clear use of SHM for some level of affinity maturation in ectotherms, there still is much to be determined of the anatomy and physiology of these selection processes in lower vertebrates. Regardless, until recently, the consensus by most immunologists was that T cells did not employ SHM at all.
Nurse sharks generate unconventional TCR incorporating IgH or IgH-like V gene segments
Nurse sharks construct distinct IgH-TCR chimeric isoforms by rearranging IgH V gene segments to a TCR constant region (C), thereby enhancing diversity of the TCR repertoire. We first identified unusual transcripts in nurse sharks that recombine IgM or IgW (as mentioned, similar to IgD) V gene segments to TCRδ (or rarely, TCRα) C regions (Criscitiello et al. 2010). The IgH V gene segments used by TCR are genetically indistinguishable from those used by BCR and consequently, are presumed to be from the conventional Ig loci (Fig. 1b). However, the lack of an assembled genome or complete Ig/TCR loci in nurse shark complicates our complete understanding of the genomic origin of these IgHV gene segments. Whether IgHV associated with TCR are located within the conventional TCRαδ locus (cis-chromosomal rearrangements), the conventional Ig locus (trans-locus rearrangements), or in a separate locus altogether (“trans” rearrangements) remains unclear, but we do know IgH clusters exist near the TCRαδ locus in nurse shark (Criscitiello et al. 2010; Venkatesh et al. 2014; Deiss et al. 2019). We recently found that nurse shark TCR utilized at least five different IgM and three different IgW V segment groups, and importantly, expression of chimeric IgHV-TCRδ chains was comparable with, or even exceeded, expression of canonical TCR chains (Ott et al. 2020). Further, sharks produce functional chimeric TCR from the IgMV-2C group which, when used by BCR, produces nonfunctional receptors due to defective Ig constant region exons. We retain the IgHV designations here when referring to these gene segments to avoid confusion. However, we acknowledge that these gene segments may require distinct names once the nurse shark genome is complete if they are indeed embedded within both Ig and TCR loci.
Partial assembly of the TCRδ locus uncovered unique Ig-like V gene segments nestled within the TCRαδ translocon that group with IgH V phylogenetically, and mRNA transcripts indicate these V gene segments (termed TCRδ-associated Ig-like V, or TAILV) are used with TCRδ (or TCRα) C regions but not with BCR C regions (Deiss et al. 2019). The presence of Ig-like TAILV within the TCRαδ locus of nurse sharks (and VHδ gene segments in a number of vertebrate lineages) suggests that T cell assimilation of both Ig and TCR V gene segments into functional TCR was the ancestral state at the genesis of the IgSF-based adaptive immune system (more below).
Complex receptors evolved convergently in sharks and mammals
Perhaps the most complex TCR isoform in sharks is the doubly rearranging NAR-TCR, composed of two V domains (each undergoing a separate VDJ recombination event) and a TCRδ C domain (Criscitiello et al. 2006) (Fig. 1c). The membrane-distal V domain (NARV domain) is closely related to IgNAR (variably called “nurse shark antigen receptor” and “new antigen receptor”), a distinct IgH isotype found only in cartilaginous fish that does not associate with light chain (Greenberg et al. 1995; Criscitiello et al. 2006). The NARV domain is supported by a membrane proximal TCRδ variable domain (STCRδV) that is assembled from distinct TCRδ V gene segments (that have lost their leader exons) rearranged to the canonical TCRδ D and J gene segments (Criscitiello et al. 2006). A draft assembly of the nurse shark TCRαδ locus identified tandem blocks of NARV V, D, and J gene segments located in a separate stretch of the TCRαδ translocon from the canonical TCRδ V-D-J gene segments, and the elephant shark confirms NAR-TCR genomically in more primitive cartilaginous fish (Venkatesh et al. 2007). Within each block, NARV VDJ are located upstream of an apparently dedicated STCR δV gene segment (Deiss et al. 2019). NAR-TCR is hypothesized to partner with TCRγ chain to form an MHC-unrestricted receptor. The resulting receptor consists of a protruding NARV domain that sits atop a base formed by the γ and δ TCR chains, with only the NARV CDRs constructing the predicted antigenbinding site of the receptor (Criscitiello et al. 2006).
The near simultaneous discovery of a unique TCR locus (TCRμ) in monotreme and marsupial mammals further blurred the distinction between B and T cell receptor components (Parra et al. 2007; Wang et al. 2011). In opossum (Monodelphis domestica), the TCRμ locus is found on a separate chromosome from conventional TCR loci and is atypically organized as tandem clusters of Vμ, Dμ, and Jμ gene segments followed by a Cμ exon (Parra et al. 2007). In addition, an exon encoding a complete V domain, with rearranged VDJ gene segments already joined together in germline DNA (Vμj), is found between the Jμ and Cμ of each cluster. TCRμ expresses two functional transmembrane isoforms (Fig. 1d). The short form, TCRμ1.0, encodes a receptor chain composed of a single Vμj domain and Cμ, forming an invariant binding site that is structurally more similar to conventional TCR (Parra et al. 2007). The long form (TCRμ2.0, the dominant isoform in peripheral lymphoid tissues) encodes a receptor chain containing two V domains and Cμ and is structurally analogous to the NAR-TCR of sharks (Parra et al. 2007). The membrane-distal V of TCRμ2.0 is formed by RAG-recombined V, D, and J gene segments that incorporate junctional diversity within the V domain, whereas the membrane-proximal V is always a (pre-joined) Vμj exon that forms an invariant V domain (Parra et al. 2007). The two V domains are linked through a mRNA splice site in the Vμj leader sequence that splices the recombined VDJ of the membrane-distal V to FR 1 of the membrane-proximal V (Parra et al. 2007). V gene segments of both variable domains (membrane-distal V domain of TCRμ2.0 and the sequence corresponding to FR1 through FR3 of Vμj in both isoforms) are phylogenetically more similar to IgH V gene segments (VH) while Cμ was derived from a TCRθ ancestor (Parra et al. 2007, 2008).
Like that of opossum, the monotreme platypus (Ornithorhynchus anatinus) TCRμ locus occurs in a separate location from conventional TCR genes, but platypus express only a single TCRμ isoform composed of two V domains that each somatically rearrange V, D, and J gene segments (Wang et al. 2011). The membrane-distal V domain (V1) rearranges two to four Dμ gene segments and adds non-template (N) nucleotides during assembly. However, while the membrane-proximal V domain (V2) incorporates both palindromic (P) and N nucleotide additions, it does not appear to use Dμ gene segments, likely because the locus encoding the V2 domain lacks D segments (Wang et al. 2011). Thus, V1 encodes longer and more junctionally diverse CDR3 than V2. As in opossum TCRμ, both V1 and V2 domains of platypus TCRμ are more similar to IgH V while Cμ is related to TCR8 (Wang et al. 2011).
The discovery of TCR homologs of similar structure in older mammalian clades to shark NAR-TCR suggests that this structure confers useful physiology and evolved multiple times in vertebrates. That NAR-TCR is found in the genome of elephant shark (an older Holocephalan cartilaginous fish than sharks and rays) yet IgNAR is not (Venkatesh et al. 2007), suggests that the T cell NAR variant may have given rise to the B cell NAR variant (Criscitiello 2014); alternatively, Holocephalans may have lost IgNAR.
VHδ gene segments discovered in sharks and most vertebrate groups
Sharks and marsupials/monotremes are not the only species to use IgH variable gene segments in their TCRδ repertoire. Functional IgH-like TCRδ V (VHδ) gene segments have been found in the genomes representing all extant gnathostome groups except teleosts and placental mammals. The Sarcopterygian coelacanth TCRαδ locus includes a track of 25 VHδ gene segments between the TCRα and TCRδ gene segments (Saha et al. 2014). In the amphibian Xenopus tropicalis, the 5’ end of the conventional TCRαδ locus encodes a separate cluster of VHδ gene segments that are expressed exclusively with a second distinct TCR δC (Parra et al. 2010). Some birds express VHδ gene segments with TCRδ as well. In the passerine zebra finch, a single VHδ gene segment is present in the TCRαδ locus and is expressed with TCR δC. However, the conventional TCRαδ locus of galliform birds (chicken, turkey, and likely duck) contains no VHδ segment. Instead, Galliformes have a second, nonsyntenic TCRδ locus containing a single VHδ-Dδ-Jδ-Cδ cluster that rearranges to form one TCRδ product (Parra et al. 2012b). The only mammal known to have functional VHδ gene segments is the monotreme platypus, which has a single VH8 gene segment located within the TCRαδ locus (Parra et al. 2012a). However, we found a single VHδ pseudogene in the TCRαδ locus of the Florida manatee (Breaux et al. 2018), suggesting that at least some eutherian mammals may have used similar gene segments at one time.
Thus, a much clearer picture is emerging of the ancestral antigen receptor locus from diverse extant vertebrates. Leading the way, the linkage of the TCR αδ locus with IgH elements is a recurring theme from cartilaginous fish to mammals. Recent work in reptiles substantiates VHδ segments in crocodilian TCR αδ loci and suggests further modeling of multiple insertion, duplication, and sometimes loss (teleosts) of B cell receptor elements in the TCR (Wang et al. 2020). In the past decade, what at first seemed to be merely curious findings of IgH in TCRδ of shark and amphibian (Criscitiello et al. 2010; Parra et al. 2010) have since spring-boarded our understanding of the natural history of the adaptive immune system. Now, a comprehensive hypothesis of the dawn of the system is supported, which includes not only the rearranging antigen receptor loci but also the MHC in a “primordial immune complex” whose components have evolved in four paralogous genomic regions since the two rounds of genome duplication in early vertebrates (Ohta et al. 2019). This genomic association is far reaching beyond Ig and TCR, to include natural killer receptors, cytokines, co-stimulation, and even the (likely) older variable lymphocyte receptors (VLR) of the jawless vertebrates, extant lamprey, and hagfish.
Somatic hypermutation augments γδ T cell receptor repertoire diversity in sharks
In addition to capitalizing on the availability of Ig V gene segments to refashion TCR γδ chains, sharks exploit traditional B cell diversifying mechanisms to expand their TCR repertoires. One such mechanism is the use of AID-catalyzed SHM to augment TCR repertoire diversity. Chen et al. (2009) reported the first evidence of targeted mutation to TCRγ V regions in the sandbar shark (Carcharhinus plumbeus). These authors sequenced the TCRγ locus and then evaluated the V region repertoire diversity using a 5′ RACE library from a single animal. Typical of TCR loci in many other vertebrates, sandbar shark TCRγ is arranged as a single translocon containing at least five V gene segments, three J gene segments, and a single C region. Expressed transcripts revealed no V segment bias for four of the five known Vs but a reduction in the use of the most 5′ (distal) V segment in the locus (Chen et al. 2009). However, comparison of cDNA clones to genomic sequences revealed a high frequency of mutation that could not be attributed to allelic variation or PCR error. Mutation patterns mirrored those of activated B cells undergoing SHM during affinity maturation, with mutation targeted to AID hotspot motifs within CDR of V gene segments (specifically CDR1), biased towards AID-favored G and C nucleotides, favoring transitions over transversions, and including both single-base and consecutive (tandem)-base changes that altered template-coded amino acids (Lee et al. 2002; Diaz et al. 1999; Chen et al. 2009, 2012). Because there was no evidence of antigen selection of mutated TCR [CDR and FR showed similar ratios of replacement (R) and silent (S) changes], Chen et al. (2012) concluded that TCRγ instead utilizes SHM to enhance repertoire diversity in γδ T cells. Research in nurse shark Ig light chains (IgL) also concluded that antigen did not drive selection except by limiting mutation to FR2, suggesting a mechanism for maintaining structural stability rather than enhanced affinity (Zhu and Hsu 2010). However subsequent work has shown significant mutation and evidence of selection in mature sharks (Iacoangeli et al. 2017).
Some mammals also hypermutate γδ TCRs
Similar analyses in both γ and δ chain of dromedary camel (Camelus dromedaries) indicated that mutation altered both chains of γδ TCR in camelids (Antonacci et al. 2011; Vaccarelli et al. 2012; Ciccarese et al. 2014). Using RT-qPCR and a 5′ RACE library, Antonacci et al. (2011) evaluated the expressed TCRδ chain repertoire of peripheral lymphoid tissues (spleen, tonsils, and blood) from a single adult camel. These transcripts were used to identify genes encoding TCRδ V gene segments in the germline. Analyses identified 13 putative germline TCRδ V gene segments belonging to 3 family groups. Comparing these germline sequences to cDNA clones revealed mutation to V regions at a rate (0.013/bp in spleen) similar to those reported in sandbar shark TCRγ and in mouse and shark IgL (see above). However, although nucleotide changes did appear to favor transitions (and included both point and tandem base changes in spleen), mutation did not target CDR over FR but instead was distributed throughout the V region (Antonacci et al. 2011). Comparison of synonymous and nonsynonymous (replacement) changes suggested (like in sandbar shark) that mutated receptors were not under antigen selection. While the authors did not report specific analyses to examine whether mutation was AID-mediated (e.g., bias to AID-favored G and C bases or targeted mutation to AID hotspot motifs), they concluded that mutation in TCRδ chain did contribute to γδ TCR repertoire diversity, but the mutations in FR and CDR did not suggest antigen selection (Antonacci et al. 2011). Analysis of camelid thymus may be needed to confirm that mutation is in the primary repertoire.
In a follow-up study, the same group reported evidence that mutations to genes encoding the TCRγ chain generate diversity within the γδ TCR repertoire (Vaccarelli et al. 2012). The group assembled and mapped the TCRγ locus from PCR products and chromosome walking fragments to identify two V-J-J-C cassettes within the TCRγ locus. While a cluster organization is atypical for TCR loci in general, this same basic cassette (V-J-J-C) structure is found in the TCRγ locus of a number of organisms (including sheep, cattle, and buffalo) and modifications to this structure are found in mice (Vernooij et al. 1993; Antonacci et al. 2007; Vaccarelli et al. 2008). An analysis of expressed transcripts from a spleen 5’ RACE library revealed targeted mutation biased towards G and C bases within AID-favored hotspot motifs. Further, although there was (again) no evidence of selection for modified receptors, the accumulation of nonconservative changes within CDR (specifically CDR2) intimated that somatic mutation contributed to the overall paratope diversity of TCRγ V regions (Vaccarelli et al. 2012). In silico structural models showed that mutation of γ or δ V regions enhances the structural stability of the γδ TCR, regardless of where (FR or CDR) these mutational changes occur within the V region (Ciccarese et al. 2014).
The presence of mutation within γδ TCR genes is not altogether surprising given the ability of γδ T cells to traverse the boundary between the innate and adaptive immune systems. Similar to αβ T cells, γδ T cells recombine V, (D), and J gene segments to create a highly specific adaptive repertoire with immunological memory (Kazen and Adams 2011). However, γδ T cells can assert an innate role in immunity as well, producing cytokines (e.g., TNFα and IFN-γ) in response to infection or tumor antigens (Gober et al. 2003; Beetz et al. 2008). In humans, γδ T cells can act as efficient antigenpresenting cells to CD8+ αβ T cells, synthesizing antigens through immunoproteasomes for cross-presentation via MHC class I (Brandes et al. 2009). Additionally, specific subsets of γδ T cells in humans (Vδ2 Tregs) express FOXP3 (forkhead/winged helix transcription factor box P3) and function as regulatory T cells, suppressing proliferation of peripheral blood mononuclear cells through the TGF-β1 signaling pathway (Casetti et al. 2009). Thus, γδ T cells combine both immediate innate-like responses to infection with on-going adaptive recognition responses [also reviewed in Kabelitz (2011)]. While some γδ TCR bind free antigen in a manner similar to BCR, some γδ TCR interact with non-classical MHC as tissue-specific receptors using restricted sets of variable and joining genes with limited junctional diversity (Allison and Garboczi 2002; Adams et al. 2005; Kazen and Adams 2011). In either case, there are some conserved binding features among γδ T cells from diverse species and tissues (Hayday and Vantourout 2020) that SHM-mediated changes to paratopes could offer flexibility to recognize new pathogens or adapt to rapidly changing ligands within restricted environments.
Could SHM be employed by αβ T cells?
While it is clear that T cells retain the same basic machinery that allows B cells to affinity mature receptors (Gellert 2002), somatically mutating αβ TCR may not provide the same benefits as to BCR or γδ TCR. Because αβ T cells are restricted to binding antigen in the context of self MHC, altering receptors that already have passed selection in the thymus could have profound consequences on receptor functionality. In fact, early studies in humans indicated that SHM in αβ T cells occurs only as a result of a diseased state (e.g., alloreactive T cell hybridomas, HIV-1, T cell lymphoma, lung, and liver tumors) (Augustin and Sim 1984; Cheynier et al. 1998; Okazaki et al. 2003; Rucci et al. 2006; Morisawa et al. 2008). Thus, while AID-mediated mutation may augment certain populations of T cells, it is clear that mutation is not likely to be beneficial. However, the assumption that αβ T cells cannot employ AID for any reason drove decades of (especially comparative) immunologists to disregard AID-driven mechanisms as an explanation for aberrations in their datasets.
Analysis of T cells from periarteriolar T cell sheath (PALS) and GC of immunized B10.A transgenic mouse spleen revealed mutation to V regions of TCR α chain (but not β chain) that was substantially higher than expected for PCR error. Further, mutation to TCRα V mirrored that of IgH V acquired from adjacent sites within the GC, suggesting a mechanism for SHM in T cells (Zheng et al. 1994). The significance of these results was questioned, citing insufficient evidence to support the claim (Bachl and Wabl 1995). However, the mutation may suggest that AID expression within splenic GC (during affinity maturation of B cells) also can impact V regions of TCRα.
In a study attempting to identify targeting elements of SHM in mice, Hackett et al. (1992) designed a rearranged TCR transgene capable of being expressed on B cells. The authors then examined cDNA transcripts of both endogenous IgH and TCR transgenes expressed on B cells to determine if TCR are targeted by SHM. Though they did observe some level of mutation (0.00017/bp) in the TCR transgenes, the frequency of mutation was minimal compared with rates observed in endogenous IgH genes (0.0021/bp), suggesting that TCR genes do not contain the required transcriptional elements for SHM (Hackett et al. 1992). In another study using Cre-ires-hCD2 (Cre) transgenic mice with a genetic reporter knocked into the AID locus, Qin et al. (2011) assessed endogenous AID production by B and T cells within spleen, lymph nodes, and Peyer’s patches. The authors found that a surprisingly large number of CD4+ memory T cells in these tissues express AID, likely resulting from T cell activation in peripheral lymphoid tissues. Activation of these T cell subsets produced a unique cytokine profile that increased with mouse age, suggesting a function in cellular aging. Though they did not examine cDNA transcripts for evidence of mutation, Qin et al. (2015) suggested that AID may play a role in T cell function or tumorigenesis. In conclusion, to date, data suggest that TCR αβ mutation in mammals seems to play little or no role in normal immune physiology.
Sharks target AID-mediated SHM to TCRα
While we found no study that specifically assesses the presence or absence of SHM in endogenous TCR, the fact that SHM is not commonly observed in mice or humans (except in diseased states) led immunologists to assume that αβ T cells cannot utilize SHM or any other receptormodifying mechanism (Kronenberg et al. 1986; Vitetta et al. 1991). The machinery for both endogenous and exogenous antigen presentation pathways seems to be shared among vertebrates from mammals to sharks, suggesting similar MHC restriction of TCR αβ (Ohta et al. 2002; Criscitiello et al. 2012, 2013). However, we recently reported evidence that nurse sharks enrich their TCR repertoire by exploiting SHM during repertoire generation in the thymus (Fig. 2). Real-time RT-qPCR and in situ hybridization expression data from nurse shark thymus confirmed AID expression in thymus at levels roughly half those observed in spleen (where B cell SHM occurs) (Ott et al. 2018). Using probes for in situ hybridization specific to either TCR αC or AID on thymus tissue, we observed a consistent “ring” pattern, where cells expressing both TCR αC and AID message surrounded a central cell expressing only TCR αC. Further, we determined that AID expression is localized to the inner cortex and medulla adjacent to the cortico-medullary junction, coincident with the location of TCRα receptor editing and thymic selection in mice (Huesmann et al. 1991; Nakagawa et al. 2012) (Fig. 3). Thus, T cells actively express AID during RAG-mediated somatic recombination of the alpha locus, permitting SHM of TCRα chain sequences while cells are being selected in the thymus (Ott et al. 2018).
Fig. 2.
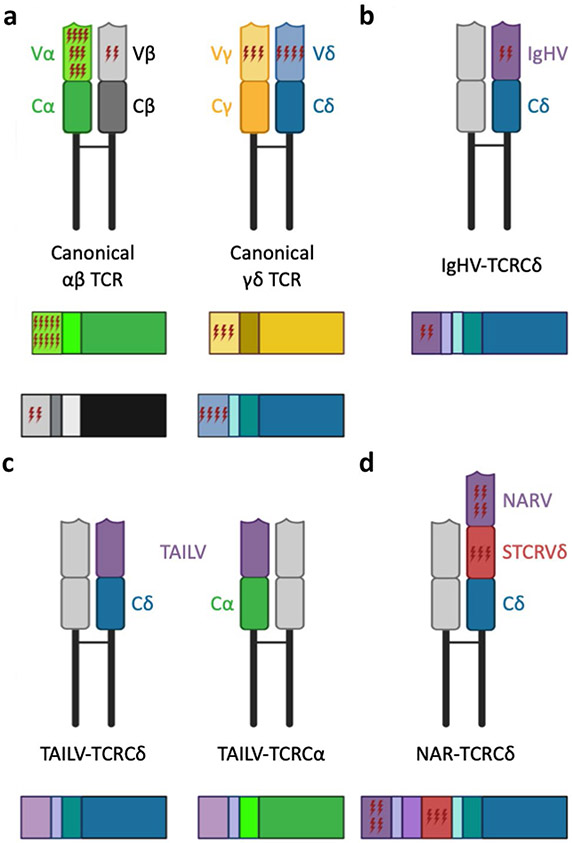
Cartoon depictions of putative assembled T cell receptors (TCR, top of each panel) and transcripts (bottom of each panel) illustrating the relative extent of somatic hypermutation (SHM) acquired by variable (V) regions of TCR chains in nurse sharks. a While V regions of all canonical TCR chains assimilate SHM, alpha chain incorporates significantly more mutation than other chains [αβTCR: alpha chain, α (green); beta chain, β (black); and γδ5TCR: gamma chain, γ (gold); delta chain, δ (blue)]; b Immunoglobulin heavy chain (IgH) V gene segments associated with TCRδ (or rarely TCRα) accumulate mutation within CDR2 regions at rates substantially lower than when used by immunoglobulin C regions; c TCR-associated Ig-like V (TAILV) gene segments, which associate with either TCRδ or TCRα C, do not appear to undergo SHM; d doubly-rearranging NAR-TCRδ are composed of two variable domains that undergo separate RAG-mediated VDJ recombination events—a membrane-distal IgNAR-like V domain (NARV, purple) supported by a membrane-proximal TCR δV domain (STCRδV, red) and associate with TCR δC incorporate few mutations to wither V domain [NAR: nurse shark (or new) antigen receptor; RAG: recombination activating genes]. Figure created with BioRender.com
We then assessed TCR transcripts for evidence of mutation and analyzed mutation patterns for similarities to AID-catalyzed mutation in affinity-matured BCR. V region sequences of BCR evolved to maximize the impacts of mutation, targeting replacement mutation to antigen binding CDR and limiting mutation to structurally important framework regions (FR) (Saini and Hershberg 2015). SHM in mouse, human, and shark BCR is biased towards G/A and C/T transitions and targeted to AID-preferred nucleotide motifs (DGYW/WRCH) (Anderson et al. 1995; Diaz et al. 1998; Lee et al. 2002; Rumfelt et al. 2002; Li et al. 2004; Odegard and Schatz 2006; Zhu and Hsu 2010). In addition to SHM-induced point mutations observed in other vertebrates, nurse shark IgH, IgL, and IgNAR sequences generate tandem substitutions of 2–5 adjacent nucleotides (Greenberg et al. 1995; Lee et al. 2002; Malecek et al. 2005; Dooley and Flajnik 2006; Dooley et al. 2006a). Though tandem mutations demonstrate a bias towards AID hotspot motifs, they do not typically favor transitions, suggesting that an additional mechanism may contribute to V region changes in nurse sharks (Zhu and Hsu 2010). Despite decades of assertions that SHM does not shape MHC-restricted αβ TCR repertoires, we identified SHM of nurse shark TCRα transcripts characteristic of AID-catalyzed SHM in shark BCR—point and tandem mutations focused on CDR, biased towards transitions, and targeted to AID motifs. Further, we detected SHM in transcripts from both thymus and peripheral lymphoid tissues, suggesting mutated receptors originated in the thymus prior to contact with foreign antigen. Together with corresponding evidence that AID expression overlaps TCRα chain rearrangement and selection in thymus, these data indicate that AID catalyzes SHM of TCRα for repertoire diversification during T cell development, implying that SHM contributes to receptor modifications that enhance selection (Ott et al. 2018).
Our discovery of AID-mediated somatic mutation in TCRα during primary lymphocyte development in thymus compelled us to examine the extent to which SHM alters the primary repertoire of other canonical (β, γ, and δ) and non-canonical (Ig or Ig-like) TCR chains. We examined transcripts from 5′ RACE cDNA libraries from nurse shark thymus to analyze mutation patterns in unconventional TCR chains and found that SHM targets TCR sequences preferentially based (generally) on the V segment used and (specifically) the C region associated with it. Despite the varying presence of AID hotspot motifs within V gene segments of all canonical and non-canonical TCR chains, only TCRα V accumulated significant mutation (Fig. 2b). Though TCR β, γ, and δ chains exhibited limited mutation, patterns paralleled those observed in BCR and TCRα of nurse sharks, with point (and tandem) mutation biased towards transitions and focused on AID hotspot motifs within CDR. In TAILV and both V domains of NAR-TCR V, the infrequent mutation we observed likely reflected the limited number of AID hotspot motifs present in sequences from these chains. Thus, AID-catalyzed mutation does not affect V segments of all chains equally. Comparing mutation between genomic V gene segments used with both alpha and delta C regions, when an alpha/delta V segment is associated with TCR αC, it acquired more than twice as many mutations as when it was associated with TCR δC regions, suggesting that, in thymus, AID displays a proclivity for mutating V regions of the TCRα chain. Even IgHV gene segments, laden with abundant AID-preferred motifs, accrued substantially lower rates of mutation than TCRα V regions associated with TCRα C regions. Further, mutation was considerably lower in IgHV associated with TCR in thymus than one would expect of the same IgHV associated with a BCR undergoing affinity maturation in spleen. The increased mutation in V regions associated with TCR αC in thymus suggests that the DNA motifs associated with this C exon are particularly important for AID targeting (Ott et al. 2020).
Comparison of human and nurse shark TCR Vα genes indicates that Vα of nurse sharks contain more AID-preferred hotspot motifs (WRCH/DGYW) per sequence than do human Vα segments, and these motifs occur 2–3× more often in shark Vα CDR than in human CDR. This suggests that, while the costs associated with somatically mutating TCR genes may outweigh the benefits for humans and mice, the same may not be true for more evolutionarily basal organisms like sharks. Sharks may be more resistant to the dangers of aberrant mutation because of their inherently slow rates of molecular mutation (10× slower than in mammals), long lifespans (> 272 years in Greenland shark), and (in many species) large body size (Martin 1999; Nielsen et al. 2016; Marra et al. 2019). Additionally, because of their considerable size and highly repetitive nature (> 50%), shark genomes may exhibit more flexibility than those of mice or humans (Stingo and Rocco 2001; Rocco et al. 2002, 2007; Hara et al. 2018). However, to realize any benefit of SHM, TCR modification would have to occur prior to or coincident with selection events in the thymus, since changes to a receptor that already passed selection could negatively affect its ability to bind self-MHC or permit binding to self-antigen.
Importance of studying immune mechanisms in non-traditional animal models
The basic components of adaptive immunity (RAG-mediated recombination of V, D, and J gene segments, B and T cell receptors, MHC class I and II, and AID-mediated somatic diversification mechanisms) are similar among extant jawed vertebrate groups, owing to the fairly recent divergence (roughly 480 Mya) of gnathostomes from their jawless ancestors (Hsu 2009; Janvier 2011; Brazeau and Friedman 2015; Flajnik 2018). Since this divergence, host immune systems evolved quite rapidly—perhaps as a consequence of rapidly evolving pathogens and influenced by varying developmental constraints, environmental adaptations, and population dynamics— ultimately permitting new, innovative features to supplant existing ones (Bailey et al. 2013). Consequently, gnathostomes evolved various accessory immune components as solutions to specific selective pressures of their environments (e.g., heavychain only antibodies of camels and sharks), and these accessory features can provide alternate views of the adaptive immune system through the window of evolution.
Compared with mice and humans, sharks have retained impressive TCR repertoire diversification strategies. Nurse sharks assemble TCR from IgM or IgW (IgD) V gene segments (from the Ig locus, TCR locus, or an altogether unique locus) or Ig-like TAILV, expanding the combinatorial potential of developing receptors (Criscitiello et al. 2010; Deiss et al.2019). Additionally, doubly rearranging NAR-TCR combines both an Ig-like (NAR) V domain with a supporting TCR V domain to create a novel receptor type (Criscitiello et al. 2006). These diversifying strategies are not limited to nurse sharks as NARTCR have definitively been found in elephant shark and Southern blotting demonstrated its presence in all tested Elasmobranch species (Criscitiello et al. 2006). In addition, despite their ancient origin and incredible diversity, all Elasmobranch species tested have preserved their vast array of antigen receptor loci, including IgM, IgW, IgNAR, four IgL isotypes, and as mentioned, NARTCR. Thus, while nurse sharks have provided the historical model for structure and function of the immune system, the molecules and mechanisms described here likely extend to all cartilaginous fish, at least the Elasmobranch sharks, skates, and rays.
Coelacanths, Xenopus, passeriform birds, and platypus all harbor Ig-like V gene segments (VHδ) in their conventional TCRαδ loci, while galliform birds house these Ig-like VHδ segments in a separate locus (Parra et al. 2010, 2012b; Parra and Miller 2012; Saha et al. 2014; Deiss et al. 2019). Marsupial and monotreme mammals acquired an additional T cell locus (TCRμ) that somatically recombines V, D, and J gene segments (or uses pre-joined segments) into a unique TCR chain with two variable domains, the most distal of which resembles IgH (Parra et al. 2007; Wang et al. 2011). Not only do nurse shark T cells borrow Ig components when recombining and assembling receptors, they derive additional diversity by pirating mechanisms traditionally used by B cells (i.e., AID-catalyzed SHM) to alter antigen binding sites. However, unlike B cell IgH that employ SHM to affinity mature antigen receptors in secondary lymphoid tissues, nurse sharks incorporate AID-catalyzed SHM in the thymus, most likely to salvage TCR in danger of failing thymic selection. Sandbar shark and dromedary camelids also have been shown to use SHM to alter V region sequences of γδ TCR (Chen et al. 2012; Ciccarese et al. 2014). More recently, reports indicate that the teleost fish Ballan wrasse (Labrus bergylta) somatically mutate both V and C regions of TCRα (Bilal et al. 2018) (curious as allelic polymorphism at TCRα C has been described in multiple other teleosts (Criscitiello et al. 2004). While these studies in sandbar sharks, Ballan wrasse, and camelids were limited to peripheral lymphoid tissues, it is possible, and we think likely that SHM-induced changes to T cells originated in the thymus of these groups as well.
Agnathan vertebrates (jawless hagfish and lamprey) evolved an alternate adaptive immune strategy to the immunoglobulin superfamily-based system of jawed vertebrates. The variable lymphocyte receptor (VLR)-based system also incorporates a tripartite adaptive defense strategy with three distinct somatically assembled receptor types. The three lineages of agnathan variable lymphocyte receptors (VLR A, B, and C) are analogous to the B cell and T cell lineages of gnathostomes (Das et al. 2015). Like B cells, VLR type B (VLRB) can be membrane-bound or secreted and functions in adaptive humoral responses (Alder et al. 2005; Pancer et al. 2005). Both VLR type A (VLRA), transcriptionally more similar to αβ T cells, and VLR type C (VLRC) transcriptionally similar to γδ T cells, occur only as a membrane-bound receptors and are predicted to function (as do T cells) in cell-mediated immune responses (Alder et al. 2005; Kasamatsu et al. 2010). However, unlike αβ T cells, neither VLRA nor VLRC seem to require antigen presentation for recognition (Deng et al. 2010). The similarities in immune defense strategies between agnathan and gnathostome vertebrates suggest that there were three lymphocyte lineages present in the vertebrate common ancestor, with discernible components of the immune system labor partitioned among them (Flajnik 2014). Hagfish and lamprey assemble VLR genes into lymphocytes using two AID homologs (CDA1 and CDA2), and CDA-mediated gene rearrangement in lampreys occurs through a serial gene conversion mechanism similar to AID-catalyzed Ig gene conversion in some birds and mammals (Rogozin et al. 2007; Guo et al. 2009). CDA1 expression occurs selectively in VLRA (and likely VLRC) lymphocytes within a thymoid (thymus like) region and orchestrates VLRA (and VLRC) gene recombination, while CDA2 expression occurs exclusively in VLRB lymphocytes and mediates VLRB gene assembly (Rogozin et al. 2007; Guo et al. 2009). Thus, there is a precedent of AID (or its homologs) being used in thymus (or thymoid organ) during primary lymphocyte diversification in vertebrates, and the use of AID during primary T cell development in nurse sharks may suggest that AID (or likely another APOBEC-family mutator)-mediated lymphocyte diversification in the earliest vertebrate ancestor, i.e., sharks retained the AID diversification mechanism present in the agnathan/gnathostome ancestor. The potential consequences of indiscriminate AID transcription (e.g., autoimmune disease, cancer) within the highly regulated, progressively compartmentalized nuclei of warm-blooded animals could have contributed to the loss of this ancestral mechanism in later vertebrates (like mice and humans). However, AID may even be capable of unheralded functions in mammals as well, such as deletions in the ultralong CDR3 of cattle IgH (Deiss et al. 2017).
Further insight into these similarities and differences in defense strategies could help elucidate the origins of lymphocyte receptors, making the study of “lower” fish immune systems ideal for comparative studies of immune evolution. It is clear that RAG often draws from B and T cell components in assembling mature variable domain encoding exons, particularly for TCR δ. It is also clear that AID activity is confined neither to B cell receptors nor the immunized post-antigen repertoire. The work reviewed here in nurse shark and other diverse vertebrates mandates a more careful mechanistic analysis of the activity of these two somatic diversification catalysts in both primary and secondary lymphoid tissues to discover the boundaries of their shaping of lymphocyte antigen receptor repertoires in health and disease.
Acknowledgments
Funding This work was supported by grants from the NIH to MFC (AI56963) and MFF (AI027877 and AI140326) and the NSF to MFC (IOS-1257829 and IOS-1656870).
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adams EJ, Chien YH, Garcia KC (2005) Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science 308:227–231. 10.1126/science.1106885 [DOI] [PubMed] [Google Scholar]
- Aghaallaei N, Bajoghli B (2018) Making thymus visible: understanding T-cell development from a new perspective. Front Immunol 9:375. 10.3389/fimmu.2018.00375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z (2005) Diversity and function of adaptive immune receptors in a jawless vertebrate. Science 310:1970–1973. 10.1126/science.1119420 [DOI] [PubMed] [Google Scholar]
- Allison TJ, Garboczi DN (2002) Structure of gammadelta T cell receptors and their recognition of non-peptide antigens. Mol Immunol 38:1051–1061. 10.1016/S0161-5890(02)00034-2 [DOI] [PubMed] [Google Scholar]
- Álvarez-Prado ÁF, Pérez-Durán P, Pérez-García A, Benguria A, Torroja C, de Yébenes VG, Ramiro AR (2018) A broad atlas of somatic hypermutation allows prediction of activation-induced deaminase targets. J Exp Med 215:761–771. 10.1084/jem.20171738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson MK, Shamblott MJ, Litman RT, Litman GW (1995) Generation of immunoglobulin light chain gene diversity in Raja erinacea is not associated with somatic rearrangement, an exception to a central paradigm of B cell immunity. J Exp Med 182:109–119. 10.1084/jem.182.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonacci R, Vaccarelli G, Di Meo GP, Piccinni B, Miccoli MC, Cribiu EP, Perucatti A, Iannuzzi L, Ciccarese S (2007) Molecular in situ hybridization analysis of sheep and goat BAC clones identifies the transcriptional orientation of T cell receptor gamma genes on chromosome 4 in Bovids. Vet Res Commun 31:977–983. 10.1007/s11259-006-0202-x [DOI] [PubMed] [Google Scholar]
- Antonacci R, Mineccia M, Lefranc MP, Ashmaoui HM, Lanave C, Piccinni B, Pesole G, Hassanane MS, Massari S, Ciccarese S (2011) Expression and genomic analyses of Camelus dromedarius T cell receptor delta (TRD) genes reveal a variable domain repertoire enlargement due to CDR3 diversification and somatic mutation. Mol Immunol 48:1384–1396. 10.1016/j.molimm.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Arakawa H, Hauschild J, Buerstedde J-M (2002) Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 295:1301–1306. 10.1126/science.1067308 [DOI] [PubMed] [Google Scholar]
- Augustin A, Sim G (1984) T-cell receptors generated via mutations are specific for various major histocompatibility antigens. Cell 39:5–12. 10.1016/0092-8674(84)90186-7 [DOI] [PubMed] [Google Scholar]
- Bachl J, Wabl M (1995) Hypermutation in T cells questioned. Nature 375:285–286. 10.1038/375285c0 [DOI] [PubMed] [Google Scholar]
- Bailey M, Christoforidou Z, Lewis M (2013) Evolution of immune systems: Specificity and autoreactivity. Autoimmun Rev 12(6):643–647. 10.1016/j.autrev.2012.10.007 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell 109:S45–S55. 10.1016/S0092-8674(02)00675-X [DOI] [PubMed] [Google Scholar]
- Beetz S, Wesch D, Marischen L, Welte S, Oberg H-H, Kabelitz D (2008) Innate immune functions of human γδ T cells. Immunobiol 213:173–182. 10.1016/j.imbio.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Bilal S, Lie KK, Sæle Ø, Hordvik I (2018) T cell receptor alpha chain genes in the teleost ballan wrasse (Labrus bergylta) are subjected to somatic hypermutation. Front Immunol 9:1101. 10.3389/fimmu.2018.01101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgulya P, Kishi H, Uematsu Y, von Boehmer H (1992) Exclusion and inclusion of alpha and beta T cell receptor alleles. Cell 69:529–537. 10.1016/0092-8674(92)90453-j [DOI] [PubMed] [Google Scholar]
- Brady BL, Steinel NC, Bassing CH (2010) Antigen receptor allelic exclusion: an update and reappraisal. J Immunol 185:3801–3808. 10.4049/jimmunol.1001158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes M, Willimann K, Bioley G, Lévy N, Eberl M, Luo M, Tampé R, Lévy F, Romero P, Moser B (2009) Cross-presenting human γδ T cells induce robust CD8+ αβ T cell responses. Proc Natl Acad Sci 106:2307–2312. 10.1073/pnas.0810059106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazeau MD, Friedman M (2015) The origin and early phylogenetic history of jawed vertebrates. Nature 520:490–497. 10.1038/nature14438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaux B, Hunter ME, Cruz-Schneider MP, Sena L, Bonde RK, Criscitiello MF (2018) The Florida manatee (Trichechus manatus latirostris) T cell receptor loci exhibit V subgroup synteny and chain-specific evolution. Dev Comp Immunol 85:71–85. 10.1016/j.dci.2018.04.007 [DOI] [PubMed] [Google Scholar]
- Casetti R, Agrati C, Wallace M, Sacchi A, Martini F, Martino A, Rinaldi A, Malkovsky M (2009) Cutting edge: TGF-β1 and IL-15 induce FOXP3+ γδ regulatory T cells in the presence of antigen stimulation. J Immunol 183:3574–3577. 10.4049/jimmunol.0901334 [DOI] [PubMed] [Google Scholar]
- Chang B, Casali P (1994) The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today 15:367–373. 10.1016/0167-5699(94)90175-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Kshirsagar S, Jensen I, Lau K, Covarrubias R, Schluter SF, Marchalonis JJ (2009) Characterization of arrangement and expression of the T cell receptor gamma locus in the sandbar shark. Proc Natl Acad Sci 106:8591–8596. 10.1073/pnas.0811283106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Bernstein H, Ranganathan P, Schluter S (2012) Somatic hypermutation of TCR γ V genes in the sandbar shark. Dev Comp Immunol 37:176–183. 10.1016/j.dci.2011.08.018 [DOI] [PubMed] [Google Scholar]
- Cheynier R, Henrichwark S, Wain Hobson S (1998) Somatic hypermutation of the T cell receptor V beta gene in microdissected splenic white pulps from HIV-1-positive patients. Eur J Immunol 28:1604–1610. 10.1002/(SICI)1521-4141(199805)28:05 [DOI] [PubMed] [Google Scholar]
- Chien YH, Iwashima M, Kaplan KB, Elliot JF, Davis MM (1987) A new T-cell receptor gene located within the alpha locus and expressed early in T-cell differentiation. Nature 327:677–682. 10.1038/327677a0 [DOI] [PubMed] [Google Scholar]
- Ciccarese S, Vaccarelli G, Lefranc MP, Tasco G, Consiglio A, Casadio R, Linguiti G, Antonacci R (2014) Characteristics of the somatic hypermutation in the Camelus dromedarius T cell receptor gamma (TRG) and delta (TRD) variable domains. Dev Comp Immunol 46:300–313. 10.1016/j.dci.2014.05.001 [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJF, Petersen-Mahrt SK, Neuberger MS (2005) Evolution of the AID/APOBEC family of polynucleotide (deoxy) cytidine deaminases. Mol Biol Evol 22:367–377. 10.1093/molbev/msi026 [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS (2007) DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol 94:37–73. 10.1016/S0065-2776(06)94002-4 [DOI] [PubMed] [Google Scholar]
- Criscitiello M, Saltis M, Flajnik M (2006) An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci 103:5036–5041. 10.1073/pnas.0507074103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello M, Flajnik M (2007) Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol 37:2683–2694. 10.1002/eji.200737263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Wermenstam NE, Pilstrom L, McKinney EC (2004) Allelic polymorphism of T-cell receptor constant domains is widespread in fishes. Immunogenetics 55:818–824. 10.1007/s00251-004-0652-7 [DOI] [PubMed] [Google Scholar]
- Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF (2010) Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol 184:6950–6960. 10.4049/jimmunol.0902774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Ohta Y, Graham MD, Eubanks JO, Chen PL, Flajnik MF (2012) Shark class II invariant chain reveals ancient conserved relationships with cathepsins and MHC class II. Dev Comp Immunol 36:521–533. 10.1016/j.dci.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Dickman MB, Samuel JE, de Figueiredo P (2013) Tripping on acid: trans-kingdom perspectives on biological acids in immunity and pathogenesis. PLOS Pathog 9:e1003402. 10.1371/journal.ppat.1003402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF (2014) Shark T cell receptors. In: Smith SL, Sim RB, Flajnik MF (eds) Immunobiology of the Shark, 1st edn. CRC Press, Boca Raton, USA. doi: 10.1201/b17773 [DOI] [Google Scholar]
- Das S, Li J, Hirano M, Sutoh Y, Herrin BR, Cooper MD (2015) Evolution of two prototypic T cell lineages. Cellular immunol 296:87–94. 10.1016/j.cellimm.2015.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss TC, Vadnais M, Wang F, Chen PL, Torkamani A, Mwangi W, Lefranc MP, Criscitiello MF, Smider VV (2017) Immunogenetic factors driving formation of ultralong VH CDR3 in Bos taurus antibodies. Cell Mol Immunol. 10.1038/cmi.2017.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiss TC, Breaux B, Ott JA, Daniel RA, Chen PL, Castro CD, Ohta Y, Flajnik MF, Criscitiello MF (2019) Ancient use of Ig variable domains contributes significantly to the TCRδ repertoire. J Immunol 203:1265–1275. 10.4049/jimmunol.1900369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Velikovsky CA, Xu G, Iyer L, Tasumi S, Kerzic M, Flajnik M, Aravind L, Pancer Z, Mariuzza R (2010) A structural basis for antigen recognition by the T cell-like lymphocytes of sea lamprey. Proc Natl Acad Sci 107:13408–13413. 10.1073/pnas.1005475107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Flajnik MF (1998) Evolution of somatic hypermutation and gene conversion in adaptive immunity. Immunol Rev 162:13–24. 10.1111/j.1600-065x.1998.tb01425.x [DOI] [PubMed] [Google Scholar]
- Diaz M, Greenberg A, Flajnik M (1998) Somatic hypermutation of the new antigen receptor gene (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci 95:14343–14348. 10.1073/pnas.95.24.14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M, Velez J, Singh M, Cerny J, Flajnik MF (1999) Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol 11:825–833. 10.1093/intimm/11.5.825 [DOI] [PubMed] [Google Scholar]
- Diaz M, Flajnik MF, Klinman N (2001) Evolution and the molecular basis of somatic hypermutation of antigen receptor genes. Phil Trans R Soc B Biol Sci 356:67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley H, Flajnik MF (2005) Shark immunity bites back: affinity maturation and memory response in the nurse shark, Ginglymostoma cirratum. Eur J Immunol 35:936–945. 10.1002/eji.200425760 [DOI] [PubMed] [Google Scholar]
- Dooley H, Flajnik MF (2006) Antibody repertoire development in cartilaginous fish. Dev Comp Immunol 30:43–56. 10.1016/j.dci.2005.06.022 [DOI] [PubMed] [Google Scholar]
- Dooley H, Stanfield RL, Brady RA, Flajnik MF (2006a) First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci 103:1846–1851. 10.1073/pnas.0508341103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley H, Stanfield RL, Brady RA, Flajnik MF (2006b) First molecular and biochemical analysis of in vivo affinity maturation in an ectothermic vertebrate. Proc Natl Acad Sci USA 103:1846–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahl SP, Coffey F, Wiest DL (2014) Origins of γδ T cell effector subsets: a riddle wrapped in an enigma. J Immunol 193:4289–4294. 10.4049/jimmunol.1401813 [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Rumfelt LL (2000) The immune system of cartilaginous fish. Curr Top Microbiol Immunol 248:249–270. 10.1007/978-3-642-59674-2_11 [DOI] [PubMed] [Google Scholar]
- Flajnik MF (2002) Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol 2:688–698. 10.1038/nri889 [DOI] [PubMed] [Google Scholar]
- Flajnik MF (2014) Re-evaluation of the Immunological Big Bang. Curr Biol 24:R1060–R1065. 10.1016/j.cub.2014.09.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF (2018) A cold-blooded view of adaptive immunity. Nat Rev Immunol 18:438–453. 10.1038/s41577-018-0003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne NRJ, Alam SM (1999) Allelic exclusion of the T cell receptor α-chain: developmental regulation of a post-translational event. Semin Immunol 11:337–347. 10.1006/smim.1999.0190 [DOI] [PubMed] [Google Scholar]
- Gellert M (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71:101–132. 10.1146/annurev.biochem.71.090501.150203 [DOI] [PubMed] [Google Scholar]
- Germain RN (2002) T-cell development and the CD4-CD8 lineage decision. Nat Rev Immunol 2:309–322. 10.1038/nri798 [DOI] [PubMed] [Google Scholar]
- Gober H-J, Kistowska M, Angman L, Jeno P, Mori L, De Libero G (2003) Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med 197:163–168. 10.1084/jem.20021500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good RA, Finstad J (1966) The Phylogenetic Development of Immune Responses and the Germinal Center System. In: Cottier H, Odartchenko N, Schindler R, Congdon CC (eds) Germinal Centers in Immune Responses. Springer-Verlag, New York Inc, University of Bern, Switzerland [Google Scholar]
- Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF (1995) A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 374:168–173. 10.1038/374168a0 [DOI] [PubMed] [Google Scholar]
- Guo P, Hirano M, Herrin BR, Li J, Yu C, Sadlonova A, Cooper MD (2009) Dual nature of the adaptive immune system in lampreys. Nature 459:796–801. 10.1038/nature08068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett J Jr, Stebbins C, Rogerson B, Davis MM, Storb U (1992) Analysis of a T cell receptor gene as a target of the somatic hypermutation mechanism. J Exp Med 176:225–231. 10.1084/jem.176.1.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y, Yamaguchi K, Onimaru K, Kadota M, Koyanagi M, Keeley SD, Tatsumi K, Tanaka K, Motone F, Kageyama Y, Nozu R, Adachi N, Nishimura O, Nakagawa R, Tanegashima C, Kiyatake I, Matsumoto R, Murakumo K, Nishida K, Terakita A, Kuratani S, Sato K, Hyodo S, Kuraku S (2018) Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat Ecol Evol 2:1761–1771. 10.1038/s41559-018-0673-5 [DOI] [PubMed] [Google Scholar]
- Hayday AC, Vantourout P (2020) The innate biologies of adaptive antigen receptors. Ann Rev Immunol 38:487–510. 10.1146/annurev-immunol-102819-023144 [DOI] [PubMed] [Google Scholar]
- Hsu E, Criscitiello MF (2006) Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J Immunol 177:2452–2462. 10.4049/jimmunol.177.4.2452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu E (2009) V(D)J Recombination: Of Mice and Sharks. Adv Exp Med Biol 650:166–179. 10.1007/978-1-4419-0296-2_14 [DOI] [PubMed] [Google Scholar]
- Hsu E (2018) Immune system receptors in vertebrates: immunoglobulins. Reference Module in Life Sciences. 10.1016/B978-0-12-809633-8.20721-8 [DOI] [Google Scholar]
- Huesmann M, Scott B, Kisielow P, von Boehmer H (1991) Kinetics and efficacy of positive selection in the thymus of normal and T cell receptor transgenic mice. Cell 66:533–540. 10.1016/0092-8674(81)90016-7 [DOI] [PubMed] [Google Scholar]
- Iacoangeli A, Lui A, Haines A, Ohta Y, Flajnik M, Hsu E (2017) Evidence for Ig light chain isotype exclusion in shark B lymphocytes suggests ordered mechanisms. J Immunol 199:1875–1885. 10.4049/jimmunol.1700762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack R, Du Pasquier L (2019) The Triumph of Individualism: Evolution of Somatically Generated Adaptive Immune Systems. Evolutionary Concepts in Immunology. Springer International Publishing, Cham. doi: 10.1007/978-3-030-18667-8_4 [DOI] [Google Scholar]
- Janvier P (2011) Comparative anatomy: all vertebrates do have vertebrae. Curr Biol 21:R661–R663. 10.1016/j.cub.2011.07.014 [DOI] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Murre C (2009) Chromatin architecture and the generation of antigen receptor diversity. Cell 138:435–448. 10.1016/j.cell.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabelitz D (2011) γδ T-cells: cross-talk between innate and adaptive immunity. Cell Mol Life Sci 68:2331. 10.1007/s00018-011-0696-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M (2010) Identification of a third variable lymphocyte receptor in the lamprey. Proc Natl Acad Sci 107:14304–14308. 10.1073/pnas.1001910107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazen AR, Adams EJ (2011) Evolution of the V, D, and J gene segments used in the primate γδ T-cell receptor reveals a dichotomy of conservation and diversity. Proc Natl Acad Sci 108:E332–E340. 10.1073/pnas.1105105108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikutani H, Inui S, Sato R, Barsumian EL, Owaki H, Yamasaki K, Kaisho T, Uchibayashi N, Hardy RR, Hirano T, Tsunasawa S, Sakiyama F, Suemura M, Kishimoto T (1986) Molecular structure of human lymphocyte receptor for immunoglobulin E. Cell 47:657–665. 10.1016/0092-8674(86)90508-8 [DOI] [PubMed] [Google Scholar]
- Kondo K, Ohigashi I, Takahama Y (2019) Thymus machinery for T-cell selection. Int Immunol 31:119–125. 10.1093/intimm/dxy081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreslavsky T, Gleimer M, Garbe AI, von Boehmer H (2010) αβ versus γδ fate choice: counting the T-cell lineages at the branch point. Immunol Rev 238:169–181. 10.1111/j.1600-65X.2010.00947.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronenberg M, Siu G, Hood LE, Shastri N (1986) The molecular genetics of the T cell antigen receptor and T cell antigen recognition. Ann Rev Immunol 4:529–591. 10.1146/annurev.iy.04.040186.002525 [DOI] [PubMed] [Google Scholar]
- Kuklina EM (2006) Revision of the antigen receptor of T-lymphocytes. Biochemistry 71:827–837. 10.1134/S0006297906080025 [DOI] [PubMed] [Google Scholar]
- Kuo TC, Schlissel MS (2009) Mechanisms controlling expression of the RAG locus during lymphocyte development. Curr Opin Immunol 21:173–178. 10.1016/j.coi.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafaille JJ, Haas W, Coutinho A, Tonegawa S (1990) Positive selection of γδ T cells. Immunol Today 11:75–78. 10.1016/0167-5699(90)90030-D [DOI] [PubMed] [Google Scholar]
- Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E (2002) Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity 16:571–582. 10.1016/s1074-7613(02)00300-x [DOI] [PubMed] [Google Scholar]
- Lefranc M-P (2014) Immunoglobulin and T cell receptor genes: IMGT and the birth and rise of immunoinformatics. Front Immunol 5:22–22. 10.3389/fimmu.2014.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc MP, Pommie C, Ruiz M, Giudicelli V, Foulquier E, Truong L, Thouvenin-Contet V, Lefranc G (2003) IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol 27:55–77. 10.1016/S0145-305X(02)00039-3 [DOI] [PubMed] [Google Scholar]
- Li Z, Woo CJ, Iglesias-Ussel MD, Ronai D, Scharff MD (2004) The generation of antibody diversity through somatic hypermutation and class switch recombination. Genes Dev 18:1–11. 10.1101/gad.1161904 [DOI] [PubMed] [Google Scholar]
- Liu M-C, Liao W-Y, Buckley KM, Yang SY, Rast JP, Fugmann SD (2018) AID/APOBEC-like cytidine deaminases are ancient innate immune mediators in invertebrates. Nat Commun 9:1948–1948. 10.1038/s41467-018-04273-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak F, Schatz DG (1996) T-cell receptor alpha locus V(D) J recombination by-products are abundant in thymocytes and mature T cells. Mol Cell Biol 16:609–618. 10.1128/mcb.16.2.609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luer C, Walsh CJ, Bodine AB, Wyffels JT, Scott TR (1995) The elasmobranch thymus: anatomical, histological, and preliminary functional characterization. J Exp Zool 273:342–354. 10.1002/jez.1402730408 [DOI] [Google Scholar]
- MacLennan ICM (1994) Germinal Centers Ann Rev Immunol 12:117–139. 10.1146/annurev.iy.12.040194.001001 [DOI] [PubMed] [Google Scholar]
- Magor BG (2015) Antibody affinity maturation in fishes-our current understanding. Biology 4:512–524. 10.3390/biology4030512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF, Hsu E (2005) Somatic hypermutation and junctional diversification at Ig heavy chain loci in the nurse shark. J Immunol 175:8105–8115. 10.4049/jimmunol.175.12.8105 [DOI] [PubMed] [Google Scholar]
- Marra NJ, Stanhope MJ, Jue NK, Wang M, Sun Q, Pavinski Bitar P, Richards VP, Komissarov A, Rayko M, Kliver S, Stanhope BJ, Winkler C, O’Brien SJ, Antunes A, Jorgensen S, Shivji MS (2019) White shark genome reveals ancient elasmobranch adaptations associated with wound healing and the maintenance of genome stability. Proc Natl Acad Sci 116:4446–4455. 10.1073/pnas.1819778116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AP (1999) Substitution rates of organelle and nuclear genes in sharks: implicating metabolic rate (again). Mol Cell Biol 16:996–1002. 10.1093/oxfordjournals.molbev.a026189 [DOI] [PubMed] [Google Scholar]
- Maul RW, Gearhart PJ (2010) Chapter six - AID and Somatic Hypermutation. In Alt FW (ed.) Adv Immunol. Academic Press; doi: 10.1016/S0065-2776(10)05006-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGargill MA, Derbinski JM, Hogquist KA (2000) Receptor editing in developing T cells. Nat Immunol 1:336–341. 10.1038/79790 [DOI] [PubMed] [Google Scholar]
- Morisawa T, Marusawa H, Ueda Y, Iwai A, Okazaki I-m, Honjo T, Chiba T (2008) Organ-specific profiles of genetic changes in cancers caused by activation-induced cytidine deaminase expression. Int J Cancer 123:2735–2740. 10.1002/ijc.23853 [DOI] [PubMed] [Google Scholar]
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T (2000) Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell 102:553–563. 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Murphy K, Weaver C (2017) Janeway’s Immunobiology, 9th edn. Garland Science, New York [Google Scholar]
- Nakagawa Y, Ohigashi I, Nitta T, Sakata M, Tanaka K, Murata S, Kanagawa O, Takahama Y (2012) Thymic nurse cells provide microenvironment for secondary T cell receptor α rearrangement in cortical thymocytes. Proc Natl Acad Sci 109:20572–20577. 10.1073/pnas.1213069109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely HR, Guo J, Flowers EM, Criscitiello MF, Flajnik MF (2018) “Double-duty” conventional dendritic cells in the amphibian Xenopus as the prototype for antigen presentation to B cells. Eur J Immunol 48:430–440. 10.1002/eji.201747260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Hedeholm RB, Heinemeier J, Bushnell PG, Christiansen JS, Olsen J, Ramsey CB, Brill RW, Simon M, Steffensen KF, Steffensen JF (2016) Eye lens radiocarbon reveals centuries of longevity in the Greenland shark (Somniosus microcephalus). Science 353:702–704. 10.1126/science.aaf1703 [DOI] [PubMed] [Google Scholar]
- Odegard VH, Schatz DG (2006) Targeting of somatic hypermutation. Nat Rev Immunol 6:573–583. 10.1038/nri1896 [DOI] [PubMed] [Google Scholar]
- Ohta Y, McKinney EC, Criscitiello MF, Flajnik MF (2002) Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages. J Immunol 168:771–781. 10.4049/jimmunol.168.2.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Flajnik M (2006) IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci 103:10723–10728. 10.1073/pnas.0601407103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Kasahara M, O’Connor TD, Flajnik MF (2019) Inferring the “Primordial Immune Complex”: origins of MHC class I and antigen receptors revealed by comparative genomics. J Immunol 203:1882–1896. 10.4049/jimmunol.1900597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki I-m, Hiai H, Kakazu N, Yamada S, Muramatsu M, Kinoshita K, Honjo T (2003) Constitutive expression of AID leads to tumorigenesis. J Exp Med 197:1173–1181. 10.1084/jem.20030275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott JA, Castro CD, Deiss TC, Ohta Y, Flajnik MF, Criscitiello MF (2018) Somatic hypermutation of T cell receptor α chain contributes to selection in nurse shark thymus. eLife 7:e28477. doi: 10.7554/eLife.28477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott JA, Harrison J, Flajnik MF, Criscitiello MF (2020) Nurse shark T cell receptors employ somatic hypermutation preferentially to alter alpha/delta variable segments associated with alpha constant region. Eur J Immunol 50:1307–1320. 10.1002/eji.201948495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD (2005) Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci 102:9224–9229. 10.1073/pnas.0503792102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Schwarz R, Deakin J, Lindblad-Toh K, Miller RD (2007) A unique T cell receptor discovered in marsupials. Proc Natl Acad Sci 104(23):9776–9781. 10.1073/pnas.0609106104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Baker ML, Hathaway J, Lopez AM, Trujillo J, Sharp A, Miller RD (2008) Comparative genomic analysis and evolution of the T cell receptor loci in the opossum Monodelphis domestica. BMC Genomics 9:111. 10.1186/1471-2164-9-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, Miller RD (2010) The dynamic TCRdelta: TCRdelta chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur J Immunol 40:2319–2329. 10.1002/eji.201040515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Lillie M, Miller RD (2012) A model for the evolution of the mammalian t-cell receptor α/δ and μ loci based on evidence from the duckbill Platypus. Mol Biol Evol 29:3205–3214. 10.1093/molbev/mss128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Miller RD (2012) Comparative analysis of the chicken TCRα/δ locus. Immunogenetics 64:641–645. 10.1007/s00251-012-0621-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Mitchell K, Dalloul RA, Miller RD (2012) A second TCRdelta locus in Galliformes uses antibody-like V domains: insight into the evolution of TCRdelta and TCRmu genes in tetrapods. J Immunol 188:3912–3919. 10.4049/jimmunol.1103521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Suzuki K, Nakata M, Chikuma S, Izumi N, Thi Huong L, Maruya M, Fagarasan S, Busslinger M, Honjo T, Nagaoka H (2011) Activation-induced cytidine deaminase expression in CD4+ T cells is associated with a unique IL-10-producing subset that increases with age. PLoS ONE 6:e29141. 10.1371/journal.pone.0029141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin T, Zhao H, Zhu H, Wang D, Du W, Hao H (2015) Immunoglobulin genomics in the prairie vole (Microtus ochrogaster). Immunol Lett 166:79–86. 10.1016/j.imlet.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Rios FM, Zimmerman LM (2015) Immunology of Reptiles. John Wiley & Sons, Ltd [Google Scholar]
- Rocco L, Morescalchi MA, Costagliola D, Stingo V (2002) Karyotype and genome characterization in four cartilaginous fishes. Gene 295:289–298. 10.1016/s0378-1119(02)00730-8 [DOI] [PubMed] [Google Scholar]
- Rocco L, Liguori I, Costagliola D, Morescalchi MA, Tinti F, Stingo V (2007) Molecular and karyological aspects of Batoidea (Chondrichthyes, Elasmobranchi) phylogeny. Gene 389:80–86. 10.1016/j.gene.2006.09.024 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z (2007) Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AIDAPOBEC family cytosine deaminase. Nat Immunol 8:647–656. 10.1038/ni1463 [DOI] [PubMed] [Google Scholar]
- Rucci F, Cattaneo L, Marrella V, Sacco MG, Sobacchi C, Lucchini F, Nicola S, Bella SD, Villa ML, Imberti L, Gentili F, Montagna C, Tiveron C, Tatangelo L, Facchetti F, Vezzoni P, Villa A (2006) Tissue-specific sensitivity to AID expression in transgenic mouse models. Gene 377:150–158. 10.1016/j.gene.2006.03.024 [DOI] [PubMed] [Google Scholar]
- Rumfelt L, McKinney E, Taylor E, Flajnik M (2002) The development of primary and secondary lymphoid tissues in the nurse shark Ginglymostoma cirratum: B-cell zones precede dendritic cell immigration and T-cell zone formation during ontogeny of the spleen. Scand J Immunol 56:130–148. 10.1046/j.1365-3083.2002.01116.x [DOI] [PubMed] [Google Scholar]
- Saha NR, Ota T, Litman GW, Hansen J, Parra Z, Hsu E, Buonocore F, Canapa A, Cheng J-F, Amemiya CT (2014) Genome complexity in the coelacanth is reflected in its adaptive immune system. J Exp Zool Part B 322:438–463. 10.1002/jez.b.22558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini J, Hershberg U (2015) B cell variable genes have evolved their codon usage to focus the targeted patterns of somatic mutation on the complementarity determining regions. Mol Immunol 65:157–167. 10.1016/j.molimm.2015.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz DG (2004) V(D)J recombination. Immunol Rev 200:5–11. 10.1111/j.0105-2896.2004.00173.x [DOI] [PubMed] [Google Scholar]
- Stingo V, Rocco L (2001) Selachian cytogenetics: a review. Genetica 111:329–347. 10.1023/a:1013747215866 [DOI] [PubMed] [Google Scholar]
- Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302:575–581. 10.1038/302575a0 [DOI] [PubMed] [Google Scholar]
- Vaccarelli G, Miccoli MC, Antonacci R, Pesole G, Ciccarese S (2008) Genomic organization and recombinational unit duplicationdriven evolution of ovine and bovine T cell receptor gamma loci. BMC Genomics 9:81. 10.1186/1471-2164-9-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaccarelli G, Antonacci R, Tasco G, Yang F, Giordano L, El Ashmaoui HM, Hassanane MS, Massari S, Casadio R, Ciccarese S (2012) Generation of diversity by somatic mutation in the Camelus dromedarius T-cell receptor gamma variable domains. Eur J Immunol 42:3416–3428. 10.1002/eji.201142176 [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh Y-H, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, Strausberg RL, Brenner S (2007) Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLOS Biol 5:e101. 10.1371/journal.pbio.0050101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V (2014) Elephant shark genome provides unique insights into gnathostome evolution. Nature 505:174–179. 10.1038/nature12826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernooij BTM, Lenstra JA, Wang K, Hood L (1993) Organization of the murine T-cell receptor γ locus. Genomics 17:566–574. 10.1006/geno.1993.1373 [DOI] [PubMed] [Google Scholar]
- Vitetta E, Berton M, Burger C, Kepron M, Wa L, Yin X (1991) Memory B and T cells. Ann Rev Immunol 9:193–217. 10.1146/annurev.iy.09.040191.001205 [DOI] [PubMed] [Google Scholar]
- Wang X, Parra ZE, Miller RD (2011) Platypus TCRmu provides insight into the origins and evolution of a uniquely mammalian TCR locus. J Immunol 187:5246–5254. 10.4049/jimmunol.1101113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huang J, Wang P, Wang R, Wang C, Yu D, Ke C, Huang T, Song Y, Bai J, Li K, Ren L, Miller RD, Han H, Zhou X, Zhao Y (2020) Analysis of the Chinese alligator TCRalpha/delta loci reveals the evolutionary pattern of atypical TCRdelta/TCRmu in tetrapods. J Immunol 205:637–647. 10.4049/jimmunol.2000257 [DOI] [PubMed] [Google Scholar]
- Zapata A, Leceta J, Barrutia MG (1981) Ultrastructure of splenic white pulp of the turtle, Mauremys caspica. Cell Tissue Res 220:845–855. 10.1007/bf00210466 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, Schatz DG (2019) Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569:79–84. 10.1038/s41586-019-1093-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng B, Xue W, Kelsoe G (1994) Locus-specific somatic hypermutation in germinal centre T cells. Nature 372:556–559. 10.1038/372556a0 [DOI] [PubMed] [Google Scholar]
- Zhu C, Hsu E (2010) Error-prone DNA repair activity during somatic hypermutation in shark B lymphocytes. J Immunol 185:5336–5347. 10.4049/jimmunol.1000779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Lee V, Finn A, Senger K, Zarrin AA, Du Pasquier L, Hsu E (2012) Origin of immunoglobulin isotype switching. Curr Biol. 10.1016/j.cub.2012.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman LM, Vogel LA, Bowden RM (2010) Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol 213:661–671. 10.1242/jeb.038315 [DOI] [PubMed] [Google Scholar]