Abstract
Background
Considering the high invasiveness and mortality of glioma as well as the unclear key genes and signaling pathways involved in the development of gliomas, there is a strong need to find potential gene biomarkers and available drugs.
Methods
Eight glioma samples and twelve control samples were analyzed on the GSE31095 datasets, and differentially expressed genes (DEGs) were obtained via the R software. The related glioma genes were further acquired from the text mining. Additionally, Venny program was used to screen out the common genes of the two gene sets and DAVID analysis was used to conduct the corresponding gene ontology analysis and cell signal pathway enrichment. We also constructed the protein interaction network of common genes through STRING, and selected the important modules for further drug-gene analysis. The existing antitumor drugs that targeted these module genes were screened to explore their efficacy in glioma treatment.
Results
The gene set obtained from text mining was intersected with the previously obtained DEGs, and 128 common genes were obtained. Through the functional enrichment analysis of the identified 128 DEGs, a hub gene module containing 25 genes was obtained. Combined with the functional terms in GSE109857 dataset, some overlap of the enriched function terms are both in GSE31095 and GSE109857. Finally, 4 antitumor drugs were identified through drug-gene interaction analysis.
Conclusions
In this study, we identified that two potential genes and their corresponding four antitumor agents could be used as targets and drugs for glioma exploration.
Introduction
Glioma is not only a very high degree of malignancy, but also a primary brain tumor with a high recurrence rate and poor prognosis, with an incidence of 3.19 cases per 100,000 person years [1]. Although some progress has been made in early diagnosis, the majority of patients are still at an advanced stage of diagnosis, resulting in extremely high rates of mortality and disability in these patients [2]. According to current medical treatment standards, even with the maximum safe resection, the rate of early recurrence after surgery is extremely high due to the inherent ability of tumor cells to infiltrate the normal brain [3]. Besides, the average overall survival time (OS) of GBM patients is only 12–18 months even after the combination of external irradiation and temozolomide combined with (TMZ) and maintenance chemotherapy, [4,5]. At present, given that gliomas are prone to relapse after treatment and have an inferior prognosis, it is necessary to strengthen the research on the pathogenesis of glioma and explore the genetic markers of glioma, so as to provide the diagnosis and treatment basis for early clinical screening and treatment.
Over the past few years, molecular diagnostics, drug target discovery and other techniques that analyze differences in gene expression have become a hot topic in clinical cancer research. A public database supported by the National Center for Biotechnology Information (NCBI), the Comprehensive Gene Expression Database (GEO), contains dozens of basic disease gene expression profile in the experiment. Currently, GEO databases are being used extensively to identify and mine key genes and underlying mechanisms involved in disease progression [6]. Text mining of biomedical literature has been recognized as a reliable hypothesis-generating method that can reveal novel associations between genes and disease occurrence [7,8]. Although a great deal of research has been carried out on glioma in recent years, the specific pathogenesis of glioma remains unclear. Therefore, we combine gene expression chips with text mining, and analyze these data through modern approach software to find clinically meaningful clues, so as to gain new perspectives, such as new diagnostic gene markers and therapeutic targets [9,10].
In this article, we downloaded the GSE31095 gene expression datasets, which included eight glioma samples and twelve normal controls, from the Gene Expression Omnibus database (GEO) and identified differentially expressed genes (DEGs) by R software (version 3.6.3) [11,12]. Meanwhile, all the glioma genes were mined from the text mining. The intersection of the gene sets obtained from DEG and text mining was analyzed via the online tool Venny to obtain the common genes, and different bioinformatics methods were further used to conduct gene ontology, signaling pathway enrichment annotation, and protein and protein interaction research on these common genes. We then validated our results on another independent GSE109857 dataset. From these data, we could find the gene markers and related pathways that might be associated with glioma, which providing new insights into the molecular mechanism of hidden gliomas.
Methods
Data collection
We abstracted the gene expression chip data GSE31095 [13] and GSE109857 from the NCBI Gene Expression Comprehensive (GEO) web resource (https://www.ncbi.nlm.nih.gov/geo/) [6,14]. The GSE31095 cohort contains eight glioma samples and twelve normal control samples, while the GSE109857 dataset includes five glioma samples and five normal control samples.
Data preprocessing
The core R package was used to process the downloaded matrix files. After normalization, the differences between glioma and the control group were determined by truncation criteria (|log2 fold change (FC)| ≥ 2, FDR < 0.05), and selected the remarkable DEGs for downstream analyses [14,15].
Text mining
Text mining was based on web services GenCLIP3 platform (http://ci.smu.edu.cn/genclip3/analysis.php/). When manipulated, GenCLIP3 was further used to retrieve all the gene names found in the existing literature relevant to the search topic [16]. We searched for the concept of glioma and screened all the genes associated with the topic from the results. The gene set obtained by text mining further intersected with the previously obtained differential gene set for the next step of analysis.
Gene ontology analysis and KEGG pathway analysis
To characterize Gene products and their functional characteristics, we used a Gene ontology (GO) approach and provided a standard vocabulary for corresponding terms. The GO terms included biological processes (BP), cellular composition (CC), and molecular function (MF), which reflected the current understanding of genes [17,18]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) database, as an open access informatic source for explaining the biological functions of organic systems, provides a large number of known biological pathways data resources, and the resources are comments for with their respective KEGG pathway of a gene or group of genes/proteins. Besides, a variety of online tools for functional and path enrichment analysis were further used to interpret the resulting intersection function and signal path analysis [19]. FDR<0.05 was considered to be statistically significant.
PPI network and module analysis
The resulting common set of genes obtained from the online database STRING, a database of 3.1 billion interactions across about 5 K organisms [20], was uploaded to the database for retrieving interacting genes [21]. Steps were as follows. The list of selected genes was firstly mapped to the STRING site to evaluate their interactions. And the genes were selected, when the PPIs comprehensive score was >0.9 and the degree of close correlation with other genes was adjusted to ≥10 [22]. After selected, the genes were constructed into a PPI network using Cytoscape visualization software [23]. MCODE was further used to classify the vital gene modules, and the related parameter standards were set by default, except k-core = 7. The genes of the selected module were finally analyzed by functional enrichment with FDR< 0.05 as the standard.
Drug-gene interaction and functional analysis of potential genes
Through drug-gene interaction, the obtained glioma genes were combined with existing drugs to analyze and explore the potential targets of glioma. Drug gene interactions database (DGIdb: https://www.dgidb.org) is an open-source web site for browsing and filtering drug-gene interactions [24]. As potential therapeutic targets, the module genes were uploaded to the drug-gene database to be match with the existing drugs to obtain the potential genes that match the drugs.
Results
DEGs identification and Text mining
Firstly, 463 DEGs were selected from glioma samples and normal controls in the GSE31095 dataset through limma package built-in R software. Then 424 upregulated genes and 39 downregulated genes were selected. Meanwhile, 528 differentially expressed genes, including 186 upregulated genes and 342 downregulated genes, were obtained by analyzing the giloma samples in the GSE109857 dataset and the normal control group. The criteria were set|log2 fold change (FC)|≥ 2 and adjusted P <0.05.
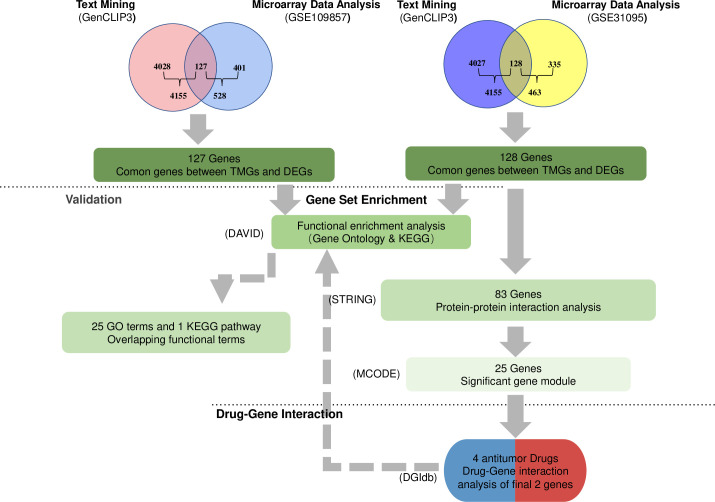
Through text mining, 4155 human genes associated with glioma. After the DEGs in the microarray data were crossed, the intersection of selected genes was obtained, and 128 genes involved in GSE31095 and 127 genes involved in GSE109857 were obtained (Fig 1).
Fig 1. The framework of data analyses.
Function and signal pathway enrichment analysis
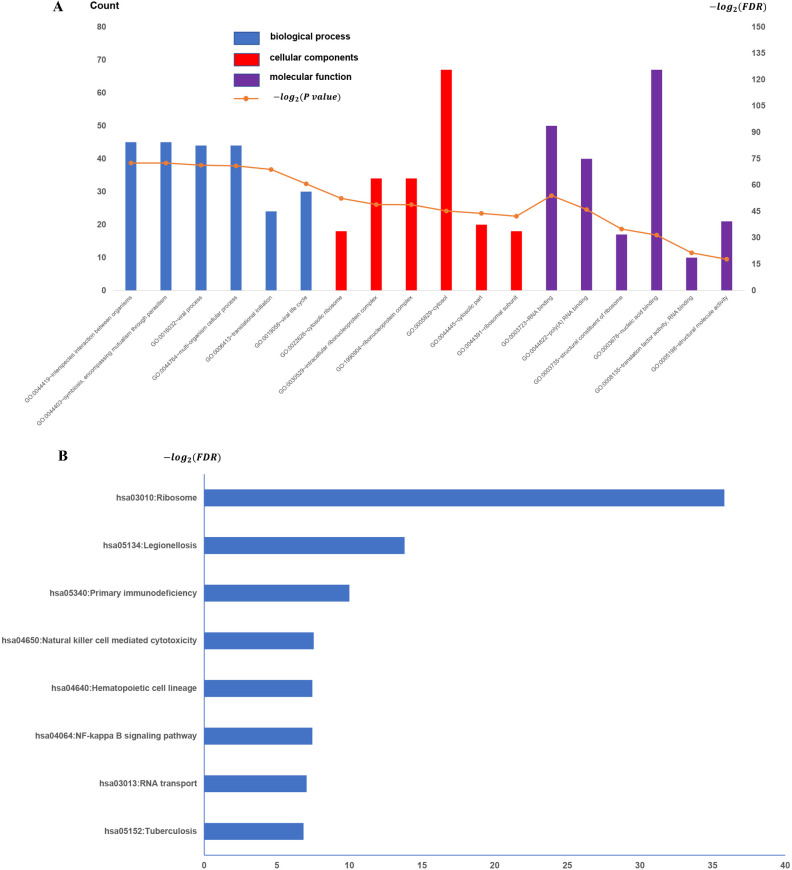
To establish the potential roles of the GSE31095 dataset DEGs, we carried out GO term analysis on the 463 genes. GO term analysis indicated that these genes were enriched for immune response (BP), inflammatory response (CC), and plasma membrane and receptor activity (MF) (Fig 2A), respectively. KEGG pathway analysis revealed 13 significantly enriched pathways. The top-5 most enriched pathways were: Tuberculosis, RNA transport, NF-kappa B signaling pathway, Hematopoietic cell lineage, and Natural killer cell-mediated cytotoxicity (Fig 2B).
Fig 2. All available significant gene ontology enrichment terms and signal pathway of the common genes from GSE31095 dataset.
(A) Top 10 GO terms. Number of gene of GO analysis was acquired from DAVID functional annotation tool. p <0.05. (B) KEGG pathway.
PPI network and module analysis
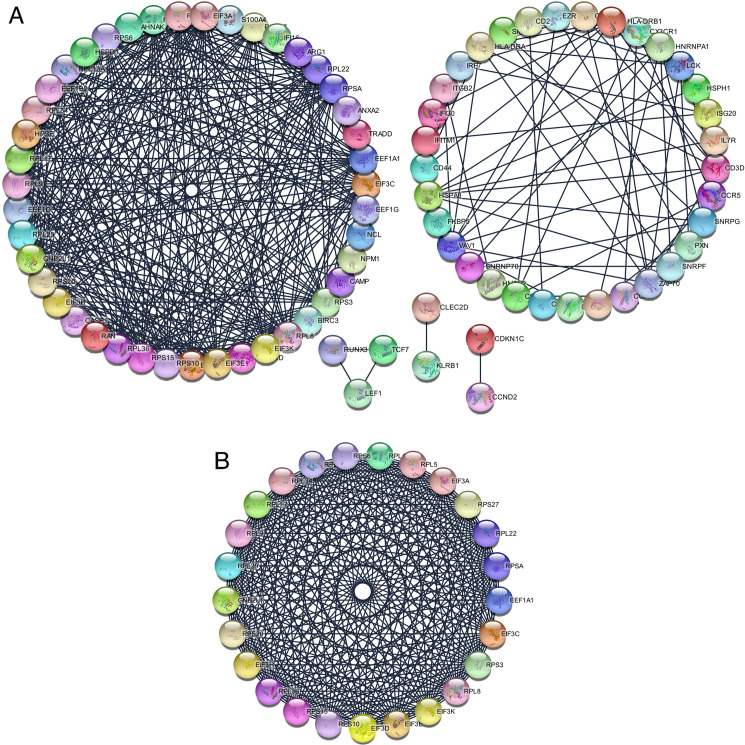
The co-genes were obtained via analyzing the STRING online database (http://string-db.org) and Cytoscape software, in which 128 genes were selected to enter the PPI network complex of co-genes with 83 nodes, 416 edges and a score of > 0.900 (highest confidence) (Fig 3A). Afterwards, based on MCODE, the highlighted modules were selected in the PPI network (25 nodes, 291 edges, Fig 3B).
Fig 3. The protein-protein interaction (PPI) networks construction and significant gene modules analysis.
(A) Based on the STRING online database, 128 common genes were filtered into common genes PPI network. (B) The most significant module from the PPI network.
Validation in GSE109857 dataset
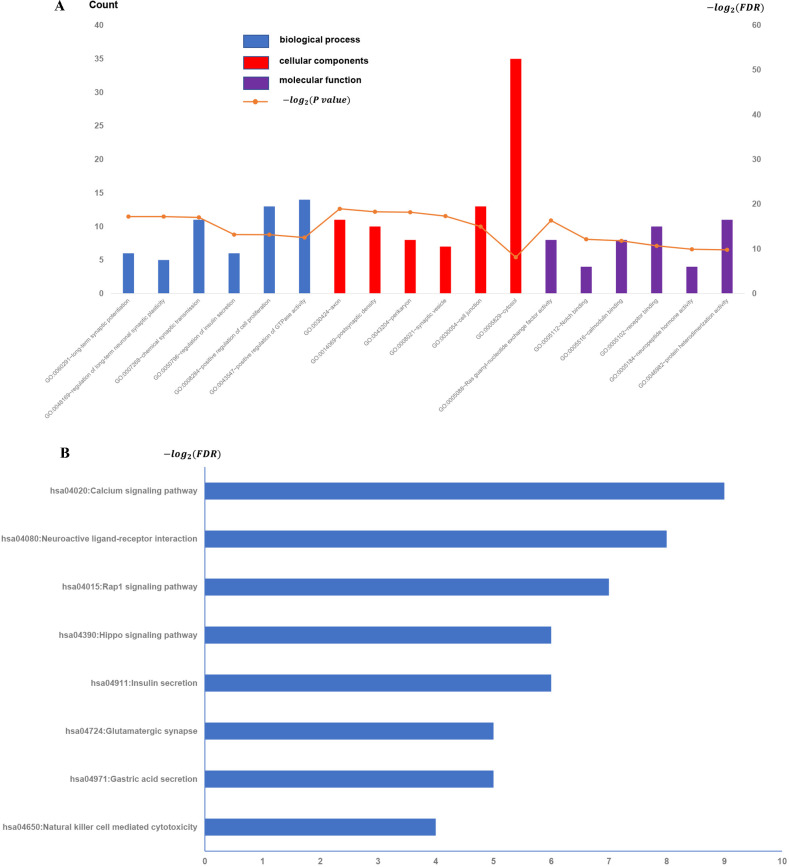
To test the reliability of the results derived from the GSE31095 dataset, we downloaded a cohort of five glioma samples and five normal control samples from another independent glioma dataset, GSE109857, and analyzed its gene expression data (Fig 4). Interesting, we found overlap of the enriched function terms between the GSE109857 and the previous GSE31095, and it is worth noting that there are 25 GO terms in the overlapping functional terms, whereas in KEGG there is only one pathway, "Natural killer cell-mediated cytotoxicity" (Table 1).
Fig 4. All available significant gene ontology enrichment terms and signal pathway of the common genes from GSE109857 dataset.
(A) Top 10 GO terms. Number of gene of GO analysis was acquired from DAVID functional annotation tool. p <0.05. (B) KEGG pathway.
Table 1. Overlap of the enriched function terms between the two datasets.
| Term | Category | Category |
|---|---|---|
| GO:0008284 | BP | positive regulation of cell proliferation |
| GO:0007417 | BP | central nervous system development |
| GO:0043065 | BP | positive regulation of apoptotic process |
| GO:0042493 | BP | response to drug |
| GO:0006468 | BP | protein phosphorylation |
| GO:0006816 | BP | calcium ion transport |
| GO:0032496 | BP | response to lipopolysaccharide |
| GO:0021537 | BP | telencephalon development |
| GO:0008285 | BP | negative regulation of cell proliferation |
| GO:0000165 | BP | MAPK cascade |
| GO:0043123 | BP | positive regulation of I-kappaB kinase/NF-kappaB signaling |
| GO:0001666 | BP | response to hypoxia |
| GO:0070374 | BP | positive regulation of ERK1 and ERK2 cascade |
| GO:0030054 | CC | cell junction |
| GO:0005576 | CC | extracellular region |
| GO:0005829 | CC | cytosol |
| GO:0043209 | CC | myelin sheath |
| GO:0009986 | CC | cell surface |
| GO:0005912 | CC | adherens junction |
| GO:0045121 | CC | membrane raft |
| GO:0005102 | MF | receptor binding |
| GO:0042802 | MF | identical protein binding |
| GO:0032403 | MF | protein complex binding |
| GO:0019899 | MF | enzyme binding |
| GO:0008092 | MF | cytoskeletal protein binding |
| hsa04650 | KEGG | Natural killer cell-mediated cytotoxicity |
GO, Gene ontology. BP. Biological processes. CC. Cellular composition. MF. Molecular function. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Drug-gene interaction and functional analysis of potential genes
Analysis of the drug-gene interaction was performed on 25 potential genes clustered in critical gene module 1. Based on the DGIdb results, there were two drugs interacted with gene EEF1A1 (eukaryotic translation elongation factor 1 alpha 1), while RPL11 (ribosomal protein L11), RPL13A (ribosomal protein L13a), RPL8 (ribosomal protein L8) and RPSA (ribosomal protein SA) were strongly associated with three different drugs, respectively. Out of these 14 drugs, only four were found to have the anti-tumor effects in glioma therapy and targeted to RPL8 and PPSA genes.
Discussion
Glioma is a deadly malignant brain tumor with strong invasiveness, vascular hyperplasia and poor prognosis [5], and lacks of effective treatment methods. Combination therapy is considered to be a promising approach to treat cancer for its effective anti-cancer effects and lower side effects. At present, although some progress has been made in multimodal treatment of glioma, including surgical removal, local irradiation and conventional chemotherapy [25], patients with glioma still have problems such as relapse and drug resistance, so the mortality rate of patients within two years after diagnosis is still very high [26].
In this regard, the candidate hub genes and signal pathways of glioma were screen out through a series of bioinformatics methods. 4155 genes related to Glioma were obtained through text mining and 428 DEGs were acquired by comparing the eight glioma samples with twelve normal control samples. After intersecting the set of genes obtained from text mining with the previously obtained DEGs, the common set of genes were got. Then, 25 hub genes were screened out by the network analysis of GO, KEGG and PPI. Finally, validation of our results using independent glioma dataset, GSE109857, verified that the expression of the some GO function and one KEGG pathway overlap with the previous data set (Table 1). Of these, 4 target RPL8 and RPSA and possess antineoplastic properties.
After validation through the GSE109857 dataset, the only overlapping KEGG term "Natural killer cell-mediated cytotoxicity" was obtained. Natural killer (NK) cells are essential lymphocytes that can kill virus-infected and cancer cells [27–29]. In recent studies, NK cells have been increasingly used in clinical trials in patients with cancer [30]. Studies have shown that NK cells release large amounts of interferon (IFN) -γ and are the main source of IFN - γ in the human body, and lack of NK cell-mediated production of IFN- γ is associated with an increased incidence of malignancy and infection [31].
RPL8 is reported to be involved in the occurrence of many diseases including osteosarcoma (OS) and also the corresponding treatment targets [32]. Besides, RPL8 regulates the protein synthesis process of Disc Degeneration (DD), suggesting that COL3A1 might be used for the diagnosis and treatment of DD [33]. A study of Swoboda et al. also showed that RPL8 antigen may be a relevant vaccine target for melanoma, glioma and breast cancer patients [34]. Since RPL8 is part of the ribosomal 60S subunit and participates in protein synthesis, RPL8 antigen is considered to be a relevant vaccine target for glioma [34].
Although Shi et al. recently have discovered that the RPSA gene might be related to the pyrazinamide (PZA) resistance in clinical Mycobacterium tuberculosis [35], some reports indicate that RPSA gene sequencing may not play a role in the detection of PZA sensitivity by molecular methods [36]. The correlation with tumors shows that RPSA can be used as a target for H2O2, and oxidized RPSA is found in clusters of specific adhesion molecules. In this study, we also found that RPSA oxidation in vitro improved the adhesion efficiency of cells to laminin [37]. Besides, RPSAs, which highly expressed in tumor cells, regulates the cell adhesion as one of its ribose in vitro functions and is directly related to metastatic potential [38,39]. Therefore, highly expressed RPSA in pancreatic cancer is reported to be closely related to the cancer invasion and metastasis due to the binding of RPSA-mediated cell adhesion laminin [40], further revealing a poor prognosis [41]. Another report further proved that RPSA regulates pancreatic cancer mainly through inhibiting caspase activity, which is a key protein of mediating apoptosis [42]. RPSA is also reported to be highly expressed in lung cancer, colorectal cancer, breast cancer and esophageal cancer, and RPSA can prevent tumor cells from autophagy in both breast cancer and esophageal cancer [43–45].
Four drugs (Puromycin targeting RPL8; Doxorubicin, Daunorubicin, Mitoxantrone targeting RPSA) were identified as potential drug candidates with antineoplastic activities and played the vital role in Glioma therapy.
Puromycin (RPL8), an old antibiotic derived from Streptomyces alboniger [46], is known that its antitumor activity is achieved by inhibiting 45S pre-ribosomal RNA and upstream binding factor (UBF) in MDA-MB-231 cells [47,48].It also has been found to induce apoptosis in breast cancer cells by insulin-like growth factor 1 (IGF-I), because it prevents the ribosomal protein generate process by causing the premature release of a polypeptide from the ribosome in malignant cells. In addition, studies have proved that puromycin can enhance its antineoplastic effect via combinating with other drugs, such as RITA or doxorubicin, which can be effectively used for wild-type P53 cancers [49].
Daunorubicin (RPSA) is a functional drug that exerts the antineoplastic effects through direct cytotoxicity and an apoptosis-inducing effect through the apoptotic signaling pathways in the cell cytoplasm and mitochondria. As a chemotherapy strategy for treating brain glioma, functionally targeted daunorubicin liposomes not only have the ability to eliminate gliomas, but also have the potential to remove glioma stem cells [50]. Meanwhile, the double-targeted daunorubicin liposomes can improve the therapeutic effect of glioma both in vitro and in vivo, and also significantly increase the transport rate of the blood-brain barrier model, up to 24.9%.
Doxorubicin (DOX) is identified as one of the most common and economic chemotherapy drugs in the treatment of malignant gliomas. However, when DOX is used alone, its clinical application is limited by its serious side effects [51,52]. Therefore, many drugs that could be combined with DOX are found in a series of subsequent studies. Among them, Gao et al., constructed a novel combination therapy to synthesize 131I-DOX-NL using two traditional drugs, DOX and 131I, which not only significantly reduced the side effects of DOX, but also effectively played an antitumor effect [53]. Besides, doxorubicin combined with dacarbazine is often used as a first-line treatment for leiomyosarcoma [54–60].
As mentioned above, the most common treatment for cancer is combination therapy [61,62], as is MTO. In previous studies, MTO has been found to be extensively used to treat metastatic, and castration-resistant prostate cancer, acute myeloid and lymphoblastic leukemias [63–68].
Up to date, the genes and drugs we have identified are only preliminarily studied in previous studies. Therefore, if further verification of its accuracy is needed, the above results need to be combined with basic experiments or computer simulations. In recent years, Chen’s professional research team has developed a computer model of miRNA-disease association prediction (MDHGI) to discover new miRNA-disease associations by integrating the predicted association probability obtained from matrix decomposition through sparse learning method [69–72]. If this model is included in biometric analysis, a broader simulation can be carried out through big data and disease data can be accurately analyzed, so as to obtain more targeted genes and targeted therapy drugs for future clinical research and treatment.
Conclusions
In summary, we analyzed a GSE31095 dataset and performed functional enrichment analysis. We then validated our approach on an independent GSE109857 dataset. Finally, 2 identified potential genes (RPL8 and RPSA) were analyzed on DGIdb and four potential antitumor drugs (Puromycin, Doxorubicin, Daunorubicin and Mitoxantrone) identified. Some of the identified genes are potential glioma biomarkers. Characterization of the identified drugs will offer more insights into potential, novel therapeutic strategies against glioma.
Acknowledgments
Thanks to Bin Zhao (Official Wechat Account: SCIPhD) of ShengXinZhuShou for English editing on the manuscript.
Data Availability
All GSE files are available from the https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31095 database.
Funding Statement
This study was funded by the National Natural Science Foundation of China, grant number 82072777 to ZW. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Dolecek. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2005–2009 (vol 14, pg 1, 2012). Neuro-Oncology. 2013;15(5):646–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou YX, Liu F, Xu QN, Wang XY. Analysis of the expression profile of Dickkopf-1 gene in human glioma and the association with tumor malignancy. Journal of Experimental & Clinical Cancer Research. 2010;29. 10.1186/1756-9966-29-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang QH, Hu BL, Hu X, Kim H, Squatrito M, Scarpace L, et al. Tumor Evolution of Glioma-Intrinsic Gene Expression Subtypes Associates with Immunological Changes in the Microenvironment (vol 32, pg 42, 2017). Cancer Cell. 2018;33(1):152. 10.1016/j.ccell.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJB, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. New England Journal of Medicine. 2005;352(10):987–96. 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 5.Meyer MA. Malignant gliomas in adults. New England Journal of Medicine. 2008;359(17):1850. [DOI] [PubMed] [Google Scholar]
- 6.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Research. 2013;41(D1):D991–D5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu S, Tranchevent L-C, De Moor B, Moreau Y. Gene prioritization and clustering by multi-view text mining. Bmc Bioinformatics. 2010;11. 10.1186/1471-2105-11-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosca E, Bertoli G, Piscitelli E, Vilardo L, Reinbold RA, Zucchi I, et al. Identification of functionally related genes using data mining and data integration: a breast cancer case study. Bmc Bioinformatics. 2009;10. 10.1186/1471-2105-10-S12-S8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo YC, Bao YH, Ma M, Yang WC. Identification of Key Candidate Genes and Pathways in Colorectal Cancer by Integrated Bioinformatical Analysis. International Journal of Molecular Sciences. 2017;18(4). 10.3390/ijms18040722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg EL. Systems biology in drug discovery and development. Drug Discovery Today. 2014;19(2):113–25. 10.1016/j.drudis.2013.10.003 [DOI] [PubMed] [Google Scholar]
- 11.Racine JS. RStudio: A Platform-Independent IDE for R and Sweave. Journal of Applied Econometrics. 2012;27(1):167–72. [Google Scholar]
- 12.Smyth GK. Limma: Linear models for microarray data. In: Gentalman R, Carey VJ, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solution Using R and Bioconductor. Statistics for Biology and Health; 2005. p. 397–420. [Google Scholar]
- 13.Hooper CM, Hawes SM, Kees UR, Gottardo NG, Dallas PB. Gene Expression Analyses of the Spatio-Temporal Relationships of Human Medulloblastoma Subgroups during Early Human Neurogenesis. Plos One. 2014;9(11). 10.1371/journal.pone.0112909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–10. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larriba Y, Rueda C, Fernandez MA, Peddada SD. Microarray Data Normalization and Robust Detection of Rhythmic Features. In: BolonCanedo V, AlonsoBetanzos A, editors. Microarray Bioinformatics. Methods in Molecular Biology. 19862019. p. 207–25. [DOI] [PubMed] [Google Scholar]
- 16.Wang J-H, Zhao L-F, Wang H-F, Wen Y-T, Jiang K-K, Mao X-M, et al. GenCLiP 3: mining human genes’ functions and regulatory networks from PubMed based on co-occurrences and natural language processing. Bioinformatics (Oxford, England). 2019. 10.1093/bioinformatics/btz807 [DOI] [PubMed] [Google Scholar]
- 17.Carbon S, Dietze H, Lewis SE, Mungall CJ, Munoz-Torres MC, Basu S, et al. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Research. 2017;45(D1):D331–D8. 10.1093/nar/gkw1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hale ML, Thapa I, Ghersi D. FunSet: an open-source software and web server for performing and displaying Gene Ontology enrichment analysis. Bmc Bioinformatics. 2019;20. 10.1186/s12859-019-2960-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4(1):44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 20.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Research. 2017;45(D1):D362–D8. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Research. 2013;41(D1):D808–D15. 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou Z, Cheng Y, Jiang YA, Liu S, Zhang M, Liu J, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. International Journal of Biological Sciences. 2018;14(2):124–36. 10.7150/ijbs.22619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4(2 Cited February 14, 2003). 10.1186/1471-2105-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffith M, Griffith OL, Coffman AC, Weible JV, McMichael JF, Spies NC, et al. DGIdb: mining the druggable genome. Nat Methods. 2013;10(12):1209. 10.1038/nmeth.2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurology. 2010;9(7):717–26. 10.1016/S1474-4422(10)70105-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rock K, McArdle O, Forde P, Dunne M, Fitzpatrick D, O’Neill B, et al. A clinical review of treatment outcomes in glioblastoma multiforme-the validation in a non-trial population of the results of a randomised Phase III clinical trial: has a more radical approach improved survival? British Journal of Radiology. 2012;85(1017):E729–E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sung PS, Jang JW. Natural Killer Cell Dysfunction in Hepatocellular Carcinoma: Pathogenesis and Clinical Implications. Int J Mol Sci. 2018;19(11). 10.3390/ijms19113648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim O, Jung MY, Hwang YK, Shin EC. Present and Future of Allogeneic Natural Killer Cell Therapy. Front Immunol. 2015;6:286. 10.3389/fimmu.2015.00286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang F, Xiao W, Tian Z. Challenges of NK cell-based immunotherapy in the new era. Front Med. 2018;12(4):440–50. 10.1007/s11684-018-0653-9 [DOI] [PubMed] [Google Scholar]
- 30.Cheng M, Chen Y, Xiao W, Sun R, Tian Z. NK cell-based immunotherapy for malignant diseases. Cell Mol Immunol. 2013;10(3):230–52. 10.1038/cmi.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton DK, Pitts-Meek S, Keshav S, Figari IS, Bradley A, Stewart TA. Multiple defects of immune cell function in mice with disrupted interferon-gamma genes. Science. 1993;259(5102):1739–42. 10.1126/science.8456300 [DOI] [PubMed] [Google Scholar]
- 32.Sun L, Li J, Yan B. Gene expression profiling analysis of osteosarcoma cell lines. Molecular Medicine Reports. 2015;12(3):4266–72. 10.3892/mmr.2015.3958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang ZD, Chen X, Zhang QL, Cai B, Chen K, Chen ZQ, et al. Dysregulated COL3A1 and RPL8, RPS16, and RPS23 in Disc Degeneration Revealed by Bioinformatics Methods. Spine. 2015;40(13):E745–E51. 10.1097/BRS.0000000000000939 [DOI] [PubMed] [Google Scholar]
- 34.Swoboda RK, Somasundaram R, Caputo L, Ochoa EM, Gimotty PA, Marincola FM, et al. Shared MHC class II-dependent melanoma ribosomal protein L8 identified by phage display. Cancer Research. 2007;67(8):3555–9. 10.1158/0008-5472.CAN-06-2763 [DOI] [PubMed] [Google Scholar]
- 35.Shi WL, Zhang XL, Jiang X, Yuan HM, Lee JS, Barry CE, et al. Pyrazinamide Inhibits Trans-Translation in Mycobacterium tuberculosis. Science. 2011;333(6049):1630–2. 10.1126/science.1208813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander DC, Ma JH, Guthrie JL, Blair J, Chedore P, Jamieson FB. Gene Sequencing for Routine Verification of Pyrazinamide Resistance in Mycobacterium tuberculosis: a Role for pncA but Not rpsA. Journal of Clinical Microbiology. 2012;50(11):3726–8. 10.1128/JCM.00620-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilas-Boas F, Bagulho A, Tenente R, Teixeira VH, Martins G, da Costa G, et al. Hydrogen peroxide regulates cell adhesion through the redox sensor RPSA. Free Radical Biology and Medicine. 2016;90:145–57. 10.1016/j.freeradbiomed.2015.11.019 [DOI] [PubMed] [Google Scholar]
- 38.Menard S, Tagliabue E, Colnaghi MI. The 67 kDa laminin receptor as a prognostic factor in human cancer. Breast Cancer Research and Treatment. 1998;52(1–3):137–45. 10.1023/a:1006171403765 [DOI] [PubMed] [Google Scholar]
- 39.Nelson J, McFerran NV, Pivato G, Chambers E, Doherty C, Steele D, et al. The 67 kDa laminin receptor: structure, function and role in disease. Bioscience Reports. 2008;28(1):33–48. 10.1042/BSR20070004 [DOI] [PubMed] [Google Scholar]
- 40.Canfield SM, Khakoo AY. The nonintegrin laminin binding protein (p67 LBP) is expressed on a subset of activated human T lymphocytes and, together with the integrin very late activation antigen-6, mediates avid cellular adherence to laminin. Journal of Immunology. 1999;163(6):3430–40. [PubMed] [Google Scholar]
- 41.Wu YH, Tan XD, Liu P, Yang YF, Huang YP, Liu XL, et al. ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Experimental Cell Research. 2019;379(1):30–47. 10.1016/j.yexcr.2019.03.022 [DOI] [PubMed] [Google Scholar]
- 42.Chetty CJ, Ferreira E, Jovanovic K, Weiss SFT. Knockdown of LRP/LR induces apoptosis in pancreatic cancer and neuroblastoma cells through activation of caspases. Experimental Cell Research. 2017;360(2):264–72. 10.1016/j.yexcr.2017.09.016 [DOI] [PubMed] [Google Scholar]
- 43.Khumalo T, Ferreira E, Jovanovic K, Veale RB, Weiss SFT. Knockdown of LRP/LR Induces Apoptosis in Breast and Oesophageal Cancer Cells. Plos One. 2015;10(10). 10.1371/journal.pone.0139584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanjuan X, Fernandez PL, Miquel R, Munoz J, Castronovo V, Menard S, et al. Overexpression of the 67-kD laminin receptor. Correlates with tumour progression in human colorectal carcinoma. Journal of Pathology. 1996;179(4):376–80. [DOI] [PubMed] [Google Scholar]
- 45.Wu MS, Tu T, Huang YC, Cao Y. Suppression subtractive hybridization identified differentially expressed genes in lung adenocarcinoma: ERGIC3 as a novel lung cancer-related gene. Bmc Cancer. 2013;13. 10.1186/1471-2407-13-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giege R, Frugier M, Rudinger J. tRNA mimics. Current Opinion in Structural Biology. 1998;8(3):286–93. 10.1016/s0959-440x(98)80060-2 [DOI] [PubMed] [Google Scholar]
- 47.Jung JH, Sohn EJ, Shin EA, Lee D, Kim B, Jung DB, et al. Melatonin Suppresses the Expression of 45S Preribosomal RNA and Upstream Binding Factor and Enhances the Antitumor Activity of Puromycin in MDA-MB-231 Breast Cancer Cells. Evidence-Based Complementary and Alternative Medicine. 2013. 10.1155/2013/879746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soderlund G, Haarhaus M, Chisalita S, Arnqvist HJ. Inhibition of puromycin-induced apoptosis in breast cancer cells by IGF-I occurs simultaneously with increased protein synthesis. Neoplasma. 2004;51(1):1–11. [PubMed] [Google Scholar]
- 49.Jung JH, Lee H, Kim JH, Sim DY, Ahn H, Kim B, et al. p53-Dependent Apoptotic Effect of Puromycin via Binding of Ribosomal Protein L5 and L11 to MDM2 and Its Combination Effect with RITA or Doxorubicin. Cancers. 2019;11(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao WY, Zhang CX, Liu L, Mu LM, Zeng F, Ju RJ, et al. Construction of Functional Targeting Daunorubicin Liposomes Used for Eliminating Brain Glioma and Glioma Stem Cells. Journal of Biomedical Nanotechnology. 2016;12(7):1404–20. 10.1166/jbn.2016.2266 [DOI] [PubMed] [Google Scholar]
- 51.Zhang YW, Shi JJ, Li YJ, Wei L. Cardiomyocyte death in doxorubicin-induced cardiotoxicity. Archivum Immunologiae Et Therapiae Experimentalis. 2009;57(6):435–45. 10.1007/s00005-009-0051-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duong HHP, Yung LYL. Synergistic co-delivery of doxorubicin and paclitaxel using multi-functional micelles for cancer treatment. International Journal of Pharmaceutics. 2013;454(1):486–95. 10.1016/j.ijpharm.2013.06.017 [DOI] [PubMed] [Google Scholar]
- 53.Gao JM, Fang L, Sun DY, Shen YM, Hu YM, Li N, et al. I-131-labeled and DOX-loaded multifunctional nanoliposomes for radiotherapy and chemotherapy in brain gliomas. Brain Research. 2020;1739. [DOI] [PubMed] [Google Scholar]
- 54.Borden EC, Amato DA, Rosenbaum C, Enterline HT, Shiraki MJ, Creech RH, et al. RANDOMIZED COMPARISON OF 3 ADRIAMYCIN REGIMENS FOR METASTATIC SOFT-TISSUE SARCOMAS. Journal of Clinical Oncology. 1987;5(6):840–50. 10.1200/JCO.1987.5.6.840 [DOI] [PubMed] [Google Scholar]
- 55.Antman K, Crowley J, Balcerzak SP, Rivkin SE, Weiss GR, Elias A, et al. AN INTERGROUP PHASE-III RANDOMIZED STUDY OF DOXORUBICIN AND DACARBAZINE WITH OR WITHOUT IFOSFAMIDE AND MESNA IN ADVANCED SOFT-TISSUE AND BONE SARCOMAS. Journal of Clinical Oncology. 1993;11(7):1276–85. 10.1200/JCO.1993.11.7.1276 [DOI] [PubMed] [Google Scholar]
- 56.Bitz U, Pink D, Busemann C, Reichardt P. Doxorubicin (Doxo) and dacarbacin (DTIC) as first-line therapy for patients (pts) with locally advanced or metastatic leiomyosarcoma (LMS) and liposarcoma (LPS). Journal of Clinical Oncology. 2011;29(15). [Google Scholar]
- 57.Zalupski M, Metch B, Balcerzak S, Fletcher WS, Chapman R, Bonnet JD, et al. PHASE-III COMPARISON OF DOXORUBICIN AND DACARBAZINE GIVEN BY BOLUS VERSUS INFUSION IN PATIENTS WITH SOFT-TISSUE SARCOMAS—A SOUTHWEST ONCOLOGY GROUP-STUDY. Jnci-Journal of the National Cancer Institute. 1991;83(13):926–32. 10.1093/jnci/83.13.926 [DOI] [PubMed] [Google Scholar]
- 58.Saiki JH, Baker LH, Rivkin SE, Shahbender S, Fletcher WS, Athens JW, et al. A USEFUL HIGH-DOSE INTERMITTENT SCHEDULE OF ADRIAMYCIN AND DTIC IN THE TREATMENT OF ADVANCED SARCOMAS. Cancer. 1986;58(10):2196–7. [DOI] [PubMed] [Google Scholar]
- 59.Judson I, Verweij J, Gelderblom H, Hartmann JT, Schoeffski P, Blay J-Y, et al. Doxorubicin alone versus intensified doxorubicin plus ifosfamide for first-line treatment of advanced or metastatic soft-tissue sarcoma: a randomised controlled phase 3 trial. Lancet Oncology. 2014;15(4):415–23. 10.1016/S1470-2045(14)70063-4 [DOI] [PubMed] [Google Scholar]
- 60.von Mehren M, Randall RL, Benjamin RS, Boles S, Bui MM, Ganjoo KN, et al. Soft Tissue Sarcoma, Version 2.2018. Journal of the National Comprehensive Cancer Network. 2018;16(5):536–63. 10.6004/jnccn.2018.0025 [DOI] [PubMed] [Google Scholar]
- 61.Batist G, Gelmon KA, Chi KN, Miller WH, Chia SKL, Mayer LD, et al. Safety, Pharmacokinetics, and Efficacy of CPX-1 Liposome Injection in Patients with Advanced Solid Tumors. Clinical Cancer Research. 2009;15(2):692–700. 10.1158/1078-0432.CCR-08-0515 [DOI] [PubMed] [Google Scholar]
- 62.Feldman EJ, Lancet JE, Kolitz JE, Ritchie EK, Roboz GJ, List AF, et al. First-In-Man Study of CPX-351: A Liposomal Carrier Containing Cytarabine and Daunorubicin in a Fixed 5:1 Molar Ratio for the Treatment of Relapsed and Refractory Acute Myeloid Leukemia. Journal of Clinical Oncology. 2011;29(8):979–85. 10.1200/JCO.2010.30.5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arseneau JC, Schoenfeld DA, Borden EC. A PHASE-II STUDY OF DIHYDROXYANTHRACENEDIONE (DHAD, MITOXANTRONE, NSC 301739) IN ADVANCED MALIGNANT-MELANOMA. Investigational New Drugs. 1986;4(1):53–6. 10.1007/BF00172017 [DOI] [PubMed] [Google Scholar]
- 64.Link KH, Sunelaitis E, Kornmann M, Schatz M, Gansauge F, Leder G, et al. Regional chemotherapy of nonresectable colorectal liver metastases with mitoxantrone, 5-fluorouracil, folinic acid, and mitomycin C may prolong survival. Cancer. 2001;92(11):2746–53. [DOI] [PubMed] [Google Scholar]
- 65.Le Deley MC, Suzan F, Cutuli B, Delaloge S, Shamsaldin A, Linassier C, et al. Anthracyclines, mitoxantrone, radiotherapy, and granulocyte colony-stimulating factor: Risk factors for leukemia and myelodysplastic syndrome after breast cancer. Journal of Clinical Oncology. 2007;25(3):292–300. 10.1200/JCO.2006.05.9048 [DOI] [PubMed] [Google Scholar]
- 66.Zinzani PL, Tani M, Pulsoni A, Gobbi M, Perotti A, De Luca S, et al. Fludarabine and mitoxantrone followed by yttrium-9 ibritumomab tiuxetan in previously untreated patients with follicular non-Hodgkin lymphoma trial: a phase II non-randomised trial (FLUMIZ). Lancet Oncology. 2008;9(4):352–8. 10.1016/S1470-2045(08)70039-1 [DOI] [PubMed] [Google Scholar]
- 67.de Bono JS, Oudard S, Ozguroglu M, Hansen S, Machiels JP, Kocak I, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010;376(9747):1147–54. 10.1016/S0140-6736(10)61389-X [DOI] [PubMed] [Google Scholar]
- 68.Schrappe M. Mitoxantrone in first-relapse paediatric ALL: the ALL R3 trial. Lancet. 2010;376(9757):1968–70. 10.1016/S0140-6736(10)62194-0 [DOI] [PubMed] [Google Scholar]
- 69.Chen X, Wang L, Qu J, Guan N-N, Li J-Q. Predicting miRNA–disease association based on inductive matrix completion. Bioinformatics. 2018; 34(24):4256–4265. 10.1093/bioinformatics/bty503 [DOI] [PubMed] [Google Scholar]
- 70.Chen X, Xie D, Zhao Q, You Z-H. MicroRNAs and complex diseases: from experimental results to computational models. Briefings in Bioinformatics. 2019;20(2):515–539. 10.1093/bib/bbx130 [DOI] [PubMed] [Google Scholar]
- 71.Chen X, Yan CC, Zhang X, You Z-H. Long non-coding RNAs and complex diseases: from experimental results to computational models. Briefings in Bioinformatics. 2017; 18(4):558–576. 10.1093/bib/bbw060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X, Yin J, Qu J, Huang L. MDHGI: Matrix Decomposition and Heterogeneous Graph Inference for miRNA-disease association prediction. PLOS Computational Biology. 2018;14(8): e1006418. 10.1371/journal.pcbi.1006418 [DOI] [PMC free article] [PubMed] [Google Scholar]