Abstract
Top-down mass spectrometry (MS) is a powerful tool for identification and comprehensive characterization of proteoforms arising from alternative splicing, sequence variation, and post-translational modifications. However, the complex dataset generated from top-down MS experiments requires multiple sequential data processing steps to successfully interpret the data for identifying and characterizing proteoforms. One critical step is the deconvolution of the complex isotopic distribution that arises from naturally occurring isotopes. Multiple algorithms are currently available to deconvolute top-down mass spectra, resulting in different deconvoluted peak lists with varied accuracy compared to true positive annotations. In this study, we have designed a machine learning strategy that can process and combine the peak lists from different deconvolution results. By optimizing clustering results, deconvolution results from THRASH, TopFD, MS-Deconv, and SNAP algorithms were combined into consensus peak lists at various thresholds using either a simple voting ensemble method or a random forest machine learning algorithm. For the random forest algorithm which had better predictive performance, the consensus peak lists on average could achieve a recall value (true positive rate) of 0.60 and a precision value (positive predictive value) of 0.78. It outperforms the single best algorithm which only achieved a recall value of 0.47, and a precision value of 0.58. This machine learning strategy enhanced the accuracy and confidence in protein identification during database search by accelerating detection of true positive peaks while filtering out false positive peaks. Thus, this method show promise in enhancing proteoform identification and characterization for high-throughput data analysis in top-down proteomics.
Keywords: Top-down mass spectrometry, machine-learning ensemble
Graphical Abstract

Introduction
Top-down mass spectrometry (MS) is a powerful tool for identification and comprehensive characterization of proteoforms, including alternative splicing, sequence variations, and post-translational modifications.1–5 One of the unique advantages of top-down MS is the ability to analyze intact proteins without proteolytic cleavage to obtain the mass spectra of various proteoforms simultaneously and subsequently fragment the proteoform to locate the site(s) of modification(s).6–7 A major challenge in top-down proteomics data analysis is the complexity of high-resolution top-down mass spectra.
The analysis of high-resolution top-down MS data requires several sequential processing steps, such as centroiding, deconvolution, proteoform identification, and quantification. Currently, many software tools have been developed to perform each step of the analysis process.8 Deconvolution is a critical step early in the analysis, as the results can significantly affect the performance of the downstream methods. In addition to the first high-resolution deconvolution software THRASH,9 other algorithms such as MS-Deconv,10 TopFD,11 pParseTD,12 and UniDec13 are also available for deconvolution of top-down MS data. Furthermore, instrument vendors also provide deconvolution algorithms such as SNAP algorithm14 by Bruker Corporation and Xtract algorithm by Thermo Scientific within their software products.
Due to the diversity of deconvolution algorithms provided to the scientific community, one potential challenge an analyst may face is the non-standardization of their parameters. Consequently, the resulting peak list from different deconvolution algorithms cannot be directly compared. Moreover, different deconvolution algorithms performed spectral deconvolution using diverse computational methods, resulting in different peak list output. For instance, THRASH9 is a subtractive peak finding routine that locates possible isotopic clusters in the spectrum by using least-squares fits to a theoretically derived isotopic abundance distributions. MS-Deconv10 is a combinatorial algorithm that uses graph theory to find the heaviest path in a largest set of potential candidate envelopes. TopFD11 is a successor to MS-Deconv which converts isotopomer envelopes to monoisotopic neutral masses after grouping top-down spectral peaks into isotopomer envelopes. The SNAP algorithm fits a function of superimposed bell curves to the peaks in order to identify the isotopic distributions (details regarding several common deconvolution algorithms were summarized in Table S1). Using a human histone dataset, Sun et al. showed that the peak list outputs among Xtract, MS-Deconv, and pParseTD had a maximum difference of 25% and 15% in recalled peak rate and recalled intensity rate, respectively.12 Last but not least, deconvolution algorithms may identify false positive peaks. The deconvolution results would need to be manually validated or corrected using software such as MASH Suite Pro,15 which can be time consuming. As a consequence of all these challenges, there is a need for the standardization of different deconvolution algorithms as well as a method that analyzes and combines results from available deconvolution algorithms.
In the machine learning community, ensemble methods (e.g. simple voting) and machine learning algorithms (e.g. random forest algorithm) have been developed to enhance the predictions of multiple distinct algorithms in order to improve the overall predictive performance.16–17 These ensemble methods and machine learning algorithms have also been employed in MS applications to improve the performance of disease diagnosis,18 to improve target protein identification,19 and to enhance de novo peptide sequence.20 In this study, by treating each deconvolution algorithm as a distinct algorithm, we propose that these ensemble methods and machine learning algorithms could be applied to combine different deconvolution results and obtain consensus peak lists. The resulting consensus peak lists should have higher accuracy, which will improve proteoform identification and mitigate manual validation efforts.
Herein, we report a novel use of machine learning strategy to combine the results from multiple deconvolution algorithms employed on high-resolution top-down MS to obtain consensus peak lists using an ensemble method and a machine learning algorithm. We compared and contrasted the predictive performance of our machine learning strategy against each deconvolution algorithm separately using a set of targeted MS data that has been annotated by an expert to obtain a “true positive” list and showed improved performance over each individual algorithm. We demonstrated that adding more deconvolution results, even results from the same algorithm with different parameters, could further improve predictive performance. Finally, we showed the utility of the consensus peak list generated by our machine learning strategy could improve downstream proteoform identification using a software tool such as MS-Align+. This machine learning strategy will be integrated into our developing software, MASH Explorer,21 a comprehensive and user-friendly tool for analyzing high-resolution top-down MS data.
Experimental Section
Mass Spectrometry Data Acquisition
The collision-induced dissociation (CID) and electron-capture dissociation (ECD) tandem MS (MS/MS) spectra from 15 sarcomeric protein proteoforms were published previously.22 The protein identification, accession number, and post-translational modifications were provided in Table S2. For data acquisition, the MS/MS data were collected on a 12 Tesla solariX Fourier transform ion cyclotron resonance (FTICR) mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with an automated chip-based nano-electrospray ionization source (Triversa NanoMate, Advion Bioscience, Ithaca, NY, USA). Details regarding sample collection, protein extraction, sample handling, experimental workflow, and instrument parameters are provided in the Supporting Information. The mass spectrometry proteomics raw data and annotations have been deposited to the ProteomeXchange Consortium via the PRIDE23 partner repository with the dataset identifier PXD018043.
Peak Extraction and Expert Annotation
Deconvoluted peaks were identified by four different algorithms, including THRASH,9 MS-Deconv,10 TopFD,11 and the SNAP algorithm from Bruker DataAnalysis14, which were available to process Bruker dataset. The peak extraction using the MASH Explorer software was executed with THRASH algorithm using a fit parameter of 60%, 70%, 80%, and 90%. The MS-Deconv algorithm was run using default parameters with a maximum charge of 30, maximum mass of 50,000, m/z error tolerance of 0.02, and an S/N ratio of 3. The TopFD deconvolution was employed using default parameters with a maximum charge of 30, MS1 S/N ratio of 3.0, precursor window size (m/z) of 3.0, maximum mass (Da) of 100,000, MS2 S/N ratio of 1.0, and an m/z error of 0.02. Using DataAnalysis available for the Bruker dataset, the deconvoluted ion list was obtained using the SNAP algorithm with a quality factor threshold of 0.1, S/N threshold of 2, relative intensity threshold (base peak) of 0.01%, absolute intensity threshold of 0, and a maximum charge state of 50. All deconvolution results were output into MSAlign format, which provides information of the monoisotopic distributions including monoisotopic mass, intensity, and charge. While this manuscript focused on data acquired from Bruker instrument, this method is applicable for datasets from other vendors if the peak information was converted to MSAlign format.
Coding Environment
Python (2.7.10) was used to generate the clusters, and R (3.6.0) was used to perform the machine learning analysis and to automate the MS-Align+ searches.
Machine Learning Strategy for Combining Multiple Deconvolution Results
A general overview of the data analysis process was provided in Figure 1. Each MSAlign file was parsed by a Python script, and the results were concatenated into one peak list which records the monoisotopic mass, charge, and source algorithm. Peaks having the same charge and similar monoisotopic mass were clustered together as the same peak. The clusters were then filtered using simple voting or machine learning methods, and the results were output into a consensus MSAlign file. Each part of the process is described in more detail below.
Figure 1. Flowchart for the machine learning strategy.

This figure shows the steps taken to combine deconvolution results into a consensus peak list using either the simple voting method or using a machine learning algorithm.
Hierarchical Clustering -
The algorithm merges the full list of deconvoluted peaks into clusters that contain the same charge and are similar in monoisotopic mass. Inspired by Robert Tibshirani’s work on ‘peak probability contrasts’,24 the method uses hierarchical clustering,25 using the difference between pairs of peaks from the log10 transformed monoisotopic mass as the distance metric. Transforming the monoisotopic mass using log removes the linear dependence of the error with mass, so a constant cutoff can be used to determine the number of clusters. A further constraint was added to ensure the charges are the same between peaks with the proposed clusters. Using Equation 1, a cutoff was determined using a user-defined threshold ppm error within the cluster, which ensured that the distances between the largest and smallest mass of the peaks within the cluster were not larger than the +/− ppm threshold. The average of the monoisotopic mass was then used as the center of the cluster. The clustering algorithms ran on each spectrum separately.
| Equation 1 |
Expert Annotation and Assignment to Clusters -
The expert annotations were obtained and verified manually using the MASH software with the embedded enhanced-THRASH algorithm at 60% fit setting.15 The peaks were manually validated by adjusting the most abundant m/z and charge state of each monoisotopic distribution. In this study, we considered expert annotated peaks as true positive peaks.
The identified clusters were annotated using the expert annotations by finding clusters that were of the same charge and within a +/−X ppm window of the expert annotated monoisotopic mass (where X is set to the same value as used in the clustering). In cases where an expert peak could be assigned to multiple clusters, we select the pair with the smallest distance between the monoisotopic mass with expert assignment that matched to multiple possible clusters as the true match. Clusters with assigned expert annotation were called expert matched peaks, and the unassigned clusters were labelled as unmatched expert annotated peaks.
Machine Learning Analysis -
The machine learning analysis was performed using the R language. For each cluster, a feature vector was generated using the features described in Table 1. We set up a machine learning task to separate expert annotated matched clusters from unmatched clusters. Precision-recall curves were estimated using leave-one-spectrum out cross-validation, where each fold estimates the probabilities of a true annotation for each of the clusters found in one spectrum using a machine learning model built from the other spectra feature vectors. Recall and precision are defined in Equation 2 and 3,
| Equation 2 |
| Equation 3 |
where a true positive is a cluster peak with an expert annotation, a false negative is an unmatched expert annotation, and a false positive is a cluster peak without an expert annotation. Recall measures the percentage of expert annotations found by the algorithm, whereas precision measures the percentage of cluster peak calls that have true annotations. We compared and contrasted the predictive performance using the random forest (randomForest R package)26 model using ntree (the number of trees used in the forest) of 100 and the remaining parameters set to their defaults.
Table 1 -.
Description of Features used in Machine Learning
| Feature Name | Data Type | Description |
|---|---|---|
| Activation | ECD/CID | Activation used to generate spectra |
| Charge | Integer | Charge of the peaks within the cluster |
| Votes | Integer | Number of deconvolution algorithms that called a peak within that cluster |
| SumIntensity | Numeric | Sum of the intensity of peaks in the cluster |
| AverageIntensity | Numeric | Average intensity of peaks in the cluster |
| MSDeconv | Boolean | MS-Deconv called this peak |
| TopFD | Boolean | TopFD called this peak |
| THRASH60 | Boolean | THRASH with 60% Fit called this peak |
| THRASH70 | Boolean | THRASH with 70% Fit called this peak |
| THRASH80 | Boolean | THRASH with 80% Fit called this peak |
| THRASH90 | Boolean | THRASH with 90% Fit called this peak |
| SNAP | Boolean | SNAP called this peak |
| PrecursorCharge | Integer | Charge of the precursor |
| AvgMass | Numeric | Average Mass of the peaks within the cluster |
| StdDev | Numeric | Standard Deviation of the mass of the peaks within the cluster |
| PrecursorMass | Numeric | Monoisotopic mass of the precursor |
| PrecursorMZ | Numeric | m/z of the precursor |
To compare the individual algorithms on the precision-recall curves, all of the true positives, true negatives (cluster peaks with no expert annotations that algorithms did not call as annotations), false positives, and false negatives results were aggregated before calculating precision and recall values. Deconvolution methods were also compared by calculating the F1 score, a metric that balances between precision and recall as defined in Equation 4. For random forest, we selected the probability threshold that maximizes the F1 score within the training dataset to make the final call on the associated test set.
| Equation 4 |
Due to the high rate of false positives within the datasets, we used precision-recall curves to visualize the accuracy of the methods. Typically, precision-recall curves have a point for recall value of 1 and precision value of 0, which indicates a machine learning algorithm that calls all clusters true peaks. However, if we count the false negatives incurred by the upstream clustering method, the curve will give a lower maximum achievable recall result. A superior performing classification algorithm would have a point (or curve) that is higher in precision and recall (i.e. more top-right) than the contrasted algorithm(s). For example, the point for one deconvolution algorithm which gives a recall of 0.50 and precision of 0.40 outperformed the point for the second deconvolution algorithm that gives a recall of 0.60 and precision of 0.50.
Cluster Filtering and Consensus Deconvolution Results -
To reduce false positive clusters, we explored two avenues of filtering. One was a simple voting heuristic that thresholds clusters based upon the number of deconvolution algorithms that called the peaks within the cluster. Another route was to apply the previously described machine learning models to assign a probability of a true expert assignment to each cluster. The clusters were filtered via thresholding upon this probability. Consensus results were output as an MSAlign file, which were processed using database search.
Database Searching -
All searches were performed using MS-Align+ v0.7.1.714327 with a fasta database file derived from a human database (Uniprot-Swissprot database, released December 2019, containing 20,367 protein sequences) for βTpm, a cynomolgus monkey database (Uniprot-Swissprot database, released January 2020, containing 77,341 protein sequences) for fsTnT5, a rat database (Uniprot-Swissprot database, release January 2020, containing 8,085 protein sequences) for αTpm, and a rhesus macaque database (Uniprot-Swissprot database, released January 2020, containing 78,285 protein sequences) for the rest of the proteins. We compared and contrasted the search results of MS-Align+ using the MSAlign file from each deconvolution algorithm, the expert annotated peaks, the simple voting method, and the random forest machine learning method.
Results and Discussion
Setting clustering ppm cutoff and clustering choices
One of the important parameters to determine for the machine learning strategy is the choice of the ppm cutoff for calling clusters. To determine the optimal cutoff for the dataset presented in this work, we evaluated four parameters: the number of clusters, the percent of peaks assigned, the cross-validated accuracy from the random forest model (the percentage of correct annotations found over the whole dataset), and the random forest’s F1 score (a measure of accuracy that is the harmonic mean of precision and recall), at multiple different cutoff levels (1 ppm, 2 ppm, 5 ppm, 10 ppm, 20 ppm, 50 ppm, 100 ppm, and 200 ppm). The results in Figure 2 demonstrated that 10 ppm was optimal for the clustering cutoff because 1) for values greater than 10 ppm clustering cutoff, there was a noticeable drop in the number of clusters (Figure 2a), 2) the percent of recalled peaks was not significantly less than that from higher ppm cutoff, while greater than that from lower ppm cutoff (Figure 2b), and 3) the accuracy and the overall accuracy measured by F1 score did not differ significantly from the optimal values in both measurements (Figure 2c and 2d).
Figure 2. Cluster cutoff performance.

Each plot is a boxplot that shows the spread of the metric measured from the 30 spectra versus different ppm cutoffs used in the hierarchical clustering step. (a) Number of clusters, (b) percent of recalled peaks versus ppm cutoff, (c) random forest accuracy versus ppm cutoff, and (d) random forest F1 score versus ppm cutoff. The black squares in the figure represent outliers in the dataset.
Figure 3. Precision-recall curves and points of the expert annotation prediction task.
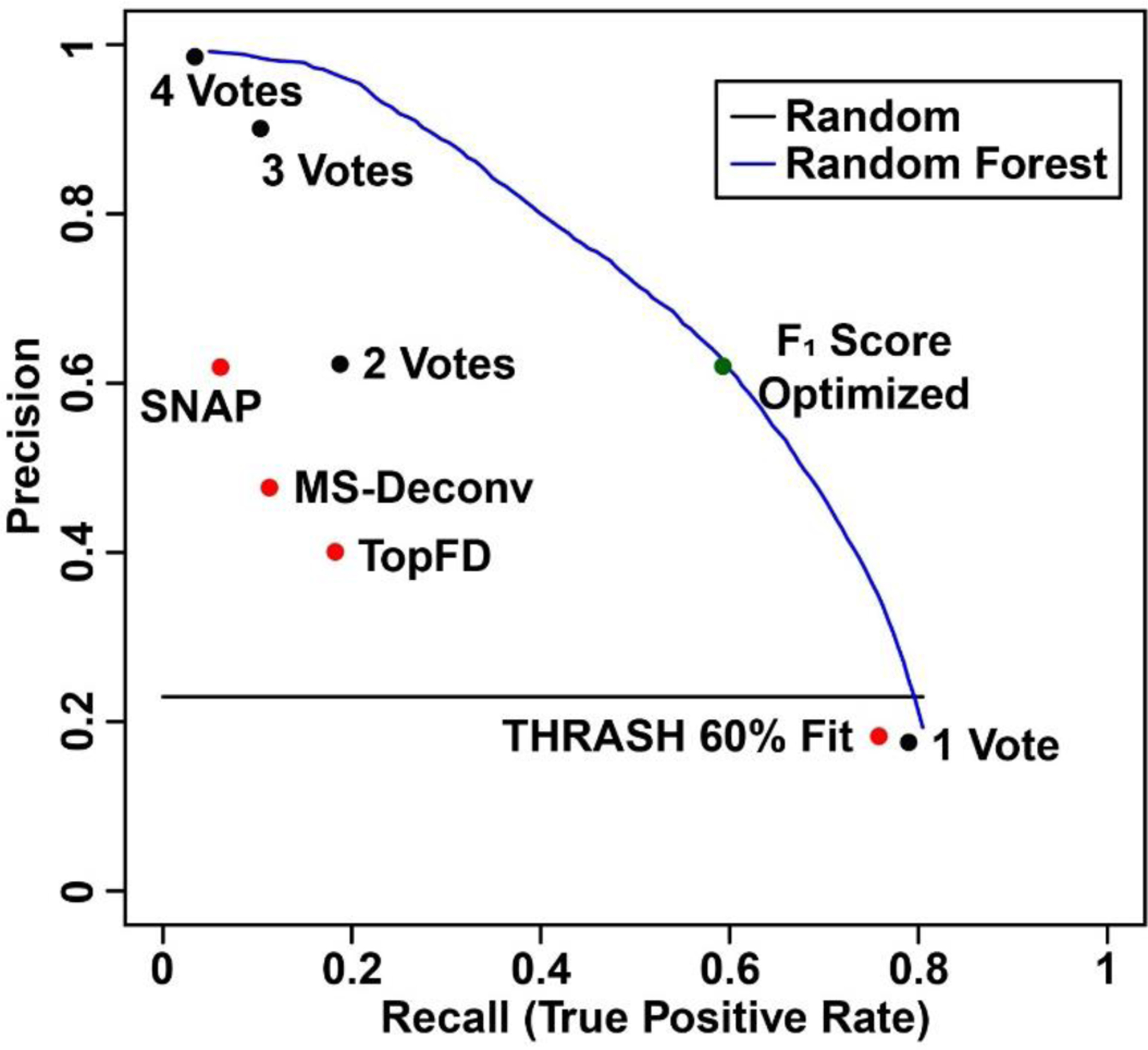
Plot displays the precision and recall performance of the deconvolution methods by themselves (red points), the simple voting (black points), and random forest (blue line). The green point represents random forest algorithm with F1 score optimized.
Many other clustering algorithms exist in the literature, including different linkage algorithms for hierarchical clustering.28 In this work, we used complete hierarchical clustering, which gives tight clusters (min/max rather than average). This is desirable for merging peaks by monoisotopic mass.
Expert annotation accuracy performance with 4-vote ensemble
After determining the optimal hierarchical clustering cutoff, the peak clusters were analyzed by ensemble/machine learning methods and individual deconvolution algorithms for comparison. In this study, we used a simple voting ensemble method which is based upon the number of unique deconvolution algorithms that called a peak within that cluster. Additionally, the random forest machine learning algorithm, which is itself an ensemble of decision trees, was utilized.26 The random forest algorithm was shown to be able to handle large datasets and exhibit excellent performance in the classification tasks.29 There are several other classification methods available, such as support vector machines30 and deep learning models.31 However, these algorithms may be difficult to tune, and deep machine learning requires numerous examples in order to learn an adequate network structure for optimizing predictive performance.
The aggregate predictive performance among individual deconvolution algorithms, the simple voting method, and the random forest machine learning algorithm are summarized in Figure 3. A majority vote (2 or more votes, Point “2 Votes”) appeared to outperform any one deconvolution method used by itself. Compared to SNAP (Point “SNAP”) and TopFD (Point “TopFD”) algorithms, a majority vote (2 or more votes) had better recall and precision, respectively. The Venn diagram between a majority vote (2 or more votes) and its overlap with expert annotation was shown in Figure S1. Although THRASH of 60% fit identified a total of 50381 peaks, 41204 of them (82%) were false positive because they were not those identified by the expert annotations. Filtering the false positive accounted for the improved accuracy in the majority vote (2 or more votes). On the other hand, this majority missed 7181 peaks from the THRASH of 60% fit, out of 12264 peaks (59%) which were expert annotated peaks, which contributes to the low recall values. To provide reference for the random forest method, we calculated the aggregate precision and recall score using a probability threshold cutoff that optimizes the F1 score on the training spectra and applied it to the corresponding test set. The aggregated precision and recall value from the random forest method shown as a green point in Figure 3 is superior to most of the methods.
Furthermore, the results suggest that the random forest algorithm could achieve superior performance for identifying clusters which are true expert annotations. To determine average metrics (precision and recall) for random forest’s performance, the probability threshold cutoff that optimizes the F1 score was determined in each training fold feature set. The probability threshold was then applied to the associated test fold feature, and the resulting performance metrics were calculated. The final precision and recall were determined by averaging the results across each testing fold. Using this process, the random forest model achieved an average recall of 0.49, a precision score of 0.69, and an F1 score of 0.55. In comparison, THRASH of 60% fit which was the best algorithm by F1 score, achieved a recall of 0.76 and precision of 0.18, with an F1 score of 0.30. Additional metrics including median, 1st and 3rd quartiles, minimum and maximum of the F1 score across the different deconvolution methods were compared, and the random forest model outperformed other algorithms (Figure S2).
A useful aspect of the random forest model is the ability to extract feature importance values. One of the metrics that the random forest can report for each feature is the Mean Decrease Accuracy, which is an estimate of the reduction in the accuracy performance of the machine learning algorithm upon permuting the values of the current feature. The features ranked at the top of the plot reduce the accuracy of the model most significantly when permuted and these features are considered to be the most important ones. In Figure S3, cluster features such as the average mass of the cluster (AvgMass), the cluster charge (Charge), and the average intensity (AvgIntensity) had the most significant impact on the model, indicating that the random forest model was learning some of the spectral features such as charge and mass ranges that contribute to a true positive cluster. Features describing characteristics of the spectrum (i.e. activation type, precursor mass, and precursor charge) had a greater influence in the performance of the random forest classifier over the simple voting model. Using the vote of each deconvolution algorithm in the random forest model also provides a way to learn the confidence in each of algorithms to determine an optimal score (THRASH 60% fit, MS-Deconv, TopFD, and SNAP). While these features did not rank high in the list, the THRASH 60% fit feature seemed to have the most effect on the model performance over the other deconvolution algorithms. This is possibly due to the number of proposed peaks that the THRASH 60% fit finds in conjunction with the other features (spectrum characteristics and cluster features) to find the best scoring clusters within all of the false positive peaks (clusters with an expert annotation).
Backward selection, which iteratively removes features in model performance optimization, is an alternative route to determine feature importance. Performing a full backward selection process with leave-one-spectrum out cross-validation, and optimizing on the median F1 score (Figure S4), we found that Charge, Precursor Charge, THRASH 60% Fit, AvgMass, and SumIntensity (five of the features shown in Table 1) can achieve the same performance as a model built from all of the features. Omitting one of these five features showed a significant decrease in the median F1 score. Moreover, features such as AvgIntensity, Votes, and SumIntensity are correlated by definition. Consequently, removal of two of these features would be sufficient for the discriminatory models.
Expert annotation accuracy performance with 7-vote ensemble
To test the hypothesis that more orthogonal deconvolution algorithms can further improve results, we generated the results using THRASH with different fit score parameters as separate deconvolution algorithms (Figure 4). Comparing the peak call results from four THRASH algorithms with different fit scores, we noticed that no result directly subsumed the peak calls of any of the others, which indicates some degrees of “orthogonality” among the different THRASH results (Figure 4a). The discordance of results from THRASH was not surprising since THRASH is heuristically finding isotope envelopes. That is, isotopic distributions found in the beginning of the THRASH algorithm can affect the peaks found later during the algorithm process. With the additional deconvolution algorithm results added to the method, our results showed an increase in the number of assignments of clusters to annotated expert peaks and an increase in the filtering performance (Figures 4b and 4c). Additionally, other metrics using the 7-vote ensemble including the number of clusters, F1 score, and the number of recalled peaks were also improved compared to those using the 4-vote ensemble (Figure S5). In comparison with the average performance as in the 4-vote ensemble, the random forest model from the 7-vote ensemble achieves an average recall or true positive rate of 0.60 and a precision score of 0.78, and an F1 score of 0.67. After calculating the recall and precision for the individual algorithms, the best algorithm (by F1 score) was found to be THRASH 90% fit, which achieved a recall of 0.47 and precision of 0.58, with an F1 score of 0.52. Since there was an increase in the number of unassigned clusters (potentially false positives) in the 7-vote ensemble (56363) vs 4-vote ensemble (45117), it suggests that the 7-vote method learned to filter out false positives more accurately than the 4-vote system (Table S2 and S3).
Figure 4. Performance comparison between 4-vote ensemble and 7-vote ensemble.
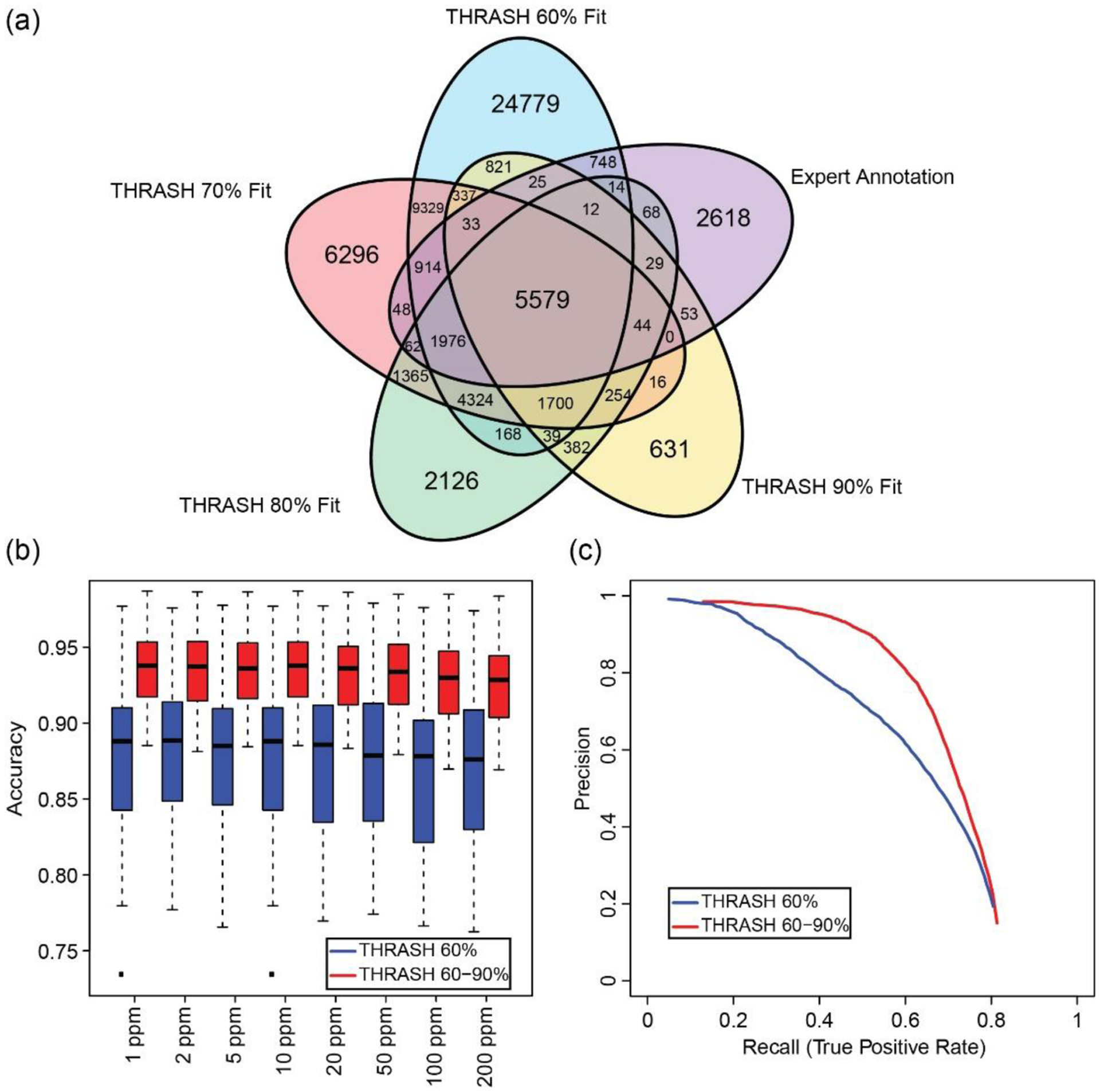
(a) Venn diagram of peaks found using THRASH with the fit parameter set at 60%, 70%, 80%, or 90%. (b) Boxplot of random forest accuracy between the 4-vote ensemble (red) and 7-vote ensemble (blue) with different cluster cutoffs. At all cluster cutoffs value, 7-vote ensemble had better performance than 4-vote ensemble. (c) Precision-recall curve using 4-vote ensemble (blue, THRASH 60%) and 7-vote ensemble (red, THRASH 60–90%). 7-vote ensemble had improved performance compared to 4-vote ensemble. The black squares in the figure represent outliers in the dataset.
In summary, adding more deconvolution algorithms has the potential of increasing the identification of peaks potentially missed by other deconvolution algorithms and improving the classification performance to filter out more false positives. Two additional deconvolution algorithms including pParseTD and UniDec which are based on online support vector machine algorithm and a Bayesian algorithm, respectively, will be ideal for continual development of this machine learning strategy due to the differences in algorithmic approaches compared to the four deconvolution algorithms used in this study. However, pParseTD currently only processes Thermo dataset, and the output peak list requires additional processing to assign charges for the isotopic distribution to locate the deconvoluted peaks in the spectrum. UniDec is optimized for native mass spectrometry where proteins and their fragment ions typically carry lower charges relative to mass compared to those in denatured conditions. Additional efforts are needed to incorporate these two algorithms into the machine learning strategy described in this study.
Unmatched clusters and missed expert annotations
When investigating the missed expert annotations (false negatives) and the unassigned clusters (false positives) from the machine learning strategy, two key observations surfaced. First, the unassigned clusters might actually be real isotopic distributions. Second, the corrected isotopic distributions may introduce a false positive and false negative calls into the analysis.
There are cases where the unassigned clusters may actually be real isotopic distributions that the manual annotator could have missed due to low abundance. These low abundance isotopic distributions might also suffer from imperfect distribution due to the noise. Figure 5a gives two examples of low abundance isotopic distributions that could be real annotations. This indicates that the method would be useful in proposing other annotations within data.
Figure 5. Example annotation of isotopic distributions.
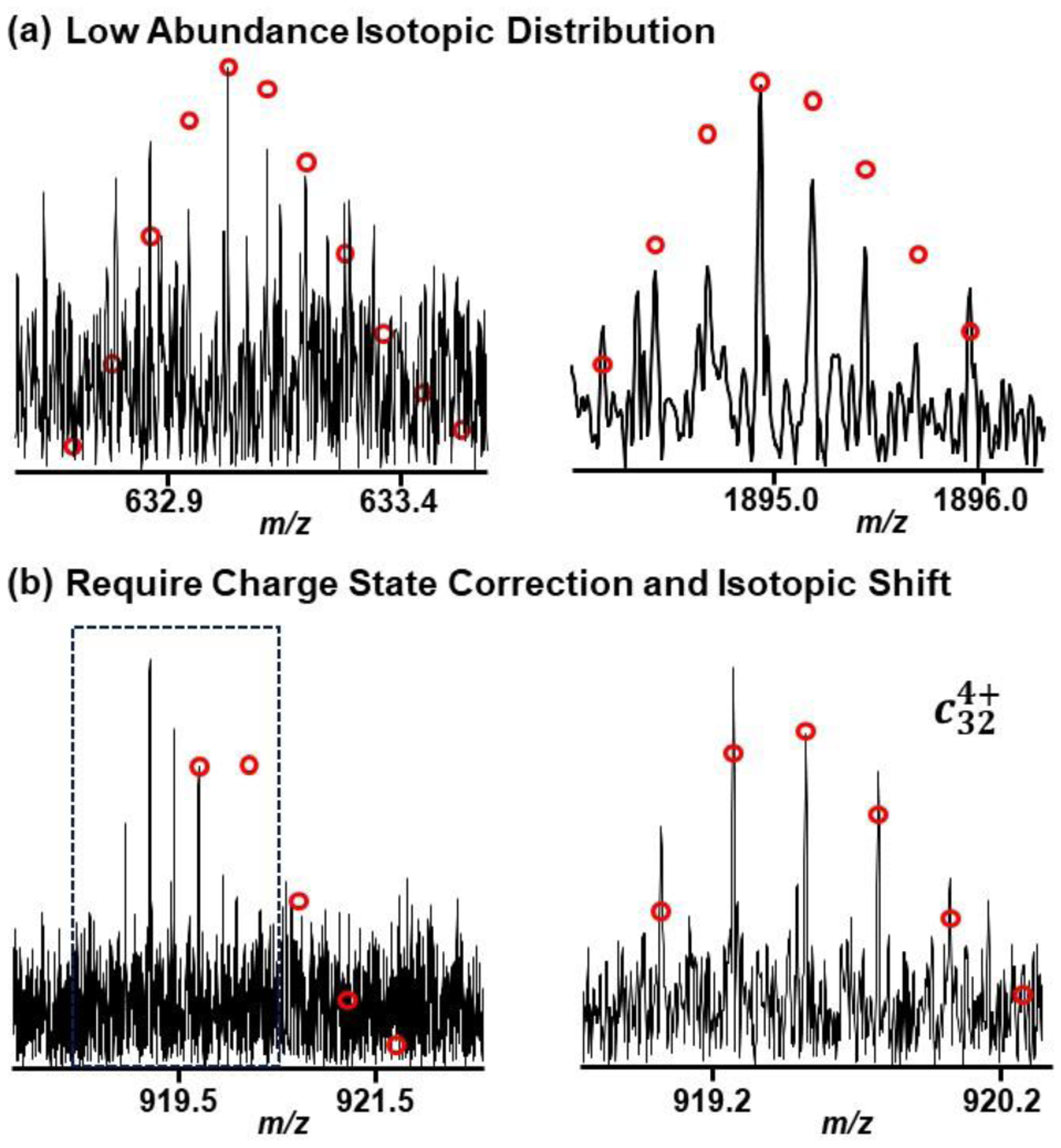
(a) Low abundance isotopic distribution that could be found by consensus peak list. These peaks were only found by the machine learning strategy. (b) Example isotopic distribution that has been manually corrected by shift the charge and monoisotopic peak.
During manual annotation and correction, there are many instances where the annotator has to correct the charge and/or peak of the most abundant mass. Figure 5b provides an example of an annotation that has been corrected by an expert annotator. Annotations that have been corrected in this way may introduce both a false positive and a false negative into the method analysis. The false positive would arise from the original peak without the correction from the deconvolution method, and the false negative would come from the corrected peak in the expert annotations.
The ability to shift the charges and most abundant mass is an area of continual research in this project, in order to identify more expert annotations without incurring more false positives. For example, generating the expert annotated results for the αTpm protein with ECD activation required the expert annotator to remove 52% (840 of 1631 peaks), adjust the charge state for 7% (109 of 1631 peaks), and shift the monoisotopic mass for 2% (38 of 1631 peaks) from the deconvoluted peaks found by THRASH 60% fit. The machine learning strategy did succeed in reducing the false positive rate but making additional modifications to identify and fix the annotations would further reduce the time spent on manual verification and peak correction.
Effects of improved deconvoluted peaks on database searching results
To investigate whether using the machine learning strategy can help with protein identification, we compared the MS-Align+ database search results from peak lists generated by different deconvolution algorithms and machine learning methods. Using the ECD spectrum of the αTpm proteoform, we evaluated and plotted the database search results using the deconvoluted results from expert annotation, TopFD, simple voting method, and random forest model (Figure 6 and Table S2). The E-value metric was utilized to evaluate the confidence of protein identification, with a lower E-value indicating high identification confidence. In the figure, the −log10 of the E-value was used for visualization instead in the y-axis, as a greater −log10(E-value) suggests higher protein identification confidence. The simple voting results were plotted by thresholding upon the number of votes. In the random forest model, the plot was generated at different thresholds of cross-validated probability of a correct expert annotation. For the 4-vote ensemble, only a small fraction of probability from simple voting and random forest model could achieve higher confidence in protein identification compared to that from expert annotations (Figure 6a). In comparison, the confidence in protein identification from the 7-vote ensemble in most majority votes from the simple voting model and most probability thresholds from the random forest model exceeded the −log10(E-value) score obtained from the expert annotations (Figure 6b). The improvement in protein identification confidence from the 4-vote ensemble to the 7-vote ensemble was also reflective of the observed increase in accuracy (in both limiting false positives while finding more peak clusters that match with an expert annotated peak, Figure 4c) when using a larger ensemble. Other proteoforms such as βTpm with CID activation and other proteins showed a similar trend in the analysis (Figure S6 and Table S2), except for a few special cases. These results indicate that some of the lower intensity isotopic distributions which were identified using the machine learning strategy could help improve the identification confidence values.
Figure 6. MS-Align+ database search results for αTpm.
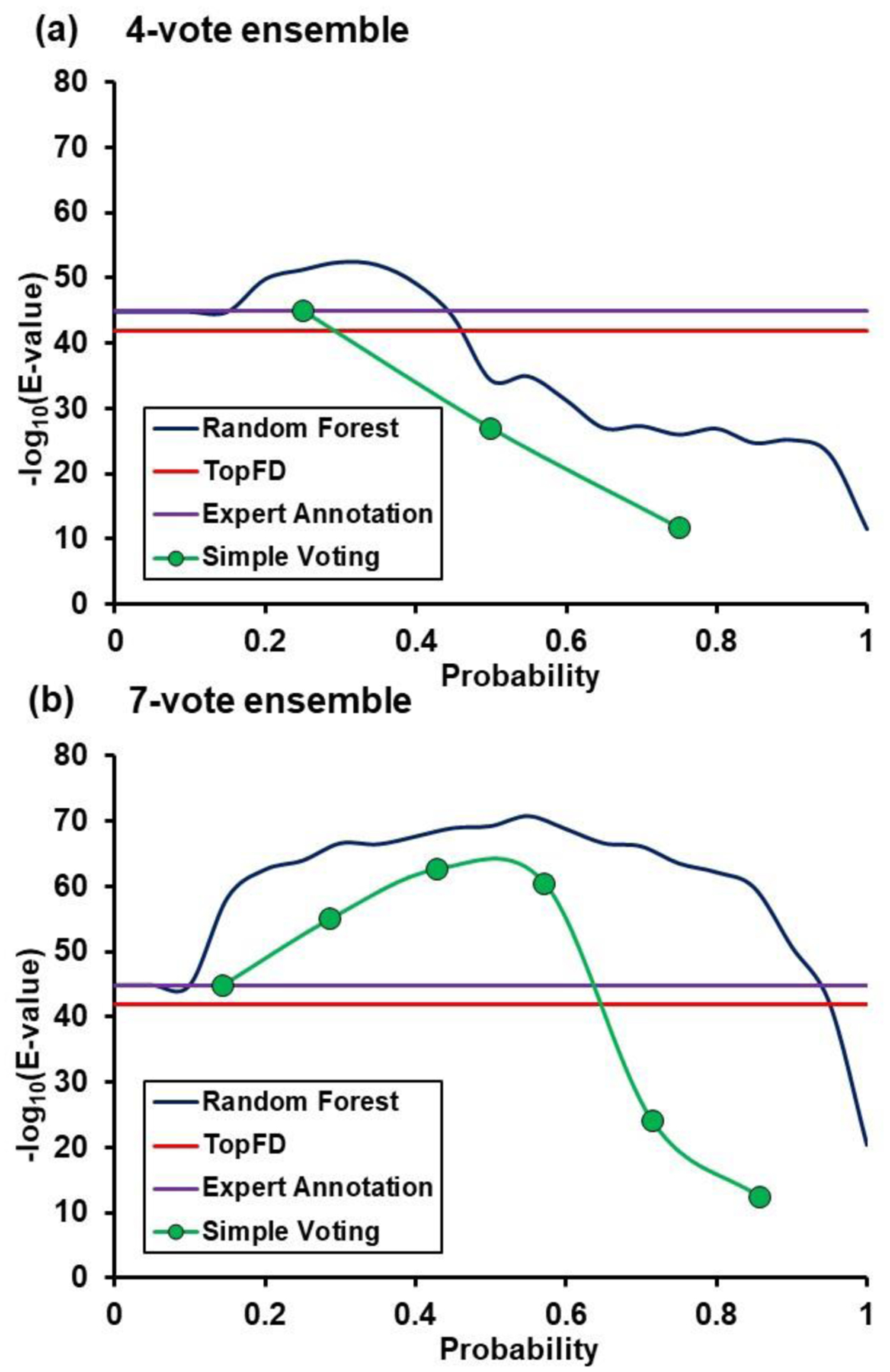
(a) 4-vote ensemble results. (b) 7-vote ensemble results. Each plot has the −log10(E-Value) for TopFD (red line), expert annotation (purple line), simple voting thresholding (#votes/max votes, green points/lines), and random forest probability thresholding (blue line).
The amount of true positive and false positive peaks that constitutes the consensus peak list has an impact on the database search when protein isoforms have a long homologous sequence. While evaluating the database search results for ssTnT ECD spectrum using generated peak lists, several isoforms were identified including A0A5K1V8N4 (Troponin T, slow skeletal muscle isoform b, correct identification), H9FC02 (Troponin T, slow skeletal muscle isoform c), A0A1D5RIQ3 (Troponin T1, slow skeletal type), and F7HR11 (Troponin T1, slow skeletal type) (Table S6). Using a sequence alignment tool, it was observed that only the N-terminal sequence has variations among these four isoforms (Figure S7). Ideally, thresholding on probability should keep the true expert annotations while reducing the number of false positives. A lower threshold would also result in the inclusion of more false positive annotations. In this particular case, simple voting method at low majority votes (less than 3 votes) yielded incorrect identification if the database search algorithm is given a set of peaks with many false positives. On the contrary, at higher thresholds for both the random forest algorithm and simple voting method, omission of true positives led to either diminishing E-value of correct identification, meaning a less confident database search result, or an incorrect identification.
For the spectrum for ssTnC protein with ECD activation, none of the single algorithms, except for THRASH of 80% fit, were able to identify the target sequence (Table S2 and S4). For the simple voting method, a majority vote (3 or more votes) could correctly find the protein. This result indicates that utilizing a consensus peak list could help identify the proteoform in spectra, even when most of the deconvolution algorithms failed to find the correct identification. If at least one algorithm can find the correct identification, theoretically the ensemble should also be able find the correct identification. Also, if there are several distinct false positive peaks (or no expert annotated peaks) from each algorithm, using a majority vote should help reduce the false positives (i.e. reduce the noise from each algorithm) to achieve a better identification rate.
Based on the database search results, both simple voting ensemble method and random forest machine learning algorithm were found to enhance both the accuracy and confidence in proteoform identification. For the simple voting ensemble method which only utilized clustering and simple voting, a majority vote (3 votes in the 7-vote ensemble) yielded the best results. In the case of random forest algorithm which required clustering and training a machine learning model, a probability threshold greater than 0.3 to 0.4 provided the optimal results.
Liquid chromatography-MS/MS data analysis
The results here are derived from targeted MS/MS data, and the machine learning strategy holds potential in improving the number of confident identifications with liquid chromatography (LC)-MS/MS runs. Further investigation needs to be done to determine whether models built using the expert annotations from MS/MS runs will improve the identification rate on a separate LC-MS/MS run, or if other annotations are needed in order to improve performance. Annotating deconvoluted peaks from spectra with confident protein identification would be a good starting point. A simple voting model would be more easily applicable for the LC-MS/MS experiment, as other machine learning algorithms may require enough annotated top-down LC-MS/MS spectra in order to develop models for performance optimization.
Integration into MASH Explorer
With the success of this project, the next step is to integrate the developed machine learning strategy as part of MASH Explorer, which provides several options for deconvolution of top-down data. Instead of deciding which deconvolution algorithm to apply, the user could run all available algorithms and automatically combine the results into a comprehensive list.
Conclusion
In summary, we have designed and demonstrated a machine learning strategy that allows for the combination of deconvolution results from multiple algorithms into an accurate consensus peak list for downstream processing. With the detection of more real isotopic distributions while filtering out false positives, the process showed promise in reducing the time spent manually validating and correcting the ion annotations in top-down MS/MS protein identification. In both simple voting ensemble method and random forest machine learning algorithm, the resulting consensus peak lists could improve on the accuracy and confidence in proteoform identification compared to a single deconvolution algorithm. This machine learning strategy shows promise for high-throughput protein identification and characterization in LC-MS/MS dataset for top-down proteomics. Integrating the tool into MASH Explorer will enable users to find more true positive deconvoluted peaks and consequently enhance the data analysis of high-resolution top-down MS dataset.
Supplementary Material
Acknowledgement
This work is dedicated to Professor John R. Yates III, the recipient of the 2019 ASMS John B. Fenn Award for a Distinguished Contribution in Mass Spectrometry. Financial support was provided by NIH R01 GM125085 (to Y. G.). Y. G. also would like to acknowledge the NIH grants, R01 GM117058 and S10 OD018475.
Footnotes
Associated Content
Skeletal Muscle Tissue Samples and Sarcomeric Protein Extraction; Offline Fraction Collection and High-Resolution MS/MS for Protein Characterization; Table S1. Summary of selected publicly available deconvolution algorithms; Table S2. Summary of protein proteoforms used for expert annotation; Table S3. Clusters classification for 4-vote ensemble; Table S4. Clusters classification for 7-vote ensemble; Table S5. Database search results for ssTnC with ECD activation from different input peak list; Table S6. Database search results for ssTnT with ECD activation from different input peak list; Figure S1. Venn diagram for the majority vote greater than two and its overlap with expert annotation (“true positive”); Figure S2. Boxplot of F1 scores using cross-validated fold results from the 4-vote ensemble comparison; Figure S3. Feature ranking for mean decrease in accuracy; Figure S4. Backward selection of features for the random forest model; Figure S5. Statistical analysis between 4-vote ensemble and 7-vote ensemble; Figure S6. E-value evaluation for βTpm spectrum with CID activation; Figure S7. Sequence comparison among four slow skeletal Troponin T isoforms.
Conflict of Interest Disclosure
The authors declare no conflict of interest.
Reference
- 1.Zhang H; Ge Y, Comprehensive Analysis of Protein Modifications by Top-Down Mass Spectrometry. Circ-Cardiovasc Gene 2011, 4 (6), 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith LM; Kelleher NL; Proteomics CTD, Proteoform: a single term describing protein complexity. Nat Methods 2013, 10 (3), 186–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai WX; Tucholski TM; Gregorich ZR; Ge Y, Top-down Proteomics: Technology Advancements and Applications to Heart Diseases. Expert Rev Proteomic 2016, 13 (8), 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen BF; Brown KA; Lin ZQ; Ge Y, Top-Down Proteomics: Ready for Prime Time? Anal Chem 2018, 90 (1), 110–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aebersold R; Agar JN; Amster IJ; Baker MS; Bertozzi CR; Boja ES; Costello CE; Cravatt BF; Fenselau C; Garcia BA; Ge Y; Gunawardena J; Hendrickson RC; Hergenrother PJ; Huber CG; Ivanov AR; Jensen ON; Jewett MC; Kelleher NL; Kiessling LL; Krogan NJ; Larsen MR; Loo JA; Loo RRO; Lundberg E; MacCoss MJ; Mallick P; Mootha VK; Mrksich M; Muir TW; Patrie SM; Pesavento JJ; Pitteri SJ; Rodriguez H; Saghatelian A; Sandoval W; Schluter H; Sechi S; Slavoff SA; Smith LM; Snyder MP; Thomas PM; Uhlen M; Van Eyk JE; Vidal M; Walt DR; White FM; Williams ER; Wohlschlager T; Wysocki VH; Yates NA; Young NL; Zhang B, How many human proteoforms are there? Nat Chem Biol 2018, 14 (3), 206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ge Y; Lawhorn BG; ElNaggar M; Strauss E; Park JH; Begley TP; McLafferty FW, Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J Am Chem Soc 2002, 124 (4), 672–678. [DOI] [PubMed] [Google Scholar]
- 7.Shaw JB; Li WZ; Holden DD; Zhang Y; Griep-Raming J; Fellers RT; Early BP; Thomas PM; Kelleher NL; Brodbelt JS, Complete Protein Characterization Using Top-Down Mass Spectrometry and Ultraviolet Photodissociation. J Am Chem Soc 2013, 135 (34), 12646–12651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaffer LV; Millikin RJ; Miller RM; Anderson LC; Fellers RT; Ge Y; Kelleher NL; LeDuc RD; Liu XW; Payne SH; Sun LL; Thomas PM; Tucholski T; Wang Z; Wu S; Wu ZJ; Yu DH; Shortreed MR; Smith LM, Identification and Quantification of Proteoforms by Mass Spectrometry. Proteomics 2019, 19 (10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horn DM; Zubarev RA; McLafferty FW, Automated reduction and interpretation of high resolution electrospray mass spectra of large molecules. J Am Soc Mass Spectr 2000, 11 (4), 320–332. [DOI] [PubMed] [Google Scholar]
- 10.Liu XW; Inbar Y; Dorrestein PC; Wynne C; Edwards N; Souda P; Whitelegge JP; Bafna V; Pevzner PA, Deconvolution and Database Search of Complex Tandem Mass Spectra of Intact Proteins. Mol Cell Proteomics 2010, 9 (12), 2772–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kou Q; Xun LK; Liu XW, TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics 2016, 32 (22), 3495–3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun RX; Luo L; Wu L; Wang RM; Zeng WF; Chi H; Liu C; He SM, pTop 1.0: A High-Accuracy and High-Efficiency Search Engine for Intact Protein Identification. Anal Chem 2016, 88 (6), 3082–3090. [DOI] [PubMed] [Google Scholar]
- 13.Marty MT; Baldwin AJ; Marklund EG; Hochberg GKA; Benesch JLP; Robinson CV, Bayesian Deconvolution of Mass and Ion Mobility Spectra: From Binary Interactions to Polydisperse Ensembles. Anal Chem 2015, 87 (8), 4370–4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Köster C, Mass spectrometry method for accurate mass determination of unknown ions. US6188064B1, 2001.
- 15.Cai WX; Guner H; Gregorich ZR; Chen AJ; Ayaz-Guner S; Peng Y; Valeja SG; Liu XW; Ge Y, MASH Suite Pro: A Comprehensive Software Tool for Top-Down Proteomics. Mol Cell Proteomics 2016, 15 (2), 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuncheva LI; Whitaker CJ, Measures of diversity in classifier ensembles and their relationship with the ensemble accuracy. Mach Learn 2003, 51 (2), 181–207. [Google Scholar]
- 17.Sollich P; Krogh A, Learning with ensembles: How over-fitting can be useful. Adv Neur In 1996, 8, 190–196. [Google Scholar]
- 18.Geurts P; Fillet M; de Seny D; Meuwis MA; Malaise M; Merville MP; Wehenkel L, Proteomic mass spectra classification using decision tree based ensemble methods. Bioinformatics 2005, 21 (14), 3138–3145. [DOI] [PubMed] [Google Scholar]
- 19.Ru XQ; Li LH; Zou Q, Incorporating Distance-Based Top-n-gram and Random Forest To Identify Electron Transport Proteins. J Proteome Res 2019, 18 (7), 2931–2939. [DOI] [PubMed] [Google Scholar]
- 20.Tran NH; Zhang XLL; Xin L; Shan BZ; Li M, De novo peptide sequencing by deep learning. P Natl Acad Sci USA 2017, 114 (31), 8247–8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIlwain SJ; Wu Z; Wenger K; Wetzel M; Tucholski T; Liu X; Sun L; Ong IM; Ge Y In MASH Explorer, a Universal, Comprehensive, and User-friendly Software Environment for Top-down Proteomics, 299322, Tuesday, Proceedings of 67th ASMS Conference on Mass Spectrometry and Allied Topics, Atlanta, GA, June 2nd - 6th, 2019. [Google Scholar]
- 22.Jin Y; Diffee GM; Colman RJ; Anderson RM; Ge Y, Top-down Mass Spectrometry of Sarcomeric Protein Post-translational Modifications from Non-human Primate Skeletal Muscle. J Am Soc Mass Spectrom 2019, 30 (12), 2460–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perez-Riverol Y; Csordas A; Bai JW; Bernal-Llinares M; Hewapathirana S; Kundu DJ; Inuganti A; Griss J; Mayer G; Eisenacher M; Perez E; Uszkoreit J; Pfeuffer J; Sachsenberg T; Yilmaz S; Tiwary S; Cox J; Audain E; Walzer M; Jarnuczak AF; Ternent T; Brazma A; Vizcaino JA, The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019, 47 (D1), D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tibshirani R; Hastie T; Narasimhan B; Soltys S; Shi GY; Koong A; Le QT, Sample classification from protein mass spectrometry, by ‘peak probability contrasts’. Bioinformatics 2004, 20 (17), 3034–3044. [DOI] [PubMed] [Google Scholar]
- 25.Virtanen P; Gommers R; Oliphant TE; Haberland M; Reddy T; Cournapeau D; Burovski E; Peterson P; Weckesser W; Bright J; al, e., SciPy 1.0--Fundamental Algorithms for Scientific Computing in Python. arXiv e-prints 2019, arXiv:1907.10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liaw A; Wiener M, Classification and Regression by randomForest. R News 2002, 2 (3), 18–22. [Google Scholar]
- 27.Liu XW; Hengel S; Wu S; Tolic N; Pasa-Tolic L; Pevzner PA, Identification of Ultramodified Proteins Using Top-Down Tandem Mass Spectra. J Proteome Res 2013, 12 (12), 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxena A; Prasad M; Gupta A; Bharill N; Patel OP; Tiwari A; Er MJ; Ding WP; Lin CT, A review of clustering techniques and developments. Neurocomputing 2017, 267, 664–681. [Google Scholar]
- 29.Zhang YY; Xin Y; Li Q; Ma JS; Li S; Lv XD; Lv WQ, Empirical study of seven data mining algorithms on different characteristics of datasets for biomedical classification applications. Biomed Eng Online 2017, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortes C; Vapnik V, Support-Vector Networks. Mach Learn 1995, 20 (3), 273–297. [Google Scholar]
- 31.Schmidhuber J, Deep learning in neural networks: An overview. Neural Networks 2015, 61, 85–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


